Abstract
The characterisation of the HIV-1 reservoir, which consists of replication-competent integrated proviruses that persist on antiretroviral therapy (ART), is made difficult by the rarity of intact proviruses relative to those that are defective. While the only conclusive test for the replication-competence of HIV-1 proviruses is carried out in cell culture, genetic characterization of genomes by near full-length (NFL) PCR and sequencing can be used to determine whether particular proviruses have insertions, deletions, or substitutions that render them defective. Proviruses that are not excluded by having such defects can be classified as genetically intact and, possibly, replication competent. Identifying and quantifying proviruses that are potentially replication-competent is important for the development of strategies towards a functional cure. However, to date, there are no programs that can be incorporated into deep-sequencing pipelines for the automated characterization and annotation of HIV genomes. Existing programs that perform this work require manual intervention, cannot be widely installed, and do not have easily adjustable settings. Here, we present HIVIntact, a python-based software tool that characterises genomic defects in NFL HIV-1 sequences, allowing putative intact genomes to be identified in-silico. Unlike other applications that assess the genetic intactness of HIV genomes, this tool can be incorporated into existing sequence-analysis pipelines and applied to large next-generation sequencing datasets.
Introduction: the need for a stand-alone HIV intactness tool that can be integrated into existing pipelines
HIV-1 replication is an error-prone process that often results in the stable integration of aberrant proviruses into the host genome [1]. To date, few consequences of integrated, defective HIV-1 genomes have been described [2]. However, integrated intact genomes are the source of high levels of viremia prior to ART initiation, persistent low-level viremia during ART, and rebound viremia when ART is interrupted. These proviruses are referred to as the HIV-1 reservoir and are the target of potential curative strategies. Analyses of near full length (NFL) proviral genomes on ART reveal that 95–99% contain some manner of defect—usually APOBEC3G-mediated hypermutation or large internal deletions up to 8.5 kb [3–6]. APOBEC-medicated hypermutation typically results in premature STOP codons in high tryptophan regions, as well as numerous missense mutations. Major defects such as hypermutation and large internal deletions are easily identifiable, but minor defects, such as packaging signal deletions, mutations in the major splice-donor site (MSD), or mutations in the rev-response element (RRE) are often observed, but less obvious. Since the vast majority of infected cells on ART contain proviruses with some combination of these lethal defects, interrogation of the HIV-1 reservoir is challenging: one must not only identify infected cells in a background of uninfected cells, but also identify those that contain intact, replication-competent proviruses.
One approach to characterising the HIV-1 reservoir without having to find infected cells is to measure and sequence residual viremia in patients on suppressive ART. Residual viremia is defined as low-level viremia below the threshold of commercial ultrasensitive viral load assays. Cells infected with replication-competent proviruses typically do not express viral RNA during treatment, but a small proportion can become activated to produce virus particles [7]. This activation explains the residual release of viruses, not suppressible by ART, which likely results in viral rebound once treatment is interrupted [8]. Although some defective proviruses could be transcribed and packaged as virions and also contribute to residual viremia [9, 10], in rare cases of persistent clonal viremia high enough to analyze, the virus is replication competent [11, 12]. Another approach to characterising the reservoir is to recover infectious virus from T cells collected from donors on ART using the quantitative viral outgrowth assay (qVOA) [13]. Although highly useful, these techniques have their drawbacks. HIV-1 plasma requires high volume samples and QVOA significantly underestimates the proportion of cells that harbor replication-competent proviruses due to inefficient latency-reversal [3, 14].
Recently, Gaebler et al. [14] and Bruner et al. [3] proposed two quantitative techniques for measuring the HIV-1 reservoir using qPCR (Q4PCR) and ddPCR (IPDA) respectively. Although these techniques can more accurately quantify the HIV-1 reservoir (reviewed in [15]), both are primer/probe-reliant and have mismatches to some donors, as recently described by Kinloch et al. [16]. Although NFL proviral sequencing mitigates some of these issues, it includes its own challenges. For one, the high genetic variation of HIV-1 necessitates the design of large primer panels and sometimes even patient-specific primers. Sanger sequencing approaches, due to their reliance on many primers, are ill-suited to accurately characterize proviral sequences [3, 5, 17–19]. The development of next-generation and third-generation sequencing approaches have overcome these challenges by being less reliant on primers. Deep sequencing is also higher throughput and less expensive than Sanger, making it better suited to NFL analyses [6, 17, 18]. Due to these improvements, it is now possible to assess the putative intactness of many proviral genomes in cells collected from donors on or off ART. However, to date, pipelines that assemble the sequencing reads of NFL HIV-1 genomes do not include a component to annotate the genomes or to infer their intactness.
Although no tools exist that can be incorporated into next-generation sequencing pipelines, there are two freely available tools for the independent bioinformatic determination of HIV-1 intactness, a web-based program called HIV-ProSeqIT [20, 21] and an R-based program called HIVSeqinR [22]. Neither of these tools are intended for high throughput analyses of HIV genomes, and neither can be easily adjusted to include or exclude more stringent checks. Here, we present HIVIntact, a command-line program written in python 3.7 that only requires MAFFT [23] and Biopython [24] to perform an intactness check, allowing ease of integration into existing deep sequencing assembly pipelines. Integration into existing pipelines allows proviral annotation and intactness inference to occur in an automated and high throughput manner, making characterization of the HIV-1 reservoir significantly more accessible. The incorporation of HIVIntact into high throughput methods for NFL proviral single-genome sequencing may constitute the most accurate method to date for measuring and characterizing the HIV-1 reservoir on ART since the NFL sequences can be assessed for minor mutations that are not detected by qPCR and ddPCR assays. Furthermore, the PCR products identified as intact by the tool can be tested in vitro for replication-competence by transfection into permissive cell lines.
Pipeline definition
Intact open reading frames (ORFs)
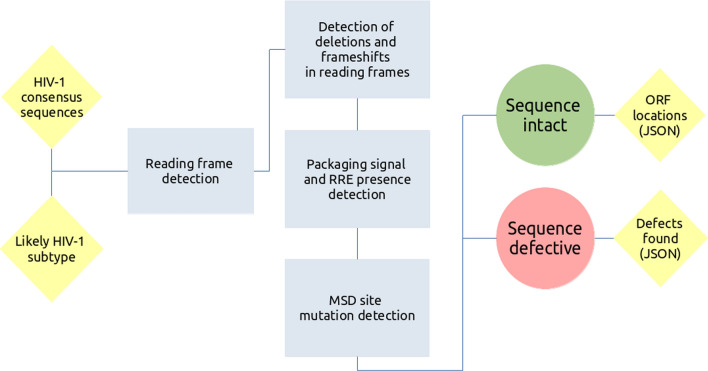
HIVIntact is invoked with a single compulsory parameter: the likely subtype of the NFL HIV-1 sequence. Reference sequences for subtypes A, B, C, D, F, G and H are available by default within the pipeline. The subtype parameter is needed to obtain the best possible estimate of the open reading frames (ORFs) and alignment of accessory genes. Each query sequence is checked for its orientation with respect to the chosen reference sequence. The NFL HIV-1 sequence is aligned in the correct orientation and is first checked for the three large ORFs (gag, pro-pol, and env) in the expected locations. If all three ORFs are present (presence being defined as the absence of a premature stop codon), the sequence is considered to have passed this first “intactness” test. The locations of the ORFs are reported in HXB2 coordinates.
Each large ORF in the candidate intact sequence is then checked for large internal deletions. Deletions amounting to up to 30 bases (consecutive or not) are permitted in gag and pol, while deletions amounting to up to 100 bases are permitted in env, in line with recommendations made in Patro et al. [17] and Shao et al. [20]. The discrepancy in allowed deletion size reflects the greater variability of the env gene in vivo. If all three ORFs contain deletions of fewer than the indicated number of bases, then the sequence is considered to have passed this phase of the intactness test.
Each large ORF is then checked for indels that introduce frameshift mutations: a combination of insertions and deletions that shifts the frame. The presence of an indel of any length in a large ORF that shifts the frame results in failure of the intactness test.
Finally, the six smaller ORFs (vif, vpr, tat, rev, vpu, nef) are also checked for “completeness”. In the case of tat and rev, each of the two exons in the ORF are checked independently. Information on ORF completeness, and the presence of indels and frameshifts within the ORF bounds, is reported. However, because it is not yet known what mutations are tolerated in these genes, this check is not considered by default in the inferred proviral intactness (it may be switched on the command line). Future studies are required to determine the effect of mutations and indels in the small ORFs, so that they can be included in inferred intactness estimations. HIVIntact is well-placed to play a role in these studies by reporting potential defects in these ORFs that may or may not contribute to intactness, which may then be tested in vitro.
Other genomic structure checks
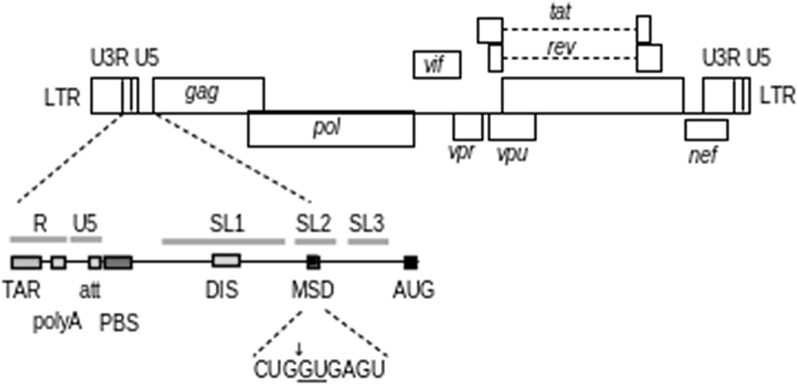
Other functional genomic structures are needed for viral replication in vivo. These include an intact packaging signal (PSI), an unmutated major splice donor (MSD) site (Fig. 1), and intact rev-response element (RRE) [25].
Fig. 1.
An illustration of the HIV-1 5′ UTR, showing major functional elements, and highlighting the major splice donor site, which must remain unmutated to produce infectious virus
The PSI locus is defined from positions 680–809 in a subtype B reference. A deletion in the PSI as small as 15 nt can render a provirus defective for replication [5]. To err on the side of caution, the defined deletion tolerance in HIVIntact for the PSI is set to 10 nt. Studies are needed to determine the most accurate tolerance for deletions in PSI. We settled on this maximum so as not to omit sequences that should be tested for replication competence in downstream analyses. Users should take note of these estimates in their reports of sequence intactness.
Deletions and mutations in the MSD (located at position 743) have also been demonstrated to render proviruses defective [5, 26]. Because no systematic study has been completed on the effect of all possible MSD mutations, we disallow any mutations in this region. However, both this check and the check for PSI intactness may be disabled by command line arguments.
The RRE locus is defined from positions 7755 to 8020 in a subtype B reference. Because truncations of the first and last 60 nt of the RRE were demonstrated to only reduce replication efficiency by 2.5-fold [27], we chose to allow tolerances in HIVIntact of 39 nt on each end of the RRE and a tolerance of 21 nt insertion/deletion adjacent the ends of the RRE.
More studies are needed to define regions of the RRE that are required for replication competence. Therefore, as with the PSI and MSD checks above, this check may be disabled at the command line by users. Of note, sequences with smaller deletions in domains III–V of the RRE are not included in the pipeline but could render the provirus defective.
Hypermutation check
An implementation of the HYPERMUT algorithm [28] is included as an additional piece of information. Sequences that fail the Fisher’s exact test for hypermutation are marked as hypermutated in the error output. However, because the intactness of a hypermutated sequence with no premature stop codons in any reading frame cannot be reliably inferred, the hypermutation status of a sequence is not counted towards overall intactness unless the hypermutation introduces a premature stop codon.
Pipeline implementation
HIVIntact is implemented as a Python 3 script, using Biopython [24] for sequence input and output. The pipeline depends on MAFFT [23] for alignment purposes but is otherwise a standalone tool. The pipeline can be installed globally, as on a high-performance computing cluster, or locally on a personal computer, using a Python package manager.
HIVIntact uses a FASTA file as input. The file should contain one or more assembled NFL HIV provirus sequences. Ideally, these assembled sequences should include coverage of the packaging signal, but a check for its presence is optional and may be switched off on the command line for shorter sequences. The pipeline, once installed, can be called using the proviral intact command, e.g.: proviral intact—subtype B sequences.fasta.
Pipeline validation
To evaluate the ability of HIVIntact to infer intactness in NFL HIV-1 sequences, we tested all sequences uploaded to the Proviral Sequence Database (PSD) as of 29 September 2020 (https://psd.cancer.gov/intro.php) [20]. The PSD is an existing curated public database of NFL HIV-1 sequences developed and maintained by the National Cancer Institute (NCI). The database contained 4870 sequences at the time of downloading. Of the total sequences, 4143 were unique. The duplicate sequences result from different single-genome sequences obtained from the same donors. Of the unique sequences, 624 were labelled intact in the database and 3519 were labelled defective.
We assessed the full set of 4143 unique sequences with HIVIntact (Table 1). The run completed on a single core of an Intel(R) Core(TM) i7-4770HQ CPU @ 2.20 GHz in 5020 s (1 h, 23 min and 39 s), equivalent to a rate of 1.2 s per sequence per core. This run rate is conducive to automation as part of a larger pipeline.
Table 1.
A comparison of intactness inference in the NCI PSD [20] with results from HIVIntact
| Inferred intactness in the PSD | 1. Reported intactness by HIVIntact (excluding small ORFs) | 2. Reported intactness by HIVIntact (including small ORFs) | |
|---|---|---|---|
| Intact | 624 | 623 | 581 |
| Defective | 3519 | 3520 | 3562 |
| Uniquely intact | 3 | 2 | 2 |
The table reports how many sequences were called intact and defective in total in the PSD, as compared against HIVIntact in two modes: (1) assessing only the three major ORFs (gag, pol, env) and (2) including intactness checks for the 6 smaller ORFs (vif, vpr, tat, rev, vpu, nef). The table also reports how many sequences were called intact uniquely by only one tool, indicating a disagreement in intactness inference
Results excluding small ORFs
We initially ran HIVIntact checking for intactness only in the three major ORFs (gag, pol and env), where defects are well known to render provirus defective. We included checks for defaults in the PSI locus, the RRE locus and the MSD. In this mode, there was very good agreement between our tool and the annotations reported in the NCI PSD.
In total, when considering large ORFs only, five sequences had discordant intactness inference between the PSD and HIVIntact. Three sequences were inferred intact in the NCI PSD but defective by HIVIntact. All three were found to have frameshifts in large ORFs, which is the only default intactness check unique to HIVIntact. Sequence ID MN090882 contained a gag frameshift, while sequences KF526323.1 and MT033880.1 both contained frameshifts in env.
Two sequences were inferred defective in the NCI PSD but intact by HIVIntact. Sequence ID MN090886 contained a large, 54-base insertion in the pol gene. The PSD considers insertions > 50 bases in pol to be defective, while HIVIntact does not currently call sequences with in-frame insertions in the three large ORFs defective. Sequence ID MK114886.1 contains a 10-base deletion in the packaging signal. HIVIntact allows up to 10 base deletions in the packaging signal, while the PSD calls nonintact when the number of deletions is greater than 8.
Results including small ORFs
We then ran HIVIntact checking for intactness in all 9 ORFS (gag, pol, env, vif, vpr, tat, rev, vpu, nef). In this mode, a further 42 sequences were called nonintact due to defects in one of the 6 small ORFs. Of these errors, 24 were in vpu, 9 in nef, 6 in tat, 2 in vif and 1 in vpr. Further research is needed to quantify which defects in these ORFs genuinely render the virus replication incompetent.
Pipeline usage
The HIVIntact pipeline and test data may be downloaded from a public GitHub repository (https://github.com/ramics/HIVIntact) under an open-source MIT license. The authors welcome feedback and contributions.
The HIVIntact output includes two FASTA files labeled intact and non-intact. The pipeline also outputs the locations of the ORFs for each sequence despite intactness and a list of defects detected. ORFs and defects are reported in standardised JSON format, allowing bioinformaticians to easily access the results using downstream software applications.
Acknowledgements
We thank John Mellors for consultation on the development of HIVIntact. We also thank the reviewers for helpful comments, which improved both the software and the paper.
Authors’ contributions
IW and MB contributed equally to the development of the pipeline and the writing of the manuscript, under guidance from WS. WS-H provided insight into conserved regions in the HIV-1 5′ UTR and produced Fig. 1. GUvZ, JMC, and MFK provided guidance on the project and wrote the manuscript. All authors read and approved the final manuscript.
Funding
Research reported in this publication was supported by the U.S. National Institutes of Health and South African Medical Research Council through its U.S.-SA Program for Collaborative Biomedical Research (National Cancer Institute Grant No. U01CA200441, awarded to MFK). The content and findings reported/ illustrated herein are the sole deduction, view and responsibility of the researchers and do not reflect the official position and sentiments of the NIH and SAMRC.
Availability of data and materials
The HIVIntact pipeline and all test data, including a snapshot of the NCI PSD and a comparison script may be downloaded from a public GitHub repository (https://github.com/ramics/proviral-intactness) under an open-source MIT license, and is available for use. The code is written in Python3 and is platform-independent.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jacobs JL, Halvas EK, Tosiano MA, Mellors JW. Persistent HIV-1 viremia on antiretroviral therapy: measurement and mechanisms. Front Microbiol. 2019;15(10):2383. doi: 10.3389/fmicb.2019.02383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katano H, Sato Y, Hoshino S, Tachikawa N, Oka S, Morishita Y, Ishida T, Watanabe T, Rom WN, Mori S, Sata T. Integration of HIV-1 caused STAT3-associated B cell lymphoma in an AIDS patient. Microbes Infect. 2007;9(14–15):1581–1589. doi: 10.1016/j.micinf.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruner KM, Murray AJ, Pollack RA, Soliman MG, Laskey SB, Capoferri AA, Lai J, Strain MC, Lada SM, Hoh R, Ho YC, Richman DD, Deeks SG, Siliciano JD, Siliciano RF. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med. 2016;22(9):1043–1049. doi: 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antar AA, Jenike KM, Jang S, Rigau DN, Reeves DB, Hoh R, Krone MR, Keruly JC, Moore RD, Schiffer JT, Nonyane BA, Hecht FM, Deeks SG, Siliciano JD, Ho YC, Siliciano RF. Longitudinal study reveals HIV-1-infected CD4+ T cell dynamics during long-term antiretroviral therapy. J Clin Invest. 2020;130(7):3543–3559. doi: 10.1172/JCI135953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, Lai J, Blankson JN, Siliciano JD, Siliciano RF. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155(3):540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hiener B, Horsburgh BA, Eden JS, Barton K, Schlub TE, Lee E, von Stockenstrom S, Odevall L, Milush JM, Liegler T, Sinclair E, Hoh R, Boritz EA, Douek D, Fromentin R, Chomont N, Deeks SG, Hecht FM, Palmer S. Identification of genetically intact HIV-1 proviruses in specific CD4+ T cells from effectively treated participants. Cell Rep. 2017;21(3):813–822. doi: 10.1016/j.celrep.2017.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiegand A, Spindler J, Hong FF, Shao W, Cyktor JC, Cillo AR, Halvas EK, Coffin JM, Mellors JW, Kearney MF. Single-cell analysis of HIV-1 transcriptional activity reveals expression of proviruses in expanded clones during ART. Proc Natl Acad Sci. 2017;114(18):E3659–E3668. doi: 10.1073/pnas.1617961114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aamer HA, McClure J, Ko D, Maenza J, Collier AC, Coombs RW, Mullins JI, Frenkel LM. Cells producing residual viremia during antiretroviral treatment appear to contribute to rebound viremia following interruption of treatment. PLoS Pathog. 2020;16(8):e1008791. doi: 10.1371/journal.ppat.1008791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imamichi H, Smith M, Adelsberger JW, Izumi T, Scrimieri F, Sherman BT, Rehm CA, Imamichi T, Pau A, Catalfamo M, Fauci AS. Defective HIV-1 proviruses produce viral proteins. Proc Natl Acad Sci. 2020;117(7):3704–3710. doi: 10.1073/pnas.1917876117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rassler S, Ramirez R, Khoury N, Skowron G, Sahu GK. Prolonged persistence of a novel replication-defective HIV-1 variant in plasma of a patient on suppressive therapy. Virol J. 2016;13(1):1–3. doi: 10.1186/s12985-016-0617-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simonetti FR, Sobolewski MD, Fyne E, Shao W, Spindler J, Hattori J, Anderson EM, Watters SA, Hill S, Wu X, Wells D. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci. 2016;113(7):1883–1888. doi: 10.1073/pnas.1522675113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halvas EK, Joseph KW, Brandt LD, Guo S, Sobolewski MD, Jacobs JL, Tumiotto C, Bui JK, Cyktor JC, Keele BF, Morse GD. HIV-1 viremia not suppressible by antiretroviral therapy can originate from large T cell clones producing infectious virus. J Clin Investig. 2020;130(11):5847–5857. doi: 10.1172/JCI138099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaebler C, Lorenzi JC, Oliveira TY, Nogueira L, Ramos V, Lu CL, Pai JA, Mendoza P, Jankovic M, Caskey M, Nussenzweig MC. Combination of quadruplex qPCR and next-generation sequencing for qualitative and quantitative analysis of the HIV-1 latent reservoir. J Exp Med. 2019;216(10):2253–2264. doi: 10.1084/jem.20190896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eriksson S, Graf EH, Dahl V, Strain MC, Yukl SA, Lysenko ES, Bosch RJ, Lai J, Chioma S, Emad F, Abdel-Mohsen M, Hoh R, Hecht F, Hunt P, Somsouk M, Wong J, Johnston R, Siliciano RF, Richman DD, O'Doherty U, Palmer S, Deeks SG, Siliciano JD. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 2013;9(2):e1003174. doi: 10.1371/journal.ppat.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdel-Mohsen M, Richman D, Siliciano RF, Nussenzweig MC, Howell BJ, Martinez-Picado J, Chomont N, Bar KJ, Xu GY, Lichterfeld M, Alcami J. Recommendations for measuring HIV reservoir size in cure-directed clinical trials. Nat Med. 2020;26(9):1339–1350. doi: 10.1038/s41591-020-1022-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinloch NN, Ren Y, Alberto WD, Dong W, Khadka P, Huang SH, Mota TM, Wilson A, Shahid A, Kirkby D, Harris M. HIV-1 diversity considerations in the application of the intact proviral DNA assay (IPDA) Nat Commun. 2021;12(1):1. doi: 10.1038/s41467-020-20314-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patro SC, Brandt LD, Bale MJ, Halvas EK, Joseph KW, Shao W, Wu X, Guo S, Murrell B, Wiegand A, Spindler J, Raley C, Hautman C, Sobolewski M, Fennessey CM, Hu WS, Luke B, Hasson JM, Niyongabo A, Capoferri AA, Keele BF, Milush J, Hoh R, Deeks SG, Maldarelli F, Hughes SH, Coffin JM, Rausch JW, Mellors JW, Kearney MF. Combined HIV-1 sequence and integration site analysis informs viral dynamics and allows reconstruction of replicating viral ancestors. Proc Natl Acad Sci USA. 2019;116(51):25891–25899. doi: 10.1073/pnas.1910334116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Einkauf KB, Lee GQ, Gao C, Sharaf R, Sun X, Hua S, Chen SM, Jiang C, Lian X, Chowdhury FZ, Rosenberg ES, Chun TW, Li JZ, Yu XG, Lichterfeld M. Intact HIV-1 proviruses accumulate at distinct chromosomal positions during prolonged antiretroviral therapy. J Clin Invest. 2019;129(3):988–998. doi: 10.1172/JCI124291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang C, Lian X, Gao C, Sun X, Einkauf KB, Chevalier JM, Chen SMY, Hua S, Rhee B, Chang K, Blackmer JE, Osborn M, Peluso MJ, Hoh R, Somsouk M, Milush J, Bertagnolli LN, Sweet SE, Varriale JA, Burbelo PD, Chun TW, Laird GM, Serrao E, Engelman AN, Carrington M, Siliciano RF, Siliciano JM, Deeks SG, Walker BD, Lichterfeld M, Yu XG. Distinct viral reservoirs in individuals with spontaneous control of HIV-1. Nature. 2020;585(7824):261–267. doi: 10.1038/s41586-020-2651-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shao W, Shan J, Hu WS, Halvas EK, Mellors JW, Coffin JM, Kearney MF. HIV proviral sequence database: a new public database for near full-length HIV proviral sequences and their meta-analyses. AIDS Res Hum Retrovir. 2020;36(1):1–3. doi: 10.1089/aid.2019.0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee GQ, Reddy K, Einkauf KB, Gounder K, Chevalier JM, Dong KL, Walker BD, Yu XG, Ndung'u T, Lichterfeld M. HIV-1 DNA sequence diversity and evolution during acute subtype C infection. Nat Commun. 2019;10(1):2737. doi: 10.1038/s41467-019-10659-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee GQ, Orlova-Fink N, Einkauf K, Chowdhury FZ, Sun X, Harrington S, Kuo HH, Hua S, Chen HR, Ouyang Z, Reddy K, Dong K, Ndung'u T, Walker BD, Rosenberg ES, Yu XG, Lichterfeld M. Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. J Clin Invest. 2017;127(7):2689–2696. doi: 10.1172/JCI93289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJ. Biopython: freely available python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25(11):1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mailler E, Bernacchi S, Marquet R, Paillart JC, Vivet-Boudou V, Smyth RP. The life-cycle of the HIV-1 Gag–RNA complex. Viruses. 2016;8(9):248. doi: 10.3390/v8090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das AT, Pasternak AO, Berkhout B. On the generation of the MSD-Ѱ class of defective HIV proviruses. Retrovirology. 2019;16(1):19. doi: 10.1186/s12977-019-0481-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Carroll IP, Thappeta Y, Fan L, Ramirez-Valdez EA, Smith S, Wang YX, Rein A. Contributions of individual domains to function of the HIV-1 Rev response element. J Virol. 2017;91(21):e00746-17. doi: 10.1128/JVI.00746-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rose PP, Korber BT. Detecting hypermutations in viral sequences with an emphasis on G→A hypermutation. Bioinformatics. 2000;16(4):400–401. doi: 10.1093/bioinformatics/16.4.400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The HIVIntact pipeline and all test data, including a snapshot of the NCI PSD and a comparison script may be downloaded from a public GitHub repository (https://github.com/ramics/proviral-intactness) under an open-source MIT license, and is available for use. The code is written in Python3 and is platform-independent.