Significance
Clathrin-coated vesicles (CCVs) are endocytic carriers responsible for the internalization of receptor-bound ligands and extracellular fluids. CCV assembly occurs by the sequential recruitment of clathrin, adaptors, and other accessory molecules that promote curvature formation. To form, CCVs need to overcome the local plasma membrane tension that varies during cell cycle, development, and cell polarization. Using quantitative fluorescence microscopy, we demonstrate that the adaptor CALM is a major determinant of CCV formation upon membrane tension increase. Since CALM is differentially expressed, our results demonstrate that competence in clathrin-mediated endocytosis is tissue specific, providing mechanistic explanation why CALM depletion strongly affects embryo development and red blood cell differentiation with minor effects in other systems.
Keywords: membrane traffic, clathrin adaptors, membrane tension, quantitative microscopy, lattice light-sheet microscopy
Abstract
The most represented components of clathrin-coated vesicles (CCVs) are clathrin triskelia and the adaptors clathrin assembly lymphoid myeloid leukemia protein (CALM) and the heterotetrameric complex AP2. Investigation of the dynamics of AP180-amino-terminal-homology (ANTH) recruitment during CCV formation has been hampered by CALM toxicity upon overexpression. We used knock-in gene editing to express a C-terminal–attached fluorescent version of CALM, while preserving its endogenous expression levels, and cutting-edge live-cell microscopy approaches to study CALM recruitment at forming CCVs. Our results demonstrate that CALM promotes vesicle completion upon membrane tension increase as a function of the amount of this adaptor present. Since the expression of adaptors, including CALM, differs among cells, our data support a model in which the efficiency of clathrin-mediated endocytosis is tissue specific and explain why CALM is essential during embryogenesis and red blood cell development.
Clathrin-mediated endocytosis (CME) controls multiple physiological activities in eukaryotic cells, including internalization of nutrients, receptors, and other membrane components. Endocytic clathrin-coated vesicles (CCVs) form by the recruitment of clathrin triskelia, adaptors, and other accessory proteins at specific plasma membrane sites (1). Biochemical experiments complemented by live-cell microscopy have allowed identification of several phases of CCV formation: initiation, growth/maturation, and completion (2). The initiation of “canonical” CCVs is defined by the coordinated recruitment of one triskelion and two heterotetrameric adaptor protein complexes 2 (AP2) at the plasma membrane (3). The majority of these nascent structures abort while a fraction can commit and grow further (4, 5). During the growth phase, the membrane invagination increases as more triskelia and adaptors are incorporated into the coat (6). Mass spectrometry analysis has shown that the AP2 and clathrin assembly lymphoid myeloid leukemia protein (CALM), also named Phosphatidylinositol-binding clathrin assembly protein (PICALM), are, together with clathrin, the most abundant protein components of CCVs (7, 8). While AP2 is a tetrameric adaptor (1), CALM is a single peptide chain that contains at its N terminus the AP180-amino-terminal-homology (ANTH) domain (9). The ANTH domain mediates CALM binding to phosphoinositides on the plasma membrane and to SNAREs that control the fusion of vesicles with specific acceptor membranes (10). CALM’s C terminus is unstructured and comprises binding motifs for clathrin, AP2, and EPS15 (11). Moreover, CALM is a driver of membrane curvature formation (12), which is critical for the construction of a CCV. Membrane curvature can be achieved only by overcoming local plasma membrane tension (13–16) that, together with the availability of CME components (17), determines the heterogeneity of clathrin structures at the membrane surface. We hypothesized that CALM recruitment can support the formation of CCVs even under mechanically stringent conditions, such as increased membrane tension. To test this, we used quantitative live-cell microscopy to study the recruitment of CALM to forming CCVs. Since CALM overexpression has dominant negative effects on intracellular clathrin distribution and CCV formation (18–20), we used gene editing to introduce protein tags at the C-terminal end of CALM so as to not perturb the N-terminal activity of CALM and to maintain its endogenous levels. Our results show that CALM functions as a major determinant of CCV formation upon membrane tension increase. Since CALM is differentially expressed among tissues, our results demonstrate that competence in CME is tissue specific and provide mechanistic explanation why CALM depletion strongly affects embryo development, red blood cell differentiation (21), and CALM polymorphisms are associated with Alzheimer's disease (22), with minor effects in other systems.
Results
Generation and Characterization of Gene-Edited SUM Cells Expressing CALM-EGFP or CALM-Halo.
CALM overexpression inhibits the internalization of EGF and transferrin and alters the distribution of clathrin in the trans-Golgi network (20). Therefore, to study CALM dynamics while avoiding artifacts due to overexpression, we used a gene-editing knock-in strategy (23), based on the CRISPR/Cas9 system (24, 25). In the quasi-diploid SUM159PT (SUM) cells (26), we inserted EGFP (enhanced green fluorescent protein) at the C terminus of CALM (SI Appendix, Fig. S1A), to avoid perturbing the CALM N-terminal domain, which is involved in curvature sensing/induction during CCV formation (12).
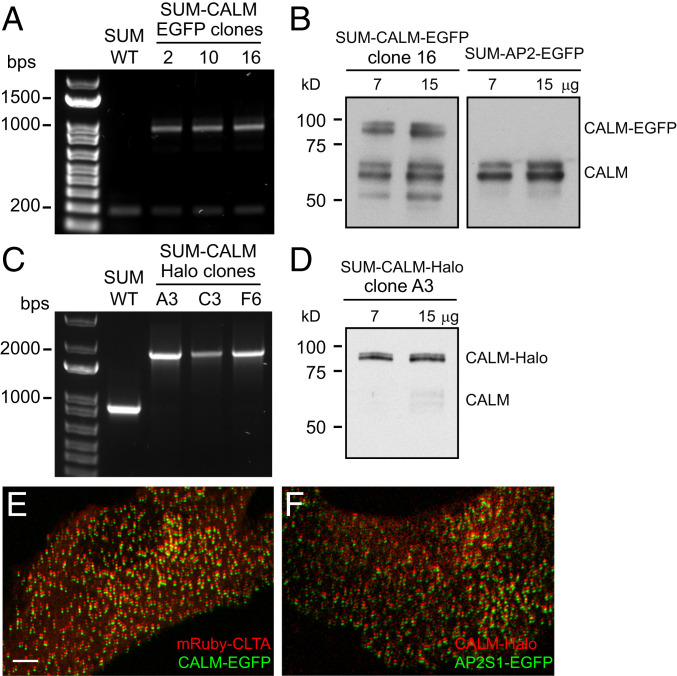
We identified several clones by genomic PCR bearing the desired insertion (SI Appendix, Fig. S1B). Biallelic integration of EGFP did occur in a few clones, suggesting complete modification. Despite the results obtained by genomic PCR, probing a protein lysate of SUM-CALM-EGFP clone 16 (one of the clones bearing modification in both alleles; SI Appendix, Fig. S1C) revealed that only a fraction of CALM was carrying the EGFP tag (SI Appendix, Fig. S1D). To exclude that this result was due to degradation of the linker used to connect CALM to the tag (23), we applied a small interfering RNA (siRNA) approach. We reasoned that if all CALM alleles were edited, every transcribed CALM-mRNA molecule would incorporate EGFP at its C terminus. Therefore, transfecting SUM-CALM-EGFP clone 16 with an siRNA specific for EGFP would result in depletion of both CALM and CALM-EGFP. Instead, EGFP siRNA treatment induced degradation of only CALM-EGFP. Conversely, when we used an siRNA specific for CALM, it induced the degradation of both protein species (SI Appendix, Fig. S1D). This result suggested that at least one allele was not modified by homologous recombination. This was confirmed by a second PCR strategy that demonstrated that the isolated SUM-CALM-EGFP clones were not completely edited (Fig. 1A). In light of these results, we concluded that SUM cells must contain at least three copies of the CALM gene. Therefore, the untagged CALM protein detected by Western blot is the wild-type form of CALM (Fig. 1B).
Fig. 1.
Development of SUM cells gene-edited to express CALM-EGFP or CALM-Halo. (A) Genomic PCR of SUM-CALM-EGFP clones 2, 10, and 16 reveals that EGFP insertion into the CALM loci is partial; SUM WT designates the parental cells. (B) Western blot analysis of cell lysates from SUM-CALM-EGFP clone 16 and SUM-AP2-EGFP (used as control) shows that two major species of CALM are detectable, that both CALM species incorporate EGFP, and that SUM-CALM-EGFP are partially edited since also the wild-type isoforms of CALM are present. (C) Genomic PCR reveals complete integration of Halo into the CALM loci in SUM-CALM-Halo clones A3, C3, and F6; SUM WT designates the parental cells. (D) Western blot analysis of cell lysates from SUM-CALM-Halo clone A3 shows that CALM is completely substituted by CALM-Halo. (E) Confocal microscopy image of the ventral surface of SUM-CALM-EGFP cells, gene-edited to express CALM-EGFP (green) and transiently transfected with mRuby-CLTB (red). (F) Confocal microscopy image of the ventral surface of SUM-CALM-Halo cells, gene-edited to express CALM-Halo (red), and transiently transfected with AP2S1-EGFP (green). To easily identify clathrin structures containing both fluorescently tagged proteins, the channels were vertically shifted by five pixels in both E and F panels. The bar in E (5 μm) is also valid for F.
To obtain completely edited cells, we designed an alternative gene-editing approach with the goal of increasing the homologous recombination efficiency. This strategy targeted exon 19 (according to https://www.ensembl.org nomenclature), the second-to-last exon in the CALM gene. We incorporated the last exon of the CALM gene in the donor template so that expression of the complete CALM protein would occur after recombination (SI Appendix, Fig. S1E). In place of EGFP, we used HaloTag, a modified enzyme that can covalently bind to a chloroalkane linker attached to a variety of molecules (27), including fluorescent dyes (28) and proteolysis targeting chimera (PROTAC) (29), resulting in a much more versatile tag than EGFP. Screening of the CALM-Halo clones by genomic PCR revealed few clones that appeared to be completely modified (SI Appendix, Fig. S1F). One such clone, SUM-CALM-Halo clone A3 (Fig. 1C), was confirmed by Western blot analysis to only express the Halo-tagged protein variant (Fig. 1D and SI Appendix, Fig. S1G). Low levels of AP180, the neuronal homolog of CALM, were detected in both wild-type SUM cells and derived clones (SI Appendix, Fig. S1H).
Using confocal microscopy, we imaged SUM-CALM-EGFP clone 16 cells transiently transfected to express clathrin light chain B labeled with monomeric Ruby, a red fluorescent protein (mRuby-CLTB), and SUM-CALM-Halo clone A3 transfected with AP2S1-EGFP and labeled with JF549-Halo ligand. Both of these experiments showed that CALM-EGFP and CALM-Halo are recruited to forming CCVs (Fig. 1 E and F, respectively). These two clones are used throughout the rest of the paper and are referred to as SUM-CALM-EGFP and SUM-CALM-Halo cells.
CCVs Formed at Ventral and Dorsal Surfaces of SUM Cells Recruit Equal Amounts of CALM with Similar Dynamics.
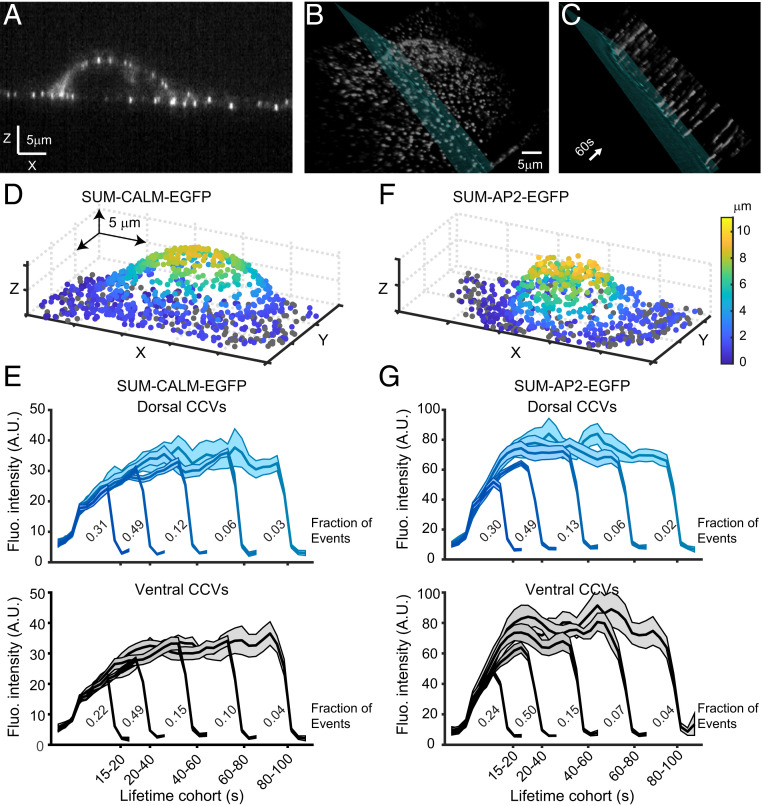
Since CALM promotes membrane curvature (12), we hypothesized that CCVs forming on the support attached (ventral) surface of cells may require more CALM than the ones forming on the dorsal surface to counteract the higher local membrane tension due to the presence of adhesion sites (14, 16, 30). To test this hypothesis, we used lattice light-sheet fluorescence microscopy (LLSFM), since this technique permits imaging the entire cell volume with unrivaled temporal resolution and significantly less photobleaching compared to other live-cell microscopy approaches (31). We imaged SUM-CALM-EGFP, and we found that CALM-EGFP spots were detected over the entire cell surface with dynamics that were consistent with CCV formation, as shown in a representative section, in the corresponding three-dimensional volume, and in the derived kymograph (Fig. 2 A–C). For a quantitative analysis of CALM recruitment, we used a single-particle tracking algorithm that efficiently detects CCV formation in the multidimensional datasets of LLSFM (25). The spatial distribution of CCVs detected over the entire cell surface of a representative SUM-CALM-EGFP is shown in Fig. 2D. The relative positions of the CCVs are color coded according to their distance from the glass surface, while CCVs forming on the ventral surface (plasma membrane-glass interface) of the cell are shown in gray. After tracking CCV formation (32) over a period of 5 min, their intensity profiles were organized into cohorts according to their lifetime (SI Appendix, Fig. S2A) and spatial localization (Fig. 2E). We found that 70% of the total CCVs were forming on the dorsal surface of the cells. When we compared the signal intensities of the respective cohorts forming on the dorsal with the ones forming on the ventral surfaces (Fig. 2 E, Top and Bottom, respectively), we did not detect a statistically significant difference, suggesting that the same amount of CALM was recruited. However, we found that short-lived CCVs (15 to 20s, first cohort) are statistically significantly less frequent on the ventral surface compared to the dorsal surface (∼10%, compare the fraction of events; p-value (pval) = 3.6 × 10−4, Kolmogorov–Smirnov test [KS-test], from seven cells as in figure legend). We reasoned that a similar difference in CCV formation would be detected in SUM cells expressing other CME markers. Therefore, we used the same approach to image SUM-AP2-EGFP cells, a SUM clone gene edited to encode EGFP at the C-terminal of AP2S1 (25). The distribution of CCVs detected on the surface of a representative SUM-AP2-EGFP cell is shown in Fig. 2F. When we compared the distributions of forming CCVs on the dorsal and ventral surfaces of the SUM-AP2-EGFP cells, we observed the same statistically significant decrease in the frequency of short-lived CCVs on the ventral compared to dorsal surface that we observed in SUM-CALM-EGFP (Fig. 2G; pval = 4.4 × 10−5 KS-test, from nine cells as in figure legend). In addition, we noticed a clear difference in the recruitment of CALM and AP2 (compare Fig. 2 E and G and SI Appendix, Fig. S2 A and B). CALM was recruited at a steadier rate over the entire lifetime of the CCVs, peaking just before uncoating, while AP2 was recruited in two phases characterized by a fast increase in intensity with a steeper slope than CALM followed by a slower rate tending to a plateau. To better appreciate this difference, we overlaid in single panels (SI Appendix, Fig. S2C) the intensity traces obtained from the first three cohorts of CALM- and AP2-labeled CCVs, which together account for 90% of the total number of the analyzed events.
Fig. 2.
LLSFM imaging of CALM and AP2 recruitment to CCVs over the entire cell surface. The data sets we acquired were composed of 120 z-stacks of 75 sequential optical sections acquired with 20 ms of exposure and spaced 250 nm apart along the detection axis that results in a total volume of 50 × 50 × 18 μm. (A–C) Example of a SUM-CALM cell imaged with LLSFM. In A, a single optical section is shown, while in B, the three-dimensional rendering of a z-stack is depicted. The kymograph generated at the intersection between the cell and the blue plane drawn in B is shown in C. (D and F) Representative three-dimensional renderings of the average positions of forming CCVs detected in SUM-CALM-EGFP (D) or SUM-AP2-EGFP (F) cells. The CCVs on the ventral surface of the cells are shown as gray dots, whereas the ones located on the dorsal surface (Top) are color-coded according to their distance from the glass coverslip (the color map on the right is valid for both D and F). (E and G) Fluorescence intensity traces of CCVs organized in lifetime cohorts. Intensities were calculated averaging data obtained from seven SUM-CALM cells (2,089 CCVs, E) and nine SUM-AP2-EGFP cells (4,456 CCVs, G). Data were organized according to the position of the CCVs on the cell surface (top plots: dorsal; lower plots: ventral) for SUM-CALM (E) and SUM-AP2-EGFP (G) cells. Productive CCVs (∼70%) were identified on the dorsal surface of both cell types. Notice the statistically significant lower frequency of CCV formation with lifetime 15 to 20 s on the ventral when compared to the dorsal surface of both CALM (pval = 3.6 × 10−4 KS-test) and AP2 (P = 4.4 × 10−5 KS-test) cells. The intensity traces in E and G are shown as mean ± SEM; the relative frequency of each lifetime cohort is reported under the respective trace (Fraction of Events).
CALM Enrichment in CCVs Is Greater than That of AP2 in SUM Cells.
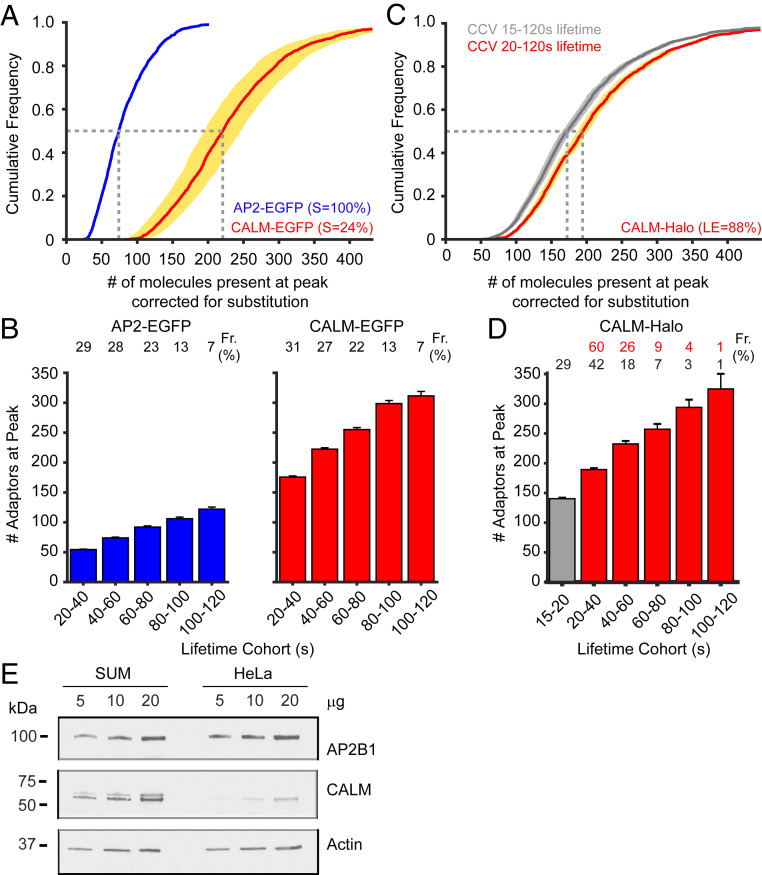
To define the number of CALM and AP2 molecules recruited during CCV formation, we used a quantitative single-molecule approach. We calibrated our spinning disk confocal microscope to the intensity of a single EGFP (33, 34) (SI Appendix, Fig. S3) and imaged CCVs over the ventral surface of SUM-CALM-EGFP and of SUM-AP2-EGFP cells. After detection and tracking of the forming vesicles (35), we expressed the maximum intensity observed per CCV as the number of fluorescently labeled adaptors (SI Appendix, Fig. S4A). Initial calculations indicated that an individual CCV contained on average ∼82 EGFP-tagged AP2 or ∼56 CALM molecules. SUM-AP2-EGFP cells are fully substituted for AP2-EGFP, meaning that the subunit sigma (AP2S1) of AP2 is completely replaced by the fluorescent chimera AP2S1-EGFP (SI Appendix, Fig. S4B). Therefore, no mathematical correction is required to estimate the total number of AP2 per CCV. However, our Western blot analyses (Fig. 1B; also refer to Fig. 4A and SI Appendix, Fig. S1 D and H) have revealed that only one-fourth of CALM molecules were fluorescently labeled in SUM-CALM-EGFP cells (24 ± 4%, mean ± SEM). Therefore, we estimated that ∼234 molecules of CALM are recruited on average per CCV (Fig. 3A), establishing greater enrichment of CALM compared to AP2 in these two models. When the data were organized in cohorts according to the lifetimes of CCVs (Fig. 3B), we noticed that the ratio of CALM to AP2 was constant across the cohorts and corresponded to 3.4 ± 0.15 (mean ± SD) CALM molecules for every AP2 that was incorporated. Previous works have estimated that a fraction of EGFP molecules is nonfluorescent, and this fraction varies from 0 to 20%. The results of these works revealed that while higher fractions of nonfluorescent EGFP were detected when chimera were transiently over-expressed (36), lower fractions were detected in stably expressing cells or in cells gene-edited to express AP2S1-EGFP (3, 25). Therefore, in our system, the fraction of immature EGFP should be negligible since EGFP maturation takes only ∼14 to 28 min at 37 °C (37, 38) while the average half-life of the proteome in mammalian cells is longer than the cell cycle (39, 40) (∼21 h for SUM159 cells). However, since we estimated the ratio of CALM to AP2, the frequency of nonfluorescent molecules should scale proportionally and consequently should not affect our conclusions.
Fig. 3.
The maximum number of AP2 or CALM molecules recruited during the formation of CCVs. The maximum number of AP2 and CALM molecules recruited to CCVs was defined as the fluorescence intensity of a given object calibrated to the intensity of a single EGFP or a single JF549-HaloTag molecule. The data were collected from three SUM-AP2-EGFP cells, three SUM-CALM-EGFP cells acquired by confocal spinning disk (100-ms exposure, 0.3 Hz), and from 10 SUM-CALM-Halo cells acquired by LLSFM (120 z-stacks of 61 sequential optical sections acquired with 35-ms exposure and spaced 300 nm apart along the detection axis, resulting in a total volume of 50 × 50 × 18.3 μm). (A) Frequency distribution of the maximum AP2 and CALM content per forming CCV (total number of CCVs: 1,743, AP2; 2,546, CALM) imaged by confocal spinning disk and corrected for the calculated substitution of 100% for AP2 and 24% for CALM. The dotted lines mark the median number of AP2 (n = 74) or CALM (n = 220) molecules recruited per CCV. The cyan and yellow areas around AP2 and CALM traces indicate the error in the estimation of the number of molecules present per CCV. (B) Distribution of the maximum number of AP2 and CALM adaptors organized in cohorts according to CCV lifetimes (mean ± SEM, data from A). The contribution of each cohort to the total number of CCVs is indicated as percentage on the top of each bar (Fr [%] = Frequency indicated as percentage). (C) Frequency distribution of the maximum CALM content per forming CCV in SUM-CALM-Halo cells (total number of analyzed CCVs: 2,603) imaged by LLSFM and corrected for the labeled efficiency (LE) of 88%. The dotted lines mark the median number of CALM molecules recruited per CCV when considering CCVs with lifetimes from 15 to 120 s (n = 172, gray distribution) or from 20 to 120 s (n = 194, red distribution). The bright gray and the yellow areas indicate the error in the estimation of the number of molecules present per CCV. (D) Distribution of the maximum number of CALM adaptors organized in cohorts according to CCV lifetimes (mean ± SEM, data from C). To easily compare this dataset with the one obtained by confocal (B), we color-coded the cohort 15 to 20 s in gray and colored the remaining cohorts in red. The contribution of each cohort to the total number of CCVs is indicated as a percentage on the top of each bar (Fr [%] = Frequency indicated as percentage). The relative contributions are in red when the 15 to 20 s cohort is excluded. (E) Lysates obtained from SUM and HeLa cells were probed with antibodies specific to AP2B1 (beta 1 subunit of AP2), CALM, and Actin by Western blotting. Comparison of band intensity between the two cell types indicates that the adaptor protein complex AP2 was present in similar amount across both cell types, while CALM expression was five times lower in HeLa cells when compared to SUM.
To confirm our quantitative data, we evaluated the number of CALM molecules recruited during the formation of CCVs in SUM-CALM-Halo cells. Although SUM-CALM-Halo are completely substituted, their fluorescence is dependent on the extent of HaloTag labeling. Therefore, we performed a competition assay to estimate the concentration of fluorescent HaloTag ligand that would result in efficient CALM-Halo labeling while avoiding nonspecific dye distribution. SUM-CALM-Halo cells were exposed to increasing concentrations of JF549-HaloTag for 1 h. After extensive washes, the cells were exposed to a saturating concentration of AlexaFluor660-HaloTag (200 nM), washed again, processed for immunofluorescence, and imaged by confocal microscopy. The competition assay suggested that incubating with 5 nM of JF549-HaloTag for 1 h resulted in specific labeling of 88 ± 5% (mean ± SEM) of CALM-Halo molecules (SI Appendix, Fig. S4C). Following this result, we imaged SUM-CALM-Halo cells labeled with 5 nM JF549-HaloTag by LLSFM. The frequency distribution of forming CCVs in SUM-CALM-Halo cells reflected the dynamics previously observed in SUM-CALM-EGFP and SUM-AP2-EGFP cells (SI Appendix, Figs. S4D and S2 A and B), suggesting that C-terminal modification of CALM results in a functional protein.
Then, we calibrated the LLSFM to the intensity of a single fluorescent molecule using a lysate obtained from SUM-CALM-Halo–labeled cells (SI Appendix, Fig. S5). The maximum intensity of CALM observed per CCV was expressed as the number of fluorescently labeled adaptors accounting for the estimated labeling efficiency of 88% (Fig. 3C). We plotted two cumulative distributions, one comprised of CCVs with a lifetime of 15 to 120 s (gray trace) and one that only accounted for CCVs with a lifetime from 20 to 120 s (red trace). We did this so that the data from the confocal experiment (Fig. 3A) could easily be referenced and compared. The combination of the higher fraction of labeled CALM molecules and faster acquisition speed allowed us to detect 5% of CCVs recruiting less than 100 CALM molecules (20- to 120-s lifetime, red trace), a fraction that increased to 9.6% when the 15- to 20-s cohort was included (gray trace). Our calculations estimated that an average of 191 CALM molecules were recruited to forming CCVs (lifetime 15 to 120 s, gray trace) and that this number rose to 212 when the 15- to 20-s cohort was excluded, resulting in ∼10% fewer molecules than the estimate we made using CALM-EGFP cells (Fig. 3 A and C, red traces). When the data were organized into cohorts (Fig. 3D), we observed that the difference between the two data sets flattened out since the number of estimated molecules in SUM-CALM-Halo cells was only 1.03 ± 0.03 (mean ± SD) times more than the number obtained using SUM-CALM-EGFP (Fig. 3 B and D). The reason for this apparent discrepancy is that the number of short-lived CCVs detected by LLSFM is more abundant than by spinning disk (CCVs with 20- to 40-s lifetime corresponds to 60% of the LLSFM data, in comparison to 31% of the spinning disk confocal data). Overall, these results suggest that both of these SUM clones have similar CALM content per CCV.
Taken together, we can conclude from these experiments that neither wild-type nor tagged CALM molecules are preferentially recruited to pits and that CALM stoichiometry is roughly three times greater than that of AP2 in CCVs of SUM cells. This result is not in agreement with previously published mass spectrometry data that suggested similar amounts of AP2 and CALM are recruited per CCV (7). The most likely explanation for this discrepancy is that SUM cells express higher levels of CALM than HeLa cells, the model used in those mass spectrometry experiments (7). Western blot analysis confirmed this hypothesis: while HeLa and SUM cells retain similar amount of AP2 content, SUM cells express roughly five times the level of CALM found in HeLa cells (Fig. 3E).
CALM Is Required for CCV Completion under High Membrane Tension.
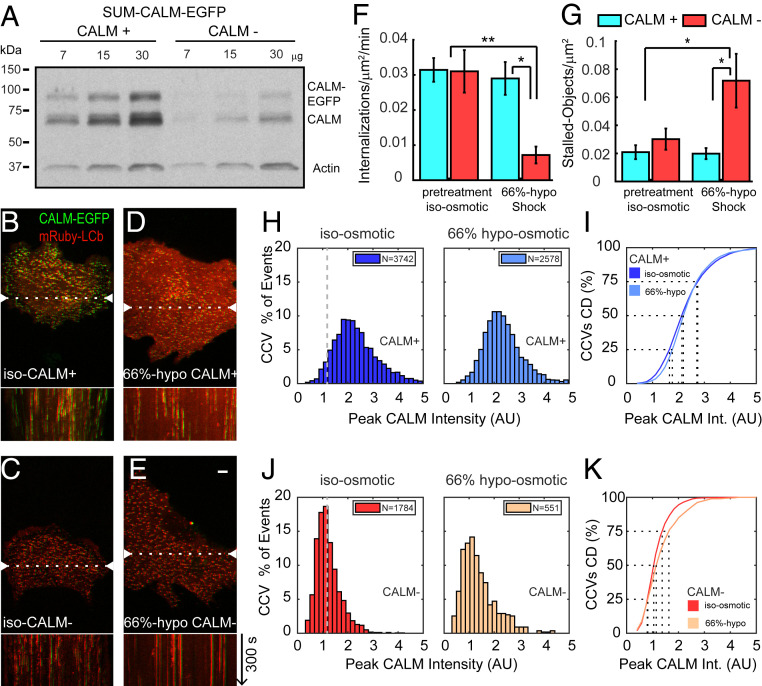
Since CALM supports generation of membrane curvature during the assembly of CCVs (12, 41) and tension on the plasma membrane decreases the internalization rate of CCVs, likely due to the additional energy required to bend the membrane (42), we hypothesized that increased membrane tension can negatively affect CME dynamics in cells expressing low levels of CALM. To test this, we developed an approach to deplete CALM by two consecutive transfections of SUM-CALM-EGFP cells with CALM siRNA separated by 48 h. In the second round of transfection we delivered, together with the siRNA oligos, a plasmid encoding for mRuby-CLTB. This approach resulted in the loss ∼80% of CALM protein over the entire cell population (Fig. 4A) and allowed us to follow forming CCVs using the clathrin signal, avoiding detection and tracking artifacts due to the lower intensity of CALM-EGFP upon siRNA-induced depletion. Spinning disk confocal time series were acquired at the ventral surface of the cells. An example of their respective kymographs of a control (CALM+) and a CALM-depleted SUM-CALM-EGFP cell (CALM-) in isotonic media is shown in Fig. 4 B and C, respectively. We did not detect a significant modification of the overall number of internalization events per surface area or an increase of stalled objects upon partial CALM depletion (Fig. 4 F and G: pretreatment bar plots).
Fig. 4.
Effect of CALM depletion and hypo-osmotic shock on CCV formation in SUM-CALM cells. (A) Western blot analysis of control (CALM+) and CALM-depleted (CALM-) SUM-CALM cells, probed with antibodies specific for CALM and Actin. Quantification of band intensities indicates that siRNA treatment depleted ∼80% of the CALM protein. (B–E) Representative frames extrapolated from time series acquired by spinning disk confocal microscopy of the ventral surface of CALM+ and CALM− cells coexpressing mRuby-CLTB exposed to isotonic (iso, B and C) or 66% hypo-osmotic media (66%-hypo, D and E). The images are shifted five pixels on the x-axis to easily visualize CALM and clathrin signals; the dotted lines indicate the plane at which we generated the kymographs. Bar (5 μm) and arrow (direction and length of kymograph) in E are valid also for B–D. (F) Analysis of CCV formation measured as number of internalizations per surface area per min (median ± SEM) in CALM+ (cyan) and CALM− (red) exposed to isotonic solution (pretreatment) or to 66%-hypo. A significant decrease in internalizations in CALM− cells was detected when exposed to 66%-hypo solution compared to CALM+ exposed to 66%-hypo (pval = 0.028) or CALM+ and CALM− cells exposed to isotonic media (pval = 2.41 × 10−4 and pval = 0.01, respectively). (G) Detection of stalled objects per cell surface area over the entire length of the movies in isotonic condition or upon exposure to 66%-hypo in CALM+ and CALM− cells. When CALM− cells were exposed to 66%-hypo solution, the number of stalled objects increased, and the differences were statistically significant when compared to CALM+ cells (pval = 0.017) or CALM− cells (pval = 0.037) exposed to isotonic solution and to CALM+ cells exposed to 66%-hypo (pval = 0.017). The legend in G is also valid for F. (H–K) Histogram distributions of the maximum CALM-EGFP intensity measured in forming CCVs observed in control SUM-CALM cells (H) or upon CALM depletion (J) and exposed to the isotonic solution are plotted in the left, while upon exposure to the 66%-hypo solution in the right. The cumulative distributions plotted in I and K correspond to the data in H and J respectively. When intensity distributions of CALM recruited in isotonic and hypotonic conditions were compared, they resulted statistically significantly different in CALM+ (pval = 5.2 × 10−6) and CALM− (pval = 3.2 × 10−6). The gray dotted lines in H and J indicate the average of CALM recruited by CCVs in CALM− cells: notice that 5% of CALM+ CCVs recruit equal or lower amount of CALM. The dotted lines in I and K indicate the CALM-EGFP intensity collected by 25, 50, or 75% of the analyzed CCVs. The data were obtained analyzing 15 CALM+ and 13 CALM− cells for the isotonic condition and 12 CALM+ and 10 CALM− cells upon exposure to 66%-hypo. P values were calculated using Student’s t test.
To determine the effects of increased membrane tension, CALM+ and CALM− cells were imaged before and after the addition of hypotonic buffers (43). We exposed the cells to two different hypotonic solutions: one in which 80% of media was replaced with water (80%-hypo) and a second in which 66% of media was replaced with water (66%-hypo). When cells were exposed to 80%-hypo, formation of CCVs was abrogated, and the fraction of persistent objects increased significantly in both CALM+ and CALM− cells, when compared to their respective controls, suggesting that CCV dynamics were strongly affected by the exposure to 80%-hypo solution, regardless of the relative expression of CALM (SI Appendix, Fig. S6 A–C).
Although exposure to 66%-hypo induced cell swelling (SI Appendix, Fig. S6D), it only partially affected CALM+ cells: no difference was visually appreciable by inspecting the kymographs (Fig. 4 B and D), and no significant decrease in the frequency of internalizations or in the number of stalled objects was observed (Fig. 4 F and G, CALM+). In contrast, we found that CALM− cells were strongly and significantly affected by the increase of membrane tension: stalled CCVs became evident in the kymographs (Fig. 4 D and E), the density of de novo CCVs decreased fourfold when compared to controls (Fig. 4F, CALM−) and stalled CCVs tripled in frequency (Fig. 4G, CALM−). These results supported our hypothesis that CALM is a major determinant in CCV formation by overcoming increases in membrane tension.
Since we observed that a small fraction of CCVs was still forming in CALM− cells exposed to 66%-hypo (Fig. 4F), we reasoned that those events could help us further establish that CALM supports CCV formation upon membrane tension increase. We hypothesized that if this was the case, more CALM molecules would be accumulated in the forming CCVs when membrane tension was increased. To this end, we plotted the amount of CALM recruited during CCV formation, expressed as maximum CALM-EGFP fluorescence intensity, in CALM+ and CALM− cells exposed to isotonic or 66%-hypo buffer. The maximum CALM fluorescence intensity measured in CCVs forming in CALM+ cells corresponded, on average, to roughly double the amount collected in CCVs forming in CALM− cells (CALM+: 2.34 ± 0.01 a.u.; CALM−: 1.21 ± 0.01 a.u. indicated as mean ± SE, Fig. 4 H and J, Right). By comparing the right panels of Fig. 4 H and J, we observed that 5% of CCVs forming in CALM+ cells collected the same or less amount of CALM recruited on average (indicated by the gray dotted line) by CALM− CCVs. This result suggested that CCVs can efficiently form in isotonic condition with roughly half of the amount of CALM recruited per CCV in CALM+ cells. Upon exposure of CALM+ cells to 66%-hypo buffer, a small but statistically significant increase in the amount of CALM recruited to forming CCVs was observed (from 2.34 ± 0.01 to 2.39 ± 0.02 a.u., P < 0.0001; Fig. 4H; also compare histograms left and right). A corresponding shift can also be observed in the cumulative distribution curve (Fig. 4I).
In CALM− cells exposed to 66%-hypo, the average amount of CALM recruited to CCVs was also significantly increased when compared to their isotonic controls (from 1.21 ± 0.01 to 1.40 ± 0.03 a.u.; P < 0.0001; Fig. 4J; also compare histograms left and right). This represents a ∼16% increase in recruited CALM in these CALM− cells compared to only a 2% increase in CALM+ cells observed under similar isotonic and 66%-hypo conditions as documented above. Even more striking, in the isotonic CALM− cells, only 7% of CCVs collected CALM corresponding to more than two fluoroscence a.u. while in 66%-hypo CALM− cells 20% of CCVs contained greater that two fluorescence a.u. (Fig. 4J; compare histograms left and right). Finally, an evident shift to the right in the cumulative distribution curve can also be observed comparing 66%-hypo to isotonic controls in CALM− cells (Fig. 4K). These results suggest that an increased amount of CALM helps counteracting membrane tension increase and facilitate CCV closure.
To further validate our findings, we designed an additional biochemical approach. We took advantage of a recently developed HaloTag ligand (Halo-PROTAC-E) (29) that induces posttranslational protein knock-down by targeting Halo-tagged chimera for proteolysis. A great advantage of this approach was that we could obtain complete CALM-Halo depletion in only 24 h (SI Appendix, Fig. S7A), while siRNA experiments often require two rounds of transfections and nearly 1 wk before obtaining silenced cells (44, 45). SUM-CALM-Halo cells depleted (CALM−) or not (CALM+) of CALM by treatment with Halo-PROTAC-E were exposed to either isotonic or hypotonic solution and probed for transferrin uptake by cytofluorometry (SI Appendix, Fig. S7B). Complete CALM depletion resulted in a significantly decrease in transferrin uptake (38% less than the untreated cells). While CALM+ transferrin internalization was not compromised by membrane tension increase, CALM− cells in hypotonic conditions internalized significantly less transferrin than CALM− cells in isotonic condition (28% less). These results support our previous conclusion that CALM promotes CCV formation upon membrane tension increase. Potentially, CALM’s contribution toward vesicle closure could be consequent to the insertion of an amphipathic helix (H0) present at the N terminus of CALM that has been shown to be involved in the regulation of CCV size (12). To evaluate this possibility, we designed a rescue experiment. SUM-CALM-Halo cells were transfected with a plasmid encoding either CALM-EGFP or a modified chimera missing the first 18 amino acids that include the H0 helix (ΔH0-CALM-EGFP). After 24 h, the cells were treated with PROTAC-E to ensure complete CALM-Halo depletion. The day after treatment, we measured transferrin uptake by cytofluorometry after exposure to either an isotonic or hypotonic solution to increase membrane tension. Interestingly, transfection of either CALM-EGFP or ΔH0-CALM-EGFP significantly rescued the phenotype by recovering transferrin internalization efficiency in cells exposed to isotonic or hypotonic solution (SI Appendix, Fig. S7B).
The results obtained suggest that CALM contributes to CCV formation since complete depletion affects transferrin uptake. Additionally, CALM-EGFP transfection could rescue the phenotype, suggesting that the effect is indeed dependent on CALM, while the presence of the H0 helix of CALM did not influence transferrin uptake in our cell system. Our findings are consistent with the hypothesis that CALM supports the formation of CCVs under increased membrane tension and that increased amounts of CALM facilitate this process.
Discussion
Over the last decades, live-cell imaging has contributed to our understanding of endocytosis integrating structural and biochemical information with real-time dynamics of the formation of endocytic cargoes (46, 47). Nevertheless, overexpression of fluorescently tagged proteins can lead to their mislocalization and data misinterpretation. This risk is negligible when the overexpressed proteins are part of a complex, since its final concentration is defined by the stoichiometry of each subunit, as demonstrated by the efficient tagging of AP2 and clathrin triskelia (3, 5, 35, 48). However, this effect is relevant when the overexpressed proteins act either independently or as oligomers, as it happens for dynamin (33, 34, 49), auxilin (50), CALM, and its homolog AP180 (20, 50). For this reason, we used a gene-editing approach to produce a cell clone expressing a fluorescent chimera of CALM without modifying its concentration. Our cell clones SUM-CALM-EGFP and SUM-CALM-Halo were fully functional and displayed similar endocytic dynamics to cells engineered to express fluorescently tagged subunits of AP2 [Fig. 2 and SI Appendix, Figs. S2 and S3, Aguet et al. (25), and Mino et al. (51)], one of the most used markers to study CCV formation (1, 46, 52). Therefore, CALM tagging by gene editing can be a useful alternative resource to study CCV dynamics.
We observed that SUM cells contain in their genome more than two copies of the adaptor protein CALM. Interestingly, copy number variations, which can involve gains or losses of genomic DNA, are unexpectedly frequent events that characterize genetic diversity in both physiological and pathological conditions (53). For example, the region of Chromosome 11 that includes CALM gene is frequently amplified, together with its transcript (54), in multiple myeloma cells, and it is associated with shorter patient survival (55). In addition, the same region is also amplified in breast cancer patients (56, 57). Therefore, the presence of more than two copies of CALM in SUM cells is not surprising even if SUM are a quasi-diploid cell line (26). These data suggest that CALM might improve the fitness of a specific subset of cancers.
During CCV formation, the recruitment of clathrin triskelia and adaptors, including CALM, drives the growth phase of the CCV (12). We observed by LLSFM that CCV dynamics are typically uniform over the entire cell surface for both CALM and AP2 (Fig. 2). However, a significant decrease in the frequency of short-lived CCVs on the ventral surface of SUM cells when compared to the dorsal surface (Fig. 2). Since lifetimes and maximum intensities are proportional to the size of the CCVs (5) and smaller CCVs have higher membrane curvature, our observation suggests that the increased membrane tension at the membrane–glass interface prevents the assembly of a fraction of small CCVs which may require higher CALM content to finish (12).
Upon partial CALM depletion, the recruitment of half of the total amount of CALM was sufficient to form CCVs in isotonic conditions but not upon membrane tension increase. Furthermore, the few CCVs forming under membrane tension increase in CALM-depleted cells recruited more CALM molecules than CCVs formed in isotonic condition in the same cells (Fig. 4). Because we lack a thorough estimation of the number of clathrin molecules, we cannot comment in absolute terms regarding the relative sizes of CCVs forming upon membrane tension increase. However, given the small footprint of CALM there is sufficient room on the CCV surface to recruit CALM molecules as needed, depending on the local membrane tension and cellular availability.
Complete CALM depletion was achieved by administering a proteolysis targeting compound to the completely edited SUM-CALM-Halo cells and resulted in a significant decrease in transferrin uptake (SI Appendix, Fig. S5). We efficiently reverted this phenotype by re-expressing CALM or its alternate version lacking the H0, N-terminal amphipathic alpha helix. This result suggests that the high concentration of CALM localized at the neck of clathrin-coated pits in SUM cells may support vesicle closure by protein–protein crowding (58) without the requirement of H0 insertion in the lipid bilayer.
However, contradictory reports are present in the literature, with CALM depletion having anywhere from minimal (45, 59) to significant (12, 44, 60) effects on transferrin uptake. Since CCV assembly is based on an extremely ductile system (61), these observed differences may reflect cell type–specific phenotypes dictated by the differential expression of CCV components (17) that may have, at least in part, overlapping functions (62). For instance, a publication that appeared while this work was in review reported that epsin can support endocytosis at high membrane tension (63). This observation together with ours suggests that epsin and CALM might have overlapping functions.
How do our experiments contribute to the general understanding of CME? In this work, we show how increasing amounts of CALM can support the formation of CCVs at higher membrane tension levels, probably promoting the closure of the CCV by narrowing the vesicle neck during budding in a dose-dependent manner. This conclusion derives from the observation that CALM is recruited until the very end of CCV assembly (Fig. 2 and SI Appendix, Fig. S2) and that static clathrin structures accumulate at the plasma membrane (Fig. 4) in CALM-depleted cells upon membrane tension increase.
Moreover, since the membrane tension exerted in our experiments is compatible with the physiological increases encountered during the late phases of the cell cycle (64), migration, and embryogenesis (42), our results can explain why CALM abrogation is lethal at early stages of mice development but produces milder effect in adult animals. Indeed, when CALM is conditionally knocked out in the hematopoietic stem cell compartment the only appreciable effect is microcytic anemia (21). In adult erythropoiesis, the differentiating cells progressively lose cell surface and their intracellular compartments, decreasing their volume and concomitantly increasing their membrane tension, that resolves when red blood cells shift their shape from round to biconcave (65). The increase in membrane tension in erythroblasts in the absence of CALM impairs CME, which is essential for iron uptake and hemoglobin production, resulting in microcytic anemia (21, 66). Finally, since CALM expression can increment more than 10 times from one tissue to another (67), our results suggest that the relative abundance of adaptors and other accessory proteins influence the efficiency of CME and can also explain why discordant results can be observed interrogating different cellular models.
In conclusion, we discovered that CALM supports CCV formation by counteracting membrane tension. This function, together with its role in imposing membrane curvature (12) and sorting a set of SNAREs (10), suggests why higher levels of this protein can protect against Alzheimer's disease (68, 69), why nonfunctional mutants can lead to alteration of selective populations of cells (60), and why it is essential for embryogenesis and erythropoiesis (21, 66).
Materials and Methods
Reagents.
Human breast cancer cells SUM159PT (SUM) and the corresponding gene edited AP2S1-EGFP (SUM-AP2-EGFP) were a kind gift of Dr. Tomas Kirchhausen (Harvard Medical School) together with the pCALM-EGFP mammalian expression vector. HeLa cells were a generous donation of Drs. Pier Paolo Di Fiore and Sara Sigismund (Istituto Europeo di Oncologia). The mammalian expression vector mRuby-CLTB was generously provided by Dr. Michael Davidson (Plasmid #55852, Addgene); Dr. Joachim Messing generously provided pUC19 vector (Plasmid #50005, Addgene) (70); the vectors for Cas9 mammalian expression were kindly contributed by Dr. Feng Zhang (Plasmids #48137 and #62988, Addgene). Mouse brain extracts were a kind gift of Dr. Kari Hoyt. Rabbit monoclonal antibodies against CALM (ab172962) and against AP2B1 (ab129168) were from Abcam as well as the rabbit polyclonal antibody against AP2S1 (ab128950) and AP180 (ab33898). These primary antibodies were used for Western blotting in conjunction with a horseradish peroxidase (HRP)-conjugated secondary (Bio-Rad). Genomic DNA was extracted using QIAamp DNA Mini Kit (Qiagen) or a DNeasy Blood & Tissue Kit, and plasmid DNA was amplified using Plasmid Miniprep or Midiprep Kits (Qiagen) or a PureLink HiPure Plasmid Filter Midiprep Kit (Thermo Fisher Scientific). Primers were obtained from IDT (Integrated DNA Technologies). Chemicals were purchased from Sigma-Aldrich unless specified otherwise.
Cell Handling.
Cells were grown at 37 °C and 5% CO2. SUM complete media consisted of F12/Glutamax (Thermo Fisher Scientific), supplemented with 5% fetal bovine serum (VWR), 100 U/mL penicillin and streptomycin (Thermo Fisher Scientific), 1 μg/mL hydrocortisone (H-4001; Sigma-Aldrich), 5 μg/mL insulin (128 to 100; Cell Applications), and 10 mM 4-(2-hydroxyethyl)-1- piperazine-ethane-sulfonic acid (Hepes), pH 7.4. For transient transfections, 4 × 10^5 SUM cells were plated in one 6-well plates well and transfected the following day with 1 μg of plasmid DNA and 3 μL of TransIT-2020 (Mirus Bio) according to the manufacturer’s instructions. Briefly, 1 μg of plasmid DNA was added to 250 μL Optimem followed by 3 μL of TransIT-2020 (Mirus Bio). After 30 min of incubation at room temperature, the mixture was added dropwise to the cells. Expression of mRuby-CLTB or AP2S1-EGFP were obtained by transient transfection. Cells were resuspended to be propagated or to be subjected to other experimental procedures by exposure to trypsin (Thermo Fisher Scientific) for 3 min at 37 °C, followed by a wash in 10 mL complete media and centrifugation at 300 g for 5 min. Cells were counted and plated as needed. Cell culture flasks and multi well plates were purchased from Greiner Bio-One.
Plasmid Modification and Assembly of Donor Templates to Promote Homologous Recombination in Knock-In Gene Editing.
Plasmids were modified using an approach based on isothermal assembly (71) and/or site directed mutagenesis using PCR amplification (72). We used the high-fidelity enzyme Phusion (New England BioLabs) to amplify the PCR products and the NEBuilder HiFi DNA Assembly kit (New England BioLabs) to ligate the products into final plasmids. Ligation products of the ligation reaction were transformed into competent DH5a Escherichia coli (5-alpha Competent E. coli) following the protocol provided by the company (New England BioLabs). For colony selection, bacteria were plated on Luria Broth agar plates supplemented with the appropriate antibiotic (ampicillin in the case of pUC19-derived vectors, kanamycin for pCALM-EGFP). Plasmids were extracted using QIAprep Miniprep Kit (Qiagen) or HiPure Plasmid Filter Midiprep Kit (Thermo Fisher Scientific) and sequenced to ensure that the correct modifications were in place.
Generation of ΔH0-CALM-EGFP vector.
The ΔH0-CALM-EGFP vector was obtained by PCR-mediated mutagenesis (72) using two specific primers (Fwd: 5′- GCC-ACCATGGGCTCTGCCGTATCCAAGAC-3′, Rev: 5′- AGAGCCCATGGTGGCGAGATC-TGAGTCCGGTAGCG-3′) that exclude the first 18 amino acids (which comprise the H0 helix, 5 to 18 aa) (12) from the original CALM-EGFP plasmid.
Generation of the templates for homologous recombination.
The templates to induce homologous recombination were obtained by two sequential cloning reactions using the isothermal assembly approach (71). The first reaction cloned into the pUC19 vector the genomic region of interest with the appropriate overlaps to promote homologous recombination. The second reaction inserted the desired modifications (e.g., protein tags). After obtaining sequence-verified vectors, we used site-directed mutagenesis to introduce silent mutations into the donor sequence corresponding to the protospacer adjacent motif (PAM) site and proximal residues of the targeting guide to prevent both the donor and the recombined locus from being further cleaved by Cas9 (73).
Generation of the CALM-EGFP donor vector.
To introduce EGFP at the C terminus of CALM, the template for homologous recombination consisted of a 2,000-bp DNA fragment around the stop codon of CALM, which was amplified and inserted into pUC19 vector (primers to linearize the puc19 vector: Fwd 5′-ATCCTCTAGAGTCGACCTG-3′, Rev 5′-TACCGAGCTCGAATTCAC-3′; primers to amplify and insert the CALM region adjacent to the last exon: 5′-gtcgactctagaggatctcgagCCTAAATCCCACAGAGGC-3′, 5′-gaattcgagctcggtactcgagCTTCTTGATGCTCTAGCC-3′; lowercase indicates the regions of homology with the linearized vector). Next, the EGFP open reading frame, preceded by a flexible linker encoding the amino acid sequence GGSGGSGGS (74), was inserted at the end of the last exon of CALM abolishing the physiological stop codon (primers to linearize the plasmid containing the C terminus CALM region: Fwd 5′-CATAAACTGTATCTGGAAAAAAAAAG-3′, Rev 5′- TAACTTGATGGAAGAAAATGG- -3′; primers to amplify the linker–EGFP sequence: Fwd 5′- ttccagatacagtttatgGGA-GGTTCTGGTGGTTCTG -3′, Rev 5′- attttcttccatcaagttaCTTGTACAGCTCGTCCATG- -3′; lowercase indicates the regions of homology with the linearized vector). A stop codon was inserted after the terminal amino acid of EGFP. To avoid the creation of a double-strand brake in the template for homologous recombination, modifications in the PAM site of the CALM-EGFP C-terminal donor were introduced by site-directed multiple plasmid mutagenesis (72) (primers: Fwd 5′-TAACTCAGTGTACTCCCATGAAGGAAATTGGGACTAGTTTAT-3′ and Rev 5′-ATG-GGAGTACACTGAGTTACTTGTAGCATTCTAATGGAAGAA-3′).
Generation of the CALM-Halo donor vector.
To modify the exon 19 region, the homologous recombination template consisted of a 1,484-bp genomic DNA fragment encapsulating CALM exon 19 and a portion of the adjacent intronic regions. This region was amplified by PCR and inserted into the pUC19 vector (primers to linearize the puc19 vector: Fwd 5′-ATCCTCTAGAGT-CGACCTG-3′, Rev 5′-TACCGAGCTCGAATTCAC-3′; primers to amplify and insert the CALM region adjacent to the exon 19: Fwd 5′-gccagtgaattcgagctcggTCTG-CTGAGAAACTCAGGCCA-3′, Rev 5′- ctgcaggtcgactctagaggAGCCTTAGAAACA-CGTCAAGAGGA-3′; lowercase indicates the regions of homology with the linearized vector). Then, DNA sequences encoding the last exon of CALM, a flexible linker (sequence GGSGGSGGS) (74) and the protein marker HaloTag (Promega) were inserted in frame following exon 19 (primers to linearize pUC19-exon19 vector: Fwd 5′-GTATTACACACTACATGTAACTG-3′ and Rev CTGTGCTCCTGATACAGG; primer to amplify linker-Halo sequence and insert last CALM exon: Fwd 5′-gccctgtatcaggagcacagATACAGTTTATGGGAGGTTC-3′ and Rev 5′-ttacatgtagtgtgtaatacctgTTAAATCTCGAGCGTCGAC-3′). A stop codon was encoded after the terminal amino acid of HaloTag. Finally, modifications in the sequence of the CALM-Halo donor vector were introduced by site-directed mutagenesis (72) to avoid the creation of double-strand brake in the template for homologous recombination (primers: Fwd 5′-GGCCCAGTT-AGCGGAGCACAGATACAGTTTATGGGAG-3′ and Rev 5′-CCGCTAACTGGGCCA-AAGGGGTTTGGAGGTCTCATG-3′).
Gene Editing of SUM Cells.
SUM159PT cells were gene edited to insert EGFP at the C terminus of CALM gene or HaloTag at the 3′ of exon 19 of CALM gene using a CRISPR/Cas9 protocol based on a free PCR strategy (24, 25). This approach was accomplished by transfecting the cells with a mixture of three components: a DNA fragment composed of a U6 promoter flanked by a specific RNA guide to target Cas9, a DNA template to promote homologous recombination (see Generation of the Templates for Homologous Recombination), and a plasmid encoding Cas9.
Generation of the DNA fragments containing the specific guides for Cas9.
A DNA fragment containing the promoter U6 and the specific DNA sequence required to produce the RNA scaffold encoding the guide for Cas9 was generated by PCR amplification using 5′- GCCGGTACCTGAGGGCCTATTTCCC-3′ Fwd primer and the Rev primer 5′-ACCTCTAGAAAAAAAGCACCGACTCGG-TGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAACNNNNNNNNNNNNNNNNNNNNCGGTGTTTCGTCC TTTCCACAAG-3′, in which the N nucleotides represent the reverse complement of the target sequences for either the 3′ untranslated region (UTR) of the CALM gene (5′-TGCTACAAG-TAACTCAGTGA-3′) or the coding sequence of exon 19 (5′-AACCCCTTTGGCCCT-GTATC-3′).
Transfection of SUM cells and clone selection.
SUM159PT cells (1 × 106) were plated in a well of a 6-well plate and transfected using Lipofectamine 3000 (Thermo Fisher Scientific) to deliver 800 ng of a plasmid to express Cas9, 2,500 ng of the donor repair template, and 150 ng of the PCR DNA fragment composed of the U6 primer and the RNA guide. The DNA components were dissolved in a total volume of 125 μL of Optimem supplemented with P3000 reagent into which 125 μL Optimem supplemented with 5 μL of Lipofectamine 3000 was added and incubated for 5 min at room temperature. The transfection mixture was then added dropwise to the cells. Identification and selection of SUM cells modified to retain EGFP or Halo in CALM loci were performed by combining fluorescence-activated cell sorting with genomic analysis by PCR (25, 34): the transfected cells were expanded to reach confluence in a T75 dish, resuspended, and sorted to enrich for EGFP- or Halo-positive cells. SUM-CALM-Halo cells were exposed for 1 h to 5 nM of JF549-HaloTag Ligand [a generous gift of Dr. Luke Lavis (75)] diluted in SUM cell media. Cells with a fluorescence signal significantly above the control were selected and expanded. PCR amplification of genomic DNA from the enriched population confirmed the presence of cells that underwent the correct homologous recombination. Clonal populations were created by diluting the enriched population and seeding one cell per well in 96-well plates. Cell clones completely edited were identified by PCR amplification of genomic DNA and verified by Western blot analysis.
Genomic PCR screening.
Genomic DNA was extracted using QuickExtract DNA Extraction Solution (Lucigen) or QIAamp DNA Mini Kit (Qiagen) or a DNeasy Blood & Tissue Kit (Qiagen). PCR reactions were performed using Hot Start Taq DNA Polymerase (New England BioLabs). The genomic PCR screening strategy to identify EGFP insertion at CALM C-terminal was based on two reactions. The first strategy used a Fwd primer designed to bind to the last exon of CALM (5′-TGA-ACACTTCTGGGCCAGTT-3′), while the second employed a FW primer complementary to EGFP (5′-GCCCTGAGCAAAGACCCCA-3′). The Rev primer for both reactions was complimentary to a region of 3′ UTR of CALM and was located downstream of the donor (5′-GACATGCCAAGCACAAAGCA-3′) to avoid the identification of false positives due to amplification of the donor plasmid. While promising CALM-EGFP clones were discovered with these strategies, a supplemental PCR screening demonstrated that the clones identified using the two original primer pairs were only partially edited. This reaction was performed using the two following primers: Fwd 5′-TGAACACTTCTGGGCCAGTT-3′ and Rev 5′-TGCTGGAAGTAAGGATTTTGCTGC-3′. To identify the insertions at the end of CALM exon19, we used a primer designed to bind in exon19 (Fwd 5′-CCTGTA-ATGACGCAACCAACCT-3′) and a second in the intronic region at the 3′ end of exon19 (Rev 5′-GTCAAAACAGTACGAGCAGAGC-3′). Amplified genomic inserts corresponding to the homologous recombination region were sequenced to confirm that the expected modification took place.
Cell Extract Preparations and Western Blotting.
Cells were detached, washed three times in phosphate-buffered saline (PBS), and solubilized at 4 °C in lysis buffer (50 mM Hepes pH 7.4, 150 mM NaCl, 15 mM MgCl2, 1 mM EGTA, 10% glycerol, and 1% Triton X-100) supplemented with a protease inhibitor mixture (Roche). Nuclei were removed from the lysate by low-speed centrifugation (800 g, 5 min, and 4 °C, Centrifuge 5424, Eppendorf). The protein concentration of the samples was determined by Bicinchoninic acid (BCA) assay (Pierce). Samples were loaded in precast sodium dodecyl sulfate polyacrylamide gels (Mini-PROTEAN TGX gels, Bio-Rad) and run at room temperature with 100 V (constant) before being transferred to polyvinylidene fluoride or nitrocellulose membranes using the Trans-Blot Turbo system (Bio-Rad) for 10 min at 2.5 A. Membranes were stained with Ponceau S solution then saturated in blocking buffer composed of Tris Buffer Saline (150 mM NaCl and 50 mM Tris pH 7.4) supplemented with 5% nonfat dry milk and 0.05% Tween20. The membranes were then incubated for 1 h with the appropriate primary antibodies diluted in blocking buffer. After three washes of 5 min apiece in blocking buffer, the membranes were incubated with 1 μg/mL of secondary antibody for 1 h. After extensive washes in Tris-buffered saline, the chemiluminescent signal was elicited by 5 min incubation with enhanced chemiluminescence substrate (ECL, Pierce, Thermo Fisher Scientific) and captured photographically on film (CL-XPosure, Thermo Fisher Scientific).
CALM Depletion by siRNA Treatment and mRuby-Clathrin Transfection in SUM Cells.
Two rounds of transfection were performed to obtain efficient silencing of the proteins of CALM or CALM-EGFP. First, 5 × 104 SUM-CALM-EGFP cells were plated in 24-well dishes. For each well, a transfection mixture containing 50 μL Optimem, 1.3 μL of 10 μM siRNA, and 0.5 μL TransIT-X2 (Mirus Bio) was prepared. After 30 min, the mixture was added dropwise to the well. After 24 h, the cells were resuspended and plated in a well of a 6-well plate. A second round of transfection was performed by adding a mixture of 250 μL Optimem, 6.5 μL of 10 μM siRNA, and 2.5 TransIT-X2 dropwise to the well. When clathrin expression was required, 2.5 μg mRuby-Clathrin plasmid was added to the second transfection mixture. The control cells were treated the same, except that scramble siRNA was administered. After 48 h, the cells were either plated on coverslips for imaging or processed for Western blot analysis to assess CALM expression. Four siRNA oligos (Dharmacon, Horizon Discovery) were tested to find the most effective: 5′-CAACAGGCAUGAUAG-GAUA-3′; 5′-GUUCAAAGAUGCCAUUAGA-3′; 5′-GUAAUGGCCUAUCCUGCUA-3′; and 5′-CAUUACAACUCAUCAUUUG-3′. In our hands, the most effective was 5′-GUAAUGGCCUAUCCUGCUA-3′. We targeted EGFP with the siRNA: 5′ GCCACA-ACGUCUAUAUCAU-3′. We used a nontargeting siRNA (5′-UGGUUUACAUGU-CGACUAA-3′) as control.
CALM Depletion by PROTAC and Rescue Evaluated by Transferrin Uptake.
In each well of a 6-well plate 4 × 105 SUM-CALM-Halo cells were placed. Two wells were transfected with p-CALM-EGFP, and two were transfected with p-ΔH0-CALM-EGFP using TransIT-2020 as described in the cell-handling section. The day after, each cell group was split across six wells of a 24-well plate to accommodate the six conditions for a complete transferrin uptake assay [isotonic solution 37 °C, hypotonic solution 37 °C, and isotonic solution 4°, each in duplicates to include or not an acid wash to exclude surface bound transferrin (34)]. The cells were then exposed to 500 nM of Halo-PROTAC-E Degrader (Medical Research Council Reagents and Services, MRC-PPU) or its solvent (dimethyl sulfoxide, DMSO) for 24 h. Afterward, the cells were processed for transferrin uptake or immunoblotting analysis. The transferrin uptake assay was performed as follows: SUM cells were washed with Hank's balanced salt solution (HBSS) and then exposed for 10 min at 37 °C to 0.5 μg/mL Transferrin-Alexa Fluor 647 (A647-Tf, Thermo Fisher Scientific), dissolved either in isotonic solution composed of minimal essential medium (MEM) alpha supplemented with 1% fetal bovine serum (FBS) or in the hypo osmotic solution composed of MEM alpha diluted with 40% of water and supplemented with 1% FBS. After 10 min of exposure to transferrin-enriched buffers, the dish containing the cells were moved on ice to block endocytosis and washed with ice-cold HBSS. To measure the amount of transferrin receptor present at rest on the cell surface, cells in other wells were exposed to the isotonic solution supplemented with A647-Tf at 4 °C. To remove the surface-bound Transferrin-A647, some wells were exposed twice to an acidic solution for 1 min (150 mM NaCl and 0.1 M glycine, pH 2.5). After two additional washes in ice cold HBSS, the cells were detached by trypsinization, collected in flow tubes containing 1 mL of media to inactivate trypsin, centrifuged at 300 g, and resuspended in MEM alpha 1% FBS at 4 °C. The samples were then quantified by flow cytometry (Gallios, Beckman Coulter), and the resulting data were analyzed by Kaluza software (Beckman Coulter). Transferrin internalization was normalized to the amount of surface-bound transferrin and to the amount of transferrin internalized by SUM-CALM-Halo cells depleted of CALM and exposed to isoosmotic solution (76). The experiment was repeated four times, and the normalized means obtained across the experiments were compared using an unpaired t test (GraphPad Prism; GraphPad Software).
Glass Coverslip Preparation and Cell Plating.
Glass coverslips (#1.5; Warner Instruments) with a diameter of 5 mm for LLSFM or 25 mm for spinning disk confocal fluorescence microscopy were cleaned by sonication (Ultrasonic Cleaner Branson 1800, Branson) in ethanol and dried at 120 °C. EGFP molecules were captured on clean coverslips to calibrate the fluorescence intensity signal (77) by spinning-disk fluorescence microscopy (33, 34). After resuspension, 5,000 to 10,000 SUM cells were plated on 5-mm diameter coverslips while 50,000 to 100,000 cells were plated on 25-mm diameter coverslips. The cells were left in the incubator for 3 h to allow complete spreading before starting imaging. SUM-CALM-Halo cells were exposed for 1 h to 5 nM of JF549-HaloTag Ligand [a generous gift of Dr. Luke Lavis (75)] diluted in SUM cell media. After an hour of washes in SUM cell media, the cells were imaged.
LLSFM Live Cell Imaging, Microscopy Calibration, and Analysis.
We imaged SUM cells with a lattice light-sheet fluorescence microscope (Intelligent Imaging Innovation). Directly derived from the instrument patented by Dr. Eric Betzig and colleagues, this system utilizes Bessel beam lattice sheet illumination via cylindrical lenses and high-speed spatial light modulator (SLM) for multicolor imaging, an annular mask array for the formation of various light sheets, and galvo mirrors to control lattice movement in x and z. It has cameras in the image and Fourier space to inspect the lattice and annular mask shapes. Assembled on an active vibration isolation table equipped with active and passive system for vibration cancellation (TMC, Ametek), it is equipped with four laser lines for sample excitation (405 nm, 300 mW, Omicron; 488 nm, 300 mW, Coherent; and 560 nm, 500 mW and 642 nm, and 500 mW, MPB Communication) and the required filter sets necessary to efficiently image two color sets at a time (Semrock). Two water immersion objectives control the illumination (28×/0.67 NA) and detection (25×/1.1 NA, expanded to reach a final 62.5× magnification). Two piezo controllers operate the translations of the stage and of the objective. The instrument is also equipped with a temperature-controlled specimen chamber 90 × 1,500 (36″ x 60″) held at 37 °C. Images are acquired by two high-speed high-resolution 2Kx2K sCMOS camera with 82%QE (ORCA-Flash4.0 V3, Hamamatsu). A Computer Work Station (Dell) is equipped with the software SlideBookTM 6 (Intelligent Imaging Innovation) that fully controls the instrument (high-speed synchronization of laser firing, SLM pattern display, galvo movements, and imaging camera readout along with the ability to de-skew and view data).
Acquisition of the Movies.
To acquire the images, we illuminated the sample with a dithered light sheet generated in the square lattice configuration (31), using an excitation NA of 0.55/0.48 and full width at half maximum (FWHM) beam length of 15 μm. SUM-CALM-EGFP clone 16 and SUM-AP2-EGFP cells were excited for 20 ms with the 488-nm laser at a power of 12 mW measured before the cylindrical lenses. The images were acquired in objective scan mode, which offers better yz resolution (78). The objectives were translated in discrete steps of 250 nm for 75 consecutive images resulting in a total imaged volume of 50 × 50 × 18.25 μm. To generate ∼5-min movies, 120 volumes were acquired every 2.8 s SUM-CALM-Halo cells were excited for 35 ms with the 561-nm laser at a power of 9 mW measured before the cylindrical lenses (500-mW laser, acousto-optic tunable filter 1 [AOTF1]). The objectives were translated in discrete steps of 300 nm for 61 consecutive images resulting in a total imaged volume of 50 × 50 × 18.3 μm. To generate ∼5-min movies, 150 volumes were acquired every 2.2 s.
LLSFM calibration.
Cytosolic extracts of SUM-CALM-Halo A3 cells were used to determine the fluorescence intensity of a single CALM-Halo molecule labeled with JF549 by single-step photobleaching under conditions of continuous LLSFM illumination (77). The cytosolic extract was diluted ∼1,000 times in PBS and then placed on top of a freshly glow-discharged glass coverslip for 5 min. The samples were subjected to continuous illumination, using 35-ms exposure and 500-mW laser power, AOTF = 100. For consistency, the volumes were collected as SUM-CALM-Halo cells were (discrete steps of 300 nm for 61 consecutive images resulting in a total imaged volume of 50 × 50 × 18.3 μm). The fluorescence intensity of diffraction-limited spots was determined as the amplitude of the fitted three-dimensional Gaussian function. The fluorescence intensity traces of continuously illuminated single fluorescent molecules were fitted using a step-fitting function (79). The mean of fluorescence intensity loss associated with a photobleaching step corresponded to the intensity of a single CALM-Halo labeled with JF549 HaloTag Ligand. To normalize the obtained single molecule intensity to the excitation used to acquire the cell data set, we calibrated the AOTF by imaging JF549 diluted in media at increasing AOTF values.
Analysis.
Forming CCVs were detected and tracked using a three-dimensional extension of cmeAnalysis (25). Given their physical size, clathrin coats appear as the LLSFM spread point function, which can be approximated to a three-dimensional Gaussian function:
where C indicates the local background, A the intensity, [μx, μy, μz] the position of the centroid of the object, and σ indicates the SD which is equal along the x and y dimensions (σxy) but differs in the z dimension (σz). We fitted fluorescent beads (100 nm in diameter, F8803 Fluospheres, Thermo Fisher Scientific, Life Technologies) to estimate σxy, σz. The averaged result was used as input in the detection function. The tracking algorithm was performed using the following parameters: tracking radius, 3 and 6 and tracking gap length, 2. Movies and three-dimensional renderings were obtained using a five-dimensional viewer (80). The lifetime distributions of CCVs detected on the dorsal and ventral surface of the cells were compared using a KStest. All the software was run in MatLab (Mathworks).
Confocal Live Cell Imaging, Fluorescence Calibration, and Analysis.
The imaging system was an Eclipse TI-E microscope (Nikon) equipped with a temperature-controlled chamber, a CSU-W1 spinning disk confocal unit (Yokogawa Electric Corporation), a 100× objective lens (Nikon CFI Plan-Apochromat Lambda, NA 1.45), an Electron Multiplying Charge Coupled Device (EMCCD) camera (iXon DU897 Ultra; Andor Technology), and 488- and 560-nm excitation lasers with 100 mW of nominal power. Image acquisition was done using NIS Elements software. All confocal movies were 5 min long, taken at 0.5 frames per second with 100 ms exposure time per channel. Cells were exposed to hypo osmotic media for 3 to 10 min before the acquisition of time series, which resulted in movies spanning from 3 to 15 min of exposure to hypo osmotic solution. We calibrated our confocal microscope to the intensity of a single EGFP as previously described (77). Briefly, free EGFP molecules were captured on clean 25-mm coverslips. These samples were then imaged with 4 s or 8 s of exposure per frame for 2 to 5 min. The resulting movies were then tracked using cmeAnalysis (77). Instances of step-like bleaching events were manually selected. Intensity of puncta was calculated as the mean intensity of a 5 × 5 pixel window around the puncta minus a background value calculated as the median intensity of the 24 pixels immediately surrounding the signal window. Detection and tracking were done using the cmeAnalysis MATLAB package (35). The parameters used were the following: numerical aperture, 1.45; magnification, 100; pixel size, 16; and marker, “egfp.” The output traces were then filtered using previously described trace-rejection algorithm (14), which rejects traces that lack either a linear intensity growth period or a linear intensity reduction period (there must be four-frame windows at the beginning and end, where the linear fit of the intensity graph is good [R2 > 0.75] and that have a positive and negative slope at the beginning and end, respectively). Moreover, we developed an algorithm for screening all clathrin fluorescent traces for true scission events. The goal thereof was to create a set of criteria that identified most events while strictly avoiding false positives, so that quantities of CALM at scission could be measured. Briefly, this algorithm scanned all traces for spans of fast, linear intensity decrease (average intensity decrease of 6% of maximum intensity per second over a 12-s period, with linear fit with R2 ≥ 0.8) that ended with a time-point at least as dim as a manually selected threshold intensity. A maximum displacement per frame of 320 nm was also imposed to preclude tracking errors. We manually verified the tracks in conditions of especially low scission rates in which a few false positives could be problematic. All two-channel tracking was done only using data from the 560-nm (clathrin) channel in order to not introduce bias to siRNA cells with lower 488 (CALM) signal. Intensity in both channels was then calculated using the pixel–sum scheme described above. Hierarchical trace clustering was done with our previously described MATLAB software createTraceLibrary (14) with 10 clusters, no minimum cluster size, and a background value of 1. Input traces were scaled to 100 time-points using linear interpolation so that the clusters would form based on trace shape independent of trace lifetime.
Supplementary Material
Acknowledgments
We thank Jack Yalowich for critical discussion and editorial help. We thank Dr. Umida Djakbarova for the acquisition of three-dimensional confocal movies that assess cell expansion upon hypotonic shock. We acknowledge the use of Microscopy, Genomics, and Flow Cytometry Shared Resources at The Ohio State University, which are supported by the National Cancer Institute (NCI, P30CA016058). We thank the Advanced Imaging Center (AIC) at Janelia Research Campus for training on their LLSFM, in particular John M Heddleston, Eric Wait, Satya Khuon, and Teng-Leong Chew from the AIC team. The AIC is jointly supported by the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation. E.C. was supported by the OSU Comprehensive Cancer Center using Pelotonia funds (Young Investigator Award, IRP46050-502339) and the Ohio Cancer Research via the McCurdy/Kimball Midwest Research Fund. C.K. was supported by NSF Faculty Early Career Development Program (Award 1751113) and NIH R01GM127526. F. C. was supported by an American-Italian Cancer Foundation Post-Doctoral Research Fellowship and by a Pelotonia Post-Doctoral Fellowship.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. M.G.G.J.F. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010438118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
Change History
June 25, 2021: The edited by line has been updated.
References
- 1.Kirchhausen T., Owen D., Harrison S. C., Molecular structure, function, and dynamics of clathrin-mediated membrane traffic. Cold Spring Harb. Perspect. Biol. 6, a016725 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mettlen M., Chen P.-H., Srinivasan S., Danuser G., Schmid S. L., Regulation of clathrin-mediated endocytosis. Annu. Rev. Biochem. 87, 871–896 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cocucci E., Aguet F., Boulant S., Kirchhausen T., The first five seconds in the life of a clathrin-coated pit. Cell 150, 495–507 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loerke D., et al., Cargo and dynamin regulate clathrin-coated pit maturation. PLoS Biol. 7, e57 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ehrlich M., et al., Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell 118, 591–605 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Traub L. M., Regarding the amazing choreography of clathrin coats. PLoS Biol. 9, e1001037 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borner G. H. H., et al., Multivariate proteomic profiling identifies novel accessory proteins of coated vesicles. J. Cell Biol. 197, 141–160 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blondeau F., et al., Tandem MS analysis of brain clathrin-coated vesicles reveals their critical involvement in synaptic vesicle recycling. Proc. Natl. Acad. Sci. U.S.A. 101, 3833–3838 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ford M. G., et al., Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science 291, 1051–1055 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Miller S. E., et al., The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM. Cell 147, 1118–1131 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moshkanbaryans L., Chan L. S., Graham M. E., The biochemical properties and functions of CALM and AP180 in clathrin mediated endocytosis. Membranes (Basel) 4, 388–413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller S. E., et al., CALM regulates clathrin-coated vesicle size and maturation by directly sensing and driving membrane curvature. Dev. Cell 33, 163–175 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boulant S., Kural C., Zeeh J. C., Ubelmann F., Kirchhausen T., Actin dynamics counteract membrane tension during clathrin-mediated endocytosis. Nat. Cell Biol. 13, 1124–1131 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferguson J. P., et al., Deciphering dynamics of clathrin-mediated endocytosis in a living organism. J. Cell Biol. 214, 347–358 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saleem M., et al., A balance between membrane elasticity and polymerization energy sets the shape of spherical clathrin coats. Nat. Commun. 6, 6249 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willy N. M., et al., Membrane mechanics govern spatiotemporal heterogeneity of endocytic clathrin coat dynamics. Mol. Biol. Cell 28, 3480–3488 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dambournet D., et al., Genome-edited human stem cells expressing fluorescently labeled endocytic markers allow quantitative analysis of clathrin-mediated endocytosis during differentiation. J. Cell Biol. 217, 3301–3311 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mettlen M., et al., Endocytic accessory proteins are functionally distinguished by their differential effects on the maturation of clathrin-coated pits. Mol. Biol. Cell 20, 3251–3260 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyerholz A., et al., Effect of clathrin assembly lymphoid myeloid leukemia protein depletion on clathrin coat formation. Traffic 6, 1225–1234 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Tebar F., Bohlander S. K., Sorkin A., Clathrin assembly lymphoid myeloid leukemia (CALM) protein: Localization in endocytic-coated pits, interactions with clathrin, and the impact of overexpression on clathrin-mediated traffic. Mol. Biol. Cell 10, 2687–2702 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishikawa Y., et al., Role of the clathrin adaptor PICALM in normal hematopoiesis and polycythemia vera pathophysiology. Haematologica 100, 439–451 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng F.-F., et al., Association of PICALM gene polymorphisms with Alzheimer’s disease: Evidence from an updated meta-analysis. Curr. Alzheimer Res. 16, 1196–1205 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Doyon J. B., et al., Rapid and efficient clathrin-mediated endocytosis revealed in genome-edited mammalian cells. Nat. Cell Biol. 13, 331–337 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ran F. A., et al., Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aguet F., et al., Membrane dynamics of dividing cells imaged by lattice light-sheet microscopy. Mol. Biol. Cell 27, 3418–3435 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forozan F., et al., Molecular cytogenetic analysis of 11 new breast cancer cell lines. Br. J. Cancer 81, 1328–1334 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Los G. V., et al., HaloTag: A novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 3, 373–382 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Zheng Q., Lavis L. D., Development of photostable fluorophores for molecular imaging. Curr. Opin. Chem. Biol. 39, 32–38 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Tovell H., et al., Rapid and reversible knockdown of endogenously tagged endosomal proteins via an optimized HaloPROTAC degrader. ACS Chem. Biol. 14, 882–892 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Batchelder E. M., Yarar D., Differential requirements for clathrin-dependent endocytosis at sites of cell-substrate adhesion. Mol. Biol. Cell 21, 3070–3079 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen B.-C. B. C., et al., Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science 346, 1257998 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaqaman K., et al., Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods 5, 695–702 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kural C., et al., Dynamics of intracellular clathrin/AP1- and clathrin/AP3-containing carriers. Cell Rep. 2, 1111–1119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cocucci E., Gaudin R., Kirchhausen T., Dynamin recruitment and membrane scission at the neck of a clathrin-coated pit. Mol. Biol. Cell 25, 3595–3609 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aguet F., Antonescu C. N., Mettlen M., Schmid S. L., Danuser G., Advances in analysis of low signal-to-noise images link dynamin and AP2 to the functions of an endocytic checkpoint. Dev. Cell 26, 279–291 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ulbrich M. H., Isacoff E. Y., Subunit counting in membrane-bound proteins. Nat. Methods 4, 319–321 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balleza E., Kim J. M., Cluzel P., Systematic characterization of maturation time of fluorescent proteins in living cells. Nat. Methods 15, 47–51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heim R., Cubitt A. B., Tsien R. Y., Improved green fluorescence. Nature 373, 663–664 (1995). [DOI] [PubMed] [Google Scholar]
- 39.Boisvert F. M., et al., A quantitative spatial proteomics analysis of proteome turnover in human cells. Mol. Cell. Proteomics 11, 011429 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alber A. B., Paquet E. R., Biserni M., Naef F., Suter D. M., Single live cell monitoring of protein turnover reveals intercellular variability and cell-cycle dependence of degradation rates. Mol. Cell 71, 1079–1091.e9 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Scott B. L., et al., Membrane bending occurs at all stages of clathrin-coat assembly and defines endocytic dynamics. Nat. Commun. 9, 419 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferguson J. P., et al., Mechanoregulation of clathrin-mediated endocytosis. J. Cell Sci. 130, 3631–3636 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dai J., Sheetz M. P., Regulation of endocytosis, exocytosis, and shape by membrane tension. Cold Spring Harb. Symp. Quant. Biol. 60, 567–571 (1995). [DOI] [PubMed] [Google Scholar]
- 44.Wang X., et al., DASC, a sensitive classifier for measuring discrete early stages in clathrin-mediated endocytosis. eLife 9, e53686 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang F., Khvorova A., Marshall W., Sorkin A., Analysis of clathrin-mediated endocytosis of epidermal growth factor receptor by RNA interference. J. Biol. Chem. 279, 16657–16661 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Mettlen M., Danuser G., Imaging and modeling the dynamics of clathrin-mediated endocytosis. Cold Spring Harb. Perspect. Biol. 6, a017038 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kural C., Kirchhausen T., Live-cell imaging of clathrin coats. Methods Enzymol. 505, 59–80 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor M. J., Perrais D., Merrifield C. J., A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 9, e1000604 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grassart A., et al., Actin and dynamin2 dynamics and interplay during clathrin-mediated endocytosis. J. Cell Biol. 205, 721–735 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao X., et al., Expression of auxilin or AP180 inhibits endocytosis by mislocalizing clathrin: Evidence for formation of nascent pits containing AP1 or AP2 but not clathrin. J. Cell Sci. 114, 353–365 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Mino R. E., Chen Z., Mettlen M., Schmid S. L., An internally eGFP-tagged α-adaptin is a fully functional and improved fiduciary marker for clathrin-coated pit dynamics. Traffic 21, 603–616 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McMahon H. T., Boucrot E., Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 12, 517–533 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Shlien A., Malkin D., Copy number variations and cancer. Genome Med. 1, 62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Largo C., et al., Identification of overexpressed genes in frequently gained/amplified chromosome regions in multiple myeloma. Haematologica 91, 184–191 (2006). [PubMed] [Google Scholar]
- 55.Gutiérrez N. C., et al., Prognostic and biologic significance of chromosomal imbalances assessed by comparative genomic hybridization in multiple myeloma. Blood 104, 2661–2666 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Cerami E., et al., The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao J., et al., Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stachowiak J. C., et al., Membrane bending by protein-protein crowding. Nat. Cell Biol. 14, 944–949 (2012). [DOI] [PubMed] [Google Scholar]
- 59.Xiao Q., et al., Role of phosphatidylinositol clathrin assembly lymphoid-myeloid leukemia (PICALM) in intracellular amyloid precursor protein (APP) processing and amyloid plaque pathogenesis. J. Biol. Chem. 287, 21279–21289 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klebig M. L., et al., Mutations in the clathrin-assembly gene Picalm are responsible for the hematopoietic and iron metabolism abnormalities in fit1 mice. Proc. Natl. Acad. Sci. U.S.A. 100, 8360–8365 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen Z., Schmid S. L., Evolving models for assembling and shaping clathrin-coated pits. J. Cell Biol. 219, e202005126 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reider A., Wendland B., Endocytic adaptors—social networking at the plasma membrane. J. Cell Sci. 124, 1613–1622 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Joseph J. G., Osorio C., Yee V., Agrawal A., Liu A. P., Complimentary action of structured and unstructured domains of epsin supports clathrin-mediated endocytosis at high tension. Commun. Biol. 3, 743 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raucher D., Sheetz M. P., Membrane expansion increases endocytosis rate during mitosis. J. Cell Biol. 144, 497–506 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H., et al., Cytoskeleton remodeling induces membrane stiffness and stability changes of maturing reticulocytes. Biophys. J. 114, 2014–2023 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Suzuki M., et al., The clathrin assembly protein PICALM is required for erythroid maturation and transferrin internalization in mice. PLoS One 7, e31854 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lonsdale J.et al.; GTEx Consortium , The genotype-tissue expression (GTEx) project. Nat. Genet. 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parikh I., Fardo D. W., Estus S., Genetics of PICALM expression and Alzheimer’s disease. PLoS One 9, e91242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thomas R. S., et al., Decreasing the expression of PICALM reduces endocytosis and the activity of β-secretase: Implications for alzheimer’s disease. BMC Neurosci. 17, 50 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Norrander J., Kempe T., Messing J., Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 26, 101–106 (1983). [DOI] [PubMed] [Google Scholar]
- 71.Gibson D. G., et al., Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 72.Liu H., Naismith J. H., An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8, 91 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cho S. W., et al., Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 24, 132–141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reddy Chichili V. P., Kumar V., Sivaraman J., Linkers in the structural biology of protein-protein interactions. Protein Sci. 22, 153–167 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grimm J. B., et al., A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 12, 244–250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bhave M., Mettlen M., Wang X., Schmid S. L., Early and nonredundant functions of dynamin isoforms in clathrin-mediated endocytosis. Mol. Biol. Cell 31, 2035–2047 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aguet F., Cocucci E., “Measuring clathrin-coated vesicle formation with single-molecule resolution” in Methods in Molecular Biology, L. E. Swan, Ed. (Humana Press, New York, NY, 2018), vol. 1847, pp. 197–216. [DOI] [PubMed] [Google Scholar]
- 78.Gao R., et al., Cortical column and whole-brain imaging with molecular contrast and nanoscale resolution. Science 363, eaau8302 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Smith D. A., A quantitative method for the detection of edges in noisy time-series. Philos. Trans. R. Soc. Lond. B Biol. Sci. 353, 1969–1981 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wait E., Winter M., Cohen A. R., Hydra image processor: 5-D GPU image analysis library with MATLAB and python wrappers. Bioinformatics 35, 5393–5395 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.