Abstract
Hypervirulent fowl adenovirus serotype 4 (hvFAdV-4) has emerged as a major pathogen of hepatitis-hydropericardium syndrome (HHS) with increased mortality in chickens, resulting in economic losses to the Chinese poultry industry since June 2015. Here, we isolated a hypervirulent FAdV-4 (hvFAdV-4) strain (designated GD616) from 25-day-old meat-type chickens with severe HHS in Guangdong Province China in June 2017. The whole genome of the strain GD616 shares high homology with those in the recently-reported hvFAdV-4 isolates in China, with natural deletions of ORF19 and ORF27. A comparative analysis of Hexon and Fiber-2 proteins revealed that 2 unique amino acid residues at positions 378 and 453 of the Fiber-2 protein might be associated with virulence due to their occurrences in all the hvFAdV-4 isolates only. To systemically evaluate the effect of age on the susceptibility of chickens to hvFAdV-4, we used this hvFAdV-4 strain to intramuscularly inoculate 7- to 180-day-old specific-pathogen-free chickens for the evaluation of pathogenicity. These results showed that the pathogenicity of the hvFAdV-4 strain GD616 to chickens exhibited age-relatedness, with younger than 59-day-old chickens showing 100% morbidity and mortality, while 180-day-old chickens still exhibited a hydropericardium syndrome-like clinicopathology with 60% morbidity and 20% mortality. These findings enrich the current available knowledge regarding the pathogenicity of the hypervirulent FAdV-4 virus in chickens with a wide range of ages, which assists with the selection of suitable-aged chickens for the evaluation of hvFAdV-4 vaccines.
Key words: hvFAdV-4, HHS, genetic property, pathogenicity, chicken age
INTRODUCTION
Hypervirulent fowl adenoviruses (hvFAdVs), which have emerged as major agents of inclusion body hepatitis-hydropericardium syndrome (IBH-HPS) and gizzard erosion, exhibit a severe damage to the poultry industry due to their contribution to enhanced mortality rates and decreased poultry growth performance (Vera-Hernández et al., 2016). FAdVs, the Aviadenovirus genus of the Adenoviridae family, are classified into 5 species (FAdV-A to E) dependent upon molecular structure, and subclassified into 12 serotypes, namely FAdV-1 to 8a and -8b to 11, according to cross-neutralization test results (Hess, 2000). After the first FAdV infection occurred in 1987 in Pakistan (Anjum et al., 1989), subsequent outbreaks have been recorded in many countries, including the USA, India, Canada, Hungary, Korea, and Japan, causing considerable economic losses to the poultry industry (Asrani et al., 1997; Hess et al., 1999; Toro et al., 1999; Jadhao et al., 2003; Kim et al., 2008; Mase et al., 2012; Rahimi and Minoosh, 2015; Zhao et al., 2015).
FAdVs are nonenveloped virions with 252 capsomers in an array of icosahedral symmetry. The genome of FAdVs is a linear, double-stranded DNA molecule with a size of approximately 43-45 kb (Griffin and Nagy, 2011; Marek et al., 2012; Zhao et al., 2015). Two separate Fiber-encoding genes noncovalently linked to the penton base in the FAdV-4 genome (Griffin and Nagy, 2011). Fiber is a structural protein located at the outermost part of the capsid for initial attachment to the host cell's receptor via the N-terminal tail that contributes to endocytosis (Shah et al., 2017). Hexon is the viral major protein which contains the neutralizing epitope, and is serotype-specific (Toogood et al., 1992; Russell, 2009). The Hexon gene sequence is directly related to the serotype of FAdVs (Niczyporuk, 2016; Li et al., 2017). Molecular analyses based on the major FAdV structural Hexon and Fiber proteins with antigenic specificity are frequently undertaken (Meulemans et al., 2001; Helena et al., 2014). The mature trimeric Hexon can bind to the phospholipids of the target cell membrane eluding the cellular receptors that form the barrier blocking viral entry (Balamurugan and Kataria, 2004; Yan et al., 2016). Moreover, they are also responsible for transportation of the FAdV-4 DNA into the nucleus via the nuclear pore complex (Yan et al., 2016).
In China, the prevalence of IBH and hepatitis-HPS (HHS) was sporadic until 2015; however, especially since May 2015, severe IBH-HPS, which is characterized by accumulated fluid in the pericardial sac and IBH, has emerged in major chicken-producing provinces of China and has caused considerable epidemics in broilers and layers. FAdV-4 infection has resulted in acute death with no overt clinical disease and high mortality rates with a range of 10% to 80% and 0% to 10% in 21- to 35-day-old broilers and younger than 140-day-old layers, respectively (Anjum et al., 1989; Balamurugan and Kataria, 2004; Asthana et al., 2013; Mittal et al., 2014). The pathological characteristics are an accumulation of straw-colored fluid in the pericardial sac (Balamurugan and Kataria, 2004), the swelling of the liver with many necrotic foci and spots, hepatocytes with nuclear inclusion bodies, and pale bone marrow (Abe et al., 1998). Here, we isolated a FAdV-4 strain (designated GD616) from 25-day-old meat-type chickens with severe HHS in Guangdong Province China in June 2017 and identified it as a hypervirulent FAdV-4 strain as evidenced by being the same genotype strain as hvFAdV-4 recently found to be prevalent in China. Previous studies have indicated that the outcome of experimentally-reproduced IBH-HHS by FAdV-4 is associated with the age of the birds (Mazaheri et al., 1998; Lim et al., 2011). Most IBH-HHS cases are observed in 21- to 35-day-old broilers, and the younger ages are associated with more severe disease consequences (Li et al., 2017). One recent study showed that the pathogenicity of FAdV-4 to geese was also dependent on age (Wei et al., 2019). Clinical disease and pericardial effusion were observed in 10-day old geese when administered subcutaneously with hvFAdV-4, whereas geese aged 20 and 30 d were not susceptible (Wei et al., 2019). However, there has been, to our knowledge, no systemic report on the effect of age on the susceptibility of chickens to hvFAdV-4. In the present study, we used this hypervirulent FAdV-4 isolate GD616 to intramuscularly inoculate 7- to 180-day-old specific-pathogen-free (SPF) chickens for the evaluation of pathogenicity and found that the pathogenicity in hypervirulent FAdV-4-inoculated chickens exhibited age-relatedness, with 180-day-old chickens having substantially low pathogenicity. The results of this study demonstrated that the pathogenicity of hypervirulent FAdV-4 is associated with the age of infected chickens, which is beneficial for the selection of suitably aged chickens for the evaluation of FAdV-4 vaccines.
MATERIALS AND METHODS
Ethics Statement
All chicken experiments were conducted according to the animal welfare guidelines of the Institutional Animal Care and Use Committee (IACUC) of the Institute of Animal Husbandry and Veterinary Medicine, Beijing Academy of Agriculture and Forestry Sciences.
Case History and Sample Collection
In June 2017, an outbreak of HPS occurred at several meat-type commercial chicken farms in Guangdong province, China. The affected chickens ranged in age from 21 to 56 d, and the morbidity and mortality rates were 80 to 100% and 50 to 80% in different chicken flocks, respectively. Necropsy was conducted on dead chickens and liver tissue samples from 5 affected birds were collected for virological and histological examination. Collected liver samples were homogenized in phosphate-buffered saline (PBS) and centrifugated, supernatant was passed through a 0.22 μM filter and stored at -80°C until use (Kaján et al., 2013).
PCR Amplification and Complete Genome Sequencing
Viral DNA from infected liver homogenates was extracted using a DNeasy Blood & Tissue Mini kit (Qiagen) in accordance with the manufacturer's instructions. The extracted DNA was resuspended in 25 μL of RNase-, DNase-, and proteinase-free water. GD616 genome sequencing was performed on the basis of the whole genome sequence of the FAdV-4-HN-15102 strain as described previously (Liu et al., 2016).
Cell Culture and Virus Isolation
A chicken hepatoma cell line (LMH) was cultured in Dulbecco's modified Eagle's medium (Gibco, 11995) with 100 mg/mL streptomycin, 100 units/mL penicillin, and 10% fetal bovine serum (FBS; Gibco, 10099-141), and incubated at 37°C in a 5% CO2 incubator. LMH cell monolayers in a 75 cm2 flask were infected with 5 mL each of 5-fold diluted liver tissue homogenates and were incubated at 37°C for 2 h followed by incubation with a maintenance medium containing 2% FBS. The cells were checked daily for cytopathic effects (CPE). The 50% tissue culture infective dose (TCID50) of the FAdV-4 strain GD616 was assayed on LMH cells as previously described (Gao et al., 2019).
Indirect Immunofluorescence and Confocal Microscopy
Monolayer LMH cells grown in chamber slides (BD) were infected with the hvFAdV-4 strain GD616 for 72 h and expression of Fiber-2 protein was detected by indirect immunofluorescence assay under confocal microscopic obversation as described recently (Yuan et al., 2021).
Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (PAGE) and Western Blotting
FAdV-4-infected LMH cells were collected and the cell lysates was extracted for determining expression of Fiber-2 protein by using Western blotting as described recently (Yuan et al., 2021).
Pathogenicity Experiments
Infection of Different hvFAdV-4 Inocula in 21-Day-Old Specific-Pathogen-Free Chickens. Seventy 21-day-old specific-pathogen-free (SPF) White Leghorn chickens (Beijing Boehringer Ingelheim Vital Biotechnology Co., Ltd, Beijing, China) were randomly assigned to 7 groups; namely, a sham-infected group (n = 10) and 6 groups of the hvFAdV-4 strain GD616-infected chickens (n = 10) at a dose of 101.5-6.5 TCID50 inoculua. The groups were observed for 9 d postinfection, and the morbidity and mortality were recorded every day.
Infection of hvFAdV-4 in Different-Aged SPF Chickens. Ten batches of SPF White Leghorn chickens aged 7 to 180 d from Beijing Boehringer Ingelheim Vital Biotechnology Co., Ltd., Beijing, China were used for further evaluation of hvFAdV-4 pathogenicity. The 10 batches of chickens corresponded to chickens aged 7, 14, 21, 28, 35, 42, 59, 80, 120, and 180 d. Each batch of 15 chickens of the same age were randomly divided into GD616-infected (n = 10) and sham-infected (n = 5) groups. All the infected chickens were intramuscularly inoculated with a dose of 0.2 mL (106.5 TCID50) of hvFAdV-4 strain GD616. All batch chickens were observed for 14 d postinfection, and the morbidity and mortality were recorded every day.
Histopathology and Immunohistochemical (IHC) Staining
Liver tissue samples of dead chickens following hvFAdV-4 infection were collected for histopathological observation and IHC staining to detect FAdV-4 viral antigens as described recently (Yuan et al., 2021).
Quantitative Real-Time Polymerase Chain Reaction (PCR)
Quantitative real-time PCR (qPCR) was performed to determine virus loads in the GD616-infected chicken sera, liver, and LMH cell culture supernatants. The forward primer (5’-TTGTTACCTCCCTCTACTTG-3’) and the reverse primer (5’-TGGACACGCTCCTATTGG-3’) were used to amply a 207 bp) fragment located in the Fiber-2 gene. Viral DNA was isolated from LMH cells, sera, and liver using a DNeasy Blood & Tissue Mini kit (Qiagen) in accordance with the manufacturer's instructions. The extracted DNA was resuspended in 25 μL of RNase-, DNase-, and proteinase-free water. The qPCR protocol followed the instructions of an IQ SYBR Green Supermix kit (Bio-Rad). The qPCR parameters consisted of total denaturation at 95°C for 3 min and 40 cycles of denaturation at 95°C for 5 s and annealing at 55°C for 30 s. Serial dilutions of a plasmid pCMV-F2 (Fiber-2 gene cloned into pCMV-HA) were used to quantitatively determine the FAdV-4 genomic copy number.
RESULTS
Isolation and Characterization of hvFAdV-4
Total DNAs in the liver tissues of dead chickens that exhibited clinical disease were extracted. A 1,440-bp FAdV-4-specific fragment (Fiber-2 gene) was amplified, and no-infections of other viruses, including Newcastle disease virus, infectious bursal disease virus, avian reovirus, avian leukosis virus, reticuloendotheliosis virus, chicken anemia virus, and Marek's disease virus, determined by PCR or RT-PCR, were present. As shown in Figure 1A, the liver homogenates-inoculated LMH cells showed severe CPE at 72 h postinouclation during cell culture passage, as compared to that of the noninfected cells. We examined FAdV-4 replication in the cultured LMH cells using double staining for Fiber-2 protein (green) and nuclei (blue) under a confocal microscope. Fiber-2 expression (green) was displayed predominantly in the cytoplasm of FAdV-4-infected cells (Figure 1B). We also determined strain GD616 replication in LMH cells by detecting the expression of the Fiber-2 protein by Western blotting at the indicated times after infection, with the peak of the Fiber-2 protein expression being at 72 h postinfection (Figure 1C).
Figure 1.
Infection of the FAdV-4 strain GD616 in cultured LMH cells. (A) Cytopathic effects in the LMH cells 72 h after FAdV-4 strain GD616 infection. (B) LMH cells following FAdV-4 infection at 72 h were fixed and incubated with a guinea pig polyclonal antibody raised against FAdV-4 Fiber-2 protein followed by a fluorescein isothiocyanate-conjugated antiguinea pig IgG antibody. LMH cells expressing Fiber-2 protein (green) and nuclear staining (blue) were merged. (C) FAdV-4-infected LMH cell lysates at the indicated times following infection were collected, resolved by SDS-PAGE, and immunoblotted. The expression level of the Fiber-2 protein was analyzed. GAPDH served as the loading control.
Genome Sequencing, Phylogenetic Tree Analysis, and Molecular Signatures of Hexon and Fiber-2 Proteins
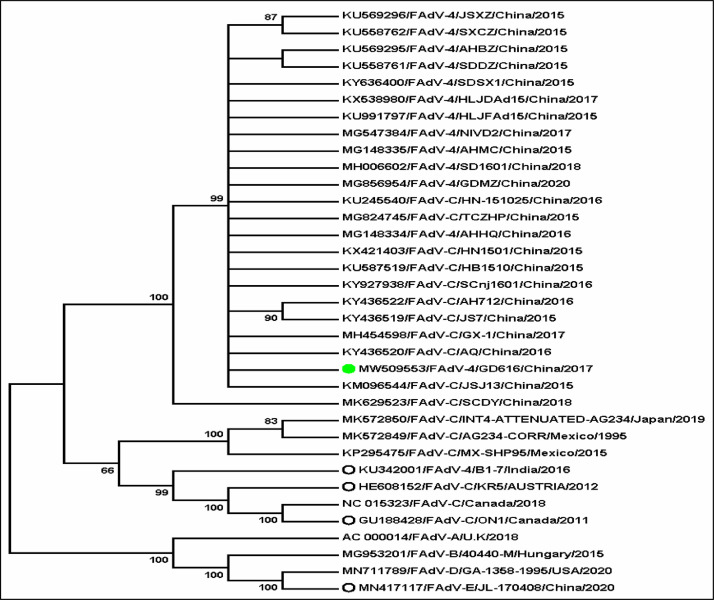
The complete genome sequence of the FAdV-4 strain GD616 from the liver samples was determined and deposited in the GenBank database under the accession number MW509553. The full genome size of strain GD616 was 44,247 bp in length. For 3 low pathogenic strains, KR5 (HE608152), ON1 (GU188428), and B1-7 (KU342001), natural deletions of ORF19, and ORF27 were observed. The strain GD616 genome possessed 99.83 to 99.95% homology with those of the reported hypervirulent FAdV-4 isolates (GDMZ, SD1601, HLJDAd15, AHMC, AHHQ, HN-151025, JSJ13, and GX-1) but exhibited 98.42 to 98.76% homology to the overseas highly pathogenic strain MX-SHP95 or low pathogenic strains (B1-7, KR5, and ON1). Phylogenetic analysis of the whole genome showed that the strain GD616 assigns as a cluster of FAdV-C viruses, while FAdV-A, FAdV-B, FAdV-D, and FAdV-E all belong to another cluster (Figure 2). The GD616 strain was further classified into a distinct group with all recently-reported hypervirulent FAdV-4 isolates in China, such as HN-151025, AHMC, GDMZ, SD1601, AHHQ, and HLJDAd15, which is different from another group consisting of the overseas highly pathogenic strain MX-SHP95 or low pathogenic strains (B1-7, KR5, and ON1) (Figure 2).
Figure 3.
Growth kinetics of the FAdV-4 strain GD616 in cultured LMH cells. (A) LMH cells were inoculated with the FAdV-4 strain GD616 at an MOI of 0.1 and harvested at the indicated times for a viral titer assay. Virus titers were expressed as TCID50/mL, and the values are shown as the mean ± SD of the results of 3 independent experiments. (B) qPCR analysis of DNA extracted from the FAdV-4 strain GD616-infected LMH cells at the indicated times postinfection. Data are means ± SD of the results of 3 independent experiments.
Figure 2.
Phylogenetic analysis of FAdV viruses based on the nucleotide sequences of the whole genomes by MAGE7. GD616 is highlighted by the listed symbol. The GenBank accession number for each FAdV sequence is indicated as the number in parenthesis.
The full Hexon and Fiber-2 genes of the FAdV-4 strain GD616 were 2,814 bp and 1,440 bp in length, and encoded for 937 and 479 amino acids, respectively. No deletions or insertions were observed as compared to those of other hvFAdV-4 strains. There were no amino acid substitutions in Hexon protein of strain GD616 when compared to that of other Chinses hypervirulent FAdV-4 strains, such as HB1510, HN/151029, NIVD2, SDSX1, HN-151025, AH-F19, and GX-1 (Table 1). Additionally, 13, 11, and 4 amino acid mutations occurred in the Hexon protein of strain GD616 compared to that of low pathogenic FAdV-4 strains KR5 (Marek et al., 2012), ON1 (Griffin and Nagy, 2011), and B1-7, respectively. Three amino acid mutations were observed at positions 193 (Q to R), 195 (E or N to Q), and 842 (G or T to A) in the Hexon protein of the GD616 strain compared to that of nonpathogenic FAdV-4 strains, but 13 amino acid substitutions in the Hexon protein of strain GD616 were observed compared to that of the highly virulent FAdV-4 strain MX-SHP95 (Vera-Hernández et al., 2016) and pathogenic strain Kr-Changnyeong (Park et al., 2017). For Fiber-2 protein, there was almost 100% identity among the recently-reported hypervirulent FAdV-4 strains in China, such as HB1510, HN/151029, NIVD2, SDSX1, HN-151025, AH-F19, GX-1, and GD616, except for unique amino acid residue A at position 219 in the Fiber-2 protein of strain GX-1 (Table 1). However, only 4 amino acid substitutions were observed at positions 261 (N or S to T), 391 (S to T), 459 (A to N), and 478 (V to L) in the Fiber-2 protein of the hvFAdV-4 isolates in China compared to those of the nonpathogenic FAdV-4 strains, but these 4 amino acid substitutions were different from those of the Fiber-2 protein in the highly pathogenic FAdV-4 isolates MX-SHP95 and Kr-Changnyeong (Table 1). However, analysis of Fiber-2 amino acid sequences revealed that 2 unique amino acid substitutions at positions T378 and A453 occurred in all highly virulent FAdV-4 isolates, regardless of whether they were the recently-reported hypervirulent FAdV-4 isolates in China or the highly pathogenic FAdV isolates MX-SHP95 and Kr-Changnyeong outside China (Table 1).
Table 1.
Comparison of variable amino acid sequences from Hexon and Fiber-2 genes among pathogenic isolates and nonpathogenic isolates.
| Genes |
Amino acids at position |
|||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hexon | Isolates | 164 | 188 | 193 | 195 | 238 | 240 | 243 | 263 | 264 | 402 | 410 | 574 | 797 | 842 | |||||||||||||||||||||||||
| GD616 | S | R | R | Q | D | T | N | I | V | A | A | I | P | A | ||||||||||||||||||||||||||
| Pathogenic isolates | HB1510 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |||||||||||||||||||||||||
| HN/151029 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | ||||||||||||||||||||||||||
| NIVD2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | ||||||||||||||||||||||||||
| SDSX1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | ||||||||||||||||||||||||||
| HN-151025 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | ||||||||||||||||||||||||||
| AH-F19 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | ||||||||||||||||||||||||||
| GX-1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | ||||||||||||||||||||||||||
| MX-SHP95 | T | . | Q | E | N | A | E | M | I | Q | T | V | A | G | ||||||||||||||||||||||||||
| Kr-Changnyeong | T | I | Q | N | N | A | E | M | I | . | T | V | A | G | ||||||||||||||||||||||||||
| Non-pathogenic strains | ON1 | T | I | Q | E | N | A | E | M | I | . | T | V | A | G | |||||||||||||||||||||||||
| KR15 | T | I | Q | E | N | A | E | M | I | . | T | . | . | G | ||||||||||||||||||||||||||
| B1-7 | . | I | Q | E | . | . | . | . | . | . | . | . | . | T | ||||||||||||||||||||||||||
| Fiber-2 | 11 | 12 | 13 | 14 | 15 | 22 | 29 | 114 | 144 | 219 | 232 | 261 | 300 | 305 | 306 | 307 | 319 | 324 | 329 | 334 | 338 | 343 | 344 | 346 | 378 | 380 | 391 | 393 | 400 | 403 | 405 | 406 | 413 | 427 | 435 | 439 | 453 | 459 | 478 | |
| GD616 | E | N | G | K | P | S | A | D | S | D | Q | T | T | A | N | A | I | V | L | A | N | L | N | A | T | T | T | P | G | E | S | I | S | I | S | E | A | N | L | |
| Pathogenic isolates | HB1510 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| HN/151029 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| NIVD2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| SDSX1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| HN-151025 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| AH-F19 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| GX-1 | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| MX-SHP95 | — | — | — | — | — | Y | P | A | A | . | . | N | . | . | H | . | . | F | . | T | T | N | S | V | . | . | S | S | A | Q | . | S | T | V | . | D | . | A | V | |
| Kr-Changnyeong | — | — | — | — | — | . | P | A | A | . | . | N | . | . | H | P | V | F | . | T | T | W | . | V | . | A | S | S | A | . | . | S | T | V | . | D | . | A | V | |
| Non-pathogenic strains | ON1 | — | — | — | — | — | . | P | . | . | G | E | S | I | S | H | P | V | F | V | T | T | N | S | . | A | A | S | . | A | Q | P | S | T | . | T | D | S | A | V |
| KR15 | . | . | . | Q | . | . | . | . | . | G | E | S | I | S | H | P | V | F | V | T | T | N | S | . | A | A | S | . | A | Q | . | S | T | . | T | D | S | A | V | |
| B1-7 | . | . | . | . | . | . | . | . | . | G | . | S | . | . | . | . | . | . | . | . | . | . | . | . | A | A | S | . | . | . | . | . | . | V | . | . | S | A | V | |
Pathogenicity of Different hvFAdV-4 Inocula in 21-Day-Old SPF Chickens
The hvFAdV-4 strain GD616 was purified by plaque test and successively passaged 10 times in LMH cells to increase viral titers. Different passages (3, 5, and 10 passages) of the GD616 strain were collected for FAdV-4 Fiber-2 gene sequencing and no nucleotide mutation was found in the Fiber-2 gene (data not shown), indicating the genetic stability of strain GD616 during continued passage in cultured LMH cells. The titer of the hvFAdV-4 strain GD616 after 10 passages was 107.7 TCID50/mL at 72 h postinfection. We adopted 6 different doses (ranging from 101.5 to 106.5 TCID50 per chicken) to intramuscularly inoculate 21-day-old SPF chickens to determine the relationship between inocula doses and pathogenicity. As expected, no clinical diseases were observed in the sham-inoculated chickens throughout the animal experiment. However, chickens inoculated with the strain GD616 regardless of different doses showed lethargy, diarrhea, and prostration. All chickens from the 106.5 TCID50-inoculated group died within 2 d postinoculation (Figure 4A). The time of symptom onset to death progressively prolonged with the decrease of inocular dose (Figure 4A). All the strain GD616-inoculated chickens died except for one chicken that survived in the 101.5 TCID50-inoculated group, which still exhibited 90% mortality, during the 9-day-observation period (Figure 4A). All the dead chickens exhibited typical clinicopathology, which was characterized by an accumulation of yellow pericardial effusion and a pale yellow, swollen, friable liver (data not shown). We determined FAdV-4 load in the sera of chickens in the 103.5 TCID50-inoculated group by qPCR. The results showed that the FAdV-4 copy number increased significantly after 3 d postinoculation until death reached the peak and thereafter declined (Figure 4B). We detected the viral load of the liver tissues in the 103.5 TCID50-inoculated group and found that the FAdV-4 genome copy number sustained an increase after inoculation and reached a peak of approximately 1010 copies/mg liver tissue at 5 d postinoculation (Figure 4C). Five randomly selected dead chicken liver tissues from the 103.5 TCID50-inoculated group or chicken liver tissues from the sham-inoculated group were subjected to hematoxylin and eosin staining and IHC staining. As shown in Figure 4D, there were many basophilic inclusion bodies (arrow c) with severe hepatic steatosis (arrow d) and infiltration of inflammatory cells (arrow e) in the liver samples of the hvFAdV-4-inoculated chickens. Heart tissue showed lymphocyte infiltration (arrow a) and myocardial Fiber-2 rupture (arrow b) (Figure 4E). As expected, liver and heart tissues from the sham-inoculated chickens were negative for the FAdV-4 antigen via IHC staining. In the liver, the inclusion bodies (arrow g) in the hepatic cells showed strong FAdV-4 antigen-positive signals (Figure 4F). For the heart tissues, only weak FAdV-4 antigen-positive signals were observed in infiltrated inflammatory cells (Figure 4F, arrow f). These results indicated that the hvFAdV-4 strain GD616 exhibited severe pathogenicity to 21-day-old SPF chickens at a dose as low as 101.5 TCID50, with the mortality rate being 90% after inoculation.
Figure 4.
Pathogenicity of the FAdV-4 strain GD616 in 21-day-old SPF chickens. (A) The survival rates in 21-day-old FAdV-4 strain GD616-infected chickens at different doses (101.5-106.5 TCID50) and sham-infected SPF chickens. (B) Total DNAs were extracted from the chicken sera (B) and liver tissues (C) of the 103.5 TCID50-inoculated group, and the FAdV-4 virus load (n = 10) was detected by qPCR. Data are shown as the mean ± SD. (D) Histological observation of heart and liver tissues from the sham- and 103.5 TCID50-inoculated chickens. The heart and liver sections of the sham-inoculated chickens exhibited no overt histopathological lesions. The hearts of the dead chickens exhibited lymphocyte infiltration (arrow a) and ruptured myocardial fibers (arrow b). The liver tissues of the dead chickens displayed a great amount of inclusion bodies (arrow c) accompanied by vacuolar degeneration (arrow d) and nuclear enlargement (arrow e). (E) Immunohistochemical staining of heart and liver tissues from the sham-inoculated and 103.5 TCID50-inoculated chickens. No staining was observed in the heart and liver tissues from sham-inoculated chickens. The heart and liver samples of FAdV-4-inoculated chickens showed many FAdV-4 antigen-positive cells (arrow f for inflammatory infiltration, arrow g for inclusion bodies, and arrow h for degenerated vacuoles).
Pathogenicity of hvFAdV-4 Strain GD616 in Different-Aged SPF Chickens
To further evaluate whether the pathogenicity of hvFAdV-4 was age-dependent in chickens, we used 106.5 TCID50/dose of the hvFAdV-4 strain GD616 to intramuscularly inoculate SPF chickens aged 7 to 180 d. For 7- to 42-day-old chickens, the morbidity and mortality rates were 100% within 5 d post-hvFAdV-4 inoculation (Figure 5A). The morbidity and mortality rates were 100% vs. 90%, 80% vs. 50%, 70% vs. 30%, and 60% vs. 20% for 59-, 80-, 120-, and 180-day-old hvFAdV-inoculated chickens, respectively (Figure 5A). The morbidity and mortality rates after hvFAdV-4 infection were inversely proportional to the age of the chickens. Only 60% morbidity and 20% mortality were observed in the 180-day-old hvFAdV-4-inocluated chickens. Figure 5B shows the mean copies of viral genomic DNA for each group of 5 randomly selected liver samples of affected or dead chickens. The maximum mean viral genomic copies (more than 108.5 copies/mg liver tissue) were observed in the 28- and 35-day-old hvFAdV-4-inoculated groups (Figure 5B), whereas the minimum mean viral genomic copies (107.0 copies/mg liver tissues) were observed in the 7- and 180-day-old hvFAdV-4-inoculated chickens (Figure 5B). The amount of viral genomic DNA from hvFAdV-4-inoculated chickens aged 21 to 80 d was approximately 107-1010 copies/mg liver tissues. In addition, the amounts of viral genomic DNA from some affected or dead chickens aged 14 d or less and 120 d or more were below 106 copies/mg liver tissue. The liver tissues from hvFAdV-4-inoculated chickens aged 21, 59, and 180 d were subjected to histological examination. Liver lesions were characterized by a large number of inclusion bodies and lipid droplets in the hepatocytes (Figure 5C). The numbers of inclusion bodies from the 180-day-old inoculated chickens were less than those of the 21-day-old inoculated chickens. IHC staining showed that a large amount of positive viral signals for the FAdV-4 antigen was widely distributed in hepatic cells (Figure 5C). The numbers of FAdV-4-positive signals in the 180-day-old inoculated chickens were significantly lower than those in the 21- and 59-day-old inoculated chickens (Figure 5C). These results indicated that the hvFAdV-4 infection-induced pathogenicity was associated with the ages of the chickens, with the maximum viral load and 100% morbidity and mortality being at 42 or less d of age, but 60% morbidity and 20% mortality were found in 180-day-old FAdV-4-infected chickens.
Figure 5.
Pathogenicity of the FAdV-4 strain GD616 in chickens of different ages. (A) The survival rates for 7-, 14-, 21-, 28-, 35-, 42-, 59-, 80-, 120-, and 180-day-old SPF chickens after GD616 infection when administered a 106.5 TCID50 dose regardless of the chicken ages. (B) The mean FAdV-4 genomic DNA copies in liver samples of the infected chickens aged 7–180 d after a 106.5 TCID50 dose of GD616 infection. (C) Liver lesions from a GD616-inoculated chicken aged 21, 59, or 180 d showed a great amount of inclusion bodies accompanied by vacuolar degeneration and nuclear enlargement. Immunohistochemical staining of the liver tissues from the FAdV-4-inoculated chickens aged 21, 59, or 180 d showed many positive cells for the FAdV-4 antigen. No histopathological lesions or FAdV-4 antigen-positive signals were found in the liver sections of the sham-inoculated chickens.
DISCUSSION
Recently, an important emerging disease with severe HHS caused by hvFAdV-4 infection has spread rapidly in many broiler and layer farms in Chinese poultry-rearing regions, resulting in severe economic losses. In the present study, a hypervirulent strain GD616 was isolated from Chinese native meat-type chickens and classified as the serotype 4 hvFAdV-4 strain on the basis of phylogenetic analysis of the whole genome (Figure 2). The whole genome sequencing of the GD616 exhibited deletions of ORF19 and ORF27, and possessed specific molecular signatures located in Hexon and Fiber-2 proteins, as observed for those of the recently-reported highly virulent FAdV-4 isolates in China (Table 1). The hypervirulent FAdV-4 isolates were highly pathogenic to 20- to 60-day-old chickens (Yin et al., 2020). In the present study, infection of chickens with the hvFAdV-4 strain GD616 showed age-relatedness, as evidenced by 100% morbidity and mortality for chickens aged 42 d or less but 60% morbidity and 20% mortality for chickens aged 180 d (Figure 5). These results suggest that the FAdV-4 isolate GD616 from local meat-type chickens was a hypervirulent FAdV serotype 4 and its pathogenicity to chickens occurs in an age-dependent manner.
The FAdV-4 strain GD616 identified in the present study possessed a common ancestor that belongs to the recently reported hypervirulent FAdV serotype 4 strains in China on the whole genome. Similar to the strain GD616, ORF19 and ORF27 deletions of nucleotides were found in almost all of the hvFAdV-4 viruses recently reported in China, including JSJ13 (Zhao et al., 2015), HLJDAd15 (Pan et al., 2017), GDMZ (Chen et al., 2019), and AH-F19 (Yin et al., 2020), but were not found in the highly virulent strain MX-SHP90 (Vera-Hernández et al., 2016) outside China or in the pathogenic strain Kr-Changnyeong or the low or nonpathogenic strains ON1, KR5 (Valle et al., 2020), and B1-7. These deletion mutant strains without ORF19 and ORF27 are emerging as novel FAdV-4 genotypes prevalent in China due to their adaption to hosts and environments (Ye et al., 2016). The ORF19 and ORF27 deletion might be responsible for viral evolution and pathogenicity (Liu et al., 2016; Ye et al., 2016; Pan et al., 2017). However, a recent study suggested that this deletion is nonessential for the enhanced pathogenicity of hvFAdV-4 isolates using the clustered regularly interspaced short palindromic repeat (CRISPR) and CRISPR-associated protein 9 system (Pan et al., 2018). By acting as the major structural proteins of the FAdV, Hexon and Fiber-2 are considered to associate with the virulence of emerging and hypervirulent FAdV-4 strains (Zhang et al., 2018). The Hexon protein of the strain GD616 showed high homology to those of the highly pathogenic strains recently isolated in China. Furthermore, the Hexon protein of the hvFAdV-4 isolates in China showed 3 amino acid substitutions at 193, 195, and 842 with respect to those of the mildly- and nonpathogenic FAdV-4 strains (Table 1). However, these 3 amino acid substitutions are different from those of the Hexon protein in the highly pathogenic FAdV-4 strains MX-SHP95 (Vera-Hernández et al., 2016) and Kr-Changnyeong (Park et al., 2017). Thus, amino acid mutations in the Hexon protein might be unrelated to hvFAdV-4 virulence. The Fiber-2 protein is associated with binding of cellular receptor and alterations in virulence (Pallister et al., 1996). D219, T300, and T380 in the Fiber-2 protein may be conserved in the pathogenic FAdV-4 strains and are associated with virulence when compared to that of nonpathogenic isolates (Mase et al., 2010; Marek et al., 2012; Vera-Hernández et al., 2016). However, A219 and T300 occur in the Fiber-2 protein of a hvFAdV-4 strain GX-1 (Ren et al., 2019) and nonpathogenic FAdV-4 strain B1-7, respectively, while A380 is located in that of the pathogenic strain Kr-Changnyeong. This indicates that D219, T300, and T380 may not contribute to FAdV-4 virulence. In the present study, we found that T261, T391, N459, and L478 in the Fiber-2 protein are conserved among the recently isolated hypervirulent FAdV-4 isolates in China (Table 1). Four amino acid substitutions (T261, T391, N459, and L478) in the Fiber-2 protein also occurred in hypervirulent FAdV-4 isolates from Guangxi, China in 2017-2019 (Rashid et al., 2020). However, these 4 amino acid substitutions are different from those of the Fiber-2 protein in the highly virulent FAdV-4 strain MX-SHP95 (Vera-Hernández et al., 2016) and the pathogenic strain Kr-Changnyeong. Further analysis of Fiber-2 amino acid sequences revealed that 2 unique amino acid substitutions at positions T378 and A453 occurred in all the highly virulent FAdV-4 isolates (Table 1). However, the involvement of the unique putative genetic marker in the virulence of highly pathogenic FAdV-4 strains must be further investigated using a reverse genetic system for the identification of the critical site required for the high virulence of FAdV-4 viruses.
In the present study, we first used 21-day-old SPF chickens to inoculate the hypervirulent FAdV-4 strain GD616 at various doses of 101.5-6.5 TCID50 and found that the morbidity and mortality of inoculated chickens at a dose of 102.5 or higher TCID50 were 100% within 7 d postinfection (Figure 4), indicating the mortality in inoculated chickens in a dose-dependent fashion. The mortablity of the strain GD616-inoculated chickens was identical to those in other hypervirulent FAdV-4 strains, such as MX-SHP95 (Vera-Hernández et al., 2016), HB1501 (Ruan et al., 2018), and GX-1 (Ren et al., 2019), when administered to 21-day-old SPF chickens with a lower dose (102.5 vs. 104.0 or 105.0 TCID50). Furthermore, the clinicopathological alterations caused by the FAdV-4 strain GD616 mainly consisted of substantially straw-colored or transparent fluid accumulated in the pericardial sac and IBH with hemorrhage or necrotic foci as observed in other highly virulent FAdV-4 isolates (Zhao et al., 2015; Li et al., 2016; Vera-Hernández et al., 2016; Pan et al., 2017; Ruan et al., 2018; Chen et al., 2019; Ren et al., 2019; Sun et al., 2019; Yin et al., 2020). These results indicated that the strain GD616 is a hypervirulent FAdV-4 virus that is highly pathogenic to chickens.
In this study, we further determined whether the pathogenicity of the hvFAdV-4 virus was related to the age of chickens and found that the pathogenicity of the hvFAdV-4 strain GD616 to chickens was dependent on their ages when administered to chickens aged 7 to 180 d with the strain GD616 at a dose of 106.5 TCID50 (Figure 5). FAdV strains instigated subclinical and clinical infections or severe diseases in chickens depending on their virulence. Hydropericardium hepatitis syndrome caused by hvFAdV-4 was mainly observed in broilers aged 21 to 35 d as well as layer and breeder pullets with mortality rates with a range of 20% to 80% (Zhao et al., 2015; Schachner et al., 2018). The 100% mortality for 35-, 98-, and 120-day-old chickens is caused by highly virulent FAdV-4 strains SD1511 (Mo et al., 2019), GDMZ (Chen et al., 2019), and AH-F19 (Yin et al., 2020), respectively. The present study further found that 60% morbidity and 20% mortality remained in the 180-day-old hvFAdV-4-inocluated chickens (Figure 5). These results enrich the current knowledge surrounding the pathogenicity of the hvFAdV-4 virus to chickens aged 180 d.
In summary, we characterized as a highly pathogenic FAdV-4 strain GD616 from a case of HPS in a 25-day-old meat-type flock affected by FAdV-4 in Guangdong, China. The whole genome of the GD616 strain shares high homology with those in the recently isolated highly virulent FAdV-4 strains in China, with natural deletions of ORF19 and ORF27. A comparative analysis revealed that 2 unique amino acid residues at positions 378 and 453 of the Fiber-2 protein might be associated with virulence due to its occurrence in all the highly virulent FAdV-4 isolates only. Animal experiments further showed the pathogenicity of the hvFAdV-4 strain GD616 to chickens dependent on days of age and 180-day-old chickens still exhibited an HPS-like clinicopathology with 60% morbidity and 20% mortality. These results enrich the current knowledge surrounding the pathogenicity of the hvFAdV-4 virus in chickens at a variety of ages, which is beneficial for the selection of suitably aged chickens for the evaluation of FAdV-4 vaccines.
Acknowledgments
ACKNOWLEDGMENTS
This work was supported by the National Key Research and Development Program of China (2016YFD0500806), China Agriculture Research System (CARS-41), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
DISCLOSURES
The authors declare no conflicts of interest.
REFERENCES
- Abe T., Nakamura K., Tojo H., Mase M., Shibahara T., Yamaguchi S., Yuasa N. Histology, immunohistochemistry, and ultrastructure of hydropericardium syndrome in adult broiler breeders and broiler chicks. Avian Dis. 1998;42:606–612. [PubMed] [Google Scholar]
- Asthana M., Chandra R., Kumar R. Hydropericardium syndrome: current state and future developments. Arch. Virol. 2013;158:921–931. doi: 10.1007/s00705-012-1570-x. [DOI] [PubMed] [Google Scholar]
- Anjum A.D., Sabri M.A., Iqbal Z. Hydropericarditis syndrome in broiler chickens in Pakistan. Vet. Rec. 1989;124:247–248. doi: 10.1136/vr.124.10.247. [DOI] [PubMed] [Google Scholar]
- Asrani R.K., Gupta V.K., Sharma S.K., Singh S.P., Katoch R.C. Hydropericardium-hepatopathy syndrome in Asian poultry. Vet. Rec. 1997;141:271. doi: 10.1136/vr.141.11.271. [DOI] [PubMed] [Google Scholar]
- Balamurugan V., Kataria J.M. The hydropericardium syndrome in poultry–a current scenario. Vet. Res. Commun. 2004;28:127–148. doi: 10.1023/b:verc.0000012115.86894.1e. [DOI] [PubMed] [Google Scholar]
- Chen Z., Shi S., Qi B., Lin S., Chen C., Zhu C., Huang Y. Hydropericardium syndrome caused by fowl adenovirus serotype 4 in replacement pullets. J. Vet. Med. Sci. 2019;81:245–251. doi: 10.1292/jvms.18-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Zhao M., Duan X., Wang Y., Cao H., Li X., Zheng S.J. Requirement of cellular protein CCT7 for the replication of fowl adenovirus serotype 4 (FAdV-4) in Leghorn male hepatocellular cells via interaction with the viral Hexon protein. Viruses. 2019;11:107. doi: 10.3390/v11020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin B.D., Nagy É. Coding potential and transcript analysis of fowl adenovirus 4: insight into upstream ORFs as common sequence features in adenoviral transcripts. J. Gen. Virol. 2011;92:1260–1272. doi: 10.1099/vir.0.030064-0. [DOI] [PubMed] [Google Scholar]
- Helena G., Krell P.J., Nagy É. Comparison of fiber gene sequences of inclusion body hepatitis (IBH) and non-IBH strains of serotype 8 and 11 fowl adenoviruses. Virus Genes. 2014;48:74–80. doi: 10.1007/s11262-013-0995-y. [DOI] [PubMed] [Google Scholar]
- Hess M. Detection and differentiation of avian adenoviruses: a review. Avian Pathol. 2000;29:195–206. doi: 10.1080/03079450050045440. [DOI] [PubMed] [Google Scholar]
- Hess M., Raue R., Prusas C. Epidemiological studies on fowl adenoviruses isolated from cases of infectious hydropericardium. Avian Pathol. 1999;28:433–439. doi: 10.1080/03079459994443. [DOI] [PubMed] [Google Scholar]
- Jadhao S.J., Deepak J.N., Kataria J.M., Kataria R.S., Verma K.C. Characterisation of fowl adenoviruses from chickens affected with infectious hydropericardium during 1994-1998 in India. Indian J. Exp. Biol. 2003;41:321–327. [PubMed] [Google Scholar]
- Kaján G.L., Kecskem´eti S., Harrach B., Benko M. Molecular typing of fowl adenoviruses, isolated in Hungary recently, reveals high diversity. Vet. Microbiol. 2013;167:357–363. doi: 10.1016/j.vetmic.2013.09.025. [DOI] [PubMed] [Google Scholar]
- Kim J.N., Seong H.B., Kim M.J., Kim J.J., Sung H.W., Mo I.P. Outbreaks of hydropericardium syndrome and molecular characterization of korean fowl adenoviral isolates. Avian Dis. 2008;52:526–530. doi: 10.1637/8178-112207-Case. [DOI] [PubMed] [Google Scholar]
- Li H., Wang J., Qiu L., Han Z., Liu S. Fowl adenovirus species C serotype 4 is attributed to the emergence of hepatitis-hydropericardium syndrome in chickens in China. Infect. Genet. Evol. 2016;45:230–241. doi: 10.1016/j.meegid.2016.09.006. [DOI] [PubMed] [Google Scholar]
- Li P.H., Zheng P.P., Zhang T.F., Wen G.Y., Shao H.B., Luo Q.P. Fowl adenovirus serotype 4: epidemiology, pathogenesis, diagnostic detection, and vaccine strategies. Poultry Sci. 2017;96:2630–2640. doi: 10.3382/ps/pex087. [DOI] [PubMed] [Google Scholar]
- Lim T.H., Lee H.J., Lee D.H., Lee Y.N., Park J.K., Youn H.N., Kim M.S., Youn H.S., Lee J.B., Park S.Y. Identification and virulence characterization of fowl adenoviruses in Korea. Avian Dis. 2011;55:554–560. doi: 10.1637/9730-032011-Reg.1. [DOI] [PubMed] [Google Scholar]
- Liu Y., Wan W., Gao D., Li Y., Yang X., Liu H., Yao H., Chen L., Wang C., Zhao J. Genetic characterization of novel fowl aviadenovirus 4 isolates from outbreaks of hepatitis-hydropericardium syndrome in broiler chickens in China. Emerg. Microbes Infect. 2016;5:e117. doi: 10.1038/emi.2016.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek A., Nolte V., Schachner A., Berger E., Schlotterer C., Hess M. Two fibre genes of nearly equal lengths are a common and distinctive feature of Fowl adenovirus C members. Vet. Microbiol. 2012;156:411–417. doi: 10.1016/j.vetmic.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Mase M., Nakamura K., Imada T. Characterization of Fowl adenovirus serotype 4 isolated from chickens with hydropericardium syndrome based on analysis of the short fiber protein gene. J. Vet. Diagn. Invest. 2010;22:218–223. doi: 10.1177/104063871002200207. [DOI] [PubMed] [Google Scholar]
- Mase M., Nakamura K., Minami F. Fowl adenoviruses isolated from chickens with inclusion body hepatitis in Japan, 2009-2010. J. Vet. Med. Sci. 2012;74:1087–1089. doi: 10.1292/jvms.11-0443. [DOI] [PubMed] [Google Scholar]
- Mazaheri A., Prusas C., Voss M., Hess M. Some strains of serotype 4 fowl adenoviruses cause inclusion body hepatitis and hydropericardium syndrome in chickens. Avian Pathol. 1998;27:269–276. doi: 10.1080/03079459808419335. [DOI] [PubMed] [Google Scholar]
- Meulemans G., Boschmans M., Van dB T.P., Decaesstecker M. Polymerase chain reaction combined with restriction enzyme analysis for detection and differentiation of fowl adenoviruses. Avian Pathol. 2001;30:655–660. doi: 10.1080/03079450120092143. [DOI] [PubMed] [Google Scholar]
- Mittal D., Jindal N., Tiwari A.K., Khokhar R.S. Characterization of fowl adenoviruses associated with hydropericardium syndrome and inclusion body hepatitis in broiler chickens. Virusdisease. 2014;25:114–119. doi: 10.1007/s13337-013-0183-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo K.K., Lyu C.F., Cao S.S., Li X., Xing G., Yan Y., Zheng X.J., Liao M., Zhou J.Y. Pathogenicity of an FAdV-4 isolate to chickens and its genomic analysis. J. Zhejiang Univ. Sci. B. 2019;20:740–752. doi: 10.1631/jzus.B1900070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niczyporuk J.S. Phylogenetic and geographic analysis of fowl adenovirus field strains isolated from poultry in Poland. Arch. Virol. 2016;161:33–42. doi: 10.1007/s00705-015-2635-4. [DOI] [PubMed] [Google Scholar]
- Pallister J., Wright P., Sheppard M. A single gene encoding the fiber is responsible for variations in virulence in the fowl adenoviruses. J. Virol. 1996;70:5115–5122. doi: 10.1128/jvi.70.8.5115-5122.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q., Liu L., Gao Y., Liu C., Qi X., Zhang Y., Wang Y., Li K., Gao L., Wang X., Cui H. Characterization of a hypervirulent fowl adenovirus 4 with the novel genotype newly prevalent in China and establishment of reproduction infection model of hydropericardium syndrome in chickens. Poul. Sci. 2017;96:1581–1588. doi: 10.3382/ps/pew431. [DOI] [PubMed] [Google Scholar]
- Pan Q., Wang J., Gao Y., Cui H., Liu C., Qi X., Zhang Y., Wang Y., Wang X. The natural large genomic deletion is unrelated to the increased virulence of the novel genotype fowl adenovirus 4 recently emerged in China. Viruses. 2018;10:494. doi: 10.3390/v10090494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.S., Lim I.S., Kim S.K., Kim T.K., Park C.K., Yeo S.G. Molecular analysis of the hexon, penton base, and fiber-2 genes of Korean fowl adenovirus serotype 4 isolates from hydropericardium syndrome-affected chickens. Virus Gen. 2017;53:111–116. doi: 10.1007/s11262-016-1393-z. [DOI] [PubMed] [Google Scholar]
- Rahimi M., Minoosh S.H.Z. Adenovirus-like inclusion body hepatitis in a flock of broiler chickens in Kermanshah province, Iran. Vet. Res. Forum. 2015;6:95–98. [PMC free article] [PubMed] [Google Scholar]
- Rashid F., Xie Z., Zhang L., Luan Y., Luo S., Deng X., Xie L., Xie Z., Fan Q. Genetic characterization of fowl aviadenovirus 4 isolates from Guangxi, China, during 2017-2019. Poult. Sci. 2020;99:4166–4173. doi: 10.1016/j.psj.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G., Wang H., Yan Y., Liu F., Huang M., Chen R. Pathogenicity of a fowl adenovirus serotype 4 isolated from chickens associated with hydropericardium-hepatitis syndrome in China. Poul. Sci. 2019;98:2765–2771. doi: 10.3382/ps/pez042. [DOI] [PubMed] [Google Scholar]
- Ruan S, Zhao J, He Z, Yang H, Zhang G. Analysis of pathogenicity and immune efficacy of fowl adenovirus serotype 4 isolates. Poult Sci. 2018;97:2647–2653. doi: 10.3382/ps/pey113. [DOI] [PubMed] [Google Scholar]
- Russell W.C. Adenoviruses: update on structure and function. J. Gen. Virol. 2009;90:1–20. doi: 10.1099/vir.0.003087-0. [DOI] [PubMed] [Google Scholar]
- Schachner A., Matos M., Grafl B., Hess M. Fowl adenovirus-induced diseases and strategies for their control–a review on the current global situation. Avian Pathol. 2018;47:1–77. doi: 10.1080/03079457.2017.1385724. [DOI] [PubMed] [Google Scholar]
- Shah M.S., Ashraf A., Khan M.I., Rahman M., Habib M., Chughtai M.I., Qureshi J.A. Fowl adenovirus: history, emergence, biology and development of a vaccine against hydropericardium syndrome. Arch. Virol. 2017;162:1833–1843. doi: 10.1007/s00705-017-3313-5. [DOI] [PubMed] [Google Scholar]
- Sun J., Zhang Y., Gao S., Yang J., Tang Y., Diao Y. Pathogenicity of fowl adenovirus serotype 4 (FAdV-4) in chickens. Infect. Genet. Evol. 2019;75 doi: 10.1016/j.meegid.2019.104017. [DOI] [PubMed] [Google Scholar]
- Toogood C.I.A., Crompton J., Hay R.T. Antipeptide antisera define neutralizing epitopes on the adenovirus hexon. J. Gen. Virol. 1992;73:1429–1435. doi: 10.1099/0022-1317-73-6-1429. [DOI] [PubMed] [Google Scholar]
- Toro H., Prusas C., Raue R., Cerda L., Geisse C., González C., Hess M. Characterization of fowl adenoviruses from outbreaks of inclusion body hepatitis/hydropericardium syndrome in Chile. Avian Dis. 1999;43:262–270. [PubMed] [Google Scholar]
- Valle F.P.D., Camba S.I., Umali D.V., Sasai K., Shirota K., Hiromitsu K., Tajima T. Molecular and pathologic characterization of avian adenovirus isolated from the oviducts of laying hens in eastern Japan. Poult. Sci. 2020;99:2459–2468. doi: 10.1016/j.psj.2019.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera-Hernández P.F., Morales-Garzón A., Cortés-Espinosa D.V., Galiote-Flores A., García-Barrera L.J., RodríguezGalindo E.T., Toscano-Contreras A., Lucio-Decanini E., Absalón A.E. Clincopathological characterization and genomic sequence differences observed in a highly virulent fowl viadenovirus serotype 4. Avian Pathol. 2016;45:73–81. doi: 10.1080/03079457.2015.1125443. [DOI] [PubMed] [Google Scholar]
- Wei Z., Liu H., Diao Y., Li X., Zhang S., Gao B., Tang Y., Hu J., Diao Y. Pathogenicity of fowl adenovirus (FAdV) serotype 4 strain SDJN in Taizhou geese. Avian Pathol. 2019;31:1–9. doi: 10.1080/03079457.2019.1625305. [DOI] [PubMed] [Google Scholar]
- Ye J., Liang G., Zhang J., Wang W., Song N., Wang P., Zheng W., Xie Q., Shao H., Wan Z., Wang C., Chen H., Gao W., Qin A. Outbreaks of serotype 4 fowl adenovirus with novel genotype, China. Emerg. Microbes. Infect. 2016;5:e50. doi: 10.1038/emi.2016.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Dong J., Wu J., Zhu R., Wang Z., Wang B., Wang L., Wang Z., Zhang H., Wu H. Interaction between hexon and L4-100K determines virus rescue and growth of hexon-chimeric recombinant Ad5 vectors. Sci. Rep. 2016;6:22464. doi: 10.1038/srep22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin D., Xue M., Yang K., Xiong X., Geng S., Tu J., Song X., Shao Y., Wang G., Qi K. Molecular characterization and pathogenicity of highly pathogenic fowl adenovirus serotype 4 isolated from laying flock with hydropericardium-hepatitis syndrome. Microb. Pathog. 2020;147 doi: 10.1016/j.micpath.2020.104381. [DOI] [PubMed] [Google Scholar]
- Zhao J., Zhong Q., Zhao Y., Hu Y.X., Zhang G.Z. Pathogenicity and complete genome characterization of fowl adenoviruses isolated from chickens associated with inclusion body hepatitis and hydropericardium syndrome in China. PLoS One. 2015;10 doi: 10.1371/journal.pone.0133073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan F., Hou L., Wei L., Quan R., Wang J., Liu H., Liu J. Fowl adenovirus serotype 4 induces hepatic steatosis via activation of liver X receptor-α. J. Virol. 2021;95 doi: 10.1128/JVI.01938-20. e01938-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu R.X., Tian K., Wang Z., Yang X., Gao D.S., Zhang Y.M., Fu J, Wang H.L., Zhao J. Fiber2 and hexon genes are closely associated with the virulence of the emerging and highly pathogenic fowl adenovirus 4. Emerg Microbes Infect. 2018;7:199. doi: 10.1038/s41426-018-0203-1. [DOI] [PMC free article] [PubMed] [Google Scholar]