Significance
Methane is a strong greenhouse gas that plays a key role in Earth’s climate. At methane seeps, large amounts of methane move upward through the seafloor, where microbial communities consume much of it. A full accounting of methane’s sources and sinks has evaded researchers—in part, perhaps, because key habitats including carbonate rock mounds have been largely neglected. We sampled seven methane seeps representing four geological settings and found that all sites had rock-hosted microbes capable of consuming methane; in lab-based incubations, some did so at the highest rates reported to date. We demonstrate several factors that help determine a sample’s methane-consuming potential and propose that carbonate rocks at methane seeps may represent a methane sink of far-reaching importance.
Keywords: methane oxidation, metabolic rates, endolith, methane seep
Abstract
At marine methane seeps, vast quantities of methane move through the shallow subseafloor, where it is largely consumed by microbial communities. This process plays an important role in global methane dynamics, but we have yet to identify all of the methane sinks in the deep sea. Here, we conducted a continental-scale survey of seven geologically diverse seafloor seeps and found that carbonate rocks from all sites host methane-oxidizing microbial communities with substantial methanotrophic potential. In laboratory-based mesocosm incubations, chimney-like carbonates from the newly described Point Dume seep off the coast of Southern California exhibited the highest rates of anaerobic methane oxidation measured to date. After a thorough analysis of physicochemical, electrical, and biological factors, we attribute this substantial metabolic activity largely to higher cell density, mineral composition, kinetic parameters including an elevated Vmax, and the presence of specific microbial lineages. Our data also suggest that other features, such as electrical conductance, rock particle size, and microbial community alpha diversity, may influence a sample’s methanotrophic potential, but these factors did not demonstrate clear patterns with respect to methane oxidation rates. Based on the apparent pervasiveness within seep carbonates of microbial communities capable of performing anaerobic oxidation of methane, as well as the frequent occurrence of carbonates at seeps, we suggest that rock-hosted methanotrophy may be an important contributor to marine methane consumption.
The anaerobic oxidation of methane (AOM) strongly modulates the emission of a potent greenhouse gas and represents a primary production pathway that mobilizes carbon, sulfur, and nitrogen on a global scale (1–3). AOM at marine methane seeps has been estimated to consume 80% of subsurface methane (2). However, the location and magnitude of methane oxidation within seep complexes are poorly constrained because the ways in which different substrate types (e.g., sediments or rocks) influence AOM have not been adequately studied.
One overlooked habitat at methane seeps is carbonate rock, which can comprise a substantial proportion of methane-perfused volume at seeps (4–9). Biomarkers and geochemical signatures suggestive of methane-oxidizing lineages—such as archaeal lipids, 16S rRNA genes, and isotopically light carbon compositions—have been detected in seep-associated carbonates (4, 10–13), but the potential role of these substrates in contemporary methane cycling has been largely neglected. Carbonate-hosted (endolithic) AOM activity has been suggested by shifts in microbial community composition (14) and explicitly demonstrated through methane oxidation rate measurements and stable isotope probing at a single location [Hydrate Ridge (15)]. Given the importance of methane in climate regulation, the presence of authigenic carbonates at seeps, and the limited but encouraging indications of rock-hosted methanotrophy to date, the extent of endolithic methane-oxidizing activity at methane seeps warranted additional study.
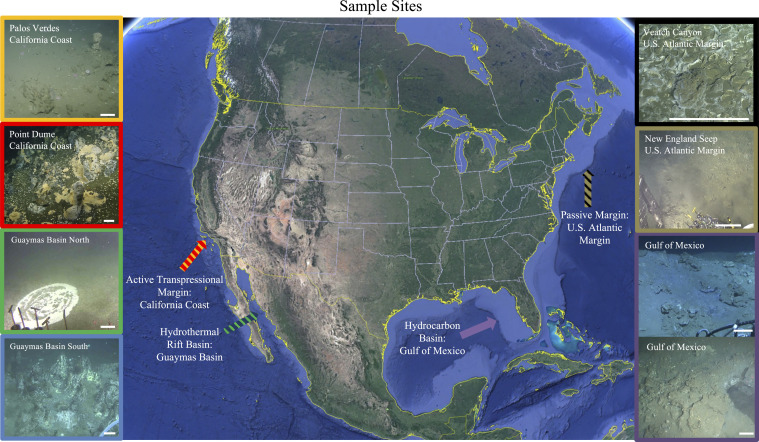
Here, we present a continental-scale survey of sediment and endolithic microbial communities and their AOM potential at a diverse set of marine methane seeps. Sampling locations included seven sites across four geological settings: 1) the northern Gulf of Mexico, where a sedimentary basin borders a carbonate platform (16); 2) two midcontinental slope submarine canyons on the US Atlantic passive margin (17); 3) two habitats in the Gulf of California’s Guaymas Basin, a heavily sedimented, organic rich hydrothermal site (18); and 4) two seep sites—including a newly discovered field of small carbonate “chimneys” near Point Dume, CA—along the Southern California coast’s active transpressional margin (19) (Fig. 1, Dataset S1, and SI Appendix, Figs. S1–S5). We quantified methane oxidation rates at methane-saturated conditions representative of many active seeps, compared our results to previously published data within a kinetic framework, and explored the likely determinants of rate differences, including microbial abundance, community composition, and mineralogical context. Our findings revealed extremely high potential rates of AOM within select carbonate structures, validating earlier research from a more limited set of samples (15) and demonstrating that endolithic methanotrophic communities are likely ubiquitous among the substantial carbonate deposits that occur within some areas of methane seepage. We posit that seep carbonates represent a habitat with attributes that can support elevated AOM rates and that carbonate-hosted AOM may play a role in greenhouse gas sequestration and the flow of methane-derived carbon into biogeochemical cycles.
Fig. 1.
An overview of the seven sites across four distinct geological settings that were sampled in this study. All scale bars are ∼25 cm; additional geographical and ground cover context is provided for each site in SI Appendix, Figs. S1–S5.
Results
Rates of AOM.
Carbonate samples from all seven sites exhibited methane oxidation when incubated in laboratory-based mesocosms (Table 1). Two approaches to short-term (<7 d) AOM rate measurements were deployed. Radiolabeled 14CH4—a commonly used, highly sensitive method (20–22)—was used to track the oxidation of methane carbon to inorganic carbon in seven triplicate sample incubations, including at least one from each geological setting. Simultaneous side-by-side experiments with CH3D, a recently developed approach that measures methane oxidation through elevated aqueous D/H ratios (23), resulted in site-specific conversion factors to link radiolabel data with D/H values (Dataset S1 and SI Appendix, Table S1). This strategy allowed us to derive apparent rates of AOM from nondestructive, higher-throughput CH3D experiments, which were ground truthed by absolute rates derived from radiolabel 14C experiments. The degree to which conversion factors apply to other samples is uncertain, but their relatively low variance measured here (1.94 ± 0.16 SD) and in the Hydrate Ridge seep sediment and carbonates (1.99 ± 0.09 SD) (23) offers confidence that apparent rates determined by CH3D experiments are reliable and robust.
Table 1.
Rates and apparent rates of AOM derived from short- and long-term incubations at atmospheric pressure and short-term incubations at high pressure (7.58 MPa)
 |
Shaded cells indicate the samples that were subjected to triplicate, side-by-side experiments with both 14CH4 and CH3D methods; the values from 14CH4 experiments are shown, and the ratios produced by these direct comparisons of methods (SI Appendix, Table S1) were used to convert the rest of the experiments (conducted with CH3D) to 14C-equivalent “apparent” rates, which are reported here and throughout this study. All data are given at 1.1 mM methane concentration values. SD, standard deviation. All rates and SDs are reported in nanomoles per cubic centimeter per day.
Most notably, carbonate-hosted communities from Point Dume (PD) chimney structures exhibited remarkably high potential rates of AOM, oxidizing up to 5,528 (±466 SD, n = 3) nmol methane ⋅ cm−3 ⋅ d−1 (as measured for the sample named PD R1). Rates from three Point Dume carbonates exceeded 1,000 nmol ⋅ cm−3 ⋅ d−1, but even the more modest values measured with samples from the other three geologic settings were substantial, ranging from ∼200 to 850 nmol ⋅ cm−3 ⋅ d−1. To directly compare our results with published rates—which have been assessed under a range of methane concentrations—all previously measured values were converted to their equivalent rate under 1.1 mM dissolved methane concentrations, assuming Michaelis–Menten kinetics and using an AOM KM of 5.65 mM (determined empirically below; see Dataset S2 and SI Appendix for conversion details). Point Dume chimney samples exhibited the highest normalized rates of methane oxidation of all previously published studies included in the survey. This maximum apparent rate from Point Dume carbonates was ∼70 times higher than the prior study of carbonate-hosted AOM from Hydrate Ridge (15), 5 times higher than rates from the most prolific marine seeps (21), and 56% higher than the highest maximum rate of AOM previously reported [from the anoxic Black Sea (24)]. Even the lowest endolithic AOM rates from several Gulf of Mexico (GoM), New England Seep, Veatch Canyon, and Guaymas Basin North (GBN) samples exceeded those of continental shelf samples and were consistent with or higher than the vast majority of previously assessed seep sediment and mud volcano samples (Tables 1 and 2).
Table 2.
Previously published AOM rates from methane seeps, mud volcanoes, and continental shelf samples
| References | Study site | AOM rates calibrated to 1.1 mM CH4 experimental conditions (nmol ⋅ cm−3 ⋅ d−1) |
| Michaelis et al. (24) | Microbial Reefs, Black Sea | 3,545 |
| Treude et al. (61) | Microbial Reefs, Black Sea | 3,001 |
| Treude et al. (21) | Seep Sediments, Hydrate Ridge | 1,065 |
| Segarra et al. (22) | Freshwater Wetlands Sediments, Coastal Georgia | 885 |
| Freshwater Wetlands Sediments, Florida Everglades | 164 | |
| Freshwater Wetlands Sediments, Maine | 248 | |
| Bowles et al. (95) | Seep Sediments, Mississippi Canyon, Gulf of Mexico | 871 |
| Seep Sediments, Guaymas Basin | 653 | |
| Seep Sediments, Monterey Bay | 51 | |
| Joye et al. (96) | Gas Hydrate Sediments, Green Canyon, Gulf of Mexico | 608 |
| Omoregie et al. (97) | Mud Volcano Sediments, Chefren, Eastern Mediterranean Sea | 494 |
| Niemann et al. (98) | Mud Volcano Sediments, Haakon Mosby, Barents Sea | 435 |
| Marlow et al. (15) | Seep Sediments, Hydrate Ridge | 193.6 |
| Seep Carbonates, Hydrate Ridge | 80.9 | |
| Nauhaus et al. (99) | Seep Sediments, Hydrate Ridge | 169 |
| Beal et al. (79) | Seep Sediments, Eel River Basin | 110 |
| Wankel et al. (100) | Brine Pool Fluids, Gulf of Mexico | 162 |
| Girguis et al. (101) | Seep Sediments, Monterey Canyon | 71.6 |
This analysis includes all known “high AOM” results reporting rates >1,000 nmol ⋅ cm−3 ⋅ d−1 under initial experimental conditions. A more complete depiction of study conditions and the rate conversion process is provided in Dataset S2 and SI Appendix.
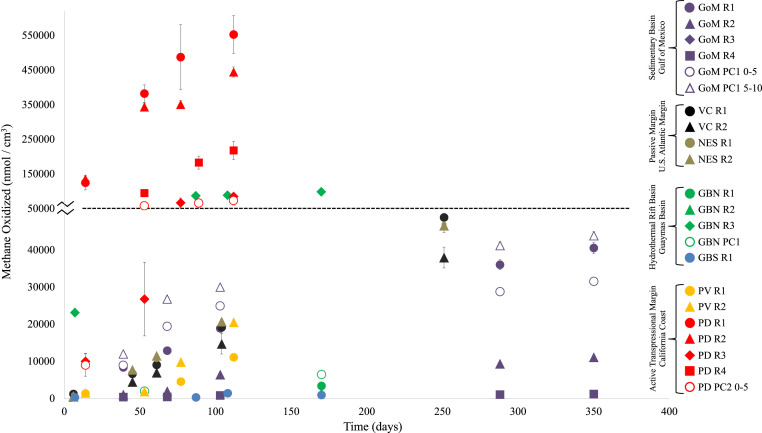
To more faithfully represent in situ conditions of seafloor methane seeps, we conducted two additional rate measurement experiments: long-term incubations at atmospheric pressure to determine how metabolic rates adjust over time with the accumulation of metabolic byproducts and short-term incubations at 7.58 MPa to evaluate the dependence of AOM on heightened pressures representative of 745 m water depth (the depth of the Point Dume seep). Over the course of 112 to 350 d at atmospheric pressure, apparent rates increased by an average of 17% over the course of the experiment, with no significant trends based on substrate (sediment versus rock) or sample site (Table 1 and Fig. 2). At elevated pressure, rocks from all four geological settings demonstrated substantially enhanced apparent rates, increasing between 13.8 (sample GBN R3) and 75.3 (sample GoM R1) times compared with the short-term experiments conducted at atmospheric pressure. Of the three sediment samples, two demonstrated pressure-induced rate increases of similar magnitudes as the carbonates (PD PC2: 26×, GBN PC1: 106.6×) though GoM PC1 0 to 5 cm was methanotrophically inactive. Point Dume chimney PD R4 exhibited an apparent AOM rate of 2.8 × 104 nmol ⋅ cm−3 ⋅ d−1 (±0.1 × 104 SD, n = 3), the highest rate of methane oxidation among the compiled studies (Tables 1 and 2).
Fig. 2.
Quantity of methane oxidized during extended laboratory-based incubation experiments. Please note the 10× shift in the y-axis scale marked by the scale break and dotted line at 50,000 nmol/cm3. Below this line, all data below 50,000 nmol/cm3 are shown with axis increments of 10,000 nmol/cm3; above this line, all data above 50,000 nmol/cm3 are shown with axis increments of 100,000 nmol/cm3. Error bars represent propagated errors from both the D/H: 14C scaling factor and, where sufficient material was available for triplicate experiments (Dataset S1), incubation SDs. Values from site-specific autoclaved control incubations were subtracted from each data point.
Kinetic, Chemical, and Physical Attributes of Seep Carbonates.
To better characterize the factors that may facilitate the remarkably high AOM rates we measured within carbonates, several key kinetic, mineralogical, and physical traits of carbonate-hosted environments were examined. Reaction kinetics reflect rate-determining ecophysiological parameters such as substrate affinity, processivity, and the relative number of reaction centers. We measured Vmax and Km values of colocated Point Dume sediment (PD PC2 0 to 5, AOM rate of 440.73 nmol ⋅ cm−3 ⋅ d−1; ±73.08 SD, n = 3) and carbonate (PD R3, 1,561.66 nmol ⋅ cm−3 ⋅ d−1; ±130.38 SD, n = 3) by quantifying apparent AOM rates with the CH3D approach at methane concentrations from 1.1 to 129 mM to populate a Michaelis–Menten curve. Sediment-based rates attained a Vmax of ∼9,500 nmol ⋅ cm−3 ⋅ d−1 with a calculated Km of 5.65 mM (SI Appendix, Fig. S6). Rock-based rates did not reach a Vmax across the range of methane concentrations tested, suggesting that carbonate-associated methane-oxidizing processes at Point Dume do not reach saturation under in situ conditions. The increasing ratio of rock- to sediment-hosted AOM with increasing methane concentrations suggests that, all other environmental factors being equal, the relative contribution of endolithic AOM (per unit volume) likely increases at methane-replete seeps in deeper waters such as the Gulf of Mexico, the US Atlantic Margin, the Guaymas Basin seeps, and others found at similar and deeper depths.
A cell’s mineralogical environment can markedly influence metabolism, from providing nutrients (25) to donating (26) and accepting (27) electrons in catabolic reactions. In this context, X-ray diffraction (XRD) analysis was performed on both carbonates and sediments to evaluate potential links between AOM rates and bulk mineralogy (SI Appendix, Table S2). Carbonate rocks from the Gulf of Mexico contained major fractions of magnesian calcite, which is consistent with subsurface formation in sulfate-depleted fluids and subsequent exposure at the seafloor (28). Multiple forms of carbonate were apparent within individual Point Dume chimneys, with aragonite or magnesian calcite predominating in different samples. The bulk mineralogy does not appear to be a first-order determinant of methane-oxidizing rates; however, the presence of specific minerals could influence metabolic relationships. Specifically, of the samples analyzed via XRD, the four that exhibited the highest rates of methane oxidation (PD R1 to 4) all contained measurable levels of pyrite. The interior fraction (>2 mm beneath the outer crust) of the highest-rate sample, PD R1, consisted of >80% pyrite. Electron microscopy further revealed abundant framboidal pyrites throughout the chimney as well as acicular crystals suggestive of aragonite fans. In some instances, pyrite framboids were also observed in direct contact with aggregations that bear a striking resemblance to consortia of anaerobic methanotrophic (ANME) Archaea and sulfate-reducing bacteria (SRB) (SI Appendix, Fig. S7).
Given the importance of extracellular electron transfer in AOM metabolism (29, 30), we wondered if the presence of iron–sulfur mineral phases could facilitate an “electron marketplace” that enables the transfer of reducing equivalents among syntrophic partners, thereby supporting elevated AOM rates. Electrical and physical properties of multiple sediment and rock samples were tested to evaluate the role of conductance and structural coherence on methane oxidation activity. Conductance values obtained from current–voltage measurements performed on the sediment and rock samples ranged from 7.32 × 10−10 Ω−1 (GoM R1) to 3 × 10−9 Ω−1 (GoM PC1 0 to 5) (SI Appendix, Table S3). Moreover, currents up to hundreds of pA were detected for modest applied voltage sweeps from −100 to 100 mV. Though no consistent relationship between conductance and AOM was observed, a comparison with previously described per-cell interfacial electron transfer rates (15 to 100 fA) (31) suggests that electron flow through both sediment and rock substrates—in addition to direct cell–cell contact—may be an important link between syntrophic partners, a phenomenon that has been observed with a number of iron-containing minerals (32). Next, we sought to establish if particle size influenced AOM rates, potentially through long-range electron transfer. Sample PD R3 was broken into four size fractions ranging from ∼400 μm to 2 cm across, and apparent rates were measured over the course of 8 d. No significant relationship between particle size and AOM rate was observed (SI Appendix, Table S4), implying that structural coherence over the spatial scale tested here is not an important rate-determining factor.
Biological Attributes of Seep Carbonates.
Having surveyed a range of abiotic factors, we next examined the abundance, composition, and arrangement of the samples’ microbial communities to determine these factors’ effects on AOM rates. Cell counts of nonincubated samples and post–long-term incubation samples revealed positive correlations between methane oxidation rates and 1) preexisting cell abundance (accounting for ∼33% of rate variance in a pairwise plot), 2) postincubation cell abundance (∼60% of rate variance), and 3) the change in cell abundance (∼61%; SI Appendix, Table S5; all values assume equivalent levels of per-cell methane oxidation).
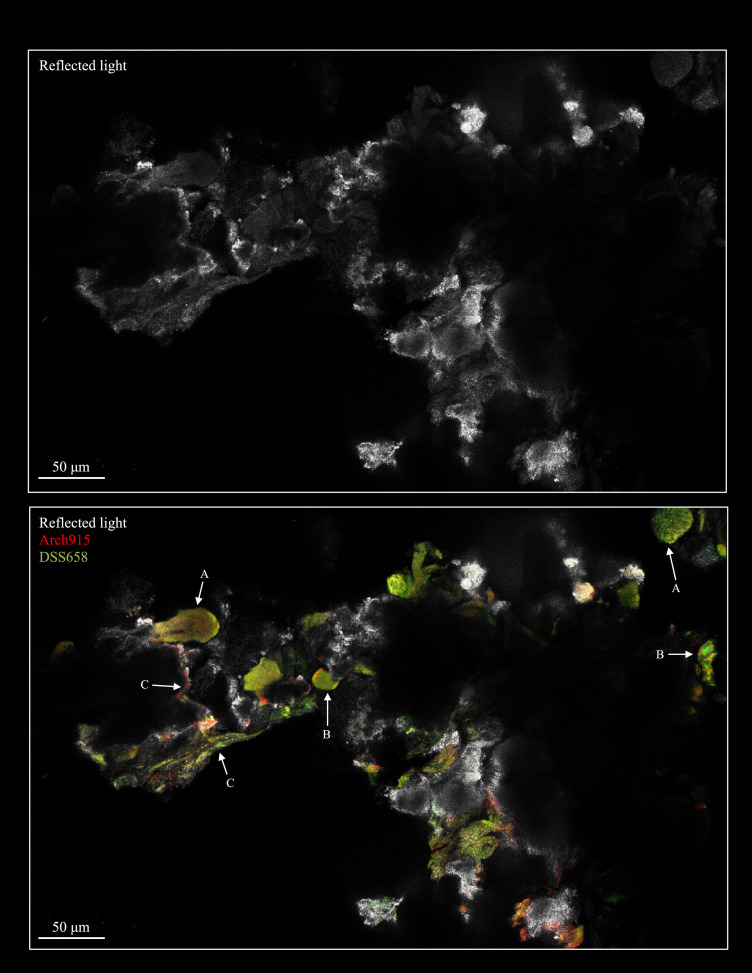
To examine the spatial relationships among microorganisms within endolithic settings, we developed a direct fluorescence in situ hybridization (FISH) imaging technique for rock fragments. This “in-place” microscopic analysis of sample PD R1 exposed dense assemblages of cells in a variety of contexts, including mixed Archaea–SRB aggregates, shell-type aggregates, and more homogenous communities coating the rock surface within pore spaces (Fig. 3 and SI Appendix, Fig. S8). The high density of surface-associated organisms—as well as the apparent diversity of cell–cell and cell–mineral spatial associations—is consistent with biotic and abiotic feedbacks that could support heightened metabolic activity. Such an intricate physical scaffold of endolithic habitats across dozens or hundreds of micrometers may retain localized redox gradients and promote electron transfer reactions that are absent in sediments. Solidified rock structures may also facilitate robust fluid flow through established conduits, providing microbial constituents with methane through upward advection and sulfate through tidal mixing (33) or hydrologic recharge (4, 34) at the base of carbonate mounds. (Notably, we observed advective fluid flow through the Point Dume chimneys, and ongoing geochemical analyses are aimed at further establishing the composition of these fluids.) Sediments, in contrast, may have more distributed channel networks with greater tortuosity and lower fluid flow rates and could be susceptible to disruption by bioturbation (35, 36). That said, endolithic communities that accelerate carbonate and metal sulfide precipitation can seemingly self-entomb (37), and the long-term implications of such processes warrant further study.
Fig. 3.
Confocal microscopy images of an intact interior portion of sample PD R1. (Top) Rock surface. (Bottom) Fluorescence signal from hybridized Arch 915 and DSS 658 FISH probes. A range of cell–cell and cell–rock arrangements were observed, including A) mixed archaeal–SRB aggregates, B) “shell-type” aggregates in which archaeal members occupy exterior spaces and SRB members comprise the interior, and C) domain-specific communities lining rock pore spaces.
To evaluate the extent to which microbial community composition explains the observed variance in AOM rates, 16S ribosomal RNA (rRNA) gene surveys were performed on 17 incubation samples at the end point of the long-term metabolic rate experiments. Although the DNA extraction procedure used herein does not distinguish between the intracellular DNA of intact cells and the extracellular DNA derived from dead cells (38), comparative studies of marine sediments report that the persistence of extracellular DNA has a negligible impact on community composition surveys (39, 40), particularly in the absence of bioturbation (as was the case in our anoxic incubations) (41). Exact sequence variants were resolved in order to observe single-nucleotide differences and improve phylogenetic comparison (42, 43). After an assessment of available computational tools, the Deblur protocol (44) produced amplicon variants most similar to sequences in the National Center for Biotechnology Information (NCBI) database (SI Appendix, Fig. S9); this approach has been shown to result in both lower overall diversity and, based on mock community controls, a more realistic representation of community identity (44). No significant correlation between methane oxidation rate and alpha diversity was apparent (SI Appendix, Fig. S10). The carbonate sample from Palos Verdes (PV R1) sample displayed the highest degree of diversity across all metrics, while the Point Dume rocks, which hosted communities exhibiting higher AOM rates in our incubations, were substantially less diverse. A similar streamlining of community diversity was observed in carbonate rocks from Hydrate Ridge (14) and Gulf of Mexico seep sediment abstracted onto artificial carbon cloth substrates that demonstrated elevated volume-specific rates (45). Nonmetric multidimensional scaling analysis examined the relationships among community compositions using distance matrices that accommodate sparse data sets (abundance-weighted Jaccard) with relative abundances (Bray–Curtis) and consider phylogenetic distances between detected organisms (weighted UniFrac) (SI Appendix, Fig. S11). Site location and mineralogy were both significant factors in sorting community relationships (permutational multivariate ANOVA P value: 0.001 to 0.036; SI Appendix, Table S6), and multiple beta diversity metrics revealed a significant link between community structure and methane oxidation rates (SI Appendix, Tables S6 and S7).
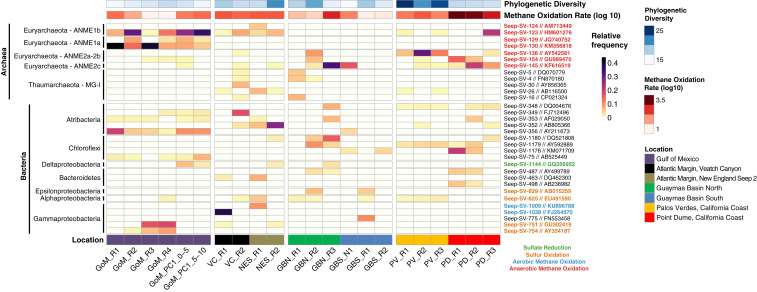
We next focused on specific high relative abundance lineages. Fig. 4 compiles alpha diversity, methane oxidation rate, and location data alongside the relative abundance of the 32 exact sequence variants that constitute at least 5% of reads in at least one sample (see Dataset S3 for all relative abundances). Among ANME, ANME-2 were most abundant in Guaymas and California Coast samples, including the high-rate PD R1 and PD R2 samples, while ANME-1 were more prevalent in lower-rate samples, particularly at Gulf of Mexico sites. Sequences from some lineages not typically associated with methane or sulfur metabolism were also detected in relatively high abundances. Five representatives of the Atribacteria candidate phylum were among the 32 high-abundance taxa, and members of the Anaerolinaceae family (in the Chloroflexi phylum) comprised >10% of communities recovered from the three highest-rate samples subjected to community analysis (PD R1, PD R2, and GBN R3).
Fig. 4.
Relative abundance heat map of the thirty-two 16S rRNA gene exact sequence variants (ESVs) that account for at least 5% of the relative abundance in at least one sample. Each row represents a distinct ESV, while columns correspond to the 22 rock or sediment samples (all are labeled at the bottom of the figure). Taxonomic groupings of the 32 ESVs and their closest hits via Basic Local Alignment Search Tool (or BLAST) are indicated on the left, and ESV labels are to the right, colored by putative metabolic category. Bars above the heat map indicate Faith’s phylogenetic diversity index (a measure of alpha diversity) and the AOM rate (on a log10 scale).
Discussion
Which Factors Determine Methane Oxidation Rates?
Our detection of pervasive endolithic AOM potential reveals carbonate rock-hosted methane consumption as an understudied component of the methane cycle. This phenomenon was observed at all of the sites we sampled—across four geological settings and seven geochemically distinct locations—and may be ubiquitous at methane seeps across the ocean that exhibit authigenic carbonate deposits. Analysis of kinetic parameters, mineralogical constituents, electrical properties, cell abundance, microbial diversity, and the presence of particular lineages pointed to multiple factors that, taken together, help explain the range of measured rates and account for the remarkable activity observed during laboratory-based incubations of Point Dume carbonates. Cell counts indicated that changes in biomass and final cell abundances showed a stronger positive correlation with eventual long-term rates than preexisting biomass. These data suggest that environmental conditions such as in situ methane concentrations played a role in priming samples for their AOM activity, but that inherent properties of the sample—for example, mineralogy or microbial community structure—were more influential.
The striking link between pyrite and higher rates is one particularly compelling avenue for further research. Framboidal pyrite, whose formation can be accelerated by sulfate reduction in the presence of reactive iron minerals (46), was also prominent in the Black Sea reefs (47), which may be the closest known analog to the poorly consolidated chimney-like structures at Point Dume (24). To further investigate pyrite prevalence in carbonates exhibiting high AOM rates, we calculated the relative abundance of putative sulfate-reducing (48–53) and sulfide-oxidizing lineages (54–58) in all samples (Dataset S4). We hypothesized that more sulfate reducers and/or fewer sulfide oxidizers could allow sulfide to accumulate and ultimately precipitate as pyrite, but no significant relationship between target lineages’ relative abundance and pyrite abundance was observed. The electrical currents across all samples—up to hundreds of picoamps compared with previous cell interface measurements in the femtoamps range (31)—suggest that the electrical properties of physical substrates may play a role in methanotrophic activity. Indeed, a similar phenomenon was observed in previous work, when seep sediment communities were transferred onto conductive carbon cloth and exhibited increased AOM rates (45).
The presence of specific lineages directly and indirectly involved with AOM may also play an important role enhancing methane oxidation rates. ANME-2, which were more prevalent in the high-rate carbonates, possess large multiheme cytochromes and surface layer proteins that may enable them to offload reducing power through direct electron transfer between consortia partners (29). ANME-1, on the other hand, were found at higher relative abundance in lower-rate samples, and these particular genotypes may lack the physiological capacity for high-throughput extracellular electron transfer (59, 60). In Black Sea carbonate reefs, zones dominated by ANME-2 exhibited lower δ13C values than those dominated by ANME-1, suggesting accelerated rates of methane oxidation and assimilation by ANME-2 (61). Atribacteria and Anaerolinaceae are not believed to be involved in the core methane-oxidizing or sulfate-reducing metabolisms, but their prevalence suggests that they may play important supporting roles. Atribacteria have been preferentially detected at marine methane seeps (62, 63) and may enter seep habitats from below through upward-migrating fluids as was demonstrated at the Gulf of Mexico seeps (64) and mud volcanoes of the Ryukyu Trench (63). Genomic analysis suggests that they perform a fermentative metabolism capable of producing acetate, CO2, and H2—potential substrates for methanogens and/or SRB (65, 66). Anaerolinaceae are commonly characterized as heterotrophs; in seep environments they have been posited to consume primary producer ANME biomass (67), but they may be more directly involved with hydrocarbon metabolism as in alkane-degrading enrichment cultures (68, 69) and Gulf of Mexico seeps (70).
Other lineage-specific attributes that could accelerate rates and account for the higher Vmax observed in Point Dume carbonate include a) increased rates of reactant uptake and product release by either syntrophic partner, b) distinct proteoforms of the enzymes performing rate-limiting reactions (methyl-coenzyme M reductase, dissimilatory sulfite reductase), c) a higher number of rate-limiting enzymes per cell, and/or d) distinct lineage-specific partnerships. Clarifying the roles of these factors will require more detailed biochemical, genomic, and proteomic studies which would likely contribute to the growing appreciation of methyl-coenzyme M reductase diversity (71–73).
Implications of Carbonate-Hosted AOM.
Existing estimates of the global extent of methane oxidation in marine settings (1, 2, 74) do not distinguish between sediment- and carbonate-hosted methanotrophy, despite the abundance and often dominance of carbonate rock at methane seeps (4–7, 75, 76). There are several important factors to consider when evaluating the influence of endolithic AOM on the global methane cycle, including the localized concentrations of metabolic reactants and products found within and around carbonates as well as the overall volume of carbonate at seeps. Some of these key parameters are not currently well constrained. Moreover, our experimental incubations were conducted under high-methane, sulfate-replete conditions that are not representative of all seep environments. For this reason, the absolute rates derived from our incubations reflect a sample’s methanotrophic potential that likely exceeds its in situ methane-oxidizing contribution. Nonetheless, by having maintained standardized experimental parameters for carbonates and sediments in our incubations, and by converting previously published results to equivalent conditions, our substrate-based and cross-study comparisons of potential AOM rates suggest that carbonate-hosted AOM could be an important methane sink in marine settings.
Carbonate rock represents a substantial proportion of the seafloor and a majority of the methane-perfused volume at many seeps (4–6, 8, 9, 77). While diffusion limitation may modulate the maximum rates of reaction in some carbonate edifices, many authigenic carbonates at seeps exhibit high porosities (4, 28, 78) and permeability values on par with those of nearby sediments (15, 33). Accordingly, reactant transport in these two environmental regimes may be relatively similar and may even be less constrained in some carbonates. Not all seeps exhibit such pervasive carbonate structures, as cation availability could limit carbonate precipitation, and only a subset of the rock volume would be expected to experience seawater concentrations of sulfate. However, nitrogen- and metal-reducing (79, 80) and low-sulfate AOM (81) as well as sulfate penetration of carbonate mounds caused by tidal forcing (33) or hydrologic recharge (4, 34) could expand the endolithic AOM habitat deep into subsurface rock habitats.
A final consideration when assessing the potential scope of endolithic AOM is the average rate enhancement in rocks compared with sediments. The rock-hosted to sediment-hosted scaling factor was derived from the short-term rates compiled in this study (Table 1). At active sites where both endolithic and sediment-based AOM was observed, substrate-specific rate values were averaged and a rock-hosted to sediment-hosted AOM ratio was determined. These ratios were 0.88 for Gulf of Mexico samples, 8.85 for Guaymas Basin North samples, and 3.47 for Point Dume samples, indicating a wide variance but, on average, higher rates associated with carbonates than sediment per unit volume under equivalent experimental conditions.
The data presented here demonstrate that carbonate samples from all seep sites examined to date, across a range of geologic settings, possess substantial anaerobic methanotrophic potential. Furthermore, some endolithic communities assessed in laboratory-based incubations exhibited the highest rates of AOM ever reported, implicating the physicochemical aspects of carbonate-hosted habitats in the promotion of enhanced methane consumption. While the full extent to which endolithic AOM influences global methane cycling remains to be established, our results demonstrate that endolithic environments may be important contributors to methane biogeochemical assessments. The consideration of methanotrophic communities in a carbonate-hosted context has implications for both mitigation of potential methane hydrate dissociation (82) and ocean acidification (83, 84). A more detailed understanding of the structural and mineralogical attributes that enable such elevated carbonate-hosted AOM rates could also help with the rational design of methane-scrubbing systems for greenhouse gas remediation or distributed biofuel production (85).
Conclusions
It is widely accepted that ANME microbial communities play a substantial role in methane consumption in marine environments, but the contribution of rock-hosted habitats is largely absent from laboratory analyses and global estimates. Following a continental-scale assessment across a range of geological settings, we show that endolithic AOM is a common phenomenon and that rates of methane consumption by microbial communities within rocks often exceeds those associated with sediments. At the newly characterized Point Dume seep off the coast of Southern California, we report chimney-like carbonate structures that oxidized methane in laboratory-based incubations at the highest rates measured to date. We identified several factors, including cell abundance, mineral composition, kinetic parameters, and the presence of specific microbial lineages, that likely play key roles in supporting elevated endolithic AOM rates. We anticipate that future studies will better characterize how these and other aspects of the system facilitate the observed rates and will clarify the contribution of carbonate-hosted AOM to marine methane budgets. Given their frequent occurrence and elevated methane-oxidizing potential, carbonate rocks at methane seeps may constitute a major marine methane sink.
Materials and Methods
Sample Recovery and Processing.
Samples were collected from the Gulf of Mexico during R/V (Research Vessel) Atlantis leg 26–12-Science Verification Cruise, the US Atlantic Margin during Atlantis leg 36, the Guaymas Basin during Atlantis leg 37–06, and the California Coast during E/V (Exploration Vessel) Nautilus cruises 073 and 084 and R/V Falkor leg 163 019. Further information on geological settings and sample collection details can be found in SI Appendix. Upon recovery from the seafloor, push cores and carbonates submerged in bottom water were transferred immediately to a 4 °C cold room and processed within several hours. Push cores were sectioned into predetermined horizons and stored at 4 °C in sterile Whirl-Pak bags within anoxic, Ar-flushed, gas-tight Mylar bags (IMPAK Corporation) until use. Carbonate rocks were rinsed to remove any adherent sediment, defaunated, and broken into smaller pieces, if needed, with ethanol-cleaned hammer and chisel. Rock pieces were placed in Mylar bags and submerged with colocalized bottom water; the bag was then sparged with Ar gas (5 min per 100 mL water volume), sealed, and stored at 4 °C.
In the laboratory, sediment and carbonate samples were transferred to new sterile bottles and Mylar bags, respectively, and submerged with bottom water from the collection site that had been pushed through a 0.22-μm Durapore filter (EMD Millipore), sparged with N2 gas, and kept overnight in an anoxic chamber. Samples were stored in the dark at 4 °C. Samples were sparged with CH4 (5 min per 100 mL water volume) 2 wk prior to rate experiment setup; this “preincubation” was performed to minimize priming effects and lag time during metabolic rate measurement experiments. These and all subsequent sample-handling steps were performed in a vinyl anoxic chamber (Coy Laboratory Products).
Metabolic Rate Measurements.
14CH4 radiotracer method.
Short-term incubations with a 14CH4 radiotracer were conducted as described in detail in ref. 21. Headspace-free incubations were set up in sterilized SVG-50 glass vials (Nichiden-Rika Glass Co. Ltd) with 10 mL sediment or carbonate and 50 mL 0.22 μm filtered anoxic seawater presparged with CH4. Gas chromatograph validation using a CH4:N2 dilution series and concentrations derived with Henry’s law (incorporating corrections for temperature and salinity) revealed an initial methane concentration of 1.35 mM. Radiolabeled methane was injected into each vial (14CH4 dissolved in seawater, with a specific activity 1.89 GBq/mmol and an activity of 12 kBq per incubation), and vials were kept in the dark at 4 °C for the duration of the incubation period. Material was then quickly transferred to a sterile 250-mL stoppered bottle prefilled with 7.5 mL 30% (weight/weight) NaOH to stop microbial activity. The flask headspace was purged with air through an 850 °C quartz tube filled with copper oxide at a rate of 30 mL/min for 30 min, combusting the unreacted 14CH4 to 14CO2. The combusted gas was routed through two sequential scintillation vials prefilled with 1 mL phenylethylamine and 7 mL 2-methyoxyethanol to which 10 mL scintillation mixture (Ultima Gold, PerkinElmer) was added. To measure 14C-labeled inorganic carbon made during the incubation period, a prefilled scintillation vial (1 mL 2.5% NaOH and 1 mL phenylethylamine) was suspended from the stopper inside the bottle, one drop antifoam and 5 mL 6 M HCl were added to the sealed container, and the bottle was left on a shaking table for 16 h at room temperature. A total of 5 mL scintillation mixture was then added. This approach has been shown to recover an average of 98% of the 14CO2 (21). Scintillation vials were measured, after a 24-h waiting period, for 10 min each with the Beckman Coulter LS 6500 scintillation counter. The rate of methane oxidation was determined by the following equation:
14CH4 represents the combusted unreacted radiolabeled methane, 14CO2 indicates the quantity of acidified oxidation product, CH4 is the initial quantity of methane in the experiment, v is the volume of sediment or carbonate rock, and t is the incubation time. Scintillation vial measurements and calculations were performed in a double-blind manner.
CH3D method.
This approach for methanotrophic rate measurements confers key scientific and procedural advantages: 1) it is nondestructive, allowing for true replication and time series measurements to be conducted on heterogeneous samples such as seep sediments and carbonates, and 2) it is safer and less logistically onerous than radioisotope work, requiring substantially less hands-on and overall time per measurement. Short-term CH3D rate experiments and carbonate fragment size tests were set up as above but with two differences: the methane sparging was done with 75% CH4 and 25% CH3D, and no 14CH4 tracer was added. For the fragment size incubations, rock was homogenized using an autoclave-sterilized mortar and pestle, and the resulting sizes of 20 fragments were determined along the longest axis by digital calipers for samples A (2.0 to 2.5 cm) and B (0.5 to 0.7 cm) and by measurement under a light microscope for samples C (∼2,000 μm) and D (∼400 μm). A negative control of autoclaved 2.0 to 2.5 cm fragments was also included. At the end of all short-term CH3D incubations, water was sampled and analyzed as detailed previously (23) unless noted otherwise below. Briefly, 1 mL incubation fluid was collected in an anoxic chamber with a sterile syringe through the stopper and pushed through a 0.22-μm filter into a 250-μL glass insert within a 1-mL gas chromatography vial. The aqueous D/H ratio of each sample was measured using a T-LWIA-45-EP liquid water isotope analyzer (Los Gatos Research), which uses off-axis integrated-cavity output spectroscopy to measure high-precision D/H and 18O/16O ratios (86). Four rounds of 10 injections per sample were performed, but only data from the last five injections were used in the analysis to avoid memory effects. Five working standards of known isotopic ratios were included: 2C (−123.7‰), 3C (−97.3‰), 4C (−51.6‰), ER2 (191.4‰), and ER3 (383.3‰); standards were remeasured every 40 to 60 injections to minimize the effects of instrumental drift. In the event of instrumental temperature or pressure flags (2.1% of all measurements, corresponding to a measurement cell temperature change of more than 0.3 °C per hour or rising pressure during the analysis), data were excluded, and D/H ratios were determined from the remaining measurements. D/H ratios were normalized to the Vienna Standard Mean Ocean Water scale using the water standards and linear interpolation/extrapolation. Using this normalized ratio, the number of activated methane molecules (processed through the first step of methane oxidation to enable D–H exchange with water but not necessarily fully oxidized to CO2) was determined. Using the 14C-based rates from the side-by-side experiments, scaling factors linking the two methods were calculated and applied to the remaining CH3D values, termed “apparent” AOM rates (SI Appendix, Table S1). Due to sample limitation, sample-specific scaling factors could not be calculated. Errors associated with the D/H: 14C scaling factors (SI Appendix, Table S1) and the CH3D rate values were propagated when calculating each CH3D-derived “apparent” rate value by adding the uncertainties in quadrature and applying the resulting error (“apparent rate error” below) to the calculated “apparent rates” of AOM:
Long-term rate experiments were prepared in 0.35-L wide-mounted, stoppered bottles (VWR International). In an anoxic chamber, 10 mL sediment or rock was added, and the bottle was filled with colocalized 0.22-μm filtered, anoxic bottom water. The residual methane was removed by N2 sparging, and gas headspace (160 mL CH3D and 80 mL N2) was added, expelling an equivalent volume of liquid through a second needle and generating a headspace of known partial pressures. The dissolved methane concentration at the start of these experiments was calculated by Henry’s law and confirmed by gas chromatography measurement to be 1.1 mM (SI Appendix). The containers were then placed upside down in the dark at 4 °C until sampling for water isotopic analysis as described above.
Custom-built 4-L titanium pressure vessels were used to 1) determine apparent rates of methane oxidation under methane-saturated conditions for several samples and 2) determine rate versus concentration curves for Point Dume sediment (PD PC2 0 to 5) and carbonate (PD R3) samples in order to evaluate reaction kinetics. In the anoxic chamber, 10 mL sediment or rock and 100 mL colocalized filtered bottom seawater were placed in Mylar bags and sealed. Outside the chamber, a small hole was made in the bag through which N2 gas was flowed at 50 mL/min for 5 min. All headspace was then removed by squeezing the bags until a drop of water emerged; 100 mL (at 1 atm pressure) of the appropriate gas mixture (SI Appendix, Table S8) was then injected using a prefilled syringe, and the bag was quickly sealed and placed in the pressure vessel in a temperature-controlled environmental room (5 °C). The 7.58 MPa target pressure (simulating ∼745 m water depth) was attained using a high-performance liquid chromatography pump (P-1536, Chrom Tech, Inc.) and regulated using a stainless-steel backpressure valve (StraVal Valve). Experiments at pressure proceeded for 8 h. At the designated end point, Mylar bags were extracted and moved to the anoxic chamber; water was collected for D/H analysis via sterile syringe through the bag and measured as described above.
For all 14CH4 and CH3D rate measurements, equipment and relevant supplies were sterilized by autoclave prior to use, and values of corresponding autoclave-sterilized control samples were subtracted. Unless otherwise noted (Dataset S1), all rate-based experiments were conducted in triplicate.
Mineralogical Analysis.
Material for XRD was subsampled from rate measurement incubations at the end of the long-term incubation experiment. Interior portions of rocks (defined as at least 3 mm from both exterior and interior surfaces) were recovered via sterile razor blade. Samples were air dried, homogenized with a sterile mortar and pestle, and run as packed powders. All samples were run on a Panalytical X’Pert3 Powder XRD using a Cu Kα source at 40 kV and 45 mA and scanned from 5 to 70° 2θ. Final scans were an average of three scans—one each with a 0, −1, and +1° wobble to confirm a lack of preferential sample orientation. HighScore Plus was used to fit and identify peaks (using a Rietveld refinement approach), and the International Center for Diffraction Data 2014 was used for sample identification. The relative quantification of phases is given in weight percent. Mineralogy as determined from XRD was constrained using elemental results obtained from scanning electron microscopy (SEM)/energy-dispersive X-ray spectroscopy performed using a Tescan VP-SEM in high vacuum mode with a backscatter detector at 20 kV.
SEM.
A preincubation subsample of rock PD R1 was dehydrated serially with 50, 80, and 100% ethanol solutions (balance deionized [DI] H2O). Interior fragments were obtained with a sterile chisel and mounted on an aluminum stub using double-sided carbon tape. Samples were then sputter coated with 10 nm Pt:Pd (80:20) (Q300T D sputtering system, Quorum Technologies) and imaged with a Zeiss Supra 55VP field emission electron microscope operating at the Harvard Center for Nanoscale Systems. Secondary electron images were obtained at a voltage of 15 kV using an Everhart–Thornley secondary electron detector. Elemental analysis was performed at 15 kV using a silicon drift detector (EDAX), and data were processed using the software Genesis.
Conductance Measurements.
Two-probe current–voltage (IV) measurements were performed on a Signatone 1160 probe station using an Agilent 4156C Precision Semiconductor Parameter Analyzer interfaced to Signatone SE-T tungsten probe tips (5-μm diameter) that were mounted on Signatone S-926 micropositioners for fine positioning. For each IV measurement, the probes were placed ∼40 μm apart on the sample. During placement and all measurements, the probe positions were monitored with an SMZ-168 stereo microscope. For IV measurements, the current response was measured during a −100 to 100 mV (and back) sweep in 20 mV steps. Each voltage step was held for 65 s before the current was measured in order to eliminate the contribution of capacitive current. The following Agilent 4156C settings were chosen to improve current sensitivity: 1 nA compliance, auto-ranging, and a long integration time (16 power line cycles). The conductance for each sample can be taken, using Ohm’s Law, as the slope of the linear regression line through the sample’s IV data points, where conductance is the inverse of the resistance.
Microscopy.
Cell counts were performed as described previously (87) with minor modifications. Representative rock and sediment samples were fixed in 2% paraformaldehyde for 1 h at room temperature and then rinsed with ddH2O and dried before homogenization with a sterile mortar and pestle. Sonication, Percoll density gradient separation, filtration, and DAPI staining were described previously (87). The cumulative volume of cell aggregates per unit volume sample was determined by counting 30 contiguous fields of view under an LSM 880 upright confocal microscope using a 60× objective lens (Zeiss). This volume was divided by a typical cell (1-μm diameter) volume and multiplied by the maximum possible spherical packing density [0.7405 (88)]. (The cell abundance within aggregates is therefore a maximum potential value; no clear differences in aggregate packing density were observed, and consistent workflow of analysis and calculation ensures that relative relationships are reliable.) Finally, the abundance of single cells observed on the filter was added.
To prepare for in-place microscopic imaging of intact microbial communities, FISH was performed directly on small pieces of rock PD R1’s interior, using buffer recipes described previously (3). The probes Arch-915 [5′-GTGCTCCCCCGCCAATTCCT-3′ (89), 5′ conjugated cyanine 3 dye] and DSS-658 [5′-TCCACTTCCCTCTCCCAT-3′ (90), 5′ conjugated 6-FAM dye] were used at final concentrations of 5 ng/μL (Biomers). A 35% formamide concentration was used in both hybridization and wash buffers. The hybridization incubation was 12 h at 46 °C, and the wash incubation was in prewarmed wash buffer for 15 min at 48 °C. Samples were then rinsed with DI water and allowed to air dry. Microscopy analysis occurred via water immersion microscopy with a LSM 880 upright confocal microscope using a 20× objective lens (Zeiss). Lasers at wavelengths 488 and 561 nm were used, and a reflectance channel recorded 80% of the 488 nm laser reflection to show rock surface features. Control experiments subjected PD R1 rock fragments to the full FISH protocol but used either the nonsense 338 probe (91) or no probes; in both cases, negligible signal was observed (SI Appendix, Fig. S8).
Microbial Community Analysis.
High-throughput sequencing of the V4 and V5 regions of the 16S rRNA gene was performed to compare the diversity and composition of the archaeal and bacterial communities from 17 sediment and rock samples (listed in Dataset S1). DNA was extracted from 0.5 g (wet weight) sample material using the PowerSoil DNA Isolation Kit (MoBio), and target regions were amplified using oligonucleotide primers 515yF (5′-GTGYCAGCMGCCGCGGTAA-3′) and 806bR (5′-GGACTACNVGGGTWTCTAAT-3′), which were modified to include the Illumina flowcell adapter sequences (92) and necessary barcodes. Merged, quality-trimmed reads were checked for chimeras using USEARCH v7.0.1090 (93) and clustered at 97% identity with UCLUST [v1.2.22q (93)] in the Quantitative Insights Into Microbial Ecology platform, QIIMEp v1.9.1 (94)]. The most abundant sequences in each cluster were selected as the representative sequences, which were then assigned taxonomy in QIIME using assign_taxonomy.py and the chimera-screened database called SILVA v128 SSURef. Details on single-nucleotide resolution approaches, diversity analyses, and phylogenetic assignments are provided in SI Appendix. Sample metadata and the SSU rRNA sequence files used in this study were submitted to the NCBI BioSample and Sequence Read Archive databases and are accessible via BioProject identifier PRJNA648152.
Supplementary Material
Acknowledgments
We thank Jenny Delaney, Dr. Aude Picard, Stephanie Connon, Stephanie Hillsgrove, Dr. Douglas Richardson, and Dr. Nicole Raineault for experimental and logistical assistance in the collection of samples and data presented here. We also thank the vehicle and vessel captains, crew, and science party members aboard the R/V Atlantis during legs AT 26-12-SVC (with the deep submergence vehicle Alvin), AT 36 (with the deep submergence vehicle Alvin and the autonomous underwater vehicle Sentry), and AT 37-06 (with DSV Alvin) and the E/V Nautilus during legs NA-073 and NA-084 (with the remotely operated vehicle Hercules). We thank the Schmidt Ocean Institute for supporting expedition FK181005, the crew of the R/V Falkor, and the pilots of the ROV SUBastian. We thank the Ocean Exploration Trust, who supported the initial expeditions to the Point Dume study site. Some analyses presented here were conducted at the Harvard University Center for Nanoscale Systems, a member of the National Nanotechnology Coordinated Infrastructure Network, which is supported by the NSF under NSF Electrical, Communications and Cyber Systems Award 1541959. We thank the Harvard Center for Biological Imaging for infrastructure and support. This work was supported by the NSF under Grants NSF-1542506 and NSF OCE-1635365 to P.R.G. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the non-US Geological Survey (USGS) author(s) and do not necessarily reflect the views of the NSF. This journal article has been peer reviewed and approved for publication consistent with USGS Fundamental Science Practices (https://pubs.usgs.gov/circ/1367/). Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the US Government. This work was also supported by NASA under Grant No. NNX17AB31G to P.R.G., issued through the Planetary Science and Technology Through Analog Research program.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006857118/-/DCSupplemental.
Data Availability
DNA sequences data have been deposited in the NCBI (PRJNA648152). All other study data are included in the article and/or supporting information.
References
- 1.Boetius A., Wenzhöfer F., Seafloor oxygen consumption fuelled by methane from cold seeps. Nat. Geosci. 6, 725 (2013). [Google Scholar]
- 2.Reeburgh W. S., Oceanic methane biogeochemistry. Chem. Rev. 107, 486–513 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Dekas A. E., Poretsky R. S., Orphan V. J., Deep-sea archaea fix and share nitrogen in methane-consuming microbial consortia. Science 326, 422–426 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Teichert B. M., Bohrmann G., Suess E., Chemoherms on Hydrate Ridge—Unique microbially-mediated carbonate build-ups growing into the water column. Palaeogeogr. Palaeoclimatol. Palaeoecol. 227, 67–85 (2005). [Google Scholar]
- 5.Klaucke I., Masson D. G., Petersen C. J., Weinrebe W., Ranero C. R., Multifrequency geoacoustic imaging of fluid escape structures offshore Costa Rica: Implications for the quantification of seep processes. Geochem. Geophys. Geosyst. 9, Q04010 (2008). [Google Scholar]
- 6.Klaucke I., Weinrebe W., Linke P., Kläschen D., Bialas J., Sidescan sonar imagery of widespread fossil and active cold seeps along the central Chilean continental margin. Geo-Mar. Lett. 32, 489–499 (2012). [Google Scholar]
- 7.Sahling H., et al., Fluid seepage at the continental margin offshore Costa Rica and southern Nicaragua. Geochem. Geophys. Geosyst. 9, Q05S05 (2008). [Google Scholar]
- 8.Jones A. T., et al., Acoustic and visual characterisation of methane-rich seabed seeps at Omakere Ridge on the Hikurangi Margin, New Zealand. Mar. Geol. 272, 154–169 (2010). [Google Scholar]
- 9.Paull C. K., Normark W. R., Ussler W. III, Caress D. W., Keaten R., Association among active seafloor deformation, mound formation, and gas hydrate growth and accumulation within the seafloor of the Santa Monica Basin, offshore California. Mar. Geol. 250, 258–275 (2008). [Google Scholar]
- 10.Aloisi G., et al., CH4-consuming microorganisms and the formation of carbonate crusts at cold seeps. Earth Planet. Sci. Lett. 203, 195–203 (2002). [Google Scholar]
- 11.Blumenberg M., Seifert R., Reitner J., Pape T., Michaelis W., Membrane lipid patterns typify distinct anaerobic methanotrophic consortia. Proc. Natl. Acad. Sci. U.S.A. 101, 11111–11116 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reitner J., et al., Concretionary methane-seep carbonates and associated microbial communities in Black Sea sediments. Palaeogeogr. Palaeoclimatol. Palaeoecol. 227, 18–30 (2005). [Google Scholar]
- 13.Yanagawa K., et al., Endolithic microbial habitats hosted in carbonate nodules currently forming within sediment at a high methane flux site in the sea of Japan. Geosciences (Basel) 9, 463 (2019). [Google Scholar]
- 14.Case D. H., et al., Methane seep carbonates host distinct, diverse, and dynamic microbial assemblages. MBio 6, e01348-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marlow J. J., et al., Carbonate-hosted methanotrophy represents an unrecognized methane sink in the deep sea. Nat. Commun. 5, 5094 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Paull C., et al., Geology of the strata exposed on the Florida Escarpment. Mar. Geol. 91, 177–194 (1990). [Google Scholar]
- 17.Skarke A., Ruppel C., Kodis M., Brothers D., Lobecker E., Widespread methane leakage from the sea floor on the northern US Atlantic margin. Nat. Geosci. 7, 657 (2014). [Google Scholar]
- 18.Teske A., et al., The Guaymas Basin hiking guide to hydrothermal mounds, chimneys, and microbial mats: Complex seafloor expressions of subsurface hydrothermal circulation. Front. Microbiol. 7, 75 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raineault N., et al., Exploration of Central California Basins, Cold Seeps, and San Juan Seamount. Oceanography (Wash. D.C.) 30, 36–37 (2017). [Google Scholar]
- 20.Alperin M. J., Reeburgh W. S., Inhibition experiments on anaerobic methane oxidation. Appl. Environ. Microbiol. 50, 940–945 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Treude T., Boetius A., Knittel K., Wallmann K., Jørgensen B. B., Anaerobic oxidation of methane above gas hydrates at Hydrate Ridge, NE Pacific Ocean. Mar. Ecol. Prog. Ser. 264, 1–14 (2003). [Google Scholar]
- 22.Segarra K. E., Comerford C., Slaughter J., Joye S. B., Impact of electron acceptor availability on the anaerobic oxidation of methane in coastal freshwater and brackish wetland sediments. Geochim. Cosmochim. Acta 115, 15–30 (2013). [Google Scholar]
- 23.Marlow J. J., et al., Monodeuterated methane, an isotopic tool to assess biological methane metabolism rates. MSphere 2, e00309–e00317 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michaelis W., et al., Microbial reefs in the Black Sea fueled by anaerobic oxidation of methane. Science 297, 1013–1015 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Valsami-Jones E., McLean J., McEldowney S., Hinrichs H., Pili A., An experimental study of bacterially induced dissolution of K-feldspar. Mineral. Mag. A 62, 1563–1564 (1998). [Google Scholar]
- 26.Lu A., et al., Growth of non-phototrophic microorganisms using solar energy through mineral photocatalysis. Nat. Commun. 3, 768 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Welch S. A., Banfield J. F., Modification of olivine surface morphology and reactivity by microbial activity during chemical weathering. Geochim. Cosmochim. Acta 66, 213–221 (2002). [Google Scholar]
- 28.Naehr T. H., et al., Authigenic carbonate formation at hydrocarbon seeps in continental margin sediments: A comparative study. Deep Sea Res. Part II Top. Stud. Oceanogr. 54, 1268–1291 (2007). [Google Scholar]
- 29.McGlynn S. E., Chadwick G. L., Kempes C. P., Orphan V. J., Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 526, 531–535 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Wegener G., Krukenberg V., Riedel D., Tegetmeyer H. E., Boetius A., Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature 526, 587–590 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Gross B. J., El-Naggar M. Y., A combined electrochemical and optical trapping platform for measuring single cell respiration rates at electrode interfaces. Rev. Sci. Instrum. 86, 064301 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Shrestha P. M., Rotaru A.-E., Plugging in or going wireless: Strategies for interspecies electron transfer. Front. Microbiol. 5, 237 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tryon M. D., Brown K. M., Torres M. E., Fluid and chemical flux in and out of sediments hosting methane hydrate deposits on Hydrate Ridge, OR, II: Hydrological processes. Earth Planet. Sci. Lett. 201, 541–557 (2002). [Google Scholar]
- 34.Paull C. K., et al., Seawater circulation through the flank of the Florida platform: Evidence and implications. Mar. Geol. 102, 265–279 (1991). [Google Scholar]
- 35.Yazdani Foshtomi M., et al., The link between microbial diversity and nitrogen cycling in marine sediments is modulated by macrofaunal bioturbation. PLoS One 10, e0130116 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng L., et al., Macrofaunal control of microbial community structure in continental margin sediments. Proc. Natl. Acad. Sci. U.S.A. 117, 15911–15922 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marlow J., Peckmann J., Orphan V., Autoendoliths: A distinct type of rock-hosted microbial life. Geobiology 13, 303–307 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Carini P., et al., Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat. Microbiol. 2, 16242 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Ramírez G. A., Jørgensen S. L., Zhao R., D’Hondt S., Minimal influence of extracellular DNA on molecular surveys of marine sedimentary communities. Front. Microbiol. 9, 2969 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Corinaldesi C., Danovaro R., Dell’Anno A., Simultaneous recovery of extracellular and intracellular DNA suitable for molecular studies from marine sediments. Appl. Environ. Microbiol. 71, 46–50 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torti A., Jørgensen B. B., Lever M. A., Preservation of microbial DNA in marine sediments: Insights from extracellular DNA pools. Environ. Microbiol. 20, 4526–4542 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Thompson L. R.et al.; Earth Microbiome Project Consortium , A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Callahan B. J., McMurdie P. J., Holmes S. P., Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 11, 2639–2643 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amir A., et al., Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2, e00191-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marlow J. J., et al., Harnessing a methane-fueled, sediment-free mixed microbial community for utilization of distributed sources of natural gas. Biotechnol. Bioeng. 115, 1450–1464 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leventhal J., An interpretation of carbon and sulfur relationships in Black Sea sediments as indicators of environments of deposition. Geochim. Cosmochim. Acta 47, 133–137 (1983). [Google Scholar]
- 47.Peckmann J., et al., Methane-derived carbonates and authigenic pyrite from the northwestern Black Sea. Mar. Geol. 177, 129–150 (2001). [Google Scholar]
- 48.Muyzer G., Stams A. J., The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 6, 441–454 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Jochum L. M., et al., Single-cell genomics reveals a diverse metabolic potential of uncultivated Desulfatiglans-related Deltaproteobacteria widely distributed in marine sediment. Front. Microbiol. 9, 2038 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rabus R., Nordhaus R., Ludwig W., Widdel F., Complete oxidation of toluene under strictly anoxic conditions by a new sulfate-reducing bacterium. Appl. Environ. Microbiol. 59, 1444–1451 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Könneke M., Kuever J., Galushko A., Jørgensen B. B., Desulfoconvexum algidum gen. nov., sp. nov., a psychrophilic sulfate-reducing bacterium isolated from a permanently cold marine sediment. Int. J. Syst. Evol. Microbiol. 63, 959–964 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Kuever J., “The family desulfarculaceae” in The Prokaryotes, Rosenberg E., DeLong E. F., Lory S., Stackebrandt E., Thompson F., Eds. (Springer-Verlag, 2014), pp. 41–44. [Google Scholar]
- 53.Greene A. C., “The family desulfuromonadaceae” in The Prokaryotes: Deltaproteobacteria and Epsilonproteobacteria, Rosenberg E., DeLong E. F., Lory S., Stackebrandt E., Thompson F., Eds. (Springer Berlin Heidelberg, 2014), pp. 143–155. [Google Scholar]
- 54.Meyer B., Imhoff J. F., Kuever J., Molecular analysis of the distribution and phylogeny of the soxB gene among sulfur-oxidizing bacteria–Evolution of the Sox sulfur oxidation enzyme system. Environ. Microbiol. 9, 2957–2977 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Pokorna D., Zabranska J., Sulfur-oxidizing bacteria in environmental technology. Biotechnol. Adv. 33, 1246–1259 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Watanabe T., Kojima H., Fukui M., Complete genomes of freshwater sulfur oxidizers Sulfuricella denitrificans skB26 and Sulfuritalea hydrogenivorans sk43H: Genetic insights into the sulfur oxidation pathway of betaproteobacteria. Syst. Appl. Microbiol. 37, 387–395 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Wirsen C. O., et al., Characterization of an autotrophic sulfide-oxidizing marine Arcobacter sp. that produces filamentous sulfur. Appl. Environ. Microbiol. 68, 316–325 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kodama Y., Watanabe K., Sulfuricurvum kujiense gen. nov., sp. nov., a facultatively anaerobic, chemolithoautotrophic, sulfur-oxidizing bacterium isolated from an underground crude-oil storage cavity. Int. J. Syst. Evol. Microbiol. 54, 2297–2300 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Timmers P. H., et al., Reverse methanogenesis and respiration in methanotrophic archaea. Archaea 2017, 1654237 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kletzin A., et al., Cytochromes c in Archaea: Distribution, maturation, cell architecture, and the special case of Ignicoccus hospitalis. Front. Microbiol. 6, 439 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Treude T., et al., Consumption of methane and CO2 by methanotrophic microbial mats from gas seeps of the anoxic Black Sea. Appl. Environ. Microbiol. 73, 2271–2283 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Inagaki F., et al., Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc. Natl. Acad. Sci. U.S.A. 103, 2815–2820 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoshino T., et al., Atribacteria from the subseafloor sedimentary biosphere disperse to the hydrosphere through submarine mud volcanoes. Front. Microbiol. 8, 1135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chakraborty A., et al., Hydrocarbon seepage in the deep seabed links subsurface and seafloor biospheres. Proc. Natl. Acad. Sci. U.S.A. 117, 11029–11037 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carr S. A., Orcutt B. N., Mandernack K. W., Spear J. R., Abundant Atribacteria in deep marine sediment from the Adélie Basin, Antarctica. Front. Microbiol. 6, 872 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nobu M. K., et al., Phylogeny and physiology of candidate phylum ‘Atribacteria’ (OP9/JS1) inferred from cultivation-independent genomics. ISME J. 10, 273–286 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wegener G., Krukenberg V., Ruff S. E., Kellermann M. Y., Knittel K., Metabolic capabilities of microorganisms involved in and associated with the anaerobic oxidation of methane. Front. Microbiol. 7, 46 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sherry A., et al., Anaerobic biodegradation of crude oil under sulphate-reducing conditions leads to only modest enrichment of recognized sulphate-reducing taxa. Int. Biodeterior. Biodegradation 81, 105–113 (2013). [Google Scholar]
- 69.Liang B., et al., Anaerolineaceae and Methanosaeta turned to be the dominant microorganisms in alkanes-dependent methanogenic culture after long-term of incubation. AMB Express 5, 117 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vigneron A., et al., Comparative metagenomics of hydrocarbon and methane seeps of the Gulf of Mexico. Sci. Rep. 7, 16015 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Evans P. N., et al., Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 350, 434–438 (2015). [DOI] [PubMed] [Google Scholar]
- 72.Vanwonterghem I., et al., Methylotrophic methanogenesis discovered in the archaeal phylum Verstraetearchaeota. Nat. Microbiol. 1, 16170 (2016). [DOI] [PubMed] [Google Scholar]
- 73.Laso-Pérez R., et al., Thermophilic archaea activate butane via alkyl-coenzyme M formation. Nature 539, 396–401 (2016). [DOI] [PubMed] [Google Scholar]
- 74.Hinrichs K.-U., Boetius A., “The anaerobic oxidation of methane: New insights in microbial ecology and biogeochemistry” in Ocean Margin Systems, Wefer G., Ed. et al. (Springer, 2002), pp. 457–477. [Google Scholar]
- 75.Dumke I., Klaucke I., Berndt C., Bialas J., Sidescan backscatter variations of cold seeps on the Hikurangi Margin (New Zealand): Indications for different stages in seep development. Geo-Mar. Lett. 34, 169–184 (2014). [Google Scholar]
- 76.Römer M., et al., Methane fluxes and carbonate deposits at a cold seep area of the Central Nile Deep Sea Fan, Eastern Mediterranean Sea. Mar. Geol. 347, 27–42 (2014). [Google Scholar]
- 77.Klaucke I., Weinrebe W., Petersen C. J., Bowden D., Temporal variability of gas seeps offshore New Zealand: Multi-frequency geoacoustic imaging of the Wairarapa area, Hikurangi margin. Mar. Geol. 272, 49–58 (2010). [Google Scholar]
- 78.Greinert J., Bohrmann G., Suess E., Gas hydrate-associated carbonates and methane-venting at Hydrate Ridge: Classification, distribution and origin of authigenic lithologies. Geophysical Monograph-American Geophysical Union 124, 99–114 (2001). [Google Scholar]
- 79.Beal E. J., House C. H., Orphan V. J., Manganese- and iron-dependent marine methane oxidation. Science 325, 184–187 (2009). [DOI] [PubMed] [Google Scholar]
- 80.Beckmann S., et al., Evidence for an expanded repertoire of electron acceptors for the anaerobic oxidation of methane in authigenic carbonates in the Atlantic and Pacific Ocean. bioRxiv [Preprint] (2020). 10.1101/2020.06.12.148429 (Accessed 14 June 2020). [DOI]
- 81.Beal E. J., Claire M. W., House C. H., High rates of anaerobic methanotrophy at low sulfate concentrations with implications for past and present methane levels. Geobiology 9, 131–139 (2011). [DOI] [PubMed] [Google Scholar]
- 82.Archer D., Buffett B., Brovkin V., Ocean methane hydrates as a slow tipping point in the global carbon cycle. Proc. Natl. Acad. Sci. U.S.A. 106, 20596–20601 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Doney S. C., Fabry V. J., Feely R. A., Kleypas J. A., Ocean acidification: The other CO2 problem. Ann. Rev. Marine Sci. 1, 169–192 (2009). [DOI] [PubMed] [Google Scholar]
- 84.Luo Y., Boudreau B. P., Mucci A., Disparate acidification and calcium carbonate desaturation of deep and shallow waters of the Arctic Ocean. Nat. Commun. 7, 12821 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Conrado R. J., Gonzalez R., Chemistry. Envisioning the bioconversion of methane to liquid fuels. Science 343, 621–623 (2014). [DOI] [PubMed] [Google Scholar]
- 86.Lis G., Wassenaar L. I., Hendry M. J., High-precision laser spectroscopy D/H and 18O/16O measurements of microliter natural water samples. Anal. Chem. 80, 287–293 (2008). [DOI] [PubMed] [Google Scholar]
- 87.Marlow J. J., et al., Microbial abundance and diversity patterns associated with sediments and carbonates from the methane seep environments of Hydrate Ridge, OR. Front. Mar. Sci. 1, 44 (2014). [Google Scholar]
- 88.Steinhaus H., Mathematical Snapshots (Courier Dover Publications, 1999). [Google Scholar]
- 89.Stahl D. A., Amann R. I., “Development and application of nucleic acid probes” in Nucleic Acid Techniques in Bacterial Systematics, Stackebrandt E., Goodfellow M., Eds. (John Wiley & Sons, 1991). [Google Scholar]
- 90.Manz W., Eisenbrecher M., Neu T. R., Szewzyk U., Abundance and spatial organization of Gram-negative sulfate-reducing bacteria in activated sludge investigated by in situ probing with specific 16S rRNA targeted oligonucleotides. FEMS Microbiol. Ecol. 25, 43–61 (1998). [Google Scholar]
- 91.Wallner G., Amann R., Beisker W., Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14, 136–143 (1993). [DOI] [PubMed] [Google Scholar]
- 92.Bates S. T., et al., Examining the global distribution of dominant archaeal populations in soil. ISME 5, 908–917 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Edgar R. C., Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010). [DOI] [PubMed] [Google Scholar]
- 94.Caporaso J. G., et al., QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bowles M., et al., Remarkable capacity for anaerobic oxidation of methane at high methane concentration. Geophys. Res. Lett. 46, 12192–12201 (2019). [Google Scholar]
- 96.Joye S., et al., The anaerobic oxidation of methane and sulfate reduction in sediments from Gulf of Mexico cold seeps. Chem. Geol. 205, 219–238 (2004). [Google Scholar]
- 97.Omoregie E., et al., Biogeochemistry and community composition of iron-and sulfur-precipitating microbial mats at the Chefren mud volcano (Nile Deep Sea Fan, Eastern Mediterranean). Appl. Environ. Microbiol. 74, 3198–3215 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Niemann H., et al., Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature 7113, 854–858 (2006). [DOI] [PubMed] [Google Scholar]
- 99.Nauhaus K., et al., In vitro demonstration of anaerobic oxidation of methane coupled to sulphate reduction in sediment from a marine gas hydrate. Environ. Microbiol., 10.1046/j.1462-2920.2002.00299.x (2002). [DOI] [PubMed] [Google Scholar]
- 100.Wankel S. D., et al., New constraints on methane fluxes and rates of anaerobic methane oxidation in a Gulf of Mexico brine pool via in situ mass spectrometry. Deep Sea Res. 2 Top. Stud. Oceanogr. 57, 2022–2029 (2010). [Google Scholar]
- 101.Girguis P. R., Orphan V. J., Hallam S. J., DeLong E. F., Growth and methane oxidation rates of anaerobic methanotrophic archaea in a continuous-flow bioreactor. Appl. Environ. Microbiol. 69, 5472–5482 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences data have been deposited in the NCBI (PRJNA648152). All other study data are included in the article and/or supporting information.