Significance
Cells have evolved multiple mechanisms restraining the usage of lower-fidelity DNA repair pathways and polymerases. This work focuses on how deregulated Replication Protein A (RPA)–single-stranded DNA (ssDNA) assembly may impact the fidelity of genome duplication and repair. The absence of Rtt105 causes lower nuclear levels of RPA and impaired RPA-ssDNA assembly, resulting in increased replication errors with large deletions or duplications and elevated usage of the low-fidelity repair pathways. We show that this mechanism is likely conserved between yeast and humans. Thus, our work could provide insights into how human cells ensure high-fidelity DNA replication and repair. Furthermore, our study could potentially offer explanations for the genetic basis of some genome arrangements in human diseases.
Keywords: Rtt105, RPA, replication fidelity, homologous recombination, genome stability
Abstract
Single-stranded DNA (ssDNA) covered with the heterotrimeric Replication Protein A (RPA) complex is a central intermediate of DNA replication and repair. How RPA is regulated to ensure the fidelity of DNA replication and repair remains poorly understood. Yeast Rtt105 is an RPA-interacting protein required for RPA nuclear import and efficient ssDNA binding. Here, we describe an important role of Rtt105 in high-fidelity DNA replication and recombination and demonstrate that these functions of Rtt105 primarily depend on its regulation of RPA. The deletion of RTT105 causes elevated spontaneous DNA mutations with large duplications or deletions mediated by microhomologies. Rtt105 is recruited to DNA double-stranded break (DSB) ends where it promotes RPA assembly and homologous recombination repair by gene conversion or break-induced replication. In contrast, Rtt105 attenuates DSB repair by the mutagenic single-strand annealing or alternative end joining pathway. Thus, Rtt105-mediated regulation of RPA promotes high-fidelity replication and recombination while suppressing repair by deleterious pathways. Finally, we show that the human RPA-interacting protein hRIP-α, a putative functional homolog of Rtt105, also stimulates RPA assembly on ssDNA, suggesting the conservation of an Rtt105-mediated mechanism.
Faithful DNA replication and repair are essential for the maintenance of genetic material (1). Even minor defects in replication or repair can cause high loads of mutations, genome instability, cancer, and other diseases (1). Deficiency in different DNA repair or replication proteins can lead to distinct mutation patterns (2–4). For example, deficiency in mismatch repair results in increased microsatellite instability, while deficiency in homologous recombination repair is often associated with tandem duplications or deletions (3–7). Sequence analysis of various cancer types has identified many distinct genome rearrangement and mutation signatures (8). However, the genetic basis for some of these signatures remains poorly understood, thus requiring further investigation in experimental models (8).
In eukaryotic cells, Replication Protein A (RPA), the major single-stranded DNA (ssDNA) binding protein complex, is essential for DNA replication, repair, and recombination (9–13). It is also crucial for the suppression of mutations and genome instability (14–17). RPA acts as a key scaffold to recruit and coordinate proteins involved in different DNA metabolic processes (14, 15, 17). As the first responder of ssDNA, RPA participates in both replication initiation and elongation (10, 12, 13). During replication or under replication stresses, the exposed ssDNA must be protected and stabilized by RPA to prevent formation of secondary structures (14, 16). RPA is also essential for DNA double-stranded break (DSB) repair by the homologous recombination (HR) pathway (18–21). During HR, the 5′-terminated strands of DSBs are initially processed by the resection machinery, generating 3′-tailed ssDNA (22). The 3′-ssDNA becomes bound by the RPA complex to activate the DNA damage checkpoint (23). RPA is subsequently replaced by the Rad51 recombinase to form a Rad51 nucleoprotein filament (19, 24). This recombinase filament catalyzes invasion of the 3′-strands at the homologous sequence to form the D-loop structure, followed by repair DNA synthesis and resolution of recombination intermediates (18, 19, 24). During HR, RPA prevents the formation of DNA secondary structures and protects 3′-ssDNA from nucleolytic degradation (25). In addition, recent work implies a role of RPA in homology recognition (26).
RPA is composed of three subunits, Rfa1, Rfa2, and Rfa3, and with a total of six oligonucleotide-binding (OB) motifs that mediate interactions with ssDNA or proteins (14, 17, 27). RPA can associate with ssDNA in different modes (28). It binds short DNA (8 to 10 nt) in an unstable mode and longer ssDNA (28 to 30 nt) in a high-affinity mode (28–31). Recent single-molecule studies revealed that RPA binding on ssDNA is highly dynamic (28, 32). It can rapidly diffuse within the bound DNA ligand and quickly exchange between the free and ssDNA-bound states (32–35). The cellular functions of RPA rely on its high ssDNA-binding affinity and its ability to interact with different proteins (28). Although RPA has a high affinity for ssDNA, recent studies have suggested that the binding of RPA on chromatin requires additional regulations (36). How RPA is regulated to ensure replication and repair fidelity remains poorly understood.
Rtt105, a protein initially identified as a regulator of the Ty1 retrotransposon, has recently been shown to interact with RPA and acts as an RPA chaperone (36). It facilitates the nuclear localization of RPA and stimulates the loading of RPA at replication forks in unperturbed conditions or under replication stresses (36). Rtt105 exhibits synthetic genetic interactions with genes encoding replisome proteins and is required for heterochromatin silencing and telomere maintenance (37). The deletion of RTT105 results in increased gross chromosomal rearrangements and reduced resistance to DNA-damaging agents (36, 38). In vitro, Rtt105 can directly stimulate RPA binding to ssDNA, likely by changing the binding mode of RPA (36).
In this study, by using a combination of genetic, biochemical, and single-molecule approaches, we demonstrate that Rtt105-dependent regulation of RPA promotes high-fidelity genome duplication and recombination while suppressing mutations and the low-fidelity repair pathways. We provide evidence that human hRIP-α, the putative functional homolog of yeast Rtt105, could regulate human RPA assembly on ssDNA in vitro. Our study unveils a layer of regulation on the maintenance of genome integrity that relies on dynamic RPA binding on ssDNA to ensure high-fidelity replication or recombination.
Results
Rtt105 Promotes DSB Repair by Gene Conversion or Break-Induced Replication.
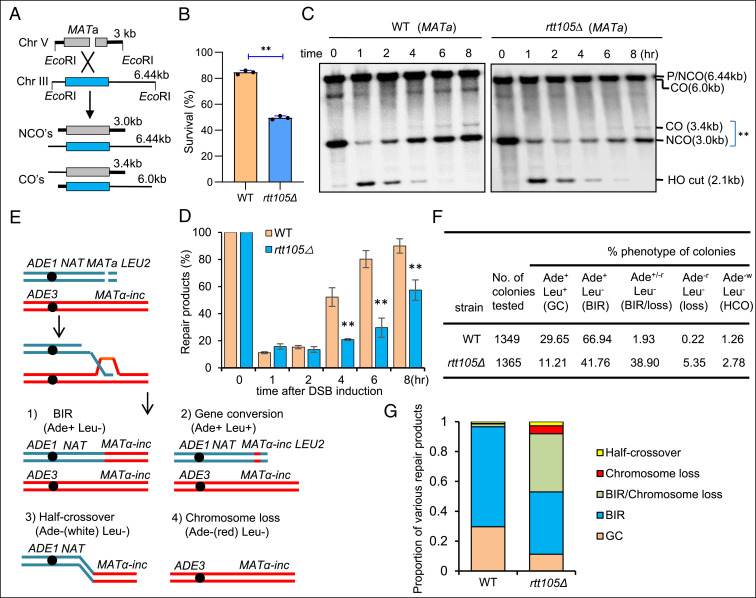
In light of the role of Rtt105 in regulating RPA (36), we aimed to define the consequences of the loss of Rtt105 on DSB repair and on spontaneous mutagenesis. First, we examined the role of Rtt105 in DSB repair by HR using an ectopic recombination system (39). In this assay, a single DSB is generated by an HO endonuclease at the MATa sequence inserted on chromosome V at the ARG5,6 locus. The DSB is repaired by HR using the homologous MATa-inc sequence located on chromosome III (Fig. 1A) (39). About 85% of the wild-type (WT) cells completed the repair and survived. The rtt105Δ mutant cells repaired the break with lower efficiency (∼50% survival) and slower kinetics as monitored by Southern blot (Fig. 1 B–D). These results indicate an important role of Rtt105 in promoting efficient DSB repair by gene conversion.
Fig. 1.
Rtt105 promotes DSB repair by gene conversion or BIR. (A) Scheme showing an ectopic recombination system. CO: crossover; NCO: noncrossover. (B) Survival rate for the WT and rtt105Δ cells repaired by ectopic recombination. (C) Southern blot analysis of the repair kinetics for indicated cells. ** marks the repair products. DSB was induced by the addition of 2% galactose to log phase cells (∼1 × 107/mL) cultured in liquid YP-Raffinose media. The samples were taken at indicated time points after DSB induction. The blot was hybridized to the MATa probe. (D) Quantification of repair products in C. The error bars in B and D represent SD from at least three independent experiments. The asterisk denotes statistical significance. **P < 0.01 (Student’s t test). (E) Diagram showing a site-specific BIR system. Repair of DSBs by different mechanisms generates distinct repair products that can be determined by following the markers on the chromosomes. (F) Repair outcomes in the WT and rtt105Δ cells. GC: gene conversion, HCO: half-crossover. Cells cultured in the preinduction liquid media were plated on YEP-Galactose media to induce DSBs. Colonies were then replica plated on Leu− or Ade− dropout media. The frequencies of BIR, half-crossover, gene conversion, and chromosome loss were calculated based on the percentage of colonies carrying markers specific for these repair outcomes. (G) Graph showing the proportion of each category of repair products in indicated strains. The statistical comparison of different repair outcomes between the WT and rtt105Δ cells in G was performed with χ2 test. P < 0.01.
Second, we tested the role of Rtt105 in break-induced replication (BIR), a unique HR mechanism for repair of one-ended DSBs that often arise during the collapse of replication forks or erosion of telomeres (40). We employed a system wherein only one end of a site-specific DSB has extensive homology to the donor sequence to coerce ∼70% of cells to use BIR to copy over 100 kilobases of chromosome III for break repair (41, 42). In most of the remaining cells (>20%), break repair occurs by gene conversion entailing the capture of the second end of the DSB that harbors short homologous sequences. BIR and other repair outcomes or chromosome loss are followed via examination of the loss of genetic markers (Fig. 1E) (41, 42). We found that BIR repair is significantly decreased and progresses with slower kinetics in rtt105Δ cells, while the gene conversion product is reduced by nearly threefold (Fig. 1 F and G and SI Appendix, Fig. S1 A–D). Notably, we observed about a 24-fold increase in chromosome loss and significant increase in sectored colonies (∼20-fold) in rtt105∆ cells, indicating partial BIR repair and partial chromosome loss (Fig. 1 F and G). Therefore, the loss of Rtt105 has a general impact on DSB repair by gene conversion or BIR.
Allelic BIR and ectopic recombination tested so far are both slow processes (8 to 10 h), involving formation of long ssDNA generated by resection and/or resulting from D-loop migration (42, 43). Long ssDNA intermediates may increase the need for RPA; therefore, we also tested repair by intrachromosomal BIR that is much faster (3 to 4 h) and likely involves much shorter ssDNA. Similarly, repair by BIR was also significantly reduced in rtt105∆ cells when compared to WT cells (SI Appendix, Fig. S2 A–C).
Consistently, the mutant cells have an impaired ability to repair the damaged chromosomes after short exposure to methyl methanesulfonate (MMS), as measured by pulsed-field gel electrophoresis (PFGE), and the mutant cells are highly sensitive to the DNA-damaging agents camptothecin (CPT) or MMS (SI Appendix, Fig. S3 A and B) (36). Therefore, Rtt105 has a broad impact on the DNA damage response and repair.
Rtt105 Suppresses Spontaneous Large Deletions, Duplications, and Chromosome Loss.
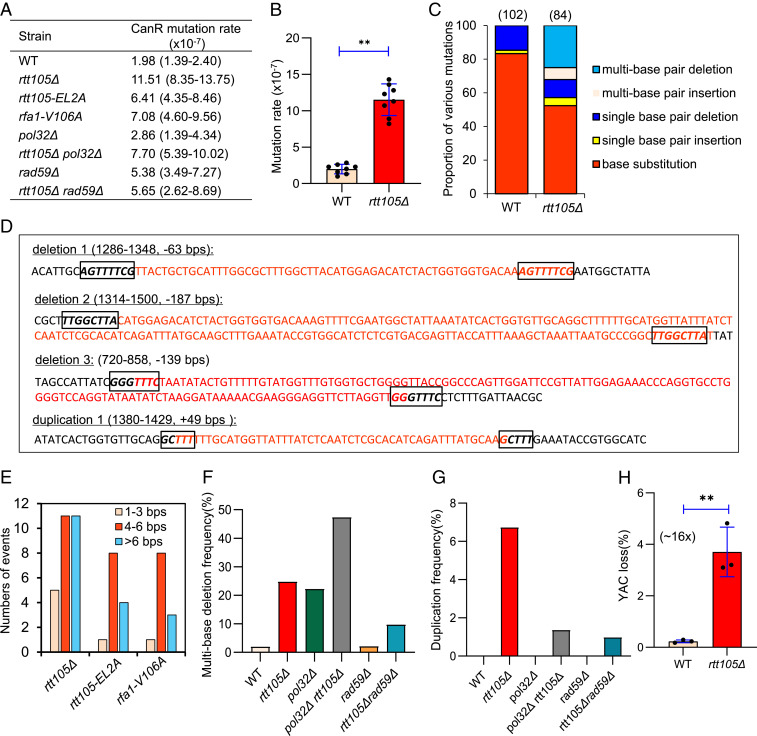
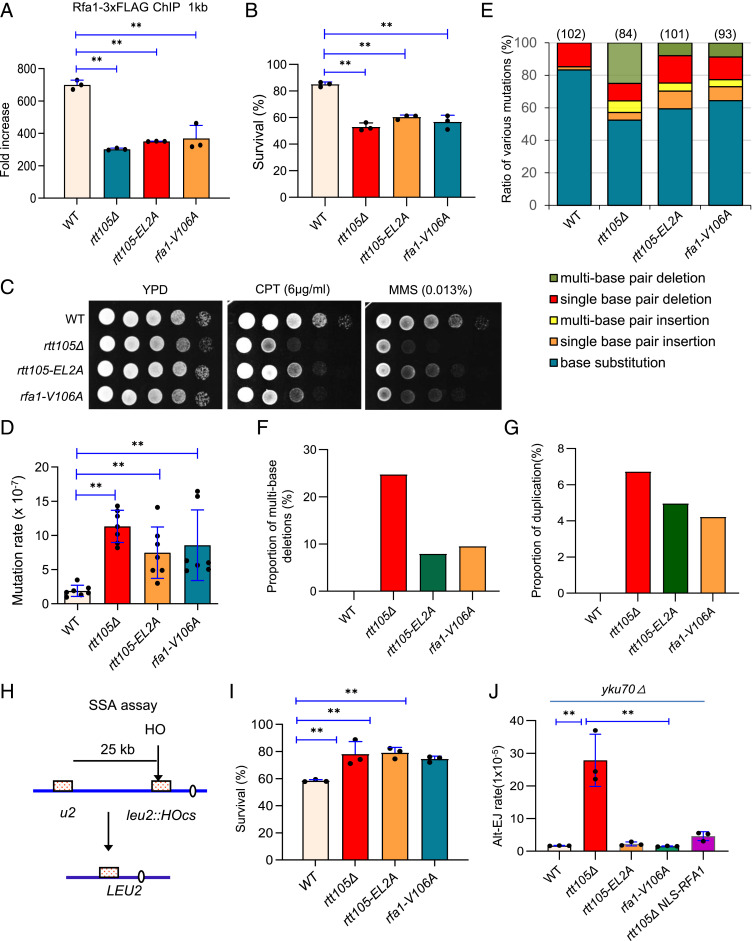
RPA plays important roles in DNA replication by preventing secondary structure formation within ssDNA produced at replication forks. To examine the fidelity of DNA replication in rtt105Δ cells, we used the CAN1 gene as a mutation reporter (44). Mutations in the CAN1 gene result in canavanine-resistant (CanR) colonies that can be selected on synthetic complete (SD) arginine-dropout plates containing canavanine. The mutation rate in the WT cells is ∼2 × 10−7, whereas it is increased to ∼12 × 10−7 (∼sixfold) in rtt105Δ cells (Fig. 2 A and B). Sequencing of the CAN1 gene from the individual CanR colony of the WT (102 colonies) or rtt105Δ (84 colonies) cells revealed that ∼83% of the mutations in WT cells were base substitutions, while the remaining are single–base pair deletions (∼15%) or rare single–base pair insertions (∼2%) (Fig. 2C). In contrast, the pattern of mutation was different in rtt105Δ cells, with ∼52.4% base substitutions, ∼10.7% single–base pair deletions, and ∼4.8% single–base pair insertions. Notably, ∼7% of mutation events in rtt105Δ cells are multi–base pair insertions (Fig. 2C). Specifically, these large insertions are all tandem duplications of DNA fragments with sizes ranging from 17 to 49 base pairs (bps) (Fig. 2D and SI Appendix, Figs. S4 and S5). Strikingly, 25% of all mutation events in rtt105Δ cells harbor large deletions of 4 to 187 bps (Fig. 2 C and D, and SI Appendix, Figs. S4 and S5). Importantly, these gross deletions or duplications did not occur in WT cells. Interestingly, they appear to occur between direct repeats with 4 to 8 bps microhomologies (Fig. 2 D and E and SI Appendix, Fig. S5).
Fig. 2.
Rtt105 promotes high-fidelity replication and suppresses chromosome loss. (A) Table listing the mutation rates for indicated strains. The values in the brackets represent 95% CIs. (B) Plots showing the rate of spontaneous mutations in the WT and rtt105Δ cells. (C) Graph showing the proportion of each category of mutation in CanR colonies derived from indicated cells. The number of CanR colonies sequenced for each strain is indicated. The statistical comparison of different mutation patterns between the WT and rtt105Δ cells in C was performed with χ2 test. P < 0.01. (D) Examples of large deletions or duplications observed in rtt105Δ cells. The deleted or duplicated DNA sequences are marked in red color. The boxed sequences in italics denote microhomologies. (E) Numbers of deletion or duplication events with indicated lengths of microhomologies in indicated cells. (F and G) Plots showing the frequency of large deletions and duplications, respectively, in indicated strains. (H) The frequency of YAC loss in the WT and mutant cells. The error bars in B and H represent SD from at least three independent experiments. **P < 0.01 (Student’s t test).
Microhomology-mediated deletion or duplication events were also seen in the rfa1 point mutants and the pol32Δ or rad27Δ deletion mutant (45–50). These events were suggested to result from polymerase slippage or DSB repair via single-strand annealing (SSA) (45–50). To test if these mechanisms contribute to the genome rearrangements in rtt105Δ cells, we first deleted RAD59, which is required for annealing of complementary single strands in SSA, in rtt105Δ cells (51, 52). We noted that large deletion events were reduced by over one-half, and the duplication events were reduced by sevenfold in this double mutant as compared to that in the rtt105Δ single mutant (Fig. 2 F and G). Thus, the repair by SSA appears to account for a majority of the duplications and over one-half of the deletions in rtt105Δ cells. POL32, a nonessential subunit of the lagging-strand polymerase Pol-δ, is important for polymerase processivity, and the absence of Pol32 increases polymerase slippage (46, 53). Consistent with previous studies, we noted that a high proportion (∼22%) of pol32Δ CanR colonies harbor deletions flanked by short repeats (Fig. 2F and SI Appendix, Fig. S5) (46, 47). However, we did not observe duplication events in pol32Δ cells. Interestingly, additional deletion of POL32 in rtt105Δ cells increased the proportion of large deletions among all mutations to ∼50%, while it did not alter overall mutation rate, suggesting that Rtt105 and Pol32 independently suppress large deletions (Fig. 2F). In contrast, the proportion of duplications was reduced by fivefold in this double mutant as compared that that of rtt105Δ cells (Fig. 2G). Together, these results suggest that ∼80% of the duplication events observed in rtt105Δ cells require highly processive Pol-δ and annealing of microhomologies by Rad59.
Finally, we tested the effect of RTT105 deletion on chromosome loss using a system that carries an extra ∼320-kb yeast artificial chromosome (YAC) (54). The deletion of RTT105 causes more than a 16-fold increase in YAC loss as compared to the WT cells (Fig. 2H). Thus, we demonstrate that Rtt105 plays important roles in suppressing chromosome loss and spontaneous mutations with the signature of microhomology-mediated large duplications or deletions. Our results are consistent with a role of Rtt105 in suppressing gross chromosomal rearrangements (38).
Rtt105 Promotes RPA and Rad51 Loading at DSB Ends.
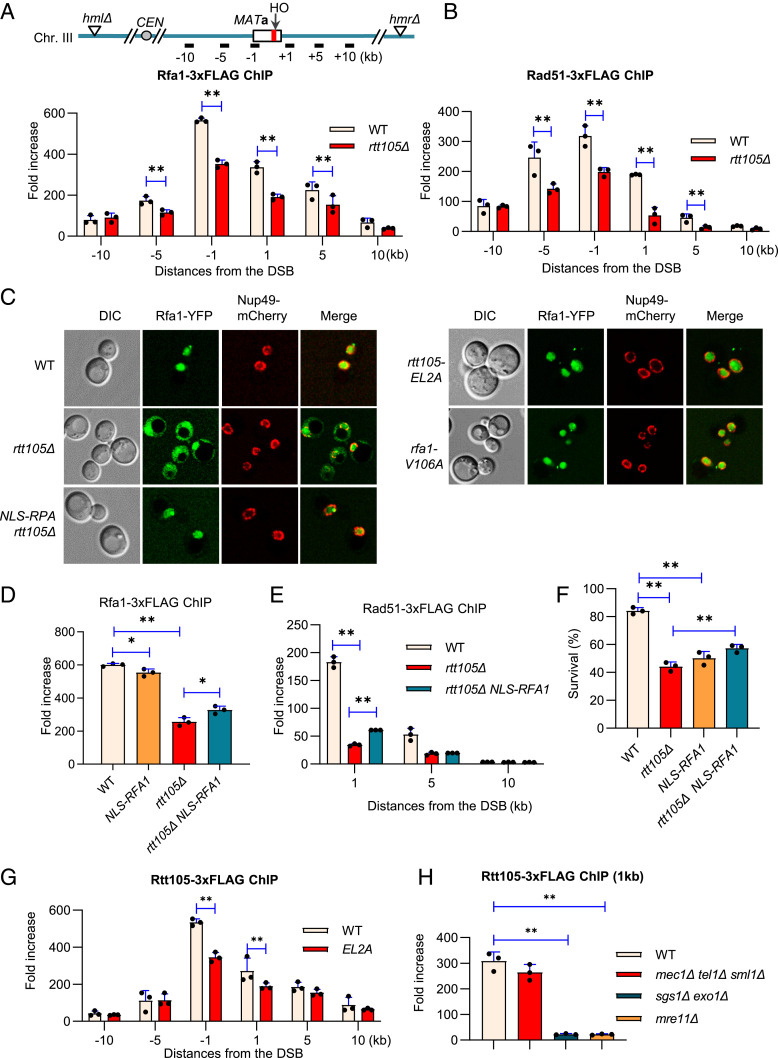
To probe how Rtt105 may affect HR, we first tested the initial step of HR, 5′-end resection, where RPA plays an important role in the Sgs1/Dna2- and Exo1-dependent pathways (25, 55, 56). We used an assay where a single nonrepairable DSB is induced per genome by HO endonuclease at the MAT locus, and the donor sequences HML and HMR are deleted (Fig. 3A) (57, 58). The Southern blot results revealed that both the initial and the long-range resection are normal in rtt105Δ cells (SI Appendix, Fig. S6 A and B). Therefore, the role of Rtt105 in RPA regulation is not important for resection. Next, we tested the recruitment of RPA and Rad51 to DSB ends by chromatin immunoprecipitation (ChIP). In the WT cells, RPA was robustly recruited and spread over 10 kb at both sides of the break. In rtt105Δ cells, the loading of RPA was significantly reduced (Fig. 3A). RPA binding helps to melt secondary structures formed on ssDNA and directly stimulates the Rad51-mediated strand exchange reaction (59). Accordingly, we noted that Rad51 loading was reduced in rtt105Δ cells (Fig. 3B). The defect is not due to any changes in RPA or Rad51 levels since they are expressed at a similar level between the WT and the mutant cells (SI Appendix, Fig. S6C). These results indicate that Rtt105 is required for proper loading of RPA and Rad51 at DSBs.
Fig. 3.
Rtt105 promotes RPA nuclear import and its loading at DSBs. (A and B) ChIP-qPCR analysis of the loading of Rfa1-3xFLAG or Rad51-3xFLAG at indicated locations 4 h following DSB induction in the WT or rtt105Δ cells. The positions of the HO cut and the qPCR primers are indicated. (C) Microscopy analysis of Rfa1-YFP subcellular localization in indicated cells. Nup49-mCherry was used to mark the nuclear membrane. (D and E) ChIP analysis of Rfa1-3xFLAG or Rad51-3xFLAG recruitment at indicated locations 4 h after DSB induction in indicated strains. (F) Survival rate of the repair by ectopic recombination in indicated strains. (G and H) ChIP analysis of Rtt105-3xFLAG recruitment 4 h following DSB induction at indicated locations or in indicated strains. The error bar denotes the SD from three independent experiments. **P < 0.01 (Student’s t test).
Defective RPA Loading in rtt105Δ Cells Is Not Solely due to Altered RPA Localization.
Rtt105 regulates the nuclear import of RPA (36, 60). Indeed, RPA is localized to the nucleus in WT cells, while it is present in both the nucleus and cytoplasm in rtt105Δ cells (Fig. 3C). The fusion of a prototypical nuclear localization signal (NLS) peptide (PKKKRKV) to the N terminus of Rfa1 in the genome fully restored RPA nuclear localization in rtt105Δ cells (Fig. 3C and SI Appendix, Fig. S7). However, it can only restore RPA loading very little at DSBs in the mutant cells (Fig. 3D). This was not due to the inability of NLS-RPA in binding ssDNA, as it loaded almost normally at the DSB in WT cells (Fig. 3D). Accordingly, Rad51 loading and HR repair remained largely defective in rtt105Δ cells with NLS-RPA (Fig. 3 E and F). These results imply that Rtt105 plays additional roles in promoting RPA loading besides facilitating its nuclear import.
Rtt105 Is Recruited to DSB Ends in a Manner Dependent on ssDNA Formation.
To test whether Rtt105 plays a direct role in HR, we examined the recruitment of Rtt105 at DSBs. We detected robust enrichment of Rtt105-3xFLAG at DSB ends that spreads up to 10 kb at each side of the break by 4 h after break induction (Fig. 3G). The recruitment of Rtt105-3xFLAG is normal in the checkpoint-deficient mec1Δ tel1Δ sml1Δ cells but is nearly abolished in mre11Δ or sgs1Δ exo1Δ mutant (Fig. 3H) that are deficient in resection. Thus, the recruitment of Rtt105 appears to rely on ssDNA formation but not checkpoint signaling. This is consistent with the result that Rtt105 directly interacts with RPA via the two residues E171 and L172 of Rtt105 (36). Indeed, disruption of their interaction by mutating both residues to alanine (rtt105-EL2A) impaired Rtt105 loading at DSBs, indicating that the recruitment of Rtt105 partially depends on its association with RPA (Fig. 3G).
Rtt105 Interacts with RPA and Stimulates the Dynamic Binding of RPA on ssDNA.
Consistent with previous studies, we found that yeast Rfa1 strongly interacts with Rtt105 but not the rtt105-EL2A mutant protein in vitro, as revealed by GST pull-down assays (SI Appendix, Fig. S8A) (36). Accordingly, Rtt105 interacts with Rfa1 in vivo, and this interaction appears to increase upon MMS treatment. As expected, the association is greatly impaired in the EL2A mutant cells (SI Appendix, Fig. S8B).
It has been shown that Rtt105 directly stimulates the binding of the yeast RPA complex (ScRPA) on ssDNA in vitro by changing the binding mode of ScRPA (36). Indeed, the addition of Rtt105 (50 nM) moderately stimulates ScRPA (5 to 50 nM) binding on ssDNA (30 nt, 20 nM) as measured by an electrophoretic mobility shift assay (EMSA), while Rtt105 itself does not bind ssDNA (SI Appendix, Fig. S8 C and D). Next, we monitored the real-time assembly of ScRPA with ssDNA by single-molecule analysis using magnetic tweezers (MT). We labeled 12.5 kilo nucleotides ssDNA molecules with digoxigenin and biotin at the 5′- and 3′-ends, respectively, and stretched ssDNA molecules using MT at 8 pN, 20 °C, and pH 7.5 (SI Appendix, Fig. S8E). As RPA-ssDNA nucleoprotein filaments are longer than naked ssDNA, we can examine the dynamic assembly of RPA on ssDNA via monitoring the elongation of ssDNA at a constant force (61, 62). For each solution condition, we calculated the average values from multiple ssDNA molecules over time-extension courses and plotted them as a time-extension curve.
As expected, neither Rtt105 nor rtt105-EL2A associates with ssDNA, as indicated by the constant extension of ssDNA before or after the addition of the protein (SI Appendix, Fig. S8F). As expected, elongation of ssDNA occurred upon the addition of ScRPA (50 nM, green) (SI Appendix, Fig. S8G). When Rtt105 (20 nM) was added together with ScRPA, we detected a faster and larger scale of elongation of ssDNA, indicating that Rtt105 enhances the speed of ScRPA assembly with ssDNA (magenta). Notably, the mutant protein EL2A (navy) was unable to do so (SI Appendix, Fig. S8G). Single exponential fitting yielded a binding rate constant (Kon) of ∼1 μM−1 ⋅ s−1 for ScRPA in our experimental condition (SI Appendix, Fig. S8H). Notably, the addition of Rtt105 increases the Kon value of ScRPA by fivefold. In contrast, the addition of rtt105-EL2A mutant protein can only marginally increase the Kon value of ScRPA (SI Appendix, Fig. S8I). Together, these results indicate that Rtt105 increases the binding speed and binding rate (Kon) of ScRPA for ssDNA in a manner dependent on their physical interaction.
Interestingly, Rtt105 can also interact with human RPA (hRPA) and stimulates its assembly on ssDNA (SI Appendix, Fig. S9 A and C). Similarly, Rtt105 interacts with the ssDNA-binding protein SSB from Escherichia coli (SI Appendix, Fig. S9B). In contrast to RPA, the assembly of E. coli SSB with ssDNA resulted in the shortening of ssDNA, as noted previously (SI Appendix, Fig. S9D) (63). The inclusion of Rtt105 further accelerated the shortening of ssDNA bound with SSB (SI Appendix, Fig. S9D), implying a stimulation effect of Rtt105 on SSB assembly with ssDNA. These results suggest the conservation of this mechanism on RPA regulation.
The Residue V106 of Rfa1 Is Essential to Mediate the RPA-Rtt105 Association.
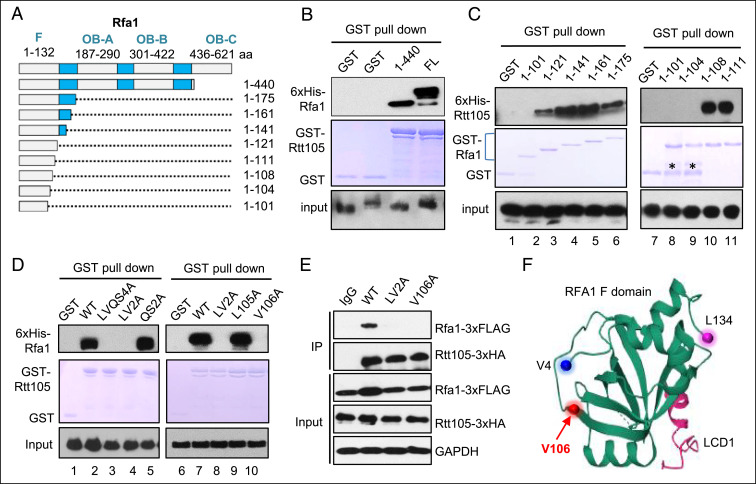
To test whether the observed phenotypes for rtt105Δ cells are due to lack of the Rtt105-mediated regulation on RPA, we sought to delineate the region in Rfa1 that mediates the RPA-Rtt105 interaction. We generated a series of GST- or His-tagged forms of truncated Rfa1 and examined their association with Rtt105 via pull-down assay. We found that the N-terminal fragment (1 to 440 amino acid) of Rfa1 is sufficient to bind Rtt105 in vitro (Fig. 4 A and B). Further analysis revealed that the N-terminal fragment harboring residues 1 to 121 or more retained the ability to bind Rtt105, while the one harboring residues 1 to 101 lost the ability (Fig. 4C, lane 1 to 6). Thus, the motif from residue 101 to 120 is critical for mediating the association. We then located the sites in a short region between residues 104 and 108 (Leucine-Valine-Glutamine-Serine [LVQS]) (Fig. 4C, lane 7 to 11). Indeed, simultaneous mutation of the four residues or the two residues LV but not QS to alanine completely ablated the association (Fig. 4D, lane 1 to 5). Finally, we found that the mutation of single amino acid, V106, to alanine is sufficient to ablate the RPA-Rtt105 association in vitro (Fig. 4D, lane 6 to 10). Consistently, the interaction was also abolished in the rtt105-V106A (V106A) mutant cells (Fig. 4E). Thus, the residue V106, which is located at the junction between a β-fold and a disordered region of the N-terminal OB-F domain of Rfa1 (Fig. 4F) (64), is essential to mediate RPA-Rtt105 interaction.
Fig. 4.
Identification of the RPA residue that mediates the interaction with Rtt105. (A) Graph showing the full-length and truncated Rfa1. The key motifs in Rfa1 are indicated. (B and D) GST pull-down assay showing the interaction between GST-Rtt105 and 6xHis-tagged full-length, truncated, or mutated Rfa1. The amount of GST-tagged proteins was indicated by Coomassie blue staining. (C) GST pull-down assay examining the association between 6xHis-Rtt105 and truncated GST-Rfa1 proteins. The amount of GST-tagged proteins was indicated by Coomassie blue staining. * denotes degraded protein fragments. The bracket marks truncated GST-Rfa1 proteins. (E) Immunoprecipitation analysis the interaction between Rtt105-3xHA and the WT or mutated Rfa1-3xFLAG. (F) A model showing the structure of the F domain of yeast Rfa1, adapted from Deshpande et al. (64). The initiation residue V4, the last residue L134, and the location of V106 are marked.
The Rtt105-RPA Interaction Facilitates RPA Loading and HR Repair.
Next, we assessed the importance of RPA-Rtt105 interaction on RPA loading. We found that RPA loading at DSBs was impaired in both rtt105-EL2A and rfa1-V106A point mutants, as seen in rtt105Δ cells (Fig. 5A). Since the absence of Rtt105 does not impair ssDNA generation (SI Appendix, Fig. S6 A and B), we reasoned that the defect of RPA loading in rtt105-EL2A or rfa1-V106A cells is due to an inability to regulate RPA binding on ssDNA. Accordingly, the repair by ectopic recombination in both point mutants was reduced to a similar level, comparable to that of rtt105Δ cells, indicating that Rtt105 promotes HR repair by regulating RPA (Fig. 5B). Notably, the impaired RPA loading or HR repair was not due to any changes in RPA localization since RPA localizes properly in the nucleus in both point mutants (Fig. 3C). Consistently, the two mutants are sensitive to CPT and MMS (Fig. 5C). In addition, DSB repair by gene conversion in the allelic BIR system was also significantly reduced in the rtt105-EL2A mutant (SI Appendix, Fig. S2D). However, the repair by BIR was slightly increased in this point mutant, which was likely resulted from decreased efficiency of the second end capture that is required to complete gene conversion.
Fig. 5.
Rtt105-mediated regulation of RPA promotes high-fidelity replication and recombination while suppressing deleterious repair pathways. (A) ChIP analysis of Rfa1-3xFLAG recruitment at 1 kb location 4 h after DSB induction in indicated strains. (B) Survival rates of DSB repair by ectopic recombination for indicated strains. (C) DNA damage sensitivity test for indicated strains at indicated drug concentrations. (D) Plot showing mutation rates for indicated strains. (E) Graph showing the ratio of each category of mutation in CanR colonies derived from indicated strains. The number of CanR colonies sequenced for each strain is indicated. (F and G) Proportion of multiple-base deletion or duplication in observed mutation events in indicated strains. (H) A diagram showing the SSA assay between two partial leu2 gene repeats separated by 25 kb. (I) Survival rates of DSB repair by SSA in indicated strains. (J) The rate of DSB repair by alt-EJ in indicated cells. The error bar denotes the SD from at least three independent experiments. **P < 0.01.
Rtt105-RPA Interaction Suppresses Large Spontaneous Deletions or Insertions.
Similarly, mutation rate is increased in both rtt105-EL2A and rfa1-V106A mutants as compared to WT cells (Figs. 2A and 5D). Notably, the CanR colonies derived from both mutants carry a high proportion of the microhomology-mediated large duplications and deletions, as seen in rtt105Δ cells (Fig. 5 E–G and SI Appendix, Figs. S4 and S5). Consistently, a similar type of gross deletion was also seen in other rfa1 mutants (45). Collectively, these phenotypes resemble that of the rtt105Δ mutant, indicating that the role of Rtt105 in avoidance of mutation, duplication, or deletion requires its direct interaction with RPA.
Rtt105-Mediated RPA Assembly on ssDNA Suppresses DSB Repair by SSA.
SSA is a conserved pathway that can repair DSBs formed between long direct repeats (65). During SSA, the ssDNA revealed by DNA end resection anneals at the repeats, leading to loss of the intervening sequence (65). RPA can suppress the annealing of complementary ssDNA and inhibit Rad52-mediated annealing (14, 17, 33, 66, 67). We tested the impact of Rtt105 and Rtt105-RPA interaction on SSA by employing an SSA assay in which two partial leu2 repeats separated by a 25-kb intervening sequence on chromosome III can anneal to repair the HO-induced DSB (Fig. 5H). Rad51, which is dispensable for SSA, was deleted to prevent repair by alternative pathways (58, 68). Approximately 54% of WT cells repaired the break and survived. Repair by SSA was higher (∼80%) in rtt105Δ cells (Fig. 5I). This is not due to any increase in resection or Rad52 loading since resection remains unchanged, while Rad52 loading is decreased in the mutant as compared to WT cells (SI Appendix, Fig. S6 A, B, and D). Similarly, an increased survival rate (∼80%) for repair by SSA was seen in the rtt105-EL2A or rfa1-V106A point mutant (Fig. 5I). Thus, the Rtt105-mediated regulation of RPA is required to promote DSB repair by gene conversion or BIR, while attenuating the deleterious SSA pathway.
Rtt105-Mediated RPA Nuclear Import Suppresses DSB Repair by Alternative End Joining.
RPA is also known to suppress DSB repair by alternative end joining (alt-EJ), a Ku-independent error-prone process involving pairing of short homologies at DSBs (33, 69, 70). We tested the effect of Rtt105 on alt-EJ in the Yku70-deficient JKM139 strain wherein the donor sequences HML and HMR are deleted. Thus, the DSB can only be repaired by the alt-EJ pathway. Notably, alt-EJ rate is increased over 30-fold when RTT105 is deleted, but it remains unaffected in the rtt105-EL2A or rfa1-V106A point mutant where RPA nuclear localization is normal. Interestingly, the elevated alt-EJ could be suppressed by forced RPA nuclear import (NLS-RPA) (Fig. 5J). Thus, proper RPA nuclear import is critical to prevent alt-EJ. Interestingly, alt-EJ and SSA exhibit distinct requirements for the Rtt105-RPA interaction. This difference could be due to the fact that SSA repair involves much longer ssDNA than alt-EJ, so it requires a higher amount or more efficient RPA loading.
Human hRIP-α Interacts with RPA and Stimulates RPA Assembly on ssDNA.
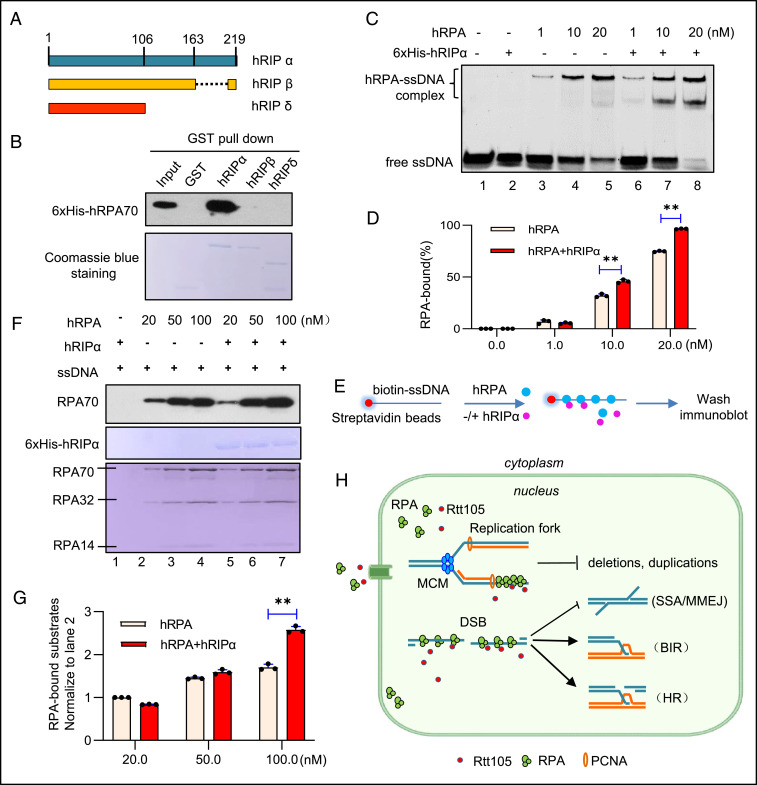
It has been suggested that the RPA-interacting protein alpha (RIP-α) in higher eukaryotes is the putative functional homolog of yeast Rtt105 (36). In Xenopus, XRIP-α is complexed with RPA and importin-β and serves as a carrier for RPA nuclear import (71). In humans, hRIP-α also functions as a nuclear transporter of RPA, and it has several splice isoforms among which hRIP-α and hRIP-β are the predominant variants (Fig. 6A) (72). We found that GST-hRIP-α but not GST-hRIP-β interacts with 6xHis-RPA70 in vitro (Fig. 6B). By an EMSA assay, we observed that the addition of human RPA (hRPA, at 1, 10, or 20 nM) caused a retarded mobility of ssDNA (20 nM) on a gel, indicating the assembly of hRPA with ssDNA (Fig. 6C, lane 3 to 5 and Fig. 6D). Notably, the inclusion of hRIP-α (20 nM) together with hRPA further stimulated hRPA assembly on ssDNA (Fig. 6C, lane 6 to 8 and Fig. 6D). As a control, hRIP-α itself does not bind ssDNA (Fig. 6C, lane 2). Consistently, in a ssDNA pull-down assay, we detected an increased amount of ssDNA-bound hRPA (at 100 nM) in the presence of hRIP-α (at 20 nM) using a biotin-labeled ssDNA (30 nt, 100 nM) coupled to streptavidin-coated magnetic beads (Fig. 6 E–G). Thus, like yeast Rtt105, hRIP-α interacts with hRPA and stimulates its assembly on ssDNA, suggesting that Rtt105 or hRIP-α–mediated regulation on RPA is conserved across species.
Fig. 6.
Human hRIP-α interacts with RPA and promotes RPA assembly on ssDNA. (A) Scheme showing the structure of different hRIP isoforms. (B) GST pull-down assay examining the interaction between 6xHis-hRPA70 and GST-tagged hRIP isoforms. GST was used as a control. hRIP isoforms used for experiments were indicated by Coomassie blue staining. (C) EMSA showing the stimulating effect of hRIP-α (20 nM) on hRPA (1, 10, or 20 nM) assembly on ssDNA (20 nM). (D) Quantitation of the RPA-bound ssDNA in C. (E) Scheme indicating a ssDNA pull-down assay. Biotin-labeled ssDNA (30 nt) coupled to streptavidin-coated magnetic beads was used to capture hRPA in the absence or presence of hRIP-α. We used 50 nM hRIP-α, 100 nM ssDNA, and a gradient amount of hRPA (20, 50, or 100 nM) for the experiment. (F) Western blot analysis of the ssDNA pull-down products from E. hRIP-α and hRPA used for the pull-down experiment were indicated by Coomassie blue staining. (G) Quantitation of the RPA-bound ssDNA in F. (H) A wok model for the roles of Rtt105 in preserving genome stability. The error bar denotes the SD from at least three independent experiments. **P < 0.01.
Discussion
High-fidelity DNA replication and repair are essential for maintenance of genome stability and avoidance of cancer. RPA plays important roles in suppressing mutations and genome instability (14–17). Here, we found that Rtt105 functions as an important regulator of RPA to promote high-fidelity replication and repair (Fig. 6H). Specifically, the deletion of RTT105 results in elevated mutations with microhomology-mediated large duplications or deletions that likely stem from replication slippage or repair by SSA (47, 49, 73). Rtt105 also stimulates RPA loading at DSBs and the repair by gene conversion or BIR, while it suppresses the deleterious SSA and alt-EJ pathway (Fig. 6H). We identified the residue within Rfa1 needed for its interaction with Rtt105 and revealed that the mutants deficient in the interaction exhibited similar phenotypes as observed in rtt105Δ cells. Interestingly, we found that Rtt105 can also interact with human RPA70 and E. coli SSB to stimulate their assemblies on ssDNA in vitro (SI Appendix, Fig. S9). Finally, we showed that hRIP-α, the putative human homolog of Rtt105, interacts with human RPA70 and stimulates hRPA assembly on ssDNA (Fig. 6 A–G). Thus, we propose that Rtt105-mediated regulation of RPA likely represents a conserved mechanism channeling ssDNA intermediates to the high-fidelity replication and repair processes.
Notably, the rtt105Δ mutant cells exhibit increased spontaneous mutations with large deletions or duplications mediated by short repeats. These types of rearrangements were also observed in cells with mutations in DNA replication genes such as RFA1, RAD27, POL3, and POL32 and were proposed to result from replication slippage or mutagenic repair by SSA (45, 47–50). In the rtt105Δ mutant, the large deletions are partially attributed to SSA since they depend on Rad59, which mediates annealing of ssDNA (51, 52). This observation is consistent with the general increase of DSB-induced SSA in rtt105Δ cells. The residual deletions are likely generated by polymerase slippage. Interestingly, Rtt105 and Pol32 appear to have an additive effect in preventing deletions, suggesting that they function by distinct mechanisms in suppressing these events.
The duplication events in rtt105Δ cells appear to be related to lagging-strand DNA synthesis and SSA repair since these events were markedly reduced when component of the lagging-strand polymerase Pol32 was eliminated or when RAD59 was deleted in rtt105Δ cells (Fig. 2 F and G). The importance of Pol32 implies the need for a highly processive polymerase for the duplications observed in rtt105Δ cells. Notably, a similar type of duplications, but with a much higher frequency, was observed in cells lacking Rad27, an Okazaki fragment–processing enzyme (48). It was proposed that during lagging-strand synthesis, Pol-δ extends DNA into the downstream Okazaki fragment and displaces it, resulting in a 5′-flap structure that needs to be cleaved by Rad27. In the absence of Rad27, unprocessed flaps are likely extended by strand displacement synthesis, resulting in short duplication of DNA sequences. These 5′-ssDNA overhangs produced can potentially be filled in by DNA synthesis and subsequently resected to expose the short repeats at the 3′-end for SSA (48). Pairing of the repeats out of register can generate either duplication or deletion events (48). RPA is known to control the coordinated actions of Dna2 and Rad27 in processing Okazaki fragments and to melt DNA secondary structures (9). Reduced RPA levels or impaired RPA assembly on ssDNA in rtt105Δ cells may directly or indirectly cause aberrant processing or displacement of Okazaki fragments or misalignment of short homologies, eventually leading to the observed duplication or deletion events. Recently, it was reported that high GC content can cause such types of deletions or duplications by polymerase slippage (73). However, we did not find any correlation between GC content and these rearrangement events (SI Appendix, Fig. S5).
RPA undergoes rapid diffusion and microscopic dissociation on ssDNA (14, 32). It was proposed that Rtt105 can change the binding mode of RPA with ssDNA (36). Here, we found that Rtt105 alters the binding rate (Kon) of RPA on ssDNA, allowing for a faster and more efficient assembly of RPA on ssDNA. Each RPA OB motif has a distinct footprint and binding dynamics on ssDNA (28). Interestingly, Rad52 appears to selectively reduce the engagement of the OB-D motif with ssDNA, providing access to the 3′-end of the occluded ssDNA (28). Here, we identified that RPA interacts with Rtt105 via the residue V106 that is located in the F domain. It remains to be determined which RPA domain is specifically modulated by Rtt105.
ssDNA is perhaps one of the most ubiquitous and important biological intermediates formed throughout the life of cells (74, 75). Due to the broad functions in DNA metabolism, RPA is tightly associated with carcinogenesis (74, 76–79). In ovarian cancer cells, RPA exhaustion leads to instability of replication forks and deficiency in nucleotide excision repair, and RPA availability represents a major determinant of cisplatin sensitivity (80). Similarly, RPA depletion or chemical inhibition impairs the survival and self-renewal of glioma cancer stem-like cells and sensitizes these cells to ionizing radiation (74). Therefore, both the nuclear import and proper binding of RPA on ssDNA are essential for avoidance of mutations or cancer. It will be of great interest to assess the roles of hRIP in regulating RPA and in preserving genome stability in normal human cells and in cancer cells. Targeting hRIP could be a potential cancer therapy strategy.
In many human cancers and germline mutations, base substitutions and large DNA duplication or deletion events have often been observed (2–4, 8, 48). The genetic basis for some of the mutations remains unknown. Our results suggest that ineffective loading or assembly of RPA on ssDNA could be a mechanism leading to the genome rearrangements accompanied by large duplications or deletions in cancer.
Materials and Methods
Yeast Strains and Plasmids.
The yeast strains used in this study are presented in SI Appendix, Table S1.
DNA Damage Sensitivity Assay.
The yeast cells were grown in yeast extract peptone dextrose (YEPD) medium overnight to saturation. Undiluted cell culture and 1/10 serial dilutions of cell cultures were spotted onto YEPD plates containing indicated concentrations of camptothecin or MMS. Plates were incubated at 30 °C for 3 d before taking pictures.
Fluorescence Microscopy.
Rfa1-YFP (yellow fluorescent protein) subcellular localization in live cells were examined using a ZEISS LSM 880 fluorescence confocal microscope carrying an Airyscan with a 63× oil immersion objective lens and a YFP or RFP (red fluorescent protein) filter. Fluorescent images were captured and processed using ZEISS Blue Lite2 software.
PFGE.
Analysis of yeast chromosome integrity by PFGE was carried out as described by Maringele et al. (81). Chromosomal DNA plugs were prepared and separated on a 1% agarose gel at 6 V/cm for 30 h (initial time = 10 s, final time = 35 s) by using the CHEF DRII apparatus (Bio-Rad).
Mutation Rate and Spectra.
The rate of accumulation of CanR mutations was determined as previously described (44). Yeast cells from single fresh colonies were plated on SC arginine-dropout plates containing 60 mg/L canavanine. Mutation rate was determined by fluctuation analysis using the median method. CanR mutation spectra were characterized by PCR amplification of the CAN1 gene from independent CanR isolates, followed by DNA sequencing.
Analysis of Ectopic Recombination, SSA, and Alt-EJ.
To test the viability of DSB repair by ectopic recombination, SSA, or alt-EJ, cells were cultured in the pre-induction medium YEP-raffinose (Yeast extract peptone raffinose) overnight to log phase. Cells were then diluted and plated on YEPD and YEP-Gal (Yeast extract peptone galactose) plates, respectively, followed by incubating at 30 °C for 3 to 5 d. Survival rate was calculated using the following formula: (Number of colonies grown on YEP-Gal)/(Number of colonies on YEPD × dilution fold) × 100%. At least three independent experiments were performed for each strain. The repair kinetics for ectopic recombination were monitored by Southern blot analysis as described (58).
Analysis of DSB Repair by BIR.
Analysis of allelic BIR was carried out as previously described (41, 42). The frequencies of BIR, gene conversion, BIR/loss, half crossovers, and chromosome loss were estimated based on the percentage of colonies carrying markers specific for these repair outcomes, as reported previously (41, 42). To analyze BIR in the H-0 system, cells were collected 0, 1, and 6 h after DSB induction with 2% galactose. Isolated DNA was digested with Bsp1286I and separated on 0.8% agarose gels. MATa-distal DNA fragment was used for Southern blot, and ACT1 fragment served as loading control probe. Repair efficiency at 6 h after DSB induction was calculated as the percentage of normalized pixel intensity of the BIR product band at 6 h compared to the normalized parental bands at 0 h. Quantitative densiometric analysis was completed with ImageQuant TL 5.2 software (GE Healthcare Life Sciences), and statistical analysis was conducted using Welch’s unpaired t test.
Analysis of 5′-End Resection by Southern Blot.
Analysis of DSB resection was analyzed using Southern blotting as reported (58).
ChIP.
Log phase yeast cells (∼1 × 107 cells/mL) grown in YEP-Raffinose medium were subject to DSB induction by the addition of 2% galactose. Chromatin DNA was sheared to an average size of ∼300 bp using a Diagenode Bioruptor. ChIP and qPCR assays were carried out as previously described (57).
Expression of Recombination Protein and GST Pull-Down Assay.
Yeast RFA1 or RTT105 gene was cloned into the vector pGEX-4T-3 or pET30a with proper restriction enzymes. The recombinant plasmid was transformed into BL21 (DE3). Protein expression and GST pull-down assay was conducted as described by Li et al. (36).
Protein Purification.
Purification of yeast RPA complex was performed as described by Binz et al. (82). The full-length human RPA composed of three subunits Rfa1, Rfa2, and Rfa3 was expressed and purified according to the protocols described previously (83). For purification of 6xHis-Rtt105 protein, please see SI Appendix, Materials and Methods for details.
EMSA.
ssDNA substrate (30 nt, 2 nM) was incubated with various amounts of ScRPA or Rtt015 for 1 h at 4 °C in 1× binding buffer (25 mM Tris HCl, pH 7.5, 5 mM MgCl2, and 5% glycerol). The reaction mixture was then loaded with 4× loading dye and resolved in either a 6% native polyacrylamide gel or 2% agarose gel in cold 0.3× Tris-boric acid-EDTA (TBE) buffer. The native PAGE or agarose gels were stained with GelRed. Signals were detected on a G-Box imager (Syngene), and band intensities were quantified by ImageJ.
Immunoprecipitation and Western Blotting.
Immunoprecipitation was carried out as previously described (57). Anti-HA or anti-FLAG antibody was purchased from MBL. Protein G-agarose beads were ordered from GE healthcare. Western blot was performed as described in SI Appendix, Materials and Methods. Anti-mouse or rabbit IgG horseradish peroxidase-conjugated secondary antibodies were ordered from Santa Cruz Biotechnology.
Single-Molecule Study.
The 12.5 k-nt ssDNA was generated by one-sided PCR, and its two ends were labeled with digoxigenin and biotin groups, respectively. In MT experiments, the digoxigenin-labeled end of a single ssDNA molecule was anchored to the anti–digoxigenin-coated glass surface in a flow cell. Then, a superparamagnetic microbead (M-270, Dynal beads) was attached to the biotin-labeled end of the anchored ssDNA molecule. A pair of permanent magnets was used to attract the microbead and thus exert a constant force to the anchored ssDNA molecule. The extension of ssDNA was determined to be the separation between the microbead and glass surface. The assembling buffer contained 100 mM NaAc, 10 mM MgAc2, 1 mM ATP, and 25 mM Tris-Ac pH 7.5. All experiments were performed at a constant force of 8 pN at 20 °C.
Supplementary Material
Acknowledgments
We thank Dr. Patrick Sung (University of Texas at San Antonio) for critical reading of the manuscript and Dr. Douglas Koshland (University of California, Berkeley) for providing the YAC system for testing chromosome loss. This research was supported by grants from the National Natural Science Foundation of China (Grant Nos. 32070573, 31872808, and 31671294) to X.C., (Grant Nos. 11704286 and 31670760) to X. Zhang, and (Grant Nos. 31725015) to Q.L., NIH Grant No. GM080600 to G.I., and Wuhan University Advanced Genetics Course Program to X.C.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2106393118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Martincorena I., Campbell P. J., Somatic mutation in cancer and normal cells. Science 349, 1483–1489 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Helleday T., Eshtad S., Nik-Zainal S., Mechanisms underlying mutational signatures in human cancers. Nat. Rev. Genet. 15, 585–598 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nik-Zainal S., et al., Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Volkova N. V., et al., Mutational signatures are jointly shaped by DNA damage and repair. Nat. Commun. 11, 2169 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalimutho M., et al., Patterns of genomic instability in breast cancer. Trends Pharmacol. Sci. 40, 198–211 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Fishel R., et al., The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell 75, 1027–1038 (1993). [DOI] [PubMed] [Google Scholar]
- 7.Strand M., Prolla T. A., Liskay R. M., Petes T. D., Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature 365, 274–276 (1993). [DOI] [PubMed] [Google Scholar]
- 8.Petljak M., et al., Characterizing mutational signatures in human cancer cell lines reveals episodic APOBEC mutagenesis. Cell 176, 1282–1294.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bae S. H., Bae K. H., Kim J. A., Seo Y. S., RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature 412, 456–461 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Dornreiter I., et al., Interaction of DNA polymerase alpha-primase with cellular replication protein A and SV40 T antigen. EMBO J. 11, 769–776 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li G. M., Mechanisms and functions of DNA mismatch repair. Cell Res. 18, 85–98 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Waga S., Stillman B., Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature 369, 207–212 (1994). [DOI] [PubMed] [Google Scholar]
- 13.Yuzhakov A., Kelman Z., Hurwitz J., O’Donnell M., Multiple competition reactions for RPA order the assembly of the DNA polymerase delta holoenzyme. EMBO J. 18, 6189–6199 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen R., Wold M. S., Replication protein A: Single-stranded DNA’s first responder: Dynamic DNA-interactions allow replication protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair. Bioessays 36, 1156–1161 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maréchal A., Zou L., RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 25, 9–23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toledo L. I., et al., ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 155, 1088–1103 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Wold M. S., Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66, 61–92 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Haber J. E., A life investigating pathways that repair broken chromosomes. Annu. Rev. Genet. 50, 1–28 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Kowalczykowski S. C., An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 7, a016410 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scully R., Panday A., Elango R., Willis N. A., DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 20, 698–714 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Symington L. S., Rothstein R., Lisby M., Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics 198, 795–835 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Symington L. S., Gautier J., Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45, 247–271 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Zou L., Elledge S. J., Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300, 1542–1548 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Lisby M., Rothstein R., Cell biology of mitotic recombination. Cold Spring Harb. Perspect. Biol. 7, a016535 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H., Lisby M., Symington L. S., RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol. Cell 50, 589–600 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crickard J. B., Moevus C. J., Kwon Y., Sung P., Greene E. C., Rad54 drives ATP hydrolysis-dependent DNA sequence alignment during homologous recombination. Cell 181, 1380–1394.e18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iftode C., Daniely Y., Borowiec J. A., Replication protein A (RPA): The eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 34, 141–180 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Pokhrel N., et al., Dynamics and selective remodeling of the DNA-binding domains of RPA. Nat. Struct. Mol. Biol. 26, 129–136 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arunkumar A. I., Stauffer M. E., Bochkareva E., Bochkarev A., Chazin W. J., Independent and coordinated functions of replication protein A tandem high affinity single-stranded DNA binding domains. J. Biol. Chem. 278, 41077–41082 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Bhat K. P., Cortez D., RPA and RAD51: Fork reversal, fork protection, and genome stability. Nat. Struct. Mol. Biol. 25, 446–453 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan J., Pavletich N. P., Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 26, 2337–2347 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma C. J., Gibb B., Kwon Y., Sung P., Greene E. C., Protein dynamics of human RPA and RAD51 on ssDNA during assembly and disassembly of the RAD51 filament. Nucleic Acids Res. 45, 749–761 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng S. K., Gibb B., de Almeida M. J., Greene E. C., Symington L. S., RPA antagonizes microhomology-mediated repair of DNA double-strand breaks. Nat. Struct. Mol. Biol. 21, 405–412 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gibb B., et al., Concentration-dependent exchange of replication protein A on single-stranded DNA revealed by single-molecule imaging. PLoS One 9, e87922 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibb B., et al., Protein dynamics during presynaptic-complex assembly on individual single-stranded DNA molecules. Nat. Struct. Mol. Biol. 21, 893–900 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li S., et al., Rtt105 functions as a chaperone for replication protein A to preserve genome stability. EMBO J. 37, e99154 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corda Y., Maestroni L., Luciano P., Najem M. Y., Géli V., Genome stability is guarded by yeast Rtt105 through multiple mechanisms. Genetics 217, iyaa035 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Putnam C. D., et al., A genetic network that suppresses genome rearrangements in Saccharomyces cerevisiae and contains defects in cancers. Nat. Commun. 7, 11256 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ira G., Malkova A., Liberi G., Foiani M., Haber J. E., Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115, 401–411 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kramara J., Osia B., Malkova A., Break-induced replication: The where, the why, and the how. Trends Genet. 34, 518–531 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deem A., et al., Defective break-induced replication leads to half-crossovers in Saccharomyces cerevisiae. Genetics 179, 1845–1860 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson M. A., et al., Pif1 helicase and Polδ promote recombination-coupled DNA synthesis via bubble migration. Nature 502, 393–396 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chung W. H., Zhu Z., Papusha A., Malkova A., Ira G., Defective resection at DNA double-strand breaks leads to de novo telomere formation and enhances gene targeting. PLoS Genet. 6, e1000948 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang M. E., Rio A. G., Nicolas A., Kolodner R. D., A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc. Natl. Acad. Sci. U.S.A. 100, 11529–11534 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen C., Umezu K., Kolodner R. D., Chromosomal rearrangements occur in S. cerevisiae rfa1 mutator mutants due to mutagenic lesions processed by double-strand-break repair. Mol. Cell 2, 9–22 (1998). [DOI] [PubMed] [Google Scholar]
- 46.Huang M. E., Rio A. G., Galibert M. D., Galibert F., Pol32, a subunit of Saccharomyces cerevisiae DNA polymerase delta, suppresses genomic deletions and is involved in the mutagenic bypass pathway. Genetics 160, 1409–1422 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kokoska R. J., Stefanovic L., DeMai J., Petes T. D., Increased rates of genomic deletions generated by mutations in the yeast gene encoding DNA polymerase delta or by decreases in the cellular levels of DNA polymerase delta. Mol. Cell. Biol. 20, 7490–7504 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tishkoff D. X., Filosi N., Gaida G. M., Kolodner R. D., A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell 88, 253–263 (1997). [DOI] [PubMed] [Google Scholar]
- 49.Tran H. T., et al., Replication slippage between distant short repeats in Saccharomyces cerevisiae depends on the direction of replication and the RAD50 and RAD52 genes. Mol. Cell. Biol. 15, 5607–5617 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weston-Hafer K., Berg D. E., Deletions in plasmid pBR322: Replication slippage involving leading and lagging strands. Genetics 127, 649–655 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davis A. P., Symington L. S., The yeast recombinational repair protein Rad59 interacts with Rad52 and stimulates single-strand annealing. Genetics 159, 515–525 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugawara N., Ira G., Haber J. E., DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell. Biol. 20, 5300–5309 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burgers P. M., Eukaryotic DNA polymerases in DNA replication and DNA repair. Chromosoma 107, 218–227 (1998). [DOI] [PubMed] [Google Scholar]
- 54.Wahba L., Amon J. D., Koshland D., Vuica-Ross M., RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol. Cell 44, 978–988 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cejka P., et al., DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature 467, 112–116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niu H., et al., Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 467, 108–111 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen X., et al., The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature 489, 576–580 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu Z., Chung W. H., Shim E. Y., Lee S. E., Ira G., Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134, 981–994 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sung P., Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 272, 28194–28197 (1997). [DOI] [PubMed] [Google Scholar]
- 60.Belanger K. D., et al., The karyopherin Kap95 and the C-termini of Rfa1, Rfa2, and Rfa3 are necessary for efficient nuclear import of functional RPA complex proteins in Saccharomyces cerevisiae. DNA Cell Biol. 30, 641–651 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Vlaminck I., et al., Torsional regulation of hRPA-induced unwinding of double-stranded DNA. Nucleic Acids Res. 38, 4133–4142 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van der Heijden T., et al., Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules. Nucleic Acids Res. 35, 5646–5657 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Naufer M. N., et al., Multiprotein E. coli SSB-ssDNA complex shows both stable binding and rapid dissociation due to interprotein interactions. Nucleic Acids Res. 49, 1532–1549 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deshpande I., et al., Structural basis of mec1-ddc2-RPA assembly and activation on single-stranded DNA at sites of damage. Mol. Cell 68, 431–445.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 65.Bhargava R., Onyango D. O., Stark J. M., Regulation of single-strand annealing and its role in genome maintenance. Trends Genet. 32, 566–575 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen R., Subramanyam S., Elcock A. H., Spies M., Wold M. S., Dynamic binding of replication protein a is required for DNA repair. Nucleic Acids Res. 44, 5758–5772 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu Y., Kantake N., Sugiyama T., Kowalczykowski S. C., Rad51 protein controls Rad52-mediated DNA annealing. J. Biol. Chem. 283, 14883–14892 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vaze M. B., et al., Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol. Cell 10, 373–385 (2002). [DOI] [PubMed] [Google Scholar]
- 69.Sfeir A., Symington L. S., Microhomology-mediated end joining: A back-up survival mechanism or dedicated pathway? Trends Biochem. Sci. 40, 701–714 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sinha S., Villarreal D., Shim E. Y., Lee S. E., Risky business: Microhomology-mediated end joining. Mutat. Res. 788, 17–24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jullien D., Görlich D., Laemmli U. K., Adachi Y., Nuclear import of RPA in Xenopus egg extracts requires a novel protein XRIPalpha but not importin alpha. EMBO J. 18, 4348–4358 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Park J., Seo T., Kim H., Choe J., Sumoylation of the novel protein hRIPbeta is involved in replication protein A deposition in PML nuclear bodies. Mol. Cell. Biol. 25, 8202–8214 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kiktev D. A., Sheng Z., Lobachev K. S., Petes T. D., GC content elevates mutation and recombination rates in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 115, E7109–E7118 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pedersen H., Anne Adanma Obara E., Elbæk K. J., Vitting-Serup K., Hamerlik P., Replication protein A (RPA) mediates radio-resistance of glioblastoma cancer stem-like cells. Int. J. Mol. Sci. 21, 1588 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zeman M. K., Cimprich K. A., Causes and consequences of replication stress. Nat. Cell Biol. 16, 2–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dai Z., Wang S., Zhang W., Yang Y., Elevated expression of RPA3 is involved in gastric cancer tumorigenesis and associated with poor patient survival. Dig. Dis. Sci. 62, 2369–2375 (2017). [DOI] [PubMed] [Google Scholar]
- 77.Levidou G., et al., Prognostic significance of replication protein A (RPA) expression levels in bladder urothelial carcinoma. BJU Int. 108, E59–E65 (2011). [DOI] [PubMed] [Google Scholar]
- 78.Tomkiel J. E., et al., Autoimmunity to the M(r) 32,000 subunit of replication protein A in breast cancer. Clin. Cancer Res. 8, 752–758 (2002). [PMC free article] [PubMed] [Google Scholar]
- 79.Xiao W., Zheng J., Zhou B., Pan L., Replication protein A 3 is associated with hepatocellular carcinoma tumorigenesis and poor patient survival. Dig. Dis. 36, 26–32 (2018). [DOI] [PubMed] [Google Scholar]
- 80.Bélanger F., et al., Replication protein A availability during DNA replication stress is a major determinant of cisplatin resistance in ovarian cancer cells. Cancer Res. 78, 5561–5573 (2018). [DOI] [PubMed] [Google Scholar]
- 81.Maringele L., Lydall D., Pulsed-field gel electrophoresis of budding yeast chromosomes. Methods Mol. Biol. 313, 65–73 (2006). [DOI] [PubMed] [Google Scholar]
- 82.Binz S. K., Dickson A. M., Haring S. J., Wold M. S., Functional assays for replication protein A (RPA). Methods Enzymol. 409, 11–38 (2006). [DOI] [PubMed] [Google Scholar]
- 83.Henricksen L. A., Umbricht C. B., Wold M. S., Recombinant replication protein A: Expression, complex formation, and functional characterization. J. Biol. Chem. 269, 11121–11132 (1994). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.