Significance
Deficiencies or defects in nuclear lamins result in cell toxicity and cause multiple human diseases. Nuclear membrane (NM) ruptures have been implicated in both cell toxicity and disease pathogenesis, but the factors that modulate susceptibility to NM ruptures are unknown. We observed low levels of LAP2β, a chromatin-tethering inner NM protein, in nuclear lamin–deficient neurons and fibroblasts with frequent NM ruptures. Further reducing LAP2β expression markedly increased NM ruptures (without affecting rupture duration), whereas increasing LAP2β virtually abolished NM ruptures—even in cells subjected to mechanical stress. The discovery that increased expression of LAP2β in nuclear lamin–deficient cells prevents NM ruptures represents an unexpected and welcome insight into factors that modulate the structural integrity of the nuclear membranes.
Keywords: nuclear lamins, nuclear membrane ruptures, lamina-associated polypeptide 2, nuclear envelope
Abstract
Defects or deficiencies in nuclear lamins cause pathology in many cell types, and recent studies have implicated nuclear membrane (NM) ruptures as a cause of cell toxicity. We previously observed NM ruptures and progressive cell death in the developing brain of lamin B1–deficient mouse embryos. We also observed frequent NM ruptures and DNA damage in nuclear lamin–deficient fibroblasts. Factors modulating susceptibility to NM ruptures remain unclear, but we noted low levels of LAP2β, a chromatin-binding inner NM protein, in fibroblasts with NM ruptures. Here, we explored the apparent link between LAP2β and NM ruptures in nuclear lamin–deficient neurons and fibroblasts, and we tested whether manipulating LAP2β expression levels would alter NM rupture frequency. In cortical plate neurons of lamin B1–deficient embryos, we observed a strong correlation between low LAP2β levels and NM ruptures. We also found low LAP2β levels and frequent NM ruptures in neurons of cultured Lmnb1−/− neurospheres. Reducing LAP2β expression in Lmnb1−/− neurons with an siRNA markedly increased the NM rupture frequency (without affecting NM rupture duration), whereas increased LAP2β expression eliminated NM ruptures and reduced DNA damage. Consistent findings were observed in nuclear lamin–deficient fibroblasts. Reduced LAP2β expression increased NM ruptures, whereas increased LAP2β expression virtually abolished NM ruptures. Increased LAP2β expression nearly abolished NM ruptures in cells subjected to mechanical stress (an intervention that increases NM ruptures). Our studies showed that increasing LAP2β expression bolsters NM integrity in nuclear lamin–deficient cells and markedly reduces NM rupture frequency.
We recently examined mouse embryonic fibroblasts (MEFs) that were homozygous for knockout mutations in the three nuclear lamin genes (Lmnb1, Lmnb2, and Lmna) (1). These “triple-knockout fibroblasts” (TKO MEFs) lacked nuclear blebs but nevertheless had frequent nuclear membrane (NM) ruptures (1). The NM ruptures were associated with increased DNA damage (1). The frequency of NM ruptures in TKO MEFs increased when cells were subjected to mechanical stress (uniaxial stretching) and decreased when cells were subjected to interventions that interfere with the transmission of cytoskeletal forces to the nucleus (1). In follow-up studies, we observed frequent NM ruptures, along with DNA damage and cell death, in migrating neurons within the cortical plate of lamin B1–deficient mouse embryos (2). Cortical plate neurons are particularly susceptible to NM ruptures because neuronal migration subjects the cell nucleus to mechanical stress (2) and because embryonic neurons do not express lamin A or lamin C (3–6). In studies of TKO MEFs and Lmnb1−/− neurons (1, 2), NM ruptures were identified by the escape of a nuclear-localized fluorescent reporter into the cytoplasm.
In our studies of NM ruptures in TKO MEFs (1), the outline of the cell nucleus was visualized with an antibody against the inner nuclear membrane protein LAP2β (lamina-associated polypeptide 2, β-isoform). LAP2β contains a transmembrane helix that anchors it to the inner nuclear membrane (7–9), and it interacts, via nucleoplasmic LEM (LAP2, emerin, and Man1) domains (10, 11), with the DNA-bridging protein BAF (barrier-to-autointegration factor). This LEM–BAF complex is thought to tether chromatin to the nuclear periphery (12). In TKO MEFs, we found, by confocal microscopy, reduced amounts of LAP2β in TKO MEFs with NM ruptures. In those cells, there were also gaps in the distribution of LAP2β along the nuclear rim (1). In TKO MEFs without NM ruptures, LAP2β was distributed evenly along the nuclear periphery (1). In our initial study of TKO MEFs (1), we focused on the impact of mechanical stress on NM ruptures and did not pursue the possibility that LAP2β expression might modulate susceptibility of cells to NM ruptures.
In the current studies, we investigated the possibility of a link between LAP2β expression and NM ruptures in nuclear lamin–deficient cells. We had two goals. The first was to investigate the potential association between abnormalities in LAP2β expression and distribution and NM ruptures—both in tissues of a nuclear lamin–deficient mouse and in a second nuclear lamin–deficient cell line. We began by studying the developing brain of Lmnb1−/− embryos. We reasoned that we would be able to assess LAP2β expression, LAP2β distribution, and NM ruptures in migrating neurons of the cortical plate (a process that subjects the nucleus to mechanical stress) and in the germinal cells of the ventricular zone (where stresses on the nucleus are low) (2). We further reasoned that Lmnb1−/− embryos would be a useful source of primary neurons for cell culture studies of NM ruptures (2).
Our second goal was to examine whether LAP2β expression levels in nuclear lamin–deficient cells are directly relevant to the susceptibility to NM ruptures. Specifically, we wanted to test the hypothesis that manipulating LAP2β expression levels would change NM rupture frequency. Initially, we were skeptical that we would find any effect of LAP2β expression levels. Earlier studies identified indirect effects of other LEM domain proteins on the repair of NM ruptures (12–16), but no one had considered the idea that altering LAP2β expression levels would influence the frequency of NM ruptures. Despite our initial skepticism, we tested whether manipulating LAP2β expression would affect the frequency of NM ruptures in TKO fibroblasts and Lmnb1−/− neurons. To our surprise, we found that reducing LAP2β expression levels markedly increased the frequency—but not the duration—of NM ruptures. Increasing LAP2β expression virtually abolished NM ruptures.
Results
NM Ruptures and LAP2β Distribution in Cortical Neurons of Lmnb1-Deficient Mouse Embryos.
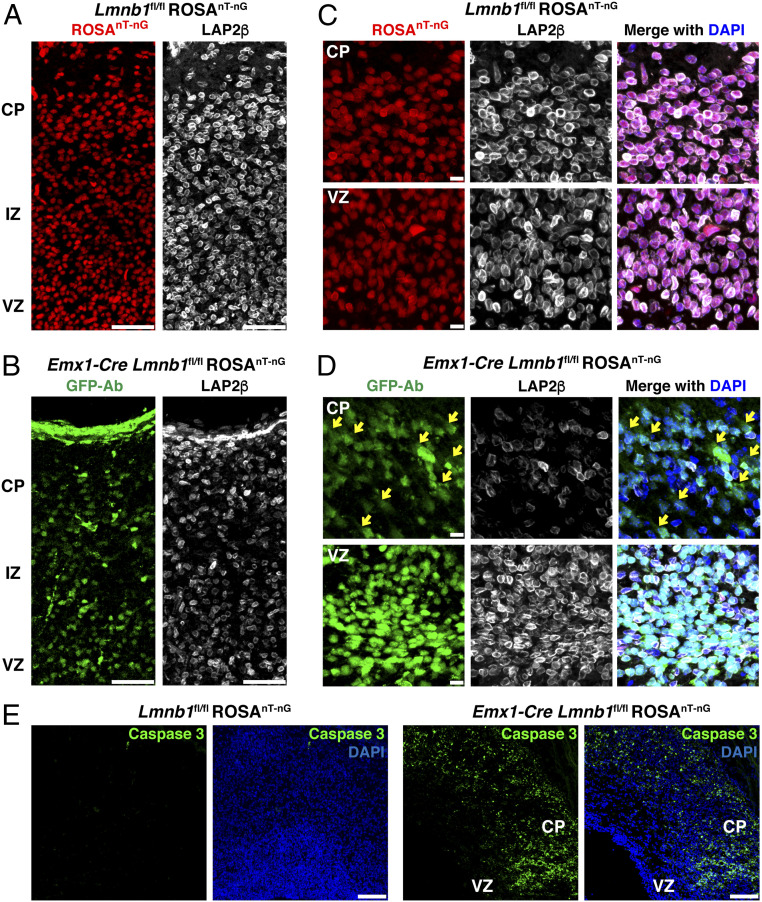
We bred Lmnb1fl/fl mouse embryos (17) harboring the forebrain-specific Emx1-Cre transgene (18) and a nuclear-targeted ROSAnT-nG transgene (2). The ROSAnT-nG transgene produces a tdTomato fluorescent reporter in the absence of Cre and GFP in the presence of Cre (Fig. 1 A and B). In embryonic day 18.5 (E18.5) Lmnb1fl/fl ROSAnT-nG embryos (where lamin B1 expression is normal), the ROSAnT-nG reporter was located in the nucleus and LAP2β was distributed evenly along the nuclear rim (Fig. 1C). In migrating neurons in the cortical plate of Emx1-Cre Lmnb1fl/fl ROSAnT-nG embryos (where lamin B1 is absent), numerous NM ruptures were present, evident by escape of the fluorescent reporter into the cytoplasm (Fig. 1D). Also, the levels of LAP2β were reduced and/or unevenly distributed along the nuclear rim (Fig. 1 B and D). In the germinal cells within the ventricular zone (which are not subjected to stresses of cell migration), we did not observe clear examples of NM rupture (the reporter was largely confined to the nucleus) (Fig. 1D). Also, LAP2β expression was robust and distributed evenly along the nuclear rim in ventricular zone cells (Fig. 1D). In cortical plate neurons of Emx1-Cre Lmnb1fl/fl ROSAnT-nG embryos, we observed increased amounts of caspase 3, a marker of apoptotic cell death (Fig. 1E). Caspase 3 staining was absent in cells of the ventricular zone (Fig. 1E).
Fig. 1.
Forebrain-specific inactivation of Lmnb1 results in reduced neuronal cell density, reduced LAP2β expression, and NM ruptures. (A and B) Fluorescence microscopy images of the cerebral cortex of E18.5 Lmnb1fl/fl ROSAnT-nG (A) and Emx1-Cre Lmnb1fl/fl ROSAnT-nG (B) mouse embryos. The ROSAnT-nG transgene produces a nuclear-targeted tdTomato reporter in the absence of Cre and a nuclear-targeted GFP in the presence of Cre. tdTomato is colored red; GFP is colored green. Sections were stained with antibodies against the inner nuclear membrane protein LAP2β (white) and GFP (green). (Scale bars, 50 μm.) (C and D) Higher-magnification images of cortical plate (CP) neurons and ventricular zone (VZ) cells in E18.5 Lmnb1fl/fl ROSAnT-nG (C) and Emx1-Cre Lmnb1fl/fl ROSAnT-nG (D) mouse embryos. DNA was stained with DAPI (blue). Yellow arrows point to NM ruptures in CP neurons in the Emx1-Cre Lmnb1fl/fl ROSAnT-nG embryo. CP neurons with NM ruptures exhibited reduced levels of LAP2β and uneven LAP2β distribution along the nuclear rim. LAP2β distribution was normal in VZ cells of Emx1-Cre Lmnb1fl/fl ROSAnT-nG embryos, and the fluorescent reporter was largely confined to the nucleus. The VZ contains dividing cells; consequently, low levels of GFP were detected in the cytoplasm of some cells. (Scale bars, 10 μm.) (E) Immunofluorescence microscopy of the cerebral cortex in E18.5 Lmnb1fl/fl ROSAnT-nG and Emx1-Cre Lmnb1fl/fl ROSAnT-nG mouse embryos after staining sections with an antibody against caspase 3 (a marker of apoptotic cell death; green). Caspase 3 was confined to CP neurons and was absent from cells of the VZ. DNA was stained with DAPI (blue). (Scale bars, 100 μm.)
In 3.5-mo-old Emx1-Cre Lmnb1fl/fl ROSAnT-nG mice, LAP2β levels in the cerebral cortex were much lower than in wild-type (WT) mice, and the LAP2β was often distributed unevenly along the nuclear rim (SI Appendix, Fig. S1 A and B).
Neurons Harvested from the Cerebral Cortex of Lmnb1−/− Embryos Have Frequent NM Ruptures and Reduced Amounts of LAP2β.
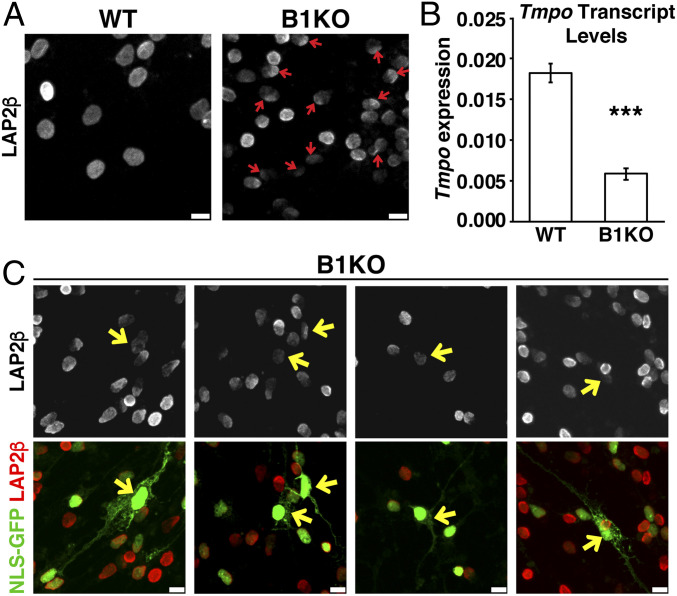
Neuronal progenitor cells from Lmnb1+/+ (WT) and Lmnb1−/− (lamin B1 knockout; B1KO) were cultured as neurospheres and allowed to differentiate. In WT neurons, there was some variation in LAP2β levels but the protein was always distributed evenly along the nuclear rim. In some B1KO neurons, LAP2β was present in reduced amounts or distributed unevenly (Fig. 2A). The reduced amounts of LAP2β in B1KO neurons may have been due, at least in part, to reduced levels of LAP2β transcripts (Fig. 2B). In B1KO neurons with a NM rupture, LAP2β staining was less intense and/or was distributed unevenly along the nuclear rim (Fig. 2C). Confocal microscopy revealed that LAP2β distribution in B1KO nuclei was polarized, with disproportionate amounts of LAP2β located on the side of the nucleus closest to the centrosome (SI Appendix, Fig. S2).
Fig. 2.
LAP2β expression in Lmnb1−/− (B1KO) embryos. (A) Immunofluorescence microscopy of Lmnb1+/+ (WT) and Lmnb1−/− (B1KO) neurons after staining cells with an antibody against LAP2β (white). Red arrows point to B1KO neurons with low levels of LAP2β and/or abnormally distributed LAP2β along the nuclear rim. (Scale bars, 10 μm.) (B) Transcript levels for Tmpo (the gene for LAP2β) in WT and B1KO neurons. Neurons had been differentiated in culture for 48 h. Transcript levels were normalized to Ppia transcripts. Bar graph shows mean ± SEM; n = 3 independent experiments. ***P < 0.0005; unpaired Student’s t test. (C) Immunofluorescence microscopy of B1KO neurons that expressed a NLS-GFP (green) after staining the cells with an antibody against LAP2β. In the Top row, LAP2β is white; in the Bottom row, LAP2β is red. Yellow arrows point to neurons with a NM rupture. LAP2β levels were low in neurons with a NM rupture. (Scale bars, 10 μm.)
In cortical plate neurons of E18.5 Emx1-Cre Lmnb1fl/fl embryos, both LAP2β and lamin B2 were distributed disproportionately to one side of the nucleus (SI Appendix, Fig. S3A). LAP2β and lamin B2 were also distributed to one side of the nucleus in cultured B1KO neurons (SI Appendix, Fig. S3B). To assess the distributions of LAP2β and lamin B2 across the nuclear area, individual nuclei were analyzed uniaxially and the fluorescence intensities of LAP2β and lamin B2 were quantified along the diameter of the cell nucleus in the direction of polarization (SI Appendix, Fig. S4A). In wild-type neurons, all nuclei had even distributions of LAP2β and lamin B2 (SI Appendix, Fig. S4B). In the majority of B1KO neurons, LAP2β and lamin B2 were polarized toward the same half of the cell nucleus (SI Appendix, Fig. S4C).
Knocking Down LAP2β Expression in B1KO Neurons Increases NM Ruptures.
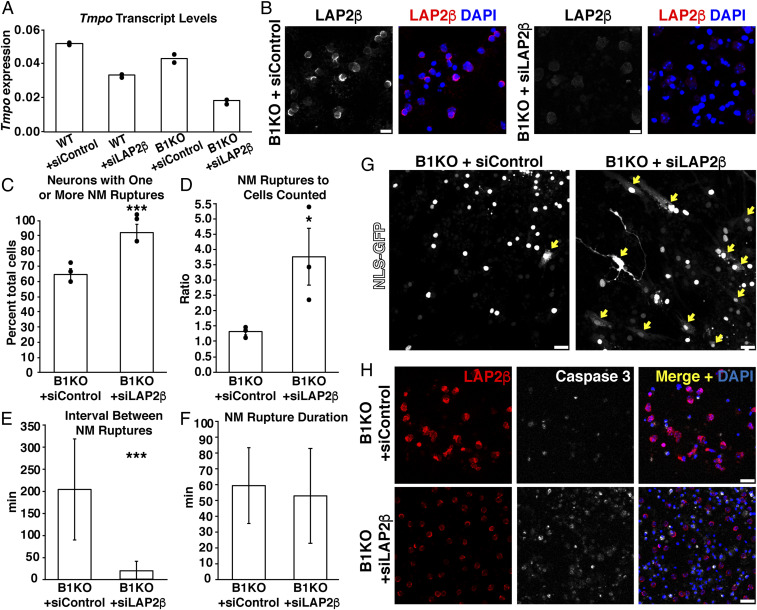
To explore the association between NM ruptures and LAP2β expression, we knocked down LAP2β expression in WT and B1KO neurons with an exon 2–specific Tmpo-specific small-interfering RNA (here designated “siLAP2β”). In parallel, neurons were also treated with a control siRNA (siControl). LAP2β transcript levels in siLAP2β-transfected neurons were reduced by ∼50% (Fig. 3A). The reduced amounts of LAP2β expression were readily detectable by immunofluorescence microscopy (Fig. 3B). Reduced LAP2β expression in B1KO neurons resulted in increased numbers of cells having at least one NM rupture during video microscopy (91.8% of 150 siLAP2β-transfected neurons imaged in three independent experiments had at least one NM rupture vs. 64.6% of 125 control siRNA–transfected neurons imaged in three independent experiments) (P < 0.0005; χ2 test) (Fig. 3C). Reduced LAP2β also increased the total number of NM ruptures (502 ruptures in 150 siLAP2β-transfected neurons vs. 167 ruptures in 125 control siRNA–transfected neurons) (P < 0.05; unpaired Student’s t test) (Fig. 3D). Of note, knocking down LAP2β expression reduced the time between NM ruptures (P < 0.0005; unpaired Student’s t test) (Fig. 3E) but had little or no effect on NM rupture duration (Fig. 3F). Increased numbers of NM ruptures were apparent by confocal microscopy (Fig. 3G). The vast majority of neurons with NM ruptures ruptured and repaired during continuous video microscopy (obviating any concerns that apoptotic cells were counted as NM ruptures). The larger numbers of NM ruptures in siLAP2β-transfected B1KO neurons were accompanied by increased apoptotic cell death (as judged by caspase 3 staining) (Fig. 3H). Reduced LAP2β expression affected NM rupture frequency only in B1KO neurons. Transfection of WT neurons with siLAP2β resulted in reduced amounts of LAP2β in the cell nucleus by immunofluorescence microscopy (SI Appendix, Fig. S5A), but we found no NM ruptures in >100 siLAP2β-transfected WT neurons during 20 h of imaging (SI Appendix, Fig. S5B). In WT cells, the ∼50% Tmpo knockdown that we achieved with the siRNA did not result in adverse phenotypes (e.g., nuclear shape abnormalities, cell death).
Fig. 3.
Reducing LAP2β expression in lamin B1–deficient (B1KO) neurons with siLAP2β (a small-interfering RNA against Tmpo) increases NM ruptures and results in more cell death. (A) Transcript levels for Tmpo in Lmnb1+/+ (WT) and B1KO neurons that had been transfected with siLAP2β or a control siRNA (siControl). Bar depicts the mean in two experiments; dots show results in two independent experiments; transcript levels were normalized to Ppia transcripts. Neurons were examined 24 h after transfections. (B) Confocal micrographs of B1KO neurons transfected with siLAP2β or siControl. Neurons were stained with an antibody against LAP2β (white in the Left; red in the merged image). DNA was stained with DAPI (blue). (Scale bars, 10 μm.) (C) Percentages of siLAP2β- or siControl-transfected B1KO neurons with a NM rupture. ***P < 0.0005; χ2 test. Mean ± SEM; n = 3 independent experiments; 20 h of imaging/experiment. Dots depict percentages in three independent experiments. In three independent experiments, a total of 150 siLAP2β-transfected neurons and 125 control siRNA–transfected neurons were analyzed. (D) Ratio of the total number of NM ruptures to the number of neurons counted. Dots depict ratios in three independent experiments. *P < 0.05; unpaired Student’s t test. In three independent experiments, a total of 150 siLAP2β-transfected neurons and 125 control siRNA–transfected neurons were analyzed. (E and F) Bar graphs depict the mean time between NM ruptures (E) and the mean duration of NM ruptures (F) in siLAP2β- or siControl-transfected B1KO neurons (mean ± SD; >35 NM ruptures analyzed/group). ***P < 0.0005; unpaired Student’s t test. (G) Confocal micrographs of siLAP2β- or siControl-transfected B1KO neurons that expressed NLS-GFP (white). Larger numbers of NM ruptures (evident by escape of GFP into the cytoplasm and/or sudden loss of GFP fluorescence within the cell nucleus; yellow arrows) were observed in siLAP2β-transfected B1KO neurons. (Scale bars, 25 μm.) (H) Confocal micrographs of siLAP2β- or siControl-transfected B1KO neurons after staining with antibodies against LAP2β (red) and caspase 3 (white); DNA was stained with DAPI (blue). (Scale bars, 20 μm.)
Overexpressing LAP2β Abolishes NM Ruptures in B1KO Neurons.
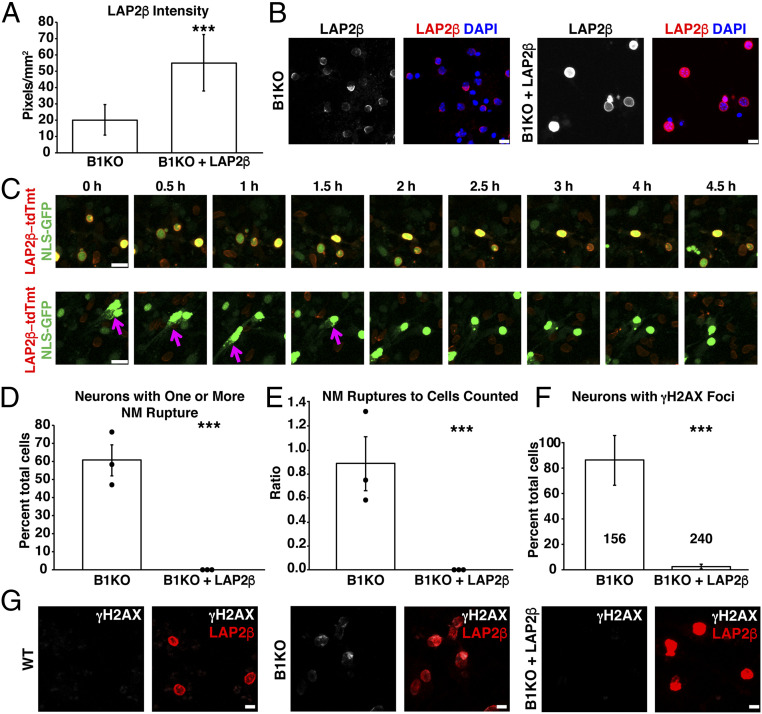
To examine the effect of increased LAP2β expression on NM ruptures, we transduced NLS-GFP–expressing B1KO neurons with a lentivirus encoding a LAP2β-tdTomato fusion protein. The transduced B1KO neurons were not subjected to drug selection; hence, some cells expressed the fusion protein while others did not. After 48 h, the cells were stained with a LAP2β-specific antibody (to assess total levels of LAP2β) and imaged by confocal microscopy. The binding of the LAP2β antibody (as judged by the ratio of LAP2β pixel intensities to nuclear area) was significantly greater in the LAP2β-tdTomato–transduced cells (n = 85) than in nontransduced cells (P < 0.0001; unpaired Student’s t test) (Fig. 4A). In the LAP2β-tdTomato–transduced cells, LAP2β staining was detectable along the entire circumference of the nucleus (Fig. 4B). In nontransduced cells, LAP2β staining was less intense and distributed unevenly along the nuclear rim (Fig. 4B). LAP2β-tdTomato expression did not alter nuclear shape.
Fig. 4.
Higher levels of LAP2β expression in differentiated B1KO neurons prevents NM ruptures. WT and B1KO neurons were transduced with a lentivirus encoding a LAP2β-tdTomato fusion protein. (A) Fluorescence intensity in nontransduced and LAP2β-tdTomato–transduced B1KO neurons. Mean ± SD ***P < 0.0001; unpaired Student’s t test. After staining neurons with an antibody against LAP2β, confocal micrographs were recorded and 10 images were analyzed by ImageJ. Fluorescence intensity for each nucleus was quantified and normalized to nuclear area (in square millimeters). (B) Immunofluorescence microscopy of nontransduced and LAP2β-tdTomato–transduced B1KO neurons. Cells were stained with an antibody against LAP2β (white on the Left; red in the merged image). DNA was stained with DAPI (blue). (Scale bars, 10 μm.) (C) Time-lapse microscopy study of NLS-GFP–expressing B1KO neurons (green) that had been incubated with a LAP2β-tdTomato lentivirus. The neurons were not subjected to drug selection; hence, some of the neurons did not express LAP2β-tdTomato (red). (Top) Micrographs of a field containing NLS-GFP–expressing B1KO neurons that were successfully transduced with LAP2β-tdTomato. NM ruptures were never observed in NLS-GFP–expressing B1KO neurons that expressed LAP2β-tdTomato. (Bottom) Confocal micrographs of another field in which several cells expressed NLS-GFP but no LAP2β-tdTomato. Neurons that expressed only NLS-GFP exhibited NM ruptures (evident by escape of GFP into the cytoplasm) (magenta arrows). (Scale bars, 20 μm.) (D) Percentages of NLS-GFP–expressing nontransduced B1KO neurons (n = 148) and LAP2β-tdTomato–transduced B1KO neurons (n = 101) that had at least one NM rupture (detected by escape of GFP into the cytoplasm). Mean ± SEM; n = 3 independent experiments; 20 h of imaging/experiment. Dots depict percentages in three independent experiments. ***P < 0.0005; χ2 test. No NM ruptures were detected in 101 LAP2β-tdTomato–transduced B1KO neurons. A total of 148 nontransduced B1KO neurons were evaluated in the three experiments. (E) Ratio of the total number of NM ruptures in nontransduced and LAP2β-tdTomato–transduced B1KO neurons divided by the total number of neurons analyzed. Mean ± SEM; n = 3 independent experiments; ***P < 0.0005; unpaired Student’s t test. No NM ruptures were observed in LAP2β-tdTomato–transduced B1KO neurons. A total of 148 nontransduced B1KO neurons were evaluated. (F) Percentages of nontransduced and LAP2β-tdTomato–transduced B1KO neurons with γH2AX foci. A total of 240 LAP2β-tdTomato–transduced B1KO neurons and 156 nontransduced B1KO neurons were analyzed. Mean ± SEM; n = 3 independent experiments. ***P < 0.0005; χ2 test. The total number of neurons scored are recorded with each bar. (G) Confocal micrographs of nontransduced and LAP2β-tdTomato–transduced B1KO neurons after staining the cells with antibodies against LAP2β (red) and the DNA damage marker γH2AX (white). (Scale bars, 10 μm.)
To assess the effect of LAP2β-tdTomato expression on NM integrity, we counted NM ruptures in nontransduced and LAP2β-tdTomato–transduced B1KO neurons (Fig. 4C and Movies S1 and S2). By live-cell microscopy, 60.6% of 148 nontransduced B1KO neurons analyzed over three independent experiments had at least one NM rupture (P < 0.0005; χ2 test) (Fig. 4D). Some of the nontransduced B1KO neurons had more than one rupture (142 NM ruptures were observed in the 148 neurons) (P < 0.0005; unpaired Student’s t test) (Fig. 4E). In contrast, no NM ruptures were detected in 101 LAP2β-tdTomato–transduced B1KO cells (Fig. 4 D and E). LAP2β-tdTomato expression protected B1KO neurons from DNA damage (Fig. 4 F and G). Only 2.4% of 240 LAP2β-tdTomato–transduced B1KO neurons had γH2AX foci in the nucleus, whereas >80% of 156 nontransduced B1KO neurons analyzed had nuclear γH2AX foci (P < 0.0005; χ2 test) (Fig. 4F). We assessed γH2AX foci in a mixed population of nontransduced and LAP2β-tdTomato–transduced B1KO neurons; γH2AX foci were rare in 240 LAP2β-tdTomato–expressing B1KO neurons but frequent in 156 nontransduced cells (Fig. 4G).
Reducing LAP2β Expression in TKO MEFs Results in More NM Ruptures and Cell Death.
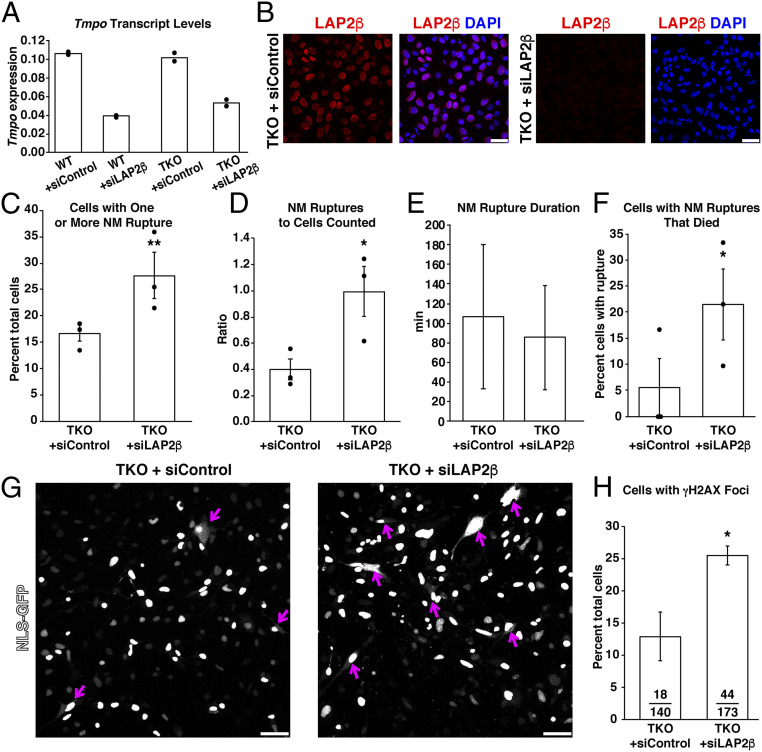
Transfection of WT and TKO MEFs with siLAP2β decreased LAP2β transcript levels by 50 to 60% (Fig. 5A), and microscopy revealed reduced amounts of LAP2β in the nucleus, consistent with the transcript levels (Fig. 5B). Reducing LAP2β expression in TKO MEFs increased the percentage of cells with NM ruptures: 27.7% of siLAP2β-transfected TKO MEFs (210 total cells counted in three independent experiments) had NM ruptures, whereas only 16.7% (140 total cells counted in three independent experiments) of control siRNA–transfected cells had a NM rupture (P = 0.009; χ2 test). Some TKO MEFs had more than one NM rupture; we detected 213 NM ruptures in 210 siLAP2β-transfected cells but only 58 ruptures in the 140 control siRNA–transfected cells (P < 0.05; unpaired Student’s t test) (Fig. 5 C and D). The duration of NM ruptures in control siRNA–transfected and siLAP2β-transfected TKO MEFs was similar (Fig. 5E). In 60 siLAP2β-transfected TKO MEFs with at least one NM rupture, 16 of the cells died during 20 h of imaging, whereas the remainder survived, only 1 of 23 control siRNA–transfected TKO MEFs with a NM rupture died (P = 0.009; χ2 test) (Fig. 5F). The increased frequency of NM ruptures in siLAP2β-transfected TKO MEFs (Fig. 5G) was accompanied by increased DNA damage (25.4% of siLAP2β-transfected TKO MEFs had nuclear γH2AX foci vs. only 12.8% of TKO MEFs transfected with the control siRNA) (Fig. 5H).
Fig. 5.
Reducing LAP2β expression in Lmnb1−/−Lmnb2−/−Lmna−/− fibroblasts (TKO) with siLAP2β (a small-interfering RNA against Tmpo) increases the frequency of NM ruptures and cell death. (A) Transcript levels for Tmpo (the gene for LAP2β) in WT and TKO MEFs transfected with siLAP2β or siControl. Means from two independent experiments; dots show results in the two independent experiments; transcript levels were normalized to Ppia. Analyses were performed 24 h after the transfection. **P < 0.005; unpaired Student’s t test. (B) Immunofluorescence micrographs of TKO MEFs transfected with siLAP2β or siControl. MEFs were stained with an antibody against LAP2β (red). DNA was stained with DAPI (blue). (Scale bars, 50 μm.) (C) Percentages of siLAP2β- and control siRNA–transfected TKO MEFs with at least one NM rupture. In 140 control siRNA–transfected TKO MEFs analyzed in three independent experiments, NM ruptures were detected in 23 cells. In 210 siLAP2β-transfected TKO MEFs, 60 cells had a NM rupture. Mean ± SEM; n = 3 independent experiments, dots show the percentages in each experiment (20 h of imaging/experiment). **P = 0.009; χ2 test. (D) Ratio of total number of total NM ruptures over the total number of MEFs evaluated. Mean ± SEM; n = 3 independent experiments, dots show the ratio in each experiment (20 h of imaging/experiment). In 140 control siRNA–transfected TKO MEFs analyzed in three experiments, 58 NM ruptures were detected. In 210 siLAP2β-transfected TKO MEFs, 213 NM ruptures were detected. *P < 0.05; unpaired Student’s t test. (E) NM rupture duration in siLAP2β- or siControl-transfected TKO MEFs (mean ± SD; >35 NM ruptures/group). (F) Percentages of siLAP2β- or control siRNA–transfected TKO MEFs that had a NM rupture and subsequently died during 20 h of imaging. Mean ± SEM; n = 3 independent experiments; dots show the percentage in each experiment. Of the 23 control siRNA–transfected TKO MEFs that exhibited a NM rupture, 1 died. Of 60 siLAP2β-transfected TKO MEFs that exhibited a NM rupture, 16 died. *P = 0.024; χ2 test. (G) Confocal micrographs of siLAP2β- or siControl-transfected TKO MEFs that expressed NLS-GFP (white). Numerous NM ruptures (magenta arrows) were observed in the siLAP2β-transfected TKO MEFs, evident by escape of GFP into the cytoplasm and/or sudden loss of GFP fluorescence within the cell nucleus. (Scale bars, 25 μm.) (H) Percentages of siLAP2β- or control siRNA–transfected TKO MEFs that exhibited nuclear γH2AX foci. Bars depict the mean from two independent experiments (140 control siRNA–transfected TKO MEFs and 173 siLAP2β-transfected TKO MEFs scored); dots depict percentages in the two experiments.
Increased LAP2β Expression in TKO MEFs Abolishes NM Ruptures and Reduces DNA Damage.
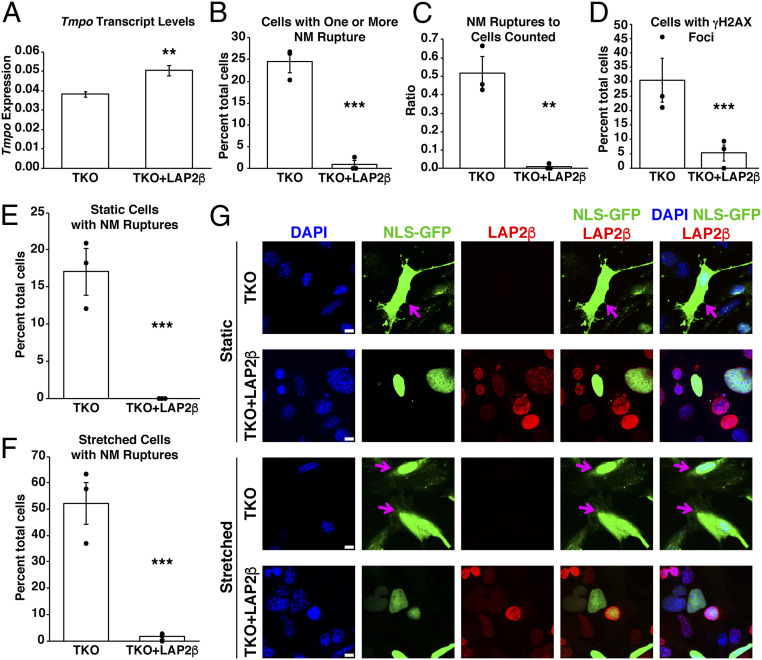
We transduced NLS-GFP–expressing TKO MEFs with the LAP2β-tdTomato lentivirus, resulting in a 32.3% increase in LAP2β transcript levels (P = 0.0079; unpaired Student’s t test) (Fig. 6A). We analyzed transduced NLS-GFP–expressing TKO MEFs with high levels of LAP2β-tdTomato expression (cells in the top 10 percentile for both GFP and tdTomato fluorescence). LAP2β-tdTomato had no effect on nuclear shape. In three independent experiments (>100 cells counted/group), ∼24.5% of nontransduced TKO MEFs exhibited one or more NM ruptures vs. only 0.85% of LAP2β-tdTomato–transduced cells (P < 0.0001; χ2 test) (Fig. 6B). We observed a total of 69 NM ruptures in 141 nontransduced TKO MEFs but only 1 rupture in 103 LAP2β-tdTomato–transduced cells (P = 0.0055; unpaired Student’s t test) (Fig. 6C). The lower number of NM ruptures in LAP2β-tdTomato–transduced TKO MEFs was accompanied by reduced DNA damage. In 50 nontransduced cells, we observed 14 cells with γH2AX foci. In 101 LAP2β-tdTomato–transduced TKO MEFs, we observed only 5 cells with γH2AX foci (P < 0.0001; χ2 test) (Fig. 6D).
Fig. 6.
Transduction of wild-type and TKO MEFs with a lentivirus encoding a LAP2β-tdTomato fusion protein prevents NM ruptures and reduces DNA damage. (A) Transcript levels for Tmpo (the gene for LAP2β) in nontransduced or LAP2β-tdTomato–transduced TKO MEFs (mean ± SEM; n = 3 independent experiments). Transcript levels were normalized to Ppia transcripts. **P = 0.0079; unpaired Student’s t test. (B) Percentages of nontransduced and LAP2β-tdTomato–transduced TKO MEFs that had a NM rupture event during 20 h of live-cell imaging. In 141 nontransduced TKO MEFs, 34 MEFs had a NM rupture. In 103 LAP2β-tdTomato–expressing MEFs, only 1 cell had a NM rupture. Mean ± SEM from three independent experiments (20 h of imaging/experiment). ***P < 0.0005; χ2 test. Black dots depict percentages in each experiment. (C) Ratio of the number of NM ruptures in TKO MEFs and LAP2β-tdTomato–transduced MEFs over the total number of cells analyzed. In 141 nontransduced TKO MEFs, there were 69 NM ruptures. In 103 LAP2β-tdTomato–expressing MEFs, there was only one NM rupture. Mean ± SEM from three independent experiments; **P = 0.0055; unpaired Student’s t test. Black dots depict ratios in each experiment. (D) Percentages of nontransduced and LAP2β-tdTomato–transduced TKO MEFs with γH2AX foci. Mean ± SEM from three independent experiments; ***P < 0.0005; χ2 test. Black dots depict ratios in each experiment. In three independent experiments, 50 nontransduced and 101 LAP2β-tdTomato–expressing MEFs were analyzed. (E) Percentages of nontransduced and LAP2β-tdTomato–transduced TKO MEFs with a NM rupture when plated on PDMS membranes under static conditions. In 172 nontransduced TKO MEFs, 29 MEFs had a NM rupture. In 195 LAP2β-tdTomato–expressing MEFs, there were no NM ruptures. Shown are mean percentages ± SEM from three independent experiments; ***P < 0.0001; χ2 test. Black dots depict percentages in each experiment. (F) Percentages of nontransduced and LAP2β-tdTomato–transduced TKO MEFs with a NM rupture when the cells were subjected to uniaxial stretching on a PDMS membrane for 2 h. In 151 nontransduced TKO MEFs, 79 MEFs had a NM rupture. Among 198 LAP2β-tdTomato–expressing MEFs, three cells had a NM rupture. Shown are mean ± SEM from three independent experiments; ***P < 0.0001; χ2 test. Black dots depict percentages in each experiment. (G) Immunofluorescence microscopy of nontransduced and LAP2β-tdTomato–transduced TKO MEFs under static and stretched conditions. The TKO MEFs expressed NLS-GFP (green) alone or NLS-GFP and LAP2β-tdTomato (red). DNA was stained with DAPI (blue). Magenta arrows point to NM ruptures. (Scale bars, 10 μm.)
The fact that increased LAP2β expression prevented NM ruptures in TKO MEFs suggested that increased LAP2β expression bolsters the integrity of nuclear membranes. If this were the case, we reasoned that LAP2β overexpression in TKO MEFs would protect against NM ruptures in the setting of mechanical stress. To test this possibility, we counted NM ruptures in nontransduced and LAP2β-tdTomato–transduced TKO MEFs under static conditions (Fig. 6E) and under uniaxial stretching (Fig. 6F). As expected (1), stretching nontransduced TKO MEFs increased the frequency of NM ruptures (52.2% of stretched cells vs. 17% of cells tested under static conditions) (Fig. 6 E–G). No NM ruptures were detected in LAP2β-tdTomato–expressing TKO MEFs under static conditions (195 cells scored) (P < 0.0001; χ2 test) (Fig. 6 E and G). When the TKO MEFs were subjected to uniaxial stretching, we observed 3 cells (from a total of 198 cells) with a NM rupture (P < 0.0001; χ2 test) (Fig. 6 F and G). Remarkably, NM rupture frequency in LAP2β-tdTomato–expressing TKO MEFs under stretching conditions (3 NM ruptures in 198 cells) was much lower than the NM rupture frequency in nontransduced TKO MEFs under static conditions (29 NM ruptures in 172 cells) (P < 0.0001; χ2 test) (Fig. 6 F and G).
Discussion
In earlier studies of TKO MEFs (1), we observed that LAP2β was present in reduced amounts or abnormally distributed in cells with NM ruptures (1, 19). Initially, we thought that the finding might be inconsequential and simply represented another example of a nuclear envelope protein whose expression pattern was altered by a deficiency of a nuclear lamin (17, 20–25). In the end, however, we investigated this finding further, motivated by the fact that the abnormality in LAP2β distribution was not observed in every TKO fibroblast but was restricted to cells with NM ruptures (1). We hypothesized that LAP2β expression might actually influence the structural integrity of nuclear membranes. Our first goal was to determine whether the association of NM ruptures with reduced LAP2β was a peculiarity of TKO MEFs. We began by studying the forebrain of lamin B1–deficient mouse embryos. We had shown previously that lamin B1 deficiency disrupts the migration of neurons to the cortical plate and reduces neuronal survival (2, 17). Neuronal migration depends on nucleokinesis, a process in which the nucleus is pulled by cytoplasmic motors into the leading edge of the cell (26, 27). In the setting of lamin B1 deficiency, we suspected that the forces of nucleokinesis might induce NM ruptures. Indeed, NM ruptures were widespread in lamin B1–deficient cortical plate neurons. Moreover, virtually all cortical plate neurons had low levels of LAP2β and/or gaps or irregularities in LAP2β distribution. In contrast, LAP2β levels and distribution were normal—and NM ruptures were absent—in the germinal cells of the ventricular zone. We also observed frequent NM ruptures, accompanied by low levels of LAP2β and/or gaps in LAP2β distribution, in cultured Lmnb1−/− neurons. Thus, the association between NM ruptures and LAP2β abnormalities was not a peculiarity of TKO MEFs.
Our second objective was to determine whether manipulating LAP2β expression would modulate the frequency of NM ruptures. In both Lmnb1−/− neurons and TKO MEFs, knocking down LAP2β expression resulted in more frequent NM ruptures and more DNA damage. Conversely, transducing Lmnb1−/− neurons and TKO MEFs with a LAP2β-tdTomato lentivirus virtually abolished NM ruptures. We suspected that higher levels of LAP2β expression bolstered the integrity of nuclear membranes. This suspicion was strongly supported by studies in which TKO MEFs were subjected to mechanical stretching. We found frequent NM ruptures in nontransduced TKO MEFs under static conditions, but the rupture frequency increased markedly when the cells were stretched. In LAP2β-tdTomato–transduced TKO MEFs, NM ruptures were abolished under static conditions and were extremely rare when the cells were stretched.
LEM domain proteins (9) have been implicated in the repair of NM ruptures (12–14, 16). Cytosolic barrier-to-autointegration factor (BAF) binds to chromatin at sites of NM ruptures and leads to rapid binding of several LEM domain proteins but not LAP2β (13, 16). LEMD2 binding to sites of NM ruptures was prolonged and led to recruitment of ESCRT-III nuclear membrane repair proteins (16). Recently, Young et al. (12) examined the duration of NM ruptures in a human osteosarcoma cell line in which lamin B1 expression had been knocked down with a shRNA. Depleting BAF—or depleting emerin or LEMD2—in those cells prolonged the duration of NM ruptures. In our studies, knocking down LAP2β expression in Lmnb1−/− neurons and TKO MEFs increased NM rupture frequency without affecting NM rupture duration. The observation that LAP2β expression levels did not alter NM rupture duration could relate to the fact that LAP2β, unlike other LEM domain proteins, did not associate with BAF at sites of NM ruptures in NIH 3T3 cells (13, 16).
The fact that LAP2β expression prevented NM ruptures in TKO MEFs in response to mechanical stretching suggested that LAP2β, either by itself or by associating with BAF and nuclear chromatin, bolsters the integrity of nuclear membranes in nuclear lamin–deficient cells. Our findings provide fresh insights into factors that modulate NM rupture frequency in nuclear lamin–deficient cells, but they also pose new questions. For example, does the protection against NM ruptures by LAP2β require its LEM domains? Would overexpression of other LEM domain proteins (e.g., MAN1, emerin, LEMD2) also reduce NM rupture frequency? Would overexpression of other NM proteins—even those without LEM domains or chromatin-binding properties—influence NM rupture frequency? All of these questions need to be addressed in future studies.
The most important and most surprising discovery in the current studies is that a LAP2β-tdTomato fusion protein prevents NM ruptures in Lmnb1−/− neurons and TKO MEFs. The Lmnb1−/− neurons express lamin B2 and extremely low levels of lamins A/C (3–5). Whether the ability of the LAP2β-tdTomato to prevent NM ruptures in Lmnb1−/− neurons depends on the expression of the remaining nuclear lamins in those cells is unknown. Also, we would point out again here [as we have in the past (28)] that the TKO MEFs that we created contained the original Lmna knockout allele from Sullivan et al. (24). That knockout allele yields low amounts of an internally truncated lamin A that lacks amino acid sequences encoded by exons 8 to 11 (29). Those sequences have been reported to mediate interactions with LEM domain proteins (29). Thus far, we have not tested whether the capacity of the LAP2β-tdTomato fusion protein to abolish NM ruptures in TKO MEFs requires the expression of the internally truncated lamin A. These issues also need to be investigated in future studies.
Materials and Methods
Mouse Studies.
Forebrain-specific Lmnb1 knockout mice harboring a ROSAnT-nG fluorescent reporter (Emx1-Cre Lmnb1fl/fl ROSAnT-nG) were generated as described (2). Mice were fed a chow diet and housed in a virus-free barrier facility with a 12-h light/dark cycle. All mouse studies were carried out according to the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (30). Animal protocols were reviewed and approved by the Animal Research Committee of the University of California, Los Angeles (UCLA).
Immunohistochemistry.
Mouse tissues were prepared for immunohistochemical studies as described (31). Adult and embryonic brains were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 2 h at room temperature, incubated in 30% sucrose in PBS at 4 °C overnight, and then frozen in optimal cutting temperature compound (Tissue-Tek, Sakura Finetek). Sections (10 μm thick) were fixed for 5 min in 4% paraformaldehyde or ice-cold methanol, followed by five dips in acetone and permeabilization with 0.1% Tween-20. Background staining with mouse antibodies was reduced with the Mouse-on-Mouse Kit (Vector Laboratories). Tissue sections were blocked with 2.5% horse serum for 1 h at room temperature and incubated overnight at 4 °C with primary antibodies at the dilutions indicated in SI Appendix, Table S2. Alexa Fluor 488– and Alexa Fluor 568–conjugated secondary antibodies (Molecular Probes, Invitrogen) were used at a 1:2,000 dilution; DyLight 649–conjugated streptavidin (Vector Laboratories) was used at 5 μg/mL. Following DAPI staining, sections were mounted with Prolong Gold antifade (Invitrogen), and images were recorded with a Zeiss LSM700 laser-scanning microscope with Plan Apochromat 20×/0.80 objective (air) or Plan Apochromat 100×/1.40 oil-immersion objectives. Images along the z axis were processed by Zen 2010 software (Zeiss).
Cell Culture Models.
Neuronal progenitor cells (NPCs) from E13.5 mouse embryos (derived from intercrosses of Lmnb1+/− mice) were used to generate Lmnb1+/+ (WT) and Lmnb1−/− (B1KO) neurospheres. Explants from the cerebral cortex were placed in Dulbecco’s Modified Eagle Medium (DMEM)/F-12 medium (Corning) and dissociated with TrypLE Select (Gibco); the NPCs were resuspended in DMEM/F-12 (Corning). Neurospheres were generated by culturing NPCs in DMEM/F-12 medium containing 2% B-27 supplement (Thermo Fisher), 100 U/mL penicillin, and 100 μg/mL streptomycin at 37 °C in 5 to 7% CO2. Neurospheres were supplemented with 3 μL of a heparin-embryonic growth factor (EGF)-fibroblast growth factor (FGF) mixture for every milliliter of medium. Briefly, FGF resuspended in PBS with 0.1% bovine serum albumin (BSA; Sigma) was mixed with EGF (Thermo Fisher) that had been resuspended in PBS containing 10% BSA. The EGF and FGF solution was diluted in DMEM/F-12 (Corning) and mixed with heparin sodium salt (Sigma-Aldrich) that had been resuspended in DMEM/F-12.
We also transduced Lmnb1+/+ and Lmnb1−/− neurons with a lentivirus encoding a mouse LAP2β-tdTomato fusion protein. To create the LAP2β-tdTomato fusion protein, the FUtdTW plasmid (#22478; Addgene) was digested with BamHI and XbaI and gel purified. A Tmpo cDNA was amplified from mouse cDNA with the CloneAMP HiFi PCR Kit (Takara Bio), forward primer 5′-ACGAGATGCCGGAGTTCCTAGAG-3′ and reverse primer 5′-CAATGC-AGCACTAACTTTACTGAGGTG-3′. The Tmpo insert was purified with UltraClean15 (Qiagen) and introduced into the FUtdTW vector with the InFusion Cloning Kit (Takara Bio). The integrity of the plasmid encoding the LAP2β-tdTomato fusion was documented by DNA sequencing. Packaging of the lentivirus and cell transductions were performed by UCLA’s Vector Core.
TKO MEFs (Lmna−/− Lmnb1−/− Lmnb2−/−) were created by Jung et al. (28) by incubating Lmna−/−Lmnb1fl/flLmnbfl/fl MEFs with a Cre adenovirus. They also generated TKO keratinocytes. Jung et al. (28) emphasized that their TKO cells were generated with the first Lmna knockout allele created by Sullivan et al. (24). Jahn et al. (29) discovered that the Lmna knockout allele yielded Lmna transcripts with an in-frame deletion of sequences derived from exons 8 to 11. This mutant Lmna transcript resulted in the production of an internally truncated lamin A protein lacking several domains important for protein interactions, including interactions of lamin A with LEM domain proteins (29). Consistent with the report by Jahn et al. (29), Jung et al. (28) documented, by Western blotting and immunohistochemistry, low levels of the internally truncated lamin A in their TKO cells. Subsequently, the properties of the TKO MEFs produced by Jung et al. (28) were studied by Chen et al. (1). As part of the latter studies, TKO MEFs were transduced with a lentiviral vector encoding a nuclear-localized GFP (NLS-GFP) (32). NLS-GFP–expressing TKO MEFs did not have nuclear blebs but displayed a high frequency of NM ruptures (1). In the current studies, we transduced the NLS-GFP–expressing TKO MEFs with the LAP2β-tdTomato lentivirus described earlier.
Neuronal Differentiation.
Cultured neurospheres were removed with polyethylene pipettes (Fisher) and pipetted into a single drop of laminin (Sigma-Aldrich) on poly-L-ornithine–coated plates. Neurospheres were allowed to settle for 30 min at 37 °C and then incubated in DMEM/F-12 containing 2% B-27 supplement, 100 U/mL penicillin, and 100 μg/mL streptomycin. Neurons that had been differentiated on coverslips were fixed with 4% paraformaldehyde in PBS or ice-cold methanol, dipped once in acetone, and permeabilized with 0.2% Triton. The fixed cells were then stained with antibodies (listed in SI Appendix, Table S2) and processed for confocal immunofluorescence microscopy (33).
Quantitative RT-PCR.
RNA was isolated from undifferentiated and differentiated neurospheres, treated with DNase I (Ambion), and reverse transcribed with random primers, oligo(dT), and SuperScript III (Invitrogen). qPCR reactions were performed on a 7900 Fast Real-Time PCR system (Applied Biosystems) with SYBR Green PCR Master Mix (Bioline). Transcript levels were determined using the comparative cycle threshold method and normalized to levels of cyclophilin A transcripts. Primers are listed in SI Appendix, Table S1.
siRNA Transfections.
In some studies, neurospheres that had been differentiated for 5 d were transfected with a MISSION esiRNA (endoribonuclease prepared siRNA) against mouse Tmpo (MilliporeSigma) (sequence shown in SI Appendix, Table S3) using the MISSION siRNA Transfection Reagent (MilliporeSigma). MEFs were allowed to adhere before being transfected with the esiRNA and transfection reagent. The Tmpo siRNA was not specific for the LAP2β isoform; it targeted exon 2 sequences that are shared by several LAP2 isoforms. We demonstrated that the siRNA significantly reduced LAP2β expression, as judged by qRT-PCR studies using exon 5 and exon 6 primers (specific for the LAP2β isoform).
Confocal Microscopy.
We prepared lamin B1–deficient neurons and nuclear lamin–deficient fibroblasts (1, 2) for laser-scanning confocal microscopy. Microscopy was used to assess LAP2β expression and distribution, to examine caspase 3 expression and γH2AX foci, and to visualize the polarization of LAP2β and lamin B2 distribution. We used live-cell video microscopy to analyze nuclear membrane ruptures. Imaging methodologies are described in more detail in SI Appendix.
Flow Cytometry.
Cells were sorted in the UCLA Jonsson Comprehensive Cancer Center (JCCC) and the Center for AIDS Research Flow Cytometry Core Facility on a FACSAria I(II) High-Speed Cell Sorter using a 488-nm blue laser (to sort for cells with NLS-GFP) and a 633-nm red laser to sort for cells transduced with the LAP2β-tdTomato lentivirus.
Cell Stretching.
Stretching MEFs on polydimethylsiloxane (PDMS) membranes was performed as described (34). Cells were seeded on flexible PDMS membranes (1 mm thick). The membranes were stretched 5 mm at 0.5 Hz for 2 h.
Statistical Analyses.
Statistical analyses were performed with GraphPad QuickCalcs (https://www.graphpad.com:443/). The percentages of cells with NM ruptures or γH2AX foci were analyzed with a χ2 test. Differences in ratios of NM ruptures over the total number of cells imaged were analyzed with an unpaired Student’s t test.
Supplementary Material
Acknowledgments
This work was supported by NIH grants HL126551 (S.G.Y.), AG047192 (L.G.F.), an NIH Ruth L. Kirschstein National Research Service Award T32GM065823 (N.Y.C.), a Whitcome Fellowship Award from UCLA’s Molecular Biology Institute (N.Y.C.), and a Vascular Biology Training Grant from the National Heart, Lung, and Blood Institute (N.Y.C.). We thank Dr. Jan Lammerding for sharing the NLS-GFP plasmid. Virus production and transduction were performed by the Integrated Molecular Technologies Core/UCLA Vector Core, which is supported by CURE/P30 DK041301. Mass cytometry was performed in the AIDS Research Flow Cytometry Core Facility, supported by NIH awards P30 CA016042 and 5P30 AI028697, the UCLA JCCC, the UCLA AIDS Institute, and the David Geffen School of Medicine at UCLA.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2107770118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Chen N. Y., et al., Fibroblasts lacking nuclear lamins do not have nuclear blebs or protrusions but nevertheless have frequent nuclear membrane ruptures. Proc. Natl. Acad. Sci. U.S.A. 115, 10100–10105 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen N. Y., et al., An absence of lamin B1 in migrating neurons causes nuclear membrane ruptures and cell death. Proc. Natl. Acad. Sci. U.S.A. 116, 25870–25879 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young S. G., Jung H. J., Coffinier C., Fong L. G., Understanding the roles of nuclear A- and B-type lamins in brain development. J. Biol. Chem. 287, 16103–16110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jung H. J., et al., Regulation of prelamin A but not lamin C by miR-9, a brain-specific microRNA. Proc. Natl. Acad. Sci. U.S.A. 109, E423–E431 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jung H. J., Lee J. M., Yang S. H., Young S. G., Fong L. G., Nuclear lamins in the brain - new insights into function and regulation. Mol. Neurobiol. 47, 290–301 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young S. G., Jung H. J., Lee J. M., Fong L. G., Nuclear lamins and neurobiology. Mol. Cell. Biol. 34, 2776–2785 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger R., et al., The characterization and localization of the mouse thymopoietin/lamina-associated polypeptide 2 gene and its alternatively spliced products. Genome Res. 6, 361–370 (1996). [DOI] [PubMed] [Google Scholar]
- 8.Foisner R., Gerace L., Integral membrane proteins of the nuclear envelope interact with lamins and chromosomes, and binding is modulated by mitotic phosphorylation. Cell 73, 1267–1279 (1993). [DOI] [PubMed] [Google Scholar]
- 9.Herrada I., et al., Purification and structural analysis of LEM-domain proteins. Methods Enzymol. 569, 43–61 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Furukawa K., LAP2 binding protein 1 (L2BP1/BAF) is a candidate mediator of LAP2-chromatin interaction. J. Cell Sci. 112, 2485–2492 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Yang L., Guan T., Gerace L., Lamin-binding fragment of LAP2 inhibits increase in nuclear volume during the cell cycle and progression into S phase. J. Cell Biol. 139, 1077–1087 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Young A. M., Gunn A. L., Hatch E. M., BAF facilitates interphase nuclear membrane repair through recruitment of nuclear transmembrane proteins. Mol. Biol. Cell 31, 1551–1560 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King M. C., Lusk C. P., Loss of nuclear envelope integrity? No probLEM-BAF has it covered. J. Cell Biol. 218, 2077–2079 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lusk C. P., Ader N. R., CHMPions of repair: Emerging perspectives on sensing and repairing the nuclear envelope barrier. Curr. Opin. Cell Biol. 64, 25–33 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thaller D. J., et al., An ESCRT-LEM protein surveillance system is poised to directly monitor the nuclear envelope and nuclear transport system. eLife 8, 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halfmann C. T., et al., Repair of nuclear ruptures requires barrier-to-autointegration factor. J. Cell Biol. 218, 2136–2149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coffinier C., et al., Deficiencies in lamin B1 and lamin B2 cause neurodevelopmental defects and distinct nuclear shape abnormalities in neurons. Mol. Biol. Cell 22, 4683–4693 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorski J. A., et al., Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J. Neurosci. 22, 6309–6314 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen N. Y., Kim P. H., Fong L. G., Young S. G., Nuclear membrane ruptures, cell death, and tissue damage in the setting of nuclear lamin deficiencies. Nucleus 11, 237–249 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo Y., Zheng Y., Lamins position the nuclear pores and centrosomes by modulating dynein. Mol. Biol. Cell 26, 3379–3389 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brachner A., Reipert S., Foisner R., Gotzmann J., LEM2 is a novel MAN1-related inner nuclear membrane protein associated with A-type lamins. J. Cell Sci. 118, 5797–5810 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Jung H. J., et al., Farnesylation of lamin B1 is important for retention of nuclear chromatin during neuronal migration. Proc. Natl. Acad. Sci. U.S.A. 110, E1923–E1932 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaughan A.et al.; Whitfield WGF , Both emerin and lamin C depend on lamin A for localization at the nuclear envelope. J. Cell Sci. 114, 2577–2590 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Sullivan T., et al., Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 147, 913–920 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fong L. G., et al., Prelamin A and lamin A appear to be dispensable in the nuclear lamina. J. Clin. Invest. 116, 743–752 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vallee R. B., Tsai J. W., The cellular roles of the lissencephaly gene LIS1, and what they tell us about brain development. Genes Dev. 20, 1384–1393 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Wynshaw-Boris A., Lissencephaly and LIS1: Insights into the molecular mechanisms of neuronal migration and development. Clin. Genet. 72, 296–304 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Jung H. J., et al., An absence of nuclear lamins in keratinocytes leads to ichthyosis, defective epidermal barrier function, and intrusion of nuclear membranes and endoplasmic reticulum into the nuclear chromatin. Mol. Cell. Biol. 34, 4534–4544 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jahn D., et al., A truncated lamin A in the Lmna -/- mouse line: Implications for the understanding of laminopathies. Nucleus 3, 463–474 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
- 31.Coffinier C., et al., Abnormal development of the cerebral cortex and cerebellum in the setting of lamin B2 deficiency. Proc. Natl. Acad. Sci. U.S.A. 107, 5076–5081 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Denais C. M., et al., Nuclear envelope rupture and repair during cancer cell migration. Science 352, 353–358 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang S. H., et al., An absence of both lamin B1 and lamin B2 in keratinocytes has no effect on cell proliferation or the development of skin and hair. Hum. Mol. Genet. 20, 3537–3544 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim P. H., et al., Disrupting the LINC complex in smooth muscle cells reduces aortic disease in a mouse model of Hutchinson-Gilford progeria syndrome. Sci. Transl. Med. 10, eaat7163 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.