Significance
The peopling of Siberia and the Americas is intriguing for archaeologists, linguists, and human geneticists, but despite significant recent developments, many details remain controversial. Here, we provide insights based on genetic diversity within Helicobacter pylori, a bacterium that infects 50% of all humans. H. pylori strains were collected from across eastern Eurasia and the Americas. Sequence analyses indicated that Siberia contains both anciently diverged and recently admixed bacteria, supporting both human persistence over the last glacial maximum and more recent human recolonization. We inferred a single migration across the Bering land bridge, accompanied by a dramatic reduction in effective population size, followed by bidirectional Holocene gene flow between Asia and the Americas.
Keywords: Helicobacter pylori, Siberia, Americas, colonization, demographic model
Abstract
The gastric bacterium Helicobacter pylori shares a coevolutionary history with humans that predates the out-of-Africa diaspora, and the geographical specificities of H. pylori populations reflect multiple well-known human migrations. We extensively sampled H. pylori from 16 ethnically diverse human populations across Siberia to help resolve whether ancient northern Eurasian populations persisted at high latitudes through the last glacial maximum and the relationships between present-day Siberians and Native Americans. A total of 556 strains were cultivated and genotyped by multilocus sequence typing, and 54 representative draft genomes were sequenced. The genetic diversity across Eurasia and the Americas was structured into three populations: hpAsia2, hpEastAsia, and hpNorthAsia. hpNorthAsia is closely related to the subpopulation hspIndigenousAmericas from Native Americans. Siberian bacteria were structured into five other subpopulations, two of which evolved through a divergence from hpAsia2 and hpNorthAsia, while three originated though Holocene admixture. The presence of both anciently diverged and recently admixed strains across Siberia support both Pleistocene persistence and Holocene recolonization. We also show that hspIndigenousAmericas is endemic in human populations across northern Eurasia. The evolutionary history of hspIndigenousAmericas was reconstructed using approximate Bayesian computation, which showed that it colonized the New World in a single migration event associated with a severe demographic bottleneck followed by low levels of recent admixture across the Bering Strait.
The gram-negative gastric bacterium Helicobacter pylori has shared an intimate coevolutionary relationship with humans for the last 100,000 y and possibly longer (1, 2). The greatest genetic diversity in both humans and H. pylori is found in Africa, the ancestral homeland of both species (3). After leaving Africa, H. pylori continued to differentiate in tandem with humans. Unlike its host, the bacterium evolved into multiple distinct geographic populations because it has high mutation and recombination rates, shorter generation times, and lower effective population sizes (4). Human populations expanding along the southern Asia coastal route carried the H. pylori precursors of hpSahul, a divergent, monophyletic population that continues to infect New Guinean Highlanders and Aboriginal Australians (5). The western and central parts of Paeleolithic Eurasia were dominated by hpAsia2, which has differentiated into hspIndia and hspLadakh (6, 7).
A Eurasian West–East divide in H. pylori is reflected by the divergence of hpEastAsia, an ancestral East Asian population, into subpopulations hspEAsia (Southeast and East Asians), hspIndigenousAmericas (formerly known as hspAmerind, native North and South Americans), and hspMaori (Austronesian speakers). However, H. pylori’s presence, diversity, and structure in northern Eurasia are still unknown. This vast region, hereafter Siberia, extends from the Ural Mountains in the west to the Pacific Ocean in the east and the Kazakh and Mongolian Steppes in the south. Siberia is inhabited by at least 41 ethnic minorities who often live in small communities of low population densities with a total of <250,000 individuals (8). Notwithstanding often harsh environmental conditions, Siberians subsist through hunter-gathering and/or pastoralism (9), and the diversity of language families spoken in the region (Uralic, Yeniseian, Turkic, Tungusic, Mongolic, Gilyak, and Chukotko-Kamchatkan) hints at a complex history of migration and isolation.
The patterns of human diversity between these ethnic groups are also largely understudied, and most genetic studies were based on traditional markers such as mitochondrial or Y chromosome DNA, whose effectiveness is compromised by relatively low mutation rates and incomplete lineage sorting (10–15). Recently, genomic studies of ancient human DNA have offered further insights, confirming that Siberia was the gateway for human migrations into northern America (16, 17) as well as into western Eurasia (18).
Two important questions remain unanswered. Firstly, although anatomically modern human hunter-gatherers first appeared in Siberia 45,000 y ago (kya) in the Upper Paleolithic (16), their continued presence in the region throughout the last glacial maximum (LGM, 26.5 to 19 kya) is controversial. One argument suggests that ancient northern Eurasian [ANE (19)] metapopulations in Siberia had already developed adaptive mechanisms that allowed them to persist throughout the LGM, at least in sheltered locations with favorable environments (20). The presence of genetically similar human remains in central Siberia at 24 kya (Ma’lta boy, MA1) and 17 kya (AG2) (17) also hints that the region may have been continuously inhabited by ANEs during the LGM. Conversely, other scientists propose that the LGM paleoclimate resulted in the complete depopulation of central Siberia, driving humans into refugia to the south where they persisted for thousands of years before repopulating northern Siberia during the Holocene (9, 21, 22). Secondly, the peopling of the Americas by anatomically modern humans has long attracted scientific attention. Recent genomic evidence suggests that a single genetically diverse human population migrated across the Bering land bridge into the Americas, followed by subsequent post-LGM gene flow (23, 24). However, the dates of colonization by these migrants, the size of their founder population, and their relationships to other human groups remain unclear.
To help answer these and other questions about human movements through Eurasia and the Americas, we undertook large-scale sampling of H. pylori from ethnically diverse aboriginal human populations across Siberia and Mongolia. Genotyping and sequence data were combined with a large reference database of Asian strains to provide an overview of the population structure of their genetic diversity and to infer the demographic and evolutionary histories of the peoples of Siberia and the Americas.
Results
From 2004 to 2006, we sampled H. pylori from 16 ethnic and multiple traditional language groups (SI Appendix, Table S1 and Fig. S1) during visits by an experienced gastroenterologist to 18 geographic locations groups in Russia and Mongolia spanning northwestern, central, northern, and eastern Siberia and Beringia. The language families sampled included Uralic, Turkic, Mongolic, Tungusic, and Chukotko-Kamchtakan as well as two Isolate languages spoken by the Ket of the Yenisei Valley and the Nivkh of Sakhalin Island. A total of 1,175 gastroendoscopic biopsies were taken from the antrum and/or corpus of the stomachs of volunteers without mishaps and with informed consent and frozen immediately in liquid nitrogen before shipping to Moscow. A total of 556 independent strains of H. pylori were cultivated from the frozen biopsies and DNA was extracted. Seven MLST (multilocus sequence typing) housekeeping genes spanning 3,406 nucleotide positions were sequenced by classical Sanger sequencing as described (4). We also extended these studies with higher resolution genomic short-read sequencing for a subset of 40 representative H. pylori strains from around the globe as well as 54 of the indigenous H. pylori strains cultured from 14 ethnicities in Siberia and the New World (Dataset S1). The short reads were assembled into draft genomic sequences in EnteroBase (25) and used to assess population structure at the genomic level.
Russians colonized Siberia centuries ago, and H. pylori from Russians usually belong to the hpEurope population (3), which evolved through Neolithic admixture (26). We were therefore not surprised that 160 of the Siberian isolates belonged to hpEurope and ignored them here because of numerous prior studies on such isolates from Europe (2–5, 27). Instead, we focus on the remaining 396 bacterial strains which were assigned to Asian populations. These Siberian MLST genotypes were combined with previously described H. pylori genotypes from other global sources (36 human populations in Asia, Oceania, and the Americas (4–7, 27–32); Dataset S2 and SI Appendix, Fig. S1) for a total of 1,002 genotypes and 1,952 polymorphic sites, which were used to elucidate the evolutionary history of H. pylori in Siberia.
The Structure of H. pylori Variation in Eastern Eurasia and the Americas.
We analyzed the genetic structure of our H. pylori MLST data set using model-free discriminant analysis of principle components (DAPC) (33) as well as the linkage model in STRUCTURE (34). Both approaches identified 10 distinct subpopulations (Fig. 1 and Table 1 and SI Appendix, Figs. S2 and S3), five of which (hspAltai, hspUral, hspSiberia1, hspSiberia2, and hspKet; Dataset S2) are first described here.
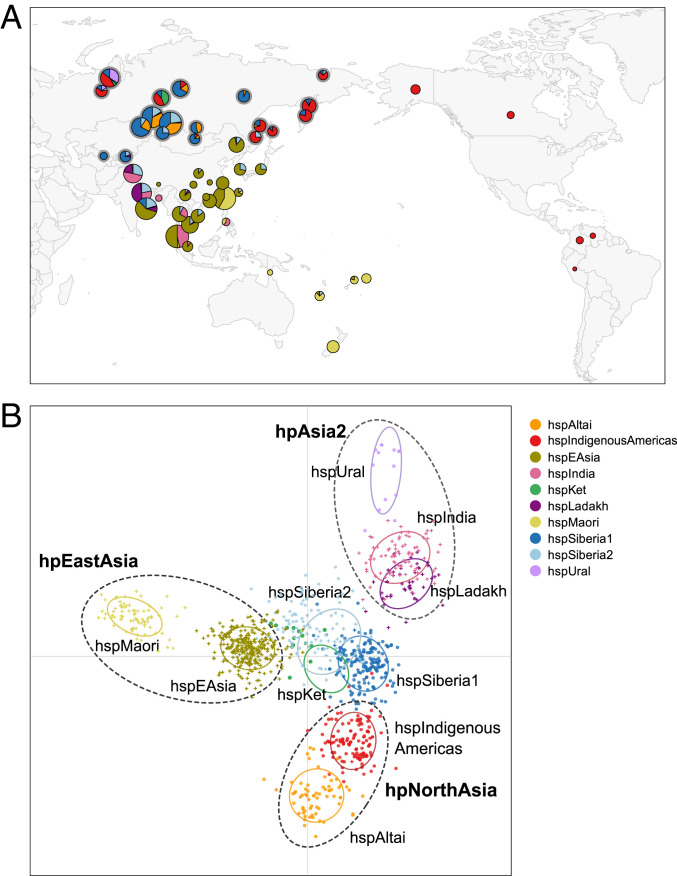
Fig. 1.
MLST genetic variation of H. pylori structured into 10 subpopulations of populations across Asia and the Americas. (A) Geographic distribution of 1,002 genotypes, color coded according to subpopulation. Circle positions indicate the sources of the bacterial isolates, and their sizes are proportional to the numbers of isolates. Newly sampled populations are denoted with a gray halo. (B) First two principal components of a DAPC analysis showing the three main H. pylori populations inhabiting Asia and the Americas: hpAsia2, hpEastAsia, and hpNorthAsia (dashed ovals; bold font). Five newly discovered subpopulations (hspAltai, hspUral, hspSiberia1, hspSiberia2, and hspKet) were endemic to northern and central Siberia. Seven subpopulations (smaller ovals) were nested within these major populations while three clustered in the DAPC space between populations. Newly isolated strains appear as full circles and reference strains as crosses. Key legend: color codes of all 10 subpopulations.
Table 1.
H. pylori’s genetic structure and geographic distribution into populations and subpopulations across Asia and the Americas
| Population | Subpopulation | Locations (ethnic groups) | # Strains | Genetic structure | |||
| MLST | Genomes | ||||||
| DAPC | STR | M-L | FS | ||||
| hpAsia2 | hspIndia | India, Nepal, Bangladesh, Philippines, Thailand | 85 | X | X | — | X |
| hspLadakh | Northern India and Nepal, Bhutan, Central Siberia (Tuvan, Kyrgyz), Yunnan | 43 | X | X | — | X | |
| hspUral | Northern Siberia (Khant, Nenet) | 16 | X | X | X | X | |
| hpEastAsia | hspEAsia | China, Vietnam, Thailand, Cambodia, Singapore, Malaysia, Bhutan, Korea and Japan | 294 | X | X | X | X |
| hspMaori | Taiwan, Philippines, Melanesia and Polynesia (Austronesian speakers) | 83 | X | X | X | X | |
| hpNorthAsia | hspIndigenous Americas | Northern Siberia (Khant, Nenet, Evenk, Ket), Central Siberia (Tuvan, Mongolian), Eastern Siberia (Nanai, Orok, Nivk), Beringia (Even, Koryak, Chukchi), the Americas (Eskimo, Athabaskan, Venezuelan, Huitoto, Peruvian) | 123 | X | X | X | X |
| hspAltai | Central Siberia (Tuvan, Tubalar, Yakut, Buryat, Mongolian), Northern Siberia (Evenk), Eastern Siberia (Orok) | 67 | X | X | X | X | |
| Potentially admixed | hspSiberia1 | Siberia (Khant, Nenet, Evenk, Ket, Uzbek, Kyrgyz, Tubalar, Tuvan, Mongolian, Buryat, Yakut, Nivk, Orok, Even, Koryak, Chukchi), Bhutan, China | 177 | X | X | INT | INT |
| hspSiberia2 | Siberia (Khant, Nenet, Kyrgyz, Tubalar, Tuvan, Mongolian, Nanai, Koryak, Chukchi), Bhutan, Nepal, India, Cambodia, Korea, China, Japan | 101 | X | X | INT | INT | |
| hspKet | Central Siberia (Ket, Nenet) | 13 | X | — | X | X | |
Hp, H. pylori population; hsp, H. pylori subpopulation; STR, analysis using the linkage model in STRUCTURE; M-L, maximum likelihood phylogenetic tree; FS, fineSTRUCTURE analysis; X denotes an identified subpopulation cluster/clade, while – denotes the opposite; and INT denotes clustering at intermediate position to existing subpopulations/clades.
hspIndia and hspLadakh from South and Southeast Asia are previously described subgroupings of hpAsia2. Our DAPC results show that hspUral from Uralic-speaking Khants and Nenets of northwestern Siberia is a third, slightly divergent hpAsia2 subpopulation (Fig. 1B and SI Appendix, Figs. S4 and S5).
hspEAsia and hspMaori (Table 1) are found predominantly in East Asia, island Southeast Asia, and the Pacific and belong to the hpEastAsia population. Previous analyses suggested that the hspIndigenousAmericas subpopulation isolated from Native Americans also belonged to hpEastAsia (4, 5). The added resolution afforded by our extensive sampling reveals this conclusion to be incorrect. We now find that hspIndigenousAmericas also included 103 other strains found in Beringia (Chukchi, Koryak, Even) along the East Siberian coast as far south as Sakhalin Island and the Lower Amur Valley and was isolated from Uralic (Khant and Nenet) and Yenisein (Ket) speakers in northwestern Siberia (Table 1). hspIndigenousAmericas was notably absent in central Siberia (Fig. 1A) except for four isolates from Evenk and one from Mongolia. The combined hspIndigenousAmericas subpopulation from Asia and the Americas is also clearly not a subgroup of hpEastAsia (Fig. 1B). Instead, hspIndigenousAmericas is most closely related to a new subpopulation, which we designated as hspAltai. hspAltai was restricted to central Siberia, where it was found with high frequencies among Turkic (Tuvan, Tubalar, Yakut) and Mongolic (Buryat, Mongolian) speakers and at lower frequencies in Evenk and Orok Tungusic groups (Fig. 1A and Table 1). hspIndigenousAmericas and hspAltai form a population that we designate hpNorthAsia, which accounted for 44% of Siberian H. pylori.
The majority of our Siberian genotypes (52%) clustered distinctly from all of these H. pylori populations and subpopulations (Fig. 1B). They formed three somewhat overlapping subpopulations (hspSiberia1, hspSiberia2, and hspKet) in the center of the triangular DAPC cluster delineated by hpEastAsia, hpNorthAsia, and hpAsia2 (Fig. 1B). hspSiberia1 was geographically widespread and was isolated from all sampled ethnicities across northern Eurasia (Fig. 1A and Table 1). hspSiberia2 was also widely distributed across Siberia, albeit at lower frequencies and from fewer ethnicities than hspSiberia1. The third new subpopulation, hspKet, was exclusively found among ethnic Kets in central Siberia (Fig. 1B and SI Appendix, Fig. S4).
We also investigated the relationship between these subpopulations on a subset of 94 complete genomes (of which 54 originated in Siberia). Maximum likelihood trees were generated from genomic single-nucleotide polymorphisms after removing recombinant regions that were detected with Gubbins (35). A total of 40 genomes from previously defined subpopulations formed subpopulation-specific monophyletic clades, except for a partial overlap of hspIndia and hspLadakh (SI Appendix, Fig. S6A), which also overlap somewhat in geographical location and in DAPC space (Fig. 1B). In a larger tree containing 54 additional Siberian genomes, hspUral and hspAltai also formed monophyletic clades (SI Appendix, Fig. S6B and Table 1). As expected, hspUral was closely associated with hspLadakh and hspIndia, while hspAltai was nested within hspIndigenousAmericas. Both trees also showed that hpNorthAsia genotypes diverged from a common ancestor and were not nested within hpEastAsia.
In contrast to these monophyletic clades, 19 hspSiberia1, hspSiberia2, and hspKet genomes formed long branches, did not cluster uniformly according to their subpopulation designations, and were intermingled to some extent with hpNorthAsia, hpEastAsia, and hpAsia2 genomes (SI Appendix, Fig. S6B). A total of 15 of those genomes had <95% bootstrap support. Together with the intermediate and overlapping positions in the DAPC plot described above, these results indicate an absence of discrete or robust structure which might possibly result from extensive mixed ancestry.
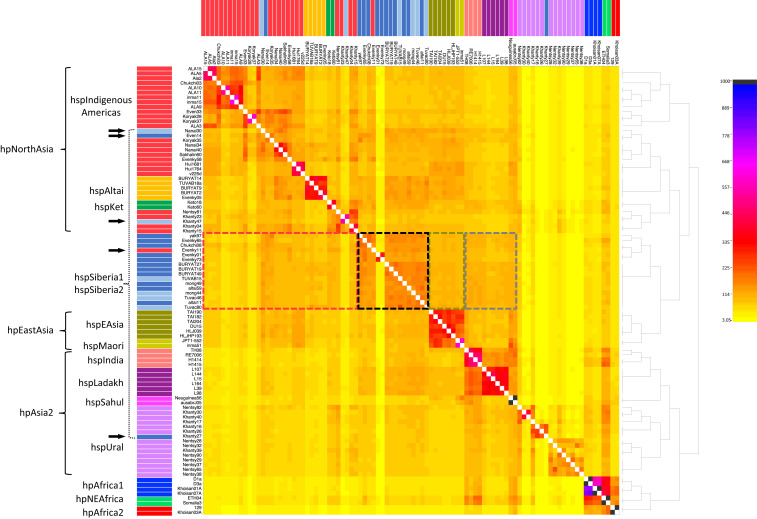
We therefore investigated the sources of ancestry of all core genomes of Siberian strains with fineSTRUCTURE (36), which has previously been used to demonstrate shared ancestry among recombining H. pylori genomes (26, 37). The coancestry matrix (Fig. 2) supported a distinct clustering of populations hpNorthAsia, hpEastAsia, and hpAsia2 and most of their subpopulations, consistent with the phylogenetic results described above (Table 1). However, the fineSTRUCTURE results were also partially inconsistent with the DAPC assignments to the subpopulations hspSiberia1 and hspSiberia2. A total of 15 genomes clustered together with hpEastAsia, but three clustered with hspIndigenousAmericas and one with hspUral (horizontal arrows, Fig. 2). In contrast, the two hspKet genomes clustered discretely with hpNorthAsia strains from Uralic-speaking Nenets and Khants, indicating a closer affinity to this population and an independent origin from other Siberian strains. Importantly, the coancestry heat map showed that strains assigned to Siberian subpopulations (dashed boxes) shared considerable proportions of their ancestry (darker shading) with each other and, to a lesser extent, with other genomes from hpNorthAsia, hpEastAsia, hspLadakh, or hspIndia (Fig. 2). Quantifying these proportions of ancestry in Siberian strains, we found that the highest levels corresponded to hpNorthAsia, hpAsia2, and hpEastAsia (SI Appendix, Table S2). The combination of these results indicates that modern Siberian subpopulations have signals of admixture between multiple ancestral populations.
Fig. 2.
Coancestry matrix of admixture among 94 globally representative H. pylori genomes obtained in fineSTRUCTURE. The heat map corresponds to the number of motifs inferred to have been imported from a donor genome (column) to a recipient genome (row) by Chromopainter. The inferred tree was generated by Bayesian clustering in fineSTRUCTURE. Strain names are displayed on the top and left of the heat map and are color coded by subpopulation as in Fig. 1. Strains belonging to DAPC subpopulations hspSiberia1 and hspSiberia2 are labeled with a dashed curly bracket. Small black horizontal arrows indicate strains not clustering according to DAPC assignment. Potential sources for the genomic ancestry of Siberian strains are highlighted on the heat map in dashed boxes and color coded: gray, hpAsia2; red, hpNorthAsia; mustard green, hpEastAsia. The black dashed box shows the levels of ancestry shared among Siberian strains.
Evolutionary Origins of Siberian Subpopulations.
We attempted to elucidate the evolutionary dynamics of H. pylori in Siberia assessing demographic simulations via approximate Bayesian computation [ABC (38); reviewed in ref. 39]. We defined sets of alternative demographic histories (“models”) that could potentially have generated the observed MLST sequence distributions. A coalescent simulator was then used to simulate DNA sequence evolution according to each of the predefined models. The simulated data were compared to the observed MLST sequences, and the model that retained the largest number of simulations at three threshold levels was chosen as best fit to the data. We also used this approach to model migration into the Americas and to date the occurrence of branch points and admixture events in the phylogenies.
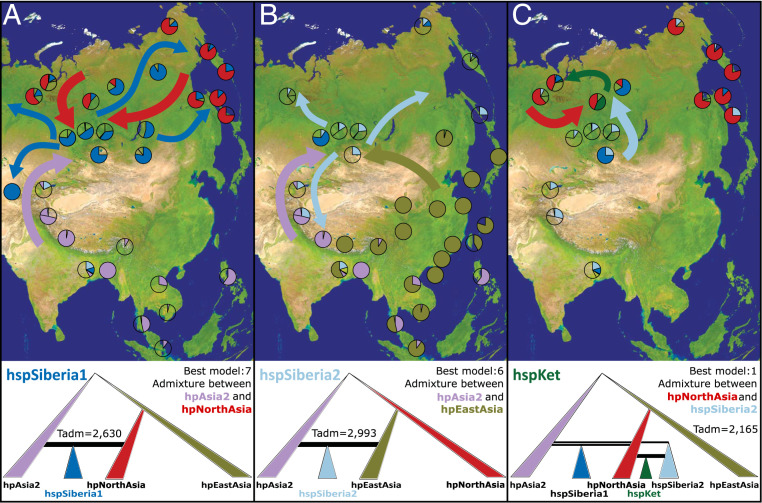
We defined the model populations on the basis of the observed genetic structure above, independent of the geographic origins of samples. The models began with H. pylori’s well-established West–East divide (40), which is reflected by the split between hpAsia2 and the common ancestor of hpEastAsia and hpNorthAsia (2, 5). We inferred the origins of hspSiberia1 and hspSiberia2, the two most common Siberian populations, by simulating a range of tree-like and admixture-based demographic models (SI Appendix, Fig. S7 and Table S3). Models one to four are tree-like; they postulate that subpopulations diverged from an ancestral population exclusively by mutation and drift. Models five to seven hypothesize that a subpopulation evolved by hybridization between two ancestral populations. The coancestry matrix (Fig. 2) and proportions of shared ancestry (SI Appendix, Table S2) suggest hpAsia2, hpNorthAsia, and hpEastAsia as potential ancestral populations, excluding hspUral, whose members are unusually divergent (Fig. 1B and SI Appendix, Figs. S4 and S5) and share fewer ancestral motifs with hspIndia and hspLadakh. A visual examination of principal component analyses (PCA) (SI Appendix, Figs. S8 and S9) indicated that the seven models were able to generate the observed data. Logistic regression showed that model posterior probabilities were stable with increasing numbers of retained simulations (SI Appendix, Table S4), and the best models yielded median posterior mutation rates of similar magnitudes to previously estimated long-term rates (1, 41). Coalescent generations were converted to years using the calibration of 1 y per generation previously established from population divergence time estimates (2, 5) and mutation rates (1). The best fit models for hspSiberia1 and hspSiberia2 indicated that both originated through admixture rather than divergence. The best model for hspSiberia1 (model seven; Fig. 3A) inferred evolution through admixture between hpAsia2 and hpNorthAsia. hpAsia2 was also one of the admixture partners for hspSiberia2, but the other partner was hpEastAsia rather than hpNorthAsia (model six, Fig. 3B). The top models chose different admixture sources but yielded similar posterior estimates of their parameter values (SI Appendix, Figs. S10 and S11 and Table S5). The admixture events were relatively recent, ∼2,630 (hspSiberia1; R2 0.51) and 2,933 (hspSiberia2; R2 0.52) y ago.
Fig. 3.
Best supported models for the evolution of newly discovered H. pylori subpopulations hspSiberia1 (A) and hspSiberia2 (B) and hspKet (C) using ABC. (A–C, Top) Sampled locations containing either source or admixed populations, the proposed migration routes of both source populations (thick arrows), and the recent colonization routes once admixture occurred (thin arrows). The transparent pie slices therefore correspond to populations that were not involved in the depicted scenario. hspSiberia1 originated through admixture between hpAsia2 and hpNorthAsia, hspSiberia2 derived via admixture between hpAsia2 and hpEastAsia, and hspKet evolved though admixture between hpNorthAsia and hspSiberia2. The median posterior estimates for admixture times (Tadm) are given for each analysis.
DAPC (Fig. 1B and SI Appendix, Fig. S4) and fineSTRUCTURE (Fig. 2) analyses suggested that hspKet might have arisen through admixture. The observation that hspKet was only resolved by DAPC at K > 9 suggested that admixture might have between preexisting admixed populations, such as hspSiberia1 or hspSiberia2, and older populations, such as hpEastAsia or hpNorthAsia. We therefore built the reconstructed demographics inferred for hspSiberia1 and hspSiberia2 into five new models to test the origins of hspKet (SI Appendix, Fig. S12 and Table S6). The best model for the evolution of hspKet was that of secondary admixture between hspSiberia2 and hpNorthAsia ∼2,165 y ago (Fig. 3C and SI Appendix, Figs. S13 and S14 and Tables S4 and S7). Thus, these analyses indicate that admixture was involved in the evolution of hspSiberia1 and hspSiberia2 as well as hspKet and also provide explanations for their equivocal clustering in phylogenetic (SI Appendix, Fig. S6B) and fineSTRUCTURE analyses (Fig. 2).
We now turn to hspAltai. This subpopulation was assigned to hpNorthAsia as was hspIndigenousAmericas (Figs. 1B and 2 and SI Appendix, Fig. S6B). We therefore used our ABC framework to estimate the divergence time of hspAltai from hpNorthAsia at ∼983 y ago (R2 = 0.70; SI Appendix, Figs. S15 and S16 and Tables S4, S8, and S9).
Migration of H. pylori from Siberia into the New World.
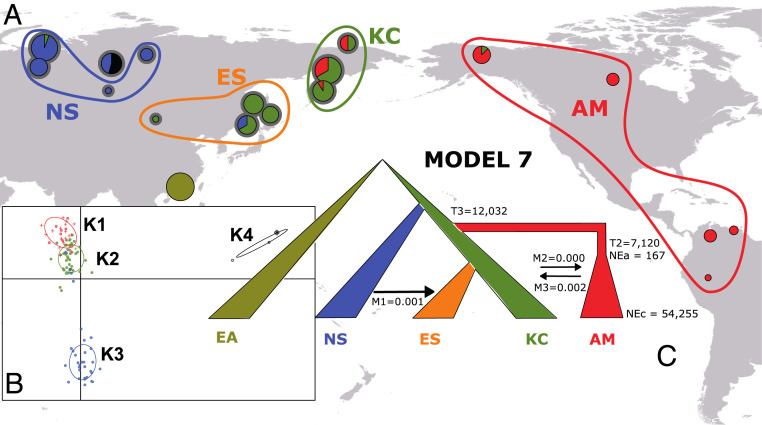
We also used ABC comparisons between multiple models to reconstruct the evolutionary history of the hpNorthAsia subpopulation hspIndigenousAmericas as it migrated from Eurasia into the Americas, but the populations used for modeling were based on geographical populations rather than genetic clusters. We restricted these analyses to hspIndigenousAmericas because no other aboriginal H. pylori subpopulation has been isolated from Native Americans. hspIndigenousAmericas genotypes were assigned to four spatial populations based on their geographic distributions across Siberia and the Americas. These were designated NS (northern Siberia; ethnic groupings: Khanty, Nenet, Tuvan, and Evenk; n = 31), ES (eastern Siberia; Nivkh, Orok, and Nanai; n = 28), KC (Kamchatka-Chukotka; Chukchi, Koryak, and Even; n = 35), and AM (Americas; Eskimo, Athabaskan, Venezuelan, Huitoto, and Peruvian; n = 19) (Fig. 4A). The geographic boundaries of the four regions correspond to geographical areas with elevated frequencies of K1 to K3, three genetic sub-subpopulations that were discerned by a DAPC analysis of hspIndigenousAmericas strains (SI Appendix, Fig. S17 and Fig. 4B). hspIndigenousAmericas from the Ket ethnic group, which comprised the K4 DAPC cluster, were excluded because they might confound the analyses due to high genetic variance (Fig. 4B) and their demonstrated history of isolation and admixture (Fig. 3C). We developed eight demographic models that potentially reconstruct the history of hspIndigenousAmericas and its colonization of the American continent, using a representative population (Hong Kong) of hpEastAsia (EA) as an outgroup (SI Appendix, Fig. S18 and Table S10). Models one to four depicted simple tree-like scenarios. Models one, three, and four were based on the underlying ingroup topology deduced from DAPC, with the initial branching of northern Siberia followed by ES, KC, and finally AM [base topology (EA(NS(ES(KC,AM))))]. Model two was based on an alternative topology, namely an early split of Native American H. pylori from Siberian bacteria. Models three and four tested whether gene flow between KC and AM occurred early (model three) or late (model four) in order to distinguish whether migrations to America preceded or succeeded the flooding of Beringia (42). Model five explicitly tested the existence of gene flow between H. pylori from ANEs (represented by NS) and Native Americans as postulated by Raghavan et al. (17). American H. pylori may also have undergone a reduction in effective population size during the flooding of Beringia; this bottleneck was simulated in models six through eight, with American H. pylori splitting from the ancestors of ES and KC. Model six excludes gene flow between Siberia and AM after the bottleneck, whereas model seven includes bidirectional gene flow across the Bering Strait in accordance with Reich et al. (23). Models six and seven both envision gene flow from NS to ES, whereas model eight envisions additional bidirectional gene flow between ES and KC in order to account for similarities in H. pylori sequences from both regions.
Fig. 4.
Reconstructing the colonization of the New World by hspIndigenousAmericas. (A) The geographic distribution of hspIndigenousAmericas subpopulations across Eurasia and the Americas. ABC was carried out on geographic populations rather than DAPC population clusters. The size of each pie represents the number of hspIndigenousAmericas strains isolated at each location, and slices represent the proportions of each DAPC-inferred populations from B, Inset. Newly sampled populations are denoted with a gray halo. (B) DAPC analysis of 123 hspIndigenousAmericas strains showing structuring into four populations. Newly sequenced strains are denoted with a filled circle, whereas reference strains are shown as crosses. An East Asian population (Hong Kong) was used as the outgroup. (C) The best supported of eight competing models with median posterior estimates. T2 is the time, in years, when the bottleneck underwent by the AM population stopped. T3 is the time, in years, when the AM population split from its common ancestor with populations ES and KC; NEa, ancestral effective population sizes; NEc, current effective population sizes; M, migration rates.
PCA confirmed that our models were able to reproduce the observed data (SI Appendix, Fig. S19), while regression analyses (SI Appendix, Table S4) supported model seven for the evolutionary history of hspIndigenousAmericas (Fig. 4C). Thus, H. pylori’s colonization of the Americas comprised the migration of a population that was ancestral to those presently inhabiting the eastern and northeastern reaches of Siberia, across Beringia and into North America ∼12 kya (95% highest posterior density [HPD] 3,105 to 22,512 y, Fig. 4C). The effective population size of the founding H. pylori was very small (ancestral effective population size = 167). This founding population subsequently expanded considerably after migrating from Siberia to a current effective population size of 54,255 (Fig. 4C and SI Appendix, Fig. S20 and Table S11).
Discussion
The Structure of hpAsia2 in the Paleolithic.
hpAsia2 was one of the first H. pylori populations to evolve outside Africa, and these bacteria infected a 5,300-y-old frozen mummy in the Italian Alps (26) and may even have been dominant in western and central Eurasia during the Paleolithic. Today, hpEurope H. pylori predominate in Europe and the surrounding areas of western Asia. However, hpAsia2 may have accompanied the first modern human hunter-gatherers into the region over 40 kya (43). Pure hpAsia2 occurs at low frequencies among Finnish and Estonian Uralic speakers, on the fringes of the range of hpEurope, and is found at higher frequencies in Central and Southeast Asia (3, 7, 44). The data presented here show that Uralic speakers in northwestern Siberia of the Khanty and Nenet ethnicities are infected by hspUral, a novel subpopulation of hpAsia2. The existence of hspUral indicates that hpAsia2 may once have been highly structured across its erstwhile Eurasian range. It also raises the possibility that hspUral might represent a microbial marker for future investigations of the westward expansion of Uralic language speakers from Siberia into northern Europe.
Equally intriguing is the propensity of hpAsia2 to form recombinant populations upon secondary contact with other H. pylori populations. In 2003, Falush et al. (4) established that hpEurope was derived from admixture between ancestral hpAsia2 and hpNEAfrica populations, the latter of which left Africa during a recent out-of-Africa migration (2). As shown here, hpAsia2 was also involved on its eastern frontiers in two further admixture events with hpNorthAsia and hpEastAsia, yielding the recombinant subpopulations hspSiberia1 and hspSiberia2, respectively. Indeed, all known recombinant H. pylori populations worldwide include hpAsia2 as one of the parent populations. Yet, other secondary contacts between H. pylori populations not involving hpAsia2 did not prevent the continued coexistence of discrete populations in Africa (hpAfrica1 and hpNEAfrica) (45) and the New World (hspIndigenousAmericas and hpEurope) (3). These contrasting historical developments may reflect the especially high fitness of hpAsia2 genes that drive the expansion of their recombinant descendants. Alternatively, the expansion of recombinant populations may have been driven by repeated Paleolithic and Holocene introductions of hpAsia2 into geographic areas containing other populations.
Our data also indicate additional admixture events for the evolution of hspKet, which currently consists of 11 isolates from Solumai. Other biopsies from Solumai yielded 11 H. pylori strains assigned to hspIndigenousAmericas and three to hspSiberia1. Our ABC analyses indicated that the evolutionary origin of hspKet was through admixture between hpNorthAsia and hspSiberia2 (model one; Fig. 3C). However, hspSiberia1 is closely related to hspSiberia2, and also to their parent populations, and we cannot definitely exclude alternative evolutionary scenarios because the four other models tested also yielded appreciable probabilities (SI Appendix, Table S4). Resolving this issue would require additional samples and higher resolution comparisons based on whole-genome sequences, which are not currently available. Due to these uncertainties, we ignored all H. pylori isolated from Kets for our reconstructions of the migrations to the Americas. Comparisons of additional samples and whole-genome sequences might elucidate additional migration events in Eurasia and to the Americas that involved hspSiberia1 and/or hspSiberia2 and their descendants.
The Peopling of Siberia.
Our reconstruction of the evolutionary history of four novel H. pylori subpopulations brings insights into past human movements in central Siberia. Three of these subpopulations (hspSiberia1, hspSiberia2, and hspKet) arose via admixture relatively recently, likely during the Holocene. We postulate that each of these evolved from recent secondary contact of populations previously isolated in LGM refugia across central or southern Siberia. This concept is also in accord with the hypothesis of a recent Holocene repopulation of northern Siberia (9, 21, 22). hspAltai arose by genetic drift from a common ancestor with hpNorthAsia; it is also likely to have differentiated in isolation near its current sources in central Siberia. We dated the split that led to hspAltai as 199 to 2,328 y ago, and its current proliferation among Turkic- and Mongolic-speaking Siberians suggests that it may have spread through the region together with recent Turkic (sixth to 16th centuries) and Mongolic (13th century) migrations (46).
The presence of other nonadmixed populations of hspIndigenousAmericas strains among northern Siberians (e.g., Khants, Nenets, Kets, and Evenks) and hspUral strains among Khants and Nenets is less compatible with expansion from refugia in southern areas. Instead, these strains may have infected ANE metapopulations that weathered the LGM at high latitudes as has also been suggested previously (17, 20, 47). We propose therefore that the peopling of Siberia may have involved both continued LGM persistence and Holocene repopulation due to the complex evolutionary dynamics underpinning the region’s human and H. pylori populations.
Peopling of the Americas.
One of the most exciting findings presented here is the existence in Siberia of hpNorthAsia, which encompasses the hspAltai and hspIndigenousAmericas subpopulations (Fig. 1). hspAltai is concentrated in central Siberia, but hspIndigenousAmericas exists throughout Siberia from west to east and throughout the Americas from north to south. Our earlier work demonstrated that hspIndigenousAmericas (at that time, hspAmerind) probably accompanied early American settlers over the Bering Strait (4), possibly even during their initial colonization of the Americas which began over 13 kya (48–50). Our understanding of the detailed Siberian genetic history that resulted in these intercontinental human migrations has been somewhat limited until recently due to a low resolution of traditional methods of human genetics and the lack of data (10–15). Recent developments have found that the human population of Beringia already contained the main East Asian and west Eurasian ancestral components common to all Native American populations before migration into North America (17, 24). The genomic distinctiveness of present-day Eskimo, Inuit, and Surui populations has been attributed to more recent Holocene bouts of gene flow across the Bering Strait (24, 51–53). The identification of hspIndigenousAmericas H. pylori and its sub-subpopulations throughout Siberia provides additional context to help reconstruct those early migrations and their sources. Our data show Siberian substructure within hspIndigenousAmericas, with discrete sub-subpopulations in the East (K2) and West (K3), which overlap in central Siberia (Fig. 4). A total of 18 H. pylori from the Americas belonged to the K1 subpopulation, as did 10 other isolates from Koryak, Chukchi, and Even ethnicities from Kamchatka and Anadyr in West Siberian Beringia. The presence of K1 hspIndigenousAmericas on both sides of the Bering Strait may reflect the ongoing interactions across the Strait between the inhabitants of the coasts of both continents (54, 55). However, the discrete geographic distributions between the other subpopulations of hspIndigenousAmericas over the entire extent of Siberia (Fig. 4) must reflect ancient patterns of isolation and supports the hypothesis of a single Pleistocene human migration into North America (23, 24).
These observations justify the use of ABC models to reconstruct H. pylori’s colonization of the Americas. We inferred that American H. pylori populations branched from Eurasian hspIndigenousAmericas prior to the diversification of East Siberian and Chukotko-Kamchatcan populations, and we dated the median time for this event to ∼12 kya, albeit with wide 95% HPDs. A single migration across the Bering land bridge that colonized both North and South America is not readily conceivable more recently than ∼11.500 kya, after the inundation of the Bering Strait (42). Although it is still debated whether Na-Dene–speaking Athabascans represent an additional source of ancestry from East Asia (23, 56), our analysis indicates that four Canadian Athabascan strains share common ancestry with the rest of American H. pylori and is in line with a hypothesis that the distinctiveness of the Na-Dene speakers evolved through an early divergence from other Native American groups shortly after the colonization of the New World followed by genetic drift (24, 57). However, our best fit model detected low levels of Holocene gene flow into AM from KC (Fig. 4C and SI Appendix, Table S11), potentially reflecting recent human migrations of Saqqaq paleo-Eskimos (52). We also detected migration in the opposite direction, which could represent back migration from the New World into northeastern Asia as previously described (23). Possibly stronger signals of American immigration from Eskimo and other sources might be obtained if additional H. pylori populations were sampled from northern Canada and Greenland.
hspIndigenousAmericas suffered a very severe demographic bottleneck during the initial stage of its colonization of North America, comprising no more than 10 to 854 effective bacteria. However, our models cannot distinguish whether this bottleneck occurred during the migration from Siberia into Beringia and/or from isolation and genetic drift after crossing the Bering Strait. Our analyses estimate an increase in population size about 7 kya (95% HPD 680 to 16,849 y), but we consider the upper limit to be 12,500 y, when the recession of Cordilleran (west) and Laurentide (east) ice sheets created a corridor for human migration (58, 59). Alternatively, the earliest North American settlers would have to have migrated south along a coastal route (49) or by boats (60).
These population analyses and dating calculations are in accord with most archaeological estimates and support the hypothesis that North and South Americans are derived from a single pre-Holocene human migration of evolutionarily ancient northern Eurasians into the Americas (23, 24). They also provide a bacterial perspective on the even earlier spread of anatomically modern humans throughout northern Eurasia.
Materials and Methods
H. pylori strains were cultured from gastric biopsies, which were de-identified after being collected endoscopically from volunteers with informed consent at 18 government hospitals or clinics in Russia and Mongolia. This study was approved by the Charité ethics committee (Ethics certificate EA1/071/07). The details of the sampling procedure, populations, and fragment and genome sequencing are given in SI Appendix. MLST sequences, primer combinations, and PCR conditions are available at https://pubmlst.org/helicobacter. We also created an EnteroBase (25) Helicobacter database (https://enterobase.warwick.ac.uk) that automatically assembles and annotates for Helicobacter draft genome sequences from Illumina short-read sequences, which are uploaded by EnteroBase users or published in short-read archives. The Helicobacter EnteroBase includes seven-gene MLST data and genomic sequences from the M.A. collection (1,871 seven-gene legacy MLST data and almost 400 genomes). The 396 Siberian MLST sequence profiles generated for this study are publicly available at http://enterobase.warwick.ac.uk/species/helicobacter/search_strains?query=workspace%3A49960. A total of 94 genomes representing the global diversity of H. pylori used in this study, and their metadata, are publicly available in the Helicobacter database at http://enterobase.warwick.ac.uk/a/49884.
The structure of genetic variation was evaluated using DAPC (33) and Structure (61) on an MLST data set from 1,002 H. pylori strains from across Asia and the Americas, including 396 previously unpublished Siberian strains. We also analyzed the structure of 94 globally representative H. pylori strains (54 from Siberia) based on draft genome sequences. Initial analyses were performed with 79 strains which belonged to both datasets (SI Appendix, Fig. S21 and Table S12). These were largely consistent but showed some discrepancies with isolates from North Asia. We therefore reconstructed a recombination-aware phylogeny by identifying and removing a variation that was assigned to a homologous recombination by Gubbins (35) and reconstructed maximum likelihood phylogenetic trees using the best substitution model for our dataset (K3P + ASC + R5) in IQ-TREE (62), with branch support determined by 1,000 μLtrafast bootstrap replicates (63). To further investigate the ancestry of Siberian genomes, we employed ChromoPainter version 2 and fineSTRUCTURE version 4 (36). Full details are provided in SI Appendix.
We used the ABC framework (38) to model both the evolutionary origins of the new subpopulations hspSiberia1, hspSiberia2, hspKet, and hspAltai as well as to reconstruct and date the migration of H. pylori into the Americas. All modeling details are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Kuvat Momynaliev and Vera Chelysheva for storing H. pylori biopsies, bacterial culture, and isolation. We thank Steffie Bernhöft and Christiana Stamer for technical assistance with PCR and sequencing. We thank Kaisa Thorell for assistance with the genome alignment and Andrinajoro Rakatoarivelo for bioinformatic support. This work was supported by a grant from the Max-Planck Society and an Investigator in Science Award to M.A. (Grant No. 202792) from the Wellcome Trust.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. D.F. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015523118/-/DCSupplemental.
Data Availability
DNA sequences data have been deposited in EnteroBase (http://enterobase.warwick.ac.uk/species/helicobacter/search_strains?query=workspace%3A49960 and http://enterobase.warwick.ac.uk/a/49884).
Change History
June 25, 2021: The license for this article has been updated.
References
- 1.Montano V., et al., Worldwide population structure, long-term demography, and local adaptation of Helicobacter pylori. Genetics 200, 947–963 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moodley Y., et al., Age of the association between Helicobacter pylori and man. PLoS Pathog. 8, e1002693 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Linz B., et al., An African origin for the intimate association between humans and Helicobacter pylori. Nature 445, 915–918 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falush D., et al., Traces of human migrations in Helicobacter pylori populations. Science 299, 1582–1585 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Moodley Y., et al., The peopling of the Pacific from a bacterial perspective. Science 323, 527–530 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wirth T., et al., Distinguishing human ethnic groups by means of sequences from Helicobacter pylori: Lessons from Ladakh. Proc. Natl. Acad. Sci. U.S.A. 101, 4746–4751 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tay C. Y., et al., Population structure of Helicobacter pylori among ethnic groups in Malaysia: Recent acquisition of the bacterium by the Malay population. BMC Microbiol. 9, 126 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rohr J. R., IWGIA Report 18 (International Work Group for Indigenous Affairs, Copenhagen, Denmark, 2014). [Google Scholar]
- 9.Pugach I., et al., The complex admixture history and recent southern origins of Siberian populations. Mol. Biol. Evol. 33, 1777–1795 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lell J. T., et al., Y chromosome polymorphisms in native American and Siberian populations: Identification of native American Y chromosome haplotypes. Hum. Genet. 100, 536–543 (1997). [DOI] [PubMed] [Google Scholar]
- 11.Starikovskaya Y. B., Sukernik R. I., Schurr T. G., Kogelnik A. M., Wallace D. C., mtDNA diversity in Chukchi and Siberian Eskimos: Implications for the genetic history of ancient Beringia and the peopling of the New World. Am. J. Hum. Genet. 63, 1473–1491 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schurr T. G., Sukernik R. I., Starikovskaya Y. B., Wallace D. C., Mitochondrial DNA variation in Koryaks and Itel’men: Population replacement in the Okhotsk Sea-Bering Sea region during the Neolithic. Am. J. Phys. Anthropol. 108, 1–39 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Derbeneva O. A., Starikovskaya E. B., Wallace D. C., Sukernik R. I., Traces of early Eurasians in the Mansi of northwest Siberia revealed by mitochondrial DNA analysis. Am. J. Hum. Genet. 70, 1009–1014 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karafet T. M., et al., Ancestral Asian source(s) of new world Y-chromosome founder haplotypes. Am. J. Hum. Genet. 64, 817–831 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derenko M. V., et al., Diversity of mitochondrial DNA lineages in south Siberia. Ann. Hum. Genet. 67, 391–411 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Fu Q., et al., Genome sequence of a 45,000-year-old modern human from western Siberia. Nature 514, 445–449 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raghavan M., et al., Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature 505, 87–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong E. H., et al., Reconstructing genetic history of Siberian and Northeastern European populations. Genome Res. 27, 1–14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lazaridis I., et al., Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuzmin Y. V., Keates S. G., Siberia and neighboring regions in the last glacial maximum: Did people occupy northern Eurasia at that time? Archaeol. Anthropol. Sci. 10, 111–124 (2018). [Google Scholar]
- 21.Goebel T., The “microblade adaptation” and recolonization of Siberia during the late upper Pleistocene. Archeol. Pap. Am. Anthropol. Assoc. 12, 15 (2002). [Google Scholar]
- 22.Graf K. E., “The good, the bad, and the ugly”: Evaluating the radiocarbon chronology of the middle and late upper paleolithic in the Enisei river valley, south-central Siberia. J. Archaeol. Sci. 36, 694–707 (2009). [Google Scholar]
- 23.Reich D., et al., Reconstructing native American population history. Nature 488, 370–374 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raghavan M., et al., POPULATION GENETICS. Genomic evidence for the Pleistocene and recent population history of Native Americans. Science 349, aab3884 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Z., Alikhan N. F., Mohamed K., Fan Y., Achtman M.; Agama Study Group , The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 30, 138–152 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maixner F., et al., The 5300-year-old Helicobacter pylori genome of the Iceman. Science 351, 162–165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Latifi-Navid S., et al., Ethnic and geographic differentiation of Helicobacter pylori within Iran. PLoS One 5, e9645 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao Y. L., et al., Core genome haplotype diversity and vacA allelic heterogeneity of Chinese Helicobacter pylori strains. Curr. Microbiol. 59, 123–129 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Guo Y., et al., Genome of Helicobacter pylori strain XZ274, an isolate from a Tibetan patient with gastric cancer in China. J. Bacteriol. 194, 4146–4147 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miftahussurur M., et al., Molecular epidemiology of Helicobacter pylori infection in Nepal: Specific ancestor root. PLoS One 10, e0134216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsunari O., et al., Rare Helicobacter pylori virulence genotypes in Bhutan. Sci. Rep. 6, 22584 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.You Y.et al., Genome sequences of three Helicobacter pylori strains isolated from atrophic gastritis and gastric ulcer patients in China. Am. Soc. Microbiol., 6314–6315 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jombart T., Devillard S., Balloux F., Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 11, 94 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Falush D., Stephens M., Pritchard J. K., Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 164, 1567–1587 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Croucher N. J., et al., Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43, e15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawson D. J., Hellenthal G., Myers S., Falush D., Inference of population structure using dense haplotype data. PLoS Genet. 8, e1002453 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yahara K., et al., Chromosome painting in silico in a bacterial species reveals fine population structure. Mol. Biol. Evol. 30, 1454–1464 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beaumont M. A., Zhang W., Balding D. J., Approximate Bayesian computation in population genetics. Genetics 162, 2025–2035 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bertorelle G., Benazzo A., Mona S., ABC as a flexible framework to estimate demography over space and time: Some cons, many pros. Mol. Ecol. 19, 2609–2625 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Achtman M., et al., Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32, 459–470 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Morelli G., et al., Microevolution of Helicobacter pylori during prolonged infection of single hosts and within families. PLoS Genet. 6, e1001036 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.England J. H., Furze M. F. A., New evidence from the western Canadian arctic archipelago for the resubmergence of Bering Strait. Quat. Res. 70, 60–67 (2008). [Google Scholar]
- 43.Moodley Y., “Helicobacter pylori: genetics, recombination, population structure and human migrations” in Helicobacter Pylori Research: From Bench to Bedside, Backert S., Yamaoka Y., Eds. (Springer Japan, 2016), pp. 3–27. [Google Scholar]
- 44.Breurec S., et al., Evolutionary history of Helicobacter pylori sequences reflect past human migrations in Southeast Asia. PLoS One 6, e22058 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nell S., et al., Recent acquisition of Helicobacter pylori by Baka pygmies. PLoS Genet. 9, e1003775 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yunusbayev B., et al., The genetic legacy of the expansion of Turkic-speaking nomads across Eurasia. PLoS Genet. 11, e1005068 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuzmin Y. V., Keates S. G., Dates are not just data: Paleolithic settlement patterns in Siberia derived from radiocarbon records. Am. Antiq. 70, 773–789 (2005). [Google Scholar]
- 48.Gilbert M. T. P., et al., DNA from pre-Clovis human coprolites in Oregon, North America. Science 320, 786–789 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Dillehay T. D., et al., Monte Verde: Seaweed, food, medicine, and the peopling of South America. Science 320, 784–786 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Waters M. R., et al., Pre-Clovis mastodon hunting 13,800 years ago at the Manis site, Washington. Science 334, 351–353 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Rasmussen M., et al., Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature 463, 757–762 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raghavan M., et al., The genetic prehistory of the New World arctic. Science 345, 1255832 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Skoglund P., et al., Genetic evidence for two founding populations of the Americas. Nature 525, 104–108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Black L. T., Some problems in interpretation of Aleut prehistory. Arctic Anthropol. 20, 49–78 (1983). [Google Scholar]
- 55.Rubicz R. C., “Evolutionary consequences of recently founded Aleut communities in the Commander and Pribilof Islands,” PhD dissertation, University of Kansas (2007).
- 56.Skoglund P., Reich D., A genomic view of the peopling of the Americas. Curr. Opin. Genet. Dev. 41, 27–35 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rasmussen M., et al., The genome of a Late Pleistocene human from a Clovis burial site in western Montana. Nature 506, 225–229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dyke A. S., Developments in Quaternary Sciences (Elsevier, 2004), vol. 2, pp. 373–424. [Google Scholar]
- 59.Gregoire L. J., Payne A. J., Valdes P. J., Deglacial rapid sea level rises caused by ice-sheet saddle collapses. Nature 487, 219–222 (2012). [DOI] [PubMed] [Google Scholar]
- 60.Erlandson J. M., Braje T. J., From Asia to the Americas by boat? Paleogeography, paleoecology, and stemmed points of the northwest Pacific. Quat. Int. 239, 28–37 (2011). [Google Scholar]
- 61.Pritchard J. K., Stephens M., Donnelly P., Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Minh B. Q., et al., IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Minh B. Q., Nguyen M. A. T., von Haeseler A., Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences data have been deposited in EnteroBase (http://enterobase.warwick.ac.uk/species/helicobacter/search_strains?query=workspace%3A49960 and http://enterobase.warwick.ac.uk/a/49884).