Abstract
Receptor-like kinases (RLKs) are key cell signaling components. The rice ARBUSCULAR RECEPTOR-LIKE KINASE 1 (OsARK1) regulates the arbuscular mycorrhizal (AM) association postarbuscule development and belongs to an undefined subfamily of RLKs. Our phylogenetic analysis revealed that ARK1 has an ancient paralogue in spermatophytes, ARK2. Single ark2 and ark1/ark2 double mutants in rice showed a nonredundant AM symbiotic function for OsARK2. Global transcriptomics identified a set of genes coregulated by the two RLKs, suggesting that OsARK1 and OsARK2 orchestrate symbiosis in a common pathway. ARK lineage proteins harbor a newly identified SPARK domain in their extracellular regions, which underwent parallel losses in ARK1 and ARK2 in monocots. This protein domain has ancient origins in streptophyte algae and defines additional overlooked groups of putative cell surface receptors.
Keywords: arbuscular mycorrhiza, SPARK receptor-like kinases, OsARK2
Land plants and arbuscular mycorrhizal (AM) fungi form an ancient and widespread nutritional mutualism. Plant–fungal reciprocal recognition in the rhizosphere is commonly followed by hyphal entry into plant roots and subsequent bidirectional nutrient exchange fostered at intracellular arbuscules. A plant-derived periarbuscular membrane (PAM) surrounds arbuscule branches as they develop, creating a potential hub for plant–fungal communication.
The receptor-like kinase (RLK) ARBUSCULAR RECEPTOR-LIKE KINASE 1 (ARK1) is required for sustenance of AM symbiosis in rice and Medicago truncatula (1, 2) and is evolutionarily conserved in genomes of AM-competent plant species (2, 3). The rice OsARK1 (LOC_Os11g26140) is a PAM RLK that regulates fungal fitness at the postarbuscule development stage (1). As RLKs tend to operate in complex signaling circuits, we aimed to identify additional components of the OsARK1 pathway.
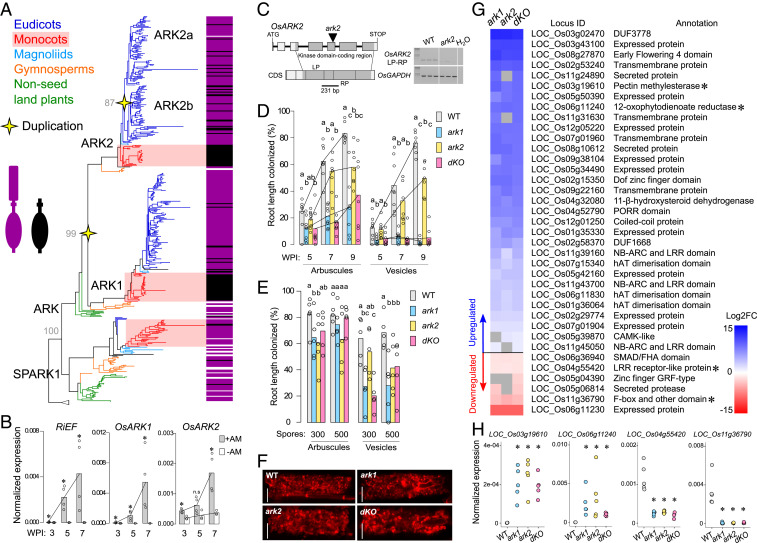
OsARK1 belongs to an uncharacterized group of land plant–specific RLKs, the Unknown Receptor Kinase-2 (URK-2) subfamily (4). In order to trace the evolutionary history of the URK-2 subfamily, we conducted a phylogenetic analysis using publicly available genomic and transcriptomic data (Fig. 1A and Dataset S1). We found that the URK-2 subfamily is composed of two members in nonseed land plants: ARK and SIMILAR PROTEIN TO ARK 1 (SPARK1). An ancient duplication in the ARK lineage gave rise to the paralogues ARK1 and ARK2 in spermatophytes. The latter experienced a further duplication early in the evolution of eudicots creating ARK2a and ARK2b, which in some cases was followed by loss of one of the paralogues. Contrary to ARK lineage genes, SPARK1 can be found in several non-AM land plants and was lost in eudicots. As in other monocots, the URK-2 subfamily in rice is composed of three members. The two paralogues OsARK1 and OsARK2 (LOC_Os04g39180) code for predicted functional RLKs that lack extracellular domains (EDs) and OsARK2 also lacks a predicted signal peptide. OsSPARK1 (LOC_Os07g12480), however, is predicted to harbor a 250-amino-acid-long ED. Occurrence of this ED was surveyed across URK-2 subfamily members, revealing that it was lost independently in ARK1 and ARK2 in monocots (Fig. 1A). This suggests that ARK1 and ARK2 follow a common evolutionary trajectory in monocots despite occurring in paralogous gene lineages that diverged early in the evolution of tracheophytes. This prompted us to functionally characterize OsARK2.
Fig. 1.
Identification and characterization of OsARK2. (A) Phylogenetic tree of the URK-2 RLK subfamily. Bootstrap values of important nodes are shown. Occurrence of the ED was surveyed for each sequence, purple denoting presence and black absence. White accounts for incomplete sequences where occurrence of the ED could not be established. A detailed tree is available in Dataset S1. (B) Comparative gene expression assay in a time course of wild-type rice. Expression levels of Rhizophagus irregularis ELONGATION FACTOR (RiEF) are included to account for increases of AM fungal biomass over time. Expression levels in inoculated (+AM) and noninoculated (−AM) roots are normalized to OsCYCLOPHILIN2. Bars represent means. Asterisks denote statistically significant differences between +AM and −AM treatments (Kruskal–Wallis and post hoc Dunn’s test P < 0.05). (C) Gene structure of OsARK2. Tos17 element is inserted between the first and second nucleotide of the codon encoding the universal Asp residue from the kinase domain catalytic loop in exon V (978 base pairs from first nucleotide of ATG). Right: RT-PCR shows no transcripts in ark2 using oligonucleotides spanning Tos17 insertion. (D) Time course colonization assay employing 250 R. irregularis spores as inoculum. (E) Colonization assay under different inoculum pressures evaluated at 6 wk postinoculation (WPI). For D and E, bars represent means and different letters denote statistically significant differences between genotypes (Kruskal–Wallis and post hoc Dunn’s test P < 0.05). (F) Confocal microscope images of fully developed arbuscules stained with wheat germ agglutinin (WGA)-Alexa Fluor 633. (Scale bar, 10 μm.) Representative images per genotype are provided. (G) Subset of DEGs from the RNA-seq assay that displayed a strict nonoverlapping expression pattern in the WT-dKO comparison. Color hue accounts for degree of up- or down-regulation compared to the WT (log2FC ≥ |1|, FDR adjusted P ≤ 0.05). Gray squares represent no expression changes in the respective genotypes. Asterisks mark genes selected for validation. (H) qRT-PCR assays confirming the pattern of expression of a subset of DEGs in an independent experiment. Expression levels are normalized to OsCYCLOPHILIN2. Asterisks denote statistically significant differences between gene expression of mutant genotypes and WT control (Kruskal–Wallis and post hoc Dunn’s test P < 0.05). FC, fold change. n.s., statistically nonsignificant.
OsARK2 transcripts have been reported to accumulate during AM symbiosis (1, 5, 6). We performed a time course gene expression assay confirming OsARK2 to be induced in AM conditions but, in contrast to OsARK1, low transcript levels of OsARK2 were also detected in noninoculated plants (Fig. 1B). We obtained an ark2 mutant allele containing a Tos17 retrotransposon element insertion in the kinase domain catalytic loop–coding region. Disruption of the OsARK2 transcript was confirmed through RT-PCR (Fig. 1C). This ark2 mutant was crossed with the previously characterized ark1-2 mutant allele (hereafter referred as ark1) (1) to generate an ark1/ark2 double knockout (dKO) line. A time course experiment revealed that the ark2 mutant had a significantly reduced AM colonization phenotype; however, arbuscules and vesicles were more abundant than in ark1 (Fig. 1D). The phenotypes of both ark1 and ark2 are plastic, as applying higher amounts of spore inoculum resulted in wild-type (WT) levels of arbuscule abundance and an even reduction of vesicles across mutant genotypes (Fig. 1E). Arbuscules were able to fully branch in ark2 akin to ark1 (Fig. 1F). The dKO largely reproduced the ark1 phenotype (Fig. 1 D–F). These observations suggest a nonredundant and nonsynergistic regulation of AM symbiosis by OsARK1 and OsARK2.
We determined the transcriptional consequences of the different mutations via RNA-sequencing (RNA-seq). To identify genotype effects, we employed 10 biological replicates per genotype and measured the correlation of arbuscule abundance to expression levels of each gene across replicates. This resulted in a filtered dataset where transcriptional responses associated solely with differential colonization levels were removed (Dataset S2). We focused on the most stringently differentially expressed genes (DEGs) whose expression levels in all 10 dKO replicates were higher or lower than all 10 wild-type replicates. This strict nonoverlapping expression was observed in a total of 31 up- and 6 down-regulated DEGs, most of which were also differentially expressed in the single mutants (Fig. 1 G and H). This convergent response suggests that OsARK1 and OsARK2 operate in a common, yet undescribed symbiotic genetic program.
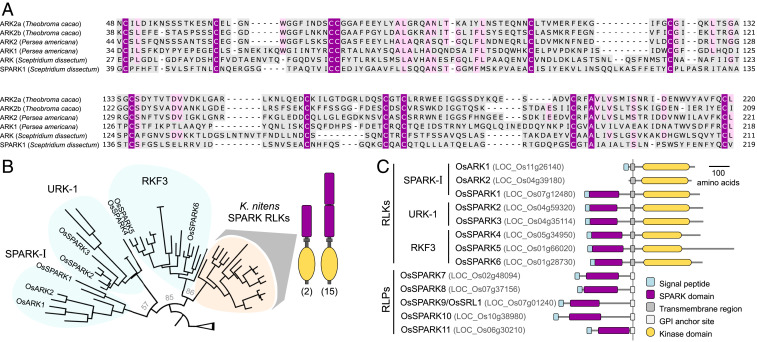
ARK1 and ARK2 in monocots are characterized by the loss of an otherwise conserved ED (Fig. 1A). This ED has not been defined and has no homology to known domains. An alignment of the EDs of all URK-2 orthologs identified in this study showed them to have a highly conserved signature arrangement of 12 cysteine residues (Fig. 2A). This newly described domain was named SPARK (Pfam ID: PF19160, available at https://pfam.xfam.org/family/PF19160). Accordingly, the URK-2 RLK subfamily was renamed SPARK-I. The SPARK domain was also found to constitute the ED of two other uncharacterized land plant RLK subfamilies; the URK-1 (Unknown Receptor Kinase-1) and RKF3 (Receptor Kinase in Flowers 3) (Fig. 2B). In addition, we detected the SPARK domain in a number of RLKs of Klebsormidium nitens, a member of a lineage of streptophyte algae sister to land plants. Surprisingly, the SPARK domain was present in more than 10% of the small repertoire of K. nitens RLKs. These RLKs form a monophyletic group and their domain architecture mostly consists in two tandemly arranged SPARK domains (Fig. 2B). In land plants, we also found the SPARK domain in predicted GPI-anchored receptor-like proteins (RLPs), including the previously characterized rice gene SEMI-ROLLED LEAF1 (OsSRL1), involved in cell wall formation (7, 8). We identified a total of 11 SPARK domain–containing proteins in rice (Fig. 2C).
Fig. 2.
The SPARK domain. (A) Amino acid sequence alignment of the SPARK domain from selected representative sequences for each member of the SPARK-I RLK subfamily. Residues colored purple are conserved in at least 80% of all sequences identified in this study. Residues in pink are conserved in at least 50% of the sequences. (B) Phylogenetic tree of SPARK domain–harboring RLK subfamilies. All sequences form the SPARK-I, URK-1, and RKF3 subfamilies from selected plant species (Physcomitrium patens, Selaginella moellendorffii, Amborella trichopoda, Arabidopsis thaliana, and rice) are included along with all 17 SPARK domain–harboring RLKs from K. nitens. Branches corresponding to rice proteins are named. Number of K. nitens RLKs having one or two SPARK domains are written below schematics. A detailed tree is available in Dataset S4. (C) Predicted protein domain architecture of rice SPARK-I subfamily members along with all rice proteins identified in this study predicted to have a SPARK domain.
In summary, we characterized OsARK2 as a RLK functioning in AM symbiosis. While previous large-scale phylogenomics studies suggested a strict association between the two RLKs and AM symbiosis (2, 3), we detected ARK2 but not ARK1 in the three orchid genomes available (Dataset S1) and ARK2 is induced during orchid mycorrhizal symbiosis (9). This may reflect a putative broader role of ARK2 compared to ARK1 in plant-fungal symbioses. Functionally characterizing the gene suite regulated by the two RLKs identified here by transcriptomics represents an avenue to resolve differential roles. In addition, the discovery of the SPARK domain provides the foundations to unravel ancestral, as yet unknown signaling pathways in the green lineage and invites further investigation into its function and the biological context allowing for its particular pattern of occurrence in symbiotic RLKs.
Materials and Methods
Molecular characterization of mutants, plant growth conditions, AM inoculation and colonization assessments were performed following previously described protocols and guidelines (1, 10). Phylogenetic analyses, gene expression assays, RNA-seq assays, and data analyses were performed as in ref. 11. Procedures are detailed in SI Appendix, Extended Methods.
Supplementary Material
Acknowledgments
We thank Rob Finn (European Bioinformatics Institute) for curating the SPARK domain into the Pfam database and James Gattward for technical assistance. H.M. was supported by the Chilean National Agency for Research and Development and Cambridge TRUST. Research in U.P.’s laboratory was supported by the Biotechnology and Biological Sciences Research Council Grant BB/P003419/1, the Engineering the Nitrogen Symbiosis for Africa project, which is funded by a grant to the University of Cambridge by the Bill & Melinda Gates Foundation, and a St. John’s College Teaching and Research grant.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2105281118/-/DCSupplemental.
Data Availability
See Dataset S1 for a detailed phylogenetic tree of Fig. 1A, Dataset S2 for RNA-seq data, Dataset S3 for a list of sequences, and Dataset S4 for a detailed phylogenetic tree of Fig. 2B. RNA-seq raw data were deposited in the National Center for Biotechnology Information gene expression omnibus (ID: GSE168162) (12). All other study data are included in the supporting information.
References
- 1.Roth R., et al., A rice Serine/Threonine receptor-like kinase regulates arbuscular mycorrhizal symbiosis at the peri-arbuscular membrane. Nat. Commun. 9, 4677 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bravo A., York T., Pumplin N., Mueller L. A., Harrison M. J., Genes conserved for arbuscular mycorrhizal symbiosis identified through phylogenomics. Nat. Plants 2, 15208 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Radhakrishnan G. V., et al., An ancestral signalling pathway is conserved in intracellular symbioses-forming plant lineages. Nat. Plants 6, 280–289 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Lehti-Shiu M. D., Shiu S. H., Diversity, classification and function of the plant protein kinase superfamily. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 2619–2639 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Güimil S., et al., Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc. Natl. Acad. Sci. U.S.A. 102, 8066–8070 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gutjahr C., et al., Transcriptome diversity among rice root types during asymbiosis and interaction with arbuscular mycorrhizal fungi. Proc. Natl. Acad. Sci. U.S.A. 112, 6754–6759 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiang J. J., Zhang G. H., Qian Q., Xue H. W., Semi-rolled leaf1 encodes a putative glycosylphosphatidylinositol-anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells. Plant Physiol. 159, 1488–1500 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W. Q., et al., CLD1/SRL1 modulates leaf rolling by affecting cell wall formation, epidermis integrity and water homeostasis in rice. Plant J. 92, 904–923 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Miura C., et al., The mycoheterotrophic symbiosis between orchids and mycorrhizal fungi possesses major components shared with mutualistic plant-mycorrhizal symbioses. Mol. Plant Microbe Interact. 31, 1032–1047 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Montero H., Choi J., Paszkowski U., Arbuscular mycorrhizal phenotyping: The dos and don’ts. New Phytol. 221, 1182–1186 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi J., et al., The negative regulator SMAX1 controls mycorrhizal symbiosis and strigolactone biosynthesis in rice. Nat. Commun. 11, 2114 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montero H., et al., A mycorrhiza-associated receptor-like kinase with an ancient origin in the green lineage. Gene Expression Omnibus (GEO). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE168162. Deposited 3 March 2021. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
See Dataset S1 for a detailed phylogenetic tree of Fig. 1A, Dataset S2 for RNA-seq data, Dataset S3 for a list of sequences, and Dataset S4 for a detailed phylogenetic tree of Fig. 2B. RNA-seq raw data were deposited in the National Center for Biotechnology Information gene expression omnibus (ID: GSE168162) (12). All other study data are included in the supporting information.