Abstract
Background
Tripidium ravennae is a cold-hardy, diploid species in the sugarcane complex (Poaceae subtribe Saccharinae) with considerable potential as a genetic resource for developing improved bioenergy and ornamental grasses. An improved understanding of the genetic regulation of reproductive processes (e.g., floral induction, inflorescence development, and seed development) will enable future applications of precision breeding and gene editing of floral and seed development. In particular, the ability to silence reproductive processes would allow for developing seedless forms of valuable but potentially invasive plants. The objective of this research was to characterize the gene expression environment of reproductive development in T. ravennae.
Results
During the early phases of inflorescence development, multiple key canonical floral integrators and pathways were identified. Annotations of type II subfamily of MADS-box transcription factors, in particular, were over-represented in the GO enrichment analyses and tests for differential expression (FDR p-value < 0.05). The differential expression of floral integrators observed in the early phases of inflorescence development diminished prior to inflorescence determinacy regulation. Differential expression analysis did not identify many unique genes at mid-inflorescence development stages, though typical biological processes involved in plant growth and development expressed abundantly. The increase in inflorescence determinacy regulatory elements and putative homeotic floral development unigenes at mid-inflorescence development coincided with the expression of multiple meiosis annotations and multicellular organism developmental processes. Analysis of seed development identified multiple unigenes involved in oxidative-reductive processes.
Conclusion
Reproduction in grasses is a dynamic system involving the sequential coordination of complex gene regulatory networks and developmental processes. This research identified differentially expressed transcripts associated with floral induction, inflorescence development, and seed development in T. ravennae. These results provide insights into the molecular regulation of reproductive development and provide a foundation for future investigations and analyses, including genome annotation, functional genomics characterization, gene family evolutionary studies, comparative genomics, and precision breeding.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-021-07641-y.
Keywords: Tripidium ravennae, Transcriptome sequencing, Differential gene expression, Next-generation sequencing, Floral transition, Flowering, Inflorescence development, Reproduction, RNA sequencing, Seed development
Background
The need for and importance of alternative energy sources becomes increasingly essential as global energy demands grow with concomitant fossil fuel reserve depletion. Bioenergy crops suitable as fuel for heat, electric power generation, and processing into cellulosic ethanol continue to attract attention as alternative fuel sources. Members of the grass family Poaceae subtribe Saccharinae, also known as the sugarcane complex, have gained attention for their broad adaptability, pest resistance, high biomass yields, and potential for perennially sequestering large amounts of carbon with few inputs on marginal lands [1–5].
The Saccharinae are diverse, spanning numerous genera include Erianthus, Miscanthus, Saccharum, and Tripidium [6–8]. Though previously placed in the genus Saccharum L. [9], then Erianthus (L.) P. Beauv [10]., Valdés and Scholz [7] transferred the four members of Old-World Erianthus into Tripidium based on both molecular and morphological features. The genus Tripidium currently circumscribes all the Old-World members of Erianthus sect. Ripidium resolving the debate over Tripidium spp. taxonomy since the partial treatment of the group by Valdés and Scholz [6, 7]. Tripidium ravennae (L.) H. Scholz (syn. E. ravennae, S. ravennae (Ravenna grass) is diploid (2n = 2x = 20) and cold-hardy to USDA Zone 5b [5]. Ravenna grass has a broad native range spanning Eastern Europe, North Africa, and Southwestern Asia, but has naturalized in several locales of the new world [10–12]. The use of T. ravennae as a landscape ornamental or on marginal lands and riparian areas for erosion control purposes [13–15] created an opportunity for escape, and it is now considered weedy in some areas [16, 17].
Conventional breeding is well suited for improving complex traits such as yield and cold-hardiness. Maren et al. [3] recently reported on new interspecific Tripidium hybrids with significantly higher biomass yields than Miscanthus ×giganteus and cold hardiness to USDA Zone 6b/7a. However, plant biotechnology has considerable potential to augment conventional breeding and make value-added improvements in elite clonally propagated cultivars without compromising other genetically complex and desirable traits. For example, silencing key reproductive processes could reduce reseeding and invasive potential of valuable bioenergy grass clones.
The role of plant reproduction in crop production and yield encouraged extensive research into the genetics of flowering among agricultural cereals such as wheat (Triticum spp.) [18–20] and barley (Hordeum vulgare) [21–23]. However, current information on the genetics and translational genomics of reproductive development in perennial grass species of the Panicoideae (including Tripidium spp.), Aristidoideae, Chloridoideae, Micrairoideae, Arundinoideae, and Danthonioideae (PACMAD) clade of the Poaceae is limited [24]. A foundation for future application of precision breeding and gene editing depends on a detailed understanding of the reproductive process’ genetic regulation. Transcriptomic and RNA-sequencing analyses allow for examining gene expression regardless of prior sequencing context and enable the identification of candidate genes for modification [25, 26]. The processes of floral initiation [27], inflorescence development [28, 29], and seed development [30, 31] involve multifaceted changes in gene expression. An extensive literature search identified minimal genetic information or gene expression analysis of T. ravennae. The aim of this study was to characterize the genetic control and differentially expressed transcripts in reproductive development pathways of the diploid perennial bioenergy grass T. ravennae.
Results
Transcriptome assembly and functional annotation
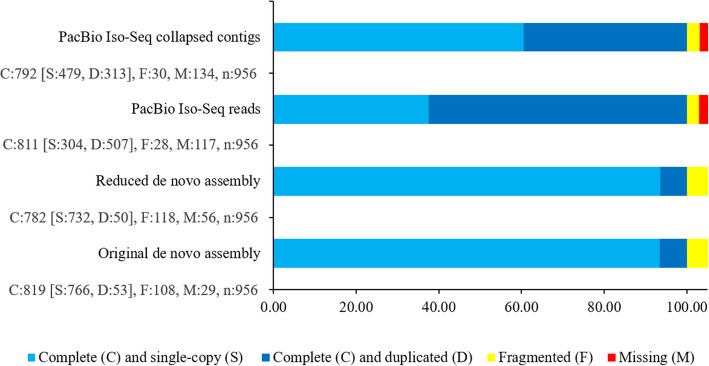
Sequencing of vegetative, developing inflorescence, floret, and seed tissues yielded 687 million raw reads (Supplemental Table 1). The primary de novo assembly utilized 15.4 million paired and quality-trimmed reads and comprised 95% of all quality-filtered reads yielding an assembly with 156,724 contigs (N50: 1265; Table 1; Supplemental Fig. 1a-c). BUSCO analysis, utilizing the 956 Plantae core set, revealed an 85.6% completion rate (Fig. 1). Alignment of the transcriptome contigs to the draft genome assembly of T. ravennae (Maren et al., in preparation) and cluster-based enrichment reduced the contig set by 68%, yielding a transcriptome assembly with 105,307 unitigs (N50: 1494; Table 1). Similarity-based clustering and genomic alignment reduced the transcript set by 33%, reducing the representation of complete conserved orthologs (BUSCO core genes) by 4.5%. The reduced transcript sets functional annotation identified 33,782 unigenes with at least one of the 130,460 annotations (Supplemental Tables 8; 9; Figs. 2, 3 and 4). Across all samples, 41,234 unigenes were expressed greater than five transcripts per million (TPM) in at least two biological samples and two-fold change (absolute value of log2 tagwise dispersion values) between two or more samples. Of those unigenes, 36,127 were transcribed and differentially expressed in at least one of the 78 pair-wise tests of differential expression.
Table 1.
Summary statistics of sequencing reads, assembly, and annotations
| Category | Valuex | ||
|---|---|---|---|
| Primary de novo Assembly |
Redundancy Reduced Assembly |
Collapsed Iso-Seq Set | |
| Raw reads | 670,892,713 | 1,463,943 | |
| Total length of raw reads (Mbp) | 206,368 | 288 | |
| Quality filtered & trimmed reads | 615,064,938 | 1,270,618 | |
| Total length of prepared reads (Mbp) | 80,819 | 146 | |
| Total Contigs | 156,724 | 105,307 | 65,696 |
| Read utilization (%) | 95 | 78 | 87 |
| Avg. length of contigs | 941 | 1058 | 2227 |
| Contig size N50 | 1265 | 1494 | 3581 |
| Min. contig length | 300 | 300 | 80 |
| Max. contig length | 21,363 | 21,363 | 9831 |
| Total nucleotides in assembly | 147,537,886 | 111,466,222 | 146,320,100 |
| Annotation Statistics | |||
| InterProScan | 156,723 (99%) | 105,307 (100%) | 48,482 (74%) |
| BLASTx (nr) hits | 85,974 (55%) | 45,192 (43%) | 59,553 (91%) |
| Uniquely aligning to genomey | 99,110 (63%) | 95,274 (90%) | 63,042 (96%) |
| Functionally annotatedz | 66,810 (43%) | 33,782 (32%) | 51,536 (78%) |
xColumns represent the three assemblies utilized in the differential gene expression analysis. The primary de novo assembly comprised of next-generation sequence reads assembled using a k-mer size of 41 and a bubble size of 350 in the Qiagen CLC Genomics Workbench 11.0. The redundancy-reduced assembly was developed via reference mapping to multiple members in the Andropogonae and further reduced by cluster enrichment with CD-Hit. The collapsed iso-seq set was developed from pooled RNA samples of the same experimental tissue and collapsed with the Cupcake TOFU pipeline
yTranscripts mapped, with GMAP, to a single genomic locus of a preliminary reference assembly with 95% coverage and identity
zUnigenes annotated with one or more gene ontology terms
Fig. 1.
Tripidium ravennae transcriptome quality assessment with BUSCO analysis. C, S, D, F, and M represent complete, single, duplicate, fragmented, and missing BUSCOs, respectively
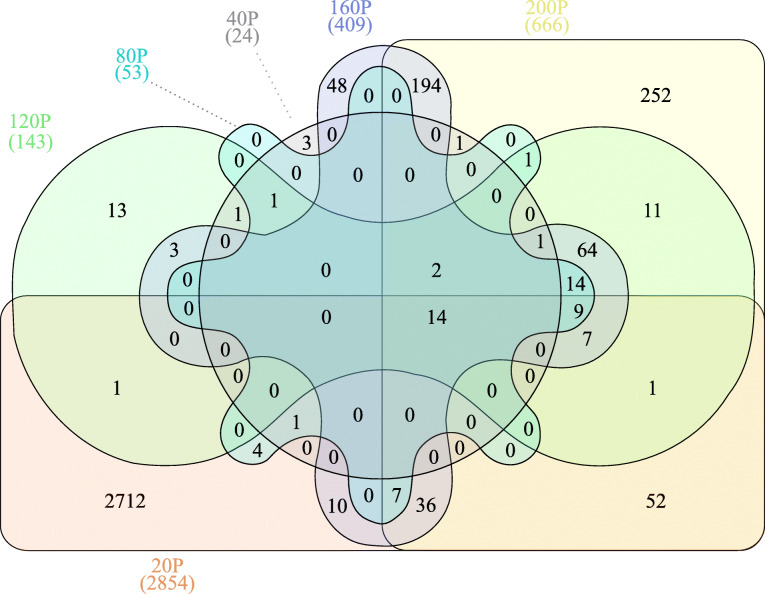
Fig. 2.
Venn diagram of differentially expressed unigenes in inflorescence development. Numbers represent unigenes with an absolute and minimum two-fold change in expression relative to controls (FDR p-value < 0.05).  Inflorescence meristem sampling at ~ 20 cm from culm base (20P).
Inflorescence meristem sampling at ~ 20 cm from culm base (20P).  Inflorescence meristem sampling at ~ 40 cm from culm base (40P).
Inflorescence meristem sampling at ~ 40 cm from culm base (40P).  Inflorescence meristem sampling at ~ 80 cm from culm base (80P).
Inflorescence meristem sampling at ~ 80 cm from culm base (80P).  Inflorescence meristem sampling at ~ 120 cm from culm base (120P).
Inflorescence meristem sampling at ~ 120 cm from culm base (120P).  Inflorescence meristem sampling at ~ 160 cm from culm base (160P).
Inflorescence meristem sampling at ~ 160 cm from culm base (160P).  Inflorescence meristem sampling at ~ 200 cm from culm base (200P)
Inflorescence meristem sampling at ~ 200 cm from culm base (200P)
Fig. 3.

Heat map of hierarchically clustered inflorescence unigenes. Absolute values of transcript per million normalized gene expression ranged from a minimum of two-fold to > 3000-fold change in expression relative to controls (FDR p < 0.05). Columns represent log2-fold expression values of all biological replicates in developing inflorescences (Table 5). Clustering utilized transcript per million (TPM) normalized expression values to iteratively calculate pair-wise manhattan distances between all clusters and joining clusters of proximity. Branch length represents the distance between clusters and reflects the similarity of expression profiles for co-expressed genes within the two clusters
Fig. 4.
Venn diagram of late spikelet and flower development. Number values represent unigenes with an absolute minimum two-fold change in gene expression (FDR p < 0.05). DEGs were derived from statistical tests within floral tissue relative to developing inflorescence controls for boot stage florets (FT), pre-anthesis florets (PAF), anther tissue (ST), anthesis florets (ANT)
Differential expression and GO enrichment of inflorescence development
Differential expression analysis of inflorescence samples identified 3463 unigenes with an absolute two-fold minimum change in expression (FDR p < 0.05; Figs. 2 and 3; Supplemental Table 5). Basic metabolic processes of carbohydrate biosynthesis, redox processes, and cell wall growth were prominent along with sexual reproduction categories in the hypergeometric annotation tests (p < 0.05). Among the unigenes involved in plant reproduction and morphogenesis, MADS-box transcription factors and other floral integrators were prominent in all inflorescence samples. Transcriptome assembly sequences were named as “Trav” followed by a seven-digit number, which have been put in parenthesis here and thereafter. MADS-box transcription factor 18 (MADS18 – Trav0007787) [32], Sorghum bicolor MADS22-X2 (SbMADS22-X2 – Trav0022274) [33], MADS-box protein SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1 – Trav0019736) [34, 35], and MADS14-X2 (Trav0004731) [32, 36], the type II subfamily of MADS-box transcription factors, were expressed at elevated transcript abundance levels in all inflorescences sampled. Transcript abundance changes were most prominent in the early phases of inflorescence development, with 2712 (69%) differentially expressed genes (DEGs) that accounted for most of the upregulated expression (Fig. 2). Early inflorescence development marked transcript abundance changes within basic developmental processes and flavonol and lignin production with CAFFEIC ACID O-METHYLTRANSFERASE (COMT – Trav0002740) [37, 38]. Several floral development genes, including multiple expansin proteins, zinc finger protein CONSTANS-like unigenes [23, 34, 35], HEADING DATE 3B-like (HD3B)-like (Trav0007479) [34], MADS47-X1 (Trav0004133), MADS50 (Trav0015948), MADS55 (Trav0005904), MADS56 (Trav0017955) [32, 33, 39–41], and GRAIN NUMBER, PLANT HEIGHT, AND HEADING DATE7 (Ghd7 – Trav0011675) [23, 42, 43] were upregulated coinciding with the transition from vegetative to reproductive growth. The abundance of DEGs diminished after the vegetative to inflorescence development shift. As inflorescences matured into the middle and later stages of development, DEG abundance increased. Unigenes relevant in floral meristem determinacy, meiosis, ovule, carpel, and stamen development increased including PARTING DANCERS isoform X1 (PTD)-X1 (Trav0016008) [44], CHROMOSOME TRANSMISSION FIDELITY 7 (CTF7 – Trav0008006) [45], POLLEN DEFECTIVE IN GUIDANCE 1 (POD1 – Trav0005801) [46, 47], SELF-PRUNING (SP – Trav0029292) [48], AGAMOUS LIKE 6-like MADS-box transcription factor (AGL6)-like (Trav0022760) [41] as well as MADS3-X1 (Trav0008160), MADS7 (Trav0000111), and MADS8 (Trav0000081) [32, 33, 39–41]. The DEG abundance substantially increased as inflorescences grew and expanded beyond the flag leaf. Unigenes involved in plant metabolic, putative floral development processes in the embryo sac, meiosis, and pollen development were identified including with ETERNAL TAPETUM 1 (EAT1 – Trav0038135) [49, 50], MEIOSIS ARRESTED AT LEPTOTENE 1 (MEL1 – Trav0018140) [50, 51], Expansin-B9 and -B11 (EXPB9 – Trav0000513 & EXPB11 – Trav0000400) [52], FLOWERING LOCUS T protein (FT – Trav0015919) [53], and floral homeotic protein AGAMOUS-like (AG)-like (Trav0011776) [54].
Throughout inflorescence development, multiple unigenes and those possessing functional annotations were revealed in the differential expression analyses. Relative to vegetative controls, inflorescence samples at the earliest stages of development had the greatest abundance of DEGs, corresponding with the floral transition (Fig. 2). The fold change in gene expression varied considerably and ranged to greater than ±8000 times the expression levels observed in the vegetative meristem control samples. Table 2 enumerates a list of genes of interest with tissue-specific expression patterns, and those whose fold changes in gene expression were most pronounced for a given stage of inflorescence development. The genes listed in Table 2 comprise a shortlist of utilitarian targets for studying gene functional genomics and gene knockout mutations that might serve to limit reproduction. This information furthermore provides the opportunity for the identification of tissue-specific promoters.
Table 2.
Unigenes upregulated with stage specific expression patterns in Tripidium inflorescence development
| Gene name | Unitig ID | E-valuex | % Idx | Fold Changey | FDR p-valuey | TPM Normalized Means | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VM | 20P | 40P | 80P | 120P | 160P | 200P | ||||||
| Inflorescence meristems at 20 cm (20P) | ||||||||||||
| Mini zinc finger protein 1 | Trav0045635 | 6.1E−28 | 85.5 | 67.9 | 2.8E−3 | 1.7 | 106.9 | 16.1 | 59.7 | 1.6 | 3.1 | 2.9 |
| Dehydrodolichyl diphosphate synthase 2 | Trav0048752 | 3.3E−93 | 89.3 | 48.8 | 7.7E−3 | 0.7 | 35.0 | 1.2 | 20.1 | 0.7 | 1.5 | 1.7 |
| n/a | Trav0022235 | n/a | n/a | 48.7 | 3.1E−3 | 9.6 | 400.4 | 13.4 | 123.6 | 9.6 | 11.8 | 13.6 |
| Transcription factor TGA4 | Trav0031073 | 3.1E−123 | 87.8 | 23.1 | 8.6E−4 | 7.5 | 153.9 | 14.9 | 51.8 | 4.9 | 5.6 | 6.5 |
| n/a | Trav0012164 | n/a | n/a | 17.2 | 4.3E−3 | 28.2 | 424.0 | 21.5 | 120.0 | 8.9 | 16.4 | 17.6 |
| Inflorescence meristems at 40 cm (40P) | ||||||||||||
| n/a | Trav0005961 | n/a | n/a | 402.5 | 2.1E−6 | 0 | 8.5 | 52.7 | 29.3 | 28.7 | 26.4 | 19.8 |
| n/a | Trav0020850 | n/a | n/a | 203.4 | 2.1E−5 | 0 | 10.4 | 26.3 | 25.2 | 24.7 | 14.4 | 8.7 |
| Inflorescence meristems at 80 cm (80P) | ||||||||||||
| MADS-box trans. factor 34 | Trav0005828 | 4.2E−103 | 86.8 | 210.4 | 5.5E−5 | 0.2 | 2.7 | 2.2 | 68.2 | 144.2 | 168.5 | 180.8 |
| Inflorescence meristems at 120 cm (120P) | ||||||||||||
| n/a | Trav0000921 | n/a | n/a | 204.8 | 5.0E−2 | 2.1 | 7.5 | 0.7 | 4.7 | 367.0 | 752.9 | 1063.1 |
| Sucrose-phosphate synthase | Trav0002204 | 0 | 93.7 | 109.1 | 1.9E−6 | 0.8 | 7.0 | 4.4 | 12.1 | 35.4 | 91.2 | 146.0 |
| MADS-box trans. factor 7 | Trav0000111 | 3.6E−171 | 92.0 | 202.0 | 1.4E−3 | 0.5 | 1.1 | 0.3 | 10.0 | 101.9 | 171.3 | 233.0 |
| Inflorescence meristems at 160 cm (160P) | ||||||||||||
| n/a | Trav0012650 | n/a | n/a | 104.4 | 5.0E−6 | 0.7 | 5.2 | 3.2 | 7.6 | 25.4 | 69.5 | 60.4 |
| Carbonic anhydrase | Trav0005356 | 5.3E−58 | 96.6 | 152.0 | 4.0E−2 | 4.7 | 24.0 | 0.4 | 3.1 | 337.7 | 573.4 | 571.9 |
| Inflorescence meristems at 200 cm (200P) | ||||||||||||
| Arginine decarboxylase 2 | Trav0036107 | 0 | 81.2 | 1472.0 | 8.2E−3 | 0.1 | 0.9 | 0.1 | 0.3 | 41.1 | 213.8 | 294.0 |
| n/a | Trav0006980 | n/a | n/a | 487.1 | 1.1E− 3 | 2.2 | 3.0 | 1.8 | 2.7 | 183.0 | 677.9 | 959.9 |
| MADS-box trans. factor 8 | Trav0000081 | 4.5E−175 | 96.2 | 471.5 | 2.7E−6 | 0.6 | 1.5 | 0.5 | 10.2 | 107.0 | 218.1 | 300.9 |
| Protein unc-13 homolog | Trav0005596 | 0 | 90.2 | 104.5 | 5.0E−3 | 1.9 | 3.8 | 1.4 | 2.6 | 61.0 | 137.7 | 174.1 |
| MADS-box trans. factor 16 | Trav0000760 | 5.5E−27 | 83.7 | 87.3 | 2.8E−4 | 1.0 | 0.4 | 0.5 | 2.9 | 40.7 | 70.5 | 85.2 |
| MADS-box trans. factor 4 | Trav0000787 | 4.6E−76 | 98.3 | 40.2 | 7.0E−6 | 3.7 | 6.3 | 3.8 | 9.9 | 41.7 | 89.3 | 139.4 |
| Sister chromatid cohesion 1 protein 1 | Trav0023214 | 0 | 88.8 | 34.2 | 4.5E− 4 | 0.5 | 1.2 | 0.5 | 2.6 | 6.6 | 15.1 | 17.8 |
xE-value and % Id derived from BLAST and InterPro Scan results and annotated utilizing BLAST2GO default annotation rules
yFold change and FDR p-values are derived from EDGE tests between the vegetative control and inflorescence development stage of each underlined subsection within the table
Differential expression and GO enrichment in floral development
Differential gene expression profiling of floret development identified 5988 unigenes with the absolute minimum two-fold change in expression (FDR p < 0.05; Figs. 4 and 5; Supplemental Table 6). Among floret development samples, DEG abundance was highest in stamen tissues (4966), followed by anthesis florets (2622), pre-anthesis florets (1970), and early development florets (698). At the earliest stage of floret development, multiple MADS-box transcription factors, including AGL6-like [41], were differentially regulated along with protein DROOPING LEAF (DL)-X1 (Trav0014582) [55] and AG-like (Trav0011776) [54]. As florets developed into pre-anthesis florets, differential gene expression analysis revealed multiple genes in floral organ morphogenesis, embryo sac, embryonic, and pollen development relative to late inflorescence development samples. DEGs implicated in floral development included guanine nucleotide-binding protein NUCLEOSTEMIN LIKE 1 (NSN1 – Trav0003163) [56, 57], SOMATIC EMBRYOGENESIS RECEPTOR KINASE 2 (SERK2 – Trav0003828) [58], FLOWERING PROMOTING FACTOR 1-LIKE 2 (FPF1)-2 (Trav0026700) [59], and BTB/POZ AND TAZ DOMAIN-CONTAINING PROTEIN 3 (BT3 – Trav0015326) [60]. Relative to late inflorescence development samples, unigenes regulating pollen integument, cellular growth, and karyogamy increased in the DEG sets in pre-anthesis florets and stamen tissues, including protein NUCLEAR FUSION DEFECTIVE 4 (NFD4)-X2 (Trav0000417) [61], and putative receptor protein CRINKLY 4 (CR4 – Trav0011689) [62].
Fig. 5.
Heat map of hierarchically clustered floral development unigenes. Absolute values of transcript per million normalized gene expression ranged from a minimum of two-fold to > 8000-fold change in expression relative to controls (FDR p < 0.05). Columns represent log2-fold expression values of all biological replicates in inflorescences at 120 cm of development (120P), developing boot stage florets (FT), pre-anthesis florets (PAF), stamens (ST), and florets at anthesis (ANT) (Table 5). Clustering utilized transcript per million (TPM) normalized expression values to iteratively calculate pair-wise Manhattan distances between all clusters and joining clusters of proximity. Branch length represents the distance between clusters and reflects the similarity of expression profiles for co-expressed genes within the two clusters
Floral tissue samples can provide detailed information to identify genes of interest for future applications of precision breeding and gene editing of reproductive processes. The utility of full-length transcripts to key floral integrators and the potential they have in the identification of multiple gene-editing mutation regions may be a key asset in precision breeding and gene editing. DEG analysis revealed tissue-specific gene expression dynamics in all floral samples. Within flower specific tissues, stamen tissues yielded the greatest abundance of upregulated differential gene expression (Fig. 4). Gene expression dynamics varied widely in stamen samples, with some genes demonstrating a 7000-fold difference in expression relative to late stage inflorescence controls. Genes of interest were amassed into a table based on tissue-specific expression and large fold changes between controls and a given floral sample (Table 3). The tissue expression specificity of these genes provides candidate genes for continued study or applied plant breeding purposes to limit reproduction in Tripidium.
Table 3.
Unigenes upregulated with stage specific expression patterns in Tripidium flower development
| Gene name | Unitig ID | E-valuex | % Idx | Fold Changey | FDR p-valuey | TPM Normalized Means | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| 120P | FT | PAF | ST | ANT | ||||||
| Stamens (ST) | ||||||||||
| Glucan endo-1,3-beta-glucosidase 8 | Trav0002923 | 0 | 92.1 | 4394.1 | 1.9E−7 | 0.1 | 208.4 | 322.4 | 749.7 | 156.3 |
| Homeobox protein ARX-like | Trav0000011 | 1.4E−112 | 79.7 | 1855.9 | 6.4E−12 | 2.1 | 903.4 | 1226.8 | 2510.5 | 678.1 |
| Pollen receptor-like kinase 3 | Trav0004991 | 0 | 82.4 | 1850.4 | 6.4E−10 | 0.2 | 94.7 | 167.5 | 331.5 | 86.5 |
| Male gametophyte-specific2 | Trav0008881 | 0 | 89.0 | 1451.8 | 1.1E−10 | 0.1 | 36.8 | 121.0 | 242.5 | 55.7 |
| Pollen receptor-like kinase 4 | Trav0008246 | 0 | 84.8 | 1286.6 | 2.8E−8 | 0.3 | 174.5 | 246.1 | 323.0 | 122.8 |
| Ethylene-responsive trans. factor ABR1 | Trav0065092 | 5.5E−33 | 74.1 | 1204.8 | 3.2E−27 | 0.1 | 4.8 | 5.1 | 208.8 | 16.4 |
| GAMETE EXPRESSED 1 | Trav0007983 | 0 | 87.8 | 898.2 | 8.4E−12 | 0.3 | 51.6 | 53.6 | 232.4 | 45.4 |
| REVEILLE 1 | Trav0051764 | 0 | 82.0 | 719.6 | 3.2E−34 | 3.4 | 19.3 | 23.5 | 1736.8 | 73.9 |
| Xylanase inhibitor protein 1 | Trav0043283 | 0 | 91.2 | 565.4 | 3.0E−13 | 2.9 | 66.0 | 68.7 | 1142.4 | 343.9 |
| Florets at anthesis (ANT) | ||||||||||
| n/a | Trav0031057 | n/a | n/a | 2120.1 | 2.2E-9 | 0.8 | 372.7 | 260.8 | 1269.9 | 1753.1 |
| 5-pentadecatri-enyl resorcinol O-methyltransferase | Trav0039042 | 1.9E−161 | 78.1 | 1345.4 | 9.4E−10 | 0.3 | 34.5 | 12.6 | 118.1 | 531.8 |
| Myosin-binding protein 1 | Trav0000816 | 0 | 78.8 | 114.9 | 7.6E−6 | 1.1 | 227.8 | 279.5 | 1010.0 | 126.2 |
| Pre-anthesis florets (PAF) | ||||||||||
| Expansin-B11 | Trav0005943 | 2.3E−39 | 93.3 | 1765.5 | 7.8E−12 | 0.2 | 80.9 | 371.5 | 206.4 | 0.2 |
| Predicted protein | Trav0003235 | 0 | 78.9 | 1600.0 | 1.4E−6 | 0.1 | 27.8 | 263.7 | 6.3 | 16.2 |
| Pollen allergen Phl p 2 | Trav0005342 | 7.9E−48 | 80.9 | 810.2 | 9.7E−10 | 0 | 31.4 | 127.6 | 71.2 | 45.8 |
| Zinc finger protein ZAT5 | Trav0002821 | 2.6E−88 | 55.8 | 485.6 | 3.9E−6 | 0.1 | 36.4 | 91.3 | 28.3 | 0.1 |
| Boot stage florets (FT) | ||||||||||
| AG-like MADS-box protein AGL66 X4 | Trav0007544 | 0 | 82.8 | 26.9 | 5.6E−3 | 0.4 | 11.8 | 22.7 | 32.7 | 9.8 |
| Retrotransposon protein, putative, unclassified | Trav0016899 | 0 | 63.8 | 24.5 | 0.04 | 0.3 | 9.8 | 11.8 | 7.5 | 17.1 |
| auxin-responsive protein SAUR71 | Trav0074745 | 6.8E−40 | 81.7 | 18.8 | 0.02 | 0.2 | 6.3 | 1.8 | 1.2 | 21.4 |
| Zn finger protein CONSTANS-LIKE 5 | Trav0016572 | 8.3E−19 | 83.0 | 11.0 | 4.3E−3 | 9.6 | 94.5 | 67.2 | 12.8 | 172.3 |
xE-value and % Id derived from BLAST and InterPro Scan results and annotated utilizing BLAST2GO default annotation rules
yFold change and FDR p-values for each gene are derived from EDGE tests between inflorescence meristem samples at 120 cm of development and each underlined subsection within the table
Differential expression and GO enrichment in seed development
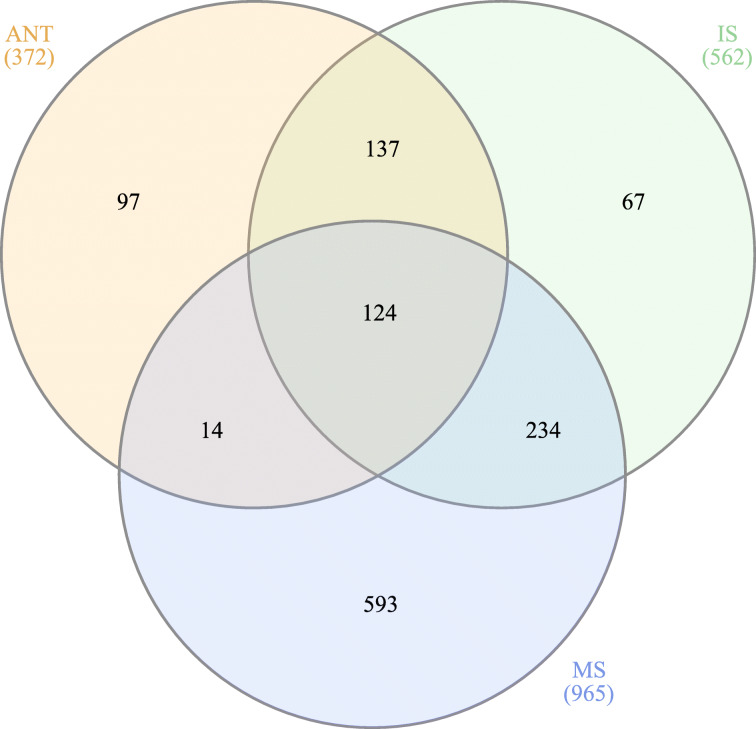
Differential gene expression profiling of seed development identified 1266 unigenes with an absolute minimum two-fold change in expression (FDR p < 0.05; Figs. 6 and 7; Supplemental Table 7). GO term enrichment analysis identified 654 terms throughout seed development. Many GO terms coincided with basic organismal processes and development. Mature seed samples had the highest number of DEGs (965) relative to immature seed (562) or anthesis florets (372). In the mature seed, several DEGs involved in multicellular organism development and seed maturation were identified. The functional annotation of these sequences indicated that they were regulatory protein VIVIPAROUS 1 (VP1 – Trav0080007) [63], WUSCHEL-related homeobox 6 (WOX6 – Trav0041173) [64], LATE EMBRYOGENESIS ABUNDANT 18 (LEA18 – Trav0122482) [65], and SUCROSE SYNTHASE 3 (SUS3 – Trav0008473) [66]. In the transition from anthesis florets to immature seed development, DEG analysis identified 80 DEGs with multiple unigenes in embryo development, seed oil-body biogenesis, and response to abscisic acid. Noteworthy annotations included OIL BODY-ASSOCIATED PROTEIN 1A (OBAP1A – Trav0053823) [67], VICILIN-like (VIC)-like (Trav0081216) [68], LATE EMBRYOGENESIS ABUNDANT EMB564 (LEA-EMB564 – Trav0096979) [65], and dehydrin Rab25-like (Rab25 – Trav0064814) [69].
Fig. 6.
Venn diagram of seed development. Number values represent unigenes with an absolute minimum two-fold change in gene expression within seed tissue relative to controls (FDR p < 0.05). DEGs derive from statistical tests within seed tissues relative to developing inflorescence controls for florets at anthesis (ANT), florets with immature seeds (IS), and florets with mature seeds (MS)
Fig. 7.
Heat map of hierarchically clustered seed development unigenes. Absolute values of transcript per million normalized gene expression ranged from a minimum of two-fold to > 8000-fold change in expression relative to controls (FDR p < 0.05). Columns represent log2-fold expression values of all biological replicates of developing florets at anthesis (ANT), immature seed (IS), and mature seed (MS) (Table 5). Clustering utilized transcript per million (TPM) normalized expression values to iteratively calculate pair-wise manhattan distances between all clusters and joining clusters of proximity. Branch length represents the distance between clusters and reflects the similarity of expression profiles for co-expressed genes within the two clusters
Seed production is a significant contributor to the successful cultivation of many plants. In developing seed samples, a variety of genes with increased expression levels involved in homeostatic regulatory roles for metal ion uptake, redox processes, heat shock, and genes known to interact under abiotic stress were found in the dataset. Fold changes in gene expression between late inflorescence stage development controls and mature seeds varied considerably and were expressed at as much as a 16,000-fold difference (Fig. 6). A shortlist of genes of interest within these analyses were amassed in Table 4 because of their tissue-specific expression pattern. Taken together, these genes represent several potential targets that may drastically limit plant reproductive characteristics in an applied precision breeding context.
Table 4.
Unigenes upregulated with stage specific expression patterns in Tripidium seed development
| Gene name | Unitig ID | E-valuex | % Idx | Fold Changey | FDR p-valuey | TPM Normalized Means | |||
|---|---|---|---|---|---|---|---|---|---|
| 200P | ANT | IS | MS | ||||||
| Mature seeds (MS) | |||||||||
| Globulin-1 S allele | Trav0051718 | 0 | 83.5 | 32,919.7 | 7.1E−7 | 0.4 | 1.4 | 7883.0 | 16,578.5 |
| Embryonic abundant protein 1 | Trav0073008 | 4.2E−41 | 95.7 | 23,575.7 | 5.8E−6 | 0.26 | 0.19 | 4424.2 | 9490.7 |
| Aquaporin TIP3–1 | Trav0101881 | 1.8E−130 | 94.0 | 14,828.8 | 8.4E−6 | 0.1 | 0.28 | 2266.5 | 3572.9 |
| Vicilin-like seed storage protein At2g28490 | Trav0121709 | 3.8E−87 | 87.7 | 9135.7 | 1.6E−5 | 0 | 0 | 628.7 | 1237.7 |
| n/a | Trav0084667 | n/a | n/a | 281.5 | 6.7E−16 | 0 | 0 | 0.2 | 40.1 |
| n/a | Trav0133404 | n/a | n/a | 276.5 | 1.4E−9 | 0 | 0 | 0 | 40.3 |
| Immature seeds (IS) | |||||||||
| Em-like protein GEA1 | Trav0097540 | 3.7E−49 | 87.0 | 21,364.3 | 1.5E−4 | 0 | 0 | 2616.3 | 4543.1 |
| Oleosin 16 kDa | Trav0094715 | 1.9E−45 | 77.2 | 7209.0 | 2.2E−5 | 0.1 | 0.2 | 1195.2 | 907.0 |
| n/a | Trav0099087 | n/a | n/a | 5148.0 | 9.4E−5 | 0 | 0 | 584.4 | 707.9 |
| Seed biotin-containing protein SBP65 | Trav0121829 | 1.7E−50 | 76.5 | 3075.0 | 9.0E−5 | 0 | 0 | 348.7 | 334.6 |
| Metal-assoc. isoprenylated plant protein 7 | Trav0143502 | 1.5E−55 | 77.0 | 2029.7 | 6.4E−4 | 0 | 0 | 230.2 | 244.4 |
| n/a | Trav0137461 | n/a | n/a | 1776.7 | 1.7E−4 | 0 | 0 | 201.9 | 212.6 |
| Globulin-2 | Trav0102378 | 7.8E−59 | 91.5 | 953.5 | 8.3E−5 | 0 | 0 | 109.6 | 96.3 |
| Cytochrome P450 99A2 | Trav0083879 | 0 | 78.5 | 842.3 | 3.7E−4 | 0 | 78.4 | 93.3 | 0.3 |
| Florets at anthesis (ANT) | |||||||||
| Berberine bridge enzyme-like 18 | Trav0097613 | 0 | 91.1 | 807.0 | 6.7E−6 | 0 | 100.3 | 36.4 | 0.3 |
| Xyloglucan endotransglucosylase/hydrolase 2 | Trav0071493 | 1.4E−74 | 77.7 | 790.4 | 7.8E−3 | 0 | 87.2 | 7.4 | 0.6 |
| n/a | Trav0083685 | n/a | n/a | 470.4 | 2.0E−3 | 0 | 47.1 | 2.8 | 0.7 |
| n/a | Trav0100871 | n/a | n/a | 418.9 | 4.9E−5 | 0 | 45.4 | 1.9 | 0 |
| Uncharacterized protein LOC8058288 | Trav0046122 | 0 | 72.6 | 363.8 | 2.5E−3 | 0.1 | 74.6 | 7.0 | 0.2 |
| n/a | Trav0081406 | n/a | n/a | 257.9 | 1.3E−4 | 0 | 26.3 | 1.1 | 0 |
| Ent-cassadiene C2-hydroxylase | Trav0106832 | 3.1E−24 | 80.5 | 186.9 | 0.01 | 0 | 21.2 | 24.0 | 0 |
xE-value and % Id derived from BLAST and InterPro Scan results and annotated utilizing BLAST2GO default annotation rules
yFold change and FDR p-values for each gene are derived from EDGE tests between inflorescence meristem samples at 200 cm of development and each underlined subsection within the table
Gene expression analysis and validation of selected candidate genes by real-time qPCR
The expression patterns between RNA-Seq data and qPCR data demonstrated a positive mean Spearman rank correlation (Fig. 8). Overall, the six genes selected for the assessment validated similar relative rank and transcript abundances throughout development. CAFFEIC ACID 3-O-METHYLTRANSFERASE (COMT - Trav0002740) is involved in the lignin biosynthetic pathway [38]. In Tripidium, unigenes designated with this annotation expressed abundantly in vegetative meristematic tissues and declined throughout inflorescence, floral, and seed development. NUCLEAR FUSION DEFECTIVE 4 (NFD4 – Trav0032568) is a gene of interest for its role in karyogamy or nuclear fusion following pollination and during female gametophyte formation [61]. Overall transcript abundance was low and increased during the critical phases of floral development. PARTING DANCERS 1 (PTD1 – Trav0016008) is essential in cross-over formation during meiosis [44]. Reports of transcriptional activity of PTD1 in rice place the highest activity in anthers and pistils at heading [44]. The notable difference in the number of possible meiocytes sampled in developing inflorescences versus those of floral development tissues likely explains some of the disparity in expression patterns between Tripidium and rice. CTF7 functions in the plant body during double-stranded break repair associated with cell division. The expression dynamics of this DNA repair associated protein expressed abundantly during active phases of inflorescence growth. RNA-DIRECTED DNA METHYLATION 4 (RDDM4 – Trav0015779) is putatively involved in the epigenetic modification system of plants [70]. Expression of RDDM4 in Tripidium was relatively consistent throughout inflorescence, diminished in anthesis floral samples, and increased in mature seeds. POLLEN DEFECTIVE IN GUIDANCE 1 (POD1 – Trav0014920) dysfunction in Arabidopsis has demonstrated major impacts on vegetative and reproductive growth [46, 47]. The expression of POD1 in Tripidium demonstrated relatively consistent expression throughout inflorescence and floral development.
Fig. 8.
Validation of RNA-Seq expression levels with qPCR. Black and red lines represent relative gene expression levels observed in the RNA-Seq and qPCR respective datasets. RNA-seq data are transcript per million (TPM) normalized means ± SEM. Relative gene expression of qPCR data utilized using BI1-like protein gene and translationally controlled tumor protein homolog TCTPH as reference genes and calculated with the 2(-ΔΔCt) algorithm. a Trav0002740 (caffeic acid 3-O-methyltransferase). b Trav0032568 (NUCLEAR FUSION DEFECTIVE 4 isoform X2). c Trav0016008 (PARTING DANCERS isoform X1). d Trav0008006 (CHROMOSOME TRANSMISSION FIDELITY 7). e Trav0015779 (RNA-DIRECTED DNA METHYLATION 4). f Trav0014920 (POLLEN DEFECTIVE IN GUIDANCE 1)
Discussion
This study aimed to scrutinize the gene regulatory environment of developing inflorescences, flowers, and seeds to maximize the knowledge base for transgenic and gene editing based plant improvements. Our approach examined both within and between-group testing to identify transcript abundance changes specific to a given stage of development.
Inflorescence development and floral transition
The abundance of differentially expressed unigenes revealed the magnitude of gene expression changes involved in floral induction. The transition from the vegetative meristem to the reproductive meristem follows an abundance of regulatory changes coinciding with the increased expression of homeobox genes, MADS-box transcription factors, and plant hormones [28]. The members of the expansin family were significantly over-represented in the hypergeometric annotation tests as well as the differential expression analysis, which identified nine different transcript annotations of expansins as well as other canonical floral induction integrators. As culms produced new vertical growth, the upregulation of multiple expansin family members validated their role in the reproductive process of Tripidium [71–73].
Many of the classic ABC(D) E model MADS-box transcription factors are essential in the fate of meristem identity and determinacy [41, 74, 75]. Within the developing inflorescence, 21 different MADS-box elements demonstrated differential expression patterns. The expression dynamics and identification of MADS18, SbMADS22-X2, MADS-box protein SOC1, and MADS14-X2 [36, 40] in inflorescence meristem samples demonstrate the conservation of roles of these floral organ identity transcription factors in Tripidium. FLOWERING LOCUS T (FT), HEADING DATE 3A (HD3A) [76], and VERNALIZATION 3 (VRN3) [77] are synonyms for florigen, now well characterized in Arabidopsis, rice, and wheat [21, 53, 78]. The most significant sequence homology of the Tripidium florigen transcript identified FT (TrFT – Trav0056419) within the transcriptome. Within inflorescence development samples, TrFT demonstrated the highest expression levels in the floral transition (20P), as reported in Sorghum bicolor [53]. As an inflorescence matures into the late stages of development, the determinacy of the meristems changes in several plant species. SELF-PRUNING (SP) is the homolog of CENTRORADIALIS and TERMINAL FLOWER 1 of Arabidopsis and functionally maintains floral meristem indeterminacy [48]. The SP expression patterning in Tripidium followed expectations with consistent expression throughout inflorescence development. The floral homeotic protein AG-like genes increased in expression in Tripidium inflorescences throughout maturation. AG-like genes are broadly involved in floral development and coordination of the floral body plan; therefore, it might have a role in Tripidium floral transition and organization. Concomitant decreases in WUSCHEL-related homeobox expression were observed as AGL increased, capitulating the relationship between these two interacting genes in Tripidium [79].
In rice and barley, their respective CONSTANS-like 9 (COL9) and CONSTANS1 (CO1) expression increases in the early weeks of plant development and diminished following the floral transition [21, 35]. As COL9 abundance represses EARLY HEADING DATE 1, delaying flowering in rice, COL9 (Trav0008717) expression diminished in Tripidium with concomitant increased HEADING DATE 3 (HD3)-like transcription from early to mid-inflorescence stages of development [21, 80, 81]. Grain number, plant height, and heading date 7 (Ghd7) is a photoperiodic responsive gene with upstream repressive effects on EHD1 and subsequently HD3A in rice [76]. The translational dynamics of Ghd7 in Tripidium support a role throughout inflorescence development [23, 42, 43, 76].
Floral and reproductive development
The identity genetics of floral development in grasses consists of several MADS-box transcription factors and their interacting proteins in articulating the floral body plan. The translation of floral homeotic class genes from the core eudicots to the monocots is tenuous due to various synonyms applied with computationally derived annotations. The road map best suited to this disambiguation lies in the consensus between expression dynamics and functional genomics. The AGL6-like MADS-box transcription factor is active in perianth and gynoecium development in rice with pleiotropic effects on lodicules, stamens, and carpels [82]. In Tripidium, the expression of AGL6-like follows similar patterns of expression in developing inflorescences and floral tissues reported in Oryza sativa [41], supporting its floral organ-specific expression and the significance of its role in floral organ specification. The role of floral organ specification and meristem determinacy are also functions of the YABBY genes DL1 and DL2 in rice carpel specification and maize [55, 83, 84]. While the tissue expression specificity between branch meristems and inflorescence meristems was not established in these experiments, the dynamic gene expression profile in inflorescence and developing floral tissues support its role in Tripidium floral determinism [55, 83, 84]. In wheat, the TaAGL6 acts on staminate floral development by working on TaAPETALA3 [85]. In Tripidium, the homeotic activity of staminate floral development could be identified by the dramatic increase in transcript abundance of floral homeotic protein AG-like in samples at 80 cm of growth.
Meiosis, embryogenesis, and caryopsis development
As inflorescences matured into the middle stages of development and beyond, transcript abundance increased. Among the changes in transcript abundance, multiple meiotic genes delineated the next phase in reproduction. Transcription factor EAT1 is a constituent of the TAPETUM DEGENERATION RETARDATION (TDR) heterodimer significant in tapetum development [86, 87]. Interestingly, the expression pattern of EAT1 follows a distribution significant in both early and post-meiosis cells [50]. This expression pattern, and our observations in Tripidium, support mid-inflorescence stage microsporogenesis. The conformity of expression patterns of the POD1 transcript to those reported by Dai et al. [46] furthermore provides evidence for the conservation of function in Tripidium. It is noteworthy that the sampling strategy employed in this study was sufficient to successfully identify unigenes of protein PTD, CTF7, and SHORTAGE IN CHIASMATA I. Differential expression analysis of meiotic unigenes in developing inflorescences highlights their significance in the reproductive process as well as the abundance of meiocytes captured here. A more comprehensive characterization of the regulatory dynamics of meiocytes involved in mega- and microsporogenesis will require additional examination in Tripidium [44, 45, 88].
The translational dynamics of floral development is foundational to plant reproduction. When the inflorescence has emerged from the flag leaf, the changes in floral morphology are apparent, but many of the morphological changes are manifestations of the developmental design laid out during inflorescence development. Many of the genes responsible for gamete production and flower development were highly expressed at the mid-inflorescence stages of development. One of the primary benefits of the sampling strategy and data analysis approaches used in this study has been to sample multiple stages of development and to compare all possible pair-wise comparisons. This has enabled the capitulation for several canonical floral genes in a novel species as well as highlighted the sequence identity of multiple novel genes.
Conclusion
Tripidium is an enigmatic plant with considerable potential as a landscape and bioenergy crop given its ornamental merit and high biomass production on marginal lands with minimal inputs. A greater understanding of gene expression throughout reproductive development provides context for gene function analysis in these basic biological processes. This research focused on the molecular genetics of plant reproduction in T. ravennae, including identifying diverse genes related to inflorescence, flower, and seed development. The expression dynamics of unigenes detailed in this study provide a guide for future biological studies on functional and comparative genomics and the development of biotechnology applications, including precision breeding.
Materials and methods
Plant material and sample collection
Vegetative and inflorescence meristems as well as floral development tissues were collected from three plants on the week of August 9th, 2017, from the North Carolina Arboretum, Asheville, NC. Inflorescence meristematic tissue from reproductive culms was collected at various heights from the ground, representing floral development progression. Inflorescences had emerged from more developed culms, but inflorescence meristems at earlier development stages were apparent within the leaf sheath of immature culms. Floral development tissues were collected at the floret boot stage, pre-anthesis, anthesis stages, and mature stamens. Spikelets containing immature and mature seeds were collected from the same plants in September and October of 2017. Culm segments, inflorescences, and developing seed samples containing target tissues were collected and immediately placed in 15 or 50 mL centrifuge tubes vials with 5–20 mL of RNAlater® Stabilization Solution (Ambion®, Life Technologies TM). Centrifuge tubes containing sample tissue were frozen in the field on a dry ice bed before transport to the laboratory and stored at − 80 °C (Table 5). Excess plant tissue was trimmed and removed or enriched under a stereomicroscope in sterile 100 mm Petri dishes containing approximately ~ 5–10 mL fresh RNAlater®. Immature floret tissue samples (FT) were purified from bulk collected inflorescence tissue before emergence from the flag leaf sheath. Pre-anthesis floret tissue samples (PAF) were purified from bulk collected inflorescence tissue that had emerged from the flag leaf sheath. The observation of spikelet expansion and glume extrusion identified PAF samples, but no evidence of anther or stigma extrusion was observed. Stamen tissue samples (ST) were purified from bulk collected floral spikelet tissues when anthers were exposed entirely outside of the glumes, and pollen was visibly dehiscing. Samples of florets at anthesis (ANT) were purified from bulk collected floral spikelet tissues by amassing florets showing stigma protrusion from the floret’s lemma and palea. Immature and mature seeds were processed by removing first, and second-order rachilla from the bulk collected tissue before tissue lysis and homogenization. Sample tissue lysis and homogenization were processed in liquid nitrogen by mortar and pestle.
Table 5.
List of samples, their codes, and corresponding sampling heights of tissues collected for RNA-seq analysis
| Tissue | Sample code | Plant Aw (cm) |
Sample code | Plant Bw (cm) |
Sample code | Plant Cw (cm) |
|---|---|---|---|---|---|---|
| Inflorescence development tissuesx | ||||||
| VM (Vegetative meristem) | VMA | 0 | VMB | 0 | VMC | 0 |
| 20P (~ 20 cm) meristem | 20PA | 16 | 20 PB | 21 | 20PC | 13 |
| 40P (~ 40 cm) meristem | 40PA | 25 | 40 PB | 40 | 40PC | 44 |
| 80P (~ 80 cm) meristem | 80PA | 60 | 80 PB | 106 | 80PC | 70 |
| 120P (~ 120 cm) meristem | 120PA | 133 | 120 PB | 121 | 120PC | 120 |
| 160P (~ 160 cm) meristem | 160PA | 170 | 160 PB | 157 | 160PC | 170 |
| 200P (~ 200 cm) meristem | 200PA | 203 | 200 PB | 193 | 200PC | 198 |
| Floral development tissuesy | ||||||
| FT (boot stage florets) | FTA | n/a | FTB | n/a | FTC | n/a |
| PAF (pre-anthesis florets) | PAFA | n/a | PAFB | n/a | PAFC | n/a |
| ANT (Florets at anthesis) | ANTA | n/a | ANTB | n/a | ANTC | n/a |
| ST (Stamens) | STA | n/a | STB | n/a | STC | n/a |
| Seed development tissuesz | ||||||
| IS (Immature seeds) | ISA | n/a | ISB | n/a | ISC | n/a |
| MS (Mature seeds) | MSA | n/a | MSB | n/a | MSC | n/a |
wMeasurements are the actual distance (cm) from the base of the culm to the inflorescence node subtending the meristem for each sample
x20P, 40P, 80P, 120P, 160P, and 200P represented six sampling groups for inflorescence development and grouped by the approximate distance from the base of the culm where those samples were collected
yFT, floral spikelet tissue excised from boot stage inflorescences. PAF, floral spikelet tissues sampled from mature inflorescences before stigma extrusion from the palea and lemma. ANT, samples at the stage of floret development where stigmas were pollen receptive and bright red. ST, samples of fresh dehiscent anther tissues
zIS, samples of floret tissue approximately one-two weeks post-anthesis. MS, mature florets
RNA isolation, library preparation, and sequencing
Total RNA was extracted from all tissues using the Spectrum® Plant Total RNA Kit (Sigma-Aldrich, Burlington MA). DNA was digested on-column with the Sigma-Aldrich DNase10 (DNASE10) kit per the manufacturer’s instructions. RNA concentration and integrity were quantitated with the QubitTM fluorimeter (Life TechnologiesTM) and the 2100 Bioanalyzer (Agilent) before library preparation, respectively. RNA samples were poly-adenylation purified, and cDNA libraries were prepared using the BiooScientific (a PerkinElmer Co.) NEXTFlex Rapid Directional RNA-Seq kit with a target insert size of 200–300 bp. Libraries were sequenced using the Illumina HiSeq 4000, 150 bp PE by Novogene (Sacramento, CA). The RNA of all samples was mixed and used to construct Pacific Biosciences Iso-Seq libraries (Protocol # 101–070-200 version 6) with three size fractions (no size selected, < 4 kb, and > 4 kb). The libraries were sequenced with four cells of a PacBio Sequel I system at NC State Genomic Sciences Laboratory.
Transcriptome assembly and functional annotation
Read quality was inspected for quality with FastQC. Trimming was conducted with CLC Genomics WB (CLC – GWB, V11.0.1, QIAGEN) to remove adapter sequences and low-quality reads (Q < 20). Multiple de novo transcriptome assemblies were constructed with the CLC – GWB using different k-mer (word size) and the bubble size of de Bruijn graph combinations and assessed for the number of contiguity and N50 (Supplemental Fig. 1a). The assembly with the lowest number of contigs but the largest N50 was selected which maximized the yield of complete BUSCOs (Supplemental Fig. 1b & c). The final assembly was mapped (GMAP, V2015-07-23, [89]) to a draft genome assembly of T. ravennae (Maren et al., Unpub.) as well as multiple reference genomic assemblies within the Andropogoneae tribe, including Sorghum bicolor [46], Saccharum officinarum [90], and Zea mays [91]. The GMAP mapping was carried out to enrich the transcriptome for plant transcripts and eliminate the transcripts of sample surface contaminants [92]. Contigs with a 95% identity and match score to two or more reference genomes were retained. The transcriptome was analysed for redundancy with CD-HIT software with 95% identity to make a nonredundant set [92, 93]. Functional annotation was carried out on a local server using BLASTx and the nr (NCBI non-redundant protein 12/2018 version) database. Searches were limited to the first 20 significant results with an E-cutoff value of 1.0E-6. Unitigs were functionally annotated utilizing default annotation rules in the BLAST2GO package [94]. The unitigs and their BLASTX results were imported into the BLAST2GO package for functional gene annotation [94]. Gene ontology (GO) term and functional annotation assignments followed InterPro scan, using the European Molecular Biology Laboratory-European Bioinformatics Institute (EMBL-EBI) database, KEGG pathway analysis [95], Rfam annotation [96], and GO mapping based characterizations online on the BLAST2GO package [97].
Differentially expressed gene (DEG) and gene ontology (GO) enrichment analysis
Quality trimmed and filtered reads from all samples were mapped to the final transcriptome assembly with default parameters in the CLC – GWB. Statistical tests for the determination of differential gene expression utilize an exact test-like generalized linear model (GLM) similar to that performed in DESeq and EdgeR [98, 99]. In developing inflorescences, the test of differential expression utilized non-flowering controls for comparison. The statistical tests for differential expression in developing floral spikelets utilized two or more pair-wise comparisons between developing inflorescences and the sample query (e.g., FT, PAF, ST, ANT). Two or more pair-wise statistical tests between inflorescence controls, floral samples, and developing seeds comprise DEG calls. Genes of interest were filtered from differentially expressed genes table in CLC-GWB using a threshold p ≤ 0.05 and a two-fold threshold change. Venn diagrams were generated from gene lists using InteractiVenn [100]. TPM normalized expression values for DEG’s presented in each Venn diagram (Figs. 2, 4, and 6) were subjected coexpression analysis utilizing the R package “MBCluster.Seq” (version 1.0) [101]. Negative binomial modeling parameters were strategically adjusted to evaluate the number of coexpression clusters. Probability estimates greater than 0.9 for each member of a pair of genes coexpressing within the same cluster, as the parameters were adjusted, were used in the progressive selection of the clustering strategy as well as their gene memberships (Supplementary Fig. 5). The final clustering utilized a reduced set of expression data containing the listing of genes from the most probable coexpressing gene set identified within the preliminary analyses. Individual clusters were analyzed independently in CLC-GWB with the hierachical clustering algorithm in the development of the heat maps presented in Figs. 3, 5, and 7.
Gene expression analysis by RT-qPCR
Bioinformatically derived differential expression statistics of inflorescence, floret, and seed development were screened for novel and putative genes in reproductive development to validate the sample set with RT-qPCR according to Zhao et al. [102]. GO enrichment by hypergeometric test (p ≤ 0.05; CLC-GWB) aided in selecting sequences from the test set for over-representation (Supplementary Table 2, 3, & 4). Unigenes were filtered for significant differential expression (FDR p ≤ 0.05) within the subset of relevant inflorescence samples. Final transcript selections were made on the unique mapping (GMAP; V2015-07-23, [89]) of the transcript to the reference genome assembly with concomitant support for gene architecture from the PacBio Iso-Seq data set. Primers were designed for each gene to maximize coverage for gene structures, which uniquely identified the isoform of interest. Multiple internal controls were selected from the RNA-seq data set by filtering the expression data set for unigenes with a minimum expression value of 200 transcripts per million (TPM), a mean value of less than 2000 TPM, and having a CV less than 0.35 [103]. Relative gene expression analysis was used in the evaluation of PCR data in the determination of gene expression values and calculated following the 2-∆∆Ct method [104].
Supplementary Information
Additional file 1: Table S1. Sequencing statistics. Figure S1a-c. Transcriptome assembly. Figure S2. Annotation statistics for primary de novo assembly. Figure S3. Annotation statistics for cluster enriched assembly. Figure S4. Annotation statistics for PB Iso-Seq sequences. Table S2. GO-term enrichment for upregulated transcripts during inflorescence development. Table S3. GO-term enrichment for upregulated transcripts during flower development. Table S4. GO-term enrichment for upregulated transcripts during seed development. Table S5. Excel workbook including summaries of DEG’s in inflorescence development. Table S6. Excel workbook including summaries of DEG’s in floral development. Table S7. Excel workbook including summaries of DEG’s in seed development. Supplemental List 1. List of FASTA formatted sequences associated with Fig. 8 and Tables 2, 3, and 4. Table S8. Table export of annotations for the cluster enriched de novo transcriptome assembly. Table S9. Table export of annotations for the collapsed Iso-seq transcript set.
Acknowledgements
The authors greatly appreciate the financial support from the North Carolina Department of Agriculture and Consumer Services, the United States Department of Agriculture (USDA), the National Institute of Food and Agriculture, and the USDA Agricultural Research Service. Additional thanks are given to the North Carolina Arboretum for providing plant material.
Abbreviations
- ANT
Florets at anthesis
- CV
Coefficient of variation
- DEG
Differentially expressed gene
- DTE
Differentially expressed transcript
- FT
Boot stage florets
- GO
Gene ontology
- IS
Florets with immature seeds
- MS
Florets with mature seeds
- PAF
Pre-anthesis florets
- RNA
Ribonucleic acid
- RNA-seq
RNA sequence
- ST
Stamens
- TPM
Transcripts per million
- VM
Vegetative meristem
- 20P
Inflorescence meristem sampled at ~ 20 cm from the base of the culm
- 40P
Inflorescence meristem sampled at ~ 40 cm from the base of the culm
- 80P
Inflorescence meristem sampled at ~ 80 cm from the base of the culm
- 120P
Inflorescence meristem sampled at ~120 cm from the base of the culm
- 160P
Inflorescence meristem sampled at ~160 cm from the base of the culm
- 200P
Inflorescence meristem sampled at ~200 cm from the base of the culm
Authors’ contributions
This research was completed as part of NM’s Ph.D. dissertation. NM, TR, DT, and HA collaborated on the design, execution, analysis, and writing of this manuscript. RA processed the raw reads for the PB Iso-Seq data set. FZ and WL collaborated on the validation of RNA-Seq expression levels with RT-qPCR. All authors reviewed and approved the final manuscript.
Funding
Bioenergy Research Initiative, North Carolina Department of Agriculture and Consumer Services, Grant/Award Numbers: 17–072-4010 and 17–078-4009. United States Department of Agriculture, National Institute of Food and Agriculture, Project No. NC02559. United States Department of Agriculture, Agricultural Research Service, Cooperative Agreement, 58–8020–9-003.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the NCBI Bioproject repository PRJNA623617, Tripidium ravennae (ID 623617) (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA623617).
Declarations
Ethics approval and consent to participate
Relevant institutional, national, and international guidelines and legislation were complied by authors to use the plant materials. Required permission to use the plant material in this study was obtained from The North Carolina Arboretum. No KEGG pathway map picture also was used in the entire manuscript and we mentioned them only with appropriate reference. The authors of KEGG pathway maps also did not believe any permission needed.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nathan Maren, Email: namaren@ncsu.edu.
Hamid Ashrafi, Email: hamidashrafi@ncsu.edu.
References
- 1.Beale CV, Long SP. Seasonal dynamics of nutrient accumulation and partitioning in the perennial C4-grasses Miscanthus ×giganteus and Spartina cynosuroides. Biomass Bioenergy. 1997;12(6):419–428. doi: 10.1016/S0961-9534(97)00016-0. [DOI] [Google Scholar]
- 2.Boehmel C, Lewandowski I, Claupein W. Comparing annual and perennial energy cropping systems with different management intensities. Agric Syst. 2008;96(1–3):224–236. doi: 10.1016/j.agsy.2007.08.004. [DOI] [Google Scholar]
- 3.Maren NA, Touchell DH, Ranney TG, Ashrafi H, Whitfield MB, Chinn M. Biomass yields, cytogenetics, fertility, and compositional analyses of novel bioenergy grass hybrids (Tripidium spp.) Glob Change Biol Bioenerg. 2020;12(5):361–373. doi: 10.1111/gcbb.12676. [DOI] [Google Scholar]
- 4.Propheter JL, Staggenborg S. Performance of annual and perennial biofuel crops: nutrient removal during the first two years. Agron J. 2010;102(2):798–805. doi: 10.2134/agronj2009.0462. [DOI] [Google Scholar]
- 5.Palmer IE, Gehl RJ, Ranney TG, Touchell D, George N. Biomass yield, nitrogen response, and nutrient uptake of perennial bioenergy grasses in North Carolina. Biomass Bioenergy. 2014;63:218–228. doi: 10.1016/j.biombioe.2014.02.016. [DOI] [Google Scholar]
- 6.Welker CAD, McKain MR, Vorontsova MS, Peichoto MC, Kellogg EA. Plastome phylogenomics of sugarcane and relatives confirms the segregation of the genus Tripidium (Poaceae: Andropogoneae) TAXON. 2019;68(2):246–267. doi: 10.1002/tax.12030. [DOI] [Google Scholar]
- 7.Valdés B, Scholz H. The euro+ med treatment of Gramineae — a generic synopsis and some new names. Willdenowia. 2006;36(2):657–669. doi: 10.3372/wi.36.36202. [DOI] [Google Scholar]
- 8.Soreng RJ, Peterson PM, Romaschenko K, Davidse G, Teisher JK, Clark LG, et al. A worldwide phylogenetic classification of the Poaceae (Gramineae) II: An update and a comparison of two 2015 classifications. J Syst Evol. 2017;55(4):259–290. doi: 10.1111/jse.12262. [DOI] [Google Scholar]
- 9.Cv L. Species plantarum [...] Tomus II. Editio Secunda: Holmiae: Impensis direct Laurentii Salvii. 1763. [Google Scholar]
- 10.Beauvois P, Joseph AMF. Essai d'une nouvelle Agrostographie: ou nouveaux genres des Graminees. Paris: Imprimerie de Fain; 1812. [Google Scholar]
- 11.Chen S, Phillips S: Saccharum Linnaeus, Sp. Pl. 1: 54. 1753. In: Poaceae. Edited by committee FoCE, vol. 22. Beijing: Science Press; 2006;576–581.
- 12.Darke R. Encyclopedia of ornamental grasses for livable landscapes. Portland: Timber Press; 2007. [Google Scholar]
- 13.Lacy B, Hoagland BW. Vascular flora of a riparian site on the Canadian River, Cleveland County, Oklahoma. Oklahoma Native Plant Record. 2006;6(1):69-79.
- 14.Lambert AM, Dudley TL, Saltonstall K. Ecology and impacts of the large-statured invasive grasses Arundo donax and Phragmites australis in North America. Invas Plant Sci Mana. 2010;3(4):489–494. doi: 10.1614/IPSM-D-10-00031.1. [DOI] [Google Scholar]
- 15.Winston RL, DesCamp W, Andreas JE, Randall CB, Milan J, Scharzlander M. Forest health technology Enterprise team, Univ Idaho extension. 2014. New invaders of the southwest. [Google Scholar]
- 16.Barkworth ME, Anderton LK, Capels KM, Long S, Piep MB. Manual of grasses for North America: University Press of Colorado. 2007. [Google Scholar]
- 17.Vincent MA, Gardener RL: Spread of the invasive Ravenna grass (Tripidium ravennae, Poaceae) in Ohio. Phytoneuron. 2016;78:1–9.
- 18.Li H, Guo J, Zhang C, Zheng W, Song Y, Wang Y. Identification of differentially expressed miRNAs between a wheat K-type cytoplasmic male sterility line and its near-isogenic restorer line. Plant Cell Physiol. 2019;60(7):1604–1618. doi: 10.1093/pcp/pcz065. [DOI] [PubMed] [Google Scholar]
- 19.Preston JC, Fjellheim S. Understanding past, and predicting future, niche transitions based on grass flowering time variation. Plant Physiol. 2020;183(3):822–839. doi: 10.1104/pp.20.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar RR, Goswami S, Sharma SK, Kala YK, Rai GK, Mishra DC, et al. Harnessing next generation sequencing in climate change: RNA-Seq analysis of heat stress-responsive genes in wheat (Triticum aestivum L.) OMICS. 2015;19(10):632–647. doi: 10.1089/omi.2015.0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Digel B, Pankin A, von Korff M. Global transcriptome profiling of developing leaf and shoot apices reveals distinct genetic and environmental control of floral transition and inflorescence development in barley. Plant Cell. 2015;27(9):2318–2334. doi: 10.1105/tpc.15.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mulki MA, Bi X, von Korff M. FLOWERING LOCUS T3 controls spikelet initiation but not floral development. Plant Physiol. 2018;178(3):1170–1186. doi: 10.1104/pp.18.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cockram J, Thiel T, Steuernagel B, Stein N, Taudien S, Bailey PC, et al. Genome dynamics explain the evolution of flowering time CCT domain gene families in the Poaceae. PLoS One. 2012;7(9):e45307. doi: 10.1371/journal.pone.0045307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tornqvist C-E, Vaillancourt B, Kim J, Buell CR, Kaeppler SM, Casler MD. Transcriptional analysis of flowering time in switchgrass. Bioenerg Res. 2017;10(3):700–713. doi: 10.1007/s12155-017-9832-9. [DOI] [Google Scholar]
- 25.Xiang LX, He D, Dong WR, Zhang YW, Shao JZ. Deep sequencing-based transcriptome profiling analysis of bacteria-challenged Lateolabrax japonicus reveals insight into the immune-relevant genes in marine fish. BMC Genomics. 2010;11(1):472. doi: 10.1186/1471-2164-11-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang Q, Ma X, Mo C, Wilson IW, Song C, Zhao H, et al. An efficient approach to finding Siraitia grosvenorii triterpene biosynthetic genes by RNA-seq and digital gene expression analysis. BMC Genomics. 2011;12(1):343. doi: 10.1186/1471-2164-12-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colasanti J, Coneva V. Mechanisms of floral induction in grasses: something borrowed, something new. Plant Physiol. 2009;149(1):56–62. doi: 10.1104/pp.108.130500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang DB, Yuan Z. Molecular control of grass inflorescence development. Annu Rev Plant Biol. 2014;65(1):553–578. doi: 10.1146/annurev-arplant-050213-040104. [DOI] [PubMed] [Google Scholar]
- 29.Whipple CJ. Grass inflorescence architecture and evolution: the origin of novel signaling centers. New Phytol. 2017;216(2):367–372. doi: 10.1111/nph.14538. [DOI] [PubMed] [Google Scholar]
- 30.Lepiniec L, Devic M, Roscoe TJ, Bouyer D, Zhou DX, Boulard C, et al. Molecular and epigenetic regulations and functions of the LAFL transcriptional regulators that control seed development. Plant Reprod. 2018;31(3):291–307. doi: 10.1007/s00497-018-0337-2. [DOI] [PubMed] [Google Scholar]
- 31.Jo L, Pelletier JM, Harada JJ. Central role of the LEAFY COTYLEDON1 transcription factor in seed development. J Integr Plant Biol. 2019;61(5):564–580. doi: 10.1111/jipb.12806. [DOI] [PubMed] [Google Scholar]
- 32.Zhao T, Holmer R, de Bruijn S, Angenent GC, van den Burg HA, Schranz ME. Phylogenomic synteny network analysis of MADS-Box transcription factor genes reveals lineage-specific transpositions, ancient tandem duplications, and deep positional conservation. Plant Cell. 2017;29(6):1278–1292. doi: 10.1105/tpc.17.00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Munster T, Wingen LU, Faigl W, Werth S, Saedler H, Theissen G. Characterization of three GLOBOSA-like MADS-box genes from maize: evidence for ancient paralogy in one class of floral homeotic B-function genes of grasses. Gene. 2001;262(1-2):1–13. doi: 10.1016/S0378-1119(00)00556-4. [DOI] [PubMed] [Google Scholar]
- 34.Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457(7229):551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 35.Li J, Gao K, Yang X, Khan WU, Guo B, Guo T, et al. Identification and characterization of the CONSTANS-like gene family and its expression profiling under light treatment in Populus. Int J Biol Macromol. 2020;161:999–1010. doi: 10.1016/j.ijbiomac.2020.06.056. [DOI] [PubMed] [Google Scholar]
- 36.Preston JC, Kellogg EA. Reconstructing the evolutionary history of paralogous APETALA1/FRUITFULL-like genes in grasses (Poaceae) Genetics. 2006;174(1):421–437. doi: 10.1534/genetics.106.057125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu X, Wu J, Luo Y, Bragg J, Anderson O, Vogel J, Gu YQ. Phylogenetic, molecular, and biochemical characterization of caffeic acid o-methyltransferase gene family in Brachypodium distachyon. Int J Plant Genomics. 2013;2013:1–12. [DOI] [PMC free article] [PubMed]
- 38.Jung JH, Altpeter F. TALEN mediated targeted mutagenesis of the caffeic acid O-methyltransferase in highly polyploid sugarcane improves cell wall composition for production of bioethanol. Plant Mol Biol. 2016;92(1-2):131–142. doi: 10.1007/s11103-016-0499-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei B, Zhang RZ, Guo JJ, Liu DM, Li AL, Fan RC, et al. Genome-wide analysis of the MADS-Box gene family in Brachypodium distachyon. PLoS One. 2014;9(1):e84781. doi: 10.1371/journal.pone.0084781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dornelas MC, Patreze CM, Angenent GC, Immink RG. MADS: the missing link between identity and growth? Trends Plant Sci. 2011;16(2):89–97. doi: 10.1016/j.tplants.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 41.Reinheimer R, Kellogg EA. Evolution of AGL6-like MADS box genes in grasses (Poaceae): ovule expression is ancient and Palea expression is new. Plant Cell. 2009;21(9):2591–2605. doi: 10.1105/tpc.109.068239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herath V. The architecture of the GhD7 promoter reveals the roles of GhD7 in growth, development and the abiotic stress response in rice. Comput Biol Chem. 2019;82:1–8. doi: 10.1016/j.compbiolchem.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 43.Yan W, Liu H, Zhou X, Li Q, Zhang J, Lu L, et al. Natural variation in Ghd7.1 plays an important role in grain yield and adaptation in rice. Cell Res. 2013;23(7):969–971. doi: 10.1038/cr.2013.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ren Y, Chen D, Li W, Zhou D, Luo T, Yuan G, et al. OsSHOC1 and OsPTD1 are essential for crossover formation during rice meiosis. Plant J. 2019;98(2):315–328. doi: 10.1111/tpj.14214. [DOI] [PubMed] [Google Scholar]
- 45.Bolaños-Villegas P, Yang X, Wang H-J, Juan C-T, Chuang M-H, Makaroff CA, et al. Arabidopsis CHROMOSOME TRANSMISSION FIDELITY 7 (AtCTF7/ECO1) is required for DNA repair, mitosis and meiosis. Plant J. 2013;75(6):927–940. doi: 10.1111/tpj.12261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dai XR, Gao XQ, Chen GH, Tang LL, Wang H, Zhang XS. ABNORMAL POLLEN TUBE GUIDANCE1, an endoplasmic reticulum-localized mannosyltransferase homolog of GLYCOSYLPHOSPHATIDYLINOSITOL10 in yeast and PHOSPHATIDYLINOSITOL GLYCAN ANCHOR BIOSYNTHESIS B in human, is required for Arabidopsis pollen tube micropylar GUIDANCE and embryo development. Plant Physiol. 2014;165(4):1544–1556. doi: 10.1104/pp.114.236133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li HJ, Xue Y, Jia DJ, Wang T, Hi DQ, Liu J, et al. POD1 regulates pollen tube guidance in response to micropylar female signaling and acts in early embryo patterning in Arabidopsis. Plant Cell. 2011;23(9):3288–3302. doi: 10.1105/tpc.111.088914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pnueli L, Carmel-Goren L, Hareven D, Gutfinger T, Alvarez J, Ganal M, et al. The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development. 1998;125(11):1979–1989. doi: 10.1242/dev.125.11.1979. [DOI] [PubMed] [Google Scholar]
- 49.Ko SS, Li MJ, Sun-Ben Ku M, Ho YC, Lin YJ, Chuang MH, et al. The bHLH142 transcription factor coordinates with TDR1 to modulate the expression of EAT1 and regulate pollen development in rice. Plant Cell. 2014;26(6):2486–2504. doi: 10.1105/tpc.114.126292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ono S, Liu H, Tsuda K, Fukai E, Tanaka K, Sasaki T, et al. EAT1 transcription factor, a non-cell-autonomous regulator of pollen production, activates meiotic small RNA biogenesis in rice anther tapetum. PLoS Genet. 2018;14(2):e1007238. doi: 10.1371/journal.pgen.1007238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nonomura K, Morohoshi A, Nakano M, Eiguchi M, Miyao A, Hirochika H, et al. A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell. 2007;19(8):2583–2594. doi: 10.1105/tpc.107.053199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kapu NUS, Cosgrove DJ. Changes in growth and cell wall extensibility of maize silks following pollination. J Exp Bot. 2010;61(14):4097–4107. doi: 10.1093/jxb/erq225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolabu TW, Zhang F, Niu LF, Kalve S, Bhatnagar-Mathur P, Muszynski MG, et al. Three FLOWERING LOCUS T-like genes function as potential florigens and mediate photoperiod response in sorghum. New Phytol. 2016;210(3):946–959. doi: 10.1111/nph.13834. [DOI] [PubMed] [Google Scholar]
- 54.Ito T, Ng KH, Lim TS, Yu H, Meyerowitz EM. The homeotic protein AGAMOUS controls late stamen development by regulating a jasmonate biosynthetic gene in Arabidopsis. Plant Cell. 2007;19(11):3516–3529. doi: 10.1105/tpc.107.055467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamaguchi T, Nagasawa N, Kawasaki S, Matsuoka M, Nagato Y, Hirano HY. The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell. 2004;16(2):500–509. doi: 10.1105/tpc.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang XM, Xie B, Zhu MS, Zhang ZM, Hong ZL. Nucleostemin-like 1 is required for embryogenesis and leaf development in Arabidopsis. Plant Mol Biol. 2012;78(1-2):31–44. doi: 10.1007/s11103-011-9840-7. [DOI] [PubMed] [Google Scholar]
- 57.Wang XM, Gingrich DK, Deng YF, Hong ZL. A nucleostemin-like GTPase required for normal apical and floral meristem development in Arabidopsis. Mol Biol Cell. 2012;23(8):1446–1456. doi: 10.1091/mbc.e11-09-0797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li D, Wang L, Wang M, Xu YY, Luo W, Liu YJ, et al. Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol J. 2009;7(8):791–806. doi: 10.1111/j.1467-7652.2009.00444.x. [DOI] [PubMed] [Google Scholar]
- 59.Ge L, Chen H, Jiang JF, Zhao Y, Xu ML, Xu YY, et al. Overexpression of OsRAA1 causes pleiotropic phenotypes in transgenic rice plants, including altered leaf, flower, and root development and root response to gravity. Plant Physiol. 2004;135(3):1502–1513. doi: 10.1104/pp.104.041996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robert HS, Quint A, Brand D, Vivian-Smith A, Offringa R. BTB and TAZ domain scaffold proteins perform a crucial function in Arabidopsis development. Plant J. 2009;58(1):109–121. doi: 10.1111/j.1365-313X.2008.03764.x. [DOI] [PubMed] [Google Scholar]
- 61.Portereiko MF, Sandaklie-Nikolova L, Lloyd A, Dever CA, Otsuga D, Drews GN. Nuclear fusion defective1 encodes the Arabidopsis RPL21M protein and is required for karyogamy during female gametophyte development and fertilization. Plant Physiol. 2006;141(3):957–965. doi: 10.1104/pp.106.079319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Becraft PW, Stinard PS, McCarty DR. CRINKLY4: a TNFR-like receptor kinase involved in maize epidermal differentiation. Science. 1996;273(5280):1406–1409. doi: 10.1126/science.273.5280.1406. [DOI] [PubMed] [Google Scholar]
- 63.Hill A, Nantel A, Rock CD, Quatrano RS. A conserved domain of the viviparous-1 gene product enhances the DNA binding activity of the bZIP protein EmBP-1 and other transcription factors. J Biol Chem. 1996;271(7):3366–3374. doi: 10.1074/jbc.271.7.3366. [DOI] [PubMed] [Google Scholar]
- 64.Mjomba FM, Zheng Y, Liu HQ, Tang WQ, Hong ZL, Wang F, et al. Homeobox is pivotal for OsWUS controlling tiller development and female fertility in rice. G3-Genes Genom Genet. 2016;6(7):2013–2021. doi: 10.1534/g3.116.028837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hundertmark M, Hincha DK. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics. 2008;9(1):118. doi: 10.1186/1471-2164-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Asano T, Kunieda N, Omura Y, Ibe H, Kawasaki T, Takano M, et al. Rice SPK, a calmodulin-like domain protein kinase, is required for storage product accumulation during seed development: phosphorylation of sucrose synthase is a possible factor. Plant Cell. 2002;14(3):619–628. doi: 10.1105/tpc.010454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lopez-Ribera I, La Paz JL, Repiso C, Garcia N, Miquel M, Hernandez ML, et al. The evolutionary conserved oil body associated protein OBAP1 participates in the regulation of oil body size. Plant Physiol. 2014;164(3):1237–1249. doi: 10.1104/pp.113.233221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sreedhar R, Tiku PK. Cupincin: a unique protease purified from rice (Oryza sativa L.) bran is a new member of the cupin superfamily. PLoS One. 2016;11(4):e0152819. doi: 10.1371/journal.pone.0152819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Close TJ. Dehydrins: a commonalty in the response of plants to dehydration and low temperature. Physiol Plant. 1997;100(2):291–296. doi: 10.1111/j.1399-3054.1997.tb04785.x. [DOI] [Google Scholar]
- 70.Kanno T, Bucher E, Daxinger L, Huettel B, Kreil DP, Breinig F, et al. RNA-directed DNA methylation and plant development require an IWR1-type transcription factor. EMBO Rep. 2010;11(1):65–71. doi: 10.1038/embor.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Che J, Yamaji N, Shen RF, Ma JF. An Al-inducible expansin gene, OsEXPA10 is involved in root cell elongation of rice. Plant J. 2016;88(1):132–142. doi: 10.1111/tpj.13237. [DOI] [PubMed] [Google Scholar]
- 72.Santiago TR, Pereira VM, de Souza WR, Steindorff AS, Cunha B, Gaspar M, et al. Genome-wide identification, characterization and expression profile analysis of expansins gene family in sugarcane (Saccharum spp.) PLoS One. 2018;13(4):e0196140. doi: 10.1371/journal.pone.0196140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Narayan JA, Dharshini S, Manoj VM, Padmanabhan TSS, Kadirvelu K, Suresha GS, et al. Isolation and characterization of water-deficit stress-responsive alpha-expansin 1 (EXPA1) gene from Saccharum complex. 3 Biotech. 2019;9(5):186. doi: 10.1007/s13205-019-1719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Malcomber ST, Kellogg EA. Heterogeneous expression patterns and separate roles of the SEPALLATA gene LEAFY HULL STERILE1 in grasses. Plant Cell. 2004;16(7):1692–1706. doi: 10.1105/tpc.021576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu C, Yang J, Box MS, Kellogg EA, Eveland AL. A dynamic co-expression map of early inflorescence development in Setaria viridis provides a resource for gene discovery and comparative genomics. Front Plant Sci. 2018;9:1309. doi: 10.3389/fpls.2018.01309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316(5827):1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- 77.Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, et al. The wheat and barley vernalization gene VRN3 is an orthologue of FT. P Natl Acad Sci USA. 2006;103(51):19581–19586. doi: 10.1073/pnas.0607142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pearce S, Vanzetti LS, Dubcovsky J. Exogenous gibberellins induce wheat spike development under short days only in the presence of VERNALIZATION1. Plant Physiol. 2013;163(3):1433–1445. doi: 10.1104/pp.113.225854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lenhard M, Bohnert A, Jurgens G, Laux T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell. 2001;105(6):805–814. doi: 10.1016/S0092-8674(01)00390-7. [DOI] [PubMed] [Google Scholar]
- 80.Danilevskaya ON, Meng X, Hou Z, Ananiev EV, Simmons CR. A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol. 2008;146(1):250–264. doi: 10.1104/pp.107.109538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, et al. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002;43(10):1096–1105. doi: 10.1093/pcp/pcf156. [DOI] [PubMed] [Google Scholar]
- 82.Duan YL, Xing Z, Diao ZJ, Xu WY, Li SP, Du XQ, et al. Characterization of Osmads6-5, a null allele, reveals that OsMADS6 is a critical regulator for early flower development in rice (Oryza sativa L.) Plant Mol Biol. 2012;80(4–5):429–442. doi: 10.1007/s11103-012-9958-2. [DOI] [PubMed] [Google Scholar]
- 83.Strable J, Vollbrecht E. Maize YABBY genes drooping leaf1 and drooping leaf2 regulate floret development and floral meristem determinacy. Development. 2019;146(6):dev171181. doi: 10.1242/dev.171181. [DOI] [PubMed] [Google Scholar]
- 84.Strable J, Wallace JG, Unger-Wallace E, Briggs S, Bradbury PJ, Buckler ES, et al. Maize YABBY genes drooping leaf1 and drooping leaf2 regulate plant architecture. Plant Cell. 2017;29(7):1622–1641. doi: 10.1105/tpc.16.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Su YL, Liu JX, Liang WQ, Dou YH, Fu RF, Li WQ, et al. Wheat AGAMOUS LIKE 6 transcription factors function in stamen development by regulating the expression of TaAPETALA3. Development. 2019;146(20):dev177527. doi: 10.1242/dev.177527. [DOI] [PubMed] [Google Scholar]
- 86.Niu N, Liang W, Yang X, Jin W, Wilson ZA, Hu J, et al. EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat Commun. 2013;4(1):1445. doi: 10.1038/ncomms2396. [DOI] [PubMed] [Google Scholar]
- 87.Ji C, Li H, Chen L, Xie M, Wang F, Chen Y, et al. A novel rice bHLH transcription factor, DTD, acts coordinately with TDR in controlling tapetum function and pollen development. Mol Plant. 2013;6(5):1715–1718. doi: 10.1093/mp/sst046. [DOI] [PubMed] [Google Scholar]
- 88.Macaisne N, Vignard J, Mercier R. SHOC1 and PTD form an XPF-ERCC1-like complex that is required for formation of class I crossovers. J Cell Sci. 2011;124(16):2687–2691. doi: 10.1242/jcs.088229. [DOI] [PubMed] [Google Scholar]
- 89.Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21(9):1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 90.Garsmeur O, Droc G, Antonise R, Grimwood J, Potier B, Aitken K, et al. A mosaic monoploid reference sequence for the highly complex genome of sugarcane. Nat Commun. 2018;9(1):2638. doi: 10.1038/s41467-018-05051-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326(5956):1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 92.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 93.Fu LM, Niu BF, Zhu ZW, Wu ST, Li WZ. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28(23):3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Conesa A, Gotz S. Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics. 2008;619832:1-12. [DOI] [PMC free article] [PubMed]
- 95.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nawrocki EP, Burge SW, Bateman A, Daub J, Eberhardt RY, Eddy SR, et al. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 2015;43(D1):D130–D137. doi: 10.1093/nar/gku1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36(10):3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Robinson MD, McCarthy DJ. Smyth GK: edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics. 2008;9(2):321–332. doi: 10.1093/biostatistics/kxm030. [DOI] [PubMed] [Google Scholar]
- 100.Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. 2015;16(1):169. doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Si Y, Liu P, Li P, Brutnell TP. Model-based clustering for RNA-seq data. Bioinformatics. 2014;30(2):197–205. doi: 10.1093/bioinformatics/btt632. [DOI] [PubMed] [Google Scholar]
- 102.Zhao F, Maren NA, Kosentka PZ, Liao YY, Lu H, Duduit JR, et al. An optimized protocol for stepwise optimization of real-time RT-PCR analysis. Hort. Res. 2021 (in review). [DOI] [PMC free article] [PubMed]
- 103.Cheng Y, Bian W, Pang X, Yu J, Ahammed GJ, Zhou G, et al. Genome-wide identification and evaluation of reference genes for quantitative RT-PCR analysis during tomato fruit development. Front Plant Sci. 2017;8:1440. doi: 10.3389/fpls.2017.01440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Sequencing statistics. Figure S1a-c. Transcriptome assembly. Figure S2. Annotation statistics for primary de novo assembly. Figure S3. Annotation statistics for cluster enriched assembly. Figure S4. Annotation statistics for PB Iso-Seq sequences. Table S2. GO-term enrichment for upregulated transcripts during inflorescence development. Table S3. GO-term enrichment for upregulated transcripts during flower development. Table S4. GO-term enrichment for upregulated transcripts during seed development. Table S5. Excel workbook including summaries of DEG’s in inflorescence development. Table S6. Excel workbook including summaries of DEG’s in floral development. Table S7. Excel workbook including summaries of DEG’s in seed development. Supplemental List 1. List of FASTA formatted sequences associated with Fig. 8 and Tables 2, 3, and 4. Table S8. Table export of annotations for the cluster enriched de novo transcriptome assembly. Table S9. Table export of annotations for the collapsed Iso-seq transcript set.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the NCBI Bioproject repository PRJNA623617, Tripidium ravennae (ID 623617) (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA623617).