Significance
As one of the most important post-translational modifications, phosphorylation is both highly abundant and dynamically regulated in cells. Characterizing subcellular phosphoproteome with high temporal resolution should shed light on their contributions to diverse cellular processes. By integrating an activatable proximity labeling enzyme with an orthogonal phosphorylation enrichment scheme, we have developed the SubMAPP strategy for mapping the phosphorylation dynamics of subcellular proteome in living systems. The high sensitivity of SubMAPP enabled the identification of phosphoproteins and phosphorylation sites in the endoplasmic reticulum (ER), revealing ER-to-mitochondria protein translocation under ER stress.
Keywords: subcellular phosphoproteome, bioorthogonal decaging, proximal labeling
Abstract
Proteome-wide profiling of protein phosphorylation has been widely used to reveal the underlying mechanism of diverse cellular signaling events. Yet, characterizing subcellular phosphoproteome with high spatial–temporal resolution has remained challenging. Herein, we developed a subcellular-specific uncaging-assisted biotinylation and mapping of phosphoproteome (SubMAPP) strategy to monitor the phosphorylation dynamics of subcellular proteome in living cells and animals. Our method capitalizes on the genetically encoded bioorthogonal decaging strategy, which enables the rapid activation of subcellular localized proximity labeling biotin ligase through either light illumination or small-molecule triggers. By further adopting an integrated orthogonal pull-down strategy with quantitative mass spectrometry, SubMAPP allowed for the investigation of subcellular phosphoproteome dynamics, revealing the altered phosphorylation patterns of endoplasmic reticulum (ER) luminal proteins under ER stress. Finally, we further expanded the scope of the SubMAPP strategy to primary neuron culture and living mice.
Phosphorylation is one of the most imperative posttranslational modifications (PTMs) that plays fundamental roles in diverse cellular processes, ranging from signal transduction to stress responses (1–3). Over the past decades, phosphoproteomic profiling techniques have been extensively developed to study various physiological and pathological processes (4, 5). Yet, these methods focused mainly on the whole-cell proteome rather than the distributions and dynamic changes of phosphoproteins at subcellular level. Therefore, spatiotemporally resolved characterization of protein phosphorylation remained highly desirable (6–9).
In recent years, several enzyme-catalyzed proximity labeling (PL) methods have been developed [e.g., APEX/APEX2 (10, 11), BioID/TurboID (12–14)] for subcellular proteomic profiling in living cells (15, 16), providing high-quality protein inventories in subcellular compartments that are difficult to isolate. However, existing PL enzymes are not readily applicable for the capture of dynamic PTM changes at subcellular level according to the following reasons: 1) the prerequisite of hydrogen peroxide (∼1 mM) by APEX would acutely affect the cellular metabolism and protein PTMs, particularly on phosphorylations (17); 2) cellular expression of TurboID would inevitably lead to pervasive biotinylation of endogenous proteins, which may perturb regular cellular processes and give rise to cytotoxicity (18–21); and 3) biotin starvation commonly adopted in BioID/TurboID assays to reduce background labeling could result in substantial stress to cellular homeostasis that alters protein PTM patterns (22). In order to address these challenges, we envision that a PL enzyme whose catalytic activity is controlled by bioorthogonal triggers can be utilized to minimize the interferences on cell physiology. Coupling with diverse PTM-enrichment protocols, the dynamic change of PTMs can be portrayed with subcellular precision (23–26).
Herein, we manufactured a bioorthogonally activatable promiscuous biotin ligase with rapid turn-on kinetics and excellent proximity labeling efficiencies in organelles including endoplasmic reticulum (ER) and mitochondria. By adopting an integrated orthogonal pull-down strategy with mass spectrometry–based quantitative proteomics, we developed a subcellular-specific uncaging-assisted biotinylation and mapping of phosphoproteome (SubMAPP) strategy to investigate ER-specific phosphoproteome and its dynamic changes upon ER stress (Fig. 1 A–C). SubMAPP has enabled the identification of 949 phosphopeptides from 500 phosphorylated proteins in the ER lumen of human embryonic kidney 293T (HEK293T) cells, which unveils phosphoproteins and phosphosites that are hardly detectable by previous methods in the ER. Finally, we expanded the application of SubMAPP toward a more complex living system including primary neuron culture and living mice.
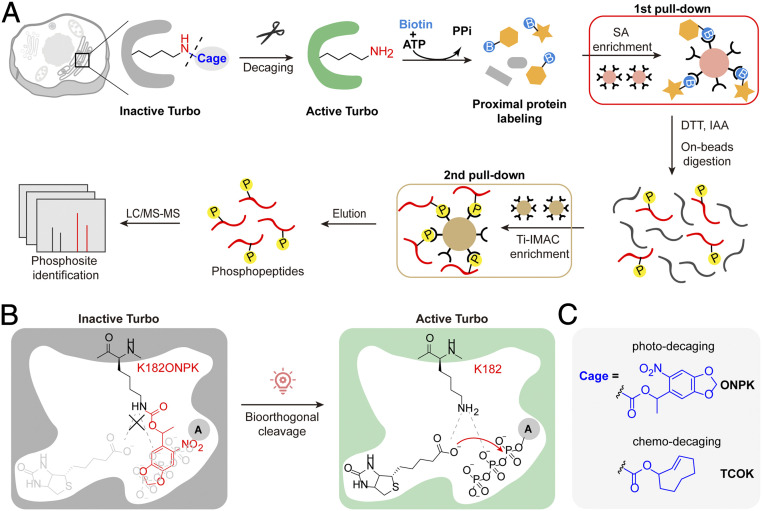
Fig. 1.
Schematic illustration of SubMAPP. (A) Subcellular targeting of bioorthogonally activatable PL enzymes, photoTurbo and chemoTurbo variants, are created by genetically replacing the catalytic lysine residue with photo-/chemo-caged lysine analogs. Temporal-gated PL enzyme activation and subcellular proteomic labeling can be achieved by in situ decaging. Biotinylated proteins subsequently undergo an orthogonal pull-down pipeline to yield digested phosphopeptides, which were subjected to quantitative MS analysis. (B) Schematic illustration of photoTurbo activation. The photo-caged lysine analog (ONPK) can be genetically and site-specifically incorporated into Turbo in place of the conserved catalytic lysine residue (K182). Upon 365 nm photolysis, photoTurbo is rapidly activated, thus triggering the enzymatic generation of biotin-AMP from biotin and ATP. (C) Chemical structures of photo-caged and chemo-caged unnatural amino acids.
Results
Design of Bioorthogonally Activatable PL Enzymes for SubMAPP.
We chose to work on the latest-generation of biotin ligase, TurboID, due to its rapid and efficient proximity-dependent biotinylation (14). Sequence alignment and structural analysis of biotin ligase variants (e.g., BioID/BioID2, Turbo, and BASU) revealed a key catalytic lysine residue (e.g., BirA-K183, Turbo-K182) that is required for the adenylation of biotin with ATP to yield the reactive intermediate, biotinyl-5′-AMP (SI Appendix, Fig. S1A) (27). By genetically replacing the K182 residue of Turbo with photo-caged lysine analog Nε-[(1-(6-Nitrobenzo[d][1,3]dioxol-5yl)ethoxy)carbonyl]-L-lysine (ONPK) via genetic code expansion (28, 29), the resulting Turbo-K182ONPK (hereafter referred to as photoTurbo) became inactive even in the presence of 100 μM biotin substrate in the culture medium (Fig. 1B and SI Appendix, Fig. S1 B and C). ONPK was chosen as opposed to other photo-caged lysine analogs (e.g., o-nitrobenzyl-oxycarbonyl-Nε-L-lysine, ONBK, SI Appendix, Fig. S1D) for its higher incorporation yield, photo-decaging efficiency, and its more biologically inert ketone by-product (SI Appendix, Fig. S1 D and E and Supplementary Note S1). Upon photolysis by 365 nm light for 5 min, ONPK can liberate the lysine side-chain and restore the enzyme activity (Fig. 1 B and C).
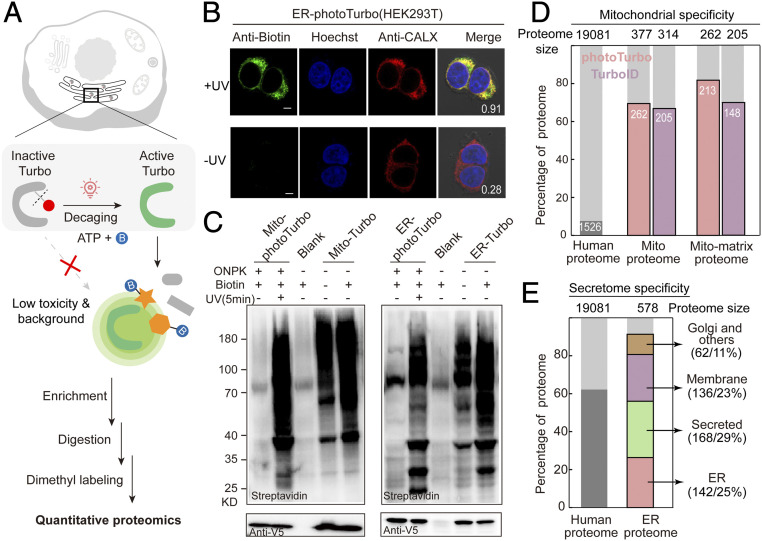
We created a HEK293T cell line stably expressing the pyrrolysine-amino acyl tRNA synthetase (PylRS) for ONPK incorporation. Utilizing this cell line, we verified localizations and labeling specificities of photoTurbo variants with fluorescence microscopy in mitochondrial matrix and ER lumen, respectively. (Fig. 2 A and B and SI Appendix, Fig. S2 A and B). We then compared the proximity-dependent labeling efficiency of these targeted photoTurbo fusions to the wild-type Turbo by Western blotting. The background biotinylation from photoTurbo prior to light illumination was almost imperceptible with the negative control omitting the enzyme and substantially lower than the sample expressing Turbo, indicating successful enzyme inactivation. Following photolysis at 365 nm for 5 min, biotinylation from decaged photoTurbo reached a similar level as that of wild-type Turbo (Fig. 2C and SI Appendix, Fig. S2C).
Fig. 2.
Engineering bioorthogonally activatable PL enzymes for SubMAPP. (A) Schematic description of photoTurbo-enabled subcellular proteomics. The light-activated photoTurbo allowed temporally gated and spatially restricted protein biotinylation that can be analyzed by MS-based quantitative proteomics. (B) Fluorescence images of HEK293T cells expressing photoTurbo in the ER lumen, with proteins biotinylated by UV illumination for 5 min. CALX: ER marker. (Scale bars: 5 µm.) Pearson's R values are shown in the corner. (C) Western blot analysis comparing the proximity-dependent protein labeling efficiency of photoTurbo with wild-type Turbo in mitochondrial matrix and endoplasmic reticulum. Cells transfected with empty vector were used as “Blank” sample. (D) MS-based identification and quantification of mitochondrial matrix proteome labeled by photoTurbo and TurboID, respectively. From left to right: the first column shows the fraction of annotated mitochondrial proteins in entire human proteome; the second and third columns compare the fractions of mitochondrial proteins in the data sets; and the fourth and fifth columns represent the fractions of mitochondrial matrix proteins in the data sets. (E) Specificity analysis of ER lumen proteome obtained via photoTurbo, with the coverage of secretion related proteins reaching 88% (508 out of 578 proteins).
We further evaluated the specificity and coverage of this photo-activatable PL enzyme with quantitative mass spectrometry (MS)-based subcellular proteomic profiling. HEK293T cells expressing organelle-targeted photoTurbo were illuminated with 365 nm light in the presence of biotin for 10 min before cell lysis, and biotinylated proteins were enriched with streptavidin-coated beads (Fig. 2A and SI Appendix, Fig. S3). Samples without UV irradiation were set as negative controls. Following on-beads digestion, enriched peptides were dimethyl labeled and analyzed by liquid chromatography–tandem mass spectrometry (LC-MS/MS). The quantitative proteomic data were filtered with a cutoff UV/−UV ratio >2. In the case of mitochondrial matrix-targeted photoTurbo, three replicated experiments covered 377 proteins with 70% specificity, which was on par with previously published mito-TurboID proteome (Fig. 2D and SI Appendix, Fig. S4 and Dataset S1) (14). For ER lumen, we identified 508 secretory pathway-related proteins (secretome proteins) with 88% specificity, including 142 ER resident proteins (28%), 33 Golgi apparatus proteins (6.5%), 168 secreted proteins (33%), and 136 membrane-bound proteins (27%) (Fig. 2E and SI Appendix, Fig. S5 and Dataset S2). Taken together, the above data demonstrated that our engineered, bioorthogonally activatable PL system exhibited both high spatial specificity and excellent coverage of subcellular proteome, while allowing precise and on-demand temporal control of enzymatic activity.
Subcellular Phosphoproteome Profiling by SubMAPP.
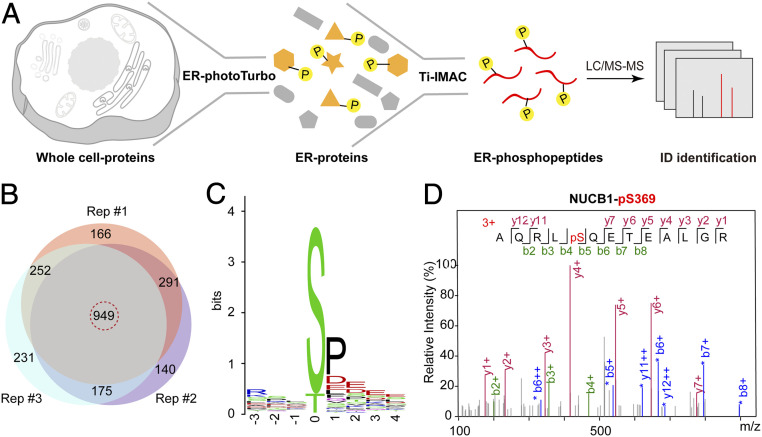
To portray the dynamic PTM of subcellular proteome, we devised a SubMAPP procedure that combined the activatable PL enzymes with phosphoproteomics. While protein phosphorylation frequently occurs in the cytoplasm, several protein kinases have been recently discovered to be responsible for phosphorylating proteins in the secretory pathway (30–33). Although this exciting discovery suggested that protein phosphorylation may occur within the ER lumen, we are currently lacking an ER-specific proteomic profiling tool to directly identify and/or verify these phosphorylation targets in living cells. Notably, previous efforts for analyzing ER phosphoproteome using ultracentrifugation were plagued with poor spatial specificity and temporal resolution (34). Alternatively, we combined ER-specific photoTurbo labeling with TiO2-mediated phosphoproteome enrichment as an orthogonal pull-down strategy: biotinylated proteins enriched from affinity purification were subjected to on-bead digestion to release phosphopeptides, which were further enriched by TiO2-coated beads and analyzed by LC-MS/MS (Figs. 1A and 3A). We have identified a total of 949 phosphopeptides from 500 phosphorylated proteins across three biological replicates in HEK293T cells (Fig. 3B and Dataset S3). Compared with our ER proteome, the specificity of ER phosphoproteome was slightly reduced to 66% (330 out of 500 proteins), with the nuclear phosphoproteins as the major source of contaminants (∼25%, SI Appendix, Fig. S6A). We therefore filtered our data set to contain only those peptides derived from secretory proteins (628 peptides from 330 proteins). Amino acid motif analysis of this curated list revealed that phosphorylation frequently occurred at the Ser residue, followed by Thr, and to a lesser extent Tyr. Notably, the majority of phosphosites were located within the S-P-E tripeptide motif (Fig. 3C), which coincided with the substrate recognition motif of Fam20C, a recently identified secretory pathway kinase (32). Our ER phosphoproteome list contains several reported Fam20C substrates such as ENPL (S306), TGO1 (S1906), and NUCB1 (S369) (Fig. 3D and SI Appendix, Fig. S6 B and C) (35). In addition, our results have unveiled phosphorylated proteins, such as ER-resident protein FKBP7 (S210), secreted protein GANAB (T440), plasma membrane protein DSG2 (S551), and HLA-B (S66 and T249) (Dataset S3 and SI Appendix, Fig. S7), which were all phosphorylated within ER lumen. Whether these proteins are phosphorylated by Fam20C, VLK, or by yet-unknown secreted kinases remain to be clarified. Taken together, SubMAPP allowed us to profile subcellular phosphoproteome, which revealed previously identified Fam20C substrates as well as kinase substrates in ER lumen.
Fig. 3.
SubMAPP-enabled subcellular phosphoproteomics. (A) Schematic description of ER phosphoproteome profiling by SubMAPP. (B) Venn diagram showing the overlap of phosphorylated peptides identified from three independent orthogonal pull-down experiments in living cells. (C) Motif analysis of secretome-related phosphopeptides. (D) MS/MS spectrum of phosphopeptide derived from NUCB1, with b- and y-ions identified by the pFind software (34).
SubMAPP for Dynamic ER Lumen Proteome and Phosphoproteome Profiling.
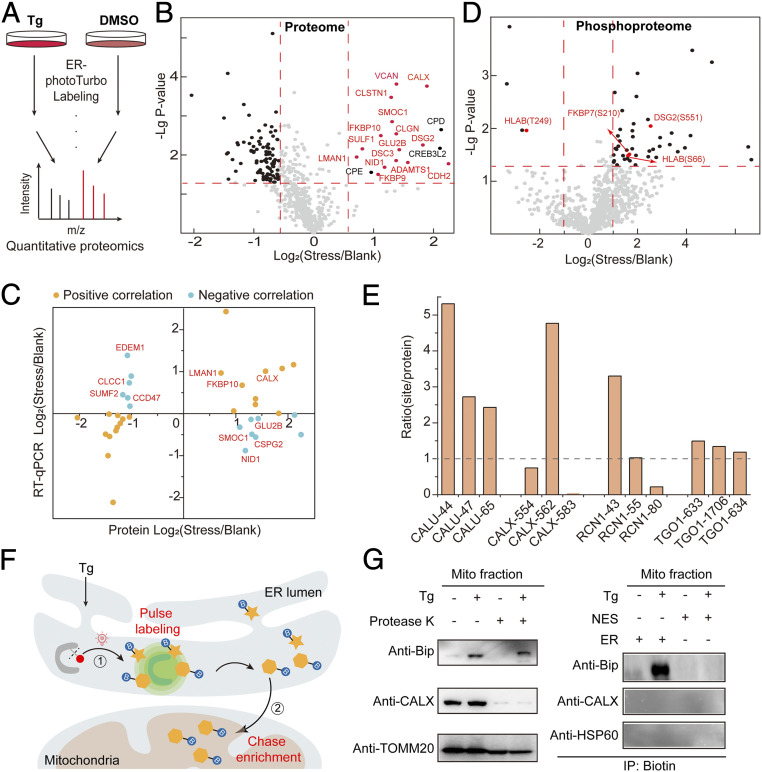
We next utilized the temporal feature of SubMAPP to monitor the dynamic changes of ER proteome and phosphoproteome upon ER stress. As an inhibitor of the sarco/endoplasmic reticulum Ca2+-ATPase (SERCA), thapsigargin (Tg) could disrupt calcium homeostasis and induce ER stress, which can activate cellular responses such as unfolded protein response (UPR) and ER associated protein degradation (ERAD) (36–38). HEK293T cells expressing ER lumen-targeted photoTurbo were treated with Tg (500 nM) or DMSO for 8 h, followed by photo-activation for ER proteome labeling (Fig. 4A). The presence of Tg-induced ER stress was verified by the elevated expression of Bip, a known ER stress marker (SI Appendix, Fig. S5A). Across three biological replicates with quantitative proteomic analysis, a total of 636 proteins were identified (Fig. 4B and Dataset S4). The characteristic, asymmetric distribution of Stress/Blank ratios indicated a global depletion of ER luminal proteins, with 93 proteins below the cutoff Stress/Blank ratio of 0.67 (P < 0.05), which may arise from ER stress-induced mRNA decay (Fig. 4B and SI Appendix, Fig. S8C) (39). Notably, against this global depletion background, 18 proteins emerged as being up-regulated (cutoff Stress/Blank ratio > 1.5, P < 0.05). Gene Ontology analysis of these up-regulated proteins revealed an overrepresentation of the following terms: protein folding, ER unfolded protein response, cell adhesion, and glycoprotein metabolic process (SI Appendix, Fig. S8B). A majority of these up-regulated proteins are calcium-binding proteins including Ca2+-binding chaperones (e.g., CALX, CALR, CLGN), protein folding enzymes (e.g., FKBP9, FKBP10) as well as proteins required for ER export (e.g., LMAN1), which may enhance the chaperone/enzyme activity and maintain ER calcium homeostasis during Tg-induced ER stress (SI Appendix, Fig. S8 A and B).
Fig. 4.
SubMAPP for dynamic subcellular proteomics and phosphoproteomics. (A) Experimental scheme for ER-photoTurbo labeling in the presence and absence of ER stress inducer, Tg. (B) Volcano plot showing significant change of secretory proteins in ER lumen under 500 nM Tg treatment. Calcium binding proteins are highlighted in red. (C) The transcription and translation correlation analysis on proteins under ER stress. (D) Volcano plot showing the change of phosphorylated peptides in ER lumen under ER stress. (E) The correction of phosphorylation level of partial phosphorylated proteins, which coincide with ER proteome under ER stress. (F) Schematic description of the photo-controlled Turbo-enabled “pulse-chase” strategy for monitoring or identification of ER-to-mitochondria translocated proteins. In brief, 1) ER proteins were first “pulse” labeled by photo-activation of Turbo upon ER stress, and 2) the mitochondria fraction was then isolated to “chase” enrich the biotinylated proteins in mitochondria. (G) Immunoprecipitation for detecting the ER-to-mitochondria translocated Bip protein under ER stress. Mitochondria fraction was isolated by mitochondria isolation kit (Abbike KTP4003) and treated with protease K before being subject to immunoblotting analysis. CALX was used as the ER marker, while TOMM20 and HSP60 were used as the mitochondrial markers.
RT-qPCR was used to examine the transcriptional change, which revealed a total of 18 up-regulated and 19 significantly down-regulated proteins by a stringent cutoff (fold-change < 0.5, P < 0.05). As expected, a positive correlation between protein abundance and transcription level was observed for most proteins, including folding factors, UPR-related proteins, and proteins involved in the extracellular matrix organization. However, a negative correlation was also detected for a subset of proteins. For instance, several proteins involved in the glycoprotein metabolism (e.g., GLU2B, SMOC1, CSPG2, NID1) with lower transcription levels were highly enriched in our proteomics data set. In contrast, several proteins involved in the ERAD pathway and glycoside catabolism (e.g., CCD47, EDEM1, SUMF2, CLCC1) had higher transcription activity but were down-regulated at the protein level (Fig. 4C and SI Appendix, Fig. S8 D and E). The discrepancies between mRNA levels and protein levels might be explained by the difference in the translation efficiency and/or the subcellular targeting efficiency.
The dynamic change of ER phosphoproteome was next investigated. From three label-free quantitative proteomic experiments (Fig. 4D and Dataset S5), we identified a total of 51 phosphosites from 42 proteins with significant changes in response to ER stress. Among these, 42 phosphopeptides from 34 secretion-related proteins were up-regulated, while another 3 phosphopeptides from 3 proteins were down-regulated (Fig. 4D). By normalizing changes in phosphorylation levels against changes in protein-levels, we found that 17 out of 24 phosphorylation sites (71%) were up-regulated under ER stress, which was likely caused by elevated kinase activity in the ER lumen (Fig. 4E and SI Appendix, Fig. S9). Interestingly, the two phosphosites in the human leukocyte antigen HLA-B, Ser66 and T249, were up- and down-regulated, respectively. Both phosphosites are located at the loop regions, which may be more accessible to the corresponding kinase (SI Appendix, Fig. S10). Since HLA-B has been associated with ER-stress in previous reports (40), our results suggested that the different phosphorylation on HLA-B may contribute to its role in ER stress.
Interestingly, while we found that the whole cell expression of a major ER chaperone BiP was up-regulated upon ER stress, its abundance remained virtually unchanged within ER lumen (SI Appendix, Fig. S11). Previous fluorescence imaging study showed that BiP was relocated to the mitochondria when cells were under ER stress (41, 42). We confirmed the presence of BiP in isolated mitochondria by Western blotting, which was insensitive to Proteinase K treatment, indicating the localization in either the mitochondrial matrix or the intermembrane space (Fig. 4G).
Given the high abundance of BiP in ER and its presence in mitochondria upon ER stress, we hypothesized that BiP might translocate from the ER lumen to the mitochondria upon Tg-induced ER stress. We thus designed a “pulse-chase” experiment to examine whether a direct ER-to-mitochondria translocation might have occurred for BiP. As shown in Fig. 4F, HEK293T cells expressing ER lumen-targeted photoTurbo were treated with 500 nM Tg for 4 h to induce ER stress, photoactivated to “pulse” biotinylate the ER luminal proteins for 10 min, and subsequently “chased” for 4 h. Mitochondria isolation and affinity purification of its biotinylated proteins were conducted at the end of the “chase” period. Indeed, BiP was only found in the ER labeled sample after Tg treatment (Fig. 4G). Taken together, our study revealed the translocation of BiP from ER lumen to mitochondria under ER stress, which further established activatable Turbo system as a spatiotemporally specific tool for studying subcellular proteome dynamics.
Photo/ChemoTurbo-Enabled Subcellular Proteomics on Neuron Culture and Living Animals.
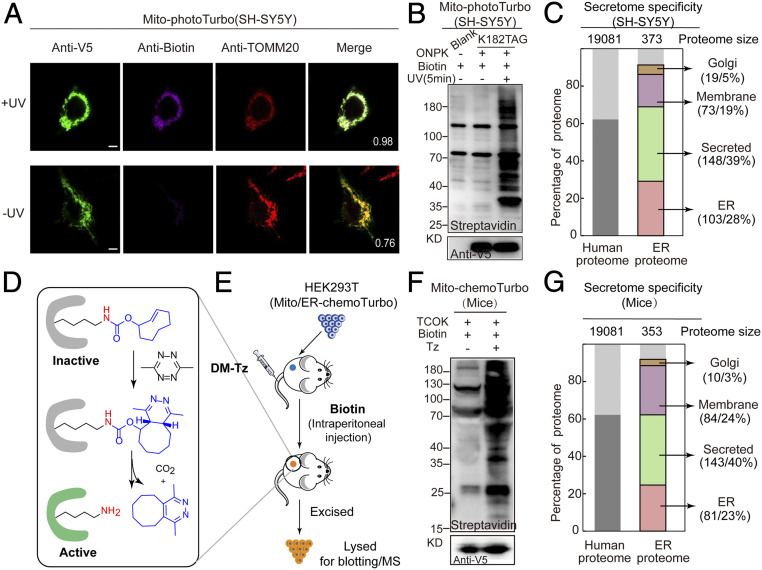
We next demonstrated the compatibility of SubMAPP in neuronal cell culture models, including the human neuroblastoma cell line SH-SY5Y and cultured rat primary neurons, which are particularly sensitive to the dysregulation of biotin metabolism (43, 44). In SH-SY5Y cells, mitochondrial matrix/ER lumen-targeted photoTurbo were expressed and allowed to specifically and efficiently label proximal proteins after 10 min irradiation (Fig. 5 A and B and SI Appendix, Fig. S12). Furthermore, we also evaluated the specificity and coverage of photoTurbo through proteomic experiments in SH-SY5Y cells. A total of 343 secretory pathway related proteins were identified with specificity up to 92% from three biological replicates (Fig.5C and Dataset S6). In cultured rat cortical neurons, photoTurbo can be expressed in the mitochondrial matrix and ER lumen. As expected, these targeted photo-Turbo variants had low biotinylation background and negligible toxicity in the presence of biotin molecules in neuron cells, which can be rapidly activated with high proximal labeling efficiency (SI Appendix, Fig. S13).
Fig. 5.
Photo/chemoTurbo-enabled subcellular proteomics on neuron culture and living mice. (A) Fluorescence imaging of photoTurbo location and the activation activity in mitochondrial matrix of SH-SY5Y. Anti-V5 (green) was used to visualize Turbo expression, purple is the biotinylation signal to verify the labeling efficiency of photo-controlled Turbo, and red is the mitochondrial marker-TOMM20. UV, 5 min. (Scale bars: 5 µm.) Pearson’s R values of biotin and TOMM20 are shown in the corner. (B) Photo-activation efficiency of photoTurbo for proximity-dependent labeling of proteins in mitochondrial matrix of SH-SY5Y. (C) MS-based identification and quantification of ER lumen proteome labeled by photoTurbo in SH-SY5Y cells. A total of 343 proteins were identified with 92% specificity (∼28% ER resident proteins, 5% Golgi apparatus proteins, 39% secreted proteins, and 19% membrane proteins). (D) Mechanism of the DM-Tz triggered IEDDA for biorthogonal decaging. (E) Schematic illustration of chemo-controlled Turbo activation in living mice. HEK293T cells expressing the mitochondria-Turbo-K182TCOK variant were subcutaneously injected into mice, followed by tail vein injection of DM-Tz and intraperitoneal injection of biotin, which led to the regeneration of active Turbo for proximal labeling. Subsequently, HEK293T cells were excised and lysed before being subject to immunoblotting analysis (F). (G) MS-based identification and quantification of ER lumen proteome labeled by chemoTurbo in mice, which identified 318 proteins (∼23% ER resident proteins, 3% Golgi apparatus proteins, 40% secreted proteins, and 24% membrane proteins).
Finally, to circumvent the limitation of photoTurbo in deep tissue or intact animals, we developed chemical-activatable Turbo (chemoTurbo) that extended the SubMAPP strategy to living mice. Through the incorporation of chemical-caged lysine analog N-(((E)-cyclooct-2-en-1-yl)-oxy)carbonyl-L-lysine (TCOK) (45, 46) at the K182 site, Turbo’s enzyme activity can be temporally blocked until being rapidly rescued by the addition of a chemical trigger, dimethyl tetrazine (DM-Tz, Fig. 5D and SI Appendix, Fig. S14A). Since chemoTurbo does not require UV illumination, it allows in situ enzyme activation in tissue or live animals. To demonstrate the efficiency of chemoTurbo in living mice, we injected HEK293T cells harboring mitochondrial matrix/ER lumen-targeted chemoTurbo subcutaneously into mice. Little biotinylation background was observed in the absence of DM-Tz. Upon the addition of DM-Tz via tail vein injection and the supplementation of biotin via intraperitoneal injection, chemoTurbo was rapidly turned on and efficiently biotinylated proteins (Fig. 5 E and F and SI Appendix, Fig. S14B). Interestingly, we further enriched the biotinylated proteins by ER lumen-targeted chemoTurbo in mice, which identified 318 secretory-related proteins with 90% specificity (Fig. 5G and Dataset S6). Taken together, these results demonstrated the capability of our SubMAPP strategy for subcellular proteome labeling in complex systems such as neurons and living animals.
Discussion
Dissecting the underlying mechanism of various protein-modulated biological processes requires comprehensive analysis of PTMs with high spatial and temporal precision (1, 2, 6). Herein, we developed a SubMAPP strategy by engineering bioorthogonally activatable PL enzymes for spatial–temporal resolved profiling of subcellular phosphoproteome in living cells. Based on the newly developed biotin ligase-TurboID (14), a spatial–temporal controlled photo/chemoTurbo was created by the genetically encoded chemical decaging strategy, which avoids cytotoxicity and PTM perturbations caused by biotin starvation, as well as the background biotinylation from endogenous biotin. The excellent spatial specificity and temporal precision allowed quantitative and comparative proteomic analysis of dynamic ER proteome under ER stress, which also offered a “pulse-chase” strategy for monitoring interorganelle protein translocation (e.g., ER to mitochondria) under living conditions.
Notably, the engineered PL enzyme could be integrated with phosphorylation-enrichment strategy and MS-based quantitative proteomics for dynamic profiling of phosphorylation changes in subcellular proteome. We profiled the phosphoproteome of ER lumen and revealed some known phosphosites of Fam20C, indicating that Fam20C may directly phosphorylate a cluster of its substrates within ER lumen (32). In addition, several phosphorylated proteins were identified by SubMAPP strategy. Through coinciding the proteome and phosphoproteome of ER lumen, we mapped the dynamic phosphorylation changes upon ER stress. Combining this strategy with affinity purification procedures for different PTMs (e.g., tyrosine phosphorylation, acetylation, and lactylation, etc.) may enable the exploration of the highly dynamic PTM processes of subcellular organelles under various physiological and/or pathological conditions.
Finally, we demonstrated the compatibility of photo/chemoTurbo-enabled SubMAPP in neuronal cell culture and living mice by immunofluorescence and MS-based proteomics. The results suggested that our strategy could be applied for spatiotemporal analysis of subcellular proteome and PTM dynamics in live neurons, whose function relies on the continual modulation of local protein synthesis and modifications (47). In addition, understanding the cellular dynamic regulation within the context of an intact mammalian organism is attractive but challenging. Our SubMAPP strategy may provide a general toolkit for spatiotemporal profiling of subcellular proteome and PTM-dynamics under in vitro and in vivo settings.
Methods
Detailed methods are available in SI Appendix.
Supplementary Material
Acknowledgments
We acknowledge support from Prof. Chu Wang, Dr. Wen Zhou, and the Proteomic Mass Spectrometry Core of the National Facilities for Protein Sciences (the Phoenix Project) at Peking University. This work was supported by the National Basic Research Program of China (Grants 2018YFA0507600, 2017YFA0503600, and 2016YFA0501500), the National Natural Science Foundation of China (Grant 21673009, 21521003, 21937001, 32088101, and 91753131), and NSF of Beijing Municipality (Grant 5182011). P.R.C. is sponsored by the Xplore Prize, and P.Z. is sponsored by Li Ge-Zhao Ning Life Science Junior Research Fellowship and Bayer Investigator Award.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025299118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Hunter T., Signaling–2000 and beyond. Cell 100, 113–127 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Blume-Jensen P., Hunter T., Oncogenic kinase signalling. Nature 411, 355–365 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Lahiry P., Torkamani A., Schork N. J., Hegele R. A., Kinase mutations in human disease: Interpreting genotype-phenotype relationships. Nat. Rev. Genet. 11, 60–74 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Sharma K., et al., Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 8, 1583–1594 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Humphrey S. J., Azimifar S. B., Mann M., High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat. Biotechnol. 33, 990–995 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Altelaar A. F. M., Munoz J., Heck A. J. R., Next-generation proteomics: Towards an integrative view of proteome dynamics. Nat. Rev. Genet. 14, 35–48 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Larance M., Lamond A. I., Multidimensional proteomics for cell biology. Nat. Rev. Mol. Cell Biol. 16, 269–280 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Lundberg E., Borner G. H. H., Spatial proteomics: A powerful discovery tool for cell biology. Nat. Rev. Mol. Cell Biol. 20, 285–302 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Zhu H., Tamura T., Hamachi I., Chemical proteomics for subcellular proteome analysis. Curr. Opin. Chem. Biol. 48, 1–7 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Rhee H. W., et al., Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328–1331 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lam S. S., et al., Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 12, 51–54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi-Rhee E., Schulman H., Cronan J. E., Promiscuous protein biotinylation by Escherichia coli biotin protein ligase. Protein Sci. 13, 3043–3050 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roux K. J., Kim D. I., Raida M., Burke B., A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Branon T. C., et al., Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin W., Cho K. F., Cavanagh P. E., Ting A. Y., Deciphering molecular interactions by proximity labeling. Nat. Methods 18, 133–143 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Y., Zou P., The evolving capabilities of enzyme-mediated proximity labeling. Curr. Opin. Chem. Biol. 60, 30–38 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Vepa S., et al., Hydrogen peroxide stimulates tyrosine phosphorylation of focal adhesion kinase in vascular endothelial cells. Am. J. Physiol. 277, L150–L158 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Pacheco-Alvarez D., et al., Paradoxical regulation of biotin utilization in brain and liver and implications for inherited multiple carboxylase deficiency. J. Biol. Chem. 279, 52312–52318 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Agrawal S., Agrawal A., Said H. M., Biotin deficiency enhances the inflammatory response of human dendritic cells. Am. J. Physiol. Cell Physiol. 311, C386–C391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solórzano-Vargas R. S., Pacheco-Alvarez D., León-Del-Río A., Holocarboxylase synthetase is an obligate participant in biotin-mediated regulation of its own expression and of biotin-dependent carboxylases mRNA levels in human cells. Proc. Natl. Acad. Sci. U.S.A. 99, 5325–5330 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.León-Del-Río A., Biotin in metabolism, gene expression, and human disease. J. Inherit. Metab. Dis. 42, 647–654 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Madsen C. T., et al., Biotin starvation causes mitochondrial protein hyperacetylation and partial rescue by the SIRT3-like deacetylase Hst4p. Nat. Commun. 6, 7726 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sletten E. M., Bertozzi C. R., Bioorthogonal chemistry: Fishing for selectivity in a sea of functionality. Angew. Chem. Int. Ed. Engl. 48, 6974–6998 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zorn J. A., Wells J. A., Turning enzymes ON with small molecules. Nat. Chem. Biol. 6, 179–188 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Li J., Chen P. R., Development and application of bond cleavage reactions in bioorthogonal chemistry. Nat. Chem. Biol. 12, 129–137 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Tarrant M. K., Cole P. A., The chemical biology of protein phosphorylation. Annu. Rev. Biochem. 78, 797–825 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sternicki L. M., Wegener K. L., Bruning J. B., Booker G. W., Polyak S. W., Mechanisms governing precise protein biotinylation. Trends Biochem. Sci. 42, 383–394 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Liu C. C., Schultz P. G., Adding new chemistries to the genetic code. Annu. Rev. Biochem. 79, 413–444 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Gautier A., et al., Genetically encoded photocontrol of protein localization in mammalian cells. J. Am. Chem. Soc. 132, 4086–4088 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Tagliabracci V. S., Pinna L. A., Dixon J. E., Secreted protein kinases. Trends Biochem. Sci. 38, 121–130 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bordoli M. R., et al., A secreted tyrosine kinase acts in the extracellular environment. Cell 158, 1033–1044 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tagliabracci V. S., et al., A single kinase generates the majority of the secreted phosphoproteome. Cell 161, 1619–1632 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J., et al., Secretory kinase Fam20C tunes endoplasmic reticulum redox state via phosphorylation of Ero1α. EMBO J. 37, e98699 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou H., et al., Analysis of the subcellular phosphoproteome using a novel phosphoproteomic reactor. J. Proteome Res. 9, 1279–1288 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Chi H., et al., Comprehensive identification of peptides in tandem mass spectra using an efficient open search engine. Nat. Biotechnol. 36, 1059–1061 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Walter P., Ron D., The unfolded protein response: From stress pathway to homeostatic regulation. Science 334, 1081–1086 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Hetz C., Chevet E., Harding H. P., Targeting the unfolded protein response in disease. Nat. Rev. Drug Discov. 12, 703–719 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Rogers T. B., Inesi G., Wade R., Lederer W. J., Use of thapsigargin to study Ca2+ homeostasis in cardiac cells. Biosci. Rep. 15, 341–349 (1995). [DOI] [PubMed] [Google Scholar]
- 39.Hollien J., Weissman J. S., Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313, 104–107 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Metin Y., Gulce S. K., The relation between ER stress and HLA-B27 misfolding. Med. Res. Innov. 1, 1–5 (2017). [Google Scholar]
- 41.Sun F.-C., et al., Localization of GRP78 to mitochondria under the unfolded protein response. Biochem. J. 396, 31–39 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li N., Zoubeidi A., Beraldi E., Gleave M. E., GRP78 regulates clusterin stability, retrotranslocation and mitochondrial localization under ER stress in prostate cancer. Oncogene 32, 1933–1942 (2013). [DOI] [PubMed] [Google Scholar]
- 43.McKay B. E., Molineux M. L., Turner R. W., Biotin is endogenously expressed in select regions of the rat central nervous system. J. Comp. Neurol. 473, 86–96 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Sedel F., Bernard D., Mock D. M., Tourbah A., Targeting demyelination and virtual hypoxia with high-dose biotin as a treatment for progressive multiple sclerosis. Neuropharmacology 110, 644–653 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Li J., Jia S., Chen P. R., Diels-Alder reaction-triggered bioorthogonal protein decaging in living cells. Nat. Chem. Biol. 10, 1003–1005 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Fan X., et al., Optimized tetrazine derivatives for rapid bioorthogonal decaging in living cells. Angew. Chem. Int. Ed. Engl. 55, 14046–14050 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Rangaraju V., Tom Dieck S., Schuman E. M., Local translation in neuronal compartments: How local is local? EMBO Rep. 18, 693–711 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.