Significance
Epstein–Barr virus (EBV) infects virtually every human by adulthood with limited symptoms. However, in the context of immune suppression, EBV can cause lymphomas. EBV-associated lymphomas have poor prognoses and limited treatment options, highlighting a need to better understand EBV-mediated tumorigenesis. One common feature of viral replication and cancer is dysregulation of metabolism. Indeed, EBV dramatically reprograms host cell metabolism, up-regulating both oxidative phosphorylation and glycolysis. In this report, we identify metabolic vulnerabilities of EBV-infected primary human B cells, EBV-positive lymphomas, and lymphomas caused by the related oncogenic herpesvirus, KSHV. Pharmacological inhibition of lactate export suppressed these lymphomas and rendered them hypersensitive to metformin and phenformin. These findings together with our mechanistic studies outline a therapeutic strategy for viral lymphomas.
Keywords: Epstein–Barr virus, viral lymphoma, lactate export, monocarboxylate transporter, cancer metabolism
Abstract
Epstein–Barr virus (EBV) is a ubiquitous herpesvirus that typically causes asymptomatic infection but can promote B lymphoid tumors in the immune suppressed. In vitro, EBV infection of primary B cells stimulates glycolysis during immortalization into lymphoblastoid cell lines (LCLs). Lactate export during glycolysis is crucial for continued proliferation of many cancer cells—part of a phenomenon known as the “Warburg effect”— and is mediated by monocarboxylate transporters (MCTs). However, the role of MCTs has yet to be studied in EBV-associated malignancies, which display Warburg-like metabolism in vitro. Here, we show that EBV infection of B lymphocytes directly promotes temporal induction of MCT1 and MCT4 through the viral proteins EBNA2 and LMP1, respectively. Functionally, MCT1 was required for early B cell proliferation, and MCT4 up-regulation promoted acquired resistance to MCT1 antagonism in LCLs. However, dual MCT1/4 inhibition led to LCL growth arrest and lactate buildup. Metabolic profiling in LCLs revealed significantly reduced oxygen consumption rates (OCRs) and NAD+/NADH ratios, contrary to previous observations of increased OCR and unaltered NAD+/NADH ratios in MCT1/4-inhibited cancer cells. Furthermore, U-13C6–glucose labeling of MCT1/4-inhibited LCLs revealed depleted glutathione pools that correlated with elevated reactive oxygen species. Finally, we found that dual MCT1/4 inhibition also sensitized LCLs to killing by the electron transport chain complex I inhibitors phenformin and metformin. These findings were extended to viral lymphomas associated with EBV and the related gammaherpesvirus KSHV, pointing at a therapeutic approach for targeting both viral lymphomas.
Epstein–Barr virus (EBV) is a ubiquitous human herpesvirus transmitted through saliva, with more than 95% of the world being infected by adulthood. Primary EBV infection is typically asymptomatic and surmounts to latency establishment in quiescent memory B cells in the peripheral blood (1). However, in immunocompromised persons, EBV reactivation can cause uncontrolled B cell proliferation, leading to various lymphoproliferative disorders and frank lymphoma. Recent evidence suggests that host responses such as the DNA damage response (DDR) and metabolic stress play a significant cell-intrinsic role in preventing EBV-driven B cell transformation (2, 3).
EBV infection of primary human peripheral blood B cells in vitro produces indefinitely proliferating lymphoblastoid cell lines (LCLs). LCLs express the Latency III growth program, where all six EBV nuclear antigens, or EBNAs, (EBNA-LP, 1, 2, 3A, 3B, and 3C) and two latent membrane proteins, or LMPs, (LMP1 and LMP2A/B) are present. This gene expression program mimics that of many EBV-associated B lymphoid cancers, making LCLs a suitable model for studying mechanisms underlying tumorigenesis (4). Prior to Latency III establishment in LCLs and shortly after EBV infection of B cells, a transient period of hyperproliferation, called Latency IIb, is induced, where only the viral EBNAs are expressed in the absence of the LMPs (5, 6). This culminates in an irreversible growth arrest in most cells caused by a persistent DDR (7). However, a very small subset (<10%) of hyperproliferating cells evade host barriers like the DDR and transition into Latency III–expressing LCLs within weeks (2).
During EBV immortalization of B cells into LCLs, increased energetic demands for continued proliferation drive an up-regulation of glycolysis and extracellular acidification rates (ECAR) (3, 8–10). While the ECAR is largely a reflection of lactate export into the extracellular environment, the magnitude and role of lactate export during EBV-driven B cell immortalization remains poorly characterized. Besides being a glycolytic waste product, lactate has recently emerged to play a key role in tumor development, growth, and metastasis (11, 12). Furthermore, intracellular lactate buildup can have dire consequences on cell homeostasis due to acidification by lactic acid, highlighting the importance of lactate export for sustained cell growth and survival (13).
Proton-coupled lactate export is driven by the plasma membrane–expressed solute carrier (SLC) family proteins monocarboxylate transporters (MCTs) 1 and 4 (SLC16A1/MCT1 and SLC16A3/MCT4). MCT1 has a much higher affinity for lactate than MCT4, making it the primary lactate transporter under normal physiological conditions (14–16). MCT4, however, is typically expressed in highly glycolytic tissues or when intracellular lactate concentrations are high, such as during aerobic glycolysis (17–19). MCT1 and MCT4 have the ability to transport other monocarboxylates such as pyruvate and ketone bodies like acetoacetate and β-hydroxybutyrate (15, 19, 20). However, these transporters are most commonly associated with their role in lactate transport across the plasma membrane, especially during tumor development, growth, and metastasis (21–23). As such, MCT1 and MCT4 act as essential buffers at the intersection of the intracellular and extracellular environments, sensing monocarboxylate concentrations and facilitating rapid transport based on the needs of the cell (24–26). Not surprisingly, MCT1 and MCT4 are commonly expressed in a wide variety of tumors, exporting lactate from glycolytic cancer cells, which can later be recycled by oxidative cells in the tumor microenvironment (27, 28).
Targeting MCT1- and MCT4-mediated lactate export has become a highly attractive therapeutic strategy in recent years as blocking lactate export can lead to cell cycle arrest and sensitize tumor cells to killing by other metabolic inhibitors (20, 22, 29, 30). MCT1 inhibitors are currently in Phase I clinical trials for a number of solid tumors and lymphomas (31). While these inhibitors have proven to effectively reduce tumor burden in preclinical models of MCT1-overexpressing cancers, they are unfortunately ineffective in MCT1/MCT4-coexpressing cancers due to the redundant role of MCT4 as a lactate exporter (32). This highlights the need for MCT4-specific inhibitors, which are unfortunately not commercially available to date. Furthermore, while inhibition of MCT1 and MCT4 universally leads to a buildup of intracellular lactate in tumor cells, the mechanisms underlying growth arrest and cell death are not fully understood. Thus, studying how MCT1 and MCT4 are regulated in diverse contexts is critical for expanding our understanding of their role not only in tumorigenesis but in cell biology and homeostasis at large.
Given their established roles in tumor development both in vitro and in vivo, we reasoned that MCT1- and MCT4-mediated lactate export might contribute to EBV-mediated B cell tumorigenesis and virus-associated lymphoma cell growth more broadly. To address this, we measured changes in lactate concentration and MCT expression through EBV-mediated B cell outgrowth and perturbed MCT1 and MCT4 function with small molecule antagonists and reverse genetic approaches. Our data identify metabolic vulnerabilities that we propose could be exploited for the treatment of viral-associated cancers.
Results
EBV Infection of Human B Cells Leads to an Increase in Lactate Accumulation, Secretion, and the Lactate Transporter MCT1.
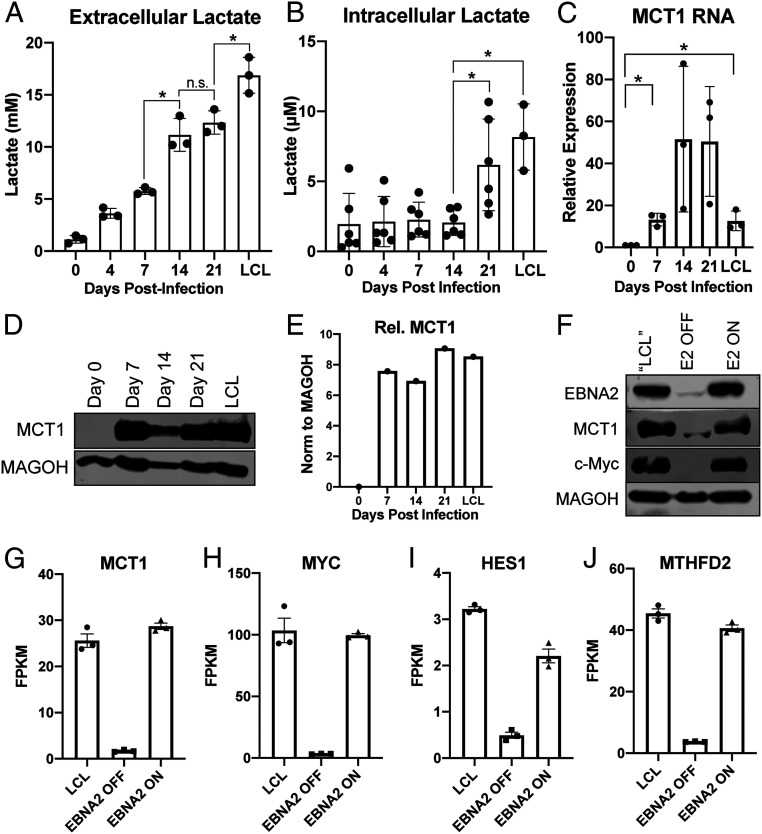
We and others have previously demonstrated that EBV infection of primary human B cells leads to up-regulation of glycolysis as evidenced by an increase in glycolytic enzymes and the ECAR (3, 10). To determine whether the increase in ECAR was indicative of an increase in lactate export from infected cells, we harvested media from EBV-infected primary human B cells at several time points following infection and measured media lactate. We found that media lactate levels increased steadily after infection as cell proliferation initiated by day 4 and through long-term outgrowth (day 35 = LCL) (Fig. 1A). However, we did not observe a concomitant rise in intracellular lactate until late times postinfection (Fig. 1B). This finding led us to assess how lactate export was controlled during early and late time points after EBV infection.
Fig. 1.
EBV infection of human B cells leads to an increase in lactate transport through MCT1. (A) Extracellular (n = 3) and (B) Intracellular (n = 6) lactate concentration during EBV-infected B cell immortalization. Extracellular concentration was obtained from the supernatant of CD19+-isolated B cells at a concentration of 1 × 106/mL and corrected to cell-free growth medium lactate concentrations. Intracellular lactate concentration is per 12,500 cells. (C) MCT1 RNA (n = 3) and (D) protein levels during EBV-infected B cell immortalization. MAGOH is a loading control used because its expression does not change during B cell outgrowth. (E) Quantitation of MCT1 protein levels from D, normalized to MAGOH. (F) Immunoblot of EBNA2, MCT1, and c-Myc in P493-6 cells at the “LCL” state with stable EBNA2 expression, EBNA2 OFF in the absence of β-estradiol, and EBNA2 ON with EBNA2 expression regained. (G–J) RNA-seq data showing MCT1, c-Myc, HES1, and MTHFD2 expression in P493-6 B cells (n = 3). FPKM = fragments per kilobase of transcript per million mapped reads. All statistical significance was determined by a paired Student’s t test, in which *P < 0.05 and n.s. (not significant) is P ≥ 0.05.
Under normal physiological conditions in most cells, lactate transport is mediated by MCT1. We found that while uninfected resting B cells had little MCT1 expressed at the RNA and protein level, infection with EBV led to an increase in MCT1 expression that persisted through LCL outgrowth (Fig. 1 C–E). MCT1 is a known transcriptional target of c-Myc, and c-Myc is transcriptionally regulated in EBV-infected cells by the EBV latency protein, EBV nuclear antigen 2 (EBNA2) (33). Thus, we hypothesized that EBNA2 drives MCT1 expression through c-Myc. To test this, we utilized the P493-6 LCL model of EBNA2 expression where an EBNA2-estrogen receptor fusion is endogenously encoded from the viral genome and thus subject to posttranslational control by β-estradiol (34). Following loss of EBNA2 upon β-estradiol withdrawal, we observed a significant decrease in MCT1 and c-Myc expression at both the messenger ribonucleic acid (mRNA) and protein levels, along with two known EBNA2 and c-Myc coregulated genes: HES1 and MTHFD2 (Fig. 1 F–J) (10, 35). Similarly, turning EBNA2 back on via addition of β-estradiol led to re-expression of all these targets.
MCT1 Inhibition Causes Growth Arrest and Intracellular Lactate Buildup in Early EBV-Infected B Cells.
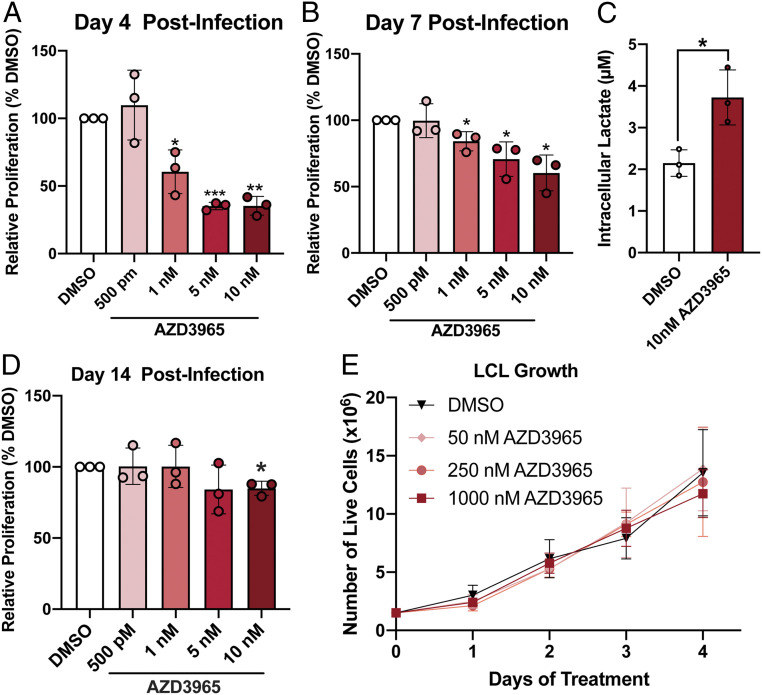
The induction of MCT1 early after EBV infection led us to interrogate its role in maintaining EBV-infected B cell proliferation given the known detrimental consequences of intracellular acidification caused by intracellular lactic acid buildup (13). To assess this, we used the MCT1 inhibitor, AZD3965, which is currently in clinical trials for MCT1-overexpressing diffuse large B cell lymphomas (DLBCLs) (31). We treated EBV-infected peripheral blood mononuclear cells (PBMCs) with AZD3965 concurrent with EBV infection and assessed CD19+ B cell proliferation by CellTrace Violet dilution. We found that treatment with AZD3965 led to a dose-dependent decrease in B cell proliferation observed at the onset of EBV-induced hyperproliferation at 4 d postinfection (d.p.i.) and further into hyperproliferation at 7 d.p.i. Importantly, this reduction in proliferation was accompanied by an increase in intracellular lactate levels and was due to a G1/S phase growth arrest, not B cell death (Fig. 2 A–C and SI Appendix, Fig. S1). However, the effect of AZD3965 on proliferation was lost by 14 d.p.i. and later in immortalized LCLs, suggesting an acquired resistance to MCT1 inhibition during B cell outgrowth (Fig. 2 D and E).
Fig. 2.
MCT1 inhibition leads to growth arrest and lactate buildup in early EBV-infected B cells. (A) Bulk PBMCs (n = 3) were treated with AZD3965 immediately following EBV infection, and CD19+ B cell proliferation was based on CellTrace Violet dilution as determined by flow cytometry 4 d later. Cell counts were bead normalized. (B) Bulk PBMCs (n = 3) were treated at 4 d postinfection and after 3 d in media. CD19+ B cell proliferation assessed at day 7 postinfection as in A. (C) CD19-purified B cells (n = 3) were treated at 4 d postinfection and intracellular lactate assessed 3 d later at day 7 postinfection. (D) Same as B, except proliferation was assessed at 14 d postinfection. (E) Growth curve of LCLs (n = 3) treated with varying concentrations of AZD3965. LCL growth was determined by daily assessment of trypan blue exclusion. Statistical significance determined was by a paired Student’s t test. *P < 0.05, **P < 0.01, and ***P < 0.001.
EBV-Infected B Cells Acquire Resistance to MCT1 Inhibition following MCT4 Up-Regulation.
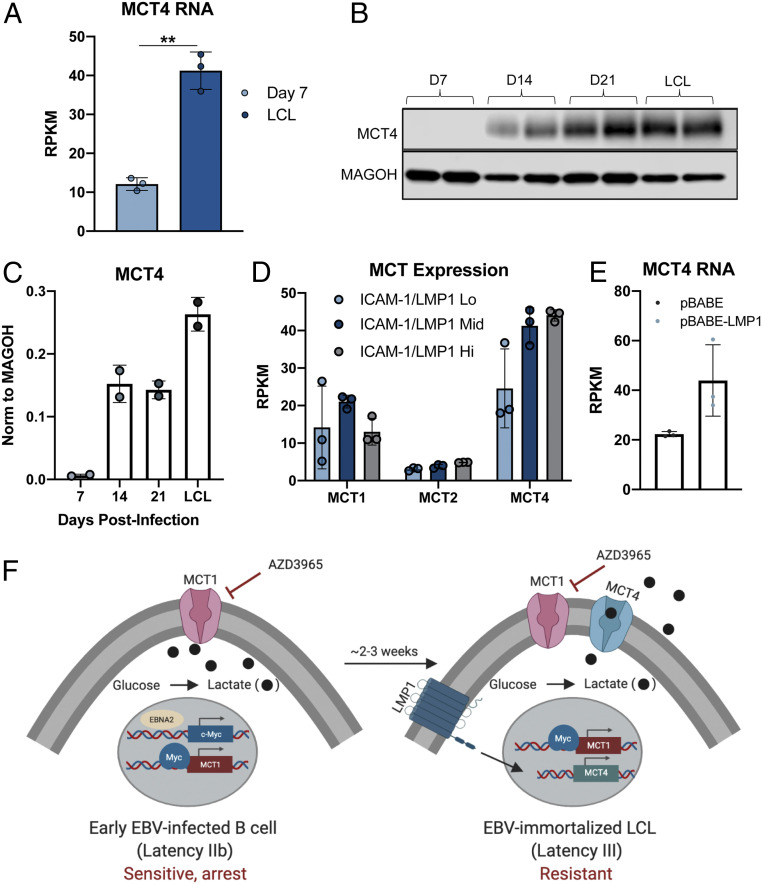
We next sought to determine the mechanism of acquired resistance to MCT1 inhibition at late times following EBV infection. Prior research indicates a role for the high-affinity lactate exporter, MCT4, in rendering highly glycolytic cancer cells resistant to MCT1 inhibition (36). We therefore queried MCT4 expression in the EBV primary B cell infection model. We found that expression of MCT4 significantly increased during the Latency IIb-to-Latency III transition in infected cells, between days 7 and 14 postinfection (Fig. 3 A–C) (37). Furthermore, LMP1 expression in LCLs correlated with MCT4 expression but not MCT1 or MCT2, which are the only other SLC-family lactate transporters expressed in EBV-infected B cells (Fig. 3D). Using a publicly available ribonucleic acid sequencing (RNA-seq) dataset, we also found MCT4 to be up-regulated upon LMP1 expression in an EBV-negative Burkitt lymphoma (BL) cell, suggesting an LMP1-regulated mechanism of MCT4 expression (Fig. 3E). Thus, the Latency IIb-to-Latency III transition and its associated expression of LMP1 leads to induction of MCT4 that could facilitate EBV-driven B cell immortalization through enabling lactate export downstream of glycolysis (Fig. 3F).
Fig. 3.
EBV-infected B cells acquire resistance to MCT1 inhibition during B cell transformation due to MCT4 induction. (A) Queried RNA-seq data of MCT4 expression in EBV-infected B cells (n = 3) at 7 d postinfection and LCL (37). RPKM = reads per kilobase of transcript per million mapped reads. (B) Western blot of MCT4 in CD19+-purified B cells during EBV-mediated B cell immortalization. MAGOH = loading control. (C) Quantification of B (n = 2). (D) MCT expression from RNA-seq of LCLs sorted by the LMP1 proxy, ICAM-1, status. (E) Queried RNA-seq data of MCT4 expression in DG75 cells (n = 3) transfected with LMP1. Statistical significance was determined by paired Student’s t test. **P < 0.01. (F) Schematic representation of MCT1 inhibitor sensitivity during EBV-mediated B lymphocyte immortalization. Created with https://biorender.com/.
Pharmacological and Genetic Perturbation of MCT1 and MCT4 Suppresses LCL Growth.
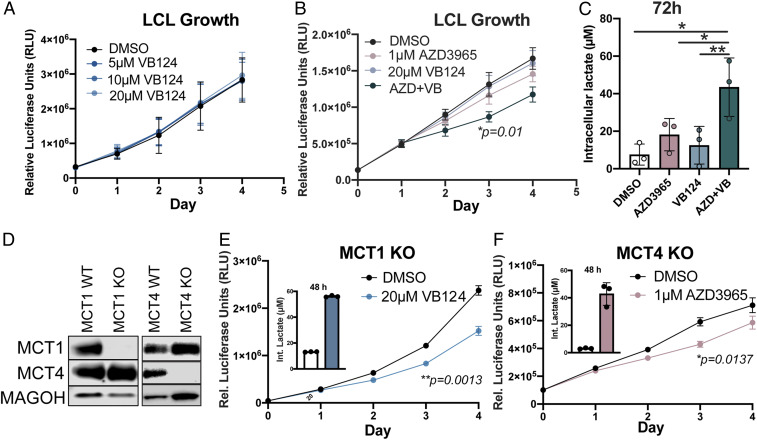
The unique up-regulation of MCT4 as a secondary means to handle elevated glycolysis and lactate accumulation in LCLs suggests that this transporter might be critical for LCL growth. To assess this, we took advantage of a recently described low-nanomolar MCT4 inhibitor, VB124, which displays >1,000-fold selectivity over MCT1 (38). Single-agent VB124 treatment did not impact LCL growth (Fig. 4A). However, this was not unexpected as LCLs also express appreciable levels of MCT1. To test whether MCT1 expression in LCLs was indeed driving VB124 resistance as well as whether MCT1 and MCT4 cooperatively promote LCL growth, we treated LCLs with both AZD3965 and VB124 and monitored cell proliferation over time. Dual MCT1/4 inhibition led to a significant decrease in LCL growth, accompanied by a fourfold increase in intracellular lactate levels (Fig. 4 B and C). Similar to the effect of MCT1 inhibition on early EBV-infected cells, dual MCT1/4 inhibition led to LCL growth arrest and not death (SI Appendix, Fig. S2).
Fig. 4.
Dual MCT1/4 inhibition reduces LCL growth and increases intracellular lactate. (A) Growth curve of LCLs (n = 3) treated with varying concentrations of VB124. Growth was determined by CellTiter Glo. (B) Growth curve of LCLs (n = 3) treated with either DMSO, 1 µM AZD3965, 20 µM VB124, or 1 µM AZD3965 + 20 µM VB124. Growth was assessed as in A. Statistical significance was determined for day 3 of treatment. (C) Intracellular lactate concentration (per 12,500 cells) at 72 h of LCLs (n = 3) treated with either DMSO, 1 µM AZD3965, 20 µM VB124, or 1 µM AZD3965 + 20 µM VB124. (D) Western blot representation of CRISPR-Cas9–mediated MCT1- and MCT4-knockout LCLs. MAGOH = loading control. (E) MCT1-knockout LCL growth (n = 3) over time and intracellular lactate concentration at 48 h following treatment with 20 µM VB124. Statistical significance was calculated at day 3. (F) MCT4-knockout LCL growth (n = 3) over time and intracellular lactate concentration at 48 h following treatment with 1 µM AZD3965. Statistical significance was calculated at day 3. Statistical significance was determined by paired Student’s t test in which *P < 0.05 and **P < 0.01.
To confirm that the reduction of LCL growth upon dual MCT1/4 inhibition was not due to off-target effects of the inhibitors, we genetically knocked out MCT1 (SLC16A1) and MCT4 (SLC16A3) using CRISPR-Cas9 (Fig. 4D). Upon loss of MCT1, LCLs became sensitive to the MCT4 inhibitor, VB124, and displayed significantly increased intracellular lactate levels similar to those observed upon inhibition of both MCT1 and MCT4 in wild-type LCLs (Fig. 4E). Similarly, knockout of MCT4 also rendered LCLs susceptible to growth arrest and increased intracellular lactate levels following MCT1 inhibition with AZD3965 (Fig. 4F). These data strongly corroborate our studies with dual pharmacological antagonism of MCT1 and MCT4 and support an important role of these molecules in LCL proliferation. Finally, treatment with exogenous L-lactate also decreased LCL growth, suggesting that the increase in intracellular lactate following MCT1/4 inhibition could be mediating the growth arrest. (SI Appendix, Fig. S3).
Dual Inhibition of MCT1/4 in LCLs Compromises Oxygen Consumption Rate and NAD+/NADH Ratios and Induces Reactive Oxygen Species.
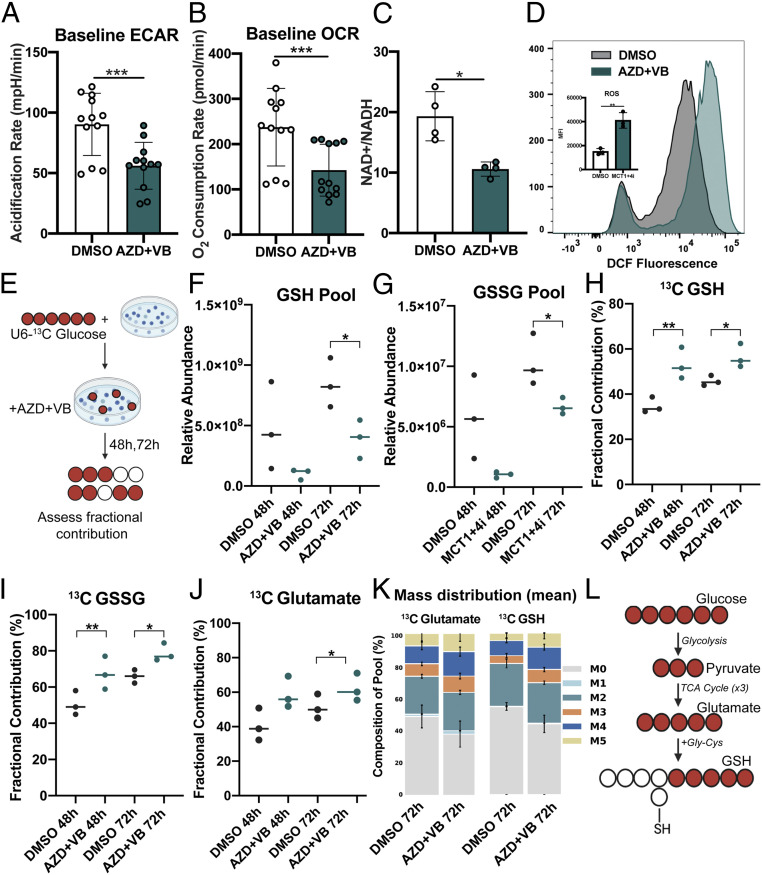
Following our observation that MCT1/4 inhibition had a significant effect on LCL growth, we became curious about underlying metabolic vulnerabilities that might be induced by dual MCT1/4 inhibition. To test this, we first measured the basal metabolism of untreated versus MCT1/4 inhibitor–treated LCLs using the Seahorse XFp Cell Energy Phenotype assay. Following 24-h treatment with 1 μM AZD3965 and 20 μM VB124, we observed a significant decrease in extracellular acidification in LCLs consistent with the role of MCT1/4 as lactate exporters (Fig. 5A). Interestingly, dual MCT1/4 inhibition also reduced the oxygen consumption rate (OCR) induced by dual MCT1/4 inhibition, suggesting a significant reduction of cellular fitness at the level of mitochondrial respiration (Fig. 5B).
Fig. 5.
Dual MCT1/4 inhibition elicits broad changes in LCL metabolism. (A) Seahorse assay showing ECAR and (B) OCR of LCLs (n = 4) in triplicate. AZD + VB = 1 µM AZD3965 + 20 µM VB124. Phenf. = 10 µM Phenformin. (C) NAD+/NADH ratio of LCLs (n = 4) treated for 48 h. (D) FACS histograms and mean fluorescence intensity quantitation of treated LCLs (n = 3) treated with 1 µM AZD3965 + VB124 and stained with 2′,7′-dichlorodihydrofluorescein diacetate (H2-DCFDA). (E) Schematic of experimental workflow for glucose labeling and inhibitor treatment in LCLs. (F–J) Metabolic analysis with U-13C6–glucose tracing of GSH or GSSG glutathione and glutamate in LCLs (n = 3) treated with either DMSO or MCT1 + MCT4 inhibitors for 48 or 72 h. Cells were cultured in the presence of U-13C6–glucose–supplemented growth medium. Fractional contribution = % 13C-labeled GSH or GSSG relative to pool size. Pool = Relative abundance of GSH or GSSH in sample, as determined by liquid chromatography-mass spectrometry (LC-MS). (K) Mass distribution of 13C-glutathione (reduced) isotopologues. (L) Schematic of flow from glucose to glutathione. All statistical significance was determined using paired Student’s t test in which *P < 0.05, **P < 0.01, and ***P < 0.001.
The production of lactate from pyruvate during glycolysis is coupled with the regeneration of NAD+ from NADH, which helps maintain the redox state of the cell. We found that MCT1/4 inhibition significantly reduced the NAD+/NADH ratio in LCLs (Fig. 5C). Given this observation, we next wondered whether MCT1/4 inhibition might be causing increased reactive oxygen species (ROS), based on the pattern of reduced OCR and NAD+/NADH ratios as well as a previous observation that inhibition of the electron transport chain (ETC) can lead to increased ROS (39). Using the H2O2-sensitive fluorescent dye DCFDA, we found that ROS levels, indeed, increased significantly upon MCT1/4 inhibition, thereby confirming increased oxidative stress (Fig. 5D).
MCT1/4 Inhibition Leads to Reduced Glutathione Pools and Increased Glucose-Derived Glutathione Synthesis.
To further understand how MCT1/4 inhibition alters EBV-infected B cell metabolism at the level of nutrient consumption, we performed 13C-glucose tracing in LCLs treated with both AZD3965 and VB124 for 48 and 72 h (Fig. 5E). We found that, correlating with the increases in ROS, MCT1/4 inhibition impacted antioxidant metabolism by reducing glutathione pool sizes (Fig. 5 F and G). We also observed an increase in glucose-labeled reduced (GSH) and oxidized (GSSG) glutathione, suggesting that glucose-driven production of glutathione could be a compensation mechanism to counteract the reduced glutathione pools and ultimately promote antioxidant activity via GSH conversion to GSSG. (Fig. 5 H and I). To further define the source of the newly synthesize glutathione, we interrogated our labeling data. Glutathione is a tripeptide comprised of cysteine, glutamate, and glycine. Although we did not observe any increases in glycine or cysteine, we did observe an increase in 13C-glutamate upon MCT1/4 inhibition as well as in the M5 isotopologues of glutamate and GSH, pointing at the tricarboxylic acid (TCA) cycle–derived glutamate downstream of pyruvate as the most probable source of glucose-labeled glutathione (Fig. 5 J–L and Datasets S1 and S2). These data confirm that dual MCT1/4 inhibition influences ROS and subsequently antioxidant metabolism by reducing glutathione pools, which likely prompts a compensatory response involving increased glutathione synthesis from glucose.
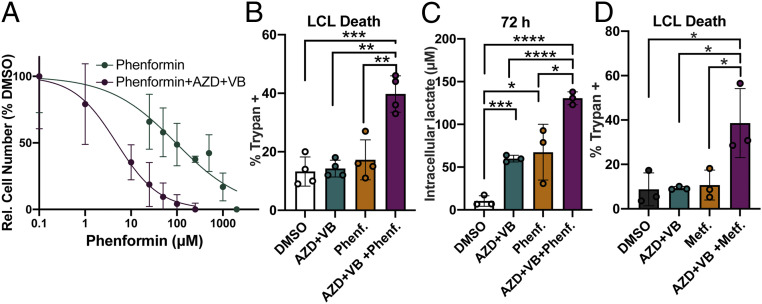
Dual MCT1/4 Inhibition Sensitizes LCLs to Killing by Phenformin and Metformin.
Given the broad effects of MCT1/4 inhibition on LCL metabolism, we wondered whether the combination of these vulnerabilities might render LCLs more susceptible to killing by ETC inhibitors, which has been observed in cancer cell lines (30, 40). Indeed, we found that inhibition of MCT1/4 with AZD3965 and VB124 increased the potency of killing by the ETC complex I inhibitor, phenformin, by 20-fold (Fig. 6 A and B). Consistently, intracellular lactate accumulated to greater levels in LCLs treated with AZD + VB and phenformin than with either AZD + VB or phenformin treatment alone (Fig. 6C). Notably, dual MCT1/4 inhibition also rendered LCLs susceptible to killing by the less-potent ETC complex I inhibitor and Food and Drug Administration (FDA)-approved antidiabetic drug metformin, suggesting this combination could be explored as a therapeutic strategy for EBV-positive tumors in the future (Fig. 6D).
Fig. 6.
MCT1/4 inhibition renders LCLs more susceptible to killing by ETC complex I inhibitors phenformin and metformin. (A) Phenformin IC50 curve generated after 72-h treatment of LCLs (n = 3) with either phenformin or phenformin plus 1 µM AZD3965 and 20 µM VB124. Relative cell number was determined by CellTiter Glo luminescence assay. Relative luminescence values (in relative luciferase units) were normalized to DMSO to obtain %DMSO. (B) LCL death (n = 3) was determined by trypan blue staining after 72 h of treatment with either DMSO, 1 µM AZD3965 + 20 µM VB124 (AZD + VB), 10 µM phenformin (Phenf.), or 1 µM AZD3965 + 20 µM VB124 + 10 µM phenformin (AZD + VB + Phenf.). (C) Intracellular lactate concentration (per 12,500 cells) at 72 h of LCLs treated as in B. (D) LCL death (n = 3) in cells treated with DMSO, 1 µM AZD3965 + 20 µM VB124 (AZD + VB), 2 mM metformin (Metf.), or 1 µM AZD3965 + 20 µM VB124 + 2 mM phenformin (AZD + VB + Metf.), was determined by trypan blue staining after 72 h. All statistical significance was determined by a paired Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
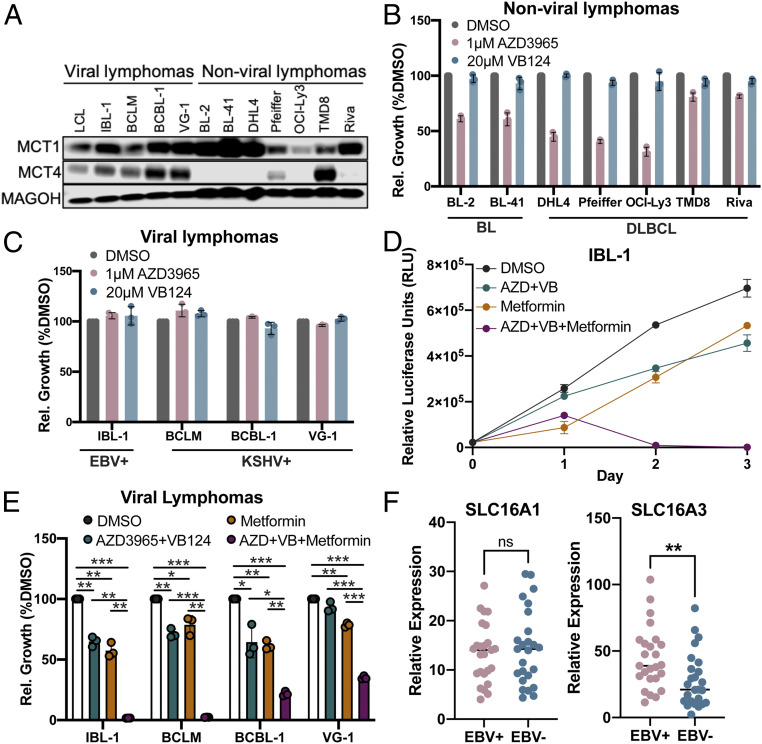
EBV+ and KSHV+ Lymphoma Cell Lines and MCT1/4+ Nonviral Lymphomas Are Hypersensitive to Killing by Metformin upon MCT1/4 Inhibition.
EBV is an etiologic factor in the development of B cell lymphomas of the immune suppressed, such as HIV-associated DLBCL and posttransplant lymphoproliferative disease (41–43). Additionally, primary effusion lymphoma (PEL) can also be driven by Kaposi sarcoma–associated herpesvirus (KSHV), a closely related oncogenic gammaherpesvirus. Therefore, we explored the role of MCT1/4 in both EBV+ and KSHV+ lymphoma cell growth. All EBV+ and KSHV+ viral lymphomas tested expressed both MCT1 and MCT4 at appreciable levels, a stark contrast to the seven nonviral BL and DLBCL lines we screened. As expected, the MCT1-high, MCT4-low nonviral lymphoma cell lines were generally sensitive to single MCT1 inhibition, consistent with previous reports (Fig. 7 A and B) (32, 44). However, the EBV+ and KSHV+ lymphoma cell lines were uniformly resistant to single MCT1 or MCT4 inhibition, similar to the single-inhibitor resistance observed in LCLs (Fig. 7C).
Fig. 7.
EBV+ and KSHV+ lymphoma cell lines are sensitized to killing by metformin upon dual MCT1/4 inhibition. (A) Western blot of MCT1 and MCT4 protein expression in viral (EBV+ or KSHV+) and nonviral (BL or DLBCL) lymphoma cell lines. MAGOH = loading control. (B) Relative cell growth (normalized to DMSO) in nonviral and, in (C), viral lymphoma cell lines. Cell growth was determined by CellTiter Glo luminescence. (D) Growth over time of the EBV + IBL-1 cell line (n = 3), after treatment with DMSO, 1 µM AZD3965 + 20 µM VB124 (AZD + VB), 2 mM Metformin (Metformin), or 1 µM AZD3965 + 20 µM VB124 +2 mM Metformin (AZD + VB + Metformin). The error bars smaller than the line symbols were automatically excluded by graphing software. (E) Relative cell growth of viral lymphoma cell lines at 48 h posttreatment as in D. IBL-1 = EBV+, KSHV− AIDS immunoblastic lymphoma, BCBL-1 = EBV−, KSHV + PEL, VG-1 = EBV−, KSHV+ PEL, BCLM = EBV−, KSHV+ PEL. Statistical significance was determined by a one-tailed paired Student’s t test where *P < 0.05, **P < 0.01, and ***P < 0.001. (F) Relative expression of SLC16A1 (MCT1) and SLC16A3 (MCT4) in EBV-positive (EBV+) and EBV-negative (EBV−) human DLBCL biopsies. RNA-seq comparisons were evaluated by Wilcoxon rank sum test; **P < 0.01, ns (not significant) P ≥ 0.05.
To determine whether dual MCT1/4 inhibition could reduce viral lymphoma cell growth, we first treated the EBV+ AIDS immunoblastic lymphoma cell line, IBL-1, which is most similar to LCLs in its Latency III state (45). IBL-1 cell growth was inhibited by dual MCT1/4 inhibition, and furthermore, these treated cells became hypersensitive to killing by metformin (Fig. 7D). We also observed increased sensitivity to metformin upon MCT1/4 inhibition of three KSHV+ PEL lines, BCLM, BCBL-1, and VG-1, suggesting that pharmacologically targeting MCT1/4 alongside metformin treatment could be explored as a novel therapeutic strategy against EBV+ and KSHV+ lymphomas (Fig. 7E). While most nonviral lymphomas were sensitive to single MCT1 inhibition, we observed that those cell lines expressing MCT4 as well as MCT1 (e.g., TMD8) became susceptible to dual MCT1/4 inhibition and metformin-induced killing similar to the viral lymphomas (SI Appendix, Fig. S4).
Our studies on the susceptibility of viral lymphomas to MCT antagonism have relied on model systems including primary B cell infection and tumor cell lines. However, to complement these mechanistic studies, we sought to characterize MCT expression in primary lymphoma tissue. Therefore, we characterized the expression of MCT1 (SLC16A1) and MCT4 (SLC16A3) in primary DLBCL tissue for which EBV baits were used to molecularly diagnose association with the virus. Consistent with our cell culture models, we found no difference in MCT1 expression between EBV-negative and EBV-positive DLBCLs but a significant increase in MCT4 among EBV+ lymphomas (Fig. 7F). These data coupled with our studies of MCT gene regulation and function in viral lymphomas support the further exploration of this metabolic pathway for clinical evaluation.
Discussion
EBV reorganizes B cell metabolic pathways to promote immortalization as a precursor to the development of lymphomas in the immune suppressed. We and others have previously described a balanced up-regulation of oxidative phosphorylation and glycolysis through EBV-mediated B cell outgrowth (3, 10). To maintain glycolytic flux during aerobic glycolysis, EBV-infected B cells must up-regulate lactate transporters. In this study, we identified an EBNA2/c-Myc–mediated up-regulation of MCT1 that was important for supporting increased glycolysis during the hyperproliferative burst early after EBV infection. However, infected cells became resistant to MCT1 antagonism between 1- and 2-wk postinfection, and we observed a strong up-regulation of MCT4. This correlated with the switch in viral latency programs in which the viral LMP1 protein promoted MCT4 expression. EBV-immortalized LCLs were sensitive to the inhibition of both MCT1 and MCT4 with specific small molecule antagonists and combined CRISPR knockout with reciprocal inhibitor, which led to their growth arrest due to a loss in the OCR, an accumulation of both lactate and ROS, and depletion of NAD+ relative to NADH. Importantly, MCT1/4 inhibition rendered LCLs vulnerable to killing by the ETC inhibitors phenformin or metformin. This lethal effect on LCLs was also manifested in lymphoma cell lines harboring EBV and also a related oncogenic gammaherpesvirus, KSHV, suggesting a strategy for targeting viral-induced cancers.
The role of MCTs has been well studied in cancer but not during viral infection. The metabolic requirements of both processes are similar, with a goal of ramping up anabolic processes to either promote proliferation or viral particle production. Latent oncogenic herpesviruses are unique in that they express specific proteins and noncoding RNAs that reprogram cellular transcription and metabolic pathways with the goal of promoting viral genome replication during host cell proliferation. For example, during KSHV lytic replication, glycolysis is required for maximal virion production, while during latent infection, 10 out of the 12 microRNAs are sufficient to induce glycolysis, thereby contributing to infected cell growth (46, 47). KSHV-associated PEL also display elevated aerobic glycolysis and fatty acid biosynthesis compared to primary B cells, highlighting the importance of these pathways in KSHV-associated tumors and another parallel between KSHV infection and tumorigenesis (48, 49). Similarly, the EBV oncoprotein LMP1 up-regulates glucose import through NF-κB signaling to promote cell survival (8). Thus, EBV and KSHV coordinately activate their infected cells in a similar manner to how cellular oncogenes cooperate to reprogram metabolism in cancer. While both EBV- and KSHV-associated lymphomas display increased ECAR and lactate export due to increased glycolysis, only a handful of studies have addressed the role of MCT1 and MCT4 in EBV+ and KSHV+ cancers (9, 50). In one study, MCT4 RNA was shown to be up-regulated in EBV-immortalized LCLs relative to mitogen-activated B cells and freshly EBV-infected B cells but mainly due to its association with the induction of hypoxia (9, 51). More indirectly, KSHV-infected PEL chemoresistance to doxorubicin and paclitaxel has been linked to expression of the MCT1/4-associated chaperone glycoprotein Emmprin, which is regulated by the KSHV-encoded latency-associated nuclear antigen in KSHV+ PEL cell lines (52, 53). Given the similarities between EBV- and KSHV-mediated reprogramming of host metabolism and based on our observation that KSHV+ PELs arrest upon treatment with both MCT1/4 inhibitors, it is therefore possible that both viruses require and/or regulate MCT1/4-mediated lactate export in similar ways. Future studies would need to elucidate whether MCT1/4-mediated lactate transport in KSHV+ lymphomas is intertwined with other aspects of metabolism like redox regulation and oxidative stress mitigation, as is the case in EBV-immortalized LCLs.
During EBV infection, the EBV master transcriptional regulator EBNA2 can both directly and indirectly influence host metabolic pathways such as one-carbon metabolism and fatty acid biosynthesis through c-Myc (10, 54). We find that concomitant induction of MCT1 through c-Myc is critical to support continued proliferation as lactate is generated through glycolysis. Interestingly, we found that early-infected cells were exquisitely sensitive to MCT1 inhibition through AZD3965 with low-nanomolar (∼1 to 10 nM) concentrations halting growth while in many other tumor cell systems, 250 nM is typically used. This suggests that MCT1-mediated lactate export could be particularly important for B cell growth in the absence of MCT4—a gene expression signature which has also been observed in germinal center B cells (32). With regards to the role of LMP1 in inducing MCT4 gene expression, it is worth noting that one of the major signaling pathways critical for LCL survival is LMP1-mediated NF-κB signaling (55). Although we were not able to identify any specific evidence for NF-κB–mediated MCT4 gene regulation, future studies could further explore how this signaling pathway could influence overall LCL metabolism. Likewise, given the analogous role of the KSHV v-FLIP protein in NF-κB activation, perhaps this molecule is involved in MCT4 up-regulation in PEL cells (56).
We were intrigued by the finding that EBV-infected B cells became resistant to MCT1 inhibition between 1 and 2 wk after infection, as this is the timeframe when infected cells switch from proliferation driven by the viral EBNA proteins (Latency IIb) to that of the EBNAs and the LMPs (Latency III) (5, 6). Given this relationship and the prior knowledge that MCT4 induction could promote resistance to MCT1 antagonism, we interrogated MCT4 up-regulation and sensitivity to a novel MCT4 inhibitor. Our data support the up-regulation of MCT4 by LMP1, which is also supported by a published data set of LMP1 expression in DG75 cells (57). Moreover, that study found that LMP1 targets dependent on the metabolic and DNA damage regulator, PARP1, were largely enriched in HIF1-α targets. In fact, their data using the PARP1 inhibitor olaparib suggests that LMP1 induction of MCT4 was PARP1 dependent. These data also nicely dovetail with that of a previous study supporting the stabilization of HIF1-α and its targets in EBV-infected cells supported a “pseudo-hypoxia” that appears to be induced by EBV (9).
The mechanism of growth arrest mediated by MCT1/4 antagonism involved a reduction in the extracellular acidification rate due to the accumulation of lactate within cells but surprisingly also a significant reduction in the OCR. Moreover, these effects were observed within 24 h following treatment, preceding cellular growth arrest, which occurs between 48 to 72 h after treatment. We were intrigued by the significant decrease in OCR upon dual MCT1/4 inhibition given that previous reports describe increased OCR upon lactate export inhibition as a means of compensating for reduced glycolysis with increased OXPHOS (20, 22, 58, 59). One possible explanation could be that MCT1/4 inhibition reduces overall cellular fitness, which is reflected in reduced ECAR and OCR. However, an alternative explanation for the reduced OCR could be due to the altered redox balance via depleted NAD+/NADH, which could inhibit NADH:ubiquinone oxidoreductase activity in complex I of the ETC as previously observed (60). In either scenario, the reduction of OCR upon selective MCT1/4 inhibition in LCLs points at an interesting vulnerability that could be exploited for therapeutic benefit in EBV-driven lymphomas in combination with ETC inhibitors like metformin. Our data are in contrast to a recent study where dual MCT1/4 inhibition in leukemia and breast cancer cell lines using the antihypertensive drug syrosingopine did not impact NAD+/NADH ratios, though the addition of metformin did alter this balance (30).
The MCT1 inhibitor AZD3965 is currently in clinical trials for various lymphomas and solid tumors (31). However, no such trials exist for MCT4, primarily due to the lack of MCT4-specific inhibitors. While pan-MCT1/4 inhibitors do exist, these also target other metabolic pathways, which could potentially cause adverse side effects (61, 62). Using the recently developed MCT4-specific inhibitor, VB124, as a tool compound, we were able to explore the therapeutic potential of MCT1/4-targeted combination therapy in viral-associated lymphomas as well as MCT1/4-double–expressing nonviral lymphomas (38). We recognize that preclinical testing of AZD3965 and VB124 in primary cells and cell lines are likely an incomplete representation of how lymphomagenesis unfolds in vivo, as contributions from the tumor microenvironment are lacking. EBV has a narrow host tropism and is unable to infect mice or other common small animal models. However, recently developed humanized mouse models could be useful in assessing how metabolic pathways like MCT1/MCT4-mediated lactate transport contribute to EBV-driven tumorigenesis in vivo (63). Furthermore, our findings that EBV+ DLBCLs display elevate MCT4 relative to nonviral DLBCLs supports the rationale for further preclinical development of this therapeutic strategy.
Materials and Methods
Cell Culture Conditions and Viruses.
Buffy coats were obtained from normal human donors through the Gulf Coast Regional Blood Center, and PBMCs were isolated by Ficoll Histopaque-1077 gradient (Sigma, H8889). B95-8 strain of EBV was produced from the B95-8 Z-HT cell line as previously described (53). Virus infections were performed in bulk by adding 50 µL filtered B95-8 supernatant to 1 × 106 PBMCs. TLR9 ligand CpG oligonucleotide (ODN 2006) was purchased from IDT and used at 2.5 μg/mL
Early-infected PBMCs and B cells were kept in Roswell Park Memorial Institute (RPMI) medium 1640 supplemented with 15% heat-inactivated fetal bovine serum (FBS) (Corning), 2 mM L-Glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen), and 0.5 µg/mL Cyclosporine A (Sigma). All LCLs and BJAB (EBV− germinal-center–derived B cell lymphoma line) cells were kept in RPMI medium 1640 supplemented with 10% heat-inactivated FBS (Corning), 2 mM L-Glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen). BJAB cells as well as the EBV− BL41 were obtained from George Mosialos, Aristotle University, Thessaloniki, Greece. Kaposi's sarcoma–associated herpesvirus-positive (KSHV+)/EBV− PEL cell lines VG-1, BCLM, and BCBL1 were provided by Dirk Dittmer, University of North Carolina, Chapel Hill, NC, or Bryan Cullen, Duke University, Durham, NC, with permission from the original authors and kept in RPMI medium 1640 supplemented with 15% FBS, 10 mM Hepes, 1 mM sodium pyruvate, and 0.05 mM β-mercaptoethanol (Sigma). The EBV-positive cell line derived from an AIDS immunoblastic lymphoma, IBL-1, was kindly provided by Ethel Cesarman, Weill Cornell Medical College, New York, NY, and kept in RPMI 1640 supplemented with 20% FBS. The DLBCL cell lines were a kind gift from Sandeep Dave, Duke University, Durham, NC. SUDHL4, Pfeiffer, and Riva were cultured in RPMI medium 1640 supplemented with 10% heat-inactivated FBS, 2 mM L-Glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. OCI-Ly3 was kept in Iscove's modified Dulbecco's medium (IMDM) supplemented with 20% heat-inactivated human serum, 2 mM L-Glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin, and TMD8 cells in minimal essential medium (MEM) supplemented with 10% FBS, 2 mM L-Glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. The EBV-negative BL2 lymphoma cell line was provided by Geoffrey Wahl, Salk Institute for Biological Studies, San Diego, CA. BL2 cells were cultured in RPMI medium 1640 containing 2 mM L-glutamine, 1 mM sodium pyruvate, 20 mM glucose, 100 U/mL penicillin, and 100 μg/mL streptomycin and supplemented with 10% heat-inactivated FBS. The P493-6 cells (a kind gift of Georg Bornkamm, Helmholtz Zentrum München, Germany) were cultured in RPMI 1640 supplemented with 10% tetracycline-free FBS (HyClone SH30070), 1 µM β-Estradiol, and 1 µg/mL Tetracycline. P493-6 cells were washed three times with β-Estradiol–free media and then cultured in the absence of β-Estradiol for 3 d to induce the EBNA2-OFF state. After washing three times, the cells were then cultured again for 3 d upon reintroducing 1 μM β-Estradiol, returning P493-6 cells to the EBNA2-ON state. All cells were cultured at 37 °C in a humidified incubator at 5% CO2.
Flow Cytometry and Sorting.
To track proliferation in EBV-infected B cells, cells were stained with CellTrace Violet (Invitrogen, C34557), a fluorescent proliferation-tracking dye. Cells were first washed in fluorescence-activated cell sorting (FACS) buffer (5% FBS in phosphate-buffered saline [PBS]), stained with the appropriate antibody for 30 min to 1 h at 4 °C in the dark, and then washed again before being analyzed on a BD FACS Canto II. Proliferating infected B cells were sorted to a pure population of CD19+/CellTrace Violet on a MoFlo Astrios Cell Sorter at the Duke Cancer Institute Flow Cytometry Shared Resource. Mouse anti-human CD19 antibody (Biolegend catalog number [Cat.] 152410) conjugated with APC was used as a surface B cell marker in flow cytometry.
Cell Growth Assays.
LCLs were seeded at a density of 3 × 105 cells/mL and treated with either dimethyl sulfoxide (DMSO), AZD3965 (Cayman Chemicals, Item No. 19912 or BioVision, Cat No. B2227), VB124 (a kind gift of Mark Parnell, Vettore LLC, San Francisco, CA), phenformin hydrochloride (Cayman Chemicals, Item 14997), or metformin hydrochloride (Cayman Chemicals, Item 13118) for 1 to 4 d. VB124 required concentrations much higher than its 50% inhibitory concentration (IC50, 8.6 nM) in serum due to high (∼99%) serum protein binding common for carboxylic acids like VB124. Drug treatments were normalized across conditions to contain 0.1% DMSO. Cell growth was assessed daily by either CellTiter Glo (Promega) or counting by trypan exclusion on an automated cell counter (Countess II, Thermo Fisher). Luminescence was measured using a microplate reader (Biotek Synergy HTX).
Gene Expression Analysis.
Total RNA was isolated from cells by using a Qiagen RNeasy kit and then reverse transcribed to generate complementary deoxyribonucleic acid (cDNA) with the High-Capacity cDNA kit (Applied Biosystems). qPCR was performed by using SYBR green (Quanta Biosciences) in an Applied Biosystems Step One Plus instrument. Primer sets include MCT1 (Forward: GACCTTGTTGGACCCCAGAG, Reverse:AGCCGACCTAAAAGTGGTGG), MCT4 (Forward: AGGTCTACCTCACCACTGGG, Reverse: CACAGGAAGACAGGGCTACC.
BrdU Staining.
Cell cycle progression was assessed by BrdU staining. Infected PBMCs treated with DMSO, Chk2 inhibitor II (EMD Millipore), or AZD3965 (Cayman Chemicals) cells were stained with CD19 and pulsed with BrdU (BD Biosciences, Cat. 552598) for 2 h following fixation and permeabilization. Afterward, cells were treated with a DNase to expose BrdU-bound epitopes and stained with a BrdU-fluorescent antibody as well as the total DNA and cell death marker 7-AAD. Then, BrdU and 7-AAD fluorescence values were acquired on a flow cytometer (FACS Canto II).
Immunoblotting.
Cells were pelleted and washed in PBS and then lysed in lithium dodecyl sulfate (LDS) Sample Buffer (NuPAGE) with complete protease inhibitors. All protein lysates were run on NuPage 4 to 12% gradient gels (LifeTechnology) and transferred to polyvinylidene difluoride (PVDF) membrane (GE Healthcare). Membranes were blocked in 5% milk in Tris-buffered saline-Tween 20 (TBST) and stained with primary antibody overnight at +4 °C, followed by a wash and staining with secondary horseradish peroxidase (HRP)-conjugated antibody for 1 h at room temperature. Antibodies include MCT1 (Abcam, Cat. ab85021, 1:1,000 dilution), MCT4 (Proteintech, Cat. 22787-1-AP, 1:1,000 dilution), LMP1 (S12; gift from Elliott Kieff, Harvard Medical School, Boston, MA), c-Myc (Santa Cruz Biotechnology, SC-764), EBNA2 (PE2; gift from Elliott Kieff), and MAGOH (Santa Cruz, SC-56724). We use MAGOH as a loading control because it does not change in expression from resting B cells through LCL outgrowth.
Generation of CRISPR/Cas9 Knockout.
The SLC16A1 (MCT1) CRISPR/Cas9 knockout LCL was generated using CRISPR-Cas9 technology. In summary Cas9-expressing LCLs [lentiCas9-enhanced green fluorescent protein (EGFP); Addgene 63592] were transduced with pLentiGuide-puro short-guide (sg)MCT1 RNA (sgRNA sequence: 5′ CACCGCCAGCCCCAGCCTCCATCTG 3′, 3′ GGTCGGGGTCGGAGGTAGACCAAA 5′), specific to the SLC16A1 gene and grown out under puromycin selection (lentiGuide-Puro; Addgene 52963). Next, sgRNA-transduced LCLs were serially diluted to obtain a clonal population and screened for MCT1 protein expression by Western blot. The clone displaying greatest reduction of MCT1 protein was then expanded and used for downstream studies.
To obtain SLC16A3 (MCT4)-knockout LCLs, LCLs were transfected with ribonucleoprotein complexes consisting of Cas9 protein duplexed with sgRNA. Briefly, to assemble the complexes, TrueCut Cas9 protein version 2 (Thermo Fisher, A36498) was incubated with both CRISPRevolution sgRNA SLC16A3 (CAACCCUCCUGGCCAUGGGA; Synthego) and sgRNA CD226 (UAGAGACAUGUUCUCGGCAA; Synthego) in a protein/sgRNA ratio of 1:3. Transfection was performed using Thermo Fisher Neon Transfection System. Loss of expression of CD226, measured by FACS, was used as a proxy to identify MCT4 knockout cells (64). CD226-negative cells were sorted on a MoFlo Astrios Cell Sorter at the Duke Cancer Institute Flow Cytometry Shared Resource, and MCT4 knockout was confirmed by Western blot.
Seahorse Analysis.
ECAR and OCR were measured using the Seahorse XFe96 extracellular flux analyzer (Agilent Technologies) Cell Energy Phenotype Test. LCLs were attached to culture plates by using Cell-Tak (BD Bioscience). ECAR and OCR were measured in Seahorse XF Base Medium supplemented with 1 mM pyruvate, 2 mM glutamine, and 10 mM glucose (Sigma-Aldrich). ECAR and OCR values were normalized to cell number. For stress measurements, ECAR and OCR were measured over time after injection of oligomycin and FCCP.
13C-Glucose Tracing.
Glucose tracing experiments were performed using U-13C6-glucose (Cambridge Isotope Laboratories CLM-1396-PK). LCLs were cultured in glucose-free RPMI supplemented with 10 mM U-13C6-glucose and 10% dialyzed FBS, seeded at a density of 300,000 LCLs/mL, and treated with either DMSO or 1 µM AZD3965 + 20 µM VB124 for 48 or 72 h. Then, cells were harvested and washed twice with ice-cold PBS, followed by metabolite extraction with ice-cold 80% methanol in deionized water and evaporation for approximately 2 h. Dried metabolites were reconstituted in 100 µL 50% acetonitrile (ACN) 50% dH2O solution. Samples were vortexed and spun down for 10 min at 17,000 g. A total of 70 µL of the supernatant was then transferred to high-performance liquid chromatography (HPLC) glass vials. A total of 10 µL of these metabolite solutions were injected per analysis. Samples were run on a Vanquish (Thermo Fisher Scientific) ultra high-performance liquid chromatography (UHPLC) system with mobile phase A (20 mM ammonium carbonate, pH = 9.7) and mobile phase B (100% ACN) at a flow rate of 150 µL/min on a SeQuant ZIC-pHILIC Polymeric column (2.1 × 150 mm 5 μm, EMD Millipore) at 35 °C. Separation was achieved with a linear gradient from 20 to 80% A in 20 min followed by a linear gradient from 80 to 20% A from 20 to 20.5 min. Then, 20% A was held from 20.5 min to 28 min. The UHPLC was coupled to a Q-Exactive (Thermo Fisher Scientific) mass analyzer running in polarity switching mode with spray voltage = 3.2kV, sheath gas = 40, aux gas = 15, sweep gas = 1, aux gas temp = 350 °C, and capillary temp = 275 °C. For both polarities, mass scan settings were kept at full scan range = (70 to 1,000), ms1 resolution = 70,000, max injection time = 250 ms, and AGC target = 1E6. MS2 data were also collected from the top three most abundant singly charged ions in each scan with normalized collision energy = 35. Each of the resulting “.RAW” files was then centroided and converted into two “.mzXML” files (one for positive scans and one for negative scans) using msconvert from ProteoWizard (65). These “.mzXML” files were imported into the MZmine 2 software package (66). Ion chromatograms were generated from MS1 spectra via the built-in Automated Data Analysis Pipeline (ADAP) chromatogram module, and peaks were detected via the ADAP wavelets algorithm (67). Peaks were aligned across all samples via the random sample consensus aligner module, gap filled, and assigned identities using an exact mass MS1(+/−15 ppm) and retention time (+/−0.5 min) search of our in-house MS1-RT database. Peak boundaries and identifications were then further refined by manual curation. Peaks were quantified by area under the curve integration and exported as comma-separated values (CSV) files. The peak areas were additionally processed via the R package AccuCor to correct for natural isotope abundance (68). Peak areas for each sample were normalized by the measured area of the internal standard trifluoromethanesulfonate (present in the extraction buffer) and by the number of cells present in the extracted sample.
Lactate and NAD+/NADH Measurements.
Intracellular and extracellular lactate concentrations were determined using the Lactate Glo (Promega) assay kit. Briefly, cells were harvested and washed in ice-cold PBS once, then immediately inactivated with 0.6 N HCl to halt metabolic activity and neutralized with 1 M Tris Base. The resulting lysate was combined at a 1:1 ratio with the lactate detection reagent and luminescence measured by a microplate reader. For intracellular lactate measurements involving early-infection time points, PBMCs were first depleted for CD3+ T cells (RosetteSep Human Granulocyte Depletion Mixture), sorted to a pure population of CD19+ B cells on a MoFlo Astrios Cell Sorter at the Duke Cancer Institute Flow Cytometry Shared Resource. Purified B cells were either treated as indicated or used as the starting cell population for time-course lactate measurements. Mouse anti-human CD19 antibody (Biolegend Cat. 152410) conjugated with APC was used as a surface B cell marker in flow cytometry.
NAD+/NADH ratios were measured using the NAD+/NADH Glo assay (Promega) according to the manufacturer’s protocol. Briefly, harvested cells were washed in PBS, lysed with Tris Base supplemented with 1% Dodecyl trimethylammonium bromide, treated with 0.4 N HCl (for NAD+), and heated (for NADH) at 60 °C for 15 min. Each sample was then added at a 1:1 ratio with a luciferin detection reagent, and NAD+ or NADH luminescence measured separately using a BioTek Synergy 2 microplate reader.
ROS Measurements.
ROS fluorescence was determined using the ROS-sensitive dye DCFDA (Cayman Chemicals Cat. 601520). Cells were pelleted and washed in the provided wash buffer and then stained in 20 µM DCFDA for 1.5 h at 37 °C protected from light. Then, cells were washed once more in the provided wash buffer and fluorescein isothiocyanate (FITC) fluorescence measured on a BD FACSCanto II flow cytometer. Pyocyanin (1 mM) was used as a positive control and N-acetylcysteine (300 mM) as a negative control.
Expression Analysis of Primary Human DLBCL Tissue.
From a previously published cohort of DLBCLs, we selected 25 cases each of EBV-positive and EBV-negative cases (69). These cases were subjected to DNA sequencing and RNA-seq. DNA sequencing included selection for the human exome as well as the EBV genome. The EBV-positive cases were verified to have an average of 42,929 DNA sequencing reads mapping to the EBV genome compared to 0 for the negative cases. We further examined the RNA-seq data from these cases for their expression of SLC16A3 and SLC16A1 genes.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant R01-CA140337 (to M.A.L.), T32GM007105-43, T32-CA009111, 1F31CA257621-01 (to E.N.B.), T32-CA009056 (to J.C.), and R01 CA215185 (to H.R.C.) as well as Grant RSG-16-111-01-MPC from the American Cancer Society (to H.R.C.), the CDI Harry Winston Fellowship award (to J.C.), and the St. Baldrick’s Foundation Fellowship award (to J.C.). Additional funding came from the Duke Center for AIDS Research, an NIH-funded program (Grant 5P30-Al064518). We would like to thank Dr. Michael Cook, Nancy Martin, Lynn Martinek, and the Duke Cancer Institute Shared Flow Cytometry core facility for invaluable assistance with flow cytometry and cell sorting. We would also like to thank Dr. Nancie McIver and Amanda Nichols of the Duke Cellular Metabolism Analysis Core for their guidance with the Seahorse XF analysis. Finally, we would like to thank Ernst Schmid of the University of California, Los Angeles, Department of Biological Chemistry for assistance with mass spectrometry and downstream analysis of isotopic tracing experiments.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. B.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022495118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Babcock G. J., Decker L. L., Volk M., Thorley-Lawson D. A., EBV persistence in memory B cells in vivo. Immunity 9, 395–404 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Nikitin P. A., et al., An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein-Barr virus transformation of primary human B cells. Cell Host Microbe 8, 510–522 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McFadden K., et al., Metabolic stress is a barrier to Epstein-Barr virus-mediated B-cell immortalization. Proc. Natl. Acad. Sci. U.S.A. 113, E782–E790 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shannon-Lowe C., Rickinson A. B., Bell A. I., Epstein-Barr virus-associated lymphomas. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160271 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Price A. M., Luftig M. A., To be or not IIb: A multi-step process for Epstein-Barr virus latency establishment and consequences for B cell tumorigenesis. PLoS Pathog. 11, e1004656 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Messinger J. E., Dai J., Stanland L. J., Price A. M., Luftig M. A., Identification of host biomarkers of Epstein-Barr virus latency IIb and latency III. mBio 10, e01006-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nikitin P. A., Price A. M., McFadden K., Yan C. M., Luftig M. A., Mitogen-induced B-cell proliferation activates Chk2-dependent G1/S cell cycle arrest. PLoS One 9, e87299 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sommermann T. G., O’Neill K., Plas D. R., Cahir-McFarland E., IKKβ and NF-κB transcription govern lymphoma cell survival through AKT-induced plasma membrane trafficking of GLUT1. Cancer Res. 71, 7291–7300 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darekar S., et al., Epstein-Barr virus immortalization of human B-cells leads to stabilization of hypoxia-induced factor 1 alpha, congruent with the Warburg effect. PLoS One 7, e42072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L. W., et al., Epstein-Barr-virus-induced one-carbon metabolism drives B cell transformation. Cell Metab. 30, 539–555.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sola-Penna M., Metabolic regulation by lactate. IUBMB Life 60, 605–608 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Sun S., Li H., Chen J., Qian Q., Lactic acid: No longer an inert and end-product of glycolysis. Physiology (Bethesda) 32, 453–463 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Chiche J., et al., In vivo pH in metabolic-defective Ras-transformed fibroblast tumors: Key role of the monocarboxylate transporter, MCT4, for inducing an alkaline intracellular pH. Int. J. Cancer 130, 1511–1520 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Halestrap A. P., Price N. T., The proton-linked monocarboxylate transporter (MCT) family: Structure, function and regulation. Biochem. J. 343, 281–299 (1999). [PMC free article] [PubMed] [Google Scholar]
- 15.Ritzhaupt A., Wood I. S., Ellis A., Hosie K. B., Shirazi-Beechey S. P., Identification and characterization of a monocarboxylate transporter (MCT1) in pig and human colon: Its potential to transport L-lactate as well as butyrate. J. Physiol. 513, 719–732 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fishbein W. N., Merezhinskaya N., Foellmer J. W., Relative distribution of three major lactate transporters in frozen human tissues and their localization in unfixed skeletal muscle. Muscle Nerve 26, 101–112 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Deuticke B., Monocarboxylate transport in erythrocytes. J. Membr. Biol. 70, 89–103 (1982). [DOI] [PubMed] [Google Scholar]
- 18.Price N. T., Jackson V. N., Halestrap A. P., Cloning and sequencing of four new mammalian monocarboxylate transporter (MCT) homologues confirms the existence of a transporter family with an ancient past. Biochem. J. 329, 321–328 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dimmer K. S., Friedrich B., Lang F., Deitmer J. W., Bröer S., The low-affinity monocarboxylate transporter MCT4 is adapted to the export of lactate in highly glycolytic cells. Biochem. J. 350, 219–227 (2000). [PMC free article] [PubMed] [Google Scholar]
- 20.Hong C. S., et al., MCT1 modulates cancer cell pyruvate export and growth of tumors that co-express MCT1 and MCT4. Cell Rep. 14, 1590–1601 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doherty J. R., Cleveland J. L., Targeting lactate metabolism for cancer therapeutics. J. Clin. Invest. 123, 3685–3692 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doherty J. R., et al., Blocking lactate export by inhibiting the Myc target MCT1 Disables glycolysis and glutathione synthesis. Cancer Res. 74, 908–920 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voss D. M., et al., Disruption of the monocarboxylate transporter-4-basigin interaction inhibits the hypoxic response, proliferation, and tumor progression. Sci. Rep. 7, 4292 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juel C., Lactate-proton cotransport in skeletal muscle. Physiol. Rev. 77, 321–358 (1997). [DOI] [PubMed] [Google Scholar]
- 25.Bonen A., Baker S. K., Hatta H., Lactate transport and lactate transporters in skeletal muscle. Can. J. Appl. Physiol. 22, 531–552 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Bosshart P. D., Kalbermatter D., Bonetti S., Fotiadis D., Mechanistic basis of L-lactate transport in the SLC16 solute carrier family. Nat. Commun. 10, 2649 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hui S., et al., Glucose feeds the TCA cycle via circulating lactate. Nature 551, 115–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitaker-Menezes D., et al., Evidence for a stromal-epithelial “lactate shuttle” in human tumors: MCT4 is a marker of oxidative stress in cancer-associated fibroblasts. Cell Cycle 10, 1772–1783 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beloueche-Babari M., et al., MCT1 inhibitor AZD3965 increases mitochondrial metabolism, facilitating combination therapy and non-invasive magnetic resonance spectroscopy. Cancer Res. 77, 5913–5924 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjamin D., et al., Dual inhibition of the lactate transporters MCT1 and MCT4 is synthetic lethal with metformin due to NAD+ depletion in cancer cells. Cell Rep. 25, 3047–3058.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.U.S. National Library of Medicine , A phase I trial of AZD3965 in patients with advanced cancer. https://clinicaltrials.gov/ct2/show/NCT01791595. Accessed 14 April 2020.
- 32.Curtis N. J., et al., Pre-clinical pharmacology of AZD3965, a selective inhibitor of MCT1: DLBCL, NHL and Burkitt’s lymphoma anti-tumor activity. Oncotarget 8, 69219–69236 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaiser C., et al., The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J. Virol. 73, 4481–4484 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pajic A., et al., Cell cycle activation by c-myc in a burkitt lymphoma model cell line. Int. J. Cancer 87, 787–793 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Maier S., et al., Cellular target genes of Epstein-Barr virus nuclear antigen 2. J. Virol. 80, 9761–9771 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polański R., et al., Activity of the monocarboxylate transporter 1 inhibitor AZD3965 in small cell lung cancer. Clin. Cancer Res. 20, 926–937 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price A. M., Messinger J. E., Luftig M. A., c-Myc represses transcription of Epstein-Barr virus latent membrane protein 1 early after primary B cell infection. J. Virol. 92, e01178–e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cluntun A. A., et al., The pyruvate-lactate axis modulates cardiac hypertrophy and heart failure. Cell Metab. 33, 629–648.e10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y., McMillan-Ward E., Kong J., Israels S. J., Gibson S. B., Mitochondrial electron-transport-chain inhibitors of complexes I and II induce autophagic cell death mediated by reactive oxygen species. J. Cell Sci. 120, 4155–4166 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Marchiq I., Le Floch R., Roux D., Simon M.-P., Pouyssegur J., Genetic disruption of lactate/H+ symporters (MCTs) and their subunit CD147/BASIGIN sensitizes glycolytic tumor cells to phenformin. Cancer Res. 75, 171–180 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Bibas M., Antinori A., EBV and HIV-related lymphoma. Mediterr. J. Hematol. Infect. Dis. 1, e2009032 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rezk S. A., Weiss L. M., Epstein-Barr virus-associated lymphoproliferative disorders. Hum. Pathol. 38, 1293–1304 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Dharnidharka V. R., et al., Post-transplant lymphoproliferative disorders. Nat. Rev. Dis. Primers 2, 15088 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Noble R. A., et al., Inhibition of monocarboxyate transporter 1 by AZD3965 as a novel therapeutic approach for diffuse large B-cell lymphoma and Burkitt lymphoma. Haematologica 102, 1247–1257 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horenstein M. G., et al., Epstein-Barr virus latent gene expression in primary effusion lymphomas containing Kaposi’s sarcoma-associated herpesvirus/human herpesvirus-8. Blood 90, 1186–1191 (1997). [PubMed] [Google Scholar]
- 46.Sanchez E. L., et al., Glycolysis, glutaminolysis, and fatty acid synthesis are required for distinct stages of Kaposi’s sarcoma-associated herpesvirus lytic replication. J. Virol. 91, e02237-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lagunoff M., Activation of cellular metabolism during latent Kaposi’s Sarcoma herpesvirus infection. Curr. Opin. Virol. 19, 45–49 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bhatt A. P., et al., Dysregulation of fatty acid synthesis and glycolysis in non-Hodgkin lymphoma. Proc. Natl. Acad. Sci. U.S.A. 109, 11818–11823 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Currie E., Schulze A., Zechner R., Walther T. C., Farese R. V. Jr, Cellular fatty acid metabolism and cancer. Cell Metab. 18, 153–161 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Delgado T., et al., Induction of the Warburg effect by Kaposi’s sarcoma herpesvirus is required for the maintenance of latently infected endothelial cells. Proc. Natl. Acad. Sci. U.S.A. 107, 10696–10701 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ullah M. S., Davies A. J., Halestrap A. P., The plasma membrane lactate transporter MCT4, but not MCT1, is up-regulated by hypoxia through a HIF-1α-dependent mechanism. J. Biol. Chem. 281, 9030–9037 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Dai L., et al., Emmprin and KSHV: New partners in viral cancer pathogenesis. Cancer Lett. 337, 161–166 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qin Z., Dai L., Slomiany M. G., Toole B. P., Parsons C., Direct activation of emmprin and associated pathogenesis by an oncogenic herpesvirus. Cancer Res. 70, 3884–3889 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang L. W., et al., Epstein-Barr virus subverts mevalonate and fatty acid pathways to promote infected B-cell proliferation and survival. PLoS Pathog. 15, e1008030 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Izumi K. M., Kaye K. M., Kieff E. D., The Epstein-Barr virus LMP1 amino acid sequence that engages tumor necrosis factor receptor associated factors is critical for primary B lymphocyte growth transformation. Proc. Natl. Acad. Sci. U.S.A. 94, 1447–1452 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matta H., Chaudhary P. M., Activation of alternative NF-kappa B pathway by human herpes virus 8-encoded Fas-associated death domain-like IL-1 beta-converting enzyme inhibitory protein (vFLIP). Proc. Natl. Acad. Sci. U.S.A. 101, 9399–9404 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hulse M., et al., Poly(ADP-ribose) polymerase 1 is necessary for coactivating hypoxia-inducible factor-1-dependent gene expression by Epstein-Barr virus latent membrane protein 1. PLoS Pathog. 14, e1007394 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xie J., et al., Beyond Warburg effect–Dual metabolic nature of cancer cells. Sci. Rep. 4, 4927 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu H., Ying M., Hu X., Lactic acidosis switches cancer cells from aerobic glycolysis back to dominant oxidative phosphorylation. Oncotarget 7, 40621–40629 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ying W., NAD+/NADH and NADP+/NADPH in cellular functions and cell death: Regulation and biological consequences. Antioxid. Redox Signal. 10, 179–206 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Park S. J., et al., An overview of MCT1 and MCT4 in GBM: Small molecule transporters with large implications. Am. J. Cancer Res. 8, 1967–1976 (2018). [PMC free article] [PubMed] [Google Scholar]
- 62.Benjamin D., et al., Syrosingopine sensitizes cancer cells to killing by metformin. Sci. Adv. 2, e1601756 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Islas-Ohlmayer M., et al., Experimental infection of NOD/SCID mice reconstituted with human CD34+ cells with Epstein-Barr virus. J. Virol. 78, 13891–13900 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grossman L., et al., Epstein-Barr virus induces adhesion receptor CD226 (DNAM-1) expression during primary B-cell transformation into lymphoblastoid cell lines. mSphere 2, e00305-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chambers M. C., et al., A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pluskal T., Castillo S., Villar-Briones A., Orešič M., MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11, 395 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Myers O. D., Sumner S. J., Li S., Barnes S., Du X., One step forward for reducing false positive and false negative compound identifications from mass spectrometry metabolomics data: New algorithms for constructing extracted ion chromatograms and detecting chromatographic peaks. Anal. Chem. 89, 8696–8703 (2017). [DOI] [PubMed] [Google Scholar]
- 68.Su X., Lu W., Rabinowitz J. D., Metabolite spectral accuracy on orbitraps. Anal. Chem. 89, 5940–5948 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reddy A., et al., Genetic and functional drivers of diffuse large B Cell lymphoma. Cell 171, 481–494.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.