Significance
Our work highlights the importance of addressing metabolite transport limitations in parallel with strain and culture optimization for production of plant-derived medicines and derivatives in engineered microbes. We show that supervised classification strategies can outperform conventional approaches when searching plant transcriptomes for metabolite transporters. Our discovery of two vacuolar tropane alkaloid (TA) exporters provides insight into the spatial organization associated with biosynthesis of these molecules. We illustrate how differences in transporter specificity and mechanisms can be leveraged to control accumulation of pathway products in an enhanced yeast-based TA production platform. By engineering this platform for production of TA derivatives using enzymes evolved for alkaloid detoxification, we reveal sources of biocatalysts for expanding biosynthetic diversity in heterologous hosts.
Keywords: tropane alkaloids, metabolic engineering, machine learning, plant natural products
Abstract
Microbial biosynthesis of plant natural products (PNPs) can facilitate access to valuable medicinal compounds and derivatives. Such efforts are challenged by metabolite transport limitations, which arise when complex plant pathways distributed across organelles and tissues are reconstructed in unicellular hosts without concomitant transport machinery. We recently reported an engineered yeast platform for production of the tropane alkaloid (TA) drugs hyoscyamine and scopolamine, in which product accumulation is limited by vacuolar transport. Here, we demonstrate that alleviation of transport limitations at multiple steps in an engineered pathway enables increased production of TAs and screening of useful derivatives. We first show that supervised classifier models trained on a tissue-delineated transcriptome from the TA-producing plant Atropa belladonna can predict TA transporters with greater efficacy than conventional regression- and clustering-based approaches. We demonstrate that two of the identified transporters, AbPUP1 and AbLP1, increase TA production in engineered yeast by facilitating vacuolar export and cellular reuptake of littorine and hyoscyamine. We incorporate four different plant transporters, cofactor regeneration mechanisms, and optimized growth conditions into our yeast platform to achieve improvements in de novo hyoscyamine and scopolamine production of over 100-fold (480 μg/L) and 7-fold (172 μg/L). Finally, we leverage computational tools for biosynthetic pathway prediction to produce two different classes of TA derivatives, nortropane alkaloids and tropane N-oxides, from simple precursors. Our work highlights the importance of cellular transport optimization in recapitulating complex PNP biosyntheses in microbial hosts and illustrates the utility of computational methods for gene discovery and expansion of heterologous biosynthetic diversity.
Microbial biosynthesis platforms are an effective strategy for synthesizing plant natural products (PNPs) that are otherwise economically, environmentally, or practically unsustainable to source directly from nature (1). Over the past two decades, high-profile microbial biosyntheses have been reported for several PNP-derived medicinal compounds of strong socioeconomic significance, such as the antimalarial artemisinin (2), opiates (3), cannabinoids (4), and tropane alkaloids (TAs) (5). Due to their genetic and metabolic tractability, microbial platforms can expand access to both natural and nonnatural PNP derivatives with unique bioactivities that may otherwise be produced in trace quantities in native plants, or which may only be accessible through derivatization using hazardous chemicals (6). The recent development of computational tools such as ATLASx (7) and BridgIT (8) for biosynthetic pathway expansion and noncanonical enzyme activity prediction can enable engineered microbial hosts to produce novel PNP derivatives de novo (9) rather than by converting expensive prederivatized precursors into cognate products (10). As efforts to engineer de novo production of PNP derivatives are often hampered by low enzyme tolerance for nonnative substrates or transformations, new approaches for optimizing pathway flux and improving accumulation of starting molecules for derivatization studies are critical for expanding chemical diversity within heterologous biosyntheses. Plant secondary metabolism exhibits complex spatial organization, as pathways for new PNPs have evolved to take advantage of the unique biochemistries found in different tissues and organelles. To date, several families of plant small-molecule transporters including adenosine triphosphate (ATP)-binding cassette (ABC) (11), multidrug and toxin extrusion (MATE) (12, 13), purine uptake permease-like (PUP) (14), and nitrate transporter 1/peptide transporter (NRT/NPF) (15) have been shown to be recruited into alkaloid biosynthesis to alleviate cellular transport limitations. Thus, recapitulating the plant transportome in the context of a microbial host may prove an effective general strategy for improving heterologous production of PNPs and their derivatives.
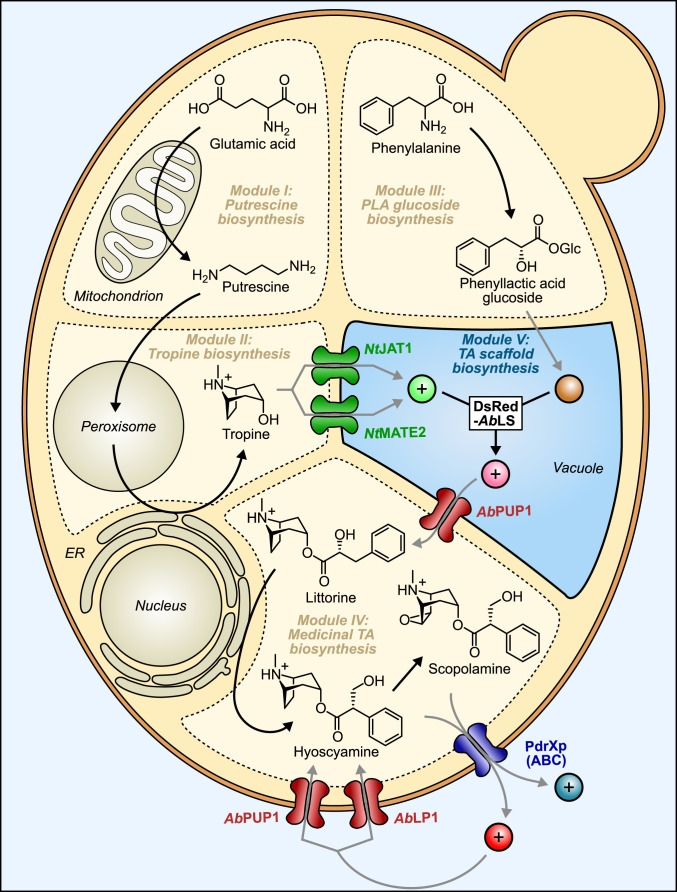
In nightshade plants (Solanaceae), biosynthesis of TAs—a class of anticholinergic molecules used to treat neuromuscular diseases (16)—is distributed across several intracellular compartments in roots, necessitating a diverse repertoire of molecular transporters. Early stages of TA production appear to have recruited enzymes from amino acid and polyamine metabolism found in the chloroplast (17), peroxisomes (18), and cytosol, and two different steps in the biosynthesis of hyoscyamine and scopolamine are catalyzed by endoplasmic reticulum (ER)–localized cytochromes P450 (19, 20). Notably, tropine and phenyllactic acid (PLA) glucoside, the acyl acceptor and donor moieties of the TA scaffold, must be transported from the cytosol across the tonoplast, where they are esterified by littorine synthase in the vacuole lumen; vacuolar littorine must then be exported back across the tonoplast into the cytosol for conversion to hyoscyamine and scopolamine (5). We recently engineered a yeast biosynthetic platform for de novo production of these medicinal TAs, which incorporated a tonoplast MATE transporter from tobacco (NtJAT1) to improve tropine import to the yeast vacuole (5). However, export of vacuolar littorine to the cytosol remains a major limitation to TA accumulation in this system (Fig. 1) particularly due to the poor intrinsic membrane permeability of protonated alkaloids in the acidic vacuole (21). As no transporters for tropane esters have yet been reported, their discovery is a key target for increasing microbial TA production.
Fig. 1.
Cellular metabolite-transport limitations in yeast engineered for de novo production of TAs. TA biosynthesis is distributed across five metabolic modules (I through V) and five cellular compartments: mitochondrion, peroxisome, ER, vacuole, and cytosol. The central esterification reaction assembling the TA scaffold from tropine and PLA glucoside is catalyzed by littorine synthase (AbLS) with N-terminally fused fluorescent protein DsRed in the vacuole. For this reaction, substrates (tropine and PLA glucoside) must be imported to the vacuole lumen, and product (littorine) must be exported to the cytosol. Principal starting materials and products are shown for each module. Black arrows represent one or more biosynthetic steps (reference SI Appendix, Fig. S3 for complete pathway); gray arrows indicate metabolite transport steps critical to pathway flux and product accumulation. Charges (“+,” positive; no symbol, uncharged) of key intermediates and products relevant to transport are indicated. Protein names: NtJAT1, Nicotiana tabacum jasmonate-inducible alkaloid transporter 1; NtMATE2, N. tabacum multidrug and toxin extrusion transporter 2; AbPUP1, A. belladonna purine uptake permease-like transporter 1; AbLP1, A. belladonna lactose permease-like transporter 1; PdrXp, yeast pleiotropic drug resistance transporters.
Comparison of gene expression across different abiotic stressors, elicitors, and/or tissues is a popular strategy for identifying novel biosynthetic gene candidates from plant transcriptomes. These coexpression approaches typically employ linear correlation analyses such as linear regression and/or hierarchical clustering based on Euclidean distance to extract gene candidates based on similarities in transcript abundance profile with those of “bait” genes with known roles in the same pathway (22). This strategy has been used recently to identify missing enzymes in the biosynthesis of monoterpene indole alkaloids (MIAs) (23), colchicine (24) and TAs (5). While biosynthetic enzymes involved in the same secondary metabolic pathway may be more likely to share linear relationships in expression profile, as they have unique organismal roles and are therefore found primarily in the same small subset of tissues (22), this assumption of linearity may not hold true for nonenzymatic genes within the same pathway. Small molecule transporters may participate in the mobilization and storage of products and precursors in plant tissues other than the primary site of biosynthesis (21), producing deviations from the linear correlations in the tissue expression profile expected of biosynthetic genes within the same pathway. This potential nonlinearity, coupled with the availability of ground-truth gene output values (i.e., genes already known to be involved in a specific pathway), makes the transporter discovery problem well-suited to nonlinear classifiers such as artificial neural networks (ANNs) (25). ANNs have been used for several gene prediction tasks in plants, including gene ontology (26), subcellular localization (27), and transcriptional regulators (28), though, to our knowledge, they have not yet been applied to identify missing genes with target activities in the context of plant biosynthetic pathways.
Here, we use a supervised learning approach to identify two TA transporters from deadly nightshade (Atropa belladonna) that alleviate cellular metabolite transport limitations when expressed in a yeast platform previously engineered for de novo TA biosynthesis (5). We first show that a simple feedforward ANN trained on A. belladonna tissue-specific transcriptome data can identify TA transporter candidates with a reduction in search space (>40,000 transcripts → 3 candidates) over 30 times better than that of conventional linear correlation strategies. We demonstrate that two of the three identified transporter candidates, AbPUP1 and AbLP1, localize to the vacuole membrane and increase hyoscyamine and scopolamine production in engineered yeast. Using growth measurements and alkaloid tolerance assays in yeast, we show that both AbPUP1 and AbLP1 transport hyoscyamine and that the former appears to be a more specific proton-dependent transporter of littorine and hyoscyamine. We combine expression of four different plant transporters to facilitate exchange of pathway intermediates between compartments with expansion of cellular reduced nicotinamide adenine dinucleotide phosphate (NADPH) availability, optimization of prototrophy, and pH tuning to develop a yeast strain capable of producing hyoscyamine and scopolamine at titers over 100-fold (480 μg/L) and 7-fold (172 μg/L) greater than our previously reported strain and growth conditions. To expand the chemical diversity of our improved TA production platform, we implement a newly reported computational tool for biosynthetic pathway prediction, ATLASx (7), and extend the engineered TA pathway to produce two different classes of derivatives, nortropane alkaloids (norTAs) and tropane N-oxides. Our results suggest a role for AbPUP1 as a key vacuolar TA exporter in the biosynthesis of these important PNPs and highlight the importance of cellular transport optimization in developing microbial platforms for production of complex PNPs and derivatives. Our work also illustrates how new computational tools can be leveraged to efficiently expand microbial biosynthesis platforms for identification and de novo production of useful PNP derivatives.
Results
Prediction of Putative TA Transporters Using Supervised Learning Models.
We first sought to identify TA transporter candidates from a publicly available A. belladonna transcriptome (29) via conventional coexpression analysis strategies (see Materials and Methods). This dataset comprised abundances for each of over 40,000 transcripts across 11 different tissues. After filtering the dataset to retain only transcripts annotated by basic local alignment search tool (BLAST) and protein family (PFAM) searches for roles in small-molecule transport, we generated linear regression models (lm) to compare the tissue-specific expression profile of each transcript with that of the bait genes littorine mutase (CYP80F1) and hyoscyamine 6β-hydroxylase/dioxygenase (H6H) (5) and then further arranged candidates by expression profile similarity via hierarchical clustering (SI Appendix, Fig. S1). Nearly 100 transporter candidates were found to coexpress (P < 0.01) and cluster with known TA biosynthetic genes.
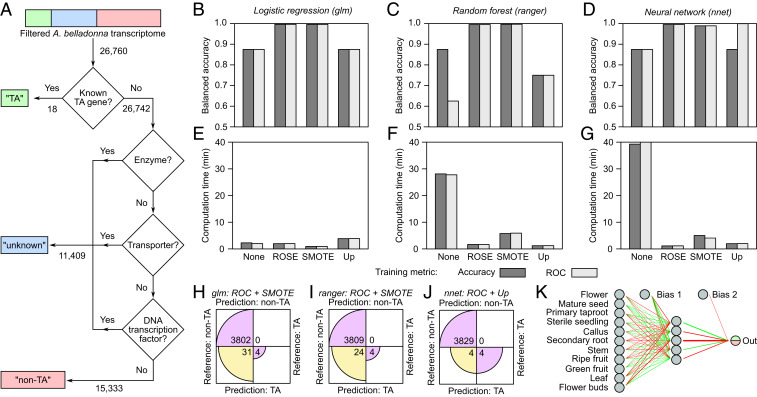
As the conventional regression/clustering analysis yielded a candidate list too large for efficient experimental validation, we examined whether classification methods could provide a greater reduction in search space. We trained and evaluated the performance of several supervised-learning models in identifying genes involved in TA biosynthesis from the same transcriptome dataset (29) (see Materials and Methods). Using BLAST functional annotations, each of the ∼27,000 transcripts that passed an initial filtering step to remove incomplete or zero-abundance transcripts was assigned one of three a priori output values depending on their involvement in TA biosynthesis (Fig. 2A); the 15,351 transcripts either known to be (denoted “TA”) or not to be involved (denoted “nonTA”) in the pathway we used for model training, cross validation, and testing. Three binary classifier models (logistic regression [LR], random forest [RF], and feedforward neural network [NN]) were trained, cross validated, and tested for prediction of TA-related genes from tissue abundance profiles using two different training objectives, four different data resampling techniques intended for highly class-imbalanced datasets, and two performance metrics (balanced accuracy and computation time). NN models showed the greatest average predictive accuracy on holdout testing data across training parameters (Fig. 2 B–D), whereas LR models required the least average computation time—although artificial resampling techniques enabled RF and NN models to be trained and tested in similar durations (Fig. 2 E–G). As all three classifier types with optimal training parameters showed comparable balanced accuracy (>0.99) and computation times (<3 min) on the testing data, we evaluated the false positive and false negative rates for each of the three optimized classifiers (Fig. 2 H–J). All three models correctly predicted 100% of “TA” genes (i.e., zero false negatives), whereas the NN model yielded far fewer false positives than either the LR or RF classifiers; 50% of transcripts predicted by the NN classifier to be involved in TA biosynthesis corresponded to known TA-related genes, in contrast to 11% and 14% for the LR and RF models, respectively. The architecture of the optimized NN model with one hidden layer of five nodes is shown in Fig. 2K (model weights are provided in Dataset S2).
Fig. 2.
Training, optimization, and testing of binary classifier models for the prediction of TA-related genes from the A. belladonna transcriptome. (A) Flowchart used for the generation of training output values for A. belladonna transcripts following dataset preprocessing. “TA”: the transcript encodes the gene product with a known role in TA biosynthesis; “nonTA”: the transcript encodes the gene product known not to be involved in TA biosynthesis; “unknown”: the transcript encodes the gene product with an unknown role in TA biosynthesis. Values indicate the number of transcripts. (B–G) Model performance in training, cross validation, and testing. Three binary classifier models were trained using one of four different oversampling methods (none: no oversampling; ROSE: random oversampling examples; SMOTE: synthetic minority oversampling technique; up, random oversampling) and one of two different performance metrics (accuracy: fraction of correctly predicted samples out of the total number of samples; ROC: area under the receiver operating characteristic curve, which plots the true positive rate versus the false positive rate). Top Row (B–D) and Bottom Row (E–G), respectively, show predictive accuracy in testing and total computation time for training, 10-fold cross validation, and testing. (H–J) Confusion matrices showing predictive performance of each of the three optimized binary classifier models on testing data. LR (H), RF (I), and NN (J) binary classifiers were trained and cross validated using the oversampling techniques and performance metrics that yielded maximum balanced accuracy and minimum computation time in B through G. Note that circular wedges are only shown to scale within each matrix row. (K) Simplified schematic of the final optimized neural network with 11-5-1 architecture (performance shown in J). Green and red lines indicate positive- and negative-weight connections, respectively, and line thickness is proportional to absolute connection weight (Dataset S2). Dual-color output neuron reflects the binary output format for predictions: green (1) = TA or red (0) = nonTA.
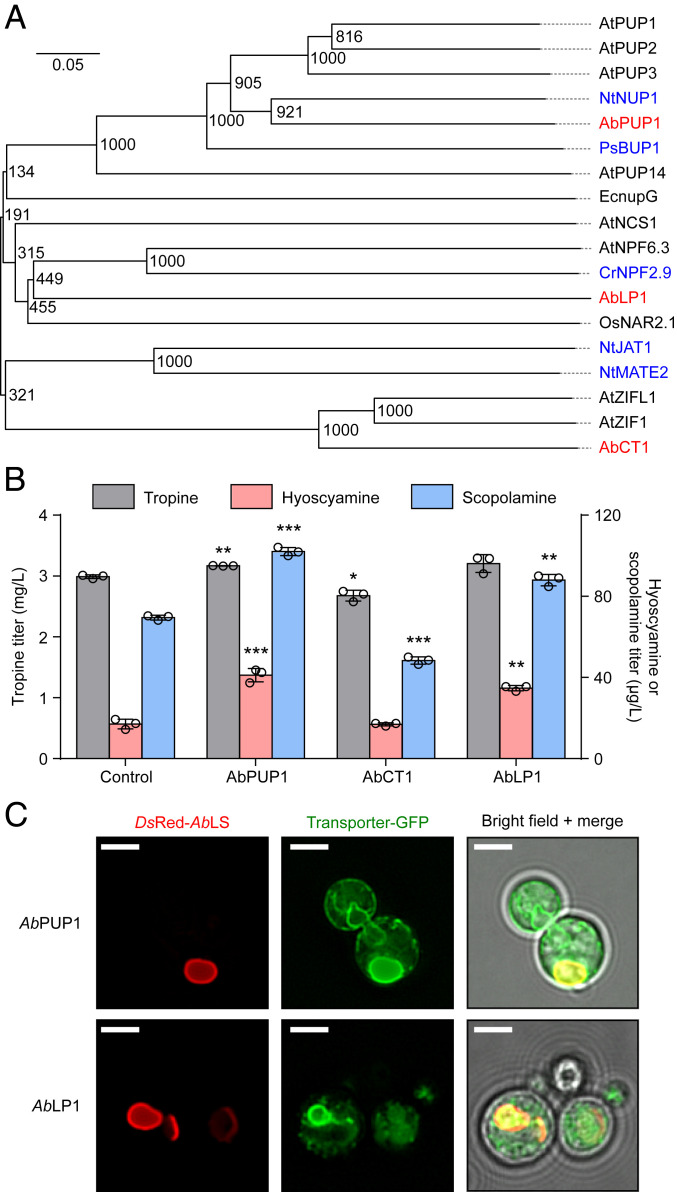
We used the trained NN classifier to identify genes with uncharacterized roles in TA biosynthesis, including TA transport. The 11,409 transcripts initially identified as encoding genes with “unknown” involvement in the TA pathway (Fig. 2A; see Materials and Methods), and which were therefore not used for model training or testing, were presented to the NN classifier, which predicted 33 transcripts to encode TA-related gene products (SI Appendix, Table S1). Consistent with localization of Solanaceae TA biosynthesis in lateral roots (30), most of the 33 genes show strong expression in secondary roots, moderate expression in primary taproots and calluses, and low expression in stems, flowers, fruits, and leaves. Three transcripts—denoted A. belladonna lactose permease-like 1 (AbLP1), carbohydrate transporter-like 1 (AbCT1), and purine uptake permease-like 1 (AbPUP1)—contained amino acid sequences annotated by homology and PFAM searches to encode organic small-molecule transporters (SI Appendix, Table S1). AbPUP1 shows a highly similar root-specific expression profile to that of known TA enzyme-coding genes, whereas AbLP1 and AbCT1 are more abundant in roots but also expressed in aerial tissues. As sequence alignments revealed missing regions in the open reading frames of some of these transcripts, we generated complete protein sequences for the putative TA transporter candidates via BLAST searches against a more complete transcriptome assembly (5) and verified them against complementary DNA (cDNA) synthesized from A. belladonna lateral root tissue (SI Appendix, Table S2). Phylogenetic analyses indicated that each of the three putative TA transporters belong to different small-molecule transporter families (Fig. 3A). AbPUP1 resembles members of the PUP family, including two transporters previously demonstrated to perform cellular import of nicotine (NtNUP1) in tobacco (31) and benzylisoquinoline alkaloids (BIAs) (PsBUP1) in opium poppy (14). In contrast, AbLP1 belongs to the NPF family, which includes a tonoplast-localized strictosidine exporter (CrNPF2.9) implicated in vinca MIA biosynthesis (15), and AbCT1, although initially annotated as a carbohydrate transporter, appears to be more closely related to vacuolar metal cation transporters of the zinc-induced facilitator family in Arabidopsis (32).
Fig. 3.
Phylogeny, heterologous activity, and subcellular localization of A. belladonna putative TA transporters. (A) Phylogenetic tree showing relationships between A. belladonna putative TA transporters (red), experimentally characterized plant alkaloid transporters (blue), and selected characterized small-molecule transporters (black). Values at each junction signify the number of bootstraps out of n = 1,000 iterations. (Scale bar, number of substitutions per amino acid site.) Species: At, A. thaliana; Nt, N. tabacum; Ps, Papaver somniferum; Ec, E. coli; Cr, Catharanthus roseus; Os, Oryzia sativa. Accession numbers for transporters are provided in SI Appendix, Table S4. (B) Effect of putative transporter expression on TA titers in engineered yeast. Transporter candidates or a BFP control were expressed from low-copy plasmids in CSY1300, and metabolites in supernatant were quantified by LC–MS/MS after 96-h growth in selective media. Data represent the mean of n = 3 biologically independent samples (open circles), and error bars show the SD. Student’s two-tailed t test: *P < 0.05, **P < 0.01, ***P < 0.001. (C) Fluorescence microscopy showing subcellular localization of TA transporter carboxyl-terminal GFP fusions in engineered yeast. AbPUP1-GFP and AbLP1-GFP were expressed from low-copy plasmids in CSY1300; microscopy and image analysis were performed as described in SI Appendix, Materials and Methods. (Scale bar, 2 μm.)
Characterization of Putative TA Transporters in Yeast.
The three TA transporter candidates were initially screened for activity in the context of an engineered biosynthetic pathway in which TA production is limited by intracellular transport. We previously engineered a yeast strain (CSY1297) for de novo biosynthesis of hyoscyamine and scopolamine by expressing 21 different enzymes and disrupting eight endogenous proteins across five metabolic modules and five subcellular compartments (5) (Fig. 1 and SI Appendix, Figs. S2 and S3). Module I, designed to increase accumulation of the TA precursor putrescine, incorporates 1) overexpression of four yeast enzymes: glutamate N-acetyltransferase (Arg2p), arginase (Car1p), ornithine decarboxylase (Spe1p), and polyamine oxidase (Fms1p); 2) a hybrid plant-bacterial pathway comprising Avena sativa arginine decarboxylase (AsADC) and Escherichia coli agmatine ureohydrolase (speB); and 3) dysregulated polyamine metabolism via disruption of genes encoding methylthioadenosine phosphorylase (Meu1p) and ornithine decarboxylase antizyme-1 (Oaz1p). Module II, designed to enable conversion of putrescine to the TA acyl acceptor tropine, incorporates 1) seven plant enzymes: putrescine N-methyltransferases from A. belladonna (AbPMT1) and Datura stramonium (DsPMT1), N-methylputrescine oxidase from Datura metel optimized for activity in yeast peroxisomes (DmMPO1ΔC-PTS1), pyrrolidine ketide synthase (AbPYKS) and tropinone synthase (AbCYP82M3) from A. belladonna, NADPH:cytochrome P450 reductase from Arabidopsis thaliana (AtATR1), and tropinone reductase 1 from D. stramonium (DsTR1); and 2) disruptions to five aldehyde dehydrogenases that divert intermediates from the pathway: Hfd1p, Ald2p, Ald3p, Ald4p, and Ald5p. Module III, designed to convert phenylalanine to the TA acyl donor phenyllactic acid (PLA) glucoside, incorporates 1) two main enzymes: phenylpyruvate reductase from Wickerhamia fluorescens (WfPPR) and uridine diphosphate (UDP)-glucosyltransferase 84A27 from A. belladonna (AbUGT); 2) overexpression of the yeast enzyme UDP-glucose pyrophosphorylase (Ugp1p) to increase UDP-glucose availability; and 3) disruption of steryl-β-glucosidase (Egh1p) to reduce PLA glucoside hydrolysis. Module IV, designed to convert the TA esterification product littorine to the downstream TAs hyoscyamine and scopolamine, incorporates three enzymes: littorine mutase from A. belladonna (AbCYP80F1), and hyoscyamine dehydrogenase (DsHDH) and H6H from D. stramonium (DsH6H). Module V comprises the central TA esterification reaction in which A. belladonna littorine synthase, previously observed to be functional in yeast only by expression in the vacuolar membrane via N-terminal fusion with the fluorescent protein DsRed (DsRed-AbLS), condenses tropine and PLA glucoside to produce littorine in the vacuole lumen. As uncharged PLA glucoside is relatively membrane-permeable, vacuolar import and export of protonated tropine and littorine, respectively, are primary limitations to flux through this Module (Fig. 1). We previously showed that the tobacco vacuolar nicotine importers NtJAT1 and NtMATE2 (33) may improve vacuolar tropine import in this pathway when expressed individually in the TA-producing strain (CSY1297) (5). However, no transporters have yet been identified to facilitate vacuolar littorine export.
We determined whether any of the three identified putative TA transporters influence TA accumulation in this yeast platform. AbPUP1, AbCT1, and AbLP1 were each expressed from low-copy plasmids in strain CSY1300, which was constructed by integrating expression cassettes for NtMATE2 and the selection marker LEU2 [previously shown to increase scopolamine accumulation (5)] into the genome of the TA production strain CSY1297, and transformants were cultured in selective media for 96 h. Liquid chromatography and tandem mass spectrometry (LC–MS/MS) analysis of culture supernatants indicated that AbPUP1 and AbLP1, but not AbCT1, increase TA production (Fig. 3B). AbPUP1 expression increased accumulation of hyoscyamine and scopolamine 2.4- and 1.5-fold (41 μg/L and 102 μg/L), respectively, relative to a blue fluorescent protein (BFP) control, while AbLP1 expression increased accumulation of these products 2.0- and 1.3-fold (35 μg/L and 88 μg/L). The lack of any substantial increase in accumulation of tropine, as we previously observed with expression of NtJAT1 and NtMATE2 (5), suggested that AbPUP1 and AbLP1 may instead transport substrates downstream of vacuolar tropine esterification.
We evaluated the subcellular localization of these transporters using fluorescence microscopy of yeast strain CSY1300 expressing AbPUP1 and AbLP1 carboxyl-terminal GFP fusions from low-copy plasmids (Fig. 3C) and of Nicotiana benthamiana leaf sections transiently expressing AbPUP1-GFP and AbLP1-GFP (SI Appendix, Fig. S4). The two transporters were observed primarily in the vacuolar membrane in both organisms, colocalizing strongly with DsRed-fused littorine synthase in the yeast vacuole membrane (Fig. 3C) and with the tonoplast marker vac-rk (34) in tobacco (SI Appendix, Fig. S4); in the latter, the localization of both transporters exhibited the transvacuolar cytoplasmic strands and vacuolar bulbs characteristic of tonoplast proteins (35–37). Collectively, these data suggested a role for AbPUP1 and AbLP1 in vacuolar substrate transport. To elucidate the structure and function of these transporters, we constructed plots of protein hydropathy and putative membrane topology using TMHMM (38) and generated homology models and gene ontology (GO) maps using C-I-TASSER (39) (SI Appendix, Figs. S5 and S6). AbPUP1 and AbLP1 comprise 10 and 12 transmembrane α-helices, respectively (SI Appendix, Fig. S5 A–D), and both transporters are predicted to be oriented in the vacuole membrane with cytosol-facing N- and carboxyl-termini (SI Appendix, Fig. S5E). For eukaryotic purine permeases, this topology is consistent with transport of substrates from the vacuole lumen (or extracellular space) into the cytosol (40). GO prediction corroborated that AbPUP1 is likely an intracellular transporter with specificity for nitrogen-containing organic compounds like purines or alkaloids (SI Appendix, Fig. S6 A–C), whereas AbLP1 showed features consistent with symport of organic substrates and cations (SI Appendix, Fig. S6 D–F).
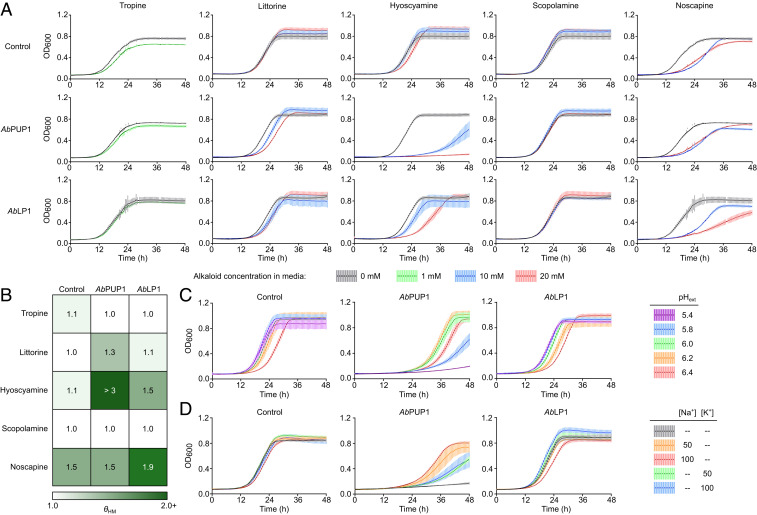
To corroborate their predicted role as vacuolar alkaloid transporters, we next characterized the substrate specificity and transport mechanism of AbPUP1 and AbLP1 via growth and toxicity assays in yeast. For these assays, we used AD12345678 (abbreviated AD1-8), a yeast strain engineered for increased sensitivity to xenobiotics due to disruptions to seven major multidrug resistance ABCs and their transcriptional regulator (41), which was previously used to study NtJAT1 substrate specificity in yeast (12). Although AbPUP1 and AbLP1 are primarily found in the yeast vacuole membrane, a fraction of cellular AbPUP1, and, to a lesser extent, AbLP1, also appears to localize to the plasma membrane (Fig. 3C). Based on the topological equivalence of the vacuole lumen and extracellular space, we reasoned that transporters exporting substrates from the vacuole to the cytosol would also be capable of importing the same extracellular substrates when situated in the plasma membrane, causing increased sensitivity to high substrate concentrations. We monitored the growth of strain AD1-8 expressing AbPUP1, AbLP1, or a BFP control from low-copy plasmids in liquid-selective media supplemented with various alkaloids at concentrations between 0 and 20 mM (Fig. 4A). Expression of either AbPUP1 or AbLP1 conferred acute sensitivity to hyoscyamine (θHM, ratio of time for half-maximal optical density at 600 nm [OD600] at 20 mM relative to at 0 mM: control, θHM ∼1.1; AbPUP1, θHM > 3; AbLP1, θHM ∼1.5) and modest sensitivity to littorine (control, θHM ∼1.0; AbPUP1, θHM ∼1.3; AbLP1, θHM ∼1.1) but not to tropine or scopolamine (Fig. 4B). AbLP1 additionally increased sensitivity to high concentrations of noscapine (θHM ∼1.5 to 1.9), a nonTA control. These data suggest that AbPUP1 and AbLP1 export vacuolar littorine and hyoscyamine to the yeast cytosol and also enable yeast to reuptake these metabolites from the extracellular space.
Fig. 4.
Elucidation of putative TA transporter substrates and transport mechanisms. Growth curve data showing (A and B) the effect of AbPUP1 and AbLP1 on alkaloid tolerance, (C) H+ dependence of TA transport by AbPUP1 and AbLP1, and (D) the effect of monovalent cations on TA transport by AbPUP1 and AbLP1 in yeast. Transporter substrate specificity (θHM) quantified as the ratio of time required for half-of-maximum OD600 increase (tHM), with maximum alkaloid concentration relative to without alkaloid supplementation (θHM = tHM,max_alk/tHM,no_alk), is shown as a heatmap in B; heatmap values were extracted from mean growth curves in A. For A through D, the yeast strain AD1-8 expressing BFP (control), AbPUP1, or AbLP1 from low-copy plasmids was cultured in selective media supplemented with (A and B) 0 to 20 mM alkaloids, (C) 10 mM hyoscyamine, and 100 mM potassium phosphate buffer, or (D) 20 mM hyoscyamine and 0 to 100 mM NaCl or KCl. Note that for A and B, tropine was supplemented in media at only 1 mM, as higher tested concentrations severely retarded the growth of AD1-8, such that growth measurements were not feasible. For D, “–” indicates no salt supplementation in excess of that already present in the yeast nitrogen base. For A, C, and D, solid lines and shaded bars, respectively, indicate the mean and SD of n = 3 biologically independent samples.
Many plant alkaloid transporters not driven by ATP hydrolysis are instead dependent on proton gradients for substrate translocation (21). We examined the proton dependence of AbPUP1- and AbLP1-mediated alkaloid sensitivity in yeast by monitoring the growth of AD1-8 expressing transporters or a BFP control in pH-buffered media supplemented with 10 mM hyoscyamine (Fig. 4C). Hyoscyamine sensitivity conferred by AbPUP1 was abrogated by increasing buffer pH, indicating that cellular hyoscyamine import (and, by equivalence, vacuolar export) by AbPUP1 occurs via a proton-dependent symport mechanism. In contrast, hyoscyamine transport by AbLP1 did not appear to be pH dependent. To determine whether transport by AbPUP1 or AbLP1 can be powered by gradients in other common symported cations, we monitored growth of the transporter-expressing strains in media supplemented with hyoscyamine and either sodium or potassium chloride (Fig. 4D). Transport by AbPUP1 was impaired by either Na+ or K+ as indicated by reduced alkaloid sensitivity at higher cation concentrations, whereas AbLP1-mediated transport appeared to be slightly enhanced at high [Na+] but not at high [K+].
Taken together, these results suggest that, although AbPUP1 and AbLP1 show similar effects on TA production in yeast and localization to the vacuolar membrane, differences in their substrate tolerances and transport mechanisms may enable them to play specialized roles in the context of TA biosynthesis and might be leveraged to modulate TA production in engineered hosts.
Yeast-Strain Engineering and Media Optimization for Increased TA Production.
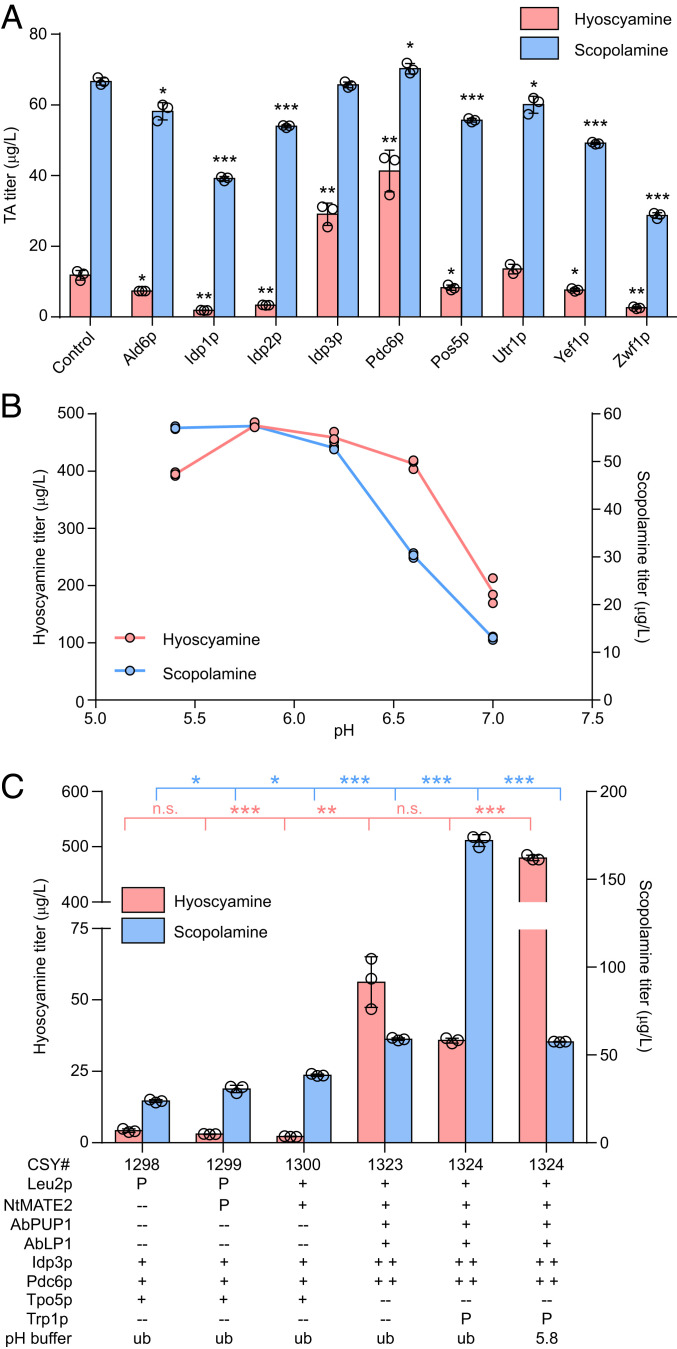
Both production of canonical TAs and exploration of TA derivatives in our engineered yeast platform are hindered by low pathway flux to hyoscyamine and scopolamine. We combined several strategies including transport engineering, cofactor regeneration, and media optimization to develop an improved TA-production platform suitable for de novo derivatization studies. We first incorporated the previously identified vacuolar tropine importers NtJAT1 and NtMATE2 (5) into a single yeast strain as a starting point for TA titer optimization. Coexpression of NtMATE2 from a low-copy plasmid with chromosomal NtJAT1 in strain CSY1299 increased scopolamine production by 29% (24 μg/L to 31 μg/L), and chromosomal expression of both transporters and the LEU2 selection marker in CSY1300 afforded a further increase of 26% (38 μg/L) (Fig. 5C).
Fig. 5.
Optimization of redox cofactor availability, intracellular transport, prototrophy, and culture conditions improve TA production in yeast. (A) Effect of NADPH-regenerating enzyme overexpression on TA titers in engineered yeast. Enzyme-coding genes or a BFP control were overexpressed from low-copy plasmids in strain CSY1300, and metabolites in the supernatant were quantified by LC–MS/MS after 96-h growth in selective media. Data represent the mean of n = 3 biologically independent samples (open circles), and error bars show the SD. (B) Effect of media pH buffering on TA titers in engineered yeast. Strain CSY1324 was cultured in selective media buffered between pH 5.4 and 7.0 with 0.1 M potassium phosphate (K2HPO4/KH2PO4), and metabolites in supernatant were quantified by LC–MS/MS after 96-h growth. Data represent the mean of n = 3 biologically independent samples (filled circles). (C) Summary of strain optimization to increase de novo production of hyoscyamine and scopolamine in engineered yeast. Strains were cultured in appropriate selective or nonselective media for 96 h before quantification of TAs in the supernatant by LC–MS/MS. Symbols: –, absence of an enzyme and/or disruption of an encoding gene; P, expression from a low-copy plasmid; +, expression from a chromosomal gene copy; ++, expression from a native gene copy plus an additional chromosomal copy; ub, unbuffered growth media; 5.8, growth media buffered to pH 5.8 with 0.1 M potassium phosphate. Data represent the mean of n = 3 biologically independent samples (open circles), and error bars show the SD. For A and C, Student’s two-tailed t test: *P < 0.05, **P < 0.01, ***P < 0.001; n.s., not significant.
Our engineered TA pathway incorporates five enzymes (AbCYP82M3, DsTR1, WfPPR, AbCYP80F1, and DsHDH) that use NADPH for electron transfer (5). As NADPH depletion by overexpressed enzymes may limit pathway flux, we determined whether expanding cellular NADPH availability could increase TA accumulation. We identified native yeast enzymes driving NADPH regeneration in the central metabolism (SI Appendix, Fig. S7) via substrate oxidation (Ald6p, Idp1p-3p, Pdc6p, and Zwf1p) or nicotinamide adenine dinucleotide [NAD(H)] phosphorylation (Pos5p, Yef1p, and Utr1p), overexpressed each corresponding gene from a low-copy plasmid in CSY1300, and measured TA accumulation in media following 96 h of growth of the transformed strains (Fig. 5A). Overexpression of peroxisomal isocitrate dehydrogenase (Idp3p) and pyruvate decarboxylase (Pdc6p) respectively increased production of hyoscyamine 2.5-fold (29 μg/L) and 3.5-fold (41 μg/L) relative to control, although no changes in scopolamine production were noted. We incorporated NADPH regeneration together with vacuolar export optimization and constructed strain CSY1323 by integrating expression cassettes for AbPUP1, AbLP1, IDP3, and PDC6 and simultaneously disrupting TPO5, which encodes a transporter responsible for Golgi-mediated exocytosis of the TA precursor putrescine (42), into the genome of CSY1300. CSY1323 produced hyoscyamine and scopolamine at titers 26-fold (56 μg/L) and 1.5-fold (59 μg/L) greater than those of CSY1300 following 96 h of growth in selective media (Fig. 5C).
We next examined the effects of amino acid prototrophy on TA production in CSY1323. We previously observed that elimination of leucine auxotrophy and enhanced NADH regeneration via LEU2 overexpression promotes scopolamine accumulation (5). As our TA-producing strains are derived from a histidine prototroph (43), we constructed strains prototrophic for uracil (Ura) and tryptophan (Trp) (the remaining two auxotrophies) by overexpressing the URA3 and TRP1 genes from low-copy plasmids in CSY1323. LC–MS/MS analysis of media supernatant from 96-h cultures of wild-type and prototrophic CSY1323 strains indicated that, whereas Ura prototrophy does not improve TA production (SI Appendix, Fig. S8), Trp prototrophy (denoted CSY1324) promotes conversion of hyoscyamine (36% decrease; 56 μg/L to 36 μg/L) to scopolamine (2.9-fold increase; 59 μg/L to 172 μg/L) (Fig. 5C).
Methods for modulating the relative accumulation of different TA products may facilitate the discovery and production of specific derivatives. As AbPUP1 transports hyoscyamine, but not scopolamine, in a proton-dependent and potassium-inhibited manner, we determined whether media pH buffering and its concomitant effect on cellular TA transport would influence the hyoscyamine–scopolamine production ratio. We measured TA production in cultures of CSY1324 following 96 h of growth in selective media buffered between 5.4 and 7.0 with 0.1 M potassium phosphate (K2HPO4/KH2PO4); in this buffer system, [H+] and [K+] respectively decrease and increase with higher pH. At all tested buffer conditions, hyoscyamine accumulation was substantially increased (≥5.3-fold) at the expense of decreased scopolamine production relative to CSY1324 grown without buffering (Fig. 5 B and C). Maximum hyoscyamine production was obtained at pH 5.8, whereas scopolamine production decreased monotonically with increasing pH (Fig. 5B). Similar trends in TA accumulation were observed in buffered cultures of CSY1323, although production of both TAs decreased with higher pH (SI Appendix, Fig. S9). Importantly, this result revealed a simple method for tuning the TA-production ratio using pH buffers: hyoscyamine accumulation is maximized during buffered growth at pH 5.8, whereas scopolamine production is maximized in unbuffered media. Under optimal conditions for each TA, hyoscyamine and scopolamine production by CSY1324 reached titers of 480 μg/L and 172 μg/L, representing improvements of 114- and 7.2-fold relative to the starting strain (CSY1298; Fig. 5C).
Collectively, these results demonstrate how conventional strain-engineering strategies—redox balancing and cofactor regeneration, auxotrophy optimization, and media buffering—can be combined with cellular-transport engineering to improve pathway flux and control the accumulation of different products in heterologous biosyntheses. For all subsequent experiments, we used our optimized TA production strain (CSY1324) and either buffered (pH 5.8) or unbuffered media to investigate derivatization of hyoscyamine or scopolamine, respectively.
De Novo Biosynthesis of Computationally Predicted Medicinal TA Derivatives in Yeast.
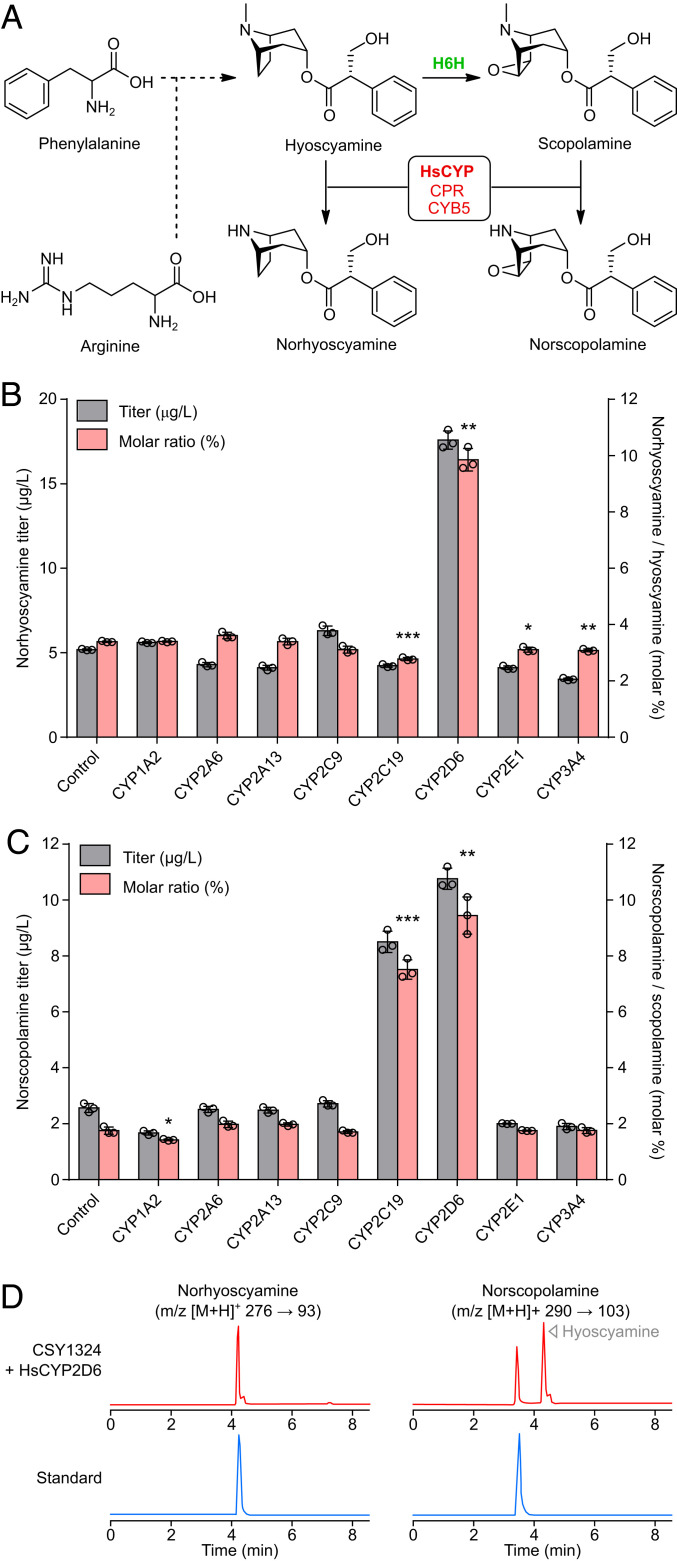
Heterologous production of PNP derivatives has been challenged by limited information on the nonnative substrate tolerance and activity of most biosynthetic enzymes (9). We applied a recently reported computational tool for in silico biosynthetic pathway expansion, ATLASx (7), to extend our engineered TA pathway for de novo production of useful derivatives in yeast. Using the ATLASx pathway prediction utility, we generated a map of all possible derivatives that might be produced in one biosynthetic step from hyoscyamine and scopolamine by known enzyme activities (SI Appendix, Fig. S10). The majority of commercial TA-derived drugs are N-functionalized derivatives of tropane esters (e.g., trospium, ipratropium, tiotropium, oxitropium, and N-butylscopolamine), since quaternary amines exhibit reduced blood–brain barrier permeability and central nervous system (CNS) toxicity (16, 30). As microbial production of secondary amine TAs might facilitate screening, discovery, and de novo biosynthesis of pharmaceutical derivatives without N-methyl groups (e.g., trospium), we searched the ATLASx-generated map for both secondary and quaternary amine TA derivatives. Two derivative classes of interest were predicted to be accessible via one-step transformations: N-demethylated norTAs and tropane N-oxides (SI Appendix, Fig. S10).
N-demethylated derivatives are byproducts of hepatic catabolism of diverse alkaloid drugs including the TA cocaine (44, 45). We reasoned that one or more human liver CYP450 enzymes (HsCYPs) might be capable of hyoscyamine and scopolamine N-demethylation in yeast (Fig. 6A). As prior attempts to express HsCYPs in yeast observed increased heterologous activity when paired with yeast NADPH:CYP450 reductase (Ncp1p) fused to human cytochrome b5 (HsCYB5A) (46), we coexpressed each of the eight HsCYPs collectively responsible for >80% of hepatic drug metabolism (44) with a Ncp1p-HsCYB5A fusion protein from low-copy plasmids (SI Appendix, Fig. S11A) in our optimized TA production strain (CSY1324). Using an extended LC–MS/MS method we developed for unambiguous identification of TAs and their derivatives (SI Appendix, Fig. S12), we analyzed the culture media of each HsCYP-expressing strain for production of norhyoscyamine (Fig. 6B) and norscopolamine (Fig. 6C) following 96 h of growth in pH-buffered or nonbuffered media, respectively. We observed low (≤5 μg/L) accumulation of both derivatives in the negative control, suggesting an existing pathway for norTA synthesis in our engineered yeast platform. Expression of HsCYP2D6 improved production of norhyoscyamine and norscopolamine over threefold (5 μg/L to 18 μg/L) and nearly fourfold (3 μg/L to 11 μg/L) in buffered (pH 5.8) and unbuffered media, respectively, while expression of HsCYP2C19 only improved production of norscopolamine in unbuffered media (3 μg/L to 9 μg/L), reflecting an increase in the norTA:TA production ratio from ≤3% to nearly 10% (Fig. 6 B–D and SI Appendix, Fig. S13). To account for potential variation in activity profile with different reductase partners, we also screened the eight HsCYPs in CSY1324 via coexpression with Ncp1p only (SI Appendix, Fig. S11 B–D) and with human NADPH:CYP450 reductase (HsPOR) fused to HsCYB5A (SI Appendix, Fig. S11 E–G). Consistent with previous results, HsCYP2D6 increased accumulation of norhyoscyamine and norscopolamine ∼3-fold (Ncp1p: 4 μg/L to 12 μg/L; HsPOR-HsCYB5A: 5 μg/L to 16 μg/L) and over 5- to 10-fold (Ncp1p: 1.2 μg/L to 6 μg/L; HsPOR-HsCYB5A: 0.8 μg/L to 9 μg/L) in buffered (pH 5.8) and unbuffered media, respectively, while expression of HsCYP2C19 improved production of norscopolamine over 7-fold (Ncp1p: 1.2 μg/L to 9 μg/L; HsPOR-HsCYB5A: 0.8 μg/L to 6 μg/L) in unbuffered media, reflecting an increase in the norTA:TA production ratio from ≤4% to up to 15% (SI Appendix, Fig. S11 B–G).
Fig. 6.
Production of norTAs in engineered yeast. (A) Simplified schematic of the TA biosynthetic pathway extended using ATLASx for de novo production of norTAs in yeast. Dashed arrows indicate abbreviated pathway steps. H6H, hyoscyamine 6β-hydroxylase/dioxygenase; HsCYP, human liver cytochrome P450; CPR, yeast NADPH-CYP450 reductase (Ncp1p); CYB5, human cytochrome b5. (B and C) Effect of human liver CYP expression on titers and the molar production ratio of (B) norhyoscyamine and (C) norscopolamine in engineered yeast. CYPs or a BFP control were coexpressed with Ncp1p fused to human cytochrome b5 (HsCYB5A) from low-copy plasmids in CSY1324, and metabolites in supernatant were quantified by LC–MS/MS after 96-h growth in selective media buffered to pH 5.8 with 0.1 M potassium phosphate (B) or unbuffered (C). Data represent the mean of n = 3 biologically independent samples (open circles), and error bars show the SD. Student’s two-tailed t test: *P < 0.05, **P < 0.01, ***P < 0.001. (D) LC–MS/MS multiple reaction monitoring chromatograms for norTAs produced de novo by CSY1324 expressing HsCYP2D6 and cultured as in B and C, or authentic chemical standards. Chromatograms are representative of n = 3 biologically independent samples.
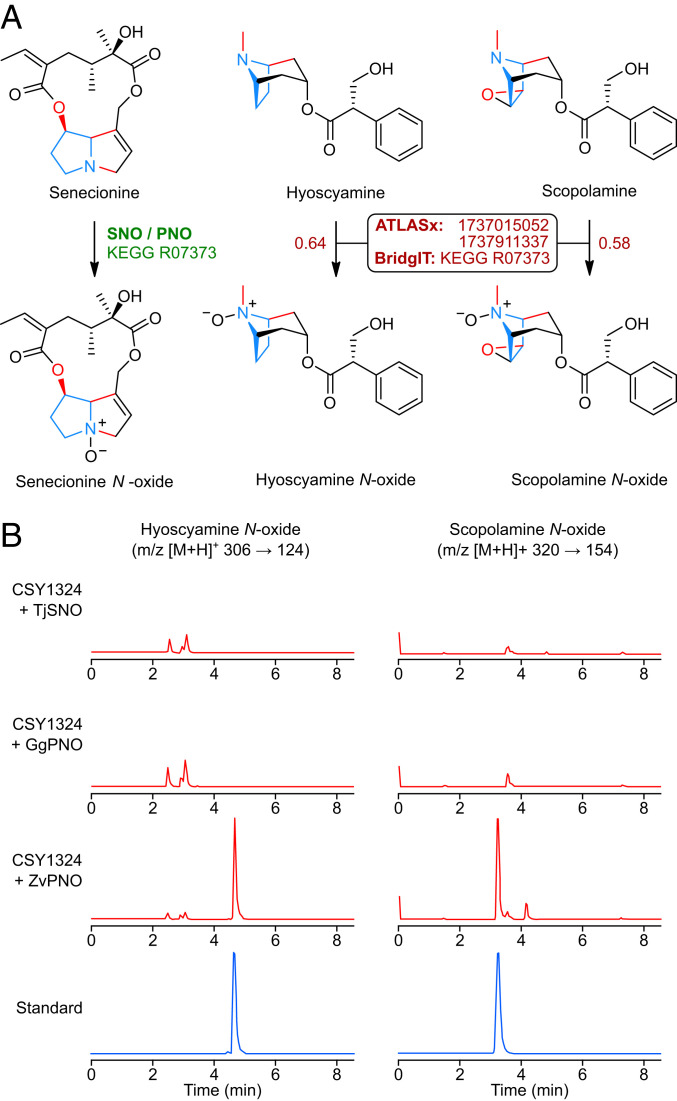
Due to their increased water solubility and lower membrane permeability compared to free bases (47), drugs derived from alkaloid N-oxides may pose a lower risk of CNS disruption. Although N-oxides are also thought to be trace byproducts of alkaloid catabolism in mammals (48), no TA N-oxygenases have yet been identified, and we did not observe N-oxide accumulation in TA-producing yeast strains expressing HsCYPs. We used BridgIT (8), a computational enzyme prediction tool integrated into the ATLASx webserver, to identify putative enzyme candidates for conversion of hyoscyamine and scopolamine to cognate N-oxides (SI Appendix, Table S3). The highest-scoring enzyme prediction for both TAs was senecionine N-oxygenase (SNO; Kyoto Encyclopedia of Genes and Genomes [KEGG] R07373, enzyme commission [EC] 1.14.13.101), an NADPH- and flavin-dependent monooxygenase responsible for detoxification and weaponization of plant pyrrolizidine alkaloids in some herbivorous insects (49). The pyrrolizidine moiety of senecionine, the canonical substrate of SNO and related pyrrolizidine N-oxygenases (PNOs), closely resembles the functional groups and atom connectivity found in the tropane ring, suggesting that these enzymes may also be capable of N-oxygenating TAs (Fig. 7A). As the substrate scope of PNOs appears to have coevolved with the dietary alkaloid diversity of their host insects (50), we selected PNO orthologs from three different species for testing in our yeast TA production platform: TjSNO from Tyria jacobaeae (cinnabar moth), a specialist feeder; GgPNO from Grammia geneura (Nevada tiger moth), a generalist herbivore of wildflowers; and ZvPNO from Zonocerus variegatus (harlequin locust), an omniherbivorous agricultural pest (51–53). We expressed each of the three candidates from low-copy plasmids in CSY1324 and analyzed the culture media by LC–MS/MS after 96 h of growth of transformed strains in pH-buffered or nonbuffered media. We observed de novo production of both hyoscyamine N-oxide (71 μg/L in buffered media, pH 5.8) and scopolamine N-oxide (14 μg/L in unbuffered media) by the strain expressing ZvPNO (Fig. 7B and SI Appendix, Fig. S14), corroborating ATLASx/BridgIT predictions that a SNO/PNO enzyme can N-oxygenate TAs.
Fig. 7.
In silico enzyme prediction using BridgIT enables de novo biosynthesis of TA N-oxides in engineered yeast. (A) Comparison of native reaction (Left) and target reactions (Center, Right) for SNO and PNO, the top-scoring enzymes predicted by BridgIT to N-oxygenate TAs. The KEGG reaction ID for native SNO/PNO transformation is shown in green; ATLASx reaction IDs and the BridgIT closest KEGG reaction match for target transformations are shown in dark red. Values beside arrows for target reactions indicate BridgIT scores. Conserved pyrrolidine core and shared functional group connectivity in substrates and products are indicated in blue and red, respectively. (B) LC–MS/MS multiple reaction monitoring chromatograms for analysis of de novo TA N-oxide production by CSY1324 expressing SNO/PNOs, or authentic chemical standards. SNO/PNO candidates from T. jacobaeae (TjSNO), G. geneura (GgPNO), and Z. variegatus (ZvPNO) were expressed from low-copy plasmids in CSY1324, and metabolites in supernatant were analyzed after 96-h growth in selective media buffered to pH 5.8 with 0.1 M potassium phosphate (for hyoscyamine N-oxide) or unbuffered (for scopolamine N-oxide). Chromatograms are representative of n = 3 biologically independent samples.
These results demonstrate that 1) two different classes of TA derivatives computationally predicted to be accessible from our engineered TA pathway—norTAs and TA N-oxides—can be synthesized de novo in yeast using enzymes with canonical roles in alkaloid detoxification (SI Appendix, Fig. S3) and that 2) optimization of pathway flux and transport are useful strategies for establishing a testing platform that can be used to rapidly screen candidate enzymes for derivatization.
Discussion
Although more sophisticated convolutional neural networks have become the current state-of-the-art for phenotype prediction from sequence or expression data (26, 28, 54), we demonstrated that a simple feedforward ANN comprising a single hidden layer could achieve a reduction in candidate search space at least comparable to—if not better than—regression- and clustering-based approaches for prediction of genes matching target activities (5, 24, 55). Our classification strategy yielded three TA transporter candidates, of which two were functional, from a transcriptome of >40,000 sequences. In contrast, the linear regression and clustering approach suggested too many candidates to efficiently screen (∼100) and was unable to identify AbLP1 as a putative TA transporter (SI Appendix, Fig. S1)—potentially because the nonzero expression of AbLP1 in aerial tissues caused its expression profile to deviate from a linear correlation with those of the more root-specific bait genes (SI Appendix, Table S1). One caveat relevant to this discrepancy in predictive power is the potential dependence of model performance on sample size: molecule transporters represent a small fraction of the transcriptome, and models were trained and evaluated on a single species-pathway dataset with a comparatively small subset of positive samples. The difference in predictive performance between the methods is likely to be diminished with a larger set of positive samples (e.g., TA-related or secondary metabolic enzymes) or if compared across multiple plant pathways. Our results demonstrate that simple neural network classifiers are powerful tools for predicting gene function within limited or small datasets—a task that is common in biosynthetic pathway discovery for natural products. The ∼50% global false-positive rate exhibited by our classifier (Fig. 2J) suggests that half of the 33 uncharacterized genes predicted to be involved in TA biosynthesis (SI Appendix, Table S1) may encode novel activities in this pathway. We noted several candidates encoding enzyme activities common to PNP biosyntheses, including a flavonoid hydroxylase, two O-methyltransferases, and several aminotransferases and 2-oxoglutarate-dependent dioxygenases. Future studies combining computational enzyme analysis, in planta gene knockouts, heterologous expression, and/or untargeted metabolomics may reveal new roles for these candidates in the biosynthesis of other tropane esters.
Phylogenetic, structural, and functional analyses of AbPUP1 and AbLP1 suggest that they may play different roles in the context of TA biosynthesis. Consistent with their evolution from disparate transporter families, AbPUP1 and AbLP1 show different substrate specificities and transport mechanisms: the former appears to be a proton symporter of littorine and hyoscyamine only, while the latter shows broader tolerance of non-TAs like noscapine and does not appear to be driven by proton gradients (SI Appendix, Fig. S5E). Considering these differences together with those observed in the tissue distribution of the transporters (SI Appendix, Table S1) and their shared vacuolar localization in yeast (Fig. 3C) and in planta (SI Appendix, Fig. S4), we propose that AbPUP1 may play a specific role in vacuolar littorine export and cytosolic hyoscyamine retention during TA biosynthesis in roots, whereas AbLP1 may facilitate TA biosynthesis as well as alkaloid transport and storage in nonroot tissues. As these hypotheses are based on characterization in heterologous hosts (yeast and tobacco), future plant studies leveraging transporter knockdown via RNA interference and/or virus-induced gene silencing in A. belladonna may elucidate the nuanced involvement of these transporters in intra- and intercellular compartmentalization of plant TA biosynthesis. Collectively, our work with plant transporters (NtJAT1, NtMATE2, AbPUP1, and AbLP1) highlights the importance of optimizing metabolite transport in parallel with pathway design, enzyme activity, and growth conditions for maximizing the performance of complex microbial biosyntheses of PNPs.
Despite our optimization of metabolite transport and pathway compartmentalization, yeast microscopy studies suggest two remaining limitations to pathway flux associated with the vacuole. First, not all of the cellular AbPUP1 and AbLP1 localized to the vacuole membrane (Fig. 3C), indicating partial mistargeting. This is often observed when membrane-associated plant proteins are expressed in yeast and may be due to limited recognition of heterologous targeting sequences and “overflow” of highly expressed proteins into other membranes (56). Optimization of vacuolar transporter trafficking may therefore enhance alkaloid transport and production. As we were unable to identify any clear tonoplast-targeting motifs in AbPUP1 or AbLP1, and, to our knowledge, no well-conserved tonoplast-targeting sequences have been identified in plants (57, 58), systematic mutagenesis and microscopy studies may reveal cryptic localization signals in AbPUP1 and AbLP1. Microscopy and functional screening of chimeric transporters with signal sequences replaced by vacuole-targeting elements from native yeast proteins (56) may identify variants with enhanced vacuolar targeting that support increased alkaloid production. Second, these transporters are necessitated by vacuolar compartmentalization of littorine synthase (AbLS; Fig. 3C). We previously demonstrated that the enzyme’s activity in yeast is only achievable as an N-terminal fluorophore-fused transmembrane enzyme (DsRed-AbLS) anchored in the vacuole membrane; the cytosol-to-vacuole secretion pathway ensures that, for this fusion construct, the enzyme domain faces toward the vacuole lumen (5). Although an inverted variant with vacuolar fluorophore and cytosolic enzyme domain would be unlikely to receive the posttranslational modifications important for this enzyme class (5), mutagenesis and directed evolution studies may identify enzyme variants that retain cytosolic activity without posttranslational modifications, obviating the need for a vacuolar step.
Optimization of yeast-strain growth conditions revealed important dependencies of alkaloid production on auxotrophies and media pH. Trp and Ura prototrophy were restored via expression of TRP1 and URA3, and neither gene products catalyze reactions with obvious links to native metabolic pathways or cofactor pools involved in TA production. However, we observed that restoration of Trp but not Ura prototrophy increased TA accumulation. We suspect that a combination of factors contributed to this discrepancy. First, as the concentration of Trp (20 mg/L) in our synthetic media is lower than that of other commercial formulations (40-200 mg/L), Trp is more likely to become limiting than Ura during growth of -Ura/-Trp auxotrophs, and restoration of Trp prototrophy via TRP1 expression provides a greater advantage to growth and protein production. Second, restoration of Trp prototrophy via TRP1 enables more robust yeast growth compared to Trp auxotrophs at temperatures lower than optimum (30 °C) (59), though the mechanism of this effect is unclear and may be linked to protein production capacity. As all yeast metabolite production in our study was carried out at 25 °C, TRP1 expression may have yielded a modest increase in growth and protein production. Third, Trp and Ura prototrophy may exert different metabolite transport effects on cells. Yeast expresses one high-affinity Trp permease (Tat2p) and a Ura permease (Fur4p) for uptake of these nutrients. Trp transport has been found to be most frequently limiting to yeast growth relative to uptake of any other nutrient (60). Moreover, Tat2p is known to be highly sensitive to changes in membrane structure and conformation (60), whereas no such sensitivity has been reported for Fur4p. Thus, changes in membrane structure caused by overexpression of several heterologous transporters may further disrupt Trp uptake and exacerbate Trp limitation, providing an advantage in growth and protein production to strains capable of endogenous Trp biosynthesis via Trp1p. Transporter–cation interactions may partially explain the observed dependence of TA production on buffer pH in engineered yeast: decreased [H+] and increased inhibitory [K+] at higher buffer pH (and relative to unbuffered growth) reduces cellular reuptake of secreted hyoscyamine, promoting its accumulation and diminishing intracellular conversion to scopolamine. These observations highlight the complex interactions between cellular physiology, central metabolism, and metabolite transport that must be considered when optimizing growth conditions for microbial cell factories.
As many commercial TA drugs are chemical derivatives of natural tropane esters (16, 30), we demonstrated that yeast can be engineered for de novo production of canonical TAs as well as two useful classes of derivatives predicted by in silico pathway exploration. Despite evidence that cocaine N-demethylation in humans is performed by HsCYP3A4 (45), we instead observed enhanced norTA production with HsCYP2D6—known to act on BIAs (44)—and HsCYP2C19 across different combinations of reductase partner enzymes, suggesting that HsCYP specificity in heterologous hosts cannot be predicted based on substrate backbone alone. We suspect that trace norTA biosynthesis without HsCYPs may be due to passage of putrescine (rather than N-methylputrescine) and subsequent nor-intermediates through the engineered pathway, which may represent a promising alternative strategy for norTA production if the tolerance of pathway enzymes for N-demethylated substrates can be enhanced.
We demonstrated that computational pathway expansion (ATLASx) and enzyme prediction (BridgIT) can be combined to accelerate the discovery and heterologous production of PNP derivatives. Production of TA N-oxides in our engineered yeast platform represents experimental validation of an enzyme computationally predicted to catalyze a native transformation on a nonnative substrate class. We detected TA N-oxygenation only by the PNO derived from the omniherbivorous Z. variegatus, consistent with its need to detoxify a broader range of dietary plant alkaloids relative to specialist herbivores (50, 53). Our results contribute to a growing body of evidence that enzymes used for PNP detoxification can be co-opted to expand biosynthetic diversity in engineered hosts (61).
Microbial synthesis of complex PNPs has been limited in part by the comparative simplicity of unicellular hosts; chemistries that might naturally be distributed across organelles and tissues in plants must be coerced into one or a few compartments in bacteria or yeast. Replicating the spatial organization and associated diverse biochemical environments of plant biosyntheses in microbial hosts is therefore an important step toward accessing the full variety of PNPs. In this work, we incorporated four different plant alkaloid transporters (NtJAT1, NtMATE2, AbPUP1, and AbLP1) to mediate the import and export of three intermediates and products at different points in the TA pathway (tropine, littorine, and hyoscyamine) across two membranes separating three cellular compartments (extracellular space, cytosol, and vacuole). Our efforts demonstrate how progressive enhancements in pathway flux afforded by addressing cellular transport limitations can synergize with those produced by strain and process optimization. As heterologous biosyntheses continue to increase in length and complexity, we anticipate that traditional design principles for metabolic pathways will need to be integrated with metabolite transport considerations and nonmetabolic information—such as the ecology or physiology of enzyme source organisms—to unlock the full potential of microbial biomanufacturing platforms.
Materials and Methods
Detailed materials and methods are provided in SI Appendix, Materials and Methods.
Transcript Coexpression Analysis via Linear Regression and Clustering.
Linear regression analysis and hierarchical clustering for the identification of TA transporter candidates was performed as described previously (5) using a custom R script described as follows. Tissue-specific abundances (fragments per kilobase of contig per million mapped reads [FPKM]) and putative protein structural and functional annotations for each of the 43,861 unique transcripts identified from the A. belladonna transcriptome were obtained from the Michigan State University (MSU) Medicinal Plant Genomics Resource (29). The transcripts were filtered for those annotated with any of the following transporter PFAM identification numbers (IDs): PF00854, PF16974, PF01554, PF00664, and PF00005; or any of the following functional annotation keywords: efflux, transporter, and ABC. In addition, any transcripts with functional annotations containing the keywords putrescine or tropinone or with locus ID 4635 [corresponding to hyoscyamine dehydrogenase (5)] were included in the filter as positive control TA-associated genes to validate clustering with bait genes. Next, mean tissue-specific expression profiles were generated for CYP80F1 and H6H as bait genes. For each of the two, lm were constructed to express the bait-gene expression profile as a linear function of each candidate gene profile, and correlation P values were extracted for each candidate (null hypothesis: slope = 0; Student’s t test on estimate of slope coefficient). The candidates identified using each of the two bait genes were pooled, and duplicates were removed. Combined P values for each candidate were computed as the sum of the log10(P values) of the correlations with each of the two bait genes. Candidates were ranked by combined P value; those with P < 0.01 were further arranged by distance from bait genes via hierarchical clustering of tissue-specific expression profiles.
Classifier Model Development and Training for TA Transporter Prediction.
All data preprocessing, model training, cross validation, and gene prediction were performed using custom R scripts (see Data Availability) and executed on an Intel Core i5 4670K processor operating at 4.1 GHz. The general computational workflow is described as follows.
Data preprocessing.
Tissue-specific abundances (FPKM) and putative protein structural and functional annotations for each of the 43,861 unique transcripts identified from the A. belladonna transcriptome were obtained from the MSU Medicinal Plant Genomics Resource (29). Low-abundance “phantom” genes with undefined (not applicable; NA) or zero abundance values across all tissues, as well as incomplete gene fragments (which are identified by the predicted function tag “gene of unknown function”), were removed. A total of 26,760 full-length transcripts passed this preprocessing filter step.
Generation of output values for model training.
Each of the 26,760 full-length transcripts in the preprocessed dataset was assigned one of three a priori output values (Fig. 2A). Transcripts with predicted functional annotations matching enzymatic activities with previously demonstrated roles in TA biosynthesis were assigned the output value “TA”; of the remaining transcripts, those whose predicted functional annotation indicated that they were not an enzyme, a transporter, or a transcription factor (i.e., no metabolic function) were given the output value “nonTA,” and the remainder (i.e., those with potential metabolic functions but without prior implication in TA biosynthesis) were given the output value “unknown.” In total, the 26,760 full-length transcripts were divided into 18 “TA,” 15,333 “nonTA,” and 11,409 “unknown”; of these, the 15,351 “TA” and “nonTA” genes were used for model training, cross validation, and testing.
Data sampling for imbalanced classification.
The minority “TA” class is represented in only 0.12% of the 15,351 training transcripts, whereas the majority “nonTA” class is found in the remaining 99.88%. This class imbalance poses a substantial challenge for supervised learning models, as conventionally trained classifiers often cannot adequately incentivize pattern learning from such a small number of minority samples (62). In this case, a model can achieve 99.88% accuracy simply by predicting that 100% of samples are “nonTA,” and therefore has no motivation to correctly identify minority-class samples. Two strategies were used to improve model training on this highly imbalanced dataset. First, in addition to a logistic regression classifier used as a baseline classifier model, two additional model types previously demonstrated to be more tolerant of imbalanced datasets—RF classifiers and NNs (62)—were trained and tested. Second, model training and cross validation were performed using each of three different resampling techniques to reduce the discrepancy between the minority and majority classes: 1) random over-sampling, in which additional copies of random minority-class samples are added to the dataset until the majority and minority classes are comparable in population; 2) synthetic minority oversampling technique (SMOTE), in which additional minority-class samples are generated by interpolating based on nearest neighbors (63); and 3) random oversampling examples (ROSE), which uses a smoothed bootstrap procedure to generate additional minority-class samples (64). Random undersampling—in which only a subset of the majority class equal in population to the minority class is used for training—was not used, since 18 samples were judged to be insufficient for adequate model training.
Model training and cross validation.
The 15,351 “TA” and “nonTA” samples were split into 75% training and 25% testing data while preserving the class distribution of the original dataset (i.e., ∼99.88% “nonTA” and 0.12% “TA”). Model training and 10-fold cross validation with on-the-fly hyperparameter tuning (grid search, 25 bootstrap samples, three levels per hyperparameter) were performed concurrently using the trainControl and train functions in the caret package (65) for each of three binary classifier models: logistic regression (glm), random forest (ranger), and neural network (nnet). For both training and testing, model performance was evaluated using two different objective functions: 1) accuracy, the fraction of correctly predicted samples out of the total number of samples; and 2) area under the receiver operating characteristic (ROC) curve, which plots the true positive rate versus the false positive rate.
For neural network (nnet) model training, three-layer networks with eleven input neurons (one for each tissue’s transcript abundance), one fully connected hidden layer of variable size, one binary output neuron (reporting as output value a single factor with two possible levels, 0 = “nonTA” and 1 = “TA” with classification threshold = 0.5), and one bias pseudoneuron were trained across three levels of each of two hyperparameters (hidden layer size: 1, 3, or 5 neurons; L2 regularization decay: 0, 10−4, 0.1) using the default logistic (sigmoid) activation function and the entropy (maximum conditional likelihood) loss function for fitting to the training set. Model weights and bias were optimized using the default Broyden–Fletcher–Goldfarb–Shanno algorithm (no batch size parameter) over 100 epochs with early termination after 10 consecutive epochs with no change in validation loss. The final optimized neural network used for prediction of TA genes, with hyperparameters selected based on the maximization of the area under the ROC curve, had a three-layer 11-5-1 architecture (11 input neurons, one hidden layer of five neurons, one binary output neuron, one bias pseudoneuron) with weights shown in Dataset S2 and was trained using L2 decay = 0.1.
Supplementary Material
Acknowledgments
We thank M. Gupta (Iowa State University) for providing the AD1-8 yeast strain; A. Cravens (Stanford University) for the yeast multiplex CRISPR–Cas9/single guide RNA plasmids (pCS3410, 3411, 3414, and 3700 to 3703); E. Carlson, W. Cody, and the Sattely laboratory (Stanford University) for providing N. benthamiana plants, Agrobacterium strain GV3101 harboring the pEAQ-mCherry binary vector, and assistance with tobacco imaging; the Stanford Cell Sciences Imaging Facility for access to microscopy equipment and training; J. Hafner, H. MohammadiPeyhani, and A. Sveshnikova (Laboratory of Computational Systems Biotechnology, École Polytechnique Fédérale de Lausanne [EPFL]) for training and suggestions on generating novel enzyme predictions using ATLASx and BridgIT; and B. Kotopka for discussions and feedback in the preparation of this manuscript. This work was supported by the NIH grant AT007886 to C.D.S., the Siebel Scholars Foundation (doctoral fellowship to P.S.), and the Natural Sciences and Engineering Research Council of Canada (doctoral postgraduate scholarship to P.S.).
Footnotes
Competing interest statement: P.S. and C.D.S. are inventors on a pending patent application; C.D.S. is a founder and CEO of Antheia, Inc.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2104460118/-/DCSupplemental.
Data Availability
Data supporting the findings of this work are available within the paper and its supporting information files. The cDNA and protein sequences of AbPUP1 and AbLP1 have been deposited in GenBank under accession nos. MW715669 (AbPUP1) and MW715670 (AbLP1). Sequences are also provided in SI Appendix, Table S2. Accession numbers for previously reported gene and protein sequences in the GenBank/UniProt databases are provided in SI Appendix, Table S4. The custom R scripts used for linear regression analysis and hierarchical clustering, binary classifier model development, and gene prediction are publicly available on the Smolke Laboratory GitHub: https://github.com/smolkelab/Supervised_TA_prediction.
References
- 1.Ozber N., Watkins J. L., Facchini P. J., Back to the plant: Overcoming roadblocks to the microbial production of pharmaceutically important plant natural products. J. Ind. Microbiol. Biotechnol. 47, 815–828 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Paddon C. J., et al., High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496, 528–532 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Galanie S., Thodey K., Trenchard I. J., Filsinger Interrante M., Smolke C. D., Complete biosynthesis of opioids in yeast. Science 349, 1095–1100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo X., et al., Complete biosynthesis of cannabinoids and their unnatural analogues in yeast. Nature 567, 123–126 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Srinivasan P., Smolke C. D., Biosynthesis of medicinal tropane alkaloids in yeast. Nature 585, 614–619 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atanasov A. G., et al., Discovery and resupply of pharmacologically active plant-derived natural products: A review. Biotechnol. Adv. 33, 1582–1614 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohammadi-Peyhani H., Hafner J., Sveshnikova A., Viterbo V., Hatzimanikatis V., ATLASx : A computational map for the exploration of biochemical space. bioRxiv [Preprint] (2021). 10.1101/2021.02.17.431583 (Accessed 19 February 2021). [DOI]
- 8.Hadadi N., MohammadiPeyhani H., Miskovic L., Seijo M., Hatzimanikatis V., Enzyme annotation for orphan and novel reactions using knowledge of substrate reactive sites. Proc. Natl. Acad. Sci. U.S.A. 116, 7298–7307 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hafner J., Payne J., MohammadiPeyhani H., Hatzimanikatis V., Smolke C., A computational workflow for the expansion of heterologous biosynthetic pathways to natural product derivatives. Nat. Commun. 12, 1760 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y., et al., Complete biosynthesis of noscapine and halogenated alkaloids in yeast. Proc. Natl. Acad. Sci. U.S.A. 115, E3922–E3931 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shitan N., et al., Involvement of CjMDR1, a plant multidrug-resistance-type ATP-binding cassette protein, in alkaloid transport in Coptis japonica. Proc. Natl. Acad. Sci. U.S.A. 100, 751–756 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morita M., et al., Vacuolar transport of nicotine is mediated by a multidrug and toxic compound extrusion (MATE) transporter in Nicotiana tabacum. Proc. Natl. Acad. Sci. U.S.A. 106, 2447–2452 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shitan N., Hayashida M., Yazaki K., Translocation and accumulation of nicotine via distinct spatio-temporal regulation of nicotine transporters in Nicotiana tabacum. Plant Signal. Behav. 10, e1035852 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dastmalchi M., et al., Purine permease-type benzylisoquinoline alkaloid transporters in opium poppy. Plant Physiol. 181, 916–933 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Payne R. M. E., et al., An NPF transporter exports a central monoterpene indole alkaloid intermediate from the vacuole. Nat. Plants 3, 16208 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grynkiewicz G., Gadzikowska M., Tropane alkaloids as medicinally useful natural products and their synthetic derivatives as new drugs. Pharmacol. Rep. 60, 439–463 (2008). [PubMed] [Google Scholar]
- 17.Borrell A., et al., Arginine decarboxylase is localized in chloroplasts. Plant Physiol. 109, 771–776 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naconsie M., Kato K., Shoji T., Hashimoto T., Molecular evolution of N-methylputrescine oxidase in tobacco. Plant Cell Physiol. 55, 436–444 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Bedewitz M. A., Jones A. D., D’Auria J. C., Barry C. S., Tropinone synthesis via an atypical polyketide synthase and P450-mediated cyclization. Nat. Commun. 9, 5281 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nasomjai P., et al., Mechanistic insights into the cytochrome P450-mediated oxidation and rearrangement of littorine in tropane alkaloid biosynthesis. ChemBioChem. 10, 2382–2393 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Shitan N., Kato K., Shoji T., Alkaloid transporters in plants. Plant Biotechnol. 31, 453–463 (2014). [Google Scholar]
- 22.Dugé de Bernonville T., Papon N., Clastre M., O’Connor S. E., Courdavault V., Identifying missing biosynthesis enzymes of plant natural products. Trends Pharmacol. Sci. 41, 142–146 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Stander E. A., et al., Identifying genes involved in alkaloid biosynthesis in vinca minor through transcriptomics and gene co-expression analysis. Biomolecules 10, 1–26 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nett R. S., Lau W., Sattely E. S., Discovery and Engineering of Colchicine Alkaloid Biosynthesis (Springer, US, 2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Denison D. G. T., Holmes C. C., Mallick B. K., Smith A. F. M., “Neural network models” in Bayesian Methods for Nonlinear Classification and Regression (John Wiley & Sons, Ltd, 2004), pp. 115–118. [Google Scholar]
- 26.Kulmanov M., Hoehndorf R., DeepGOPlus: Improved protein function prediction from sequence. Bioinformatics 36, 422–429 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Almagro Armenteros J. J., et al., SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423 (2019). [DOI] [PubMed] [Google Scholar]
- 28.MacLean D., A convolutional neural network for predicting transcriptional regulators of genes in Arabidopsis transcriptome data reveals classification based on positive regulatory interactions. bioRxiv [Preprint] (2019). 10.1101/618926 (Accessed 27 February 2021). [DOI]
- 29.Michigan State University , Medicinal Plant Genomics Resource. http://medicinalplantgenomics.msu.edu/. Accessed 27 February 2020.
- 30.Kohnen-Johannsen K. L., Kayser O., Tropane alkaloids: Chemistry, pharmacology, biosynthesis and production. Molecules 24, 1–23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hildreth S. B., et al., Tobacco nicotine uptake permease (NUP1) affects alkaloid metabolism. Proc. Natl. Acad. Sci. U.S.A. 108, 18179–18184 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haydon M. J., Cobbett C. S., A novel major facilitator superfamily protein at the tonoplast influences zinc tolerance and accumulation in Arabidopsis. Plant Physiol. 143, 1705–1719 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Brito Francisco R., Martinoia E., The vacuolar transportome of plant specialized metabolites. Plant Cell Physiol. 59, 1326–1336 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Nelson B. K., Cai X., Nebenführ A., A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Al-Harrasi I., et al., A novel tonoplast Na+/H+ antiporter gene from date palm (PdNHX6) confers enhanced salt tolerance response in Arabidopsis. Plant Cell Rep. 39, 1079–1093 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Peng Q., et al., Functional analysis reveals the regulatory role of PpTST1 encoding tonoplast sugar transporter in sugar accumulation of peach fruit. Int. J. Mol. Sci. 21, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yadav A. K., et al., A rice tonoplastic calcium exchanger, OsCCX2 mediates Ca2+/cation transport in yeast. Sci. Rep. 5, 17117 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. L., Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305, 567–580 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Zheng W., et al., Folding non-homology proteins by coupling deep-learning contact maps with I-TASSER assembly simulations. in press. [DOI] [PMC free article] [PubMed]
- 40.Alguel Y., et al., Structure of eukaryotic purine/H(+) symporter UapA suggests a role for homodimerization in transport activity. Nat. Commun. 7, 11336 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Decottignies A., et al., ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J. Biol. Chem. 273, 12612–12622 (1998). [DOI] [PubMed] [Google Scholar]
- 42.Tachihara K., Uemura T., Kashiwagi K., Igarashi K., Excretion of putrescine and spermidine by the protein encoded by YKL174c (TPO5) in Saccharomyces cerevisiae. J. Biol. Chem. 280, 12637–12642 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Srinivasan P., Smolke C. D., Engineering a microbial biosynthesis platform for de novo production of tropane alkaloids. Nat. Commun. 10, 3634 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zanger U. M., Schwab M., Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 138, 103–141 (2013). [DOI] [PubMed] [Google Scholar]
- 45.LeDuc B. W., et al., Norcocaine and N-hydroxynorcocaine formation in human liver microsomes: Role of cytochrome P-450 3A4. Pharmacology 46, 294–300 (1993). [DOI] [PubMed] [Google Scholar]
- 46.Inui H., Maeda A., Ohkawa H., Molecular characterization of specifically active recombinant fused enzymes consisting of CYP3A4, NADPH-cytochrome P450 oxidoreductase, and cytochrome b5. Biochemistry 46, 10213–10221 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Nowak M., Selmar D., Cellular distribution of alkaloids and their translocation via phloem and xylem: The importance of compartment pH. Plant Biol. 18, 879–882 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Phillipson J. D., Handa S. S., Gorrod J. W., Metabolic N-oxidation of atropine, hyoscine and the corresponding nor-alkaloids by guinea-pig liver microsomal preparations. J. Pharm. Pharmacol. 28, 687–691 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hartmann T., Ober D., “Defense by pyrrolizidine alkaloids: Developed by plants and recruited by insects” in Induced Plant Resistance to Herbivory, Schaller A., Ed. (Springer, Dordrecht, The Netherlands, 2008), pp. 213–231. [Google Scholar]
- 50.Macel M., Attract and deter: A dual role for pyrrolizidine alkaloids in plant-insect interactions. Phytochem. Rev. 10, 75–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naumann C., Hartmann T., Ober D., Evolutionary recruitment of a flavin-dependent monooxygenase for the detoxification of host plant-acquired pyrrolizidine alkaloids in the alkaloid-defended arctiid moth Tyria jacobaeae. Proc. Natl. Acad. Sci. U.S.A. 99, 6085–6090 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sehlmeyer S., et al., Flavin-dependent monooxygenases as a detoxification mechanism in insects: New insights from the arctiids (lepidoptera). PLoS One 5, e10435 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang L., Beuerle T., Timbilla J., Ober D., Independent recruitment of a flavin-dependent monooxygenase for safe accumulation of sequestered pyrrolizidine alkaloids in grasshoppers and moths. PLoS One 7, e31796 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kotopka B. J., Smolke C. D., Model-driven generation of artificial yeast promoters. Nat. Commun. 11, 2113 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lau W., Sattely E. S., Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone. Science 349, 1224–1228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feyder S., De Craene J. O., Bär S., Bertazzi D. L., Friant S., Membrane trafficking in the yeast Saccharomyces cerevisiae model. Int. J. Mol. Sci. 16, 1509–1525 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Komarova N. Y., Meier S., Meier A., Grotemeyer M. S., Rentsch D., Determinants for Arabidopsis peptide transporter targeting to the tonoplast or plasma membrane. Traffic 13, 1090–1105 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Wolfenstetter S., Wirsching P., Dotzauer D., Schneider S., Sauer N., Routes to the tonoplast: The sorting of tonoplast transporters in Arabidopsis mesophyll protoplasts. Plant Cell 24, 215–232 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leng G., Song K., Watch out for your TRP1 marker: The effect of TRP1 gene on the growth at high and low temperatures in budding yeast. FEMS Microbiol. Lett. 363, 1–2 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Abe F., Iida H., Pressure-induced differential regulation of the two tryptophan permeases Tat1 and Tat2 by ubiquitin ligase Rsp5 and its binding proteins, Bul1 and Bul2. Mol. Cell. Biol. 23, 7566–7584 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sheludko Y. V., Volk J., Brandt W., Warzecha H., Expanding the diversity of plant monoterpenoid indole alkaloids employing human cytochrome P450 3A4. ChemBioChem 21, 1976–1980 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson J. M., Khoshgoftaar T. M., Survey on deep learning with class imbalance. J. Big Data 6, 27 (2019). [Google Scholar]
- 63.Chawla N. V., Bowyer K. W., Hall L. O., Kegelmeyer W. P., SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 16, 321–357 (2002). [Google Scholar]
- 64.Menardi G., Torelli N., Training and assessing classification rules with imbalanced data. Data Min. Knowl. Discovery 28, 92–122 (2014). [Google Scholar]
- 65.Kuhn M., Building predictive models in R using the caret package. J. Stat. Softw. 28, 1–26 (2008).27774042 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this work are available within the paper and its supporting information files. The cDNA and protein sequences of AbPUP1 and AbLP1 have been deposited in GenBank under accession nos. MW715669 (AbPUP1) and MW715670 (AbLP1). Sequences are also provided in SI Appendix, Table S2. Accession numbers for previously reported gene and protein sequences in the GenBank/UniProt databases are provided in SI Appendix, Table S4. The custom R scripts used for linear regression analysis and hierarchical clustering, binary classifier model development, and gene prediction are publicly available on the Smolke Laboratory GitHub: https://github.com/smolkelab/Supervised_TA_prediction.