Significance
Fever is a common symptom of infections and inflammation. We observed CD8+ T cells exposed to febrile-range temperature during activation, augmented mitochondrial mass, and metabolic activity. An adoptive transfer of these cells into tumor-bearing mice led to enhanced antitumor responses, suggesting that exposure to febrile temperature during activation contributes to the development of robust T cell responses. We identified that mitochondrial translation is integral to the enhanced mitochondrial mass and function of CD8+ T cells exposed to febrile temperature. Collectively, this research raises the possibility that fever may be beneficial for optimizing metabolic and functional responses in CD8+ T cells.
Keywords: T cell, metabolism, immunology, fever, mitochondria
Abstract
Fever can provide a survival advantage during infection. Metabolic processes are sensitive to environmental conditions, but the effect of fever on T cell metabolism is not well characterized. We show that in activated CD8+ T cells, exposure to febrile temperature (39 °C) augmented metabolic activity and T cell effector functions, despite having a limited effect on proliferation or activation marker expression. Transcriptional profiling revealed an up-regulation of mitochondrial pathways, which was consistent with increased mass and metabolism observed in T cells exposed to 39 °C. Through in vitro and in vivo models, we determined that mitochondrial translation is integral to the enhanced metabolic activity and function of CD8+ T cells exposed to febrile temperature. Transiently exposing donor lymphocytes to 39 °C prior to infusion in a myeloid leukemia mouse model conferred enhanced therapeutic efficacy, raising the possibility that exposure of T cells to febrile temperatures could have clinical potential.
Fever is a metabolically intensive and complex physiological process that often occurs in response to infection. While fever can directly alter bacterial and viral replication, it is also apparent that fever can beneficially alter host immunity (1). Fever induction usually occurs rapidly following infection and coincides with the ramping up of immune responses. Innate immune cells primarily drive fever, which in turn can have multiple effects on the adaptive arm of the immune system. For example, fever can induce T cell migration, Th subset polarization, and enhanced cytokine production (2–5).
Upon activation, CD8+ T cells undergo a well-characterized transition from quiescent naïve T cells into fully activated effector T (TE) cells that produces cytokines and cytotoxic molecules important for controlling pathogens or tumor growth. During this transition, substantial molecular and metabolic reprogramming occurs to support and enhance the required proliferative and effector functions of the cell (6). As many metabolic processes and cellular organelles are sensitive to temperature changes, we hypothesized that febrile temperatures could impact this early metabolic reprogramming and lead to functional changes in activated CD8+ T cells.
To test this, we utilized methods to induce febrile range temperature without the confounding effects of classical pyretic inflammatory mediators. We observed that CD8+ T cells exposed to 39 °C during activation had augmented glycolytic metabolism and mitochondrial oxidative phosphorylation (OXPHOS). An adoptive transfer of these cells into tumor-bearing mice led to enhanced antitumor responses, suggesting that exposure to febrile temperature during activation is advantageous to developing robust and protective T cell responses. Through genetic and pharmacological approaches, we identified that mitochondrial translation is integral to the enhanced mitochondrial mass and function of CD8+ T cells exposed to febrile temperature.
Results
Febrile Temperature Augments CD8+ TE Cell Cytokine Production and Glucose Metabolism In Vitro.
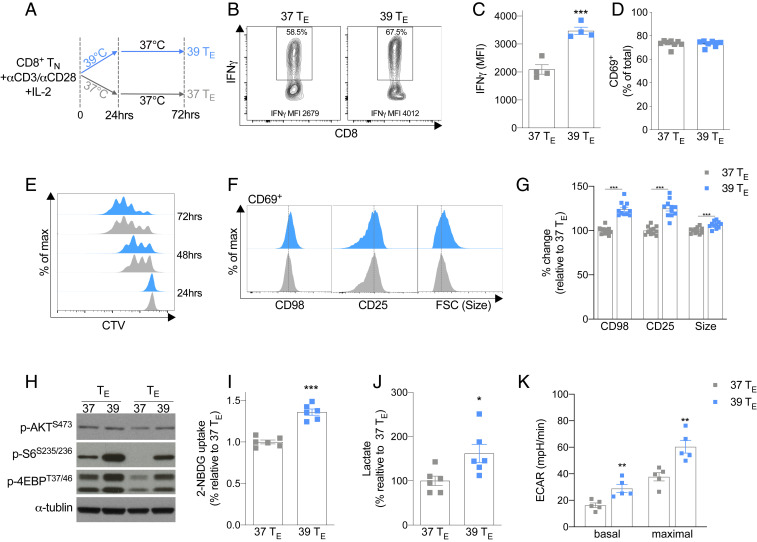
Fever typically occurs acutely after exposure to pyretic stimuli such as bacterial products or in response to prostaglandins or cytokines, such as PGE2, TNF, and IL-1β (3, 7). We reasoned that in the context of infectious disease, this time point would likely coincide with the very early activation of T cells. To replicate this situation in vitro, we activated purified naïve CD8+ T cells with anti-CD3/CD28 at 37 °C (37 °C TE) or 39 °C (39 °C TE) (Fig. 1A). To mimic the transient nature of fever, 24 h after stimulation, 39 °C TE cells were returned to 37 °C for the remainder of the culture period. Despite the cells only being exposed to 39 °C for the first 24 h, when restimulated 3 d after activation, they produced more IFN-γ (Fig. 1 B and C), consistent with previous reports (3). We observed no difference in the expression of the early activation marker CD69 between 37 and 39 °C TE cells 24 h after activation (Fig. 1D), suggesting the observed changes in IFN-γ expression on day 3 were not simply due to an increased percentage of cells becoming activated during exposure to 39 °C. Despite exhibiting a similar proliferative profile between 24 and 72 h after activation (Fig. 1E and SI Appendix, Fig. S1A), 39 °C TE cells had increased CD98 and CD25 expression and an increase in cell size 24 h after activation when compared with 37 °C TE cells (Fig. 1 F and G). This effect, however, was only seen in activated T cells, as T cells that were not expressing the early activation marker CD69 failed to show increased cell size or enhanced expression of CD98 or CD25 (SI Appendix, Fig. S1 B and C). Increased cell size and enhanced CD98 expression often occur in conjunction with the augmentation of anabolic processes and glycolytic metabolism within T cells (6). Consistent with this concept, we observed increased levels of phosphorylated mTORC1 targets 4E-BP (p4E-BP) and ribosomal protein S6 (p-S6) (Fig. 1H), an enhanced acquisition of the fluorescently labeled glucose analog 2-NBDG (Fig. 1I) and a concomitant increase in lactate production (Fig. 1J) in the 39 °C TE cells. The levels of p-AKTS473, a phosphorylation site associated with mTORC2 signaling, did not markedly change (Fig. 1H). Using extracellular flux analysis, we also observed increases in both basal extracellular acidification (ECAR) and maximal glycolytic response in 39 °C TE cells (Fig. 1K and SI Appendix, Fig. S1D). Collectively, these results indicated that TE cells exposed to febrile temperatures have an altered metabolic profile consistent with enhanced anabolism and increased glucose metabolism.
Fig. 1.
Exposure to 39 °C during activation promotes CD8+ TE cell function and anabolic metabolism. (A) Naïve CD8+ T cells were activated with anti-CD3/CD28 at 37 or 39 °C for 24 h and then subsequently cultured at 37 °C. (B and C) Representative histogram and bar graph of mean fluorescent intensity (MFI) of IFN-γ production assessed 72 h postactivation. (D) Percent of CD69+ cells 24 h after activation. (E) Representative histogram of proliferation as assessed by CellTrace Violet (CTV) 24 to 72 h after activation. (F and G) Representative histogram and bar graphs of the MFI of CD69+ cells, surface-expressed CD25 and CD98, and forward scatter (FSC) cell size 24 h postactivation. (H) Representative Western blots of two biological replicates showing phosphorylated (p) AKT, S6, and 4E-BP1 24 h postactivation. (I) Potential glucose uptake measured by 2-NBDG uptake 24 h postactivation. (J) Lactate production 24 h postactivation. (K) ECAR measured at baseline and at the maximal rate following addition of rotenone and antimycin. n ≥ 3 biological replicates/group as indicated by individual data points and shown as mean ± SEM (except for the Western blots, n = 2). *P < 0.05, **P < 0.01, ***P < 0.001.
Febrile Temperature Increases Mitochondrial Mass and Activity in CD8+ TE Cells.
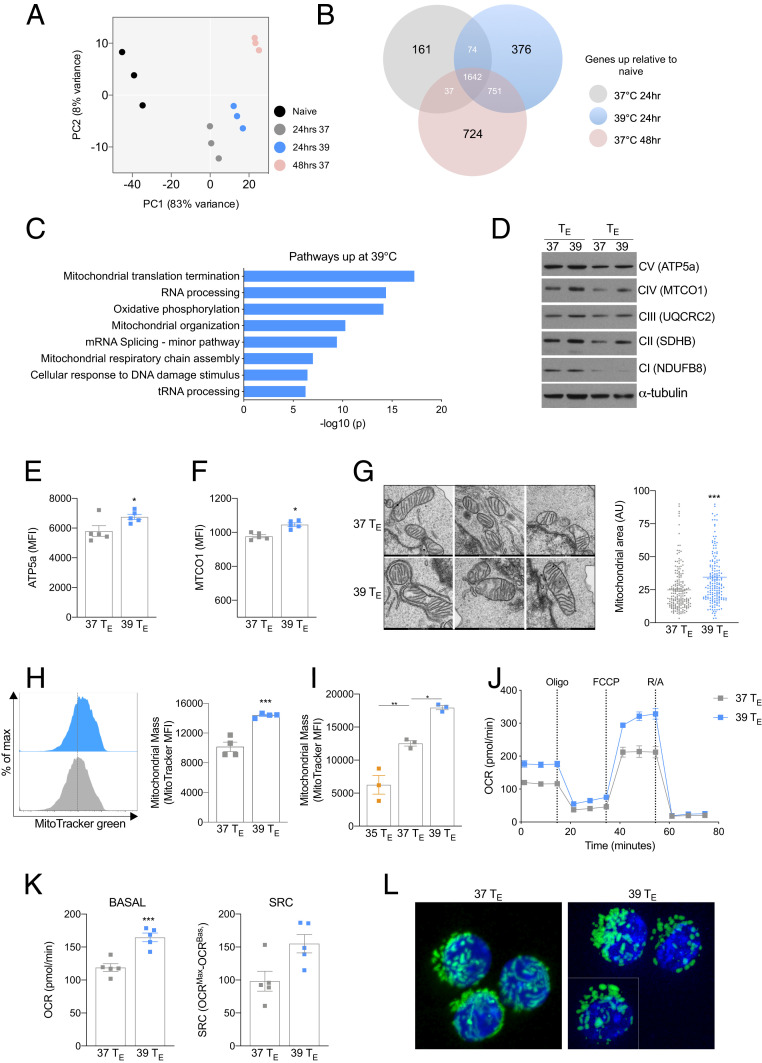
As enhanced anabolic metabolism is a characteristic of highly active TE cells (6), we reasoned that the metabolic changes in 39 °C TE cells could simply be due to a more rapid transition to an effector phenotype. Indeed, a small but significant increase in T-bet and Eomesodermin (EOMES) was detected in the 39 °C TE cells 24 h after activation (SI Appendix, Fig. S2A). To explore this, we conducted RNA sequencing (RNA-seq) analysis of 37 and 39 °C TE cells 24 h after activation and compared their transcriptional profiles with naïve T cells and 37 °C TE cells cultured for 48 h. Principal component analysis revealed that all activated TE cell populations had substantially distinct gene expression profiles compared with naïve T cells (Fig. 2A). Consistent with the phenotypic changes shown in Fig. 1, 39 °C TE cells had an altered transcriptional profile compared with 37 °C TE cells at 24 h, which, however, was also distinct from 37 °C TE cells cultured for 48 h (Fig. 2A), indicating that 39 °C TE cells did not simply transition to a 48 h–like phenotype early. To explore these data further, we identified the expression of transcripts that were significantly increased in any of the groups relative to that of naïve T cells (Fig. 2B). As expected, there was substantial overlap between all three activated populations; however, 376 genes were specifically up-regulated in the 39 °C TE cells (Fig. 2B), suggesting that exposure to 39 °C induced a distinct transcriptional profile in CD8+ TE cells. We performed pathway analysis on these 376 genes and found that four of the six most significantly enriched pathways were related to mitochondrial processes (Fig. 2C). As OXPHOS was one of the enriched pathways, we assessed expression of OXPHOS machinery proteins (Fig. 2D) and observed increases in nuclear-encoded succinate dehydrogenase (SDH) B and ATP5a (Fig. 2 D and E) and mitochondria-encoded cytochrome c oxidase subunit I (MTCO1) in 39 °C TE cells (Fig. 2 D and F). When we assessed electron micrograph images of 39 °C TE cells, we observed an increase in mitochondrial area compared with 37 °C TE cells (Fig. 2G), suggesting that in addition to an increase in mitochondrial protein expression, the total mitochondria mass was also likely enhanced. To confirm this, we measured total mitochondrial mass using MitoTracker Green (Fig. 2H) or expression of the mitochondrial import protein TOMM20 (SI Appendix, Fig. S2B) and detected an increase in mitochondrial mass within 39 °C TE cells. The observed increase in mitochondrial mass was sensitive to temperature, as cells activated at 35 °C had lower mitochondrial mass compared with both 37 and 39 °C TE cells (Fig. 2I). As 39 °C TE cells exhibited increased glycolysis (Fig. 1K), we cultured cells in reduced glucose media (1 mM glucose) or with the glycolysis inhibitor 2 deoxy-glucose (2DG) and found the trend of increased mitochondrial mass, while less pronounced, was preserved (SI Appendix, Fig. S2 C and D). To confirm that the increase in mitochondrial mass within 39 °C TE cells also occurred following T cell receptor (TCR) activation with antigen, we stimulated naïve CD8+ OT-I cells (which express a TCR specific for the OVA peptide, SIINFEKL) with bone marrow–derived dendritic cells loaded with the SIINFEKL peptide (SI Appendix, Fig. S2E) and also observed greater MitoTracker Green staining. Together, these results demonstrated that febrile temperature increases mitochondrial mass in CD8+ TE cells.
Fig. 2.
Exposure to 39 °C augments mitochondrial metabolism. (A) Principal component analysis plot of RNA expression in naïve 37 or 39 °C CD8+ TE cells. (B) Gene expression of significantly increased transcripts relative to naïve cells. (C) Top eight up-regulated pathways (Metascape gene analysis) in 39 °C TE cells using data derived from the same experiments as A and B. (D) Western blot of two biological replicates of mitochondrial respiratory components 24 h postactivation. (E and F) Mean fluorescent expression (MFI) of ATP5a and MTCO1 expression 24 h postactivation measured by flow cytometry. (G) Representative electron microscope images and mitochondrial area measured in arbitrary units (AU) from electron micrographs of cells 24 h postactivation. Each point represents an individual averaged mitochondrial area. (H) Representative histogram and bar graph of mitochondrial mass as measured by MitoTracker Green 24 h postactivation. (I) Mitochondrial mass measured by MitoTracker Green MFI 24 h postactivation at 35, 37, or 39 °C. (J) A representative plot of OCR at baseline and following administration of oligomycin (oligo), fluoro-carbonyl cyanide phenylhydrazone (FCCP), and rotenone and antimycin (R/A) of T cells 24 h postactivation. (K) Basal oxygen consumption and SRC 24 h postactivation. (L) Confocal images of mitochondria (green) in CD8+ OT-I PhAM GFP cells counterstained with nuclear stain DAPI (blue) in 37 or 39 °C TE cells 24 h postactivation. The white dividing line indicates an overlaid cropped image. (I) For all experiments, n ≥ 3 biological replicates/group and shown as mean ± SEM (except for L, n = 1). *P < 0.05, **P < 0.01, ***P < 0.001.
Previous reports have shown that increased temperatures can enhance plasma membrane fluidity in T cells, allowing for enhanced microdomain clustering and amplified signaling downstream of the TCR (3, 8). However, we still observed the effects of increased mitochondrial mass when we bypassed TCR signaling by activating the T cells with phorbol 12-myristate 13-acetate (PMA) and ionomycin (IONO) (SI Appendix, Fig. S2F) or by chemically increasing membrane fluidity with ethanol (8) (SI Appendix, Fig. S2G), indicating that enhanced mitochondrial mass within 39 °C TE cells is not simply caused by amplified TCR signaling. Further supporting this concept, increasing membrane fluidity by incubating naïve T cells at 39 °C prior to (but not during) activation did not induce increased mitochondrial mass (SI Appendix, Fig. S2H).
As cellular thermogenesis can be regulated through expression of mitochondrial uncoupling proteins (UCP), we assessed the protein expression of UCP1 and UCP2 but observed no difference between 37 and 39 °C TE cells (SI Appendix, Fig. S2I). The changes in integrin expression and CD4+ T cell polarization during exposure to febrile range temperature have previously been found to be dependent on increased HSP90 expression (2, 4). To test if increases in mitochondrial mass was also dependent on HSP90, we incubated cells with an HSP90 inhibitor (HSP90i) at concentrations previously used in T cells (2). We observed no significant difference in mitochondrial mass in 39 °C TE cells treated with the HSP90i, suggesting that increased mitochondrial mass in this context is not dependent on enhanced HSP90 expression (SI Appendix, Fig. S2J).
Consistent with increased mitochondrial mass in 39 °C TE cells, we observed enhanced basal oxygen consumption rates (OCR), although we did not observe a significant increase in spare respiratory capacity (SRC) 24 h after activation (Fig. 2 J and K). These results combined with the observed increase in ECAR (Fig. 1K) indicated that 39 °C TE cells are more metabolically active 24 h after activation. Increased basal OCR, but not SRC or ECAR, were sustained 72 h after activation (SI Appendix, Fig. S2 K–N), despite the 39 °C TE cells only being transiently exposed to 39 °C during the first 24 h of activation. This suggested that transient exposure to increased temperature can result in prolonged changes in T cell mitochondrial activity. Consistent with this concept, when we incubated 37 and 39 °C TE cells with IL-15 on days 3 to 6 of culture (SI Appendix, Fig. S2O) to induce in vitro memory cells (37 and 39 °C TM), we observed enhanced OCR and ECAR in the 39 °C TM cells following restimulation with PMA and IONO (SI Appendix, Fig. S2P) and increased IFN-γ production after stimulation with anti-CD3/CD28 (SI Appendix, Fig. S2Q).
A fused mitochondrial morphology has been associated with enhanced mitochondrial OXPHOS after T cell activation (9, 10). We wondered if the enhanced OCR in 39 °C TE cells could also be correlated to a more fused mitochondrial phenotype. Despite the increased OCR, 39 °C TE cells did not exhibit a more fused mitochondrial morphology when compared with 37 °C TE cells (Fig. 2L) 24 h after activation, suggesting that increased mitochondrial content, rather than changes in mitochondrial morphology, contributed to the enhanced OCR.
A Nonpyrogenic Model of Whole-Body Heating Enhances Mitochondrial Mass and Metabolism in CD8+ T Cells In Vivo.
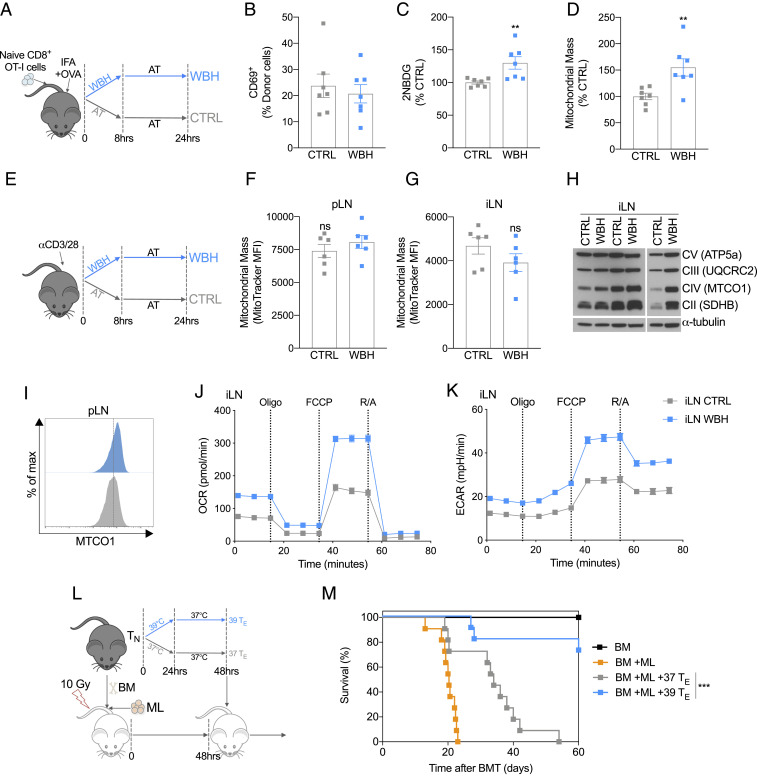
To determine if similar changes in T cell metabolism occur in vivo in response to febrile temperatures, we utilized a method of whole-body heating (WBH) (3) that increases and maintains the core body temperature of mice in a controlled manner (Fig. 3A and SI Appendix, Fig. S3A). WBH allowed us to explore the effect increased body temperature has on activated T cells without the confounding influence of infection or exogenous pyrogens that induce innate immune cell cytokines and the febrile response (3). We intravenously injected naïve CD8+ transgenic OT-I T cells into congenic recipient mice and then activated these cells by injecting OVA emulsified in sterile mineral oil (incomplete Freund's adjuvant, IFA) subcutaneously into the hind flanks. We then either exposed the mice to WBH for 8 h or kept the mice in comparable conditions (CTRL) at ambient temperature (AT) (Fig. 3A). After 24 h, we harvested the cells from the inguinal lymph nodes (iLN) that were proximal to the injection site. Similar to activated T cells exposed to 39 °C in vitro, no significant difference in the percentage of T cells expressing CD69 was observed between WBH and CTRL groups (Fig. 3B). However, there was a significant increase in both 2-NBDG uptake and MitoTracker Green staining in activated CD8+ T cells exposed to WBH (Fig. 3 C and D).
Fig. 3.
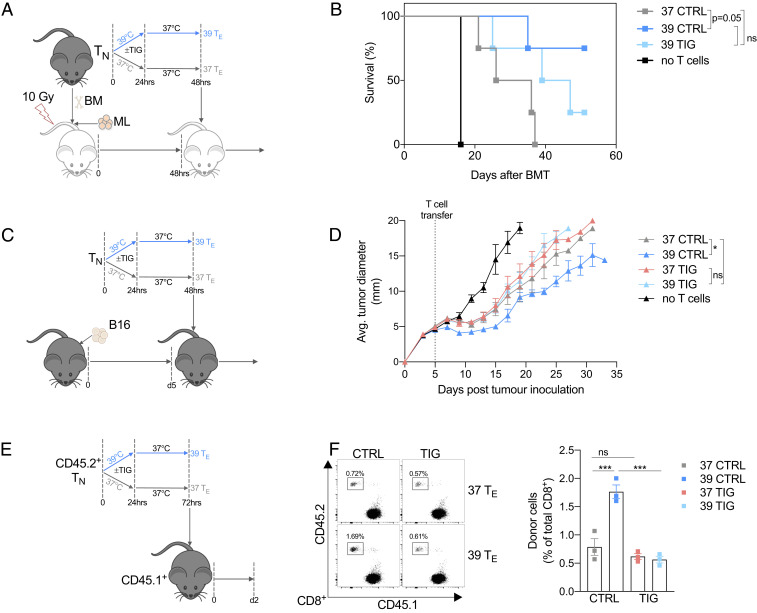
Exposure to 39 °C improves T cell metabolism and function in vivo. (A) Naïve OT-I T cells were injected (i.v) into congenic hosts, and then mice were injected s.c. in the hind flanks with OVA peptide emulsified in IFA. Mice were then either subjected to WBH to raise their temperature to 39 °C or were treated to CTRL at AT. After 8 h, all mice were housed at AT, and then cells were harvested from the mice at the 24-h timepoint. (B) Activation state of donor OT-I cells measured as percent CD69+. (C) Cells from the iLN were incubated ex vivo in the presence of 2-NBDG to measure glucose uptake. (D) Cells from the iLN were incubated with MitoTracker Green to assess mitochondrial mass. (A–D) Data from two independent experiments, n = 7 mice/group. (E) To induce a polyclonal T cell response, anti-CD3/CD28 was injected into the hind footpad of mice. Mice were then either subjected to WBH for 8 h or treated to CTRL at AT. Cells were harvested from the mice 24 h after anti-CD3/CD28 injection. (F) CD8+ cells were isolated from the popliteal LN and (G) iLN , and the mitochondrial mass was measured by MitoTracker Green. Data from two independent experiments, n = 6 mice/group. (H) Western blot of mitochondrial respiratory chain components in CD8+ T cells isolated from iLN 24 h after injection of anti-CD3/CD28 (each column represents cells pooled from two mice). (I) Representative histogram of MTCO1 expression in CD8+ T cells from iLN 24 h after injection of anti-CD3/CD28 measured by flow cytometry (each condition represents cells pooled from three mice). (J) OCR and (K) ECAR at baseline and following administration of oligomycin (oligo), fluoro-carbonyl cyanide phenylhydrazone (FCCP), and rotenone and antimycin (R/A) of CD8+ T cell from iLN 24 h after activation (each group represents cells pooled from three mice). Data from one of two independent experiments. (L) To model ML, irradiated recipient mice were injected with allogeneic BM cells and leukemic (ML: WEHI 3B) cells. After 2 d, 37 or 39 °C TE cells were injected, and survival was monitored. (M). Survival of mice following BMT and leukemia induction. Data from two independent experiments, n = 11 mice/group. **P < 0.01, ***P < 0.001.
To confirm that these effects were T cell intrinsic, we utilized a second in vivo model where we directly activated endogenous nontransgenic, polyclonal T cells by injecting anti-CD3/CD28 antibodies into the hind footpad of mice (Fig. 3E). We reasoned that this approach would allow us to directly activate T cells in the animal while minimizing any extenuating effects that increased temperature could have on other physiological processes, such as differences in migration or antigen presentation (4, 11). Mice were then either subjected to WBH or maintained at AT for 8 h (Fig. 3E). The following day, T cells were isolated from the draining lymph nodes (LN) and analyzed. In contrast to the previous experiments using OT-I cells (Fig. 3 A–D), activation with anti-CD3/CD28 in vivo did not induce a significant change in MitoTracker Green staining in CD8+ T cells from either the popliteal LN (pLN) (Fig. 3F) or the iLN (Fig. 3G) within WBH mice. We speculate that this discrepancy in MitoTracker Green staining between models may have to do with the intensity of activation induced, as almost 100% of the isolated T cells were CD69+ in the anti-CD3/CD28 model (SI Appendix, Fig. S3B) compared with ∼20% of the cells in the OT-I model (Fig. 3B). It is likely that cross-linking TCRs with high concentrations of anti-CD3 in vivo induces the release of inflammatory mediators and cytokines, which could result in a supraphysiological response. However, despite this, when we assessed protein expression of mitochondrial OXPHOS proteins, we observed increased expression in T cells isolated from WBH mice (Fig. 3 H and I). This suggested to us that despite not observing changes in MitoTracker Green staining within this context, WBH still impacted activated T cell mitochondria. Consistent with this, when we conducted extracellular flux analysis on CD8+ T cells isolated from anti-CD3/CD28–treated mice, we observed increases in both basal OCR and ECAR in the WBH group compared with the CTRL group (Fig. 3 J and K and SI Appendix, Fig. S3 C and D). Collectively, these results indicated that exposure to febrile temperature alone, without infection, induces enhanced mitochondrial activity in activated CD8+ T cells in vivo.
CD8+ TE Cells Exposed to Febrile Temperature Confer Better Protection in a Mouse Model of Myeloid Leukemia.
There is an accumulating body of literature demonstrating a strong link between robust mitochondrial metabolism and T cell function (6, 10, 12, 13). To test whether the enhanced mitochondrial properties evident in TE cells after exposure to increased temperatures translated into better functional properties in these cells, we employed a mouse model of myeloid leukemia (ML). This model is analogous to a treatment strategy commonly used in patients with specific hematological malignancies (14). In this treatment, allogeneic hematopoietic cell transplantation (HCT) is followed by donor lymphocyte infusions (DLI) to induce a robust graft-versus-leukemia response. To mimic this in mice, irradiated recipient mice were injected with allogeneic bone marrow (BM) cells and leukemic (ML: WEHI 3B) cells (Fig. 3L). After 2 d, 37 or 39 °C TE cells, that had been previously activated in vitro for 2 d, were injected into the leukemic mice. All mice that received a BM transplant (BMT) but no ML cells survived, whereas all mice that received BMT and ML died within 3 wk (Fig. 3M). The infusion of donor 37 °C TE cells following BMT prolonged the survival of mice with ML, indicating that the DLI of TE cells provides some benefit in promoting anti-leukemic responses, although 90% of these mice still died 40 d after leukemia induction (Fig. 3M). Remarkably, mice that received DLI of 39 °C TE cell following BMT and ML had substantially improved rates of survival when compared with those that received donor 37 °C TE cells, with 80% of the mice alive after 60 d (Fig. 3M). These results demonstrate that exposing T cells to febrile temperature prior to adoptive transfer fosters a more durable T cell response with enduring functional capacity.
Febrile Temperature Promotes Mitochondrial Translation in CD8+ TE Cells.
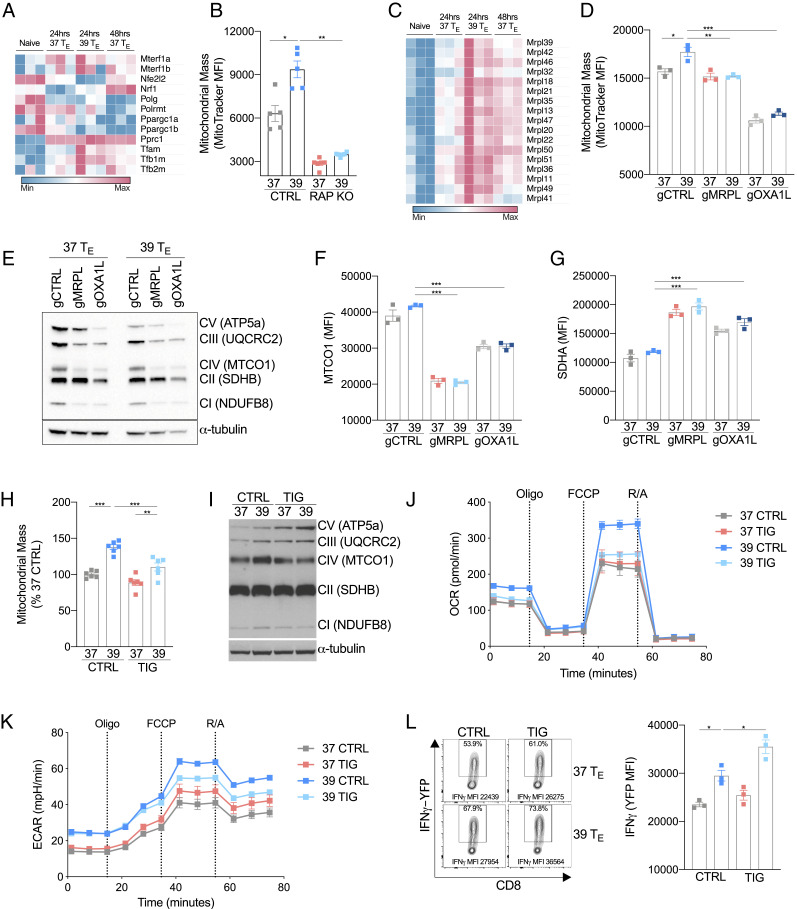
To determine the mechanistic basis of the enhanced functional capacity of TE cells exposed to increased temperature, we focused our attention on what was driving the enhanced mitochondrial mass and metabolism in 39 °C TE cells. Mitochondrial biogenesis involves a complex coordination of both nuclear and mitochondrially associated genes (15). We assessed the expression of a number of genes involved in mitochondrial transcription, and although there was a trend toward increased expression in some key genes such as TFAM, TFB1M, and TFB2M (15), other genes closely associated with mitochondrial biogenesis, like PGC-1α, were not increased on either the RNA or protein level (Fig. 4A and SI Appendix, Fig. S4A). We also failed to observe a clear pattern of augmented mitochondrially derived transcripts in the 39 °C TE cells (SI Appendix, Fig. S4B). This suggested to us that the changes in mitochondrial mass observed in 39 °C TE cells were influenced by additional factors beyond enhanced mitochondrial transcription. Since 39 °C TE cells exhibited increased mTORC1 signaling, and it has previously been shown that mTOR signaling can influence mitochondrial biogenesis through the regulation of nucleus-encoded mitochondria-related messenger RNAs (mRNAs) (16, 17), we assessed the role of mTOR in inducing mitochondrial mass. We compared mitochondrial mass in RAPTOR-deficient 37 °C TE cells with that of 39 °C TE cells. Consistent with previous literature, mitochondrial mass was significantly reduced in RAPTOR-deficient cells, with a reduction observed in both 37 and 39 °C TE cells (Fig. 4B). This indicates the importance for mTOR-related translation in supporting mitochondrial biogenesis. A similar effect was also observed in WT cells when the activity of mTOR was inhibited with rapamycin (SI Appendix, Fig. S4C).
Fig. 4.
Mitochondrial translation is enhanced by febrile temperature. (A) Heatmap showing mRNA expression of genes associated with mitochondrial biogenesis in naïve or activated CD8+ T cells. (B) MFI of MitoTracker Green 24 h postactivation in wild-type (CTRL) and RAPTOR-deficient (RAP KO) CD8+ T cells. (C) Heatmap showing mRNA expression of a selection of genes that encode mitochondrial ribosomal proteins (MRPL) in naïve or activated CD8+ T cells. (D) MFI of MitoTracker Green of CD8+ T cells targeted with CRISPR/Cas9 guides against MRPL39 (gMRPL), OXA1L (gOXA1L), or a nontargeting control (gCTRL) 24 h postactivation. (E) Western blot of mitochondrial respiratory components and (F and G) flow cytometry analysis of MTCO1 and SDHA in cells targeted with CRISPR/Cas9 guides against MRPL39 (gMRPL), OXA1L (gOXA1L), or a nontargeting control (gCTRL) 24 h postactivation. (H) Cells were treated with TIG (a mitochondrial-specific translation inhibitor) or CTRL during activation, and MitoTracker Green was measured 24 h postactivation. (I) Representative Western blot of mitochondrial respiratory components in CTRL- or TIG-treated groups. (J and K) OCR and ECAR at baseline and following administration of oligomycin (oligo), fluoro-carbonyl cyanide phenylhydrazone (FCCP), and rotenone and antimycin (R/A) of T cells 24 h after activation grouped from three replicates per condition. (L) Representative histogram and bar graph showing mean fluorescent expression (MFI) of IFN-γ production measured as expression of IFN-γ-YFP in CD8+ cells isolated from IFN-γ reporter (GREAT) mice and assessed 72 h postactivation. n ≥ 3 biological replicates/group as indicated by individual data points and shown as mean ± SEM (except E, n = 1). *P < 0.05, **P < 0.01, ***P < 0.001.
As mitochondrial biogenesis involves coordinated expression of both nuclear and mitochondrially associated proteins (15) and the pathway analysis of transcripts enhanced in 39 °C TE revealed an enrichment of processes involved in mitochondrial translation (Fig. 2C), we next focused our attention specifically on mitochondria translation. We observed a pattern of increased gene expression of mitochondrial ribosomal subunits (mrpl genes) in 39 °C TE cells (Fig. 4C), suggesting that mitochondria-specific protein translation, in addition to cytoplasmic translation, could have a role in enhancing mitochondrial processes in 39 °C TE cells. To further explore this, we impaired mitochondrial translation using CRISPR/Cas9 to delete the mitochondrial large 39s ribosomal subunit MRPL39 (SI Appendix, Fig. S4D) or the mitochondrial ribosome–associated insertase, OXA1L. We observed decreased MitoTracker Green staining at comparable levels in both 37 and 39 °C TE, indicating that mitochondrial translation contributes to changes in total mitochondrial mass and that defective mitochondrial translation inhibits the increase of mitochondrial mass in 39 °C TE cells (Fig. 4D). Consistent with the importance of mitochondrial translation for all activated T cells, a prolonged culture time of 72 h led to a more pronounced inhibition of mitochondrial mass in both 37 and 39 °C TE cells (SI Appendix, Fig. S4E). As expected, within 24 h of activation, mitochondrial respiratory components were impaired in MRPL39- and OXA1L-targeted groups, with a marked loss of mitochondrially translated MTCO1 (Fig. 4E). When we specifically compared the protein expression of MTCO1 with nuclear-encoded SDHA by flow cytometry, the loss of MRPL39 or OXA1L selectively impaired the expression of MTCO1 but enhanced SDHA expression (Fig. 4 F and G), indicating that although mitochondrial translation was affected and total mitochondrial mass decreased, nuclear-encoded mitochondrial proteins could still be effectively translated. Collectively, these results suggest that mitochondrial translation contributes to enhanced mitochondrial mass in 39 °C TE cells and that impairing mitochondrial translation can attenuate the enhanced expression of mitochondrially translated proteins in 39 °C TE cells.
To support these findings, we cultured 37 and 39 °C TE cells with a submaximal concentration (SI Appendix, Fig. S4F) of the antibiotic tigecycline (TIG; 0.5 μM), an inhibitor of mitochondrial translation. TIG treatment limited the increased mitochondrial mass (Fig. 4H) without impairing cytoplasmic protein synthesis (SI Appendix, Fig. S4G). Consistent with this, pharmacological impairment of mitochondrial translation limited the expression of mitochondrially transcribed MTCO1 but not non-mitochondrially derived components of the ETC in 39 °C TE cells (Fig. 4I). When oxygen consumption was assessed, TIG-treated 39 °C TE cells looked comparatively similar to 37 °C TE control cells, despite basal extracellular acidification remaining similar to untreated 39 °C TE cells (Fig. 4 J and K). These data suggest that the temperature-induced enhancement of mitochondrial translation contributes to increased mitochondrial oxygen consumption in 39 °C TE cells and that these processes are independent from the parallel increase in extracellular acidification/aerobic glycolysis. It is likely that TIG does not significantly impact 37 °C TE cells in this context due to the short time frame of drug exposure and the choice of a lower concentration of drug than that which is typically used to fully inhibit translation (17). Interestingly, in addition to having little effect on the 39 °C TE cell ECAR, TIG treatment did not impair IFN-γ production but rather increased IFN-γ mean fluorescent expression (MFI) in both 37 and 39 °C TE cells (Fig. 4L). Collectively, these results indicate that enhanced mitochondrial translation contributes to increased mitochondrial oxygen consumption in 39 °C TE cells, and this effect occurs independently from other physiological process such as enhanced aerobic glycolysis or cytokine production.
Protective Antitumor Effects of CD8+ TE Cells Exposed to Febrile Temperature Are Lost when Mitochondrial Translation Is Impaired.
We next set out to determine if enhanced mitochondrial translation was responsible for the increased functional capacity of 39 °C TE cells. We treated 39 °C TE cells with TIG and tested their anticancer function using the ML model described in Fig. 3L (Fig. 5A). Consistent with what we had previously observed (Fig. 3M), mice injected with 39 °C TE cells exhibited superior survival kinetics compared with mice injected with 37 °C TE cells (Fig. 5B). TIG treatment, however, impaired the efficacy of 39 °C TE cells (Fig. 5B).
Fig. 5.
Superior antitumor activity of 39 °C primed T cells is dependent on enhanced mitochondrial translation. (A and B) Cultures of 37 or 39 TE cells were incubated with TIG or CTRL for 48 h then adoptively transferred into leukemic mice (using the same model as Fig. 3I). n = 4 per group (except no T cell group n = 1). (C and D) To model melanoma, recipient mice were injected s.c. with B16 melanoma cells in the flank. After 5 d, 37 or 39 °C TE cells previously cultured with TIG or CTRL for 72 h were injected i.v., and survival was monitored. n = 4 per group and shown as mean ± SEM. (E) Cultures of 37 or 39 °C TE cells were incubated with TIG or CTRL and then adoptively transferred into congenic recipient mice; 2 d later, donor cells were isolated. (F) Representative histogram and bar graph showing percentage of donor cells isolated from the LNs of recipient mice. n = 3 per group shown as mean ± SEM. *P < 0.05, ***P < 0.001.
To further confirm this effect, we employed a second aggressive tumor model using a subcutaneous B16-OVA melanoma model. We activated naïve OT-I T cells at 37 or 39 °C for 24 h and then 1 d later transferred these cells into tumor-bearing mice (Fig. 5C). Despite not clearing the tumors, 39 °C TE cells controlled tumor growth more effectively compared with 37 °C TE cells, an effect that was lost in 39 °C TE cells treated in vitro with TIG (Fig. 5D). To directly show that febrile temperature-enhanced mitochondrial translation supports T cell persistence and viability in adoptive cell therapy models, we used the same culture technique but this time transferred the cells into healthy mice (Fig. 5E). After 2 days, we could recover substantially more 39 °C TE than 37 °C TE cells from the LN and spleen of these mice, and TIG abrogated this effect (Fig. 5E and SI Appendix, Fig. S5SA). Collectively, these results suggest that mitochondrial translation and/or enhanced oxygen consumption contributes beneficially to the greater antitumor responses and in vivo survival of 39 °C TE cells.
Discussion
The functional outcome of a T cell response is intimately linked to correct metabolic reprogramming. We observed that activation of CD8+ T cells at 39 °C promotes metabolic reprogramming and a phenotype characterized by enhanced mitochondrial mass and OXPHOS. Furthermore, we demonstrate that this mitochondrial phenotype is dependent on mitochondrial translation and leads to enhanced antitumor responses when T cells are adoptedly transferred into tumor-bearing mice.
Fever is a physiological response to infection that is conserved across a number of species (1). It is a metabolically intensive process, requiring substantial additional energy consumption to sustain increased body temperature, which suggests it must have a strong biological benefit to be maintained during evolution (1). In the context of infection, early T cell activation events will often coincide with a febrile response. The enhancement of T cell function through febrile range temperature may provide a layer of temporal regulation, whereby the T cells activated during fever are likely to be responding to disease-related antigens, and an enhanced response would be desirable. In addition to our current findings, it has previously been demonstrated that febrile range temperature can enhance T cell migration and subset polarization and effector T cell function (2–5), which suggests fever beneficially impacts multiple cellular processes in T cells.
The antitumor benefits of cells exposed to 39 °C appear to be dependent on mitochondrial metabolism, as enhanced mitochondrial translation was necessary for the optimal antitumor response of 39 °C TE cells. We do not rule out that increased cytokine production or aerobic glycolysis may also be advantageous to 39 TE cells in vivo antitumor effects. However, as exposure to TIG in vitro led to decreased mitochondrial translation and OXPHOS but not glycolysis or IFN-γ production, we infer that enhanced glycolysis or IFN-γ are not solely sufficient to drive the 39 °C TE cells antitumor responses. We have previously observed that robust mitochondrial metabolism improves survival and antitumor responses in vivo (9, 18). Consistent with this, durable remission in patents with FLT3-ITD–driven acute ML correlated with enhanced mitochondrial function in CD8+ T cells following HCT and sorafenib treatment (19).
Mitochondrial remodeling and biogenesis occur within the first 24 h after T cell activation and is crucial for both optimal function of TE cells and the subsequent development of memory T cell populations (6, 10, 12, 13, 17, 20, 21). It has also been previously shown that aerobic glycolysis and engagement of mTORC1 signaling is important for nutrient signaling and the development of proliferative and effector responses (22–24). It appears that glycolytic and mitochondrial metabolic pathways work in concert following T cell activation to provide the correct metabolic reprogramming needed for optimal effector T cell development and function. We observed enhanced mTORC1 signaling and aerobic glycolysis during exposure to 39 °C, but unlike the increased mitochondrial mass and OCR, the enhancement of ECAR was not sustained after the cells were transitioned from 39 °C back to 37 °C. When we assessed memory T cell responses in vitro, we observed that 39 °C TM cells had substantially enhanced metabolic responses to restimulation as well as increased cytokine production. This suggests that transient exposure to increased temperature can result in lasting changes in T cell mitochondrial metabolism.
It has been shown that febrile range temperatures enhance plasma membrane fluidity in T cells, resulting in enhanced microdomain clustering and amplified signaling downstream of the TCR (3, 8). Although signaling strength can have a role in enhancing mitochondrial mass, as increased mitochondrial mass is far more pronounced in activated T cells, it does not appear to be the sole driver. Increased mitochondrial mass was still observed in 39 °C TE cells when TCR signaling was bypassed using PMA/IONO, suggesting that factors independent of TCR signaling are involved.
The relationship between mitochondrial translation and increased mitochondrial activity is not unique to 39 °C TE cells and it is well documented that upon activation, T cells undergo mitochondrial biogenesis and require mitochondrial translation for correct polarization or function (10, 13, 17, 20, 21, 25). However, in experiments that targeted mitochondrial translation, it was evident that 39 °C TE cells were more sensitive to mitochondrial translation impairment than 37 °C TE cells. This is consistent with the idea that the increased mitochondrial mass and OXPHOS detected in 39 °C TE cells would require enhanced levels of mitochondrial translation. Additionally, transcriptional analysis highlighted a number of mitochondrially associated pathways that were up-regulated in 39 °C TE cells, indicating that both transcription and translational differences were present.
The mitochondrial genome encodes less than 1% of the total mitochondrial proteins, and the formation of mitochondria requires a finely orchestrated assembly of both nuclear and mitochondrial subunits (26, 27). How exactly this is coordinated and what specific signals drive the cross-talk between these two translational pathways is not well understood (27). We observed increases in both the nuclear-encoded mitochondrial protein SDHA in addition to mitochondrial-encoded protein MTCO1, suggesting that temperature increase leads to a coordination of both mitochondrial and cytoplasmic protein expression. The interplay between cytoplasmic and mitochondrial translation in orchestrating increases in mitochondrial mass are evident by the fact that both inhibition of mitochondrial translation and the loss of mTORC1-dependent cytoplasmic translation substantially reduced mitochondrial mass in TE cells. It has previously been shown that in addition to impairing translation of mitochondrially targeted proteins, inhibition or loss of mTORC1 signaling results in decreased expression of the mitochondrial transcription factor TFAM, genes associated with OXPHOS, and mitochondrial translation, which suggests increased mTORC1 in 39 °C TE cells could have a role in driving increased mitochondrial biogenesis (16, 17).
Nuclear-encoded mitochondrial proteins that are translated in the cytosol require active transport into the mitochondria with the assistance of chaperones such as heat-shock proteins (HSP) and the translocases of the outer membrane complexes (27). HSP family members have been documented to increase following febrile range temperatures in T cells, and HSP90 expression is required for both enhanced integrin expression and for the polarization of CD4+ T cells to a TH17 phenotype observed at febrile temperatures (2, 4). Surprisingly, we did not observe a significant decrease in mitochondrial mass when HSP90 was inhibited. The role for these chaperones in mitochondrial mass and function is complex; for example, the inhibition of HSP70 in tumor cells can impair mitochondrial processes (28), and conversely, the overexpression of HSP70 has been shown to enhances aerobic glycolysis but impair OXPHOS (29). It has also been observed that HSP90 inhibition can lead to increased mitochondrial mass (30). Although identifying the specific role these HSP have in altering mitochondrial function is beyond the scope of this study, our results suggest HSP90 was not directly responsible for increased mitochondrial mass in 39 °C TE cells. Despite this result, we do not preclude the potential that these chaperones could be important in influencing other aspects of T cell metabolism when exposed to higher temperatures. Likewise, we do not rule out the possibility that transcription factors may influence the phenotype of 39 °C TE cells. We did observe an increase in T-bet and EOMES expression in 39 °C TE cells, which may correlate with their increased effector potential and persistence (31).
The increase in mitochondrial mass from cells incubated at 39 °C compared with 37 or 35 °C indicates that there is a gradient of expression that is sensitive to temperature. The attenuated mitochondrial mass observed at 35 °C relative to 37 and 39 °C suggests that T cells activated in cooler situations, such as in exposed peripheral tissues or during hypothermia, could lead to a suboptimal metabolic phenotype. Like many biological processes, there is often a thermal performance curve associated with metabolic processes in which metabolic activity rises relative to temperature to an optimal level and then declines once an excessive temperature is reached (32). In the current study, we did not explore temperatures higher than 39 °C, as we wanted to avoid the complexities of classic heat-shock responses and unfolded protein responses; however, we speculate that it is likely within T cells that a similar curve could exist for mitochondrial processes, and a “sweet spot” for early activated T cells could be within the febrile temperature range.
It has recently been suggested that mitochondria reach much higher temperatures within cells (33). If this effect were to occur in T cells, it is likely to be enhanced by increased metabolic activity following activation. Interestingly, a recent study assessing protein melting temperature across cell types and species noted the stability of proteins from the mitochondrial electron transport chain were particularly stable upon increased temperature, and this effect was more pronounced in primary T cells compared with other human cell types (34). This raises the as yet untested possibility that T cell mitochondrial metabolism may be particularly tolerant to increased temperature, an attribute that could be beneficial for optimal functioning in fever states. In a broader context, there are other situations where fluctuations in body temperature occur, for example, during intense exercise. Repeated exposure of human skeletal muscle to increased temperature can enhance electron transport chain complex expression and respiration (35), suggesting that the effects of mild heating on mitochondrial processes can occur both in skeletal muscles as well as T cells.
It has previously been demonstrated that tumor-infiltrating T cells exhibit a loss of mitochondrial function and mass, an effect which is characterized by chronic AKT signaling and progressive loss of PGC-1α (13). Enhancing PGC-1α expression through the attenuation of AKT signaling or retroviral overexpression of PGC-1α in T cells increases mitochondrial mass in CD8+ T cells, and an adoptive transfer of PGC-1α–overexpressing T cells into tumor-bearing mice enhances subsequent survival (12, 13). Our current findings are consistent with these previous studies indicating the importance of robust mitochondrial metabolism in T cell antitumor responses. Within our experiments, we did not observe changes in AKT signaling or PGC-1α expression in 39 °C TE cells, suggesting that increases in mitochondrial mass in response to febrile range temperature occur through a separate mechanism. Collectively, however, this raises the possibility that there may be therapeutic value in enhancing PGC-1α expression in conjunction with increased culture temperature to induce an additive or synergistic increase in mitochondrial biogenesis and OXPHOS. Such studies could be particularly beneficial to explore in the context of adoptive T cell treatments such as during the manufacturing of chimeric antigen receptor T cell therapies.
The augmentation of mitochondrial mass and OXPHOS induced by febrile range temperatures could also be particularly beneficial during infectious diseases or following vaccinations in which robust and sustained T cell responses are required. In these situations, refraining from administering antipyretics and letting a fever “run its course” could be advantageous. Conversely, these same attributes may be detrimental in situations in which prolonged or hyperactive immune responses occur, and aggressively suppressing fever or even inhibiting T cell mitochondrial translation with TIG or other mitochondrial ribosome–targeting antibiotics may be beneficial. Adding further complexity to these scenarios, the whole-body metabolic demands of sustaining fever in some circumstances could in itself be beneficial or detrimental to an organism’s ability to respond to inflammatory stimuli (36).
Collectively, this research highlights the role temperature can have in influencing metabolic reprogramming of CD8+ T cells. Even transient exposure to febrile range temperature during activation can lead to sustained changes in OXPHOS and enhanced mitochondrial mass, which supports robust antitumor T cell responses. This research highlights a potentially beneficial role for fever in optimizing metabolic and functional responses in CD8+ T cells.
Materials and Methods
Mice.
C57BL/6J (JAX:000664), PhAM (JAX:018397), major histocompatibility complex class I–restricted OVA specific TCR OT-I transgenic mice (JAX:003831), Great IFN-γ reporter (JAX:017580), CD45.1 C57BL/6J (congenic B6.SJL-Ptprca Pepcb/BoyJ; JAX:002014), or Thy1.1 C57BL/6J congenic (JAX:000406) mouse strains were maintained at the Max Planck Institute for Immunobiology and Epigenetics and cared for according to the Institutional Animal Use and Care guidelines. C57BL/6 (H-2Kb) and BALB/c (H-2Kd) mice used for ML experiments were maintained at the animal facility of Freiburg University Medical Center and cared for according to the Institutional Animal Use and Care guidelines.
WBH Experiments.
We implanted emperature transponders (IPTT-300; Bio Medic Data Systems) subcutaneously in the dorsal region of isoflurane anesthetized mice 1 wk prior to experimentation. Age-matched C57BL/6 mice were randomly divided into WBH or control groups prior to experimentation. Temperature was measured remotely, prior to and every half an hour following initiation of WBH using a handheld transponder reader (Bio Medic Data Systems). WBH mice were placed in a mouse-warming cabinet (Scanbar 48-VS-III), and the cabinet temperature was adjusted to maintain the body temperature of mice at 39 °C. After 8 h, mice were returned to AT and kept in standard housing conditions. Mice within the control group were kept in and treated with comparable conditions at an AT. All mice had normal access to food and water throughout the duration of the experiments. For experiments using the adoptive transfer of OT-I cells, naïve CD8+ OT-I T cells (3 × 106 cells) were injected intravenously (i.v.), and 50 μg endotoxin free Ovalbumin (Invivogen) emulsified in 100 μL IFA (Sigma) was injected subcutaneously (s.c.) into the hind flanks of age- and sex-matched congenic mice. Mice were randomly allocated to WBH or control conditions for 8 h and then returned to standard housing conditions. LNs draining the immunization site were isolated 24 h after the initiation of the experiment and analyzed by flow cytometry. For experiments using in vivo α-CD3/α-CD28, age- and sex-matched isoflurane anesthetized C57BL/6 mice were injected in hind footpads with 50 μg α-CD3 and 5 μg α-CD28 (per mouse) (both BioXcell). Mice were randomly allocated to WBH or control conditions for 8 h and then returned to standard housing conditions. LNs draining the immunization site were isolated 24 h after the initiation of the experiment and analyzed by flow cytometry.
In Vivo Tumor and Survival Experiments.
For the ML model, BALB/c recipients were transplanted with 1 × 104 ML (WEHI-3B) cells and 5 × 106 allogeneic C57/BL6 donor BM cells i.v. after lethal irradiation with 10 Gy split into two equal doses 4 h apart. Naïve CD8+ T cells isolated from the same donor C57/BL6 were activated in vitro with α-CD3/CD28 ± TIG at 39 or 37 °C for 24 h and were then cultured for an additional 24 h at 37 °C. A total of 2 × 105 C57/BL6 d2 donor TE cells were introduced i.v. on day 2 following initial transplantation. For the B16 melanoma model, mice were shaved and injected s.c. into the right flank with 1 × 106 B16-OVA cells in 100 µL phosphate-buffered saline (PBS) while under isoflurane anesthesia. On day 5, 4 × 106 OT-I TE cells that had previously been activated in vitro with α-CD3/CD28 ± TIG at 39 or 37 °C for 24 h and then cultured for an additional 24 h at 37 °C were transferred i.v. into mice randomized from littermate cages. Tumor growth measurements were assessed by multiple investigators. To detect in vivo T cell survival, 1 × 106 cells that had previously been activated in vitro with α-CD3/CD28 ± TIG were transferred i.v. into congenic recipient mice. After 2 d, mice were euthanized, and the spleen and LNs were isolated.
Cell Culture.
Naïve CD8+ T cells were isolated from the spleen and LNs from 6- to 12-wk-old mice using a negative selection naïve CD8 T cell kit (STEMCELL Technologies) according to the manufacturer´s protocol. Unless otherwise stated, isolated T cells (1 × 106/mL) were activated using plate bound α-CD3 (5 μg/mL) and soluble α-CD28 (0.5 μg/mL) in T cell media (TCM; 1640 Roswell Park Memorial Institute [RPMI] medium with 10% fatal calf serum, 4 mM L-glutamine, 1% penicillin/streptomycin, and 55 μM beta-mercaptoethanol) supplemented with 100 U/mL rhIL-2 (Peprotech). Cells were cultured at 37 or 39 °C as indicated in humidified incubators with 5% CO2 and atmospheric oxygen for 24 h following activation. To induce in vitro memory T cells (TM), naïve CD8+ T cells were activated with anti-CD3/CD28 at 37 or 39 °C for 24 h and then subsequently cultured in rhIL-2 (100 U/mL) at 37 °C for 2 d followed by three additional days cultured in mIL-15 (100 U/mL; Peprotech). In experiments using drug treatments, cells were incubated with 500 nM TIG (Sigma), 20 nM Rapamycin (LC Laboratories), 0.05% ethanol (Sigma), 2 mM 2DG (Sigma), or 200 nM NMS-E973 (Seleckchem) for 24 h or as indicated. Vehicle controls consisted of equivalent amounts of dimethyl sulfoxide ≥ 1:5,000 (Sigma), except for ethanol (no vehicle). For activation by PMA/IONO, 50 ng/mL PMA + 500 ng/mL IONO (both Sigma) were used. For bone marrow–derived dendritic cell activation, C57BL/6 BM was cultured in the presence of 20 ng/mL granulocyte-macrophage colony-stimulating factor (Peprotech) for 9 d, then stimulated overnight with 100 ng/mL lipopolysaccharides (Sigma), pulsed with 1 μg/mL SIINFEKL for 1 h, and then washed twice before naïve OT-I T cells (1 × 106/mL) were added and cocultured for 24 h.
Flow Cytometry.
Fluorochrome-conjugated antibodies were purchased from eBioscience, BD Bioscience, Cell Signaling, or BioLegend. Staining was performed in 1% fetal bovine serum/PBS, and dead cells were excluded with the LIVE/DEAD Fixable Dead Cell Stain Kits (Thermo Scientific). For intracellular cytokine staining of IFN-y, cells were restimulated with 50 ng/mL PMA + 500 ng/mL IONO (all Sigma) for 4 h in the presence of Brefeldin A prior to fixation using Cytofix/Cytoperm (BD Bioscience). To detect IFN-y in TM cells, the cells were restimulated overnight with α-CD28 (5 μg/mL) and α-CD28 (0.5 μg/mL), and Brefeldin A was added to the last 4 h of culture prior to fixation. For detection of other intracellular proteins, cells were stained with Fixable LIVE/DEAD, and cell surface markers were then fixed with Intracellular Fixation and Permeabilization buffers (eBioscience) or following manufacturer’s instructions (MitoBiogenesis Flow Cytometry Kit: Abcam). CellTrace Violet (Invitrogen) staining was performed according to the manufacturer’s instructions (Invitrogen). MitoTracker staining was performed according to the manufacturer’s instructions (Invitrogen) using a 50 nM final concentration. Protein synthesis was analyzed using O-propargyl-puromycin (Invitrogen) according to the manufacturer’s instructions. To assess glucose uptake, cells were incubated at 37 °C for 10 min, in medium without glucose, in the presence of 500 nM 2-NBDG (Invitrogen) and washed twice before analysis. Cells were detected using Fortessa, LSR2 (both BD Bioscience), or Cytoflex (Beckman Coulter) flow cytometers and analyzed using FlowJo software (FlowJo).
Confocal and Electron Microscopy.
For confocal microscopy, activated OT-I Pham CD8+ T cells were transferred to glass bottom dishes (MatTek) coated with fibronectin (Sigma) in complete medium containing IL-2 and assessed using a Zeiss spinning disk confocal with an Evolve (electron-multiplying charge coupled device ) camera. During collection, cells were maintained in a humidified incubation chamber (Tokai Hit) at 37 °C with 5% CO2. Images were processed using Imaris software. For electron microscope imaging, 2 × 106 T cells were fixed in 2.5% glutaraldehyde in 100 mM sodium cacodylate washed in cacodylate buffer. After dehydration, samples were embedded in Eponate 12 resin (Ted Pella), and sections were cut. Images were acquired using a FEI Tecnai 12 Transmission electron microscope equipped with a TIETZ digital camera. The mitochondrial area analysis was performed blinded and was measured using ImageJ software (NIH).
Metabolic Profiling.
ECAR and OCR were measured using a Seahorse XF 96 Bioanalyser (Seahorse Bioscience). For cultured cells, 2 × 105 T cells (or 1.5 × 105 for day 3 activated cells) per well were used. For ex vivo samples, CD8+ cells were isolated (STEMCELL Technologies) from mice and plated at 2 × 105 cells per well. Cells were spun onto poly-D-lysine–coated seahorse 96-well plates and preincubated at 37 °C in the absence of CO2. OCR and ECAR were measured in XF media (nonbuffered RPMI 1640 containing 25 mM glucose, 2 mM L-glutamine, and 1 mM sodium pyruvate) under basal conditions and in response to 1 μM oligomycin, 1.5 μM fluoro-carbonyl cyanide phenylhydrazone, and 100 nM rotenone + 1 μM antimycin A (all Sigma). For the restimulation of TM cells, PMA (50 ng/mL) and IONO (500 ng/mL) were injected in port A.
Lactate was measured in cell culture supernatants with the Cedex Bio Analyzer (Roche).
Western Blotting.
For Western blot analysis, cells were washed with ice cold PBS and lysed in 1 × Cell Signaling lysis buffer [20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA, 1 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid), 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, and 1 μg/mL leupeptin], supplemented with 1 mM phenylmethylsulfonyl fluoride. The samples were frozen and thawed three times followed by centrifugation at 20,000 × g for 10 min at 4 °C. The cleared protein extracts were denatured with lithium dodecyl sulfate loading buffer at room temperature (mitochondrial complexes) or for 10 min at 70 °C (all other targets) and loaded onto precast bis-Tris protein gels (Invitrogen). The proteins were transferred onto nitrocellulose membranes using the iBLOT2 system following the manufacturers protocols (Thermo Fisher). The membranes were blocked with 5% weight/volume milk and 0.1% Tween-20 in tris-buffered saline (TBS) and incubated with the appropriate antibodies in 5% weight/volume bovine serum albumin in TBS with 0.1% Tween-20 overnight at 4 °C. All primary antibody incubations were followed by an incubation with secondary horseradish peroxidase–conjugated antibody (Pierce) in 5% milk and 0.1% Tween-20 in TBS and visualized using SuperSignal West Pico or Femto Chemiluminescent Substrate (Pierce). The membranes were developed using film or a ChemiDoc XRs transilluminator (Bio-Rad).
RNA-seq.
RNA isolations were done by using the RNeasy kit (Qiagen) as per manufacturer’s instructions and quantified using Qubit 2.0 (Thermo Fisher Scientific). The libraries were prepared using the TruSeq stranded mRNA kit (Illumina) and sequenced in a HISeq 3000 (Illumina) by the Deep-Sequencing Facility at the Max Planck Institute for Immunobiology and Epigenetics. Sequenced libraries were processed with the Galaxy platform and deepTools for quality control (37), Spliced Transcripts Alignment to a Reference (STAR) (38) for trimming and mapping, and featureCounts (39) to quantify mapped reads. Raw mapped read counts were processed in R (Lucent Technologies) with DESeq2 (40) to determine differentially expressed genes and generate normalized read counts, which were visualized using Morpheus (Broad Institute). Gene ontology analysis was performed using the Metascape online platform (41).
CRISPR-Cas9.
Guide RNAs (gRNAs) were designed and synthesized by Integrated DNA Technologies (IDT). The duplexes of two separate guides per target gene were prepared by annealing (5 min; 98 °C) equimolar concentrations of CRISPR gRNAs and trans-activating crispr RNAs and then incubating (20 min; room temperature) with Alt-R S.p. Cas9 Nuclease V3 (IDT). For the delivery of ribonucleoprotein (RNP) complexes, naïve CD8+ T cells were washed in PBS and mixed with RNP complexes and electroporation enhancer P4 Primary Cell Buffer (Lonza) immediately prior to electroporation (4D-Nucleofector; Lonza-Program DS137). Per electroporation, 60 pmol of Cas9 Nuclease and 180 pmol of annealed gRNAs were used. Electroporated cells were recovered in T cell medium for 3 to 5 d in TCM with rIL-7 (Peprotech) prior to activation with α-CD3/α-CD28.
Statistical Analysis.
Statistical analysis was performed using Prism 8 software (GraphPad), and the results are represented as mean ± SEM unless otherwise indicated. The comparisons for two groups were calculated using unpaired two-tailed Student’s t tests, and the comparisons of more than two groups were calculated using one-way or two-way ANOVA with Bonferroni’s multiple comparison test. Statistical significance: *P < 0.05; **P < 0.01; ***P < 0.001.
Supplementary Material
Acknowledgments
We thank Fabian Hässler, Marco Cavallari, Andrea Quintana, Raima Kyle, John Sutherland, and Joerg Buescher for providing technical assistance. We are also grateful to the Electron Microscopy Laboratory at the University of Padova and the Imaging and Flow Cytometry facilities at the Max Planck Institute of Immunobiology and Epigenetics for technical support. This work was supported by the Max Planck Society. R.Z. was supported by SFB1160 TP B09, European Research Council Consolidator Grant GvHDCure Proposal 681012, Deutsche Krebshilfe (Grant 70113473), and the Jose-Carreras Leukemia Foundation (Grant DJCLS 01R/2019). M.V. and M.C. were supported by Alexander von Humboldt Postdoctoral Fellowships. M.A.S. received funding from the Swiss National Science Foundation. D.E.S was supported by the German Research Foundation (DFG) SFB1160 and the DFG under Germany’s Excellence Strategy (Centre for Integrative Biological Signalling Studies EXC-2189 Project 390939984).
Footnotes
Competing interest statement: E.L.P. is a Scientific Advisory Board member of ImmunoMet Therapeutics, and E.J.P. and E.L.P. are founders of Rheos Medicines.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023752118/-/DCSupplemental.
Data Availability
RNA-seq data can be retrieved from the Gene Expression Omnibus repository, https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE171245). All other study data are included in the article and/or SI Appendix.
References
- 1.Evans S. S., Repasky E. A., Fisher D. T., Fever and the thermal regulation of immunity: The immune system feels the heat. Nat. Rev. Immunol. 15, 335–349 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X., et al., Febrile temperature critically controls the differentiation and pathogenicity of T helper 17 cells. Immunity 52, 328–341.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Mace T. A., et al., Differentiation of CD8+ T cells into effector cells is enhanced by physiological range hyperthermia. J. Leukoc. Biol. 90, 951–962 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin C., et al., Fever promotes T lymphocyte trafficking via a thermal sensory pathway involving heat shock protein 90 and α4 integrins. Immunity 50, 137–151.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Umar D., et al., Febrile temperature change modulates CD4 T cell differentiation via a TRPV channel-regulated Notch-dependent pathway. Proc. Natl. Acad. Sci. U.S.A. 117, 22357–22366 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geltink R. I. K., Kyle R. L., Pearce E. L., Unraveling the complex interplay between T cell metabolism and function. Annu. Rev. Immunol. 36, 461–488 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mace T. A., Zhong L., Kokolus K. M., Repasky E. A., Effector CD8+ T cell IFN-γ production and cytotoxicity are enhanced by mild hyperthermia. Int. J. Hyperthermia 28, 9–18 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zynda E. R., et al., A role for the thermal environment in defining co-stimulation requirements for CD4(+) T cell activation. Cell Cycle 14, 2340–2354 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buck M. D., et al., Mitochondrial dynamics controls T cell fate through metabolic programming. Cell 166, 63–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein Geltink R. I., et al., Mitochondrial priming by CD28. Cell 171, 385–397.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Basu S., Srivastava P. K., Fever-like temperature induces maturation of dendritic cells through induction of hsp90. Int. Immunol. 15, 1053–1061 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Dumauthioz N., et al., Enforced PGC-1α expression promotes CD8 T cell fitness, memory formation and antitumor immunity. Cell. Mol. Immunol. (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scharping N. E., et al., The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T cell metabolic insufficiency and dysfunction. Immunity 45, 374–388 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orti G., et al., Donor lymphocyte infusions in AML and MDS: Enhancing the graft-versus-leukemia effect. Exp. Hematol. 48, 1–11 (2017). [DOI] [PubMed] [Google Scholar]
- 15.D’Souza A. R., Minczuk M., Mitochondrial transcription and translation: Overview. Essays Biochem. 62, 309–320 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morita M., et al., mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 18, 698–711 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Tan H., et al., Integrative proteomics and phosphoproteomics profiling reveals dynamic signaling networks and bioenergetics pathways underlying T cell activation. Immunity 46, 488–503 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Windt G. J., et al., Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36, 68–78 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathew N. R., et al., Sorafenib promotes graft-versus-leukemia activity in mice and humans through IL-15 production in FLT3-ITD-mutant leukemia cells. Nat. Med. 24, 282–291 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ron-Harel N., et al., Mitochondrial biogenesis and proteome remodeling promote one-carbon metabolism for T cell activation. Cell Metab. 24, 104–117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer M., et al., Early effector maturation of naïve human CD8+ T cells requires mitochondrial biogenesis. Eur. J. Immunol. 48, 1632–1643 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Menk A. V., et al., Early TCR signaling induces rapid aerobic glycolysis enabling distinct acute T cell effector functions. Cell Rep. 22, 1509–1521 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang C.-H., et al., Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153, 1239–1251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pollizzi K. N., Powell J. D., Regulation of T cells by mTOR: The known knowns and the known unknowns. Trends Immunol. 36, 13–20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Almeida L., et al., Ribosome-targeting antibiotics impair T cell effector function and ameliorate autoimmunity by blocking mitochondrial protein synthesis. Immunity 54, 68–83.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Couvillion M. T., Soto I. C., Shipkovenska G., Churchman L. S., Synchronized mitochondrial and cytosolic translation programs. Nature 533, 499–503 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dennerlein S., Wang C., Rehling P., Plasticity of mitochondrial translation. Trends Cell Biol. 27, 712–721 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Leu J. I., et al., Inhibition of stress-inducible HSP70 impairs mitochondrial proteostasis and function. Oncotarget 8, 45656–45669 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L., Schumann U., Liu Y., Prokopchuk O., Steinacker J. M., Heat shock protein 70 (Hsp70) inhibits oxidative phosphorylation and compensates ATP balance through enhanced glycolytic activity. J. Appl. Physiol. (1985) 113, 1669–1676 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Margineantu D. H., Emerson C. B., Diaz D., Hockenbery D. M., Hsp90 inhibition decreases mitochondrial protein turnover. PLoS One 2, e1066 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaech S. M., Cui W., Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12, 749–761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulte P. M., The effects of temperature on aerobic metabolism: Towards a mechanistic understanding of the responses of ectotherms to a changing environment. J. Exp. Biol. 218, 1856–1866 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Chrétien D., et al., Mitochondria are physiologically maintained at close to 50 °C. PLoS Biol. 16, e2003992 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jarzab A., et al., Meltome atlas-thermal proteome stability across the tree of life. Nat. Methods 17, 495–503 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Hafen P. S., Preece C. N., Sorensen J. R., Hancock C. R., Hyldahl R. D., Repeated exposure to heat stress induces mitochondrial adaptation in human skeletal muscle. J Appl Physiol (1985) 125, 1447–1455 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Ganeshan K., et al., Energetic trade-offs and hypometabolic states promote disease tolerance. Cell 177, 399–413.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramírez F., et al., deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dobin A., et al., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liao Y., Smyth G. K., Shi W., featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Y., et al., Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data can be retrieved from the Gene Expression Omnibus repository, https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE171245). All other study data are included in the article and/or SI Appendix.