Significance
The pituitary is our master endocrine gland. Local damage and aging present important threats. We started to decrypt the ill-defined regulation of the gland’s stem cells, typically dormant but acutely activated upon damage. Single-cell transcriptomics uncovered interleukin-6 as a pituitary stem cell activator upon local damage, corroborated in vivo and in vitro using stem cell–derived organoids. This competence extinguishes at aging, concurrent with a raised inflammatory state in the older gland (inflammaging). However, the aging pituitary’s stem cells retain intrinsic activatability, resurfacing once released from their impeding tissue milieu. Our insights may instigate tactics to refrain the pituitary from aging or rejuvenate the aging gland. The single-cell transcriptomic database provides a powerful resource to decipher pituitary damage and aging.
Keywords: pituitary, stem cells, interleukin-6, aging, organoids
Abstract
Stem cells in the adult pituitary are quiescent yet show acute activation upon tissue injury. The molecular mechanisms underlying this reaction are completely unknown. We applied single-cell transcriptomics to start unraveling the acute pituitary stem cell activation process as occurring upon targeted endocrine cell–ablation damage. This stem cell reaction was contrasted with the aging (middle-aged) pituitary, known to have lost damage-repair capacity. Stem cells in the aging pituitary show regressed proliferative activation upon injury and diminished in vitro organoid formation. Single-cell RNA sequencing uncovered interleukin-6 (IL-6) as being up-regulated upon damage, however only in young but not aging pituitary. Administering IL-6 to young mice promptly triggered pituitary stem cell proliferation, while blocking IL-6 or associated signaling pathways inhibited such reaction to damage. By contrast, IL-6 did not generate a pituitary stem cell activation response in aging mice, coinciding with elevated basal IL-6 levels and raised inflammatory state in the aging gland (inflammaging). Intriguingly, in vitro stem cell activation by IL-6 was discerned in organoid culture not only from young but also from aging pituitary, indicating that the aging gland’s stem cells retain intrinsic activatability in vivo, likely impeded by the prevailing inflammatory tissue milieu. Importantly, IL-6 supplementation strongly enhanced the growth capability of pituitary stem cell organoids, thereby expanding their potential as an experimental model. Our study identifies IL-6 as a pituitary stem cell activator upon local damage, a competence quenched at aging, concomitant with raised IL-6/inflammatory levels in the older gland. These insights may open the way to interfering with pituitary aging.
The pituitary gland is the key orchestrator of the endocrine system, translating central and peripheral inputs into strictly regulated hormone outputs. Consequently, this master gland steers a multitude of fundamental processes including body growth, metabolism, sexual development, reproduction, and stress management. To exert this crucial function, the pituitary encompasses specialized hormone-producing cell types, mainly located in the anterior lobe of the gland (anterior pituitary [AP]) and comprising somatotropes (producing growth hormone [GH]), lactotropes (prolactin [PRL]), corticotropes (adrenocorticotropic hormone [ACTH]), gonadotropes (luteinizing hormone [LH] and/or follicle-stimulating hormone [FSH]), and thyrotropes (thyroid-stimulating hormone [TSH]). On top, the pituitary contains a population of stem cells, essentially marked by sex-determining region Y-box 2 (SOX2) expression (1–4). The physiological role of the pituitary stem cells remains poorly understood (5, 6). Transgenic SOX2 lineage tracing revealed involvement in adult gland homeostasis and adaptation to altered endocrine demands, although the contributions were not large (3, 4). Progressively, the picture emerges that adult pituitary stem cells are mainly dormant and not predominantly involved in tissue homeostasis and physiological remodeling (7–9), reminiscent of findings in other tissues with comparable low turnover rates [such as muscle and liver (10, 11)]. Yet, in the case of damage in the adult pituitary, the resident stem cell compartment shows swift activation (12, 13). More in particular, following diphtheria toxin (DT)-triggered endocrine cell–ablation injury in the GHCre/iDTR mouse model (expressing the Cre recombinase under control of the Gh promoter, as well as the Cre-inducible DT receptor [iDTR)], the pituitary stem cell population is promptly activated, displaying enhanced proliferative activity and expansion (12). Substantial regeneration of the obliterated somatotropes is eventually observed after 5 to 6 mo (12). Surprisingly, this regenerative competence of the gland rapidly drops with aging, because it is not observed anymore when mice reach middle age [from 8 to 10 mo of age (14)].
Virtually nothing is known about the molecular machinery driving the quiescent pituitary stem cells into activation (as observed following local injury) or about how this route may change at advancing age. In other tissues, it was found that stem cells undergo an intrinsic aging process [such as in the hematopoietic system (15)] or regress in functionality because of the aging tissue milieu [as described in muscle and brain (16–18)]. Here, we started to tackle this query by interrogating young adult and aging (middle aged), damaged and undamaged pituitary (specifically, its major endocrine AP lobe) using single-cell transcriptomics. We focused on the prompt stem cell reaction that occurs immediately upon the DT-induced local damage in the GHCre/iDTR model and substantiated transcriptomic findings using in vivo and in vitro exploration. To achieve the latter, we applied our recently developed mouse AP-derived organoid model (19). These organoids originate from the (SOX2+) stem cells and maintain the pituitary stem cell phenotype in culture. Moreover, growth characteristics reflect the stem cells’ activation (as observed following damage and at neonatal age), and organoid-based findings were found reliably translatable to the in vivo situation (19). Hence, this organoid system provides an interesting in vitro pituitary stem cell biology/activation readout tool.
In the present study, we identified interleukin-6 (IL-6) to be up-regulated in the young pituitary following tissue injury and uncovered its pituitary stem cell–activating competence, which, however, is quenched at aging concomitant with a raised inflammatory nature in the older gland. When released from the in situ microenvironment through organoid culturing, the aging pituitary’s stem cells regain activatability. These insights may be harnessed to combat pituitary aging and concomitant regenerative decline.
Results
Acute Proliferative Activation of the Pituitary Stem Cells upon Local Injury Subsides at Aging.
To controllably inflict injury in the pituitary, we used our previously designed GHCre/iDTR transgenic mouse model (12) (SI Appendix, Extended Methods). GHCre/+;R26iDTR/+ mice (further referred to as GHCre/iDTR) and control GH+/+;R26iDTR/+ animals (further referred to as -/iDTR) were injected with DT for 3 d, causing local tissue damage in the GHCre/iDTR pituitary by inducing apoptosis in endocrine cells [in particular, somatotropes and lactotropes (12)]. In accord with our previous findings (12), the resident SOX2+ stem cells of the young adult pituitary (8- to 12-wk-old mice, further referred to as “young”) show an acute increase in proliferative activity (as assessed by Ki67 immunostaining; Fig. 1A) and ensuing expansion of SOX2+ cell number (SI Appendix, Fig. S1A). This prompt pituitary stem cell activation is significantly lower at older age (10- to 15-mo-old, middle-aged mice, further referred to as “aging”) (Fig. 1A and SI Appendix, Fig. S1A). In addition, the aging basal pituitary houses a reduced number of SOX2+ stem cells when compared to the young gland (SI Appendix, Fig. S1A), consistent with prior findings (14).
Fig. 1.
Pituitary stem cell activation following tissue injury subsides at aging. (A) Immunofluorescence staining of SOX2 (magenta) and Ki67 (green) in basal (undamaged) and damaged AP-derived cytospin samples of young and aging mice. Nuclei are labeled with Hoechst33342 (blue). The arrowheads indicate double-immunopositive cells (Scale bar, 50 μm). The bar graphs show the proportion of SOX2+Ki67+ cells in SOX2+ cells (mean ± SEM) and the fold change in absolute SOX2+Ki67+ cell number (mean ± SEM) after damage (i.e., relative to undamaged AP, set as 1 [dashed line]) (for calculation of absolute cell numbers, see SI Appendix, Extended Methods). The data points represent biological replicates. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001. (B) Organoid formation efficiency from undamaged and damaged, young, and aging AP. Representative bright-field pictures of organoid cultures are shown (passage 0, P0). (Scale bar, 500 μm.) The bar graphs indicate number of organoids developed and fold change in organoid number after pituitary damage (relative to undamaged AP) (mean ± SEM). The data points represent biological replicates. *P ≤ 0.05; **P ≤ 0.01. (C) Percentage of organoid-initiating SOX2+ cells per well of 10,000 seeded AP cells from the conditions as indicated. The bar graphs show mean ± SEM, and the data points represent biological replicates. *P ≤ 0.05.
Recently, we established an in vitro organoid model starting from mouse pituitary (particularly, from the AP), which recapitulates biology and activation of the pituitary stem cells (19). The organoids develop from the SOX2+ stem cells as shown before for young AP (19) and demonstrated here for aging pituitary using SOX2eGFP/+ reporter-mouse AP giving rise to only enhanced green fluorescent protein (eGFP)+ organoids (SI Appendix, Fig. S1B). In addition, when starting from a mixture of cells from SOX2eGFP/+ and nonfluorescent wild-type (WT) aging AP, the developing organoids are either entirely fluorescent or nonfluorescent (SI Appendix, Fig. S1B), thereby supporting a clonal origin. Moreover, the organoids display a pituitary stemness phenotype in culture, both from young AP (see ref. 19) and from aging gland (SI Appendix, Fig. S1C), expressing known pituitary stem cell markers [SOX2, E-cadherin, and cytokeratin-8/18 (19)] and showing absence of hormone-secretory granules and presence of microvilli, similar to the pituitary stem cells as present in the cleft-lining marginal zone (MZ) (SI Appendix, Fig. S1D). Using this model system as pituitary stem cell biology and activation readout (19), we observed that primary organoid formation efficiency (referred to as passage 0 or P0) from the aging pituitary is lower than from the young gland and that the increase in organoid development capacity upon damage is less pronounced at the older age (Fig. 1B), both findings in line with the in vivo observations of regressed stem cell number and inferior activation response in aging versus young pituitary. Because the endocrine cell ablation in the damaged pituitary, together with the expansion of the SOX2+ cell population, entails that a higher absolute number of SOX2+ stem cells is seeded per well (i.e., per 10,000 AP cells) from damaged than from undamaged gland, we determined the number of organoid-initiating SOX2+ stem cells by normalizing for the calculated SOX2+ cell numbers seeded (SI Appendix, Extended Methods). Similar conclusions were reached, showing an increase upon injury at both ages (indicative of stem cell activation) but again less prominent at older age (Fig. 1C).
Single-Cell Transcriptomics Uncovers IL-6 as Up-Regulated in Young Pituitary upon Damage.
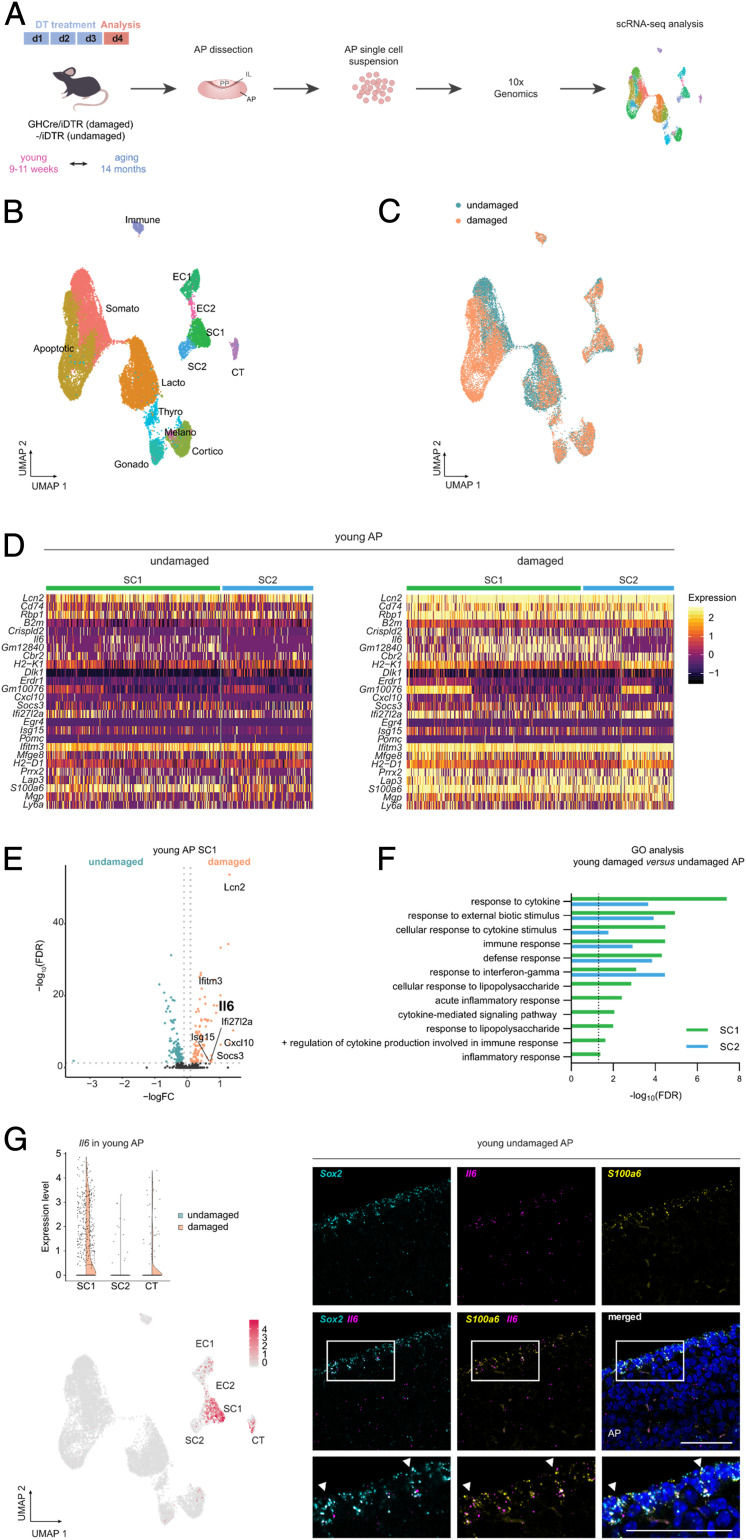
To search in detail for molecular underpinnings of the damage-induced pituitary stem cell response and of the subsided reaction in the aging gland, we applied single-cell transcriptomics to the different pituitary conditions (i.e., young and aging and damaged and undamaged AP; Fig. 2A). After filtering out dead and low-quality cells, potential doublets, and “background” (ambient) RNA (SI Appendix, Fig. S2A and Extended Methods), applied collectively on all single-cell RNA sequencing (scRNA-seq) data obtained in this study (i.e., from young and aging and damaged and undamaged AP, in total yielding 26,115 good-quality cells), unsupervised clustering and visualization using Uniform Manifold Approximation and Projection [UMAP (20)] were performed (SI Appendix, Fig. S2B). Subsequent superposition of canonical lineage markers exposed all known pituitary hormone–producing cell populations (Fig. 2B, Dataset S1, and SI Appendix, Fig. S2B). In addition, a connective tissue cluster (annotated in analogy to ref. 21 and indicated with CT) and immune cell cluster were distinguished, as well as an endothelial and stem cell population, both subdivided in two subclusters (Fig. 2B). The endothelial cell subcluster 1 (EC1) shows more expression of mature endothelial cell markers than EC2 (Dataset S1 and SI Appendix, Fig. S2C), and a first basic mining of the stem cell population revealed that the subclusters (referred to as SC1 and SC2) differ in expression levels of several (pituitary) stemness markers (Dataset S1 and SI Appendix, Fig. S2D). Finally, a population of dying (apoptotic) cells was identified, being the result of the DT-induced apoptotic process in the GHCre/iDTR (damaged) AP, also included in the unsupervised clustering of the aggregate data (Fig. 2 A–C). Of note, a small cluster of melanotrope cells (housed in the intermediate lobe [IL] of the pituitary) was also observed (Fig. 2B and SI Appendix, Fig. S2B), likely representing some limited IL tissue still attached to the AP after the latter’s isolation from the mouse. Our cell-type categorization outcome, as described above, was validated by performing the clustering based on regulon activity (i.e., transcription factors taken together with their positively regulated target genes) instead of based on differential gene expression (as in Fig. 2B), by applying “single-cell regulatory network inference” [SCENIC (22)]. This alternative approach resulted in an analogous cell-type categorization pattern (SI Appendix, Fig. S2E). Moreover, integrating recently published pituitary scRNA-seq data of comparable mouse age, gender, and strain [i.e., young WT C57/Bl6 male (21, 23)] with our equivalent dataset showed prominent correlation (SI Appendix, Fig. S2F).
Fig. 2.
Single-cell transcriptomic profiling reveals up-regulation of IL-6 in young pituitary following tissue injury. (A) Experimental schematic for the scRNA-seq analysis. DT, diphtheria toxin; AP, anterior pituitary; IL, intermediate lobe; PP, posterior pituitary. The mouse icon was obtained from http://freepik.com. (B) UMAP plot of the annotated cell clusters in the aggregate AP samples (i.e., collective single-cell transcriptome datasets from young and aging, undamaged and damaged AP). Somato, somatotropes; Lacto, lactotropes; Cortico, corticotropes; Gonado, gonadotropes; Thyro, thyrotropes; Melano, melanotropes; SC1 and SC2, stem cell cluster 1 and 2; EC1 and EC2, endothelial cell cluster 1 and 2; Immune, immune cells; CT, connective tissue cells; Apoptotic, apoptotic cells. (C) UMAP plot of undamaged and damaged AP (young and aging combined). (D) Heat maps displaying the scaled expression of inflammatory response genes in the stem cell clusters SC1 and SC2 of young undamaged and damaged AP. (E) Volcano plot displaying DEGs in SC1 of young AP. The colored dots represent significantly up- (orange) and down- (green) regulated genes in damaged versus undamaged AP. A selection of genes (as mentioned in the section Single-Cell Transcriptomics Uncovers IL-6 as Up-Regulated in Young Pituitary upon Damage) is indicated, and Il6 is highlighted. (F) Significant (false discovery rate [FDR] ≤ 0.05) DEG-associated GO terms linked with inflammatory processes enriched in SC1 and SC2 of young damaged versus undamaged AP. (G) Violin plot (Top) and projection on UMAP diagram (Bottom) of Il6 expression in young undamaged and damaged AP with indication of relevant cell clusters. Triple RNAscope in situ hybridization analysis of young (undamaged) AP for Sox2 (cyan), Il6 (magenta), and S100a6 (yellow). The boxed areas are magnified. The nuclei are stained with DAPI (blue). The arrowheads indicate cells with colocalized expression. (Scale bar, 50 μm.)
Looking deeper into the transcriptomic data of the stem cell population (Dataset S1), we detected, in addition to the well-known pituitary stem cell markers [Sox2, Sox9, Cdh1, Krt8, and Krt18; SI Appendix, Fig. S2D (19)], a number of interesting genes which we validated by in situ immunostaining analysis. Tacstd2 (alias Trop2), found in stem cells of certain other tissues (24, 25), is within the AP stem cell population particularly expressed in SC1 (Dataset S1 and SI Appendix, Fig. S2G). In situ, TACSTD2/TROP2 protein expression was observed in the cleft-lining MZ stem cell compartment where it coincides with SOX2 (SI Appendix, Fig. S2G). Interestingly, TACSTD2 was not detected in the SOX2+ cell groups in the AP parenchyma (SI Appendix, Fig. S2G), thereby providing an appealing marker to distinguish MZ from parenchymal stem cells (1, 2, 7). Of note, Tacstd2 is also observed in the corticotrope cell cluster (Dataset S1 and SI Appendix, Fig. S2G). In agreement, TACSTD2 protein was detected in certain ACTH+ cells, mainly located at the transition area between AP and IL [the so-called “wedges” (26)] (SI Appendix, Fig. S2G). Furthermore, the core Hippo pathway component Yap1 was found highly expressed in the stem cell population (both SC1 and SC2; Dataset S1 and SI Appendix, Fig. S2H). In analogy, nuclear YAP+ signal is present in SOX2+ stem cells (both in the MZ and parenchyma) (SI Appendix, Fig. S2H), thereby expanding our previous findings (14) and confirming former studies that identified Hippo pathway activity in pituitary stem cells as particularly studied during embryonic and neonatal development (27, 28). Yap1 expression is also seen in the connective tissue cluster and in the endothelial cell clusters (Dataset S1 and SI Appendix, Fig. S2H), which is consistent with YAP reported in pituitary endothelial cells (27). Finally, our scRNA-seq exploration revealed high and specific expression of Cyp2f2 in the pituitary stem cell population (both SC1 and SC2; Dataset S1 and SI Appendix, Fig. S2I), in agreement with another recent pituitary scRNA-seq study (21). Here, we validated this expression and found that CYP2F2 is indeed localized in SOX2+ stem cells (SI Appendix, Fig. S2I).
As described above, the pituitary stem cell population acutely reacts to local tissue damage, predominantly in the young gland. To search for molecular mechanisms underlying this acute injury response, we contrasted the stem cell transcriptomes of young, damaged AP with undamaged gland using differentially expressed gene (DEG) and gene ontology (GO) analyses. Among the top DEGs, we found multiple inflammatory-related genes (e.g., Cxcl10, Ifi27l2a, Ifitm3, Il6, Lcn2, and Socs3) that were up-regulated following injury (Fig. 2 D and E and Dataset S2). Accordingly, cytokine-/inflammatory response–related GO terms were enriched in the stem cell clusters upon damage (Fig. 2F and Dataset S3). Interestingly, within this context, the cytokine Il6 was found highly up-regulated, particularly in subcluster SC1 (Fig. 2 D, E, and G and Dataset S2). Il6 up-regulation was also readily detected by RT-qPCR analysis in damaged versus undamaged AP (SI Appendix, Fig. S2J). Further scRNA-seq scrutiny showed that Il6 expression is not only present in SC1 but also in the connective tissue cluster in which it also rises upon damage (Fig. 2G and Dataset S2). To validate the scRNA-seq expression pattern, we performed RNAscope in situ hybridization for Il6, Sox2, and S100a6, a gene highly expressed in both stem cell and connective tissue clusters (Dataset S1 and SI Appendix, Fig. S2K). This in situ examination showed cellular overlap of the messenger RNA signals (Fig. 2G). Both stem cell and connective (supportive) tissue cells belong to the so-called folliculostellate (FS) cell group of the pituitary, a heterogeneous cell population in the past designated as local IL-6 source, and in rat marked by S100-β (29–31). In analogy, S100A6 immunoreactivity is found in SOX2+ stem cells of mouse pituitary as well as in some non-SOX2+ cells (SI Appendix, Fig. S2K). Finally, Il6 gene expression is also detected in the endothelial cell population by scRNA-seq mining (Fig. 2G and Dataset S1), in situ also supported by RNAscope analysis showing Il6 signal in a number of cells expressing the endothelial cell–specific plasmalemma vesicle–associated protein (Plvap) (SI Appendix, Fig. S2L).
IL-6 Acts as a Pituitary Stem Cell–Activating Factor at Young Age.
Since IL-6 has been shown to activate stem cells in certain other tissues when up-regulated upon local damage [such as in muscle and intestine; (32–34)], we addressed the question whether the cytokine may also act as pituitary stem cell–activating factor.
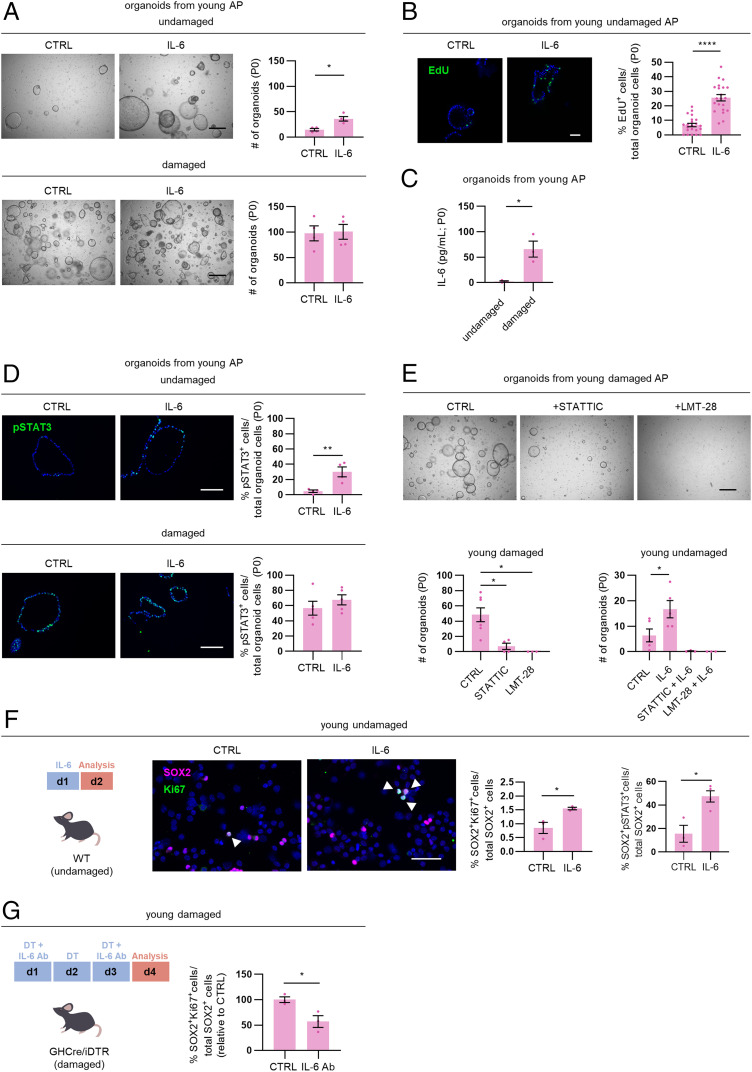
Adding IL-6 to organoid culture augments organoid formation efficiency from undamaged (young) AP (Fig. 3A), concomitant with proliferative activation of the organoid-driving stem cells (as analyzed by EdU incorporation; Fig. 3B). In contrast, IL-6 does not further enhance organoid formation from the damaged (young) gland (Fig. 3A) in which the stem cells are already activated and endogenous IL-6 levels elevated (see Fig. 1 and 2) and as observed in the supernatant of starting organoid cultures (P0, day 3 of culture) (Fig. 3C).
Fig. 3.
IL-6 acts as pituitary stem cell–activating factor at young age. (A) Organoid development from undamaged and damaged young AP in the absence (control, CTRL) or presence of IL-6 (P0). (Scale bar, 500 µm.) The bar graphs show number of organoids formed (mean ± SEM). The data points represent biological replicates. *P ≤ 0.05. (B) Proliferative activity of organoids grown in the absence (CTRL) or presence of IL-6 (P0), as assessed by EdU incorporation (green). Nuclei are stained with Hoechst33342 (blue). (Scale bar, 50 µm.) The bar graph shows percentage of EdU+ cells in organoids (mean ± SEM). The data points represent individual organoids from three biological replicates. ****P ≤ 0.0001. (C) IL-6 protein levels in supernatant of organoid cultures (P0, day 3 of culture) from young undamaged and damaged AP. The bar graph shows mean ± SEM, and the data points represent biological replicates. *P ≤ 0.05. (D) Immunofluorescence staining of pSTAT3 (green) in young AP organoids grown in the absence (CTRL) or presence of IL-6 (P0). Nuclei are stained with Hoechst33342 (blue). (Scale bar, 100 µm.) The bar graphs show percentage of pSTAT3+ cells in organoids (mean ± SEM). The data points represent biological replicates. **P ≤ 0.01. (E) Organoid development from young, damaged AP treated as indicated (P0; Scale bar, 500 µm). The bar graphs show number of organoids formed (mean ± SEM) from young damaged and undamaged AP under conditions as indicated. The data points represent biological replicates. *P ≤ 0.05. (F) Time schedule of in vivo treatment with IL-6. Immunofluorescence staining of SOX2 (magenta) and Ki67 (green) in basal (undamaged) AP-derived cytospin samples of young WT mice, in vivo injected with IL-6 or vehicle (CTRL). Nuclei are labeled with Hoechst33342 (blue). The arrowheads indicate double-immunopositive cells (Scale bar, 50 μm). The bar graphs show percentage of SOX2+Ki67+ or SOX2+pSTAT3+ cells in SOX2+ cells (mean ± SEM), and the data points represent biological replicates. *P ≤ 0.05. (G) Time schedule of in vivo treatment with anti–IL-6 antibody (IL-6 Ab). The bar graph shows percent change (relative to CTRL with mean set to 100%) of SOX2+Ki67+ cells in SOX2+ cells (determined using cytospin samples) in the AP of young mice subjected to DT-induced damage infliction and simultaneously treated with IL-6 Ab (mean ± SEM). The data points represent biological replicates. *P ≤ 0.05.
The JAK/STAT pathway is a key downstream mediator of IL-6 signaling (35). IL-6 indeed augments the number of phosphorylated STAT3 (phospho-STAT3 or pSTAT3)-immunopositive cells in organoids from undamaged (young) AP but not from damaged gland in which the pSTAT3+ status is already high without IL-6 (Fig. 3D), advocating that the JAK/STAT pathway is activated in stem cells following the DT-induced tissue injury, as also supported by intense Stat3 regulon activity in the damaged AP SC1 (SI Appendix, Fig. S3A). Adding the STAT3 inhibitor STATTIC to AP cells from damaged gland largely blocks organoid formation, indicating the importance of JAK/STAT signaling in this stem cell–driven process (Fig. 3E). Furthermore, supplementation of LMT-28, an antagonist of the IL-6 coreceptor gp130 (36), abolishes organoid formation (Fig. 3E). Along the same line, adding STATTIC and LMT-28 to undamaged AP cells counteracts the formation of organoids, with the stimulatory effect of IL-6 no longer being observed (Fig. 3E). Exposure of fully grown organoids to LMT-28 results in a decrease in pSTAT3+ cells and proliferative activity and an increase in apoptosis (as analyzed by cleaved caspase 3 immunostaining; SI Appendix, Fig. S3B), supporting that gp130/pSTAT3-mediated signaling is needed for stem cell proliferation and survival in the organoids. Taken together, organoid readout scrutiny indicates that IL-6 can act as pituitary stem cell activator and reveals the importance of the IL-6–associated JAK/STAT and gp130 signaling pathways in pituitary stem cell behavior.
To inspect whether IL-6 acts similarly in vivo, young WT mice were intraperitoneally (i.p.) injected with the cytokine, and the effect on pituitary stem cell–proliferative activity was analyzed. The proportion of proliferating SOX2+ stem cells is significantly elevated following IL-6 treatment, concomitant with an increase in pSTAT3+ cells in the SOX2+ cell population (Fig. 3F), together convincingly extrapolating the in vitro organoid-based findings to in vivo. Moreover, IL-6 injection generated a pituitary stem cell activation response in IL-6 knockout (KO; IL-6−/−) mice, whereas a general inflammatory condition [as induced by CpG oligodeoxynucleotide (CpG) injection (37)] did not (SI Appendix, Fig. S3C), demonstrating the specificity of the effect by IL-6 (independent of a general inflammatory reaction if any). Of note, CpG-induced inflammation in WT mice triggers a pituitary stem cell–proliferative reaction comparable to IL-6 (SI Appendix, Fig. S3C), in line with the up-regulated systemic IL-6 levels as reported to occur in this model (37). Intriguingly, the SOX2+ stem cell population is not different in the IL-6 KO pituitary regarding number and quiescent (low proliferative) status (SI Appendix, Fig. S3D). However, primary organoid formation from IL-6 KO AP is reduced (versus WT AP), and IL-6 KO organoids are not passageable (SI Appendix, Fig. S3E), indicating that endogenous IL-6 is important for these stem cell activities.
Finally, to determine whether IL-6, being up-regulated in the pituitary after damage, is involved in the injury-induced stem cell activation, we applied anti–IL-6 antibody during the acute DT-triggered damage infliction in GHCre/iDTR mice (Fig. 3G). The stem cell–proliferative reaction is significantly reduced (Fig. 3G), coinciding with lowered pSTAT3+ cells in the AP (SI Appendix, Fig. S3F). Similarly, in vivo LMT-28 administration during damage infliction reduces the proportion of proliferating stem cells and pSTAT3+ cells in the AP (SI Appendix, Fig. S3G).
Taken all together, our data show that IL-6 is up-regulated in young pituitary upon tissue damage and can act as pituitary stem cell–activating factor. In addition, they provide evidence that IL-6 is involved in the early stem cell activation reaction to injury.
IL-6 Does Not Activate Stem Cells in the Aging Pituitary, Which Is Typified by an Elevated IL-6/Inflammatory Phenotype.
In clear contrast to the observations in young mice, i.p. injection of IL-6 in aging animals does not trigger a stem cell–proliferative activation response (Fig. 4A). Intriguingly, basal Il6 expression level was found higher in aging versus young (undamaged) AP (Fig. 4B), in particular in the stem cell subcluster SC1 (Fig. 4C and Dataset S4). DEG and GO analysis of the stem cell transcriptomes identified up-regulation of cytokine-/inflammatory response–related terms and genes in the aging versus young (undamaged) pituitary SC1 (Fig. 4 C–E and Datasets S3 and S4) and even more broadly in the whole AP (Fig. 4E and Datasets S3 and S4). In analogy, gene set enrichment analysis applied to the single-cell transcriptomic dataset revealed a striking enrichment of the “inflammatory response” hallmark in aging versus young (undamaged) AP and in particular also in its SC1 (Fig. 4F). Also, the “IL-6/JAK/STAT3 signaling” hallmark is significantly enriched in aging versus young AP and stem cell population (Fig. 4F). In accordance, pSTAT3+ cells are more abundant in the aging pituitary (SI Appendix, Fig. S4A). Together, these findings indicate that the aging pituitary, including its stem cells, displays a basally higher IL-6/inflammatory status than the young gland, which may explain the absence of a stem cell activation reaction in the older AP upon IL-6 administration (Fig. 4A) and the inferior stem cell reaction to injury (Fig. 1A). In support, injection of IL-6 does not further elevate the number of pSTAT3+ cells in the aging gland (SI Appendix, Fig. S4B). Moreover, inflicting pituitary damage in aging mice does not significantly increase Il6 expression levels any further in the gland (SI Appendix, Fig. S4C) or its SC1 and connective tissue cluster (Dataset S5 and SI Appendix, Fig. S4D). A raised inflammatory nature at aging has also been found to occur in other organs, epitomized in the concept of inflammaging, which states that a chronic low-grade inflammation gradually develops at progressing age, not only at the systemic but also organ level (38, 39), proposed to underlie deteriorating organ and stem cell functionality during aging (40–43). Regarding systemic signs of inflammaging, increased IL-6 level is the most clear and supported marker (38, 39). In agreement, we found significantly up-regulated IL-6 plasma levels in aging mice when compared to young animals (Fig. 4G). Numbers of immune cells in the pituitary, encompassing resident, and infiltrated cells are not altered in the aging animals (SI Appendix, Fig. S4E), similar to findings in the spleen (SI Appendix, Fig. S4F), thereby suggesting that the inflammaging process may still be subtle in the middle-aged (∼1-y-old) animals when compared to elderly mice (∼2 y old) in which macrophage infiltration has been reported in certain organs (44, 45). Along the same line, plasma levels of (pro)inflammatory cytokines other than IL-6, of which specifically TNF-α has been reported to be up-regulated in elderly mice in some studies (38, 39), are not significantly changed (yet) in the middle-aged mice analyzed here (SI Appendix, Fig. S4G).
Fig. 4.
IL-6 does not activate stem cells in aging pituitary, which displays an elevated IL-6/inflammatory phenotype. (A) Time schedule of in vivo treatment with IL-6. Immunofluorescence staining of SOX2 (magenta) and Ki67 (green) in basal (undamaged) AP-derived cytospin samples of aging WT mice, in vivo injected with IL-6 or vehicle (CTRL). Nuclei are labeled with Hoechst33342 (blue). The arrowheads indicate double-immunopositive cells. (Scale bar, 50 μm.) The bar graph shows percentage of SOX2+Ki67+ cells in SOX2+ cells (mean ± SEM), and the data points represent biological replicates. (B) Il6 gene expression in aging versus young basal (undamaged) AP as determined by RT-qPCR. The bar graph shows fold change in aging AP relative to young (mean ± SEM), and the data points represent biological replicates. **P ≤ 0.01. (C) Volcano plot displaying DEGs in SC1 in young and aging undamaged AP. The colored dots represent significantly up- (blue) and down- (pink) regulated genes in aging versus young AP. A selection of genes is indicated, and Il6 is highlighted. (D) Significant (FDR ≤ 0.05) DEG-associated GO terms linked with inflammatory processes enriched in the stem cell cluster SC1 of aging versus young (undamaged) AP. (E) Heat map displaying the fold change (presented as −logFC) of inflammatory response genes up- (blue) and down- (pink) regulated at aging. (F) Gene set enrichment analysis of “inflammatory response” and “IL-6/JAK/STAT3” hallmarks in aging compared to young (undamaged) AP and its SC1 and SC2, visualized as normalized enrichment score (NES). **FDR ≤ 0.01; ****FDR ≤ 0.0001; ns, nonsignificant. (G) Systemic plasma levels of IL-6 in young and aging WT (undamaged) mice. The bar graph shows mean ± SEM, and the data points represent biological replicates. *P ≤ 0.05.
Taken together, our findings provide evidence that aging pituitary displays a raised IL-6/inflammatory phenotype, which may underlie the declined stem cell activation upon injury or IL-6 exposure at aging.
Activatability of Aging Pituitary Stem Cells Resurfaces in Organoid Culture.
Unexpectedly, in contrast to the absence of a stem cell–activating effect by IL-6 in the aging gland in vivo (Fig. 4A), we observed that IL-6 is able to increase the formation and proliferative activity of organoids from aging (undamaged) pituitary in vitro (Fig. 5 A and B), concomitant with an elevation of pSTAT3+ cells (SI Appendix, Fig. S5A). We hypothesized that the elevated inflammatory status may swiftly disappear in culture when the stem cells are released from their old in vivo (micro)environment. In support, as opposed to the up-regulated IL-6/inflammatory response genes and hallmarks in aging pituitary and its stem cell clusters (Fig. 4 E and F and Datasets S3 and S4), expression of Il6 and inflammatory response genes is not different anymore between aging and young pituitary stem cells once cultured in vitro in organoid conditions (analyzed at day 14 of P0 organoid culture; Fig. 5C).
Fig. 5.
Aging pituitary’s stem cells regain activatability to IL-6 in organoid culture. (A) Organoid development from undamaged and damaged aging AP in the absence (CTRL) or presence of IL-6 (P0). (Scale bar, 500 µm.) The bar graphs show number of organoids formed (mean ± SEM). The data points represent biological replicates. *P ≤ 0.05. (B) Proliferative activity of organoids grown in the absence (CTRL) or presence of IL-6 (P0), as assessed by EdU incorporation (green). Nuclei are stained with Hoechst33342 (blue). (Scale bar, 50 µm.) The bar graph shows percentage of EdU+ cells in organoids (mean ± SEM). The data points represent individual organoids from three biological replicates. **P ≤ 0.01. (C) Expression levels of Il6/inflammatory response genes in organoid culture (P0, day 14) from aging and young (undamaged) AP as determined by RT-qPCR (mean ± SEM). The data points represent biological replicates. (D) Bar graphs showing the (maximum) number of AP organoid passage at present reached, grown in the absence (CTRL) or presence of IL-6 (mean ± SEM). The data points represent biological replicates. *P ≤ 0.05; **P ≤ 0.01; ****P ≤ 0.0001. (E) Il6 gene expression levels in organoids from young and aging (damaged) AP at consecutive passages (each at day 14 of culture) as determined by RT-qPCR. The bars show mean ± SEM, and the data points represent biological replicates. *P ≤ 0.05; **P ≤ 0.01.
Finally, in view of its pituitary stem cell–activating competence and importance for organoid culture as found in IL-6 KO conditions, we tested whether addition of IL-6 to organoid cultures prolongs their yet-limited expandability (19). Indeed, administration of IL-6 significantly increased organoid passageability from both young and aging, damaged and undamaged pituitary (Fig. 5D). Endogenous IL-6 expression and production was found to substantially decline during organoid culturing in subsequent passages (Fig. 5E and SI Appendix, Fig. S5B), plausibly underlying the before-limited expandability in the absence of exogenous IL-6 supplementation. After long-term passaging, organoids maintain their morphological and pituitary stem cell phenotype (as shown for damaged AP; SI Appendix, Fig. S5C).
Taken together, the aging pituitary’s stem cells retain intrinsic activation capability, which resurfaces in vitro when liberated from the plausibly impeding IL-6/inflammatory stress in vivo. Moreover, achieving robust long-term expansion empowers the applicability of the organoid model system toward extensive exploration of pituitary stem cell biology and activation.
Discussion
In the present study, we searched for molecular mechanisms underlying the acute activation of adult pituitary stem cells upon local tissue injury, at present entirely unknown, and looked for differences with the aging gland, reported before to have lost damage-repair capacity (14). By applying single-cell transcriptomic profiling, we tracked down IL-6 as a factor that has the capacity to bring pituitary stem cells into activation mode. Back in 1989, we made the intriguing observation that a cytokine known for expression and function in the immune system (i.e., IL-6) was also expressed in an endocrine organ [i.e., the pituitary gland (30, 46)]. IL-6 was found to be produced by the so-called FS cell population, which represents a yet ill-defined, heterogeneous cell-type assembly in the pituitary, proposed to encompass paracrine-regulatory cells, physically supportive (connective tissue) cells, immune-associated cells, and more recently, stem cells (1, 2, 29, 31). Now, 30 y later, our scRNA-seq interrogation eventually confirmed and refined the pituitary IL-6 source. The up-regulation of IL-6 upon injury in the young gland, occurring particularly in the stem cell and connective tissue subsets, proposes a role, paracrine and/or autocrine, for these specific cell subpopulations in the injury-triggered stem cell activation (summarized in SI Appendix, Fig. S5D). In vitro, IL-6 was found to activate the pituitary stem cells, resulting in more efficient organoid development, a recently developed tool to probe pituitary stem cell biology and activation (19). In vivo, IL-6 triggered acute pituitary stem cell activation in the young gland, while blockade of IL-6 or associated signaling pathways strongly reduced the stem cell reaction at injury, together providing evidence that IL-6 is involved in the acute activation process of the quiescent pituitary stem cells in response to local tissue damage. Of note, IL-6 does not seem to be involved in the stem cell phenotype of the homeostatic gland, which is not changed in IL-6 KO mice, not illogical given the highly quiescent state of the stem cells in the basal gland, not in need of IL-6 action. It should be remarked that damage still induced some proliferative stem cell activation in the aging pituitary in vivo, while IL-6 injection did not (Figs. 1A and 4A) and that the proliferative activation reached in young mice after damage appeared higher than after IL-6 injection (Figs. 1A and 3F). One explanation may be that local IL-6 levels in young pituitary after damage are higher than achieved after i.p. IL-6 injection. Furthermore, still other factors may additionally be involved in the stem cell activation process after injury (SI Appendix, Fig. S5D). Our scRNA-seq resource now provides an invaluable means to elucidate this molecular machinery in depth, including the search for the upstream activators of IL-6 expression during damage.
Excitingly, our study demonstrates that the stem cells of aging pituitary regain activatability when removed from their in vivo tissue milieu. Hence, receded in vivo responsiveness and reaction to injury is not an intrinsic aging process of the pituitary stem cells but may rather be imposed by an oppressive (inflammatory) microenvironment. Also in certain other tissues, it has been shown that stem cells retain their functional capacities at aging, which is repressed by the environment (16–18, 47). Substituting the old milieu for a younger equivalent restored stem cell functionality in these tissues (18, 38, 47, 48). Taken together, we advance the concept (as summarized in SI Appendix, Fig. S5D) that the raised inflammatory environment in the aging pituitary, indicative of inflammaging, presents a roadblock for full activation of the resident stem cells upon injury. Or in other words, the prevailing IL-6/inflammatory milieu in the aging pituitary sets a threshold that is hard to surpass for unfolding an adequate acute reaction when challenged by injury. In the end, the subsided acute reaction of the stem cells may contribute to the absence of the later regeneration in aging pituitary upon cell-ablation injury (14). Indeed, it has been reported in other tissues that acute activation of the resident stem cells by IL-6 following insult represents a first essential step toward eventual repair (34, 49, 50). Definite evidence for an in vivo role of IL-6 in eventual pituitary regeneration awaits extensive and comprehensive scrutiny of GHCre/iDTR mice on the IL-6 KO background, ideally in a conditional (temporospatial), pituitary (stem/connective cell)-specific manner. Interestingly, it has been found that anti-inflammatory intervention can restore regenerative capacity at aging in certain tissues [such as skin, muscle, liver, and gut (40–43)], an appealing path for future pituitary aging research. Finally, the present study strongly enlarges the applicability of our recently developed pituitary organoid model by effectively extending organoid expandability using IL-6, thus compensating for the decline of endogenous IL-6 production, which could be due to the disappearance of in situ stimulatory factors.
In conclusion, we identified and characterized IL-6 as pituitary stem cell activator, a competence quenched at aging concurrent with a raised IL-6/inflammatory stress level in the gland. Still, aging pituitary stem cells retain intrinsic stemness properties and show activatability when released from their in vivo microenvironment. These insights may be instrumental to find strategies for restraining the master endocrine pituitary gland from aging or for rejuvenating a burdened old gland. And more in general, our single-cell transcriptome database provides a rich source to search for processes underlying pituitary aging, whose understanding is currently poor, and for potential therapeutic targets. In the end, IL-6 and inflammaging may represent appealing candidates.
Methods
Mice and In Vivo Treatments.
GHCre/iDTR and control (-/iDTR) mice were injected with DT and pituitaries (damaged and undamaged, respectively) collected (Fig. 2A). Young and/or aging mice were treated with IL-6, anti–IL-6 antibody, CpG, or LMT-28 according to the indicated or described schedules. Further details are provided in SI Appendix.
scRNA-Seq.
Damaged and undamaged AP from young and aging mice were dissociated into single cells (51, 52) and subjected to scRNA-seq analyses using 10x Genomics (Fig. 2A), according to manufacturer instructions. Libraries were sequenced and downstream analysis was performed in R using Seurat (53). Gene regulatory networks (regulons) were determined using SCENIC (22) in Python (pySCENIC). More details are given in SI Appendix.
Pituitary Organoids.
AP cells were plated in a drop of 70% Matrigel/30% serum-free defined medium (Thermo Fisher Scientific), and pituitary organoid culture medium (19) was added. After growth (10 to 14 d), organoids were dissociated into small fragments, which were reseeded in Matrigel drops for passaging. More details are provided in SI Appendix.
Immunostaining.
Whole pituitary and organoids were fixed, and sections were subjected to immunofluorescence staining (for antibodies, see SI Appendix, Table S1) or transmission electron microscopy (SI Appendix). Immunopositive-cell quantification and EdU labeling in organoids are described in SI Appendix.
RNAscope In Situ Hybridization.
Whole pituitary was fixed, and sections were subjected to RNAscope analysis according to the manufacturer’s recommendations (Advanced Cell Diagnostics). More details are provided in SI Appendix.
Gene Expression Analysis by RT-qPCR.
RNA was reverse-transcribed and subjected to qRT-PCR as previously described (19) using primers as listed in SI Appendix, Table S2. Further details are given in SI Appendix.
IL-6 Measurement.
IL-6 concentration was measured in organoid culture supernatant and mouse plasma using Meso Scale Discovery kits according to the manufacturer’s protocol (SI Appendix).
Statistical Analysis.
Statistical analysis was performed using GraphPad Prism as described in detail in SI Appendix. Statistical significance was defined as P ≤ 0.05.
Supplementary Material
Acknowledgments
We thank Y. Van Goethem and V. Vanslembrouck for valuable technical help. We thank the Vlaams Instituut voor Biotechnologie (VIB) Nucleomics Core (in particular Rekin’s Janky) and Katholieke Universiteit (KU) Leuven Genomics Core (particularly Álvaro Cortés Calabuig) for their expert assistance in scRNA-seq analysis, as well as Thomas Van Brussel and Bram Boeckx (D.L.’s group, KU Leuven) for technical and bioinformatical support in scRNA-seq experiments, respectively. The computational resources used for scRNA-seq analysis were provided by the Vlaams Supercomputer Centrum, managed by the Fonds Wetenschappelijk Onderzoek (FWO)–Vlaanderen. We are also grateful to the Imaging Core (VIB, KU Leuven) and the Cell and Tissue Imaging Cluster (KU Leuven) for the use of microscopes and the Center for Brain and Disease Research Histology unit (VIB, KU Leuven) for the use of histology equipment. We acknowledge the use of the Electron Microscopy Platform (VIB, KU Leuven) and the Institute of Development, Aging and Cancer (Tohoku University, Sendai, Japan) for transmission electron microscopy. This work was supported by several grants from the Bijzonder Onderzoeksfonds KU Leuven and from Fonds voor Wetenschappelijk Onderzoek (FWO)–Vlaanderen awarded to the principal investigators. A.V. (Grant No. 1141717N), E.L. (Grant No. 11A3320N), B.C. (Grant No. 11W9215N), A.J. (Grant No. 1158318N), and C.N. (Grant No. 1S14218N) are supported by a PhD Fellowship from the FWO/FWO-Strategisch Basisonderzoek (SB).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2100052118/-/DCSupplemental.
Data Availability
scRNA-seq data have been deposited in ArrayExpress (E-MTAB-10021). All other study data are included in the article and/or supporting information.
References
- 1.Fauquier T., Rizzoti K., Dattani M., Lovell-Badge R., Robinson I. C. A. F., SOX2-expressing progenitor cells generate all of the major cell types in the adult mouse pituitary gland. Proc. Natl. Acad. Sci. U.S.A. 105, 2907–2912 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen J., et al., Pituitary progenitor cells tracked down by side population dissection. Stem Cells 27, 1182–1195 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Rizzoti K., Akiyama H., Lovell-Badge R., Mobilized adult pituitary stem cells contribute to endocrine regeneration in response to physiological demand. Cell Stem Cell 13, 419–432 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andoniadou C. L., et al., Sox2(+) stem/progenitor cells in the adult mouse pituitary support organ homeostasis and have tumor-inducing potential. Cell Stem Cell 13, 433–445 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Vankelecom H., Chen J., Pituitary stem cells: Where do we stand? Mol. Cell. Endocrinol. 385, 2–17 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Cox B., Roose H., Vennekens A., Vankelecom H., Pituitary stem cell regulation: Who is pulling the strings? J. Endocrinol. 234, R135–R158 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Vankelecom H., “Pituitary stem cells: Quest for hidden functions” in Stem Cells in Neuroendocrinology, Pfaff D., Christen Y., Eds. (Springer International Publishing, 2016), pp. 81–101. [PubMed] [Google Scholar]
- 8.Zhu X., Tollkuhn J., Taylor H., Rosenfeld M. G., Notch-dependent pituitary SOX2+ stem cells exhibit a timed functional extinction in regulation of the postnatal gland. Stem Cell Reports 5, 1196–1209 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roose H., et al., Major depletion of SOX2+ stem cells in the adult pituitary is not restored which does not affect hormonal cell homeostasis and remodelling. Sci. Rep. 7, 16940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brack A. S., Rando T. A., Tissue-specific stem cells: Lessons from the skeletal muscle satellite cell. Cell Stem Cell 10, 504–514 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyajima A., Tanaka M., Itoh T., Stem/progenitor cells in liver development, homeostasis, regeneration, and reprogramming. Cell Stem Cell 14, 561–574 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Fu Q., et al., The adult pituitary shows stem/progenitor cell activation in response to injury and is capable of regeneration. Endocrinology 153, 3224–3235 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Fu Q., Vankelecom H., Regenerative capacity of the adult pituitary: Multiple mechanisms of lactotrope restoration after transgenic ablation. Stem Cells Dev. 21, 3245–3257 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Willems C., et al., Regeneration in the pituitary after cell-ablation injury: Time-related aspects and molecular analysis. Endocrinology 157, 705–721 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Rossi D. J., et al., Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc. Natl. Acad. Sci. U.S.A. 102, 9194–9199 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schultz M. B., Sinclair D. A., When stem cells grow old: Phenotypes and mechanisms of stem cell aging. Development 143, 3–14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domingues-Faria C., Vasson M. P., Goncalves-Mendes N., Boirie Y., Walrand S., Skeletal muscle regeneration and impact of aging and nutrition. Ageing Res. Rev. 26, 22–36 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Segel M., et al., Niche stiffness underlies the ageing of central nervous system progenitor cells. Nature 573, 130–134 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cox B., et al., Organoids from pituitary as a novel research model toward pituitary stem cell exploration. J. Endocrinol. 240, 287–308 (2019). [DOI] [PubMed] [Google Scholar]
- 20.McInnes L., Healy J., Melville J., UMAP: Uniform manifold approximation and projection for dimension reduction. arXiv [Preprint] (2018). https://arxiv.org/abs/1802.03426#:∼:text=UMAP%20 (Accessed 18 September 2020).
- 21.Cheung L. Y. M., et al., Single-cell RNA sequencing reveals novel markers of male pituitary stem cells and hormone-producing cell types. Endocrinology 159, 3910–3924 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aibar S., et al., SCENIC: Single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayran A., et al., Pioneer and nonpioneer factor cooperation drives lineage specific chromatin opening. Nat. Commun. 10, 3807 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez Vallone V., et al., Trop2 marks transient gastric fetal epithelium and adult regenerating cells after epithelial damage. Development 143, 1452–1463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldstein A. S., et al., Trop2 identifies a subpopulation of murine and human prostate basal cells with stem cell characteristics. Proc. Natl. Acad. Sci. U.S.A. 105, 20882–20887 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gremeaux L., Fu Q., Chen J., Vankelecom H., Activated phenotype of the pituitary stem/progenitor cell compartment during the early-postnatal maturation phase of the gland. Stem Cells Dev. 21, 801–813 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Lodge E. J., Russell J. P., Patist A. L., Francis-West P., Andoniadou C. L., Expression analysis of the Hippo cascade indicates a role in pituitary stem cell development. Front. Physiol. 7, 114 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lodge E. J., et al., Homeostatic and tumourigenic activity of SOX2+ pituitary stem cells is controlled by the LATS/YAP/TAZ cascade. eLife 8, 1–26 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vankelecom H., Pituitary stem cells drop their mask. Curr. Stem Cell Res. Ther. 7, 36–71 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Vankelecom H., Carmeliet P., Van Damme J., Billiau A., Denef C., Production of interleukin-6 by folliculo-stellate cells of the anterior pituitary gland in a histiotypic cell aggregate culture system. Neuroendocrinology 49, 102–106 (1989). [DOI] [PubMed] [Google Scholar]
- 31.Allaerts W., Vankelecom H., History and perspectives of pituitary folliculo-stellate cell research. Eur. J. Endocrinol. 153, 1–12 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Serrano A. L., Baeza-Raja B., Perdiguero E., Jardí M., Muñoz-Cánoves P., Interleukin-6 is an essential regulator of satellite cell-mediated skeletal muscle hypertrophy. Cell Metab. 7, 33–44 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Muñoz-Cánoves P., Scheele C., Pedersen B. K., Serrano A. L., Interleukin-6 myokine signaling in skeletal muscle: A double-edged sword? FEBS J. 280, 4131–4148 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuhn K. A., Manieri N. A., Liu T. C., Stappenbeck T. S., IL-6 stimulates intestinal epithelial proliferation and repair after injury. PLoS One 9, e114195 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rose-John S., Interleukin-6 family cytokines. Cold Spring Harb. Perspect. Biol. 10, 1–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong S.-S., et al., A novel small-molecule inhibitor targeting the IL-6 receptor β subunit, glycoprotein 130. J. Immunol. 195, 237–245 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Behrens E. M., et al., Repeated TLR9 stimulation results in macrophage activation syndrome-like disease in mice. J. Clin. Invest. 121, 2264–2277 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franceschi C., Campisi J., Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J. Gerontol. A Biol. Sci. Med. Sci. 69, S4–S9 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Franceschi C., et al., Inflamm-aging. An evolutionary perspective on immunosenescence. Ann. N. Y. Acad. Sci. 908, 244–254 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Neves J., Sousa-Victor P., Regulation of inflammation as an anti-aging intervention. FEBS J. 287, 43–52 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Oh J., et al., Age-associated NF-κB signaling in myofibers alters the satellite cell niche and re-strains muscle stem cell function. Aging (Albany NY) 8, 2871–2896 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jurk D., et al., Chronic inflammation induces telomere dysfunction and accelerates ageing in mice. Nat. Commun. 2, 4172 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doles J., Storer M., Cozzuto L., Roma G., Keyes W. M., Age-associated inflammation inhibits epidermal stem cell function. Genes Dev. 26, 2144–2153 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Büttner R., et al., Inflammaging impairs peripheral nerve maintenance and regeneration. Aging Cell 17, e12833 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu R., Sampathkumar N. K., Benayoun B. A., Measuring phagocytosis in bone marrow-derived macrophages and peritoneal macrophages with aging. Methods. Mol. Biol. 2144, 161–170 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vankelecom H., et al., Immunocytochemical evidence that S-100-positive cells of the mouse anterior pituitary contain interleukin-6 immunoreactivity. J. Histochem. Cytochem. 41, 151–156 (1993). [DOI] [PubMed] [Google Scholar]
- 47.Conboy I. M., et al., Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature 433, 760–764 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Ahmed A. S. I., Sheng M. H., Wasnik S., Baylink D. J., Lau K. W., Effect of aging on stem cells. World J. Exp. Med. 7, 1–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tadokoro T., et al., IL-6/STAT3 promotes regeneration of airway ciliated cells from basal stem cells. Proc. Natl. Acad. Sci. U.S.A. 111, E3641–E3649 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Galun E., Rose-John S., “The regenerative activity of Interleukin-6” in Tissue-Protective Cytokines: Methods and Protocols, Ghezzi P., Cerami A., Eds. (Humana Press, Totowa, NJ, 2013), pp. 59–77. [DOI] [PubMed] [Google Scholar]
- 51.Denef C., Hautekeete E., De Wolf A., Vanderschueren B., Pituitary basophils from immature male and female rats: Distribution of gonadotrophs and thyrotrophs as studied by unit gravity sedimentation. Endocrinology 103, 724–735 (1978). [DOI] [PubMed] [Google Scholar]
- 52.Van der Schueren B., Denef C., Cassiman J.-J., Ultrastructural and functional characteristics of rat pituitary cell aggregates. Endocrinology 110, 513–523 (1982). [DOI] [PubMed] [Google Scholar]
- 53.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
scRNA-seq data have been deposited in ArrayExpress (E-MTAB-10021). All other study data are included in the article and/or supporting information.