Significance
Clostridioides difficile is the leading cause of healthcare-associated diarrhea worldwide following antibiotic treatment. Consequently, there is medical need for novel antibacterial agents acting against C. difficile that leave the resident microbiota unharmed. The development of such narrow-spectrum antibiotics requires precise knowledge of the mechanisms that fine-tune gene expression to orchestrate the genomic output at each locus in the genome. We address this issue by defining the global transcriptome architecture of C. difficile including noncoding regulatory elements, many of which are expressed during gut colonization. Our analysis of these regulators provides evidence for the global function of Hfq in sRNA binding and stabilization in a gram-positive bacterium.
Keywords: small RNA, Hfq, differential RNA-seq, C. difficile, transcript termination site
Abstract
The gram-positive human pathogen Clostridioides difficile has emerged as the leading cause of antibiotic-associated diarrhea. However, little is known about the bacterium’s transcriptome architecture and mechanisms of posttranscriptional control. Here, we have applied transcription start site and termination mapping to generate a single-nucleotide–resolution RNA map of C. difficile 5′ and 3′ untranslated regions, operon structures, and noncoding regulators, including 42 sRNAs. Our results indicate functionality of many conserved riboswitches and predict cis-regulatory RNA elements upstream of multidrug resistance (MDR)-type ATP-binding cassette (ABC) transporters and transcriptional regulators. Despite growing evidence for a role of Hfq in RNA-based gene regulation in C. difficile, the functions of Hfq-based posttranscriptional regulatory networks in gram-positive pathogens remain controversial. Using Hfq immunoprecipitation followed by sequencing of bound RNA species (RIP-seq), we identify a large cohort of transcripts bound by Hfq and show that absence of Hfq affects transcript stabilities and steady-state levels. We demonstrate sRNA expression during intestinal colonization by C. difficile and identify infection-related signals impacting its expression. As a proof of concept, we show that the utilization of the abundant intestinal metabolite ethanolamine is regulated by the Hfq-dependent sRNA CDIF630nc_085. Overall, our study lays the foundation for understanding clostridial riboregulation with implications for the infection process and provides evidence for a global role of Hfq in posttranscriptional regulation in a gram-positive bacterium.
Antibiotic-resistant bacteria are a major global threat to human health, endangering our ability to perform a range of modern medical interventions. The obligate anaerobe, spore-forming Clostridioides difficile has become the leading cause of antibiotic-associated diarrhea over the past two decades (1). Increasing numbers of multiresistant clinical isolates (2) and recurrent infections are key challenges in the treatment of C. difficile infections (CDI), often leaving fecal microbiota transfer as the only clinical option (3, 4).
The clinical challenges posed by CDI have prompted much effort to understand how this pathogen regulates virulence in response to environmental conditions. As a result, there is a comprehensive body of literature focusing on toxin production and sporulation control by several global metabolic regulators including CcpA, CodY, Rex, and PrdR (5, 6). Furthermore, several specialized and general sigma factors including TcdR (6), Spo0A (7), SigD (8, 9), SigH (10), and SigB (11–13) are linked to virulence and metabolism, although the exact molecular mechanisms often remain unknown. Most of this knowledge has been accumulated through detailed studies of individual genes and promoters, whereas RNA-sequencing (RNA-seq)–based annotations of the global transcriptome architecture, which have accelerated research in the gram-positive pathogens Listeria (14), Staphylococcus (15), and Streptococcus (16), have only recently become available for C. difficile (17).
This paucity of global knowledge about RNA output in C. difficile readily extends to posttranscriptional control of gene expression. The bacterium is of particular scientific interest, being the only gram-positive species thus far in which deletion of hfq seems to have a large impact on gene expression and bacterial physiology (18–20). Specifically, deletion of hfq increases sporulation (21), a crucial pathogenic feature of this bacterium that enables transmission between hosts. In gram-negative bacteria, Hfq commonly exerts global posttranscriptional control by facilitating short base-pairing interactions of small regulatory RNAs (sRNAs) with trans-encoded messenger RNAs (mRNAs) (22, 23), but its role in gram-positive bacteria remains controversial. For example, functional studies on Hfq in Bacillus subtilis, a model bacterium for Firmicutes, revealed in vivo association with a subset of sRNA (24), but comparative analyses of a wild-type and hfq knockout strain revealed only moderate effects on sRNA and mRNA transcript levels (25) and the absence of any significant growth defect (26), which lead to the conclusion that Hfq plays a minor role in posttranscriptional regulation in B. subtilis.
Previous efforts using in silico methods (27) and RNA-seq (28) have predicted >100 sRNA candidates in C. difficile and identified widespread RNA binding by Hfq (20), which suggests the existence of a large posttranscriptional network in C. difficile. However, under which conditions these sRNAs are expressed, which targets they regulate, and whether they depend on Hfq remain fundamental open questions.
Another important feature of posttranscriptional control in C. difficile are cis-regulatory RNA elements. Pioneering work has deciphered the function of genetic switches in the 5′ untranslated region (UTR) of the flgB operon, containing the early-stage flagellar genes, and the cell wall protein–encoding gene cwpV (29–31). Moreover, cyclic di-GMP–responsive riboswitches were shown to regulate biofilm formation and toxin production (9, 32–34). However, other members of the many different riboswitch classes have not been systematically identified. Combined with the nascent stage of sRNA biology in C. difficile, this argues that global approaches are needed to understand the full scope of posttranscriptional regulation in this important human pathogen.
In the present study, we applied recently developed methods of bacterial RNA biology (35, 36) to construct a global atlas of transcriptional and posttranscriptional control in C. difficile. We provide an example for a gram-positive bacterium with Hfq-dependent sRNA-based gene regulation and identify the ethanolamine utilization pathway as a target of sRNA-mediated regulation in C. difficile. Overall, we provide evidence for extensive Hfq-dependent posttranscriptional regulation and lay the foundation for future mechanistic studies of RNA-based gene regulation in C. difficile.
Results
High-Resolution Transcriptome Maps of C. difficile 630.
For generating high-resolution transcriptome maps of C. difficile, we chose the reference strain 630 (DSM 27543, CP010905.2). Being widely used by the C. difficile community, this strain offers the most comprehensive genome annotation. Using two global RNA-seq approaches, we analyzed RNA samples from three different conditions: late-exponential and early stationary growth in tryptone-yeast (TY) broth and late-exponential growth in brain heart infusion (BHI) broth (Fig. 1A). The resulting genome-wide maps provide single-nucleotide–resolution transcriptional start sites (TSSs) and termination sites (TTSs) (Fig. 1B). We have used this information to annotate 5′ and 3′ UTRs and operon structures, to correct previous open reading frame (ORF) annotations, to add previously overlooked small genes, and to annotate sRNA loci. Inspired by other gene expression databases such as SalCom (37), AcinetoCom (38), and Theta-Base (39), we have launched the interactive web browser “Clost-Base” (https://www.helmholtz-hiri.de/en/datasets/clostridium) for our transcriptome data. This online resource for C. difficile allows visualization of the transcriptomic data in the context of annotated coding and noncoding genes as well as transcript features (e.g., TSS) that we have experimentally determined in this work. The browser enables search queries and retrieval of primary sequences for any annotated feature in the database to serve as a resource for the C. difficile community.
Fig. 1.
Global RNA-seq approaches for high-resolution transcriptome mapping. (A) Sequencing samples were generated from strain C. difficile 630 grown to late-exponential and stationary phase in tryptone-yeast broth, as well as late-exponential phase in BHI broth. The error bars show SD of three biological replicates. (B) Read profiles generated with dRNA-seq and RNAtag-seq allow the annotation of TSSs and TTSs. For dRNA-seq, one fraction of total RNA is treated with terminator exonuclease (TEX+), which specifically degrades processed transcripts carrying a 5′-P or 5′OH. The other fraction remains untreated. This differential treatment results in a relative read enrichment for primary transcripts (5′-PPP) in the TEX+-treated libraries allowing the identification of TSSs. For RNAtag-seq, adapters for sequencing are ligated to RNA 3′ ends, thereby capturing 3′ ends of transcripts. (C) Benchmarking of dRNA-seq approach. Two experimentally determined growth phase–dependent TSSs for spo0A are consistent with dRNA-seq–based identification of spo0A-associated TSSs. Abbreviations: LE, late-exponential growth phase; ST, stationary growth phase; CDS, coding sequence; TSS, transcriptional start site. (D) Venn diagram showing distribution of TSSs among classes. TSS classification is based on expression strength and genomic location: primary (P), secondary (S), internal (I), antisense (A), and orphan (O).
Genome-wide Annotation of TSSs.
Differential RNA-seq (dRNA-seq) (40, 41) was performed to capture 5′ ends of transcripts. In brief, one-half of the sample remains untreated (Tex−) to capture both, primary (5′- PPP) and processed (5′-P or 5′OH) 5′ ends of transcripts; the other half is treated with terminator exonuclease (Tex+), leading to specific degradation of processed (5′-P or 5′OH) RNAs, thus enriching primary transcripts and enabling TSS annotation (Fig. 1B). Conversely, relative read enrichment in the Tex− complementary DNA (cDNA) libraries indicate RNA processing sites (Fig. 1B). The C. difficile 630 genome comprises annotations for 3,778 coding sequences (CDS) as well as transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), and the housekeeping RNAs 6S RNA, RNaseP, tmRNA, and SRP. By identifying 2,293 TSSs, we were able to define transcriptional units of individual genes and polycistronic operons for approximately one-half of the genome (Dataset S1).
Benchmarking our global data with previous studies of individual genes (10, 30, 42), our dRNA-seq results are consistent with published TSSs of sigH, cwpV, cwp19, and spo0A (Fig. 1C). For annotation purposes, we assigned TSSs to one of five classes according to their genomic location and expression level: pTSS (primary TSS of a gene or operon), sTSS (secondary TSS showing lower expression level compared to pTSS for the same gene or operon), iTSS (internal TSS located inside a gene), aTSS (antisense to a gene within 100-nt distance), and oTSS (orphan TSS, no nearby gene) (Fig. 1D). Naturally, some of these TSS annotations overlap (e.g., among the 1,627 pTSS, 126 are located within a gene (iTSS), and 46 are transcribed antisense (aTSS) to a gene). However, in contrast to other bacteria where antisense transcription is a pervasive transcriptome feature (43), it only accounts for ∼6% of TSS events in C. difficile.
Internal TSSs were abundant and accounted for 20% of all start sites, many of which seem to uncouple downstream genes within operons. However, they may also indicate misannotated translational start codons or protein isoforms. One example is the putative hydrolase (CDIF630_02227) encoded between ermB1 and ermB2 of the erythromycin resistance cassette, which belongs to a conserved family of ATPases involved in plasmid partitioning. In addition to a very weakly expressed primary TSS upstream of the annotated start codon, we detected a strongly expressed iTSS located 14 nt inside the coding sequence. Manual checking for start codons (AUG, UUG, and GUG) and a ribosome-binding site (RBS) downstream of the iTSS identified a start codon 43 nt downstream of the existing ORF annotation, placing an RBS in optimal distance to the start codon and generating a 5′ UTR length of 29 nt (SI Appendix, Fig. S1A). This annotation is in line with earlier studies in Streptococcus pyogenes, which suggested this internal start codon is used in vivo (44) and accordingly used the shorter variant to solve the respective protein crystal structure (PDB ID: 2oze). Similarly, we propose reannotations in eight additional cases, including the spaFEGRK operon encoding an antibiotic/multidrug–family ATP-binding cassette (ABC) transport system (Dataset S2).
Promoter Architectures.
C. difficile encodes 14 sigma factors, among them, consensus sequences have been proposed for the vegetative SigA (28), the general stress response SigB (11–13), the major transition phase SigH (10), and the sporulation-specific SigK (45) sigma factor, mostly based on comparative transcriptome analyses of respective deletion strains. In addition, a recently published strategy for genome-wide identification of promoter sequences, which combined TSS mapping and positional weight matrices based on transcriptome profiles of sigma factor mutants, expanded several existing sigma factor regulons and defined novel regulons, including one for SigL (17).
Here, we applied multiple expectation maximizations for motif elicitation (MEME)-based searches upstream of all TSS leading to the confirmation of published consencus sequences for SigA, SigB, SigH, and SigK (SI Appendix, Fig. S1B and Datasets S1 and S3). In line with recent data (17), the majority of detected TSS (1,188/2,293) were associated with a SigA-type promoter (Dataset S3), including the known TSSs for sigH and clnR (10, 46, 47). Similarly, we identify many genes previously predicted to be regulated by SigH (10, 17). Furthermore, we identified 52 genes associated with a SigK promoter signature, including known genes of the SigK regulon such as sleC and cdeC (48). Comparing our promoter predictions with previous studies showed partial overlaps which we attribute to differential computational strategies for promoter assignment as well as growth conditions used (e.g., sigK promoter mutant analyses were performed in sporulation conditions using specific sporulation medium). Overall, our results add confidence to existing regulon predictions and extend the list of high-confidence candidates that have been repeatedly identified to be associated with a certain sigma factor (indicated in Dataset S3).
We further identified 39 genes with alternative TSSs that were associated with two different promoter sequences. For example, RecA, which controls the DNA damage response by homologous recombinational repair of damaged DNA, is associated with a SigA- and SigB-type promoter supporting its role in stress responses (SI Appendix, Fig. S1C).
Global Mapping of Transcript Ends.
To map TTSs, we adopted the RNAtag-seq protocol (49), a technique utilizing initial adapter ligation to exposed RNA 3′ ends. This approach yielded 2,042 experimentally determined TTSs, which were assigned to the following classes: 3′ UTR (downstream of CDS or noncoding RNA), 5′ UTR (between the TSS and the start codon of CDS), CDS (within a coding sequence), orphan (downstream of an orphan TSS), and CRISPR (associated with a CRISPR array) (Fig. 2A).
Fig. 2.
Global features associated with transcription termination sites (TTSs). (A) Classification of TTS based on their genomic location: 3′ UTR (downstream of CDS or noncoding gene), 5′ UTR (within the 5′ UTR of a coding sequence), CDS (within coding sequence), orphan (downstream of orphan TSS), and CRISPR (associated with CRISPR array). The numbers indicate the amount of annotated TTSs for each class. (B) Consensus motif associated with 3′ UTR–located TTSs downstream of coding sequences was identified using the MEME suite. The identified motif is located within 15 bp downstream of detected 3′ UTR TTS positions. (C) Calculation of average GC content (%) at each indicated position relative to 3′ UTR–located TTSs reveals a spike in GC content immediately upstream of the TTS, indicating a stable stem structure. (D) Frequencies of 3′ UTR lengths based on 1,741 detected 3′ UTR TTSs. (E) Reads in the RNAtag-seq libraries reveal overlapping TTSs for the convergently transcribed genes tcdA and tcdC.
The majority of TTSs mapped to the region downstream of CDSs. Analysis of primary sequences and secondary structures around these TTSs reveals a downstream poly-U tail motif for the majority (75%) of them (Fig. 2B and Dataset S4). In addition, a decrease in folding energy (SI Appendix, Fig. S2A) and a spike in guanine-cytosine (GC) content at preceding TTS positions (Fig. 2C) indicate stable stem–loop structures, pointing to an important role for intrinsic transcription termination in C. difficile.
Our genome-wide map of 3′ UTRs (Dataset S4) reveals that only ∼7% of 3′ UTR TTSs are in close distance to a preceding stop codon (<20 nt), whereas 42% of them are >100 nt away (Fig. 2D). This contrasts a recent study in B. subtilis that revealed a large fraction of intrinsic terminators overlapping with coding sequences (50). The ability of these terminators to retain efficient transcription termination activity was assigned to runaway transcription of the RNA polymerase, leading to insensitivity to translation-based transcriptional regulation. We explored the original publication, in which a variety of genomes were analyzed in silico, with respect to the presence and location of intrinsic terminators. Reanalysis of the original dataset for C. difficile confirms our experimental results that intrinsic terminators rarely overlap with stop codons (SI Appendix, Fig. S2B), suggesting that runaway transcription is not a determining feature of gene expression in C. difficile.
We further detected many overlapping termination events between convergently transcribed genes, such as for the toxin gene tcdA (toxA in CP010905.2 which was used in this study) and the negative regulator of toxin gene expression tcdC (Fig. 2E), which is also a pervasive transcriptome feature in Escherichia coli (51). Overall, our UTR annotations revealed an abundance of overlapping transcripts between convergently transcribed genes (246 gene pairs or 32.06% of transcripts with TTSs), whereas overlaps between divergently transcribed genes were rare (8 or 1.17% of transcripts with TSSs) (Dataset S8).
Annotation of Small ORFs and Operons.
High-resolution transcriptome maps allow the identification of ORFs and operon structures that have been overlooked in automated genome annotations. This includes so called small ORFs (sORFs) of usually 50 amino acids or less, an emerging class of bacterial genes with an unfolding spectrum of biological functions (52, 53). In many cases, they are predicted to be membrane proteins, containing an alpha-helical transmembrane domain (54). Focusing on oTSS in particular, we searched for novel ORFs based on the following criteria: 1) presence of a start and stop codon, 2) presence of an RBS within 15 base pairs (bp) upstream of the start codon, and 3) sequence conservation in other Clostridioides strains. Based on this approach, we identified 12 sORF candidates, seven of which are predicted to contain a transmembrane helix (Dataset S2). Among the identified candidates, six are toxins and part of previously identified C. difficile type-I toxin–antitoxin systems (55, 56). Among the remaining sORFs, four are high-confidence candidates; one being a conjugal transfer protein with annotation in other C. difficile strains; two candidates that each have a Shine–tDalgarno (SD) sequence and a predicted α-helical transmembrane domain; and one sORF associated with a 160-nt long 5′ UTR region harboring a c-di-GMP-I riboswitch.
Transcriptome studies have revealed operon structures more complex than previously anticipated (51, 57, 58), but predicted operons in C. difficile are still largely based on computational inference from other bacterial species (59). Here, our transcriptome-based annotation predicts 440 operons (Dataset S1), including 64 previously unknown operons such as the dicistronic transcript comprising spermine/spermidine acetyltransferase bltD and CDIF630_01360, a putative N-acetyltransferase. Furthermore, we identify 109 suboperon structures which, in 29 cases, uncouple the last or the last two genes from upstream genes within an operon. One example is acpS-ndoA, in which the last two genes encoding a type-II toxin–antitoxin system (ndoA and ndoA1) are uncoupled from the full operon.
5′ UTRs and Associated Regulatory Elements.
Bacterial 5′ UTRs can influence gene expression, usually through embedded cis-regulatory elements such as riboswitches, RNA thermometers, and genetic switches. Similar to B. subtilis and Listeria monocytogenes, the majority of experimentally mapped 5′ UTR lengths ranged from 20 to 60 nt (Fig. 3A and Dataset S1) (14, 40, 60). A likely RBS (aGGAGg) was detected in ∼90% of these 5′ UTRs (Fig. 3 A, Inset). Only six mRNAs appeared leaderless, having a 5′ UTR of <10 nt, including the spoVAE gene within the tricistronic spoVACDE operon (61).
Fig. 3.
Discovery of cis-regulatory elements in 5′ UTR regions. (A) Frequencies of 5′ UTR lengths based on 1,646 primary and secondary TSSs. The red bars indicate six leaderless mRNAs with a 5′ UTR length of <10 nt. The Inset shows the predicted SD sequence motif of C. difficile 630. (B) Classification of PTT events in 5′ UTRs of mRNAs. The majority is associated with conserved RNA families. The remaining 19 PTT events lack homology to known riboregulators and are classified putative regulators. (C) Putative L20 leader in the 5′ UTR of the infC operon. (Top) Predicted secondary structure for CDIF630nc_018 using RNAfold. Nucleotides are colored according to base-pairing probabilities. (Bottom) Schematic overview of the infC operon organization including 5′ UTR region and expression analysis of CDIF630nc_018 by Northern blot using a radioactively labeled DNA probe. Total RNA was extracted at midexponential (ME), late-exponential (LE), and stationary (ST) growth phases from C. difficile 630 grown in BHI, TY, and TY supplemented with 0.5% glucose (TYG) media.
Many genes (561) were associated with surprisingly long 5′ UTRs of >100 nt. Among those having a very long 5′ UTR of >300 nt, we identified 77 Rfam-predicted riboswitch candidates (62) (Fig. 3B). Premature transcription termination (PTT) was evident for 57 of them, indicating their ON state in which transcription of the parental gene is repressed (Dataset S5). Interestingly, we detected a speF riboswitch previously only documented in gram-negative alphaproteobacteria associated with CDIF630_01955, encoding a methyltransferase domain protein. As before, the associated termination site suggests that the riboswitch is functional, which is further supported by a 220-nt Northern blot signal (SI Appendix, Fig. S3).
We further detected 17 putative PTT events in 5′ UTRs lacking similarities to conserved riboregulators. A potential generation from mRNA processing is unlikely in these cases, since the characteristic read enrichment in the untreated (TEX−) cDNA library, usually associated with processing sites, is missing. One such PTT event is located in the 5′ UTR of the infC-rpmI-rplT operon whose expression is controlled through an L20 leader via transcription attenuation in B. subtilis (63, 64). This autoregulatory structure is also found in other low-GC gram-positive bacteria (Rfam family RF00558) but does not seem to be conserved on the primary sequence level in C. difficile. Nevertheless, secondary structure prediction reveals extensive interactions between distant bases that is reminiscent of the antiterminator conformation of the L20 leader described in B. subtilis (Fig. 3C). Furthermore, PTT-associated genes encoded phosphotransferase systems (bglF, bglF1, bglG3, and bglG4) known to be regulated by antiterminator proteins in B. subtilis (65), transcriptional regulators (CDIF630_00097, CDIF630_02384, and CDIF630_02922), multidrug resistance (MDR)-type ABC transporters (CDIF630_03083, CDIF630_02847, and CDIF630_03664), and aroF (Dataset S5). Northern blot validation for a selection of candidates confirmed the RNA-seq–predicted transcript sizes and revealed larger bands in several cases that are likely corresponding to the full-length parental gene or its degradation products (SI Appendix, Fig. S3).
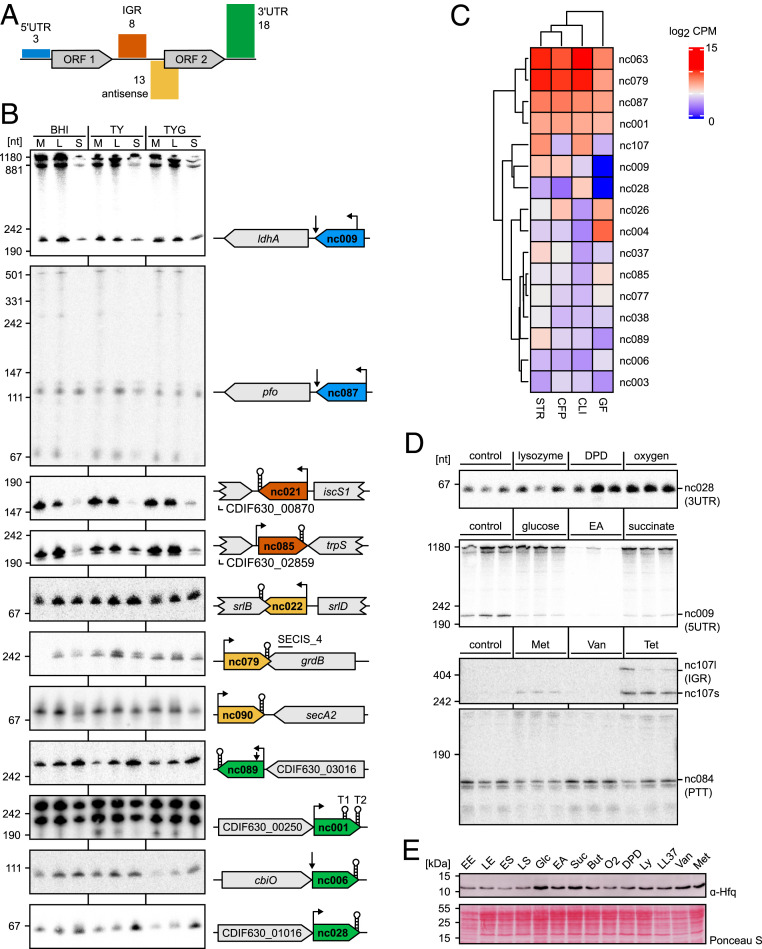
The sRNA Landscape of C. difficile in Changing Environmental Conditions and during Infection.
Our primary transcriptome analysis identified 42 transcripts that lacked an internal ORF, qualifying them as potential sRNAs (Dataset S5). A classification based on their genomic location (Fig. 4A) revealed the largest group to be 3′ UTR–derived sRNAs (18 3′ UTR), followed by those encoded cis-antisense either to a gene, a 5′/3′ UTR, or another sRNA (13 AS). In comparison, only few sRNAs were located in intergenic regions (8 IGR) or derived from the 5′ UTR of mRNAs (3 5′ UTR). Sequence conservation beyond C. difficile was extremely rare (SI Appendix, Fig. S4A and Dataset S7), whereas most are highly conserved among clinical strains of C. difficile covering five major genomic clades and diverse toxin repertoires (SI Appendix, Fig. S4B) (66). Some of the sRNAs that were absent in a subset of strains were either transposon-associated sRNAs (CDIF630nc_004 and CDIF630nc_069) or located on a prophage (CDIF630nc_095). Of note, this high level of sequence conservation within the species generally extended to noncoding 5′ and 3′ UTR regions (92% of 5′ UTRs with >90% identity and 90% of 3′ UTRs with >90% identity, Dataset S9).
Fig. 4.
C. difficile 630 sRNA landscape in changing environmental conditions. (A) Classification of annotated sRNA candidates based on their genomic location. The numbers indicate amount of annotated sRNAs for each class. (B) Expression profiles of representative candidates for each sRNA class. Total RNA was extracted at midexponential (ME), late-exponential (LE), and stationary (ST) growth phases from C. difficile 630 grown in TY, TY supplemented with 0.5% glucose (TYG), and BHI medium and analyzed by Northern blot using radioactively labeled DNA probes. (C) Heatmap of log2 CPM values for newly annotated sRNAs during C. difficile 630 intestinal colonization of different antibiotic-treated mouse models (Streptomycin, STR; Cefoperazone, CFP; Clindamycin, CLI) and exGerm-free mice (GF). Published data were taken from ref. 67 and reanalyzed using the updated genome annotation. Only sRNAs with a CPM value ≥50 were considered to be expressed. (D) Expression profiles of sRNAs that are differentially expressed in response to various nutritional and stress conditions. Total RNA was extracted from midexponential TY cultures of C. difficile 630 WT exposed to TY (control), lysozyme, iron limitation (2,2′-Dipyridine, DPD), oxygen, glucose, ethanolamine (EA), succinate, metronidazole (Met), vancomycin (Van), and tetracycline (Tet) for 20 min and analyzed by Northern blot using radioactively labeled DNA probes. (E) Western blot analysis of Hfq protein levels across different growth phases and growth media. Equal optical density (OD) units of total cell lysates of C. difficile 630 WT were loaded. Western blot membranes were incubated with anti-Hfq antibody. Ponceau S staining of the blotting membrane served as loading control. Total cell extracts of equal OD units were prepared from C. difficile 630 WT grown in TY medium to early exponential (EE), late-exponential (LE), early stationary (ES), and late stationary (LS) phase and from bacteria grown to late-exponential phase in TY medium supplemented with glucose (Glc), ethanolamine (EA), succinate (Suc), butyrate (But), 2,2′-Dipyridine (DPD), lysozyme (Ly), LL-37, vancomycin (Van), metronidazole (Met), or in 1% oxygen atmosphere (O2). Precise concentrations used are listed under SI Appendix, Materials and Methods.
Northern blot validation revealed expression throughout exponential growth for most sRNA candidates (Fig. 4B). About one-half of them are down-regulated after entry into stationary phase, while three candidates (CDIF630nc_028, CDIF630nc_089, and CDIF630nc_105) showed a marked accumulation. In agreement with the expression profile, CDIF630nc_028 has a predicted promoter sequence for SigB, while CDIF630nc_089 is associated with a SigH promoter. Moreover, Northern blot validation of sRNA expression in the hypervirulent RT027 isolate R20291 revealed identical expression profiles in both strains for the selected candidates (SI Appendix, Fig. S5).
To gain insight into sRNA activities during infection, we took advantage of published in vivo transcriptome data from C. difficile–infected ex-germ–free mice as well as conventional mice pretreated with different antibiotics (67). Reanalysis of the data revealed expression of 16 sRNAs (cutoff = 50 counts per million mapped reads [CPM]), including the most broadly conserved sRNAs CDIF630nc_001, CDIF630nc_026, and CDIF630nc_079 as well as the SigB- and SigH-associated CDIF630nc_028 and CDIF630nc_089, respectively (Fig. 4C and Dataset S10). To identify infection-related signals regulating sRNA expression, we exposed C. difficile to a panel of nutritional and stress conditions including intestinal metabolites (glucose, ethanolamine, succinate, and butyrate), oxygen stress, iron limitation, as well as antimicrobial compounds (lysozyme, LL-37, vancomycin, metronidazole, and tetracycline). Within this panel, we observed regulation for several sRNAs or PTTs (Fig. 4D), e.g., SigB-regulated CDIF630nc_028 responded to iron limitation and oxygen stress, suggesting a potential role during infection. To complement the picture, we also quantified native protein levels of the sRNA chaperone Hfq during growth in these conditions (Fig. 4E). We observed similar Hfq levels not only in different growth phases but also under a variety of nutritional and stress conditions, demonstrating that Hfq is constitutively produced in changing environmental conditions.
C. difficile Hfq Is a Global RNA Chaperone.
In many gram-negative model organisms, sRNAs require the RNA chaperone Hfq to facilitate their interaction with target mRNAs (68). However, whether Hfq functions as a central RNA-binding protein (RBP) in C. difficile remains unknown. Therefore, we performed Hfq immunoprecipitation followed by sequencing of bound RNA species (RIP-seq) in a strain expressing C-terminally 3×FLAG-tagged hfq (Hfq-FLAG) under its native promoter to draft the spectrum of Hfq-bound RNAs in vivo. Bacteria were grown to midexponential, late-exponential, and stationary phase (SI Appendix, Fig. S6A) and subjected to the RIP-seq protocol (SI Appendix, Fig. S6B). Western blot validation using monoclonal FLAG antibody confirmed expression of Hfq throughout all growth phases and specific enrichment of Hfq-FLAG from C. difficile 630 lysate (SI Appendix, Fig. S6C). In addition, copurification of Hfq-bound sRNAs was confirmed by Northern blot analysis of lysates and eluate fractions from Hfq-FLAG and Hfq-ctrl cultures (SI Appendix, Fig. S6C).
The majority of RNA species coimmunoprecipitating with Hfq-3×FLAG were mRNAs (Fig. 5A and Dataset S6). Hfq bound mostly to the 5′ and 3′ UTRs of target mRNAs, and of note, many enriched 5′ UTRs harbored a riboswitch or PTT (66/124 5′ UTRs) (Fig. 5A) as recently observed in an independent Hfq RIP-seq analysis (20). Many mRNAs showed growth phase–dependent association with Hfq (SI Appendix, Fig. S7 B and C) and mainly included genes involved in the biosynthesis of amino acids, membrane transport, signal transduction, and translation. Most interestingly, we also found several mRNAs encoding proteins involved in sporulation (spo0A, spoVB, oppB, sigG, and sspA2), toxin production (tcdE), motility (flgB, fliJ, flhB, and cheW1), quorum sensing (luxS and agrB), the cell wall protein–encoding cwpV, as well as several adhesins potentially involved in biofilm formation (CDIF630_03429, CDIF630_01459, and CDIF630_03096) (SI Appendix, Fig. S7C and Dataset S6).
Fig. 5.
The spectrum of Hfq-associated RNA ligands and Hfq impact on RNA stability. (A) Pie chart for Hfq RIP-seq showing the relative amount of Hfq-associated sequences mapping to different RNA classes. Hfq RIP-seq was performed on C. difficile 630 WT and C-terminally 3xFLAG-tagged hfq expressed from a plasmid, grown to midexponential, late-exponential, and early stationary phases in TY medium in two independent experiments. (B) Bar chart showing the fraction of sRNAs for each class that were bound by Hfq in vivo. (C) Hfq acts as a stabilizing factor for its ncRNA ligands in vivo. (Top) Total RNA was extracted from WT, Δhfq, and the complemented hfq mutant (hfqC) in late-exponential phase after addition of rifampicin at the indicated time points and analyzed by Northern blotting using radioactively labeled DNA probes. The Hfq-independent sRNA nc009 served as negative control. The representative of three independent Northern blots is shown. Abbreviations: RSW (riboswitch), AS (antisense), IGR (intergenic).
The enrichment of many 5′ UTRs could suggest that Hfq is recognizing the SD or ATG sequence region, but lack of clear Hfq peaks at SD or start codon sites does not sufficiently support this assumption at this point. In addition, the enrichment of many 3′ UTRs and of 5′ UTRs harboring a riboswitch or PTT event could point to a potential role of terminator hairpins in Hfq binding. An RNA motif search using CMfinder identified an RNA structure reminiscent of a terminator hairpin that was enriched in Hfq-bound transcripts, albeit nucleotide identity was low (SI Appendix, Fig. S7A).
Importantly, many predicted sRNAs associated with Hfq, including the previously identified Hfq-binding sRNA CDIF630nc_070 (also known as Rcd1, Fig. 5A and Dataset S6) (18, 28). In addition, all but one type-I antitoxin transcript were enriched (Fig. 5A and Dataset S6), similar to a recent RIP-seq analysis of Hfq in C. difficile (20). Classifying Hfq-associated sRNAs with respect to their genomic location revealed that the majority of cis-antisense–encoded sRNAs in C. difficile were bound by Hfq, as well as approximately one-half of all intergenic, 5′ UTR–, and 3′ UTR–encoded sRNAs (Fig. 5B). Additionaly, many sRNAs were bound across all three growth phases, with CDIF630nc_070 and CDIF630nc_079 being the two top-enriched sRNAs (SI Appendix, Fig. S6D and Dataset S6). However, a few sRNAs displayed growth phase–dependent association with Hfq, such as CDIF630nc_090, which was only enriched in stationary phase (SI Appendix, Fig. S6D).
Hfq Acts as a Stabilizing Factor for its noncoding RNA Ligands.
One major function of gram-negative Hfq homologs is the stabilization of their sRNA ligands, which appears to be the exception in gram-positive bacteria (69–71). To test if transcript stabilities of Hfq-associating noncoding RNAs (ncRNAs) were affected by Hfq, we generated a hfq deletion (Δhfq) in C. difficile 630 using a homologous recombination vector we developed for effective counterselection on standard laboratory media using the tightly regulated E. coli endoribonuclease mazF (for details, see SI Appendix). To complement hfq in cis (hfqC), we established an additional vector system which enables efficient insertion of DNA by homologous recombination between pyrE (CD630_01870) and CD630_01880.
C. difficile wild type (WT), Δhfq, and hfqC strains were grown to late-exponential phase, and rifampicin was added to arrest transcription. Northern blotting of RNA samples collected at several timepoints after rifampicin addition was performed to quantify transcript levels for several sRNAs as well as a Hfq-bound riboswitch and type-I antitoxin. Remarkably, half-lives and, in some cases, the steady-state levels of all analyzed Hfq-associated sRNAs (CDIF630nc_006, CDIF630nc_085, and CDIF630nc_022) were decreased, an effect that could be restored in the hfqC strain (Fig. 5C and SI Appendix, Fig. S8 A and B) and that was not observed for the Hfq-independent sRNA CDIF630nc_009. In addition, half-lives were also reduced for the tested type-I antitoxin as well as the c-di-GMP-II riboswitch, suggesting a potential dual function for the latter cis-acting regulator (SI Appendix, Fig. S8B).
A Role of CDIF630nc_085 in the Regulation of Ethanolamine Utilization.
To gain insight into sRNA functions, we chose the bona fide intergenic sRNA CDIF630nc_085 (short nc085) (Fig. 4B) for further characterization because its expression during intestinal colonization (Fig. 4C) suggested a potential virulence function. Secondary structure prediction showed a moderately folded structure with an extended stem loop at the 5′ end, two hair pins in the center of the sRNA, and a Rho-independent terminator at the 3′ end (Fig. 6A). To identify potential mRNA targets of nc085, we pulse expressed the sRNA from an anhydrotetracycline-inducible plasmid in a ΔCDIF630nc_085 background (Fig. 6B) and determined differentially expressed genes using RNA-seq analysis (Fig. 6C and Dataset S11). We observed many sporulation-associated genes to be down-regulated upon nc085 expression (Dataset S11). In addition, gene cluster analysis revealed ABC transporters and two-component systems as the largest group of regulated genes (SI Appendix, Fig. S9B). Searching for base-pairing regions within the 5′ UTRs of differentially expressed genes identified two potential seed regions within nc085 (Fig. 6A, SR1 and SR2), both complementarity to several target mRNAs (SI Appendix, Fig. S9A).
Fig. 6.
CDIF630nc_085 regulates ethanolamine utilization in C. difficile. (A) Predicted secondary structure for CDIF630nc_085 (nc085) using RNAfold. Nucleotides are colored according to base-pairing probabilities. Two seed regions predicted to interact with differentially expressed mRNA targets are located in an extended small stem loop (seed region 1) and a single-stranded stretch (seed region 2) in the 3′ region of the sRNA, respectively. (B) Northern blot validation of nc085 pulse expression in a nc085 deletion strain. sRNA expression was transiently induced for 20 min by addition of 100 ng/mL anhydrotetracycline (ATc) to late-exponential cultures of C. difficile WT, Δnc085 (−), Δnc085 harboring the empty pRPF185 backbone (EV), and Δnc085 harboring the pulse-expression vector carrying nc085 (PE) under the control of an ATc-inducible promoter. (C) RNA was extracted from ATc-induced cultures harboring the empty vector (EV) or the pulse-expression plasmid (PE) and analyzed by RNA-seq. The genes that were differentially expressed upon pulse expression of nc085 (false discovery rate < = 0.1, log2 fold change [log2FC] ≥ 1, or log2FC < = −1) are colored according to their transcript class. (D) Read profiles from untreated (Tex−), Tex-treated (Tex+), and RNAtag-seq libraries mapping to the eut region. TSSs were identified upstream of eutS and eutV. An RNA duplex formed by nc085 with the start codon region of eutV was predicted using IntaRNA. The red letters indicate the start codon. (E) In-line probing of 0.2 pmol of 32P-labeled nc085 in the absence (lane 3) or presence of either 0.2 pmol (lane 4), 2 pmol (lane 5), 20 pmol (lane 6), or 200 pmol (lane 7) eutV target region. Partially RNase T1– (lane 1; T1) or alkali-digested (lane 2; OH) nc085 served as ladders. Seed 1 (red) denotes the seed region that is protected from cleavage changes in the presence of the target. A representative image of two independent experiments is shown. (F) Growth of C. difficile 630 WT, Δhfq, and Δnc085 in CDMM supplemented with either glucose (5 mM) or ethanolamine (50 mM). Growth experiments were performed in two independent experiments with three biological replicates each (n = 6). The error bars show SEM. Bottom table: listed are the calculated doubling times (DT) ± SD as well as the maximum optical densities (OD600) and the timepoint when they were reached during the experiment, for each strain and each growth condition. Due to the complex, diauxic growth behavior of all strains in the analyzed growth conditions and doubling times were calculated only for the first exponential phase.
Among them was eutV, which is part of a two-component system encoded by the eutVW operon, a positive regulator of ethanolamine utilization. Our transcriptome data identified low-level constitutive expression across all growth conditions and a TSS upstream of the eutVW operon (Fig. 6D, data only shown for late-exponential TY condition). Moreover, the predicted base-pairing region within the eutV 5′ UTR comprised the start codon site of eutV (Fig. 6D).
To test the prediction experimentally, we performed in-line probing of nc085 with or without the eutV 5′ UTR. Although high concentrations of the target mRNA were required, changes in the protection from cleavage in the presence of eutV were observed (Fig. 6E, lane 7) and provided support for the computationally predicted base-pairing region in nc085. Considering the location of the seed region within a hairpin structure, this interaction may need to be facilitated by Hfq in vivo. To explore the impact of nc085 on ethanolamine utilization, we compared growth of C. difficile WT, Δnc085, and the Δhfq mutant in C. difficile minimal medium (CDMM) supplemented either with glucose or ethanolamine. Cultivation in glucose-supplemented medium resulted in identical growth for WT and Δnc085, whereas Δhfq had an extended lag phase and grew to lower maximum optical densities. In contrast, when grown in CDMM supplemented with ethanolamine, both Δnc085 and Δhfq exhibited accelerated growth in comparison to the WT (Fig. 6F). Specifically, both strains entered exponential growth faster and reached their maximum optical densities at earlier timepoints (Fig. 6 F, Bottom). Together, these data identify a regulatory function of nc085 in the utilization of the abundant intestinal carbon and nitrogen source ethanolamine, which was shown to impact pathogenicity of C. difficile (72).
Discussion
In the present study, we have applied global approaches of bacterial RNA biology (35) to capture the transcriptome architecture of C. difficile and unravel the scope of posttranscriptional regulation in this important human pathogen.
A Gram-Positive Hfq Homolog that Impacts sRNA Stabilities and Function.
It is undisputed that sRNAs are central players in the regulation of physiological and virulence pathways in gram-positive bacteria including L. monocytogenes, B. subtilis, and Staphylococcus species. For example, the sRNA RsaE, which is conserved in the order Bacillales, has been shown to coordinate central carbon and amino acid metabolism in Staphylococcus aureus (73, 74) and also to promote biofilm formation in Staphylococcus epidermidis by supporting extracellular DNA release and the production of polysaccharide-intercellular adhesin (75). However, the function of Hfq in facilitating the interactions between sRNA regulators and their target mRNAs in gram-positive bacteria remains elusive since most of the known sRNAs seem to exert their regulatory activity independent of this global RBP (76).
In the present study, our characterization of Hfq–sRNA interactions in C. difficile not only revealed a large network of Hfq-binding sRNAs along with potential target mRNAs but also provides clear evidence for the impact of Hfq on sRNA steady-state levels and stabilities in vivo. Our data align with initial studies using a Hfq-depleted C. difficile strain that identified pleiotropic effects of Hfq depletion on gene expression using microarray analysis (18). In line with these observations, absence of Hfq impacts growth on various carbon and nitrogen sources, as shown in our study, and results in increased sensitivity to stresses, sporulation rates, and biofilm formation (18). Taken together, the regulatory activities of Hfq in C. difficile strongly resemble the situation in gram-negative model organisms such as E. coli and Salmonella enterica and open avenues for the study of sRNA-based gene regulation in this important pathogen.
Ethanolamine Utilization Is Governed by sRNA Regulation in Gram-Positive Pathogens.
We have taken a first step toward the characterization of sRNA-mRNA interactions in C. difficile by identifying the ethanolamine utilization pathway to be regulated by the intergenic sRNA nc085. Ethanolamine is an abundant intestinal nutrient that is exploited by both gram-negative and gram-positive enteric pathogens, including C. difficile, during intestinal colonization (72, 77–79). Interestingly, the ethanolamine utilization (eut) gene cluster is regulated by a small regulatory RNA in both Enterococcus faecalis (EutX) and L. monocytogenes (Rli55). In both species, the sRNA contains a classic AdoCbl-responsive riboswitch in its 5′ portion that allows expression of the full-length sRNA only in the absence of vitamin B12 (80, 81). This full-length transcript harbors a dual-hairpin structure in its 3′ part that serves as a binding platform for the ethanolamine-specific, RNA-binding response regulator EutV (82), thereby sequestering EutV from binding to its target sequences in the 5′ UTRs of eut genes. An in silico search for an AdoCbl riboswitch failed to identify such a conserved element within nc085, and secondary structures of the three sRNAs show a less-complex folding pattern for nc085 that is missing the dual hairpin for EutV binding. Overall, this suggests that the mechanism of eut gene regulation by nc085 differs from EutX and Rli55. Currently, down-regulation of the eutVW operon upon nc085 pulse expression together with a predicted target site comprising the start codon of eutV suggests a classical mode-of-action whereby nc085 binding interferes with translation initiation. Consequently, deleting nc085 should relieve the repression and increase the pool of EutVW that can sense ethanolamine and activate the eut gene cluster. Aligning with this model, we observed early onset of growth for the nc085 deletion strain in CDMM with ethanolamine as the sole carbon source (Fig. 6F).
Overall, our findings further confirm that ethanolamine utilization is a conserved target of sRNA-based regulation in gram-positive pathogens.
Materials and Methods
Comprehensive descriptions of all materials and methods that were used in this study are provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank the Core Unit SysMed at the University of Würzburg for technical support with RIP-seq and RNAtag-seq data generation. This work was supported by the Interdisciplinary Center for Clinical Research (IZKF) at the University of Würzburg (Project No. Z-6). We also acknowledge the recombinant protein expression facility of the Rudolf Virchow Center for the expression and purification of Hfq. We thank the Vogel Stiftung Dr. Eckernkamp for supporting F.P. with a Dr. Eckernkamp Fellowship. J.S. was supported by the German Research Foundation (DFG Grant No. FA 1113/2-1). J.A.K. was supported by the Wellcome Trust (Grant No. 204877/Z/16/Z).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2103579118/-/DCSupplemental.
Data Availability
All RNA-sequencing data are available at the National Center for Biotechnology Information Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo) under the accession number GSE155167. Plasmids pKJAK112, pJAK184, and pJAK080 have been deposited with Addgene (167279–167281).
References
- 1.Smits W. K., Lyras D., Lacy D. B., Wilcox M. H., Kuijper E. J., Clostridium difficile infection. Nat. Rev. Dis. Primers 2, 16020 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peng Z., et al., Update on antimicrobial resistance in Clostridium difficile: Resistance mechanisms and antimicrobial susceptibility testing. J. Clin. Microbiol. 55, 1998–2008 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khanna S., Gerding D. N., Current and future trends in Clostridioides (Clostridium) difficile infection management. Anaerobe 58, 95–102 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Guery B., Galperine T., Barbut F., Clostridioides difficile: Diagnosis and treatments. BMJ 366, l4609 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Bouillaut L., Dubois T., Sonenshein A. L., Dupuy B., Integration of metabolism and virulence in Clostridium difficile. Res. Microbiol. 166, 375–383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin-Verstraete I., Peltier J., Dupuy B., The regulatory networks that control Clostridium difficile toxin synthesis. Toxins (Basel) 8, 153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pettit L. J., et al., Functional genomics reveals that Clostridium difficile Spo0A coordinates sporulation, virulence and metabolism. BMC Genomics 15, 160 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El Meouche I., et al., Characterization of the SigD regulon of C. difficile and its positive control of toxin production through the regulation of tcdR. PLoS One 8, e83748 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKee R. W., Mangalea M. R., Purcell E. B., Borchardt E. K., Tamayo R., The second messenger cyclic Di-GMP regulates Clostridium difficile toxin production by controlling expression of sigD. J. Bacteriol. 195, 5174–5185 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saujet L., Monot M., Dupuy B., Soutourina O., Martin-Verstraete I., The key sigma factor of transition phase, SigH, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile. J. Bacteriol. 193, 3186–3196 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kint N., et al., The alternative sigma factor σB plays a crucial role in adaptive strategies of Clostridium difficile during gut infection. Environ. Microbiol. 19, 1933–1958 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Kint N., et al., The σB signalling activation pathway in the enteropathogen Clostridioides difficile. Environ. Microbiol. 21, 2852–2870 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Boekhoud I. M., Michel A.-M., Corver J., Jahn D., Smits W. K., Redefining the clostridioides difficile σB regulon: σB activates genes involved in detoxifying radicals that can result from the exposure to antimicrobials and hydrogen peroxide. mSphere 5, e00728-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wurtzel O., et al., Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol. Syst. Biol. 8, 583 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mäder U., et al., Staphylococcus aureus transcriptome architecture: From laboratory to infection-mimicking conditions. PLoS Genet. 12, e1005962 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warrier I., et al., The transcriptional landscape of Streptococcus pneumoniae TIGR4 reveals a complex operon architecture and abundant riboregulation critical for growth and virulence. PLoS Pathog. 14, e1007461 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soutourina O., et al., Genome-wide transcription start site mapping and promoter assignments to a sigma factor in the human enteropathogen Clostridioides difficile. Front. Microbiol. 11, 1939 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boudry P., et al., Pleiotropic role of the RNA chaperone protein Hfq in the human pathogen Clostridium difficile. J. Bacteriol. 196, 3234–3248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caillet J., Gracia C., Fontaine F., Hajnsdorf E., Clostridium difficile Hfq can replace Escherichia coli Hfq for most of its function. RNA 20, 1567–1578 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boudry P., et al., Identification of RNAs bound by Hfq reveals widespread RNA partners and a sporulation regulator in the human pathogen Clostridioides difficile. RNA Biol., 10.1080/15476286.2021.1882180 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maikova A., Kreis V., Boutserin A., Severinov K., Soutourina O., Using an endogenous CRISPR-cas system for genome editing in the human pathogen Clostridium difficile. Appl. Environ. Microbiol. 85, e01416-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmqvist E., Vogel J., RNA-binding proteins in bacteria. Nat. Rev. Microbiol. 16, 601–615 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Kavita K., de Mets F., Gottesman S., New aspects of RNA-based regulation by Hfq and its partner sRNAs. Curr. Opin. Microbiol. 42, 53–61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dambach M., Irnov I., Winkler W. C., Association of RNAs with Bacillus subtilis Hfq. PLoS One 8, e55156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hämmerle H., et al., Impact of Hfq on the Bacillus subtilis transcriptome. PLoS One 9, e98661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rochat T., et al., Tracking the elusive function of Bacillus subtilis Hfq. PLoS One 10, e0124977 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y., Indurthi D. C., Jones S. W., Papoutsakis E. T., Small RNAs in the genus Clostridium. mBio 2, e00340-10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soutourina O. A., et al., Genome-wide identification of regulatory RNAs in the human pathogen Clostridium difficile. PLoS Genet. 9, e1003493 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anjuwon-Foster B. R., Tamayo R., A genetic switch controls the production of flagella and toxins in Clostridium difficile. PLoS Genet. 13, e1006701 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emerson J. E., et al., A novel genetic switch controls phase variable expression of CwpV, a Clostridium difficile cell wall protein. Mol. Microbiol. 74, 541–556 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trzilova D., Anjuwon-Foster B. R., Torres Rivera D., Tamayo R., Rho factor mediates flagellum and toxin phase variation and impacts virulence in Clostridioides difficile. PLoS Pathog. 16, e1008708 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKee R. W., Harvest C. K., Tamayo R., Cyclic diguanylate regulates virulence factor genes via multiple riboswitches in Clostridium difficile. mSphere 3, e00423-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bordeleau E., Fortier L. C., Malouin F., Burrus V., c-di-GMP turn-over in Clostridium difficile is controlled by a plethora of diguanylate cyclases and phosphodiesterases. PLoS Genet. 7, e1002039 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peltier J., et al., Cyclic diGMP regulates production of sortase substrates of Clostridium difficile and their surface exposure through ZmpI protease-mediated cleavage. J. Biol. Chem. 290, 24453–24469 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hör J., Gorski S. A., Vogel J., Bacterial RNA biology on a genome scale. Mol. Cell 70, 785–799 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Westermann A. J., Vogel J., Cross-species RNA-seq for deciphering host-microbe interactions. Nat. Rev. Genet. 22, 361–378 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Kröger C., et al., An infection-relevant transcriptomic compendium for Salmonella enterica Serovar Typhimurium. Cell Host Microbe 14, 683–695 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Kröger C., et al., The primary transcriptome, small RNAs and regulation of antimicrobial resistance in Acinetobacter baumannii ATCC 17978. Nucleic Acids Res. 46, 9684–9698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryan D., Jenniches L., Reichardt S., Barquist L., Westermann A. J., A high-resolution transcriptome map identifies small RNA regulation of metabolism in the gut microbe Bacteroides thetaiotaomicron. Nat. Commun. 11, 3557 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma C. M., et al., The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464, 250–255 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Sharma C. M., Vogel J., Differential RNA-seq: The approach behind and the biological insight gained. Curr. Opin. Microbiol. 19, 97–105 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Wydau-Dematteis S., et al., Cwp19 is a novel lytic transglycosylase involved in stationary-phase autolysis resulting in toxin release in Clostridium difficile. mBio 9, e00648-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wade J. T., Grainger D. C., Pervasive transcription: Illuminating the dark matter of bacterial transcriptomes. Nat. Rev. Microbiol. 12, 647–653 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Pratto F., et al., Streptococcus pyogenes pSM19035 requires dynamic assembly of ATP-bound ParA and ParB on parS DNA during plasmid segregation. Nucleic Acids Res. 36, 3676–3689 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saujet L., et al., Genome-wide analysis of cell type-specific gene transcription during spore formation in Clostridium difficile. PLoS Genet. 9, e1003756 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Serrano M., et al., A recombination directionality factor controls the cell type-specific activation of σK and the fidelity of spore development in Clostridium difficile. PLoS Genet. 12, e1006312 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woods E. C., Edwards A. N., Childress K. O., Jones J. B., McBride S. M., The C. difficile clnRAB operon initiates adaptations to the host environment in response to LL-37. PLoS Pathog. 14, e1007153 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pishdadian K., Fimlaid K. A., Shen A., SpoIIID-mediated regulation of σK function during Clostridium difficile sporulation. Mol. Microbiol. 95, 189–208 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shishkin A. A., et al., Simultaneous generation of many RNA-seq libraries in a single reaction. Nat. Methods 12, 323–325 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson G. E., Lalanne J. B., Peters M. L., Li G. W., Functionally uncoupled transcription-translation in Bacillus subtilis. Nature 585, 124–128 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ju X., Li D., Liu S., Full-length RNA profiling reveals pervasive bidirectional transcription terminators in bacteria. Nat. Microbiol. 4, 1907–1918 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hemm M. R., Weaver J., Storz G., Escherichia coli small proteome. Ecosal Plus 9, 10.1128/ecosalplus.ESP-0031-2019 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garai P., Blanc-Potard A., Uncovering small membrane proteins in pathogenic bacteria: Regulatory functions and therapeutic potential. Mol. Microbiol. 114, 710–720 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Storz G., Wolf Y. I., Ramamurthi K. S., Small proteins can no longer be ignored. Annu. Rev. Biochem. 83, 753–777 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maikova A., et al., Discovery of new type I toxin-antitoxin systems adjacent to CRISPR arrays in Clostridium difficile. Nucleic Acids Res. 46, 4733–4751 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soutourina O., Type I., Type I toxin-antitoxin systems in clostridia. Toxins (Basel) 11, 253 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yan B., Boitano M., Clark T. A., Ettwiller L., SMRT-Cappable-seq reveals complex operon variants in bacteria. Nat. Commun. 9, 3676 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sáenz-Lahoya S., et al., Noncontiguous operon is a genetic organization for coordinating bacterial gene expression. Proc. Natl. Acad. Sci. U.S.A. 116, 1733–1738 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karp P. D., et al., The BioCyc collection of microbial genomes and metabolic pathways. Brief. Bioinform. 20, 1085–1093 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Irnov I., Sharma C. M., Vogel J., Winkler W. C., Identification of regulatory RNAs in Bacillus subtilis. Nucleic Acids Res. 38, 6637–6651 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Donnelly M. L., Fimlaid K. A., Shen A., Characterization of Clostridium difficile spores lacking either SpoVAC or dipicolinic acid synthetase. J. Bacteriol. 198, 1694–1707 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kalvari I., et al., Non-coding RNA analysis using the Rfam database. Curr. Protoc. Bioinformatics 62, e51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choonee N., Even S., Zig L., Putzer H., Ribosomal protein L20 controls expression of the Bacillus subtilis infC operon via a transcription attenuation mechanism. Nucleic Acids Res. 35, 1578–1588 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Babina A. M., Parker D. J., Li G. W., Meyer M. M., Fitness advantages conferred by the L20-interacting RNA cis-regulator of ribosomal protein synthesis in Bacillus subtilis. RNA 24, 1133–1143 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fujita Y., Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci. Biotechnol. Biochem. 73, 245–259 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Riedel T., et al., High metabolic versatility of different toxigenic and non-toxigenic Clostridioides difficile isolates. Int. J. Med. Microbiol. 307, 311–320 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Jenior M. L., Leslie J. L., Young V. B., Schloss P. D., Clostridium difficile colonizes alternative nutrient niches during infection across distinct murine gut microbiomes. mSystems 2, e00063-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hör J., Matera G., Vogel J., Gottesman S., Storz G., Trans-acting small RNAs and their effects on gene expression in Escherichia coli and Salmonella enterica. Ecosal Plus 9, 10.1128/ecosalplus.ESP-0030-2019 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ul Haq I., Müller P., Brantl S., Intermolecular communication in Bacillus subtilis: RNA-RNA, RNA-protein and small protein-protein interactions. Front. Mol. Biosci. 7, 178 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nielsen J. S., et al., Defining a role for hfq in gram-positive bacteria: Evidence for hfq-dependent antisense regulation in Listeria monocytogenes. Nucleic Acids Res. 38, 907–919 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Christiansen J. K., et al., Identification of small Hfq-binding RNAs in Listeria monocytogenes. RNA 12, 1383–1396 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nawrocki K. L., Wetzel D., Jones J. B., Woods E. C., McBride S. M., Ethanolamine is a valuable nutrient source that impacts Clostridium difficile pathogenesis. Environ. Microbiol. 20, 1419–1435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bohn C., et al., Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism. Nucleic Acids Res. 38, 6620–6636 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rochat T., et al., The conserved regulatory RNA RsaE down-regulates the arginine degradation pathway in Staphylococcus aureus. Nucleic Acids Res. 46, 8803–8816 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schoenfelder S. M. K., et al., The small non-coding RNA RsaE influences extracellular matrix composition in Staphylococcus epidermidis biofilm communities. PLoS Pathog. 15, e1007618 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pitman S., Cho K. H., The mechanisms of virulence regulation by small noncoding RNAs in low GC gram-positive pathogens. Int. J. Mol. Sci. 16, 29797–29814 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bertin Y., et al., Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ. Microbiol. 13, 365–377 (2011). [DOI] [PubMed] [Google Scholar]
- 78.Kaval K. G., et al., Loss of ethanolamine utilization in Enterococcus faecalis increases gastrointestinal tract colonization. mBio 9, e00790-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thiennimitr P., et al., Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl. Acad. Sci. U.S.A. 108, 17480–17485 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.DebRoy S., et al., Riboswitches. A riboswitch-containing sRNA controls gene expression by sequestration of a response regulator. Science 345, 937–940 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mellin J. R., et al., Riboswitches. Sequestration of a two-component response regulator by a riboswitch-regulated noncoding RNA. Science 345, 940–943 (2014). [DOI] [PubMed] [Google Scholar]
- 82.Del Papa M. F., Perego M., Ethanolamine activates a sensor histidine kinase regulating its utilization in Enterococcus faecalis. J. Bacteriol. 190, 7147–7156 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-sequencing data are available at the National Center for Biotechnology Information Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo) under the accession number GSE155167. Plasmids pKJAK112, pJAK184, and pJAK080 have been deposited with Addgene (167279–167281).