Significance
Poly(rC)-binding protein 1 (PCBP1) regulates multiple cellular processes; many studies have focused on its role in tumor formation and metastasis through its capacity to bind nucleic acids. PCBP1 binds to DNA and RNA and mediates interactions with other proteins that affect transcription and messenger RNA processing. PCBP1 is also an iron chaperone. It binds cytosolic iron as an Fe–glutathione complex and controls the chemical reactivity, sensing, and trafficking of iron in the cell. How are these distinct activities coordinated in a single protein? We identify the iron binding sites on PCBP1 and show that iron and RNA binding occur independently at separate sites. Iron- and RNA-binding activity separately function to prevent DNA damage and prevent cell death.
Keywords: hnRNP E1, ferritin, BolA2, DNA damage, RNA-binding protein
Abstract
Poly(rC)-binding protein (PCBP1) is a multifunctional adaptor protein that can coordinate single-stranded nucleic acids and iron–glutathione complexes, altering the processing and transfer of these ligands through interactions with other proteins. Multiple phenotypes are ascribed to cells lacking PCBP1, but the relative contribution of RNA, DNA, or iron chaperone activity is not consistently clear. Here, we report the identification of amino acid residues required for iron coordination on each structural domain of PCBP1 and confirm the requirement of iron coordination for binding target proteins BolA2 and ferritin. We further construct PCBP1 variants that lack either nucleic acid– or iron-binding activity and examine their functions in human cells and mouse tissues depleted of endogenous PCBP1. We find that these activities are separable and independently confer essential functions. While iron chaperone activity controls cell cycle progression and suppression of DNA damage, RNA/DNA-binding activity maintains cell viability in both cultured cell and mouse models. The coevolution of RNA/DNA binding and iron chaperone activities on a single protein may prove advantageous for nucleic acid processing that depends on enzymes with iron cofactors.
“Moonlighting” is a term for multifunctional proteins that are composed of a single polypeptide chain and exhibit multiple biological functions that are not due to gene fusions, alternative splicing, or multiple proteolytic fragments (1). The earliest identified multifunctional proteins were crystallins: Protein components of the crystallin lens of the eye that were found to be identical to enzymes of intermediate metabolism [e.g., lactate dehydrogenase in ducks (2)]. There are hundreds of known multifunctional proteins and they are a highly diverse group with multiple evolutionary origins; they may exhibit related or unrelated functions.
Poly(rC)-binding protein 1 (PCBP1) is a multifunctional protein that coordinates both RNA and iron. PCBP1 was initially identified as a component of heterogeneous nuclear ribonucleoprotein complexes and is also known as hnRNP E1 (3–5). PCBP1 binds to cytosine-rich motifs in RNA and single-stranded DNA and can affect multiple steps in messenger RNA (mRNA) processing, stability, and translation, as well as transcription, which ultimately results in altered levels of gene expression. This protein can simultaneously coordinate an RNA species and interact with other proteins that mediate the processing of the oligonucleotide. PCBP1 is proposed to function as a tumor suppressor gene, as it is significantly down-regulated in many types of cancer, and elevated expression may be associated with reduced epithelial–mesenchymal transition and metastatic potential (6). The PCBP1 locus also gives rise to a long noncoding RNA (PCBP1-AS1) that extends 462 bp upstream of the PCBP1 coding sequence and also appears to have a role in cancer progression. Elevated expression of PCBP1-AS1 was associated with metastasis in hepatocellular carcinoma (6), relapse in Hodgkin lymphoma (6, 7), but improved survival in glioma (8).
More recently, PCBP1 was independently identified as an iron chaperone in mammalian cells (9, 10). It binds iron from the chemically reactive, labile iron pool of the cytosol and delivers it to ferritin, the iron storage protein, and other iron-dependent enzymes via direct, metal-mediated protein–protein interactions. Target proteins for the iron chaperone include the nonheme iron enzymes, which contain mono- and dinuclear iron centers (11–13). PCBP1 forms an intermediate iron chaperone complex with BolA2, a component of the [2Fe-2S] cluster chaperone complex comprised of monothiol glutaredoxin Glrx3 and BolA2 (14–16). PCBP1 complexes with BolA2 via a bridging Fe(II) ligand, which is needed for the assembly of the [2Fe-2S] on Glrx3-Bola2 (17).
The PCBP family of hnRNPs is structurally composed of three hnRNP K-homology (KH) domains, which are ancient, conserved, and RNA-binding modules. Structure–function analysis of the third KH domain of PCBP1 revealed that Fe(II) is coordinated on KH3 by cysteine and glutamate residues and the free thiol of a noncovalently bound molecule of reduced glutathione (GSH) (17). These residues are physically separated from the RNA binding site previously characterized on KH domains (18). This separation raised the possibility that the RNA- and iron-binding activities of PCBP1 could be examined independently. Here, we report the identification of iron binding sites on each of the KH domains of PCBP1 and the construction of PCBP1 variants that lack either oligonucleotide- or iron-binding activities. Expression of these variants in cells and mouse livers depleted of wild-type PCBP1 (P1-WT) reveals the independent, essential activities of the iron chaperone and hnRNP.
Results
Binding of Oligonucleotide and Iron on Separate Faces of PCBP1.
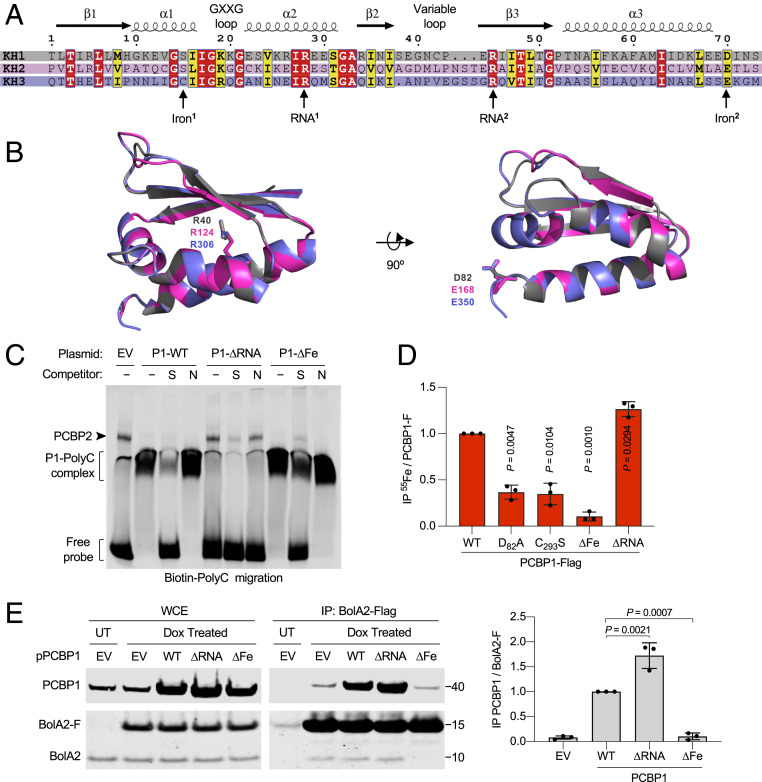
To separately examine the oligonucleotide- and iron-binding activities of PCBP1, we constructed variants containing amino acid substitutions in each KH domain that were designed to abrogate either RNA or iron coordination. Amino acid sequence alignment of individual KH domains of PCBP1 revealed >25% sequence similarity. Structural studies have revealed that two conserved arginine residues (RNA1 and RNA2) are involved in hydrogen bond formation and specific recognition of the three cytosine residues of the C-rich nucleic acid target sequence (Fig. 1A) (18). Examination of the superimposed structures of individual KH domains also revealed conserved orientation of arginine (RNA1) side chains (Fig. 1B). Recent mutagenesis analysis of the KH3 domain revealed that cysteine (Iron1) and glutamate (Iron2) residues are required for iron coordination (17). Although the cysteine residue is not conserved on KH1 or KH2, the glutamate (Iron2) residue is conserved on KH1 and KH2 domains (Fig. 1 A and B). We observed that residues involved in ligating oligonucleotides and iron are located on different surfaces of the PCBP1 KH domains, supporting the idea that these activities could be separated. Using site-directed mutagenesis, we constructed full-length PCBP1 variants that encode alanine substitutions for either RNA1- (∆RNA: R40A/R124A/R306A) or putative Iron2- (∆Fe: D82A/E168A/E350A) binding residues.
Fig. 1.
Identification of RNA- and iron-binding residues of PCBP1. (A) Sequence alignment of the three KH domains from PCBP1 illustrating secondary structure characteristics. (B) Structural comparison of three KH domains of PCBP1. Superimposed ribbon structures of KH1 (gray), KH2 (pink), and KH3 (purple) domains illustrating conserved ligands for RNA-binding (RNA1 residues: R40/R124/R306) and iron-binding (Iron2 residues: D82/E168/E350) residues as sticks. (C) Specific binding of PCBP1 variants to a poly(C) oligonucleotide probe. Biotin-labeled poly(C) oligonucleotide probe was mixed with HEK293 cell lysate transfected with EV or PCBP1-Flag (P1) variants (WT, ∆Fe: D82A/E168A/E350A, and ∆RNA: R40A/R124A/R306A) and separated by nondenaturing polyacrylamide gel electrophoresis. Specific [S, unlabeled poly(C)] or nonspecific [N, unlabeled poly(A)] oligonucleotide competitors were added, and migration of the biotin-labeled probe was analyzed by immunoblot using streptavidin-labeled antibody. (D) Requirement of iron-binding ligands for 55Fe binding to PCBP1. HEK293 cells were transfected with PCBP1-Flag WT, KH1 mutant (D82A), KH3 mutant (C293S), ∆Fe mutant, or ∆RNA mutant plasmids and labeled with 55FeCl3 for 16 h. Whole-cell extracts (WCE) and anti-Flag immune complexes were analyzed by immunoblot and scintillation counting. Specific (PCBP1-F) 55Fe binding was normalized to whole-cell 55Fe content and expressed relative to WT control. (E) Interaction of iron-binding PCBP1 variants and BolA2 in cells. Tetracycline-inducible stable cell lines expressing BolA2-Flag (BolA2-F) were transfected with plasmid (p), EV, or PCBP1 variants as indicated; treated overnight with buffer (UT) or doxycycline (Dox); and performed using anti-Flag IP. WCE and immune complexes were analyzed by immunoblot using antibodies for PCBP1 or BolA2 (Left), and the relative ratio of coprecipitated PCBP1 to BolA2-F in IP was quantified (Right). Data are means ± SD of n = 3 independent experiments. Significant P values, as determined by one-way ANOVA and unpaired t tests with Welch’s correction, are shown.
To investigate the ligand specificity of the PCBP1 variants, we measured the binding activity of a biotin-labeled, poly(C) oligonucleotide probe using an electrophoretic mobility-shift assay (EMSA) coupled with quantitative Western blotting (Fig. 1C and SI Appendix, Fig. S1). Human embryonic kidney 293 (HEK293) cells were transfected with plasmids encoding wild-type (WT) and variant PCBP1. Lysates from cells overexpressing P1-WT exhibited a high amount of PCBP1–poly(C) complex formation, compared with lysate from cells transfected with control vector (empty vector [EV]). The specificity of the PCBP1–poly(C) complex was confirmed by its disappearance when a 10-fold excess of unlabeled poly(C) oligo was added as a specific competitor (S), but presence when a 10-fold excess of control poly(A) oligo was added as a nonspecific competitor (N). In contrast, lysate from cells expressing the RNA-binding mutant (P1-∆RNA) exhibited very low amounts of protein–poly(C) complex formation. Lysate from cells expressing the PCBP1 iron-binding mutant (P1-∆Fe) showed levels of protein–poly(C) complex similar to P1-WT lysate. These data suggest that the PCBP1 RNA-binding mutant has lost poly(C) probe–binding activity and the PCBP1 iron-binding mutant has retained WT levels of poly(C) probe–binding activity.
Our previous study demonstrated that PCBP1 can bind cytosolic iron as an Fe(II)–GSH complex (17). To evaluate the iron-binding activity of PCBP1 variants, we transfected cells with plasmid-encoding PCBP1-Flag variants as indicated, labeled cells with 55FeCl3, and measured the amount of iron coprecipitating with PCBP1 (Fig. 1D). All variants were expressed at similar levels in cells (SI Appendix, Fig. S2). As previously observed, we detected 55Fe coprecipitating in P1-WT–Flag complexes isolated from cells. The D82A and C293S variants contain single mutations in the KH1 (D28A) or KH3 (C293S) domains, and each mutation lowered the level of bound iron to less than half that of WT PCBP1. These data suggest that while KH1 and KH3 contribute significantly to iron binding, the KH2 domain may not independently contribute to iron binding in cells. This observation is consistent with previous studies indicating that KH2 does not exhibit an iron- and GSH-dependent interaction with BolA2 (17). However, the ∆Fe iron-binding mutant exhibited a 90% reduction in the amount of bound 55Fe, compared with WT-P1, indicating that these three amino acid substitutions resulted in a PCBP1 variant without significant iron-binding activity. In contrast, we measured a 1.3-fold higher amount of 55Fe bound to the ∆RNA mutant, compared with WT-P1. These results identified critical iron-binding ligands of PCBP1 that are required for the formation of PCBP1–Fe complexes. Furthermore, they indicated that the iron- and RNA-binding activities of PCBP1 were separable and suggested that iron binding was enhanced in the absence of RNA-binding activity.
In human cells, PCBP1 and BolA2 form a complex in the presence of iron and GSH, and iron-binding residues on KH3 are required to form a KH3–BolA2 complex. To test whether the ∆RNA or ∆Fe variants of PCBP1 were able to form complexes with BolA2, we overexpressed the untagged PCBP1 variants in BolA2-Flag–inducible cell lines and measured the amount of PCBP1 coprecipitating with BolA2-Flag (Fig. 1E). Both the WT and ∆RNA mutant of PCBP1 were readily detectable in complex with BolA2-Flag, with the ∆RNA mutant consistently associating with BolA2 in higher amounts than WT. In contrast, BolA2-Flag failed to form a complex with the ∆Fe mutant of PCBP1, as the coprecipitating signal was not greater than the level of the EV control. These data suggest that iron-binding activity of PCBP1 is necessary for complex formation with BolA2. Collectively, it also suggests that the binding of one ligand may influence the coordination of the other.
Coordination of Iron and GSH on PCBP1 Necessary for Ferritin Binding and Iron Storage.
PCBP1 functions as a ferritin iron chaperone in the cytosol of mammalian cells by directly interacting with and facilitating iron accumulation into ferritin (9, 19). Whether PCBP1 requires iron-binding activity or other factors such as GSH to bind or load iron into ferritin remains to be determined. We transfected cells with plasmids encoding PCBP1-Flag variants or a control plasmid, treated cells with 100 µM ferric ammonium citrate (FAC) to induce endogenous ferritin, and measured the amount of ferritin coprecipitating with PCBP1 (Fig. 2A and SI Appendix, Fig. S3A). Ferritin was readily detectable in immune complexes from cells expressing WT-P1–Flag but not from cells containing an EV, confirming the specificity of the interaction. The amount of ferritin coprecipitating with the ∆RNA variant was modestly diminished (<20% reduction), compared with WT-P1–Flag, which likely reflects the lower level of expression of the ∆RNA variant in stable cell lines. In contrast, the amount of ferritin associated with the ∆Fe variant was markedly reduced (90% reduction), compared with WT-P1–Flag. These results indicate that the iron-binding activity of PCBP1 is required for ferritin interaction in cells.
Fig. 2.
Depletion of GSH impairs PCBP1-dependent iron incorporation into ferritin. (A) Interaction of endogenous ferritin with iron-binding PCBP1 variants in cells. HEK293 cells were transfected with plasmids (p), EV, or PCBP1-Flag (PCBP1-F) variants as indicated; treated overnight with buffer (UT) or 100 µM FAC; and performed using anti-Flag IP. Whole-cell extracts (WCE) and immune complexes were analyzed by immunoblot using antibodies for ferritin or Flag (Left), and the relative ratio of coprecipitated ferritin to PCBP1-F in IP was quantified (Right). (B) Requirement of GSH and iron for ferritin–PCBP1 complex stabilization. Tetracycline-inducible stable cell lines expressing PCBP1-F were treated with 100 µM FAC for 24 h and induced overnight with UT or doxycycline (Dox). Cells were lysed in IP UT ± 5 mM GSH or 100 μM chelator mix (desferrioxamine B [Dfo]/bathophenanthroline disulfonic acid [Bps]), as indicated. WCE and anti-Flag IPs were analyzed by immunoblot using antibodies for ferritin and Flag tag (Left) with quantitation (Right). (C) GSH depletion alters 55Fe incorporation into ferritin. Tetracycline-inducible stable cell lines expressing PCBP1-Flag were grown in control (NT) or 100 µM butathione sulfoximine (BSO) medium for 48 h. Cells were treated overnight with 10 µM FAC and UT or Dox (P1OX). Cells were labeled with 10 µM reduced 55Fe in serum-free medium for 2 h. (Left) WCE were analyzed by immunoblot using antibodies for ferritin or PCBP1, and accumulation of 55Fe in ferritin was analyzed by native gel electrophoresis and phosphorimaging. (Right) Relative ratio of 55Fe-ferritin content was quantified. Quantitative data are means ± SD of n = 3 to 4 independent experiments. Significant P values (P < 0.05), as determined by one-way ANOVA and unpaired t tests with Welch’s correction, are shown.
To identify ligands involved in complex formation between PCBP1 and ferritin, we induced ferritin expression and performed PCBP1-Flag coimmunoprecipitation (IP) in the presence or absence of GSH and/or iron. The ferritin–PCBP1 complex was stable to IP when isolated in buffer containing 5 mM GSH (Fig. 2B). In contrast, the addition of iron chelators and/or omission of GSH destabilized the complex, resulting in a 70 to 80% decrease in ferritin–PCBP1. These data suggest that both iron and GSH are required for stable ferritin–PCBP1 complex formation. Depletion of cellular GSH can induce an iron-dependent cell death pathway known as ferroptosis (20). The identification of GSH as a critical ligand for coordinating the cytosolic labile iron pool and for coordinating iron on PCBP1 suggests that GSH is directly involved in cellular iron trafficking. We tested whether GSH was involved in ferritin iron loading in human cells by depleting cellular GSH, loading cells with 55Fe, and measuring the amount of 55Fe loaded into endogenous cytosolic ferritin. HEK293 cells were treated with buthionine sulfoximine or buffer, and 90% depletion of cellular GSH was confirmed (SI Appendix, Fig. S3C). Interestingly, depletion of GSH tended to increase ferritin protein levels by 1.5-fold (SI Appendix, Fig. S3E), which may reflect changes in the sensing of the labile iron pool. To account for changes in ferritin expression, we examined 55Fe loading into ferritin with (Fig. 2C) and without (SI Appendix, Fig. S3D) normalization to ferritin protein. Cells depleted of GSH exhibited a 50% reduction in the amount of 55Fe incorporated into ferritin when compared with control cells, and the overexpression of PCBP1 did not restore the 55Fe ferritin–ferritin protein ratios to WT levels (Fig. 2C). The total amount of 55Fe loaded into ferritin was slightly increased in cells expressing induced levels of PCBP1, compared with control cells expressing endogenous PCBP1 with or without GSH depletion (Fig. 2C and SI Appendix, Fig. S3D), suggesting that PCBP1 overexpression produced small increases in 55Fe loading into ferritin. Although iron coordination on PCBP1 is GSH dependent, cells with 90% depletion of GSH still contain GSH at concentrations in the 200 to 500 μM range. At this level of GSH, Fe–GSH coordination by PCBP1 is likely to occur and could contribute to ferritin iron loading. These changes in ferritin iron loading were not due to lowered levels of intracellular 55Fe, because these levels did not change significantly when GSH was depleted. Collectively, these data suggest that both iron and GSH are required for stable PCBP1–ferritin complex formation and efficient iron incorporation into ferritin.
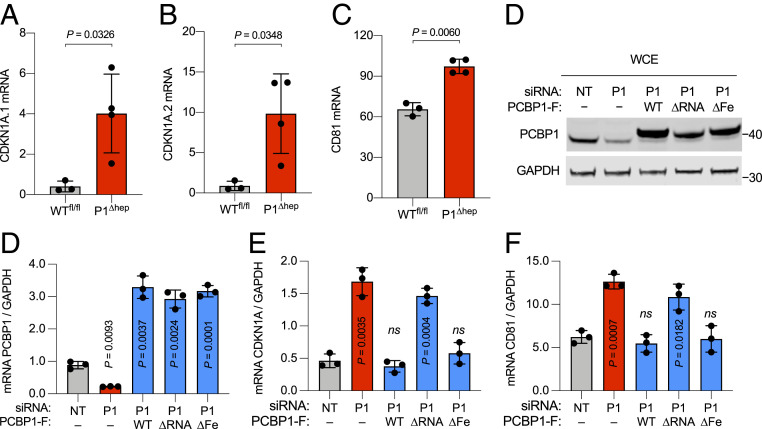
WT and ∆Fe PCBP1 Decrease Levels of mRNA Targets CDKN1A and CD81 but ∆RNA Does Not.
PCBP1 regulates the expression of a number of cellular and viral mRNAs at the levels of splicing, stability, and translation. Two of the mRNAs specifically bound and destabilized by PCBP1 are transcripts encoding the cyclin-dependent kinase inhibitor 1A (CDKN1A or p21WAF) and the transmembrane receptor CD81 (TAPA-1) (21, 22). By RNA sequence analysis, mice lacking PCBP1 in hepatocytes exhibited elevated levels of CDKN1A and CD81 transcripts (Fig. 3 A–C). We analyzed the mRNA destabilization activities of PCBP1 variants in stable cell lines using a combination of small interfering RNA (siRNA) depletion and inducible expression of PCBP1 variants. Efficient depletion of endogenous PCBP1 and expression of PCBP1 variants were confirmed by qPCR and immunoblot analyses (Fig. 3 D and E). Transcript levels of CDKN1A and CD81 were increased by 3- and 2.5-fold, respectively, in PCBP1-depleted HEK293 cells (Fig. 3 F and G). Induction of PCBP1 variants WT and ∆Fe reduced the mRNA levels of CDKN1A and CD81 to the level of nontargeting (NT) control. In contrast, induction of PCBP1 ∆RNA mutant had no effect on mRNA levels of CDKN1A and CD81, compared with cells lacking PCBP1. These results indicate that the RNA-binding activity of PCBP1 is required to regulate CDKN1A and CD81 transcript levels in cells. These results also suggest that the ∆Fe mutant of PCBP1 retained RNA-binding activity and was functionally active in HEK293 cells.
Fig. 3.
PCBP1 regulates Cdkn1A and CD81 transcript levels. (A–C) Induction of Cdkn1A and CD81 transcripts in PCBP1-deleted mouse livers were determined by RNA sequencing, expressed as reads per kilobase of transcript per million reads mapped. (D–G) RNA-binding activity of PCBP1 regulates Cdkn1A and CD81 transcript levels. Tetracycline-inducible stable cell lines expressing EV (−) or PCBP1 variants were transfected with NT or 3′UTR-PCBP1 (P1) siRNA and variant expression induced for 72 h. (D) Protein levels were analyzed by immunoblot using antibodies against PCBP1 and GAPDH. The transcripts’ abundance of Pcbp1 (E), Cdkn1A (F), and CD81 (G) were analyzed by real-time PCR (RT-PCR), normalized to GAPDH, and are shown as relative to NT-controlled siRNA-treated cells. Data are means ± SD of n = 3 or 4 independent experiments. Significant P values, as determined by one-way ANOVA and unpaired t tests with Welch’s correction, are shown. ns, not significant.
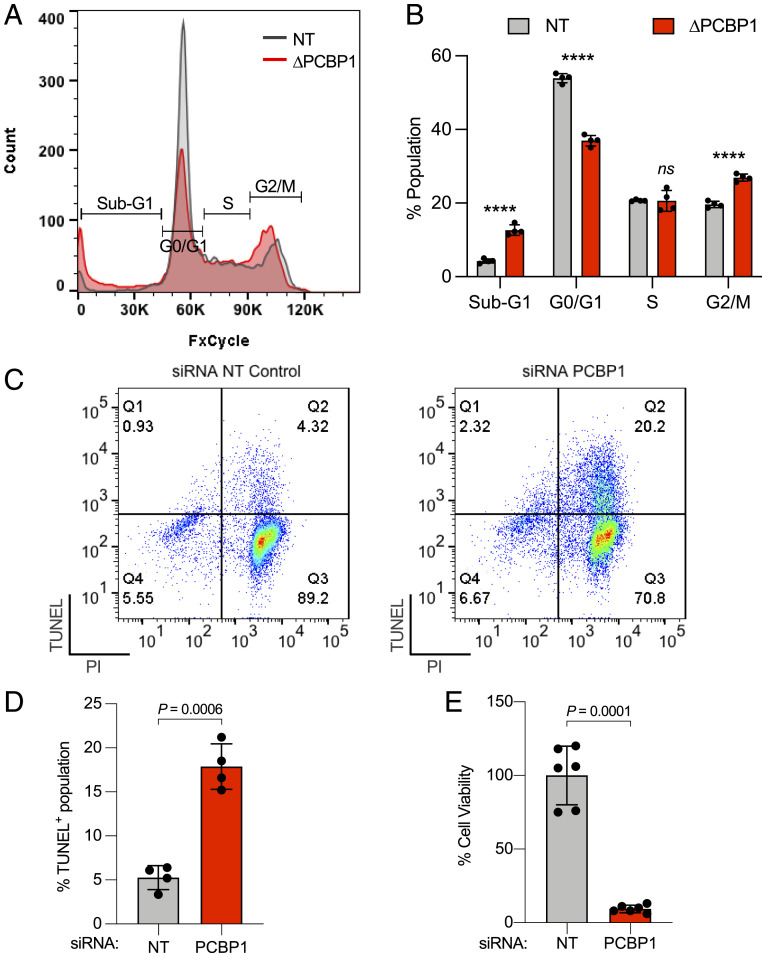
Depletion of PCBP1 Is Associated with Cell Cycle Delay, DNA Damage, and Loss of Viability in HEK Cells.
The observed increase in CDKN1A transcripts in PCBP1-depleted cells suggested that PCBP1 might influence cell cycle progression. To test this possibility, PCBP1 was depleted from HEK293 cells and cell cycle progression was evaluated by DNA staining and flow cytometry (Fig. 4 A and B). In comparison with NT siRNA transfection, we observed no significant change in cell density at 3-d posttransfection of PCBP1 siRNAs. However, flow cytometry revealed that PCBP1 depletion induced a cell cycle delay at the G2-M phase and promoted the appearance of a sub-G1 population, which contained less than a full complement of DNA. This sub-G1 induction was accompanied by a reciprocal decrease in cells in the G1 phase when PCBP1-depleted cells were compared with the NT control cells.
Fig. 4.
Depletion of PCBP1 alters cell cycle progression, induces DNA damage, and reduces cell viability. (A and B) Depletion of PCBP1 affects cell cycle progression. HEK293 cells were transfected with NT or PCBP1 (P1) siRNA for 48 h. Cells were fixed, stained, and analyzed by flow cytometry. Histogram showing sub-G1, G0/G1, S, and G2/M phase distribution (A) and quantitation (B) based on DNA content. (C and D) Depletion of PCBP1 induces DNA damage in cells. HEK293 cells were transfected with NT control or PCBP1 (∆PCBP1) siRNA for 48 h. Cells were fixed stained with TUNEL/propidium iodide (PI). (C) Representative image of flow cytometric dot plot analyses of cells after TUNEL/PI staining. Q1–Q4, quadrant 1–4. (D) Composite levels of TUNEL+ cells. (E) Depletion of PCBP1 reduces cell viability. HEK293 cells were transfected with NT or PCBP1 siRNA for 72 h. Cell viability was assessed using cell counting kit eight assays in duplicates. All data are means ± SD of n = 3 to 4 independent experiments. Significant P values, as determined by two-way ANOVA and Šídák’s multiple comparisons test, are shown in B. ****P < 0.0001, ns, not significant. For D and E, P values were determined by unpaired t test with Welch’s correction.
The expanded sub-G1 population in PCBP1-depleted cells corresponds to cells with DNA fragmentation and chromosome loss. We used the terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay to detect DNA strand breaks and fragmentation in PCBP1-depleted cells (Fig. 4 C and D). This assay revealed a fourfold increase in TUNEL+ cells when PCBP1 was depleted, compared with control cells. Finally, we measured cell viability following siRNA treatment and found a 90% reduction in viability of PCBP1-depleted cells when compared with NT controls (Fig. 4E). Thus, PCBP1 is essential for maintaining DNA integrity and viability in HEK293 cells.
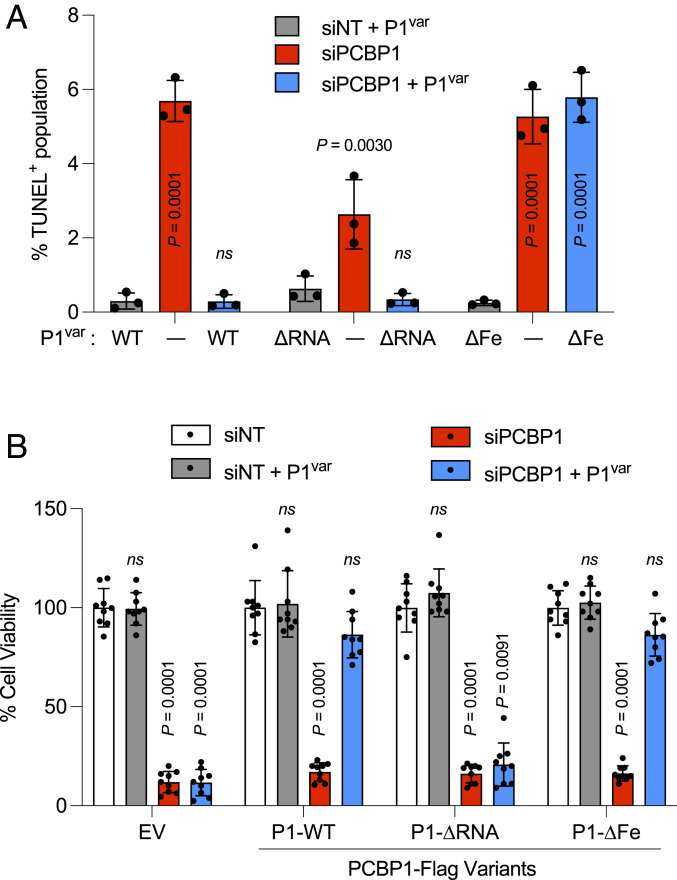
Suppression of DNA Damage and Restoration of Viability by Different PCBP1 Activities in HEK Cells.
To determine whether PCBP1 variants can suppress the DNA damage or loss of viability measured in PCBP1-depleted HEK293 cells, we again used a combination of siRNA-mediated depletion of endogenous PCBP1 and inducible expression of PCBP1 variants in stable cell lines. Again, the depletion of endogenous PCBP1 was associated with a G2-M delay and 4 to 15-fold increase in the frequency of TUNEL+ cells (Fig. 5A and SI Appendix, Fig. S5). This G2-M delay and increase in TUNEL+ cells was suppressed to control levels in PCBP1-depleted cells that were inducibly expressing the WT and ∆RNA variants but remained elevated in cells expressing the ∆Fe variant. In contrast, when cell viability was measured (Fig. 5B), the WT and ∆Fe variants restored viability, but the ∆RNA variant did not. Thus, the capacity of PCBP1 to suppress DNA damage and delay cell cycle and the capacity to support viability were due to separate and independent iron- and RNA-binding activities, respectively.
Fig. 5.
RNA- or iron-binding activity of PCBP1 are associated with different phenotypes in PCBP1-depleted cells. (A) Iron-binding activity of PCBP1 is required to suppress DNA damage in PCBP1-depleted cells. Tetracycline-inducible stable cell lines expressing PCBP1-Flag variants of WT, RNA-binding mutant (∆RNA), or iron-binding mutant (∆Fe) were transfected with NT or 3′UTR-PCBP1 siRNA and treated with (+P1var) or without (−) doxycycline to induce PCBP1 variants (P1var) for 48 h. Cells were fixed and stained with TUNEL/propidium iodide. The bar graph shows the percentage of TUNEL+ cells in each group. (B) RNA-binding activity of PCBP1 is required for cell viability in PCBP1-depleted cells. Tetracycline-inducible stable cell lines expressing EV or PCBP1-Flag variants (P1var) of WT, RNA-binding mutant (∆RNA), or iron-binding mutant (∆Fe) were transfected with NT or 3′UTR-PCBP1 siRNA and treated with doxycycline (+P1var) for 72 h, as indicated. Cell viability was assessed using cell counting kit eight assays in triplicates. All data are means ± SD of n = 3 independent experiments. Significant P values, as determined by two-way ANOVA and Dunnett’s multiple comparisons test, are shown. ns, not significant.
Suppression of Steatosis, DNA Damage, and Hepatocyte Death by Different PCBP1 Activities in Mouse Liver.
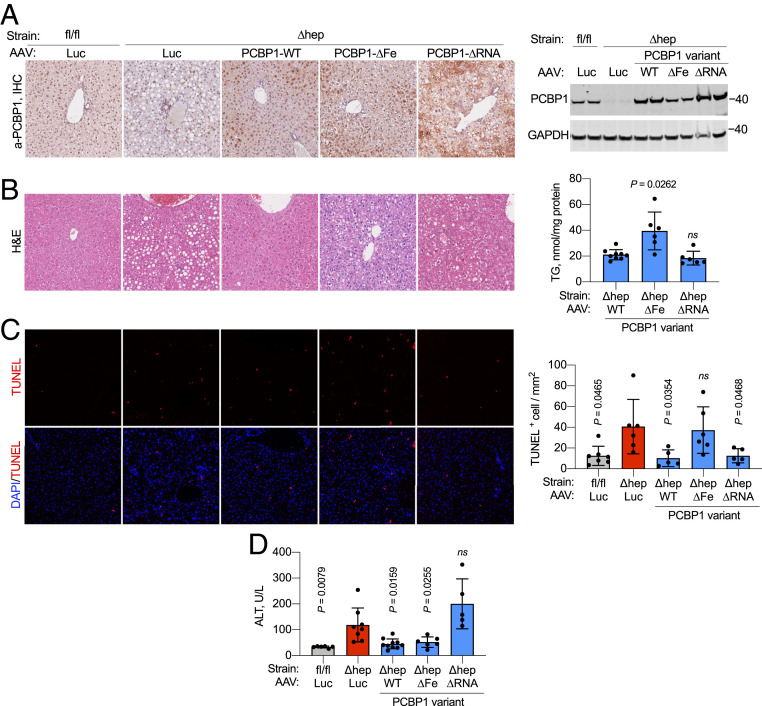
Mice with conditional deletion of PCBP1 in hepatocytes and cholangiocytes of the liver exhibit steatosis and ferroptotic cell death in hepatocytes due in part to unchaperoned iron, which catalyzes the formation of reactive oxygen species and oxidatively damaged lipids (13, 23). Whether loss of the RNA-binding activity of PCBP1 or loss of the PCBP1-adjacent lncRNA contributes to these phenotypes is not known. We constructed adeno-associated virus (AAV) serotype 8 particles to transduce hepatocytes and express PCBP1 variants under the control of a hepatocyte-specific promoter. Adult (8 wk) mice, WT or deleted for PCBP1 in the liver (PCBP1fl/fl and PCBP1 ∆hep mice, respectively), were injected with AAVs and then placed on purified synthetic diets supplemented with 48 ppm ferric iron (normal iron diet) for 16 to 18 d prior to analysis. Expression of PCBP1 variants was confirmed by immunohistochemistry, immunoblotting, and qPCR (Fig. 6A and SI Appendix, Fig. S6). By immunohistochemistry (Fig. 6 A, Left), endogenous PCBP1 was uniformly expressed across the liver lobule. AAV-transduced expression of PCBP1 variants tended to be lower in periportal zones, as previously reported (24), but was detected at levels at or above endogenous PCBP1 throughout the liver. By immunoblotting (Fig. 6 A, Right), each of the PCBP1 variants was expressed at levels at or above the level of endogenous PCBP1, although the PCBP1-∆Fe variant was consistently detected at lower levels than the WT or ∆RNA variants. Steatosis was apparent in hematoxylin- and eosin-stained sections of PCBP1 ∆hep livers transduced with either control (luciferase) or iron-binding mutant (PCBP1-∆Fe) AAVs (Fig. 6 B, Left), but steatosis was reduced in livers transduced with WT and ∆RNA variants. Measurement of triglyceride levels in transduced PCBP1 ∆hep livers (Fig. 6 B, Right) confirmed that transduction with the ∆Fe variant failed to suppress triglyceride levels, while transduction with the WT and ∆RNA variants effectively lowered lipid levels. The reduction in triglycerides with WT and ∆RNA variants also suggested that the PCBP1-associated lncRNA, which was deleted in the PCBP1 ∆hep mice and not restored by AAV transduction, was not required to suppress steatosis.
Fig. 6.
RNA- and iron-binding activities of PCBP1 are associated with different phenotypes in PCBP1-depleted hepatocytes. (A) Transduction of AAV constructs with PCBP1 WT, RNA-binding mutant (∆RNA), or iron-binding mutant (∆Fe) in PCBP1 WT (fl/fl) and deleted (∆hep) livers. (Left) Representative immunohistochemistry showing the distribution of PCBP1 in AAV-transduced livers. AAV-luciferase (Luc) was used as negative control. Shown is a representative immunoblot of PCBP1 in liver lysates from PCBP1fl/fl and PCBP1 ∆hep mice injected with the indicated AAV constructs. (Right) GAPDH used as a loading control. (B) Persistence of steatosis in livers transduced with PCBP1 ∆Fe. (Left) Hematoxylin and eosin (H&E) staining of PCBP1fl/fl and PCBP1 ∆hep mice injected with AAVs. (Right) Total acylglycerols (TG) in livers of PCBP1∆hep mice injected with the PCBP1 variant AAVs. (C) Increased TUNEL+ cells in liver from PCBP1 ∆hep mice and PCBP1 ∆hep mice expressing ∆Fe mutant. Representative images of liver sections stained with TUNEL and DAPI (Left) with quantitation (Right). (D) RNA-binding activity of PCBP1 is required to suppress hepatocyte cell death. Plasma ALT levels as units per liter (U/L) in PCBP1fl/fl and PCBP1 ∆hep mice transduced with AAVs. Data represent means ± SD; P values calculated using one-way ANOVA and unpaired t test with Welch’s correction. ns, not significant. See also SI Appendix, Fig. S6.
We next examined whether PCBP1-deleted livers exhibited DNA damage similar to that of PCBP1-depleted HEK cells. TUNEL staining of PCBP1fl/fl and PCBP1 ∆hep livers demonstrated a threefold increase in TUNEL+ cells in livers deleted for PCBP1, confirming that DNA damage was also occurring in the PCBP1 ∆hep livers. Transduced expression of WT and ∆RNA variants suppressed the DNA damage, but expression of the ∆Fe variant failed to lower the number of TUNEL+ cells. These data indicated that the iron chaperone activity of PCBP1 was critical in suppressing DNA strand breaks in both cultured cells and mouse liver. Furthermore, the appearance of DNA damage could not be attributed to loss of PCBP1-mediated RNA or DNA binding. Finally, we measured plasma levels of alanine aminotransferase (ALT), a liver-specific enzyme that is released into the blood by dying hepatocytes and is elevated in PCBP1 ∆hep mice. Surprisingly, transduction with the PCBP1 WT and ∆Fe variants lowered ALT levels to those of PCBP1fl/fl mice, but the ∆RNA variant failed to lower the ALT levels. These results, similar to the viability data in HEK cells, indicated that DNA strand breaks did not contribute to hepatocyte cell death in PCBP1 ∆hep livers and suggests that the RNA-binding activity of PCBP1 is required for WT levels of viability in both mouse hepatocytes and HEK cells.
Discussion
We present data demonstrating that the iron chaperone and oligonucleotide-binding activities of PCBP1 are separable and individually account for phenotypes observed in genetic-loss-of-function models. Initial studies of the in vitro iron-binding activity of PCBP1 indicated a stoichiometry of 3 Fe(II):1 protomer with micromolar affinity but did not determine individual iron binding sites (9). More recently, cell-based and in vitro studies determined that the KH3 domain of PCBP1 binds iron as an Fe(II)–GSH complex (17). Individual amino acid residues needed for coordination of the Fe(II) (Cys293 and Glu350) and GSH (Asn301, Glu304, and Arg346) ligands were identified on KH3. Based on structural homology modeling, we now identify potential iron-coordinating residues Asp82 and Glu168 on KH1 and KH2, respectively. The PCBP1 variant, in which the conserved Asp and Glu residues on each KH domain are mutated, completely lacked iron-, BolaA2-, and ferritin-coordination activities, confirming that the iron-binding sites on each KH domain were effectively disrupted and that iron coordination was necessary for BolA2 and ferritin binding. We also demonstrated that GSH depletion interfered with PCBP1–ferritin interaction and efficient iron loading, further supporting the role of GSH in the coordination of the cytosolic labile iron pool.
Iron Chaperone and RNA-Binding Activities Are Separable yet Independently Essential.
As expected, we determined that mutation of a single critical arginine residue on each KH domain of PCBP1 completely abrogated RNA-binding activity in vitro and in cells. Perhaps less expected was the finding that RNA- and iron-binding activities are completely separable, and the ∆RNA variant has levels of iron chaperone activity at or better than the P1-WT. Similarly, the ∆Fe variant has RNA-binding activity at the level of P1-WT. The increases in iron binding by the RNA mutant and RNA binding by the iron mutant suggest that protein–protein interactions, mediated by binding the iron or RNA ligands, could impair binding by the other ligand. Thus, the effective iron-binding capacity of the PCBP1 RNA mutant is increased in the absence of competing RNA-mediated interactions. The analogous condition likely increased RNA-binding activity in the PCBP1 iron mutant.
PCBP1 directly interacts with and destabilizes CDKN1a mRNA, which encodes a cyclin-dependent kinase inhibitor that regulates the cell cycle and the response to DNA damage. PCBP1-depleted cells exhibited alterations in cell cycle progression and increased DNA damage. Surprisingly, the RNA-binding activity of PCBP1 was not required to correct these alterations. Furthermore, RNA-binding activity was also not required to suppress the DNA damage in PCBP1-depleted mouse livers. Instead, the iron chaperone activity was required in both models. These data raise the possibility that some phenotypes attributed to PCBP1 (e.g., tumor suppressor activity) may involve the iron chaperone functions, as well as its nucleic acid–binding activity. PCBP1-depleted HEK cells exhibited a profound loss of viability. Although PCBP1 RNA-binding activity was not required to suppress DNA damage, this activity was needed to maintain viability of HEK cells and to suppress the release of ALT, a marker of hepatocyte turnover, in the PCBP1-depleted mouse livers. Clearly, this mixture of loss-of-function phenotypes suggests a complex set of essential functions for PCBP1.
Repeated Evolution of Iron-Binding and Nucleic Acid–Binding Regulatory Activities in a Single Protein.
PCBP1 is not the first example of an iron-binding protein that also binds to nucleic acid and regulates gene expression. Iron regulatory protein 1 (IRP1) is a well-characterized example of a protein that binds specifically to mRNA structures and affects the translation or stability of transcripts encoding proteins of iron homeostasis. Incorporation of a [4Fe-4S] cluster in the RNA binding site converts IRP1 to the enzyme cytosolic aconitase (25). A second example is the nuclear coactivator 4 (NCOA4) transcription factor, which associates with MCM7 in the nucleus to control replication (26). NCOA4 is also a cargo receptor in the cytosol that binds ferritin and mediates its autophagy (27, 28). NCOA4 is itself regulated by iron binding, as the iron-bound NCOA4 undergoes ubiquitin-mediated degradation (29, 30). We add to this list PCBP1, which controls iron trafficking in the cytosol and patterns of gene expression through RNA and DNA binding. Why would these iron-binding activities evolve in proteins associated with nucleic acids, which are sensitive to iron-mediated damage? Perhaps iron binding offers a level of protection against iron-mediated oxidative damage. Interestingly, several components of the DNA replication and repair machinery require coordination of a [4Fe-4S] cluster for full activity (31) and iron cofactors are required for transfer RNA, ribosomal RNA, and nucleobase modifications (32, 33). The capacity of PCBP1 to bind nucleic acid and iron may enhance the assembly or repair of iron cofactors in DNA- and RNA-modifying enzymes.
Materials and Methods
Detailed materials and methods may be found in SI Appendix, Supplementary Materials and Methods.
Plasmids, Cell Lines, and Media.
Multiple substitutions were introduced into plasmids pcDNA5-PCBP1 and pcDNA5-Flag-PCBP1 to generate RNA-binding mutant (∆RNA: R40A/R124A/R306A), iron-binding mutant (∆Fe: D82A/E168A/E350A), and D82A or C293S variants. All plasmids were confirmed by DNA sequencing. HEK293 cells were grown in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum, 50 U/mL penicillin G, and 50 μg/mL streptomycin. Tetracycline-inducible Flp-In T-Rex 293 cell lines were constructed according to the manufacturer’s instructions using pcDNA5-Flag-PCBP1 variants. Where indicated, doxycycline was used at 1 μg/mL, or iron was used as FAC. Protein depletion by siRNA was performed using Stealth siRNA according to manufacturer’s protocol, as described (17).
Electrophoretic Mobility-Shift Assays.
HEK293 cells transfected with indicated plasmids were lysed in IP buffer and analyzed by EMSA and Western blotting with anti-FLAG antibody. EMSA contained 10 μg cell lysate and 0.5 µM biotin-poly(C) probe with or without 5 µM unlabeled poly(C) probe as S or 5 μM poly(A) probe as N. Reactions were separated on native gel, transferred to PVDF, and biotin-labeled oligonucleotides detected with IR800-labeled streptavidin.
55Fe Labeling, IPs, GSH Depletion, and Ferritin–55Fe Incorporation and Immunoblotting.
For iron labeling, HEK293 cells were treated with 2 to 5 μM 55FeCl3 for 16 h. IP was performed, and retained 55Fe was quantitated by liquid scintillation counting. Immunoblotting was performed, as previously described (17). Where indicated, GSH was removed from IP buffer or iron chelators desferrioxamine B and bathophenanthroline disulfonic acid added at 100 μM. For cellular GSH depletion, HEK293 cell lines were grown in medium supplemented with 100 µM buthionine sulphoximine for 48 h and doxycycline for 24 h. For ferritin-55Fe, labeling cells in serum-free medium were treated with reduced 55Fe (10 µM 55FeCl3 + 100 µM ascorbic acid) for 2 h. Cells were lysed and proteins separated on native gels. 55Fe ferritin in dried gels was quantified by phosphorimaging.
Assays.
RNA extraction and real-time PCR were performed, as described. For cell cycle analyses, HEK293 cells were transfected with siRNA for 48 h, fixed in 4% paraformaldehyde, stained with FxCycle Violet ready flow reagent, and analyzed by flow cytometry. For TUNEL assay, HEK cell lines, transfected with siRNAs and expressing PCBP1 variants, were fixed and analyzed using the APO-BrdU TUNEL Assay Kit (Invitrogen) and flow cytometry. Cell viability was measured using the cell counting kit 8 method.
Animal Studies.
All animal study protocols were reviewed and approved by the National Institute of Diabetes and Digestive and Kidney Diseases Animal Care and Use Committee and performed in compliance with NIH guidelines for the humane care of animals. The 8-wk-old male mice with genotypes PCBP1fl/fl (WT) and Tg (Alb-Cre) PCBP1fl/fl were injected via tail vein with AAV serotype 8 constructs expressing PCBP1 variants under the control of the thyroxine-binding globulin promoter. Mice were placed on purified diets (Envigo TD.80394) supplemented with 48 ppm iron for 16 to 18 d prior to euthanasia and analysis. Plasma was collected and analyzed for ALT; liver samples were collected and analyzed by real-time PCR, quantitative Western blotting, and immunohistochemistry, as described (23). TUNEL assay on fixed liver sections was performed using In Situ Cell Death Detection Kit, TMR-Red (Roche) according to manufacture instructions. TUNEL+ nuclei were counted in the whole section and expressed as TUNEL+ cells per square millimeter.
Quantification and Statistical Analysis.
Data were analyzed using Prism 9 (GraphPad) and reported as means ± SD. Outliers were identified using the integrated robust regression and outlier removal method and excluded. Differences among groups were determined by Brown–Forsythe and Welch’s one-way ANOVA test followed by unpaired Student’s t test with Welch’s correction for unequal variance. Where indicated, two-way ANOVA followed by Dunnett’s or Šídák’s tests for multiple comparisons were performed.
Supplementary Material
Acknowledgments
We thank Ankit Saxena of the Flow Cytometry Core Facility of the National Heart, Lung, and Blood Institute; J. Reese of the Advanced Light Microscopy and Image Analysis Core; and O. Gavrilova of the Mouse Metabolism Core of the National Institutes of Diabetes and Digestive and Kidney Diseases, NIH. These studies were supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2104666118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Jeffery C. J., Protein moonlighting: What is it, and why is it important? Philos. Trans. R. Soc. Lond. B Biol. Sci. 373, 20160523 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wistow G. J., Mulders J. W., de Jong W. W., The enzyme lactate dehydrogenase as a structural protein in avian and crocodilian lenses. Nature 326, 622–624 (1987). [DOI] [PubMed] [Google Scholar]
- 3.Dreyfuss G., Philipson L., Mattaj I. W., Ribonucleoprotein particles in cellular processes. J. Cell Biol. 106, 1419–1425 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grelet S., Howe P. H., hnRNP E1 at the crossroads of translational regulation of epithelial-mesenchymal transition. J. Cancer Metastasis Treat. 5, 16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X., et al., Multilevel regulation and molecular mechanism of poly (rC)-binding protein 1 in cancer. FASEB J. 34, 15647–15658 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Luo T., et al., lncRNA PCBP1-AS1 aggravates the progression of hepatocellular carcinoma via regulating PCBP1/PRL-3/AKT pathway. Cancer Manag. Res. 12, 5395–5408 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang Y., et al., Construction of relapse-related lncRNA-mediated ceRNA networks in Hodgkin lymphoma. Arch. Med. Sci. 16, 1411–1418 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luan F., et al., An autophagy-related long non-coding RNA signature for glioma. FEBS Open Bio 9, 653–667 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi H., Bencze K. Z., Stemmler T. L., Philpott C. C., A cytosolic iron chaperone that delivers iron to ferritin. Science 320, 1207–1210 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Philpott C. C., Ryu M.-S., Frey A., Patel S., Cytosolic iron chaperones: Proteins delivering iron cofactors in the cytosol of mammalian cells. J. Biol. Chem. 292, 12764–12771 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frey A. G., et al., Iron chaperones PCBP1 and PCBP2 mediate the metallation of the dinuclear iron enzyme deoxyhypusine hydroxylase. Proc. Natl. Acad. Sci. U.S.A. 111, 8031–8036 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nandal A., et al., Activation of the HIF prolyl hydroxylase by the iron chaperones PCBP1 and PCBP2. Cell Metab. 14, 647–657 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Philpott C. C., Patel S. J., Protchenko O., Management versus miscues in the cytosolic labile iron pool: The varied functions of iron chaperones. Biochim. Biophys. Acta Mol. Cell Res. 1867, 118830 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banci L., Camponeschi F., Ciofi-Baffoni S., Muzzioli R., Elucidating the molecular function of human BOLA2 in GRX3-dependent anamorsin maturation pathway. J. Am. Chem. Soc. 137, 16133–16143 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Frey A. G., Palenchar D. J., Wildemann J. D., Philpott C. C., A Glutaredoxin·BolA complex serves as an iron-sulfur cluster chaperone for the cytosolic cluster assembly machinery. J. Biol. Chem. 291, 22344–22356 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H., Mapolelo D. T., Randeniya S., Johnson M. K., Outten C. E., Human glutaredoxin 3 forms [2Fe-2S]-bridged complexes with human BolA2. Biochemistry 51, 1687–1696 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel S. J., et al., A PCBP1-BolA2 chaperone complex delivers iron for cytosolic [2Fe-2S] cluster assembly. Nat. Chem. Biol. 15, 872–881 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sidiqi M., et al., Formation of an alphaCP1-KH3 complex with UC-rich RNA. Eur. Biophys. J. 34, 423–429 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Ryu M.-S., Zhang D., Protchenko O., Shakoury-Elizeh M., Philpott C. C., PCBP1 and NCOA4 regulate erythroid iron storage and heme biosynthesis. J. Clin. Invest. 127, 1786–1797 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stoyanovsky D. A., et al., Iron catalysis of lipid peroxidation in ferroptosis: Regulated enzymatic or random free radical reaction? Free Radic. Biol. Med. 133, 153–161 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waggoner S. A., Johannes G. J., Liebhaber S. A., Depletion of the poly(C)-binding proteins alphaCP1 and alphaCP2 from K562 cells leads to p53-independent induction of cyclin-dependent kinase inhibitor (CDKN1A) and G1 arrest. J. Biol. Chem. 284, 9039–9049 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trifillis P., Day N., Kiledjian M., Finding the right RNA: Identification of cellular mRNA substrates for RNA-binding proteins. RNA 5, 1071–1082 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Protchenko O., et al., Iron chaperone poly rC binding protein 1 protects mouse liver from lipid peroxidation and steatosis. Hepatology 73, 1176–1193 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bell P., et al., Inverse zonation of hepatocyte transduction with AAV vectors between mice and non-human primates. Mol. Genet. Metab. 104, 395–403 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rouault T. A., Maio N., Biogenesis and functions of mammalian iron-sulfur proteins in the regulation of iron homeostasis and pivotal metabolic pathways. J. Biol. Chem. 292, 12744–12753 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bellelli R., et al., NCOA4 transcriptional coactivator inhibits activation of DNA replication origins. Mol. Cell 55, 123–137 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Dowdle W. E., et al., Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat. Cell Biol. 16, 1069–1079 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Mancias J. D., Wang X., Gygi S. P., Harper J. W., Kimmelman A. C., Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 509, 105–109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mancias J. D., et al., Ferritinophagy via NCOA4 is required for erythropoiesis and is regulated by iron dependent HERC2-mediated proteolysis. eLife 4, e10308 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryu M.-S., Duck K. A., Philpott C. C., Ferritin iron regulators, PCBP1 and NCOA4, respond to cellular iron status in developing red cells. Blood Cells Mol. Dis. 69, 75–81 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barton J. K., Silva R. M. B., O’Brien E., Redox chemistry in the genome: Emergence of the [4Fe4S] cofactor in repair and replication. Annu. Rev. Biochem. 88, 163–190 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fedeles B. I., Singh V., Delaney J. C., Li D., Essigmann J. M., The AlkB family of Fe(II)/α-Ketoglutarate-dependent dioxygenases: Repairing nucleic acid alkylation damage and beyond. J. Biol. Chem. 290, 20734–20742 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kimura S., Suzuki T., Iron-sulfur proteins responsible for RNA modifications. Biochim. Biophys. Acta 1853, 1272–1283 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.