Significance
We have found that bacterial inhibition of host TAK1 inflammatory signaling elicits an alternative host defense mechanism involving production of mitochondrial reactive oxygen species through caspase 8 and RIPK3. This finding allows a reinterpretation of mouse phenotypes harboring tissue-specific gene deletion of Tak1, many of which die from tissue damage previously ascribed to impaired TAK1-dependent tissue homeostasis. We suggest that these phenotypes arise from misrecognition of compromised TAK1 as pathogen invasion.
Keywords: TAK1, ROS, mitochondria, intracellular bacteria
Abstract
Mitogen-activated protein kinase kinase kinase 7 (MAP3K7), known as TAK1, is an intracellular signaling intermediate of inflammatory responses. However, a series of mouse Tak1 gene deletion analyses have revealed that ablation of TAK1 does not prevent but rather elicits inflammation, which is accompanied by elevation of reactive oxygen species (ROS). This has been considered a consequence of impaired TAK1-dependent maintenance of tissue integrity. Contrary to this view, here we propose that TAK1 inhibition–induced ROS are an active cellular process that targets intracellular bacteria. Intracellular bacterial effector proteins such as Yersinia’s outer membrane protein YopJ are known to inhibit TAK1 to circumvent the inflammatory host responses. We found that such TAK1 inhibition induces mitochondrial-derived ROS, which effectively destroys intracellular bacteria. Two cell death–signaling molecules, caspase 8 and RIPK3, cooperatively participate in TAK1 inhibition–induced ROS and blockade of intracellular bacterial growth. Our results reveal a previously unrecognized host defense mechanism, which is initiated by host recognition of pathogen-induced impairment in a host protein, TAK1, but not directly of pathogens.
Bacterial infection activates inflammatory cellular responses upon recognition of bacterial moieties through cell surface and intracellular pattern recognition receptors (PRRs), Toll-like, and Nod-like receptors (1). Mitogen-activated protein kinase kinase kinase 7 (MAP3K7), known as TAK1, is a major intracellular mediator of proinflammatory gene expression through activating two signaling cascades, MAPK-AP1 and IκB kinase-NF-κB (2). Through these pathways, TAK1 renders a transcriptional reprograming toward an inflammatory state. TAK1 is ubiquitously expressed not only in immune cells but also in all cell types that have been tested (2). To delineate the role of TAK1 in each tissue, others and we have characterized a variety of tissue-specific Tak1-deficient mice. The striking finding from these analyses is that ablation of TAK1 elicits inflammation in many tissues such as the epidermis (3) and the intestinal epithelium (4) despite a well-established TAK1’s role as an activator of inflammation. Inflammation in Tak1-deficient tissues exhibits two major characteristics, namely, cell death through two cell death pathways, caspase 8–induced apoptosis and RIPK3-induced necroptosis, and elevation of reactive oxygen species (ROS) (2, 5, 6).
Pathogenic bacteria such as Salmonella enterica, Listeria monocytogenes, and several Yersinia species including Y. pseudotuberculosis, Y. enterocolitica, and Y. pestis are invasive intracellular bacteria, which actively enter into and colonize the host cells. To combat such bacteria, host cells activate inflammatory signaling pathways and xenophagy (7). To evade the host strategy, invasive bacteria express a variety of proteins to manipulate the host defense mechanisms, which are referred to as bacterial effector proteins. Inflammatory signaling pathways, particularly MAPK and NF-κB pathways, are prominent targets of bacterial effectors to diminish the inflammatory responses. A Yersinia effector protein, YopJ, inhibits TAK1 activity through acetylation of TAK1’s activation loop, which results in blockade of MAPK and NF-κB activation (8). However, recent studies have discovered that YopJ’s TAK1 inhibition elicits macrophage death through pyroptosis (9, 10), which could be an alternative host defense mechanism to fight back such bacteria. This prompted us to connect our finding of TAK1 ablation–induced ROS with a host defense mechanism. We have for a long time thought that TAK1 ablation induces ROS because TAK1 plays a critical role in cellular redox homeostasis. However, ROS may be intentionally up-regulated when cells sense TAK1 inhibition. We hypothesize that TAK1 inhibition–induced ROS provide an additional layer of the host defense to disrupt intracellular bacteria.
Results
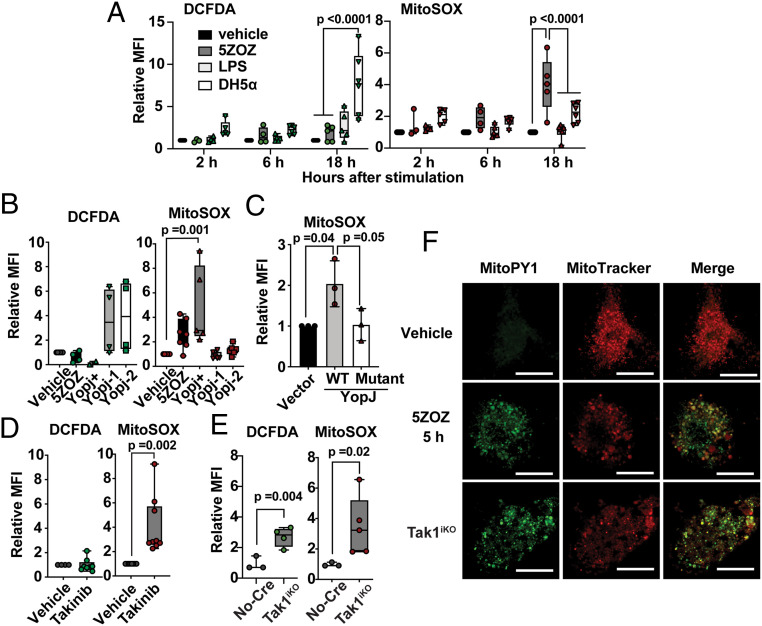
We first defined the source of TAK1 inhibition–induced ROS. The pharmacological TAK1 inhibitor 5Z-7oxozaenol (5ZOZ) (11) was used, as it was reported to elicit cellular responses identical to YopJ (9, 10). Bone marrow–derived macrophages (BMDMs) were treated with 5ZOZ, and ROS levels were determined using ROS dyes; CM-H2DCFDA, a general ROS and nitric oxide reactive chemical (12, 13); and MitoSOX, a mitochondrial ROS dye (13). As macrophages are known to elevate production of ROS and nitric oxides through assembly of NADPH oxidase 2 and transcriptional activation of nitric oxide synthase in response to bacterial moieties through PRRs (14), we also treated cells with the bacterial moiety lipopolysaccharide (LPS) and the noninvasive bacteria Escherichia coli DH5-α to compare the characteristics between bacterial moiety–driven and TAK1 inhibition–driven ROS. Consistent with previous notions (14), noninvasive bacteria and LPS up-regulated cytosolic ROS that were detectable preferentially by CM-H2DCFDA but less reactive with MitoSOX in BMDMs (Fig. 1A and SI Appendix, Fig. S1A). We found that, unlike bacteria moiety–induced ROS, TAK1 inhibition–induced ROS were readily detectable by MitoSOX but only marginally by CM-H2DCFDA. We also confirmed that Y. enterocolitica, which expresses YopJ, up-regulated mitochondrial ROS but not cytosolic ROS (Fig. 1B). In contrast, two yopJ-deficient Y. enterocolitica strains lacked an ability to elevate mitochondrial ROS but profoundly up-regulated cytosolic ROS (Fig. 1B). yopJ-deficient Y. enterocolitica strains activated TAK1’s downstream MAPKs, p38 and JNK, which could lead to transcriptional activation of ROS and nitric oxide generating enzymes, while the YopJ-expressing strain was incapable of activating them (SI Appendix, Fig. S1B). To verify that YopJ inhibition of TAK1 is accountable for mitochondrial ROS, we examined ROS in HeLa cells expressing wild-type or acetyltransferase-dead mutant YopJ protein instead of bacterial infection (Fig. 1C). As HeLa cells do not express one of the TNF signaling components, receptor-interacting protein kinase 3 (RIPK3) (15), we used HeLa cells stably expressing RIPK3 (HeLa-RIPK3) (16). TAK1 inhibition alone was less effective in elevating ROS, but cotreatment with TNF highly up-regulated mitochondrial ROS in HeLa-RIPK3 cells (SI Appendix, Fig. S1 C and D). Only wild-type YopJ but not the acetyltransferase-dead mutant was capable of inducing mitochondrial ROS (Fig. 1C). Thus, YopJ inhibition of TAK1 is the cause of elevated mitochondrial ROS. Collectively, TAK1 inhibition elicits mitochondrial ROS elevation, whereas PRR signaling pathways up-regulate cytosolic ROS.
Fig. 1.
Mitochondrial ROS are induced by TAK1 inhibition but not by PRR stimulation. (A) BMDMs were treated with 300 nM 5ZOZ, 100 ng/mL LPS, or E. coli DH5-α. DH5-α number was used at 100-fold of the BMDM number. Cells were harvested at 2, 6, and 18 h post treatment and were incubated with CM-H2DCFDA (DCFDA) and Sytox Red (cell viability dye) or with MitoSox and Sytox Green (cell viability dye). The median fluorescence intensity (MFI) of live cells was determined. Data include all separately acquired data points in BMDMs from three different animals are shown as fold-inductions relative to vehicle-treated samples. (B) Yersinia strains expressing YopJ (YopJ+) and two strains with no yopJ gene (YopJ-1 and YopJ-2) were infected into BMDMs, and ROS levels were determined at 18 h post infection. (C) Wild-type YopJ and acetyltransferase-dead mutant YopJ (YopJ mutant) were expressed in HeLa-RIPK3 cells. At 24 h post transfection, cells were treated with 50 ng/mL TNF, and ROS levels were analyzed only in transfected and live cells at 48 h post transfection. (D) BMDMs were treated with 10 µM Takinib for 18 h, and ROS levels were analyzed. (E) No-Cre control Tak1flox/flox and inducible Tak1-deficient (Tak1iKO) BMDMs were treated with the Cre inducer (4OHT) for 5 d. All data points acquired form different animal-derived BMDMs are shown. (F) BMDMs were treated with vehicle or 300 nM 5ZOZ for 5 h (Top and Middle). Tak1iKO BMDMs were treated with 4OHT for 5 d (Bottom). Mitochondrial ROS dye MitoPYI and MitoTracker Red staining are shown. (Scale bars, 20 µm.) One-way ANOVA, multiple comparisons, Tukey test (A–C), and unpaired Student’s’ t test (D and E).
To verify this unique mitochondrial ROS elevation, we used other means to disrupt TAK1: another pharmacological TAK1 inhibitor, Takinib (17), and Tak1 gene deletion. Unlike 5ZOZ, Takinib was incapable of inhibiting TAK1 autophosphorylation (17) (SI Appendix, Fig. S1E). Nevertheless, 5ZOZ, Takinib, and YopJ blocked TAK1-dependent p38 and JNK activation (SI Appendix, Fig. S1E). Takinib also elevated mitochondrial ROS in BMDMs (Fig. 1D). Tak1 gene deletion in BMDMs using the inducible Rosa26.Cre ERT system (18) depleted TAK1 protein at 4 to 5 d after treatment of the CreERT activator, 4-hydroxytamoxifen (4OHT) (19, 20). Similar to TAK1 inhibition, elevation of mitochondrial ROS was observed in Tak1-deficient BMDMs (Fig. 1E), while cytosolic ROS were also elevated in TAK1 depletion (Fig. 1E). These cytosolic ROS may be consequences of stress conditions mediated by mitochondrial ROS, since TAK1 protein depletion gradually occurred after gene deletion over a 3- to 5-d period (19). To further verify the relationship between TAK1 ablation–induced ROS and mitochondria, another mitochondria-directed ROS dye, MitoPY1 (21), was used together with a mitochondrial marker, MitoTracker Red. With 5ZOZ treatment or Tak1 gene deletion, MitoPY1-positive puncta were found to largely overlap with mitochondria (Fig. 1F). Collectively, we conclude that TAK1 ablation induces mitochondrial ROS.
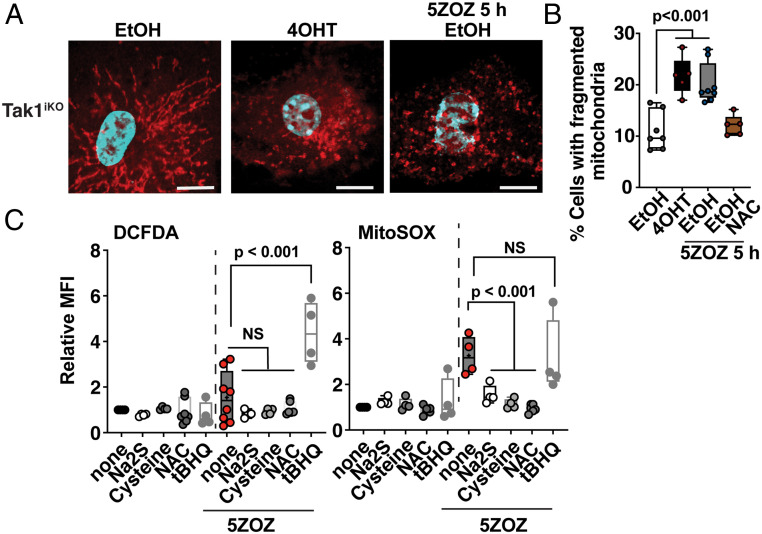
Mitochondria are constantly generating ROS as byproducts of respiration. Disruption of normal mitochondrial electron transport processes is known to up-regulate production of ROS. Such disruption is often associated with mitochondrial morphological changes, particularly mitochondrial fragmentation (22, 23). As MitoTracker staining in Fig. 1F seemed to indicate mitochondrial morphological changes, we quantified the frequency of altered mitochondrial morphology with TOM20 staining (Fig. 2A). While filamentous mitochondria were commonly observed in wild-type BMDMs, small donut-shaped mitochondria were increased by Tak1 gene deletion or with TAK1 inhibition (Fig. 2 A and B). These results indicate that TAK1 inhibition alters normal mitochondrial homeostasis that might be the cause of elevated ROS. If TAK1 inhibition up-regulates ROS through impaired mitochondrial respiration, restoring mitochondrial respiration should reduce the level of mitochondrial ROS. We chose to use sodium sulfide (a precursor of hydrogen sulfide) that is a stimulator of mitochondrial respiration by donating electrons to complex III in the electron transport chain (24, 25). Although hydrogen sulfide is toxic to cells at high concentrations by inhibiting cytochrome c oxidase, it facilitates electron flux by supplying electrons through sulfide quinone oxidoreductase at lower concentrations (26). Hydrogen sulfide is known to play a protective role on mitochondrial metabolism under stress conditions (24, 25). We found that sodium sulfide effectively blunted TAK1 inhibition–induced ROS (Fig. 2C). Hydrogen sulfide is generated from cysteine by several cytoplasmic enzymes (e.g., cystathionine γ-lyase) (27). N-acetyl cysteine (NAC) is a pharmacological precursor of cysteine. Both cysteine and NAC were found to be effective in blocking TAK1 inhibition–induced ROS (Fig. 2C). We are aware that cysteine and NAC are precursors of glutathione, which is the major endogenous ROS scavenger in cells. Thus, one might predict that they act through glutathione but not hydrogen sulfide. However, Dick’s group has recently determined that NAC is a modulator of mitochondria through hydrogen sulfide but does not act as a general ROS scavenger (27). Consistent with their finding, NAC was an ineffective ROS scavenger for bacterial moiety– and an oxidative agent tert-butylhydroperoxide (tBHP)–induced ROS (SI Appendix, Fig. S2). In contrast, a simple free radical scavenger, tert-butylhydroquinone (tBHQ), was capable of reducing bacterial moiety– and tBHP-induced ROS (SI Appendix, Fig. S2), while it did not work as a reducer of TAK1 inhibition–induced mitochondrial ROS (Fig. 2C and SI Appendix, Fig. S2C). When used together with 5ZOZ, tBHQ rather profoundly up-regulated ROS (Fig. 2C and SI Appendix, Fig. S2C), which may be associated with pro-oxidant actions of tBHQ’s reaction products (28). These results demonstrate that facilitating mitochondrial electron flux but not simple free radical scavenging is effective in blocking TAK1 inhibition–induced ROS, suggesting that TAK1 inhibition impairs mitochondria respiration resulting in elevated ROS production.
Fig. 2.
TAK1 inhibition impairs mitochondria, which is restored by hydrogen sulfide but not by general ROS scavenger. (A) Tak1iKO BMDMs were treated with vehicle (ethanol, EtOH) or with 4OHT for 5 d. Some EtOH-treated cells were also treated with 300 nM 5ZOZ for 5 h before the fixation. Mitochondrial morphology was visualized with anti-TOM20 immunofluorescence staining. (B) Quantification of mitochondrial morphology changes at 5 d post EtOH or 4OHT treatment. Some EtOH-treated cells were treated with 300 nM 5ZOZ and 3 mM NAC for 5 h. (C) Different types of ROS modulators; modulators of mitochondrial electron flux, sodium sulfide (0.4 mM); cysteine (3 mM); NAC (3 mM); or a general free radical scavenger, tBHQ (20 µM), were simultaneously treated with 300 nM 5ZOZ in BMDMs for 18 h. Cells were analyzed with CM-H2DCFDA (DCFDA) and Sytox Red or with MitoSox and Sytox Green. MFIs of live cells relative to that in vehicle-treated cells (none) are shown. Effectiveness of tBHQ was verified in separate experiments shown in SI Appendix, Fig. S2. One-way ANOVA, multiple comparisons, and Tukey test; N.S., not significant (B and C).
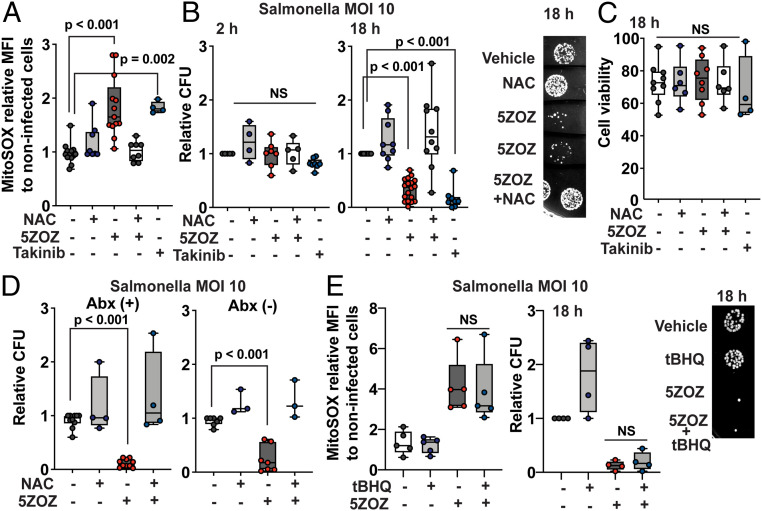
We next examined whether mitochondrial ROS prevent intracellular bacterial growth. S. enterica serovar Typhimurium LT-2, hereafter referred to as Salmonella, was used as a model intracellular bacterial strain. In contrast to highly virulent Salmonella strains such as SL1344 (29), the LT-2 strain is nonvirulent in mice (30). We found that LT-2 persistently colonized BMDMs (SI Appendix, Fig. S3 A and B). Salmonella produces the acetyltransferase AvrA that is also inhibitory to the host innate immune signaling (31). However, unlike YopJ, AvrA does not have the ability to inhibit TAK1 (32). Consistently, Salmonella infection alone did not up-regulate mitochondrial ROS, as shown in Fig. 3A and SI Appendix, Fig. S3C. We confirmed that neither 5ZOZ nor Takinib had any direct inhibitory effect on Salmonella growth (SI Appendix, Fig. S3C). To monitor only intracellular bacterial growth, cells were treated with gentamicin, which cannot penetrate the mammalian plasma membrane. The Salmonella infection frequencies in BMDMs were 70 to 80% regardless of TAK1 inhibition (SI Appendix, Fig. S3 A and B). Treatment with 5ZOZ up-regulated mitochondrial ROS in BMDMs infected with Salmonella (Fig. 3A and SI Appendix, Fig. S3D). We utilized NAC to reduce mitochondrial ROS, as sodium sulfide is toxic to Salmonella, while NAC did not directly affect Salmonella proliferation (SI Appendix, Fig. S3E). At 2 h post infection, we found that the numbers of Salmonella that invaded into macrophages were not altered by TAK1 inhibition (Fig. 3B). At 18 h, the viable intracellular Salmonella number was profoundly lower in the presence of TAK1 inhibitor than that in untreated macrophages, which was effectively restored by NAC treatment (Fig. 3B). We note here that TAK1 gene deletion gradually kills BMDMs as the TAK1 protein level declines over 5 d, while the mouse macrophage cell line RAW264.7 dies within 1 d with 5ZOZ treatment (33). However, we did not observe profound cell death in BMDMs under the current experimental conditions (Fig. 3C). The determinant of TAK1 inhibition–induced macrophage death is not yet fully clear as discussed also in an earlier study (9). Furthermore, removal of extracellular gentamicin after 2 h treatment did not affect the effectiveness of TAK1 inhibition in Salmonella growth (Fig. 3D). This indicates that the reduced Salmonella number is not due to Salmonella exposure to extracellular gentamicin after macrophage death. The general ROS scavenger tBHQ had little effects on reducing mitochondrial ROS and on restoring intracellular Salmonella growth (Fig. 3E). Collectively, we conclude that TAK1 inhibition–induced mitochondrial ROS are the cause of reduced intracellular Salmonella.
Fig. 3.
TAK1 inhibition blocks intracellular Salmonella growth through mitochondrial ROS. (A and B) BMDMs were incubated with Salmonella, MOI of 10, for 30 min, and extracellular Salmonella was killed by gentamicin treatment. TAK1 inhibitors, 300 nM 5ZOZ and 10 µM Takinib, and 3 mM NAC were added to the medium when starting gentamicin treatment. Cells were analyzed at 18 h post infection for ROS (A). The median intensity of live cells relative to that in noninfected cells with vehicle treatment is shown. Intracellular bacteria numbers were determined at 2 and 18 h post infection (B). Representative Salmonella colonies from cell lysate at 18 h are shown (B, Right). (C) Cell viability was assessed by SytoxGreen staining at 18 h post infection. (D) BMDMs were infected with Salmonella using the same procedure of A and B. At 2 h of gentamicin treatment, culture medium was changed to the same medium with gentamicin (left graph) or to a medium without antibiotics (right graph). (E) BMDMs were infected with Salmonella using the same procedure of A and B. A general ROS scavenger, 20 µM tBHQ, was cotreated with 300 nM 5ZOZ. Representative Salmonella colonies from cell lysate at 18 h post infection are shown (Right). One-way ANOVA, multiple comparisons, and Tukey test; N.S., not significant.
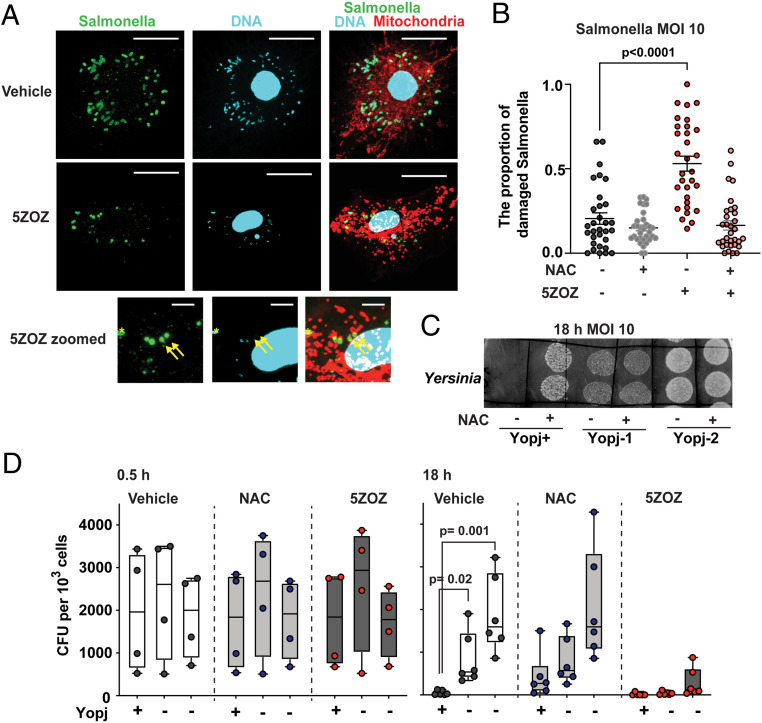
We further visually observed intracellular Salmonella in the presence and absence of 5ZOZ (Fig. 4A). The antibody against Salmonella did not have any nonspecific staining (SI Appendix, Fig. S4A). Whereas rod-shaped Salmonella cells containing small DNA particles were observed in untreated BMDMs, 5ZOZ treatment elevated the number of disintegrated Salmonella, many of which exhibited loss of DNA (Fig. 4A and SI Appendix, Fig. S4B, yellow arrows). The number of such damaged intracellular Salmonella was increased by 5ZOZ, and it was restored by NAC treatment (Fig. 4B). Intracellular Salmonella is known to reside in vacuoles, known as Salmonella-containing vacuoles (SCV). We observed that intracellular Salmonella was partially colocalized with the SCV marker, lamp1, while Salmonella was also found in the cytoplasm (SI Appendix, Fig. S4C). Both cytoplasmic and SCV-residing Salmonella may be targets for mitochondrial ROS, as an earlier study demonstrated that mitochondrial ROS are delivered to intracellular vacuoles through mitochondria-derived vesicles (34). We also found that cytoplasmic gram-positive bacterium, L. monocytogenes, was effectively blocked by TAK1 inhibition (SI Appendix, Fig. S4 D and E). Collectively, TAK1 inhibition–induced mitochondrial ROS destroy intracellular bacteria in the cytoplasm and probably also in the bacteria-harboring vacuoles.
Fig. 4.
Intracellular Salmonella and Yersinia. (A and B) TAK1 inhibition destroys intracellular Salmonella. BMDMs were infected with Salmonella, MOI of 10, and were treated with 300 nM 5ZOZ and/or 3 mM NAC. Cells were incubated with MitoTracker Red at 18 h post infection. Intracellular Salmonella was visualized by immunofluorescence staining of anti-Salmonella. Bottom shows enlarged pictures of the 5ZOZ-treated cell. Yellow arrows indicate damaged Salmonella having no DNA. Yellow asterisk indicates an intact Salmonella cell. (Scale bars, 20 µm in Top and Middle; 5 µm in Bottom.) Quantification of the proportion of damaged intracellular Salmonella is shown in B. Each data point represents one BMDM cell. Data from 30 cells per each treatment in three independent animal-derived BMDMs are shown. (C and D) One YopJ-expressing and two yopJ-deficient Y. enterocolitica strains were infected in BMDMs (MOI of 10). Bacteria colonies of two technical replicate spots from intracellular Yersinia at 18 h post infection with and without 3 mM NAC are shown (C). The bacteria number was quantified at the initial invasion, 30 min (D, left graphs), and at the intracellular maintenance/proliferation phase, 18 h (D, Right graphs). One-way ANOVA, multiple comparisons, and Tukey test (B); unpaired two-way Student’s’ t test (D).
We next examined whether bacterial TAK1 inhibition results in reducing colonization in host cells. We used YopJ-expressing and yopJ-deficient Yersinia strains. YopJ-expressing Yersinia poorly colonized BMDMs compared with yopJ-deficient strains (Fig. 4 C and D). NAC promoted YopJ-expressing Yersinia growth, whereas NAC was less effective in promoting YopJ-negative Yersinia growth (Fig. 4 C and D). These indicate that YopJ-induced TAK1 inhibition blocks intracellular Yersinia growth through mitochondrial ROS.
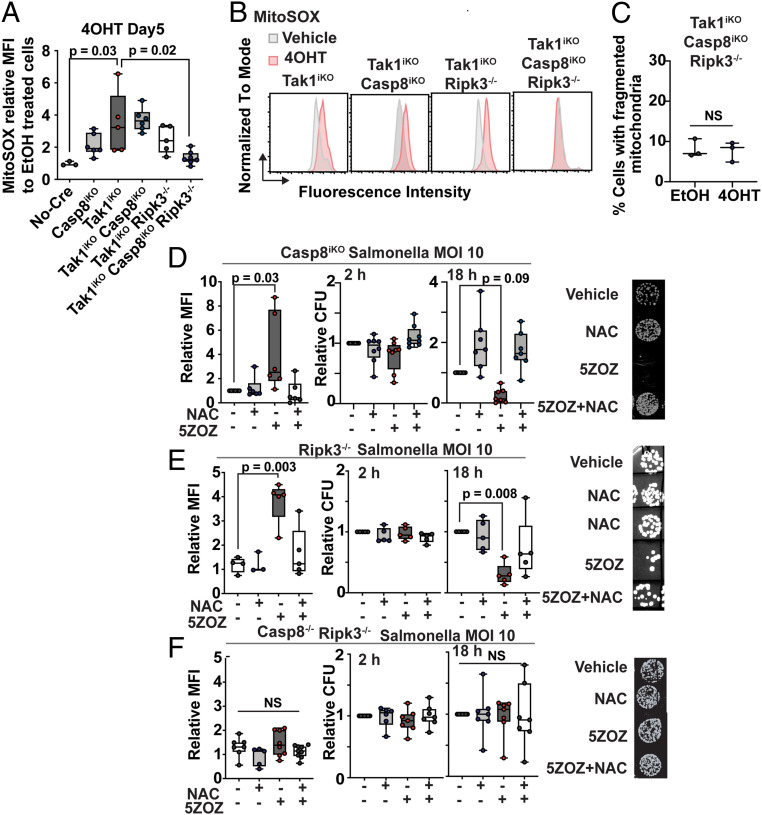
Finally, we examined the involvement of cell death–signaling molecules in mitochondrial ROS and blockade of intracellular Salmonella growth. TAK1 inhibition or Tak1 gene deletion ultimately causes cell death in which both caspase 8 and RIPK3 pathways participate (2, 9, 10, 33). To determine whether they are also mediators of mitochondrial ROS, we generated BMDMs harboring single, double, and triple deletion of Tak1, Casp8, and Ripk3. Like Tak1, Casp8 single-knockout mice are lethal during embryogenesis (35), and therefore, we used the inducible Cre system for Tak1 and Casp8 gene deletions. The Casp8 deletion alone moderately up-regulated mitochondrial ROS at 5 d after the initiation of gene deletion (Fig. 5 A and B). Tak1 deficiency–induced mitochondrial ROS was not reduced by additional single deletions of either Casp8 or Ripk3 (Fig. 5 A and B). However, triple deletion of Tak1, Casp8, and Ripk3 exhibited no increase of mitochondrial ROS (Fig. 5 A and B), in which we did not observe any increase in mitochondrial fragmentation (Fig. 5C). These demonstrate that both caspase 8 and RIPK3 participate in TAK1 inhibition–induced mitochondrial ROS. Intracellular Salmonella growth was examined in BMDMs harboring Casp8 and Ripk3 single and double deletions (Fig. 5 D–F). TAK1 inhibition up-regulated mitochondrial ROS and was still capable of blocking intracellular Salmonella growth in Casp8 or Ripk3 single-deficient BMDMs (Fig. 5 D and E). However, TAK1 inhibition–induced mitochondrial ROS and blockade of Salmonella growth were completely abolished in Casp8 and Ripk3 double-deficient BMDMs (Fig. 5F). These results demonstrate that caspase 8 and RIPK3 cell death–signaling pathways are critical in mediating TAK1 inhibition–induced mitochondrial ROS to block intracellular bacterial growth. We note here that autocrine TNF is partially accountable for Tak1 deficiency–induced macrophage death (33). Thus, TNF may be a mediator of these events. However, we found that gene deletion of the major TNF receptor, Tnfr1, did not effectively blunt TAK1 inhibition–induced mitochondrial ROS or blockade of intracellular Salmonella growth (SI Appendix, Fig. S5). Thus, in this context, PRRs but not TNF may be the predominant mediators of caspase 8 and RIPK3 pathways.
Fig. 5.
Caspase 8 and RIPK3 cooperatively mediate Tak1 deficiency–induced mitochondrial ROS and blockade of intracellular Salmonella growth. (A) BMDMs were incubated with 4OHT or vehicle (ethanol) for 5 d. Relative MFIs of MitoSOX in live cells relative to vehicle-treated BMDMs are shown. (B) Representative flow cytometric data in A. (C) Tak1iKO Casp8iKO Ripk3−/− BMDMs were incubated with 4OHT or vehicle for 5 d. Mitochondrial fragmentation was quantified as described in Fig. 2B. (D–F) BMDMs were infected with Salmonella (MOI 10) for 30 min. Extracellular Salmonella was killed by gentamicin. Relative MFIs of MitoSOX in live cells relative to vehicle-treated BMDMs at 18 h post infection are shown (left graph). Intracellular Salmonella numbers were determined at 2 h and 18 h post infection (middle and right graphs). Representative Salmonella colonies are shown (Right). One-way ANOVA, multiple comparisons, and Tukey test; N.S., not significant.
Discussion
Beside their prominent role as energy powerhouses, mitochondria play pleiotropic roles in innate immunity and cell death (36). Mitochondria have also been emerging as means for directly attacking invasive pathogens through releasing mitochondrial vesicles (34, 37) and itaconate (38). Our current study further revealed that mitochondria are actively modulated to produce ROS in response to bacterial inhibition of the host inflammatory signaling pathway. We have determined that caspase 8 and RIPK3 cooperatively participate in TAK1 inhibition–induced mitochondrial ROS. Caspase 8 not only induces apoptosis but also cleaves the pore-forming protein gasdermin D leading to pyroptosis, and RIPK3 induces another pore formation by MLKL oligomeriztion leading to necroptosis (39). TAK1 ablation indeed ultimately kill macrophages (33). Cell killing could provide an additional layer of host defense by eliminating pathogen colonizing niches. However, it alone might be inefficient to block pathogen spreading as many bacterial pathogens colonize both intracellular and extracellular environments. Our study provides evidence of host’s sophisticated strategy in that host cells kill intracellular pathogens before killing themselves to prevent pathogen spreading.
TAK1 is a major signaling intermediate of inflammatory responses. However, mouse studies of tissue-specific Tak1 gene deletion have shown that loss of TAK1 does not alleviate inflammation in many tissues. On the contrary, Tak1 gene deletion itself elicits inflammation and cell death through ROS even during embryogenesis, in which limited inflammatory stimuli are present. We propose that this puzzling phenotype is the revelation of the previously unrecognized host defense mechanism to block intracellular pathogen colonization in response to TAK1 inhibition. Engineered Tak1 gene deletion (TAK1 loss) is misinterpreted as pathogen attack in the tissues, which elevates mitochondrial ROS leading to inflammation and ultimately cell death.
Besides Tak1, gene deletion of other signaling intermediates associated with inflammatory responses such as IκB kinases (Ikbkb, Ikbkg) and receptor-interacting protein kinase 1 (Ripk1) also causes embryonic lethality in the mouse models (40–45). Furthermore, inflammation-associated cell death pathway molecules, Casp8 and Ripk3, also cause mouse embryonic lethality when deleted or mutated to destroy the catalytic activity, respectively (46, 47). Embryonic lethality has been understood as indication of their functional importance during embryogenesis. However, there is little evidence that active inflammatory pathways are required during embryogenesis. Given our results, we propose that the embryonic lethality caused by loss of signaling molecules in inflammatory and cell death pathways may be due to misinterpretation of gene manipulations as pathogen invasion. These genes are likely dispensable for embryogenesis; however, mammalian cells may have evolved the mechanism to exert alternative inflammatory responses when these inflammatory signaling molecules are disrupted. This alternative inflammatory mechanism leads to severe tissue damage resulting in animal mortality. It is noteworthy that the phenotypes (tissue damages) of mice harboring some of these gene deletions are remarkably similar even though their functional roles are totally different. A typical example for such similarities is the phenotypes of Casp8 and Tak1 gene deletion (46, 48). While caspase 8 is an apoptosis mediator, TAK1 is an activator of inflammatory transcription factors. However, the germline gene deletion of either gene results in embryonic lethality around embryonic day 9 to 10 exhibiting similar vascular defects. Furthermore, epidermal- and intestinal epithelium–specific gene deletion of Casp8 or Tak1 exhibit almost identical tissue damages (3, 4, 49–51). The phenotype similarities may be explained if either gene deletion is interpreted as pathogen invasion to activate the same alternative inflammatory mechanism.
Bacterial effectors attack a variety of host defense mechanisms including the small G protein RHO, which plays critical roles in cytoskeleton rearrangement processes, such as phagocytosis, in response to pathogen invasion (52). Shao’s group discovered that bacterial inhibition of RHO activates pyrin-inflammasome, which is an alternative host defense to fight back the pathogens inhibiting RHO-dependent host defense (53). This mechanism was thought to be somewhat similar to evolution in plant innate immunity in which host recognizes consequences of the pathogen’s virulence factors in the host but not directly pathogen-derived molecules. This type of evolution in the immune system is referred to as “guard theory” (54). Our finding of TAK1 inhibition–induced mitochondrial ROS may be a second example of such a category of host defense mechanisms in mammals.
Materials and Methods
BMDMs and Bacteria.
Bone marrow cells from wild-type, Tak1iKO [Rosa26-CreERT (18) Tak1flox/flox (55)], Ripk3−/− (56), Casp8iKO [Casp8flox/flox (35) Rosa26.CreERT], Casp8−/− (spontaneously acquired from Casp8flox/flox) Ripk3−/−, Tnrfr1-/(57), and littermate or age-matched no-Cre control mice were isolated. The genotypes were determined by PCR and were confirmed by Western blotting. The mouse care and the procedure of bone marrow isolation were conducted with the approval of the North Carolina State University Institutional Animal Care and Use Committee. Bone marrow cells were isolated with a standard method and were cultured in macrophage media; Dulbecco’s modified Eagle’s medium supplemented with 10% bovine growth serum (HyClone), 50 I.U./mL penicillin–streptomycin, and 30% L929 conditioned media at 37 °C with 5% CO2. After 3-d culture, fully differentiated BMDMs were replated and treated with 0.3 μM 4OHT or vehicle (ethanol) alone for 5 d to achieve complete Tak1 gene deletion. S. enterica Typhimurium LT2, L. monocytogenes serotype 1/2b strain 2011L-2858, implicated in a major outbreak of listeriosis via contaminated cantaloupe in 2011 (58), Y. enterocolitica WA, which express YopJ (59), Y. enterocolitica [ATCC 9610, SK 3181 (60)], which are not expressing YopJ, and E. coli strain DH5-α were used in this study. Salmonella and E. coli were culture in Luria–Bertani (LB) broth, and Listeria and Yersinia were cultured in brain heart infusion (BHI) broth (BD Bioscience) at 30 °C.
Reagents.
TAK1 protein kinase inhibitors, 300 nM 5ZOZ (11), and 10 µM Takinib (17) were used. Sodium sulfide (0.4 mM), 3 mM cysteine, 3 mM NAC, 1 mM tBHP, and 20 µM tBHQ (Sigma-Aldrich) were also used. To measure the level of ROS, 8 µM CM-H2DCFDA (Thermo Fisher Scientific), 1.25 µM MitoSOX (Thermo Fisher Scientific), and 10 µM MitoPY1 (TOCRIS) were used. To monitor cell viability and to visualize mitochondria, 30 nM Sytox Green, 10 nM Sytox Red, and 500 nM MitoTracker Red CMXRos (Thermo Fisher Scientific) were used, respectively. Antibodies against TOM20 (FL-145, Santa Cruz), Salmonella (Salmonella group antigen, Bio-Rad), Myc (9E10, Santa Cruz), and green fluorescent protein (GFP; GT859, GeneTex), phospho-TAK1(Thr187) (4536, Cell Signaling), TAK1 (61), phospho-JNK (9251, Cell Signaling), JNK (sc-571, Santa Cruz), phospho-p38 (9211, Cell Signaling), p38 (sc-7972, Santa Cruz), and β-actin (AC-15, Sigma-Aldrich) were used. LPS from Salmonella Minnesota (Sigma-Aldrich), gentamicin sulfate (VWR), and 4OHT (Sigma-Aldrich) were also used.
ROS Analysis.
BMDMs were washed with phosphate-buffered saline (PBS) and were detached from culture plates with gentle scraping. Cells were precipitated with centrifugation at 5,000 g for 1 min were incubated with mixtures of 8 µM CM-H2DCFDA and 10 nM Sytox Red or 1.25 µM MitoSOX and 30 nM Sytox Green in PBS for 30 min at room temperature. Live (Sytox dye-negative) cells were gated, and the median fluorescence intensities (MFIs) were determined with a flow cytometer (Accuri C6 Plus, BD Bioscience) and FlowJo software (BC Bioscience). For microscopic ROS analysis, cells were cultured in chambered coverslips (IBID). At 16 h posttreatment, 10 µM MitoPYI was added to the medium, which was followed by a 30-min incubation with 500 nM MitoTracker Red. Images were captured with FV3000 confocal microscopy (Olympus) with 60× oil objective lens (UPLXAPO60XO) and Fluoview software (FV30S-SW). To determine the role of acetyltransferase of YopJ, HeLa-RIPK3 (16) were used. HeLa-RIPK3 were cultured with Dulbecco’s modified Eagle’s medium supplemented with 10% bovine growth serum (HyClone) and were cotransfected with an enhanced green fluorescent protein–expressing vector and mammalian expression vectors for Myc-tagged empty, Myc-tagged YopJ, or Myc-tagged acetyltransferase-dead mutant YopJ (32) at the ratio of 1:10 using TransIT-X2 (Mirus Bio). At this ratio of the plasmids, we anticipated that most GFP-positive cells expressed wild-type or mutant YopJ. At 24 h post transfection, cells were treated with vehicle or 50 ng/mL human TNF (Peprotech). Cells were harvested at 48 h post transfection and were incubated with a mixture of 1.25 µM MitoSOX and 10 nM Sytox Red. Live (Sytox Red–negative) GFP-expressing cells were gated, and the MFI with a fluorescence compensation was determined with flow cytometry (Accuri C6 Plus, BD Bioscience) and FlowJo software (BC Bioscience).
Western Blotting.
BMDMs and HeLa-RIPK3 cells were lysed in extraction buffer (20 mM Hepes [pH 7.4], 150 mM NaCl, 12.5 mM β-glycerophosphate, 1.5 mM MgCl2, 2 mM EGTA, 10 mM NaF, 2 mM DTT, 1 mM Na3VO4, 1 mM PMSF, 20 ΜM aprotinin, and 0.5% Triton X-100). The cell extracts were resolved using sodium dodecyl sulphate–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Thermo Fisher Scientific). The membranes were immunoblotted, and the bound antibodies were visualized with horseradish peroxidase–conjugated antibodies using the WesternSure PREMIUM Chemiluminescent Substrate (LI-COR Biosciences).
Immunofluorescence Staining of BMDMs.
BMDMs were seeded on glass coverslips (#1.5) in 6-well plates and were incubated with MitoTracker Red in PBS for 30 min prior to 4% paraformaldehyde fixation for 10 min at room temperature. Fixed cells were permeabilized with methanol for 10 min at −30 °C and were incubated with a blocking buffer, PBS containing 5% goat serum for 30 min at room temperature. The cells were incubated with anti-TOM20 (1:200) and/or anti-Salmonella (1:1,000) antibodies followed by incubation with anti-rabbit IgG conjugated with Alexa 594 or Alexa 488, respectively (1:1,000, Thermo Fisher Scientific). Cells were then counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich). The coverslips were mounted with 50% glycerol and were observed by a fluorescence microscope (model BX41; Olympus) with a 20× objective lens (UPLXAPO20X) or by a confocal microscope (FV3000, Olympus) with a 60× oil objective (UPLXAPO60XO). For quantification of mitochondrial morphology, eight or more images each in samples from three different mice were randomly captured with a 20× objective lens and analyzed. Cells having greater than 50% area of mitochondria showing fragmented morphology were counted as cells with fragmented mitochondria. For quantification of intracellular Salmonella, Z-stack (0.6 µm slice interval) images with the 60× objective lens, covering entire cells, were captured. Randomly picked 30 cells per each treatment from samples from three different mice (10 cells per each animal) were used. Salmonella cells without any associated DAPI staining were counted as damaged bacteria. For quantification of the proportion of Salmonella-infected cells, five or more images were randomly captured with 10× objective lens for each treatment from three different mice. The proportion of cells with positive Salmonella staining of each image was registered as a data point.
Bacterial Infection.
Bacteria were precultured overnight and were inoculated into fresh culture broth at 1:10 dilution. After 1.5-h incubation at 30 °C, bacterial numbers were enumerated by optical density, and bacteria were suspended in PBS after centrifugation at 5,000 g for 10 min. BMDMs were seeded on 6-well plates for ROS analysis or 12-well plates for bacterial colony number analysis 1 to 3 d before infection. When cells reached to the confluency level at 80 to 100%, the culture medium was changed to one without any antibiotics, and the cell numbers per wells were counted. The cells were incubated with the indicated multiplicity of infection (MOI) of Salmonella and Listeria for 30 min or of Yersinia for 2 h at 37 °C in 5% CO2. The cells were then washed with sterile PBS and incubated in a medium supplemented with gentamicin (100 μg/mL) for 2 h with the indicated chemicals. After 2-h incubation, the culture medium was changed to a standard medium supplemented with 50 I.U./mL penicillin–streptomycin together with the indicated chemicals. To determine the initial invasion efficiency, at 2 h for Salmonella and Listeria or at 30 min for Yersinia after starting the gentamicin treatment, the cells were washed with sterile PBS and lysed in PBS containing 0.2% Triton X-100. To determine the intracellular bacterial growth, cells were washed with sterile PBS and lysed in PBS containing 0.2% Triton X-100 at 18 h post infection. Serial dilutions of the lysates were spotted (10 μL/spot) on agar plates (LB agar for Salmonella; BHI agar for Listeria and Yersinia) for enumeration of intracellular bacteria. To determine YopJ and 5ZOZ inhibition of TAK1, BMDMs were infected with Yersinia stains: WA, 9610, YE, with an MOI of 10 for 2 h at 37 °C in 5% CO2. Cells were treated with gentamicin (100 g/mL) for 30 min, then with 5ZOZ for 1 h. Cells were lysed in extraction buffer and analyzed by immunoblotting.
Statistical Analysis.
All experiments were conducted in BMDMs isolated from at least three mice as indicated. The results are confirmed by at least three separately performed experiments. The box and whisker graphs represent the mean (middle line), the 25th and 75th percentiles (box), the minimum and maximal date points (whisker), and all data points. Differences between experimental groups were assessed for significance by using the one-way ANOVA with Tukey multiple comparisons test, the unpaired Student’s t test (two-tailed) with equal distributions, the simple linear regression test, or the one sample t and Wilcoxon test as indicated in the figure legends.
Supplementary Material
Acknowledgments
We thank Dr. Akira for Tak1-floxed mice, Drs. Dixit and Newton for Ripk3−/−, Drs. Hakem and Green for Casp8-floxed mice, Drs. Neish and Jones for YopJ vectors, and North Carolina State University facilities (partially supported by National Institute of Environmental Health Sciences, Center for Human Health and the Environment P30ES025128). This work was supported by NIH R01GM112986 (J.N.-T.) and US Department of Agriculture National Institute of Food and Agriculture Award 2018-07464 (Grant 12687985) (S.K.). NIH fellowship F31GM135975 (W.L.-P.) and Training Grant T32 ES007046 (W.L.-P. and Y.S.) partially supported graduate students’ work.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023647118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Kawai T., Akira S., Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34, 637–650 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Mihaly S. R., Ninomiya-Tsuji J., Morioka S., TAK1 control of cell death. Cell Death Differ. 21, 1667–1676 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Omori E., et al., TAK1 is a master regulator of epidermal homeostasis involving skin inflammation and apoptosis. J. Biol. Chem. 281, 19610–19617 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kajino-Sakamoto R., et al., Enterocyte-derived TAK1 signaling prevents epithelium apoptosis and the development of ileitis and colitis. J. Immunol. 181, 1143–1152 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kajino-Sakamoto R., et al., TGF-β-activated kinase 1 signaling maintains intestinal integrity by preventing accumulation of reactive oxygen species in the intestinal epithelium. J. Immunol. 185, 4729–4737 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Omori E., Morioka S., Matsumoto K., Ninomiya-Tsuji J., TAK1 regulates reactive oxygen species and cell death in keratinocytes, which is essential for skin integrity. J. Biol. Chem. 283, 26161–26168 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang J., Brumell J. H., Bacteria-autophagy interplay: A battle for survival. Nat. Rev. Microbiol. 12, 101–114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paquette N., et al., Serine/threonine acetylation of TGFβ-activated kinase (TAK1) by Yersinia pestis YopJ inhibits innate immune signaling. Proc. Natl. Acad. Sci. U.S.A. 109, 12710–12715 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarhan J., et al., Caspase-8 induces cleavage of gasdermin D to elicit pyroptosis during Yersinia infection. Proc. Natl. Acad. Sci. U.S.A. 115, E10888–E10897 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orning P., et al., Pathogen blockade of TAK1 triggers caspase-8-dependent cleavage of gasdermin D and cell death. Science 362, 1064–1069 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ninomiya-Tsuji J., et al., A resorcylic acid lactone, 5Z-7-oxozeaenol, prevents inflammation by inhibiting the catalytic activity of TAK1 MAPK kinase kinase. J. Biol. Chem. 278, 18485–18490 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Kalyanaraman B., et al., Measuring reactive oxygen and nitrogen species with fluorescent probes: Challenges and limitations. Free Radic. Biol. Med. 52, 1–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dikalov S. I., Harrison D. G., Methods for detection of mitochondrial and cellular reactive oxygen species. Antioxid. Redox Signal. 20, 372–382 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mizel S. B., Honko A. N., Moors M. A., Smith P. S., West A. P., Induction of macrophage nitric oxide production by Gram-negative flagellin involves signaling via heteromeric Toll-like receptor 5/Toll-like receptor 4 complexes. J. Immunol. 170, 6217–6223 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Sun L., et al., Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 148, 213–227 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Sai K., Parsons C., House J. S., Kathariou S., Ninomiya-Tsuji J., Necroptosis mediators RIPK3 and MLKL suppress intracellular Listeria replication independently of host cell killing. J. Cell Biol. 218, 1994–2005 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Totzke J., et al., Takinib, a selective TAK1 inhibitor, broadens the therapeutic efficacy of TNF-α inhibition for cancer and autoimmune disease. Cell Chem. Biol. 24, 1029–1039.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Badea T. C., Wang Y., Nathans J., A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J. Neurosci. 23, 2314–2322 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mihaly S. R., Morioka S., Ninomiya-Tsuji J., Takaesu G., Activated macrophage survival is coordinated by TAK1 binding proteins. PLoS One 9, e94982 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mihaly S. R., Sakamachi Y., Ninomiya-Tsuji J., Morioka S., Noncanonical cell death program independent of caspase activation cascade and necroptotic modules is elicited by loss of TGFβ-activated kinase 1. Sci. Rep. 7, 2918 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dickinson B. C., Chang C. J., A targetable fluorescent probe for imaging hydrogen peroxide in the mitochondria of living cells. J. Am. Chem. Soc. 130, 9638–9639 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyazono Y., et al., Uncoupled mitochondria quickly shorten along their long axis to form indented spheroids, instead of rings, in a fission-independent manner. Sci. Rep. 8, 350 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmad T., et al., Computational classification of mitochondrial shapes reflects stress and redox state. Cell Death Dis. 4, e461 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olson K. R., Mitochondrial adaptations to utilize hydrogen sulfide for energy and signaling. J. Comp. Physiol. B 182, 881–897 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Fu M., et al., Hydrogen sulfide (H2S) metabolism in mitochondria and its regulatory role in energy production. Proc. Natl. Acad. Sci. U.S.A. 109, 2943–2948 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szabo C., et al., Regulation of mitochondrial bioenergetic function by hydrogen sulfide. Part I. Biochemical and physiological mechanisms. Br. J. Pharmacol. 171, 2099–2122 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ezeriņa D., Takano Y., Hanaoka K., Urano Y., Dick T. P., N-acetyl cysteine functions as a fast-acting antioxidant by triggering intracellular H2S and sulfane sulfur production. Cell Chem. Biol. 25, 447–459.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carocho M., Ferreira I. C., A review on antioxidants, prooxidants and related controversy: Natural and synthetic compounds, screening and analysis methodologies and future perspectives. Food Chem. Toxicol. 51, 15–25 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Branchu P., Bawn M., Kingsley R. A., Genome variation and molecular epidemiology of Salmonella enterica serovar typhimurium pathovariants. Infect. Immun. 86, e00079-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swords W. E., Cannon B. M., Benjamin W. H. Jr, Avirulence of LT2 strains of Salmonella typhimurium results from a defective rpoS gene. Infect. Immun. 65, 2451–2453 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reddick L. E., Alto N. M., Bacteria fighting back: How pathogens target and subvert the host innate immune system. Mol. Cell 54, 321–328 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones R. M., et al., Salmonella AvrA coordinates suppression of host immune and apoptotic defenses via JNK pathway blockade. Cell Host Microbe 3, 233–244 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Sakamachi Y., et al., TAK1 regulates resident macrophages by protecting lysosomal integrity. Cell Death Dis. 8, e2598 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abuaita B. H., Schultz T. L., O’Riordan M. X., Mitochondria-derived vesicles deliver antimicrobial reactive oxygen species to control phagosome-localized Staphylococcus aureus. Cell Host Microbe 24, 625–636.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salmena L., et al., Essential role for caspase 8 in T-cell homeostasis and T-cell-mediated immunity. Genes Dev. 17, 883–895 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinberg S. E., Sena L. A., Chandel N. S., Mitochondria in the regulation of innate and adaptive immunity. Immunity 42, 406–417 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deo P., et al., Mitochondrial dysfunction caused by outer membrane vesicles from Gram-negative bacteria activates intrinsic apoptosis and inflammation. Nat. Microbiol. 5, 1418–1427 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Chen M., et al., Itaconate is an effector of a Rab GTPase cell-autonomous host defense pathway against Salmonella. Science 369, 450–455 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samir P., Malireddi R. K. S., Kanneganti T. D., The PANoptosome: A deadly protein complex driving pyroptosis, apoptosis, and necroptosis (PANoptosis). Front. Cell. Infect. Microbiol. 10, 238 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Q., Van Antwerp D., Mercurio F., Lee K. F., Verma I. M., Severe liver degeneration in mice lacking the IkappaB kinase 2 gene. Science 284, 321–325 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Pasparakis M., et al., TNF-mediated inflammatory skin disease in mice with epidermis-specific deletion of IKK2. Nature 417, 861–866 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Egan L. J., et al., IkappaB-kinasebeta-dependent NF-kappaB activation provides radioprotection to the intestinal epithelium. Proc. Natl. Acad. Sci. U.S.A. 101, 2452–2457 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidt-Supprian M., et al., NEMO/IKK gamma-deficient mice model incontinentia pigmenti. Mol. Cell 5, 981–992 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Makris C., et al., Female mice heterozygous for IKK gamma/NEMO deficiencies develop a dermatopathy similar to the human X-linked disorder incontinentia pigmenti. Mol. Cell 5, 969–979 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Kelliher M. A., et al., The death domain kinase RIP mediates the TNF-induced NF-kappaB signal. Immunity 8, 297–303 (1998). [DOI] [PubMed] [Google Scholar]
- 46.Varfolomeev E. E., et al., Targeted disruption of the mouse Caspase 8 gene ablates cell death induction by the TNF receptors, Fas/Apo1, and DR3 and is lethal prenatally. Immunity 9, 267–276 (1998). [DOI] [PubMed] [Google Scholar]
- 47.Newton K., et al., Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science 343, 1357–1360 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Jadrich J. L., O’Connor M. B., Coucouvanis E., The TGF beta activated kinase TAK1 regulates vascular development in vivo. Development 133, 1529–1541 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Kovalenko A., et al., Caspase-8 deficiency in epidermal keratinocytes triggers an inflammatory skin disease. J. Exp. Med. 206, 2161–2177 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Günther C., et al., Caspase-8 regulates TNF-α-induced epithelial necroptosis and terminal ileitis. Nature 477, 335–339 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simmons A. N., Kajino-Sakamoto R., Ninomiya-Tsuji J., TAK1 regulates Paneth cell integrity partly through blocking necroptosis. Cell Death Dis. 7, e2196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jank T., Aktories K., Structure and mode of action of clostridial glucosylating toxins: The ABCD model. Trends Microbiol. 16, 222–229 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Xu H., et al., Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature 513, 237–241 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Jones J. D., Dangl J. L., The plant immune system. Nature 444, 323–329 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Sato S., et al., Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat. Immunol. 6, 1087–1095 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Newton K., Sun X., Dixit V. M., Kinase RIP3 is dispensable for normal NF-kappa Bs, signaling by the B-cell and T-cell receptors, tumor necrosis factor receptor 1, and Toll-like receptors 2 and 4. Mol. Cell. Biol. 24, 1464–1469 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pfeffer K., et al., Mice deficient for the 55 kd tumor necrosis factor receptor are resistant to endotoxic shock, yet succumb to L. monocytogenes infection. Cell 73, 457–467 (1993). [DOI] [PubMed] [Google Scholar]
- 58.McCollum J. T., et al., Multistate outbreak of listeriosis associated with cantaloupe. N. Engl. J. Med. 369, 944–953 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Johnson S. L., et al., Thirty-two complete genome assemblies of nine Yersinia species, including Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica. Genome Announc. 3, e00148-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Azizoglu R. O., Kathariou S., Impact of growth temperature and agar versus liquid media on freeze-thaw tolerance of Yersinia enterocolitica. Foodborne Pathog. Dis. 7, 1125–1128 (2010). [DOI] [PubMed] [Google Scholar]
- 61.Ninomiya-Tsuji J., et al., The kinase TAK1 can activate the NIK-I kappaB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature 398, 252–256 (1999). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.