Significance
In a typical prokaryotic cell, hundreds of metabolites are present and interconverted simultaneously, some serving as allosteric regulators for unrelated reactions or accidental substrates for promiscuous enzymes or being downright reactive and toxic. Although certain spatial separation of prokaryotic cells has long been proposed, only few examples are known. In many bacteria, the key carboxylase of the Calvin–Benson cycle is located in carboxysomes to concentrate CO2 and deplete O2, while other enzymes of the cycle are in the cytoplasm. The archaeon Ignicoccus hospitalis exhibits a two-membrane structure with an energized outer membrane. Here, we show that CO2 fixation in this organism takes place in a peripheric cytoplasmic compartment, being separated from information processing occurring in the central cytoplasmic compartment.
Keywords: Archaea, compartmentalization, CO2 fixation, immunogold labeling
Abstract
The prokaryotic cell is traditionally seen as a “bag of enzymes,” yet its organization is much more complex than in this simplified view. By now, various microcompartments encapsulating metabolic enzymes or pathways are known for Bacteria. These microcompartments are usually small, encapsulating and concentrating only a few enzymes, thus protecting the cell from toxic intermediates or preventing unwanted side reactions. The hyperthermophilic, strictly anaerobic Crenarchaeon Ignicoccus hospitalis is an extraordinary organism possessing two membranes, an inner and an energized outer membrane. The outer membrane (termed here outer cytoplasmic membrane) harbors enzymes involved in proton gradient generation and ATP synthesis. These two membranes are separated by an intermembrane compartment, whose function is unknown. Major information processes like DNA replication, RNA synthesis, and protein biosynthesis are located inside the “cytoplasm” or central cytoplasmic compartment. Here, we show by immunogold labeling of ultrathin sections that enzymes involved in autotrophic CO2 assimilation are located in the intermembrane compartment that we name (now) a peripheric cytoplasmic compartment. This separation may protect DNA and RNA from reactive aldehydes arising in the I. hospitalis carbon metabolism. This compartmentalization of metabolic pathways and information processes is unprecedented in the prokaryotic world, representing a unique example of spatiofunctional compartmentalization in the second domain of life.
Compartmentalization is one of the distinguishing features of eukaryotic cells, which contain membrane-bound organelles in order to perform their specific functions more efficiently, like photosynthesis and CO2 fixation in plants. Here, autotrophic CO2 assimilation proceeds via the Calvin–Benson cycle in the stroma of chloroplasts. Evolutionary ancestors of chloroplasts, the cyanobacteria, contain the key carboxylating enzyme of this cycle, ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO), in specifically insulated proteinaceous microcompartments, carboxysomes, to enrich both the carboxylase and its substrate CO2 and to exclude competing O2 at the active site of RubisCO (1). Other enzymes involved in autotrophy are located in the cytoplasm. Besides the Calvin–Benson cycle, autotrophic prokaryotes have developed at least six alternative ways of CO2 fixation (2–4). The recently discovered autotrophic cycle, the dicarboxylate/4-hydroxybutyrate (DC/HB) cycle (Fig. 1A), functions in the anaerobic hyperthermophilic Crenarchaeota of the orders Desulfurococcales and Thermoproteales, for example, Ignicoccus hospitalis, Thermoproteus neutrophilus, and Pyrolobus fumarii (2, 5–9). In the carbon fixation phase of this pathway, one molecule acetyl-CoA, one molecule CO2, and one molecule bicarbonate are converted to oxaloacetate via pyruvate synthase and phosphoenolpyruvate (PEP) carboxylase reactions. In the reduction phase, oxaloacetate is reduced to succinyl-CoA and then to 4-hydroxybutyrate, a name-giving intermediate of the cycle. Finally, in the acetyl-CoA regeneration phase of the cycle, 4-hydroxybutyrate is activated to the corresponding CoA-ester, dehydrated to crotonyl-CoA, and converted to two molecules of acetyl-CoA via β-oxidation reactions.
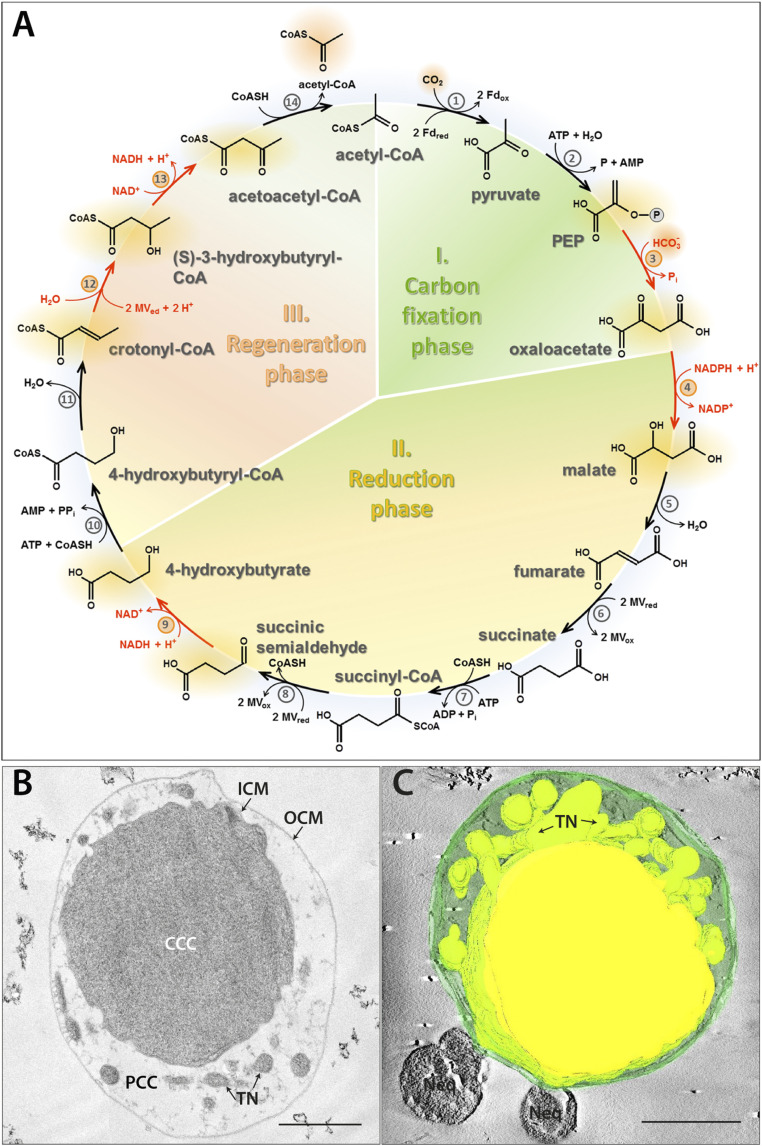
Fig. 1.
The DC/HB cycle (adapted from ref. 3) (A), ultrastructure (B), and 3D model (C) of I. hospitalis. Enzymes: 1) pyruvate synthase, 2) pyruvate:water dikinase, 3) PEP carboxylase, 4) malate dehydrogenase, 5) fumarate hydratase, 6) fumarate reductase (natural electron donor is not known), 7) succinyl-CoA synthetase, 8) succinyl-CoA reductase (natural electron acceptor is not known), 9) succinic semialdehyde reductase, 10) 4-hydroxybutyrate-CoA ligase, 11) 4-hydroxybutyryl-CoA dehydratase, 12) crotonyl-CoA hydratase, 13) (S)-3-hydroxybutyryl-CoA dehydrogenase, and 14) acetoacetyl-CoA β-ketothiolase. Fd, ferredoxin; MV, methyl viologen. The enzymes studied in this work are highlighted in red. Ultrathin section (B) and 3D model of a semithin section (C) of a cryo-fixed, freeze-substituted, Epon-embedded cell. CCC (central cytoplasmic compartment), PCC (peripheric cytoplasmic compartment), ICM (inner cytoplasmic membrane), TN (tubular network), OCM (outer cytoplasmic membrane), and Neq (Nanoarchaeum equitans). 3D model highlights the TN originating from the CCC. (Scale bars, 500 nm.)
The DC/HB cycle has first been discovered in the hyperthermophilic autotrophic sulfur-reducing archaeon Ignicoccus hospitalis (6) that grows under anaerobic conditions with molecular hydrogen and sulfur at T = 73 to 98 °C (Topt 90 °C) (10). Apart from the unusual carbon assimilation pathway, Ignicoccus cells exhibit an extraordinary two-membrane ultrastructure (Fig. 1 B and C). The outer cellular membrane (not to be confused with the outer membrane of gram-negative bacteria) encases the intermembrane compartment, while the inner membrane surrounds a modified cytoplasm. Here, we suggest naming these compartments peripheric and central cytoplasmic compartment (CC), respectively, that are enclosed by outer and inner cytoplasmic membranes (CM). The peripheric CC contains a complex tubular network, derived from the central CC (10–13). The spatial compartmentalization goes along with a functional compartmentalization, as energy-conserving ATP synthase and H2:sulfur oxidoreductase are located in the outer CM, while the major energy-consuming anabolic steps, DNA replication, RNA synthesis, and translation, take place in the central CC (14). Here, we present the subcellular localization of four enzymes of the DC/HB cycle and the biochemical characterization of three of them. Our study shows that autotrophic CO2 assimilation in I. hospitalis proceeds in the peripheric CC, thus demonstrating another unique feature of I. hospitalis: the spatial separation of major anabolic processes in I. hospitalis, namely DNA replication, RNA synthesis, and protein biosynthesis from inorganic carbon fixation.
Results
Biochemical Characterization of Enzymes of the CO2 Fixation Pathway in I. hospitalis.
To study the localization of the DC/HB cycle, we chose four enzymes (Fig. 1A, highlighted in red) involved in different phases of the cycle: PEP carboxylase (reaction three; carbon fixation phase), malate dehydrogenase (reaction four; reduction phase) and succinic semialdehyde reductase (reaction nine; reduction phase), and bifunctional crotonyl-CoA hydratase/(S)-3-hydroxybutyryl-CoA dehydrogenase (reactions 12 and 13; acetyl-CoA regeneration phase). While the genes for three of the enzymes had been identified in our previous studies (Igni_0341 for PEP carboxylase, Igni_1263 for malate dehydrogenase, and Igni_1058 for crotonyl-CoA hydratase/(S)-3-hydroxybutyryl-CoA dehydrogenase) (6, 15), their function was not confirmed biochemically. Furthermore, the gene encoding succinic semialdehyde reductase was not known. To identify the corresponding gene, we searched the genome of I. hospitalis for various alcohol dehydrogenases and identified igni_0132, which is present in all sequenced autotrophic members of the Desulfurococcales (SI Appendix, Fig. S1). The synthesized gene was expressed in Escherichia coli, and the recombinant protein was purified and characterized. Indeed, the enzyme showed succinic semialdehyde reductase activity and was highly specific for its substrate, succinic semialdehyde (KM 75 µM, Vmax 500 U ⋅ mg−1 protein at 65 °C; Table 1). Igni_0132 had only very little (if any) activity with other tested aldehydes (SI Appendix, Table S1), confirming its characterization as succinic semialdehyde reductase from the DC/HB cycle. Interestingly, the enzyme accepted both NADH and NADPH as the substrates, with a preference for NADH (catalytic efficiencies kcat/KM of 9.2 and 1.6 s−1 ⋅ µM−1, respectively; Table 1). The genes for PEP carboxylase, malate dehydrogenase, and bifunctional crotonyl-CoA/(S)-3-hydroxybutyryl-CoA dehydrogenase (igni_0341, igni_1263, and igni_1058, respectively) were synthesized and expressed in E. coli, and the corresponding proteins were purified and checked for activity. Heterologous expression of PEP carboxylase Igni_0341 led to the production of an insoluble and inactive protein despite all our attempts to optimize expression or refold the protein from inclusion bodies. Nevertheless, the identity of the PEP carboxylase gene was unambiguous (6), and the purified protein could be used for the production of antibodies (see Subcellular Localization of Enzymes of the CO2 Fixation Pathway in I. hospitalis). Igni_1263 catalyzed both the NADPH- and NADH-dependent reduction of oxaloacetate to malate, with a preference for NADPH (kcat/KM of 77.9 and 9.6 s−1 ⋅ µM−1, respectively; Table 1); no activity was detected with other tested substrates (SI Appendix, Table S2). Crotonyl-CoA and (S)-3-hydroxybutyryl-CoA dehydrogenase activities were shown for Igni_1058, as the enzyme converted both crotonyl-CoA and (S)-3-hydroxybutyryl-CoA into acetoacetyl-CoA. The enzyme was specific for NAD and acted on the (S)-stereoisomer of 3-hydroxybutyryl-CoA as the substrate, whereas no activity was shown with either NADP or (R)-3-hydroxybutyryl-CoA (Table 1).
Table 1.
Enzymes of the DC/HB cycle characterized in this study
| Enzyme/Substrate | Gene | Assay temp. [°C] | Vmax [U ⋅ mg−1 protein] | KM [µM] | kcat/KM [s−1 ⋅ µM−1] * | |
| Measured | extrapolated at 90 °C | |||||
| Malate dehydrogenase | Igni_1263 | 45 | ||||
| Oxaloacetate | 628 ± 20 | 14,200 ± 450 | 270 ± 28 | 32.2 | ||
| NADPH | 621 ± 61 | 14,000 ± 1,380 | 110 ± 26 | 77.9 | ||
| NADH | 109 ± 6 | 2,460 ± 140 | 157 ± 21 | 9.6 | ||
| Succinic semialdehyde reductase | Igni_0132 | 65 | ||||
| Succinic semialdehyde | 504 ± 21 | 2,850 ± 120 | 75 ± 8 | 30.5 | ||
| NADPH | 14 ± 1 | 80 ± 6 | 41 ± 10 | 1.6 | ||
| NADH | 497 ± 44 | 2,810 ± 250 | 246 ± 43 | 9.2 | ||
| Crotonyl-CoA hydratase/ (S)-3-hydroxybutyryl-CoA dehydrogenase | Igni_1058 | 50 | ||||
| Crotonyl-CoA (NAD reduction) | 6.1 ± 0.2 | 98 ± 3 | 71 ± 8 | 1.8 | ||
| (S)-3-Hydroxybutyryl-CoA (NAD reduction) | 7.3 ± 0.3 | 117 ± 5 | 86 ± 13 | 1.8 | ||
| NAD [with (S)-3-hydroxybutyryl-CoA] | 5.5 ± 0.1 | 88 ± 2 | 15 ± 2 | 7.6 | ||
| PEP carboxylase† | Igni_0341† | NA† | NA† | NA† | NA† | NA† |
NA, not applicable. The Vmax values are normalized to 90 °C based on the assumption that a 10 °C rise in temperature doubles the reaction rate. The determination of KM and Vmax values was at least performed two times.
kcat /KM was calculated for the activities at 90 °C.
PEP carboxylase was produced in the insoluble form and thus was not active.
Subcellular Localization of Enzymes of the CO2 Fixation Pathway in I. hospitalis.
To determine the localization of the studied enzymes in I. hospitalis cells, we performed transmission electron microscopy in combination with immunogold labeling on ultrathin sections of resin-embedded cells and additionally on thawed cryosections, so-called “Tokuyasu sections” (16). Cryosection labeling was used as an additional control experiment because it is considered to be more sensitive than room temperature labeling. Room temperature labeling in contrast is superior in ultrastructural preservation (17). Since there is currently no genetic system available for the Ignicoccus species, we raised antibodies against recombinant proteins and assured specific binding of antibodies to target proteins in advance by Western blot analysis following sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE). Indeed, antibodies recognized targeted proteins at the expected sizes, and only one specific band for each protein was detected by Western blot analysis using crude extracts of I. hospitalis cells (SI Appendix, Fig. S2).
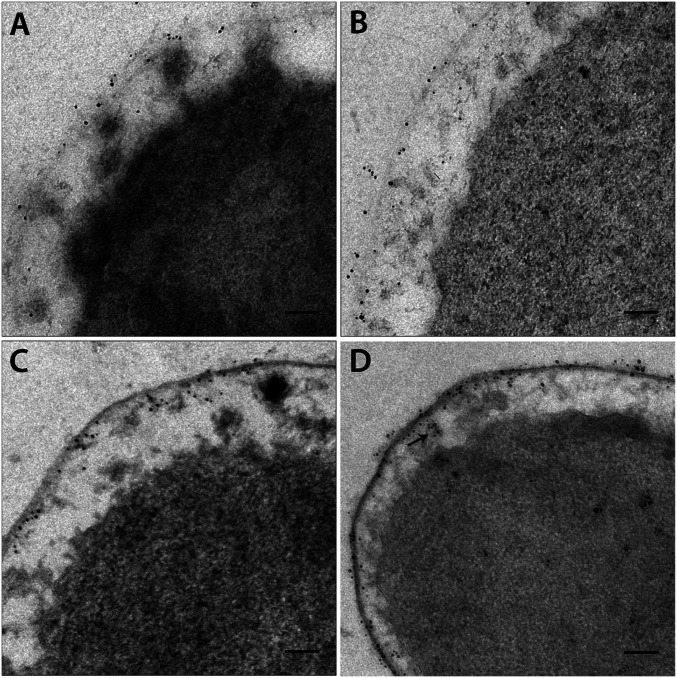
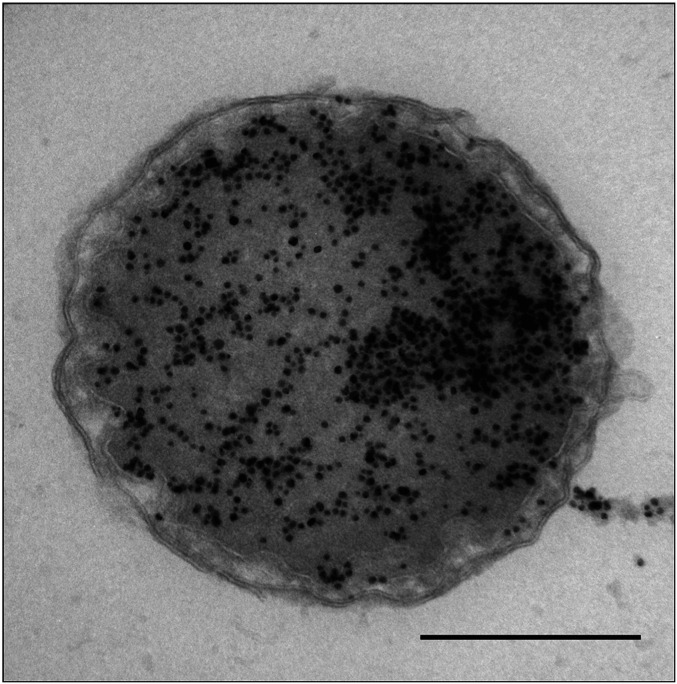
Using immunolabeling on ultrathin-sectioned I. hospitalis cells with antibodies directed against PEP carboxylase, malate dehydrogenase, succinic semialdehyde reductase, and crotonyl-CoA hydratase/(S)-3-hydroxybutyryl-CoA dehydrogenase, we found an almost exclusive localization of these enzymes in the peripheric CC in close vicinity of the outer CM. While less than ∼5% of gold particles were detected in the central CC (Fig. 2 and SI Appendix, Fig. S3), they were observed frequently within the tubular network in the peripheric CC (Fig. 2D and SI Appendix, Fig. S3D). In control experiments without primary antibodies, only few gold particles could be detected in the background. Labeling of thawed cryosections confirmed these results (SI Appendix, Fig. S4). Due to the higher sensitivity of antibody binding on Tokuyasu sections, a higher amount of gold particles was distributed across the cells, but we still found 90 to 95% of labeling in the peripheric CC. In contrast, gold particles were exclusively detected in the central CC of I. hospitalis after immunogold labeling with commercially available antibodies directed against double-stranded DNA (dsDNA) on plastic and on thawed cryosections (Fig. 3).
Fig. 2.
Localization of enzymes involved in the DC/HB cycle (resin sections). (A–D) Image sections of SI Appendix, Fig. S3. Localization of PEP carboxylase (A), malate dehydrogenase (B), succinic semialdehyde reductase (C), and crotonyl-CoA hydratase/(S)-3-hydroxybutyryl-CoA dehydrogenase (D). The black arrow in D indicates gold particles in protrusions derived from the central CC. (Scale bars, 100 nm.)
Fig. 3.
Localization of DNA (cryosection). I. hospitalis cell is labeled with an anti-DNA antibody according to Tokuyasu (16). (Scale bar, 500 nm.)
Discussion
Structural Separation of Central Carbon Metabolism from Information Processing.
Initially, the cell structure of Ignicoccus was compared to the structure of gram-negative bacteria. Indeed, the peripheric CC stains only weakly used standard preparations for electron microscopy. From this, it was assumed that this compartment hardly contains any cellular material and, thus, resembles the periplasm of gram-negative bacteria with the outer membrane containing a porin-like protein Imp1227 (18). Furthermore, cell appendages of Ignicoccus (first identified as flagella and later shown to be adhesion structures; refer to refs. 19 and 20) are anchored in the inner CM, while the central CC contains ribosomes and DNA and, thus, resembles the cytoplasm of gram-negative bacteria. Therefore, the identification of ATP synthase and H2:sulfur oxidoreductase in the outer CM (14) was surprising and led to the reannotation of the “periplasm” as an intermembrane compartment. Here, we show that at least one essential metabolic pathway, the autotrophic DC/HB cycle, is located in the peripheric CC as well, therefore suggesting that the central and peripheric CC are together analogous (and probably homologous) to a typical cytoplasm of a prokaryotic cell. Indeed, studied enzymes belong to different phases of the pathway, and intensive transport of intermediates of the cycle between central and peripheric CC seems to be unlikely. Therefore, we conclude that the whole CO2 fixation pathway of I. hospitalis takes place in the peripheric CC. That makes sense, as CO2 fixation requires high amounts of both ATP and reducing equivalents in the form of NAD(P)H and reduced ferredoxin that are generated in the peripheric CC (14). Although the studied enzymes of the DC/HB cycle were found in close vicinity of the outer CM, this association does not appear to be tight, as they were not identified in previous proteomics studies of the membrane-associated proteins (21, 22), and we were able to produce three of them in an active and soluble form. Interestingly, acetyl-CoA synthetase activating acetate to acetyl-CoA is also located in the peripheric CC (22), suggesting that other reactions of the central carbon metabolism, namely gluconeogenesis starting from PEP and pentose phosphate synthesis, may also take place in the peripheric CC. Nevertheless, three costly anabolic processes, that is, DNA replication, RNA synthesis, and protein biosynthesis, are located in the central CC, therefore being spatially separated from both energy conservation and inorganic carbon assimilation.
What could be the reason for the separation of central carbon metabolism from information processing in Ignicoccus cells? The separation seems problematic in a first view, as it requires intensive transport of ATP and AMP across the inner membrane of Ignicoccus cells. Nevertheless, this separation may give an additional protection for information processing from reactive compounds arising in the metabolism of I. hospitalis. Succinic semialdehyde (an intermediate of the DC/HB cycle) and methylglyoxal (arising from gluconeogenic intermediates like triosephosphates) belong to reactive carbonyl species, mutagenic compounds damaging DNA (23). Methylglyoxal poses a serious problem for hyperthermophiles, as it is inevitably produced from triosephosphates with a high rate at elevated temperatures. Indeed, triosephosphates have a half-life time of 4 min at 80 °C, being converted to methylglyoxal (24). Gluconeogenesis is tightly integrated in the autotrophic pathway, requires reductants, and is therefore probably also located in the peripheric CC. The spatial separation of DNA from the reactions that contain (or produce) mutagenic compounds would be beneficial for a hyperthermophile. Nevertheless, other hyperthermophiles have a classical cell structure and are still capable to cope with the methylglyoxal stress, even using this compound in biosynthetic reactions (25). The usage of a bifunctional fructose 1,6-bisphosphate aldolase/phosphatase ensuring a quick removal of triosephosphates and preventing their conversion to methylglyoxal is also an important adaptation to hyperthermophily (26). This enzyme is present in Ignicoccus (26). We may add that the mechanism in how these hyperthermophiles cope with the methylglyoxal stress is, however, neither systematically studied nor understood (27). The unusual cell biology may be an additional/alternative mechanism to protect information processing from reactive metabolites. Interestingly, I. hospitalis is able to survive high doses of ionizing radiation (up to ∼19 kGy), highlighting its extraordinarily high genome stability (28).
Comparison of the Ignicoccus Peripheric CC with Other Prokaryotic Compartments.
Bacterial microcompartments (carboxysomes and metabolosomes) and anammoxosomes specifically encapsulate metabolic reactions and pathways. Carboxysomes contain only two enzymes and serve as a carbon-concentrating mechanism, while metabolosomes encapsulate enzymes involved in catabolism of a variety of organic compounds (1, 29). The key feature of bacterial microcompartments is the selective permeability of their shell protecting the cell from toxic intermediates of the pathways, preventing side reactions, increasing enzyme concentration and stability, and/or enhancing flux through the pathway (1, 30). In comparison to the I. hospitalis peripheric CC comprising roughly half of the cell volume, microcompartments in Bacteria are usually relatively small in their dimension and encapsulate only a few enzymes or short metabolic pathways. In contrast, anammoxosomes comprise the largest compartment of the cell of anammox bacteria (Planctomycetes). They are dedicated to energy conservation, as their stable ladderane lipid bilayer membranes protect the cell against the reactive intermediate hydrazine produced during anaerobic ammonia oxidation. Still, the anammox pathway is relatively short (31). On the contrary, the peripheric CC of I. hospitalis resides between two membranes differing in chemical structure (a tetraether monolayer in the inner CM and archaeol in the outer CM, refer to refs. 18 and 32) and encapsulates ATP conservation (14), acetyl-CoA synthesis (22), CO2 fixation via a pathway consisting of 14 reactions (Fig. 1A), and potentially other metabolic pathways. It also contains a tubular network and a filamentous matrix presumably functioning as a cytoskeleton within this compartment (13, 21, 33). This is unprecedented in the microbial world and demonstrates that the I. hospitalis peripheric CC is strikingly different from other known cellular structures, including the periplasm of gram-negative bacteria. Actually, the bacterial periplasm occupies usually only ∼10% of the total cell volume (34) and does not contain ATP, therefore being hardly suitable for biosynthetic reactions. Instead, it contains soluble proteins involved in trafficking, protein folding, and the degradation of proteins in the absence of ATP (35). It is surrounded by a peptidoglycan layer anchored to the outer membrane.
Transportation Routes and Targeting Signals.
The two-membrane structure of Ignicoccus requires intensive protein and metabolite transport between the central and peripheric CC. There is growing support for the participation of cytoplasmic protrusions in protein transportation/translocation in I. hospitalis. The usage of electron tomography techniques offered a comprehensive three-dimensional (3D) view on a dynamic tubular network system in I. hospitalis, canceling our former concept of a vesicular transportation system (Fig. 1C) (13). Furthermore, pore-like, cylindrical-shaped structures were detected underneath the outer CM. These structures possibly connect the tube-like extensions/central CC with the outer CM, potentially serving as transport routes between the central and peripheric CC and even the outer CM. Consequently, we frequently detected gold particles in these “cytoplasmic extensions.” Similarly, immunogold labeling located subunits of the ATP synthase to these protrusions as well (14). This route may also contribute to import metabolic intermediate products of the carbon metabolism into the central CC (13).
In all domains of life, the Sec and Tat pathways requiring a specific signal sequence are the main pathways responsible for protein transport across membranes (36, 37). In I. hospitalis, several components of the Sec pathway and the Tat system were identified in the genome and the proteome (38, 39), and the corresponding signal peptides were found in some of its proteins (39). Nevertheless, such signal peptides could not be identified in any of the known enzyme of the DC/HB cycle, with the exception of crotonyl-CoA hydratase/3-hydroxybutyryl-CoA dehydrogenase (SI Appendix, Fig. S5). Interestingly, a putative signal sequence in this enzyme resembled the eukaryotic sequences that require a suitable leader peptidase. However, this leader sequence was missing not only in homologous crotonyl-CoA hydratase/3-hydroxybutyryl-CoA dehydrogenases from autotrophic Desulfurococcales that possess only one membrane but also in the I. islandicus protein (SI Appendix, Fig. S6). As I. islandicus has the same two-membrane cellular structure as I. hospitalis (33), it seems unlikely that this putative signal peptide is essential for the enzyme trafficking. The incapability of the used signal peptide prediction tools to predict signal sequences in the proteins studied here or investigated in the past (Ihomp, ATPase) might be due to exceptional signal peptide pattern or the usage of an unknown secretion system. Furthermore, protein import pathways might be dependent on internal targeting signals as it was described for mitochondrial membrane proteins (40).
Structural and Functional Compartmentalization in Other Archaea?
The two-membrane structure of I. hospitalis cells was first regarded as a unique feature of Ignicoccus cells. However, an outer membrane as part of the cell envelope was also shown for other Archaea like Methanomassiliicoccus luminyensis, Candidatus Altiarchaeum hamiconnexum, and members of the proposed archaeal phylum Parvarchaeota, suggesting its widespread occurrence among Archaea (41–44). Whether these structures result in comparable functional compartmentalization as found in I. hospitalis remains an open question. So far, the compartmentalization of Ignicoccus cells combined with their functional separation of metabolic and anabolic pathways is unprecedented in the prokaryotic world.
Materials and Methods
Strains and Cultivation Conditions.
I. hospitalis was cultivated anaerobically in modified 1/2 SME medium [synthetic sea water (10, 11)] containing elemental sulfur and a gas phase consisting of H2/CO2 (250 kPa; 80:20 volume/volume). Serum bottles were inoculated with 0.2 mL logarithmically grown preparatory culture and incubated at 90 °C while shaking (50 rpm).
E. coli cells (BL21 Star [DE3], Thermo Fisher Scientific) were grown at 37 °C in lysogeny broth (LB) medium. The incubation was carried out while shaking (200 rpm), and ampicillin was added to the medium to a final concentration of 100 μg ⋅ mL−1.
Gene Synthesis.
The genes igni_0341 (PEP carboxylase), igni_1263 (malate dehydrogenase), igni_0132 (succinic semialdehyde reductase), and igni_1058 (crotonyl-CoA hydratase/(S)-3-hydroxybutyryl-CoA dehydrogenase) were synthesized by Invitrogen GeneArt (Thermo Fisher Scientific). The sequences were optimized for the expression in E. coli, and the sequence encoding C-terminal polyhistidine tag (6 × histidine) was integrated in front of the stop codon (SI Appendix, Table S3). The synthesized genes were cloned in the expression vector pRSETA_A185 (Thermo Fisher Scientific).
Heterologous Expression of I. hospitalis Genes in E. coli.
Recombinant I. hospitalis enzymes were produced in E. coli strain BL21 Star (DE3) (Thermo Fisher Scientific). For gene expression, the cells transformed with the corresponding plasmids were grown in 2 l LB medium containing ampicillin (100 µg/mL) at 37 °C while shaking (200 rpm). Expression was induced at optical density (OD600nm) of 0.6 with 1 mM isopropyl-β-D-thiogalactopyranoside. After induction, the temperature was lowered to 20 °C, and cells were harvested after additional growth for 4 h and stored at −20 °C until use.
Preparation of Cell Extracts.
Frozen cell pellets containing heterologously produced his6-tagged proteins Igni_0341, Igni_1263, Igni_0132, and Igni_1058 were thawed on ice and resuspended in 5 mL lysis-equilibration buffer (50 mM NaH2PO4 [pH 8] and 300 mM NaCl) per 1 g pelleted cells. Lysozyme was added to the resuspended cells to a final concentration of 1 mg/mL, and cells were then incubated on ice for 30 min while stirring the solution. The solution was sonicated on ice (HD 2070, Bandelin; 10 times for 15 s with cooling periods of 30 s). After sonication, deoxyribonuclease I (5 µg/mL) was added, and the cell suspension was incubated on ice for an additional 15 min under stirring. Finally, samples were centrifuged at 10,000 g for 30 min at 4 °C. The supernatant and pellet were separated in different tubes and further analyzed via SDS-PAGE. In case proteins were found in the supernatant (Igni_1263 and Igni_0132), the supernatant was used for protein purification under native conditions. In case proteins were detected in the pellet (Igni_0341 and Igni_1058), proteins were purified from the pellets under denaturing conditions.
Purification of Heterologously Expressed Proteins.
The heterologously produced his6-tagged proteins Igni_0341, Igni_1263, Igni_0132, and Igni_1058 were purified by affinity chromatography using Protino Ni-TED Resin (Macherey-Nagel GmbH) following the manufacturers’ instructions.
Purification of Igni_1263 and Igni_0132.
Heat precipitation (60 °C for 15 min) was used as an initial purification step after cell disruption, followed by centrifugation at 13,000 g for 15 min at 4 °C. The corresponding supernatants were directly incubated with Ni-TED resin in a 15-mL centrifugation tube, and the suspension was mixed on an orbital shaker for 15 min at 4 °C. The suspension was then centrifuged at 500 g for 1 min (4 °C), and the supernatant was discarded. The resin was washed in 1 mL lysis-equilibration buffer and transferred to a 2-mL reaction tube. The washing was repeated three times. The elution of proteins was performed by adding 200 µl lysis-equilibration buffer containing 500 mM imidazole (elution buffer). The suspension was incubated for 5 min in the elution buffer and centrifuged at 500 g for 5 min. This step was repeated two times. The combined elution fractions (containing purified enzymes) were mixed with glycerol (1:1) and stored at −20 °C.
Purification and Refolding of Igni_0341 and Igni_1058.
As recombinant Igni_0341 and Igni_1058 were insoluble, the disruption of inclusion bodies and protein purification were performed under denaturing conditions. For the solubilization of inclusion bodies, cell pellets were washed in 10 mL lysis-equilibration buffer per 1 g cell and centrifuged at 10,000 g for 30 min at 4 °C, and the supernatants were discarded. The resulting pellets containing inclusion bodies were then resuspended in 2 mL denaturing lysis-equilibration buffer containing 50 mM NaH2PO4 (pH 8), 300 mM NaCl, and 8 M urea per 1 g of pelleted cells. The inclusion bodies were sonicated (HD 2070, Bandelin) on ice 10 times for 15 s with a cooling periods of 30 s and were further dissolved by stirring on ice for 60 min. To remove insoluble materials, the dissolved inclusion bodies were centrifuged at 10,000 g for 30 min at room temperature. The supernatant was then transferred to a clean reaction tube and incubated with Ni-TED resin in a 15-mL centrifugation tube. The suspension was mixed on an orbital shaker for 15 min at 4 °C. The suspension was then centrifuged at 500 g for 1 min (4 °C), and the supernatant was discarded. The resin was washed in 1 mL denaturing lysis-equilibration buffer and transferred to a 2-mL reaction tube. Washing was repeated three times. The elution of proteins was performed by adding 200 µl denaturing lysis-equilibration buffer containing 500 mM imidazole (denaturing elution buffer). The suspension was incubated for 5 min in the denaturing elution buffer and centrifuged at 500 g for 5 min. This step was repeated two times.
The proteins purified under denaturing conditions were dialyzed stepwise in 500 mL buffer containing 5 mM Hepes (pH 7.5) and (at first) 2 M urea, and then 1 M urea, and no urea in a final step. For dialysis, Slide-A-Lyzer Dialysis Cassettes (10K, MWCO, Thermo Fisher Scientific) were filled with 1 mL purified enzyme. Buffers were exchanged at least three times, and each step was performed for 2 h. Dialyzed enzymes were finally mixed with glycerol (1:1) and stored at −20 °C.
Analysis of Purified Proteins.
Purified protein fractions were analyzed by SDS-PAGE and Western blotting. Western blot analysis was performed using mouse-anti-polyhistidine primary antibodies (dilution 1:500) and secondary goat-anti-mouse antibodies conjugated with horseradish peroxidase (dilution: 1:20,000). The identity of the purified recombinant proteins was confirmed using in-gel digestion by trypsin followed by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) tandem mass spectrometry (MS/MS) analysis (SI Appendix, Table S4). MALDI analysis was performed at the institute of Biochemistry (University of Regensburg) with a 4700 Proteomics Analyzer. The evaluation of data was accomplished with the software tool Mascot (45) (Matrix Science LTD.).
Enzyme Assays.
Enzyme assays were performed spectrophotometrically in 0.5-mL glass cuvettes (Hellma) at 365 nm (εNADH = 3.4 cm−1 ⋅ mM−1, εNADPH = 3.5 cm−1 ⋅ mM−1) (46). Unless otherwise indicated, malate dehydrogenase was measured at 45 °C, succinic semialdehyde reductase at 65 °C, and crotonyl-CoA hydratase/(S)-3-hydroxybutyryl-CoA dehydrogenase at 50 °C. For determination of KM and Vmax values, the software tool GraphPad Prism 6 (Statcon) was used.
Malate dehydrogenase was measured spectrophotometrically as oxaloacetate-dependent NAD(P)H oxidation in the reaction mixture containing 100 mM 3-(N-morpholino)propanesulfonic acid (MOPS) (pH 7), 5 mM MgCl2, 5 mM dithiothreitol (DTT), 0.3 mM NAD(P)H, and recombinant purified enzyme. The reaction was started by the addition of oxaloacetate (3 mM). For the determination of KM and Vmax values, the concentration of one substrate was varied, while the other substrates were kept at saturating concentrations (oxaloacetate: 0.01 to 2 mM, NADH/NADPH: 0.01 to 0.5 mM). For the determination of the substrate specificity of Igni_1263, pyruvate (10 mM), acetaldehyde (10 mM), propionaldehyde (10 mM), or butyraldehyde (10 mM) were added instead of oxaloacetate.
Succinic semialdehyde reductase was measured as succinic semialdehyde-dependent NAD(P)H oxidation in the reaction mixture containing 100 mM MOPS (pH 7), 5 mM MgCl2, 5 mM DTT, 0.5 mM NAD(P)H, and purified recombinant protein. The reaction was started by the addition of succinic semialdehyde (0.2 mM). For the determination of KM and Vmax values, the reaction mixture contained different concentrations of substrates or cosubstrates (succinic semialdehyde: 0.005 to 0.5 mM, NADH/NADPH: 0.005 to 0.5 mM). The reverse reactions were measured at 65 °C in the reaction mixture containing 100 mM Tris/HCl (pH 8.5), 5 mM MgCl2, 5 mM DTT, 5 mM MgCl2, 5 mM NAD, and purified recombinant protein. Assays were started with 10 mM 4-hydroxybutyrate or 5 mM 3-hydroxypropionate, respectively. For the determination of the substrate specificity of Igni_0132, acetaldehyde (10 mM), propionaldehyde (10 mM), or butyraldehyde (10 mM) were added instead of oxaloacetate. The malonic semialdehyde reductase activity of Igni_0132 was measured at 50 °C in a reaction mixture containing 100 mM MOPS (pH 7.8), 5 mM MgCl2, 5 mM DTT, 0.5 mM NADPH, 5 mM malonyl-CoA, and 2 U ⋅ ml−1 of recombinant malonyl-CoA reductase from Sulfolobus tokodai (47, 48). For the formation of malonic semialdehyde, the mixture was incubated for 10 min at room temperature. Afterward, the reaction was started by adding a recombinant I. hospitalis enzyme.
Crotonyl-CoA hydratase/(S)-3-hydroxybutyryl-CoA dehydrogenase activity was measured at 50 °C in a reaction mixture containing 100 mM Tris/HCl (pH 9.0), 5 mM NAD, 5 mM DTT, and purified recombinant protein. The reaction was started by the addition of 3 mM crotonyl-CoA or (S)-3-hydroxybutyryl-CoA, respectively. For the determination of KM and Vmax values, the reaction mixture contained different concentrations of substrates or cosubstrates (crotonyl-CoA: 0.05 to 1 mM, (S)-3-hydroxybutyryl-CoA: 0.05 to 1 mM, NAD(P): 0.01 to 1 mM).
Electron Microscopy.
For concentrating cells, they were enriched on ultrafiltration membranes as described previously (13). Afterward, cells were either prepared for room temperature sectioning or for cryosectioning (16). For room temperature sectioning, concentrated cells were applied on gold-plated specimen carriers and high-pressure frozen in an EM-PACT2 (Leica). Samples were freeze substituted in an EM AFS2 device (Leica) containing a substitution solution composed of 5% H2O, 0.5% (weight/volume) uranyl acetate, 0.5% (volume/volume) glutaraldehyde, and 94.5% acetone. Cells were either embedded in Epon 812 substitute resin (Fluka Chemie AG) or Lowicryl HM20 (Polysciences Inc.).
For freeze substitution and Epon embedding, we used the following protocol: −90 °C for 8 h, raising to −60 °C within 6 h; −60 °C for 8 h, raising to −30 °C within 4 h; and −30 °C for 4 h, raising to 0 °C within 4 h. Afterward, samples were washed three times with ice-cold acetone and infiltrated in acetone/Epon (2 + 1) for 1 h, acetone/Epon (2 + 1) for 1 h, acetone/Epon (1 + 1) for 2 h, acetone/Epon (1 + 2) for 20 h (all at 22 °C), and in freshly prepared Epon for 2 h at 30 °C. The polymerization of Epon was performed at 60 °C for 2 d. For freeze substitution followed by Lowicryl embedding, we used the following protocol: −90 °C for 8 h, raising to −60 °C within 1 h; −60 °C for 6 h, raising to −40 °C within 1 h; and −40 °C for 7 h. After washing cells three times with acetone, cells were infiltrated with Lowicryl HM20 in acetone/Lowicryl (2 + 1) for 2 h, acetone/Lowicryl (1 + 1) for 2 h, acetone/Lowicryl (1 + 2) for 2 h, and freshly prepared Lowicryl for 16 h at −40 °C. Lowicryl was polymerized under ultraviolet light at −40 °C for 2.5 d. For cryosectioning, cells were enriched via filtration and immediately fixed in 2% formaldehyde and 0.1% glutaraldehyde in phosphate-buffered saline buffer (PBS buffer, pH 7.0) for 30 min at room temperature. To remove aldehydes, fixed samples were centrifuged at 6,000 g for 15 min and washed with PBS buffer. Pellets were then covered in a droplet of 10% gelatin, gently mixed, and immediately transferred onto ice. After 15 min incubation on ice, gelatin-embedded cell pellets were cut into small cubes and infiltrated in 2.3 M sucrose overnight on a rotary incubator at 4 °C. The following day, infiltrated blocks were mounted on aluminum pins and frozen in liquid nitrogen.
Room temperature sections were cut with a UC7 μLtramicrotome, and cryosections were obtained from a UC7-cryo ultramicrotome (both obtained from Leica).
Transmission electron microscopy was performed using a 120 kV Philips CM12 (Thermo Fisher–FEI) equipped with a LaB6 cathode and a charge-coupled device slow-scan camera (TEM-1024, TVIPS) and alternatively used a JEM-2100F (JEOL GmbH) equipped with a field emission gun operated at 200 kV and a TemCam-F416 CMOS camera (TVIPS GmbH). The software EM-Menu 4.0 (TVIPS GmbH) was used for data acquisition and documentation. Micrographs were processed using ImageJ (49) and Photoshop CS5 (Adobe). The segmentation and 3D reconstruction of a semithin section with a nominal thickness of 200 nm was performed with Amira (Thermo Fisher Scientific) as described previously (13).
Antibodies.
Prior to immunization, purified proteins were in-gel trypsin digested and prepared for MALDI-TOF analysis. For immunization in rabbits, purified recombinant proteins were provided by Davids Biotechnologie GmbH. Preimmune sera and test sera were requested in advance and were analyzed by Western blotting and immunogold labeling. We generated antibodies against the proteins Igni_0341, Igni_1263, Igni_0132, and Igni_1058. The obtained antisera were used for Western blotting and immunogold labeling. In addition, we used commercially available antibodies from mice directed against dsDNA (Abcam) and mouse anti-polyhistidine tag antibodies (Sigma-Aldrich). As a secondary antibody for immunogold labeling, we used a goat anti-rabbit or a goat anti-mouse antibody coupled with ultrasmall gold particles (Aurion). For Western blot analysis, we used anti-rabbit and anti-mouse secondary antibodies linked to horseradish peroxidase (Sigma-Aldrich).
Immunogold Labeling.
Immunogold labeling was performed on ultrathin sections of resin-embedded cells and on thawed cryosections (16, 50). For immunogold labeling, primary antibodies were diluted from 1:100 to 1:1,000. Detection of the primary antibody was carried out with secondary antibodies linked to ultrasmall gold particles. Ultrasmall gold particles were afterward silver enhanced using R-Gent SE-EM silver enhancement reagents (Aurion).
Western Blot Analysis.
For Western blot analysis, proteins were transferred from a 10% polyacrylamide gel to a polyvinylidene membrane (pore size 0.45 µm) by semidry blotting (51). Primary antibodies were used in dilutions of 1:10,000 to 1:50,000; secondary antibodies were diluted from 1:10,000 to 1:50,000. Detection was carried out with Western Lightning Chemiluminescent Reagent (PerkinElmer) as stated by the manufacturer. The visualization of the peroxidase reaction was done with a Fusion FX 7 Imager (Vilber Lourmat).
Bioinformatics.
ProtParam (52) was used to calculate general features including molecular masses of expressed proteins. The Basic Local Alignment Search Tool analyses were carried out using the National Center for Biotechnology Information’s (NCBI) protein BLAST (BLASTP) (53) and the Joint Genome Institute’s Integrated Microbial Genomes system (54). To find signal peptides, Phobius and SignalP 4.1 were applied on sequences (55, 56). Sequence alignments for the crotonyl-CoA hydratase/(S)-3-hydroxybutyryl-CoA dehydrogenase of I. hospitalis and other members of the Desulfurococcales were obtained by using T-Coffee (57). Evolutionary analyses were conducted in Molecular Evolutionary Genetic Analysis X (58). To create the tree shown in SI Appendix, Fig. S1, a standard NCBI BLASTP search with Igni_0132 as a query was performed, and the first 100 hits were analyzed. The evolutionary history was inferred by using the maximum likelihood method and Jones–Taylor–Thornton (JTT) matrix-based model (59). The tree with the highest log likelihood (−30,059.92) is shown. The initial tree(s) for the heuristic search were obtained automatically by applying the Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the JTT model and then selecting the topology with superior log likelihood value. There was a total of 506 positions in the final dataset.
Supplementary Material
Acknowledgments
We thank Y. D. Stierhof, Tübingen for scientific support and for cryosectioning and G. Fuchs, Freiburg for discussions during this work and for his critical reading of the manuscript. The work of I.A.B. was funded by the Deutsche Forschungsgemeinschaft (BE 4822/5-1). R.R. and H.H. were also funded by the Deutsche Forschungsgemeinschaft (HU703/2).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022114118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Kerfeld C. A., Aussignargues C., Zarzycki J., Cai F., Sutter M., Bacterial microcompartments. Nat. Rev. Microbiol. 16, 277–290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuchs G., Alternative pathways of carbon dioxide fixation: Insights into the early evolution of life? Annu. Rev. Microbiol. 65, 631–658 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Berg I. A., Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl. Environ. Microbiol. 77, 1925–1936 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sánchez-Andrea I., et al., The reductive glycine pathway allows autotrophic growth of Desulfovibrio desulfuricans. Nat. Commun. 11, 5090 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jahn U., Huber H., Eisenreich W., Hügler M., Fuchs G., Insights into the autotrophic CO2 fixation pathway of the archaeon Ignicoccus hospitalis: Comprehensive analysis of the central carbon metabolism. J. Bacteriol. 189, 4108–4119 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huber H., et al., A dicarboxylate/4-hydroxybutyrate autotrophic carbon assimilation cycle in the hyperthermophilic Archaeum Ignicoccus hospitalis. Proc. Natl. Acad. Sci. U.S.A. 105, 7851–7856 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramos-Vera W. H., Berg I. A., Fuchs G., Autotrophic carbon dioxide assimilation in Thermoproteales revisited. J. Bacteriol. 191, 4286–4297 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berg I. A., Ramos-Vera W. H., Petri A., Huber H., Fuchs G., Study of the distribution of autotrophic CO2 fixation cycles in Crenarchaeota. Microbiology (Reading) 156, 256–269 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Berg I. A., et al., Autotrophic carbon fixation in archaea. Nat. Rev. Microbiol. 8, 447–460 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Paper W., et al., Ignicoccus hospitalis sp. nov., the host of ‘Nanoarchaeum equitans’. Int. J. Syst. Evol. Microbiol. 57, 803–808 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Huber H., et al., Ignicoccus gen. nov., a novel genus of hyperthermophilic, chemolithoautotrophic Archaea, represented by two new species, Ignicoccus islandicus sp nov and Ignicoccus pacificus sp nov. and Ignicoccus pacificus sp. nov. Int. J. Syst. Evol. Microbiol. 50, 2093–2100 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Huber H., Küper U., Daxer S., Rachel R., The unusual cell biology of the hyperthermophilic Crenarchaeon Ignicoccus hospitalis. Antonie van Leeuwenhoek 102, 203–219 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Heimerl T., et al., A complex endomembrane system in the archaeon Ignicoccus hospitalis tapped by Nanoarchaeum equitans. Front. Microbiol. 8, 1072 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Küper U., Meyer C., Müller V., Rachel R., Huber H., Energized outer membrane and spatial separation of metabolic processes in the hyperthermophilic Archaeon Ignicoccus hospitalis. Proc. Natl. Acad. Sci. U.S.A. 107, 3152–3156 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramos-Vera W. H., Weiss M., Strittmatter E., Kockelkorn D., Fuchs G., Identification of missing genes and enzymes for autotrophic carbon fixation in crenarchaeota. J. Bacteriol. 193, 1201–1211 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tokuyasu K. T., A technique for ultracryotomy of cell suspensions and tissues. J. Cell Biol. 57, 551–565 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kellenberger E., Dürrenberger M., Villiger W., Carlemalm E., Wurtz M., The efficiency of immunolabel on Lowicryl sections compared to theoretical predictions. J. Histochem. Cytochem. 35, 959–969 (1987). [DOI] [PubMed] [Google Scholar]
- 18.Burghardt T., Näther D. J., Junglas B., Huber H., Rachel R., The dominating outer membrane protein of the hyperthermophilic Archaeum Ignicoccus hospitalis: A novel pore-forming complex. Mol. Microbiol. 63, 166–176 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Meyer C., Heimerl T., Wirth R., Klingl A., Rachel R., The Iho670 fibers of Ignicoccus hospitalis are anchored in the cell by a spherical structure located beneath the inner membrane. J. Bacteriol. 196, 3807–3815 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braun T., et al., Archaeal flagellin combines a bacterial type IV pilin domain with an Ig-like domain. Proc. Natl. Acad. Sci. U.S.A. 113, 10352–10357 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burghardt T., et al., Insight into the proteome of the hyperthermophilic crenarchaeon Ignicoccus hospitalis: The major cytosolic and membrane proteins. Arch. Microbiol. 190, 379–394 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayer F., et al., AMP-forming acetyl coenzyme A synthetase in the outermost membrane of the hyperthermophilic crenarchaeon Ignicoccus hospitalis. J. Bacteriol. 194, 1572–1581 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang J. H., Oxidative damage of DNA induced by methylglyoxal in vitro. Toxicol. Lett. 145, 181–187 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Imanaka H., Fukui T., Atomi H., Imanaka T., Gene cloning and characterization of fructose-1,6-bisphosphate aldolase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Biosci. Bioeng. 94, 237–243 (2002). [DOI] [PubMed] [Google Scholar]
- 25.White R. H., Xu H., Methylglyoxal is an intermediate in the biosynthesis of 6-deoxy-5-ketofructose-1-phosphate: A precursor for aromatic amino acid biosynthesis in Methanocaldococcus jannaschii. Biochemistry 45, 12366–12379 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Say R. F., Fuchs G., Fructose 1,6-bisphosphate aldolase/phosphatase may be an ancestral gluconeogenic enzyme. Nature 464, 1077–1081 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Szwergold B. S., Maillard reactions in hyperthermophilic archaea: Implications for better understanding of non-enzymatic glycation in biology. Rejuvenation Res. 16, 259–272 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Koschnitzki D., et al., Questioning the radiation limits of life: Ignicoccus hospitalis between replication and VBNC. Arch. Microbiol. 203, 1299–1308 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brinsmade S. R., Paldon T., Escalante-Semerena J. C., Minimal functions and physiological conditions required for growth of Salmonella enterica on ethanolamine in the absence of the metabolosome. J. Bacteriol. 187, 8039–8046 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jakobson C. M., Tullman-Ercek D., Slininger M. F., Mangan N. M., A systems-level model reveals that 1,2-Propanediol utilization microcompartments enhance pathway flux through intermediate sequestration. PLoS Comput. Biol. 13, e1005525 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Teeseling M. C., Neumann S., van Niftrik L., The anammoxosome organelle is crucial for the energy metabolism of anaerobic ammonium oxidizing bacteria. J. Mol. Microbiol. Biotechnol. 23, 104–117 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Jahn U., Summons R., Sturt H., Grosjean E., Huber H., Composition of the lipids of Nanoarchaeum equitans and their origin from its host Ignicoccus sp. strain KIN4/I. Arch. Microbiol. 182, 404–413 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Rachel R., Wyschkony I., Riehl S., Huber H., The ultrastructure of Ignicoccus: Evidence for a novel outer membrane and for intracellular vesicle budding in an archaeon. Archaea 1, 9–18 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klingl A., S-layer and cytoplasmic membrane–Exceptions from the typical archaeal cell wall with a focus on double membranes. Front. Microbiol. 5, 624 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruiz N., Kahne D., Silhavy T. J., Advances in understanding bacterial outer-membrane biogenesis. Nat. Rev. Microbiol. 4, 57–66 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Pohlschröder M., Giménez M. I., Jarrell K. F., Protein transport in Archaea: Sec and twin arginine translocation pathways. Curr. Opin. Microbiol. 8, 713–719 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Natale P., Brüser T., Driessen A. J. M., Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane–Distinct translocases and mechanisms. Biochim. Biophys. Acta 1778, 1735–1756 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Podar M., et al., A genomic analysis of the archaeal system Ignicoccus hospitalis-Nanoarchaeum equitans. Genome Biol. 9, R158 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giannone R. J., et al., Proteomic characterization of cellular and molecular processes that enable the Nanoarchaeum equitans–Ignicoccus hospitalis relationship. PLoS One 6, e22942 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmidt O., Pfanner N., Meisinger C., Mitochondrial protein import: From proteomics to functional mechanisms. Nat. Rev. Mol. Cell Biol. 11, 655–667 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Probst A. J., et al., Biology of a widespread uncultivated archaeon that contributes to carbon fixation in the subsurface. Nat. Commun. 5, 5497 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Comolli L. R., Baker B. J., Downing K. H., Siegerist C. E., Banfield J. F., Three-dimensional analysis of the structure and ecology of a novel, ultra-small archaeon. ISME J. 3, 159–167 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Dridi B., Fardeau M. L., Ollivier B., Raoult D., Drancourt M., Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int. J. Syst. Evol. Microbiol. 62, 1902–1907 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Paul K., Nonoh J. O., Mikulski L., Brune A., “Methanoplasmatales,” Thermoplasmatales-related archaea in termite guts and other environments, are the seventh order of methanogens. Appl. Environ. Microbiol. 78, 8245–8253 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S., Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 (1999). [DOI] [PubMed] [Google Scholar]
- 46.Bergmeyer H. U., Neue Werte für die molaren Extinktions-Koeffizienten von NADH und NADPH zum Gebrauch im Routine-Laboratorium. Z. Klin. Chem. Klin. Biochem. 13, 507–508 (1975). [PubMed] [Google Scholar]
- 47.Alber B., et al., Malonyl-coenzyme A reductase in the modified 3-hydroxypropionate cycle for autotrophic carbon fixation in archaeal Metallosphaera and Sulfolobus spp. J. Bacteriol. 188, 8551–8559 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Otte J., Mall A., Schubert D. M., Könneke M., Berg I. A., Malonic semialdehyde reductase from the archaeon Nitrosopumilus maritimus is involved in the autotrophic 3-hydroxypropionate/4-hydroxybutyrate cycle. Appl. Environ. Microbiol. 81, 1700–1707 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flechsler J., et al., 2D and 3D immunogold localization on (epoxy) ultrathin sections with and without osmium tetroxide. Microsc. Res. Tech. 83, 691–705 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Towbin H., Staehelin T., Gordon J., Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. U.S.A. 76, 4350–4354 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gasteiger E., et al., “Protein identification and analysis tools on the ExPASy server” in The Proteomics Protocols Handbook, Walker J. M., Ed. (Springer Protocols Handbooks, Humana Press, Totowa, NJ, 2005), pp. 571–607. [Google Scholar]
- 53.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 54.Markowitz V. M., et al., IMG/M 4 version of the integrated metagenome comparative analysis system. Nucleic Acids Res. 42, D568–D573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Käll L., Krogh A., Sonnhammer E. L., A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338, 1027–1036 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Petersen T. N., Brunak S., von Heijne G., Nielsen H., Signal P., SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Di Tommaso P., et al., T-Coffee: A web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39, W13–W17 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar S., Stecher G., Li M., Knyaz C., Tamura K., MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jones D. T., Taylor W. R., Thornton J. M., The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282 (1992). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.