Abstract
Therapeutic antibody discovery using synthetic diversity has been proved productive, especially for target proteins not suitable for traditional animal immunization-based antibody discovery approaches. Recently, many lines of evidences suggest that the quality of synthetic diversity design limits the development success of synthetic antibody hits. The aim of our study is to understand the quality limitation and to properly address the challenges with a better design. Using VH3–23 as a model framework, we observed and quantitatively mapped CDR-H3 loop length-dependent usage of human IGHJ4 and IGHJ6 germline genes in the natural human immune repertoire. Skewed usage of DH2-JH6 and DH3-JH6 rearrangements was quantitatively determined in a CDR-H3 length-dependent manner in natural human antibodies with long CDR-H3 loops. Structural modeling suggests choices of JH help to stabilize antibody CDR-H3 loop and JH only partially contributes to the paratope. Our observations shed light on the design of next-generation synthetic diversity with improved probability of success.
Keywords: CDR-H3, diversity, synthetic antibody library, JH4, JH6
STATEMENT OF SIGNIFICANCE
Therapeutic antibody discovery using synthetic diversity has been proved productive. The quality of diversity design limits the developability of synthetic hits. Here, we quantitatively determined the CDR-H3 length-dependent usage of human DH/JH germline genes and the resulting spatial paratope landscape, which sheds light on rational design of synthetic diversity with improved probability of success.
INTRODUCTION
Therapeutic antibodies can be discovered via in vivo, in vitro or in silico approaches [1, 2]. In vivo approach relies on the immunization of wild type or transgenic animals carrying human antibody gene segments, while in vitro approach employs the selection power by display technologies to pan large and diverse antibody libraries [3]. Both approaches have been proved very successful and have generated best-selling antibody therapeutics such as Keytruda and Humira, respectively [4, 5]. Recent advances in artificial intelligence and machine learning also facilitated in silico rational antibody design [1].
In vitro selection of synthetic antibody libraries complements in vivo immunization-based approaches by providing unique, non-natural antibody paratopes and escaping the limitations of self-tolerance [6, 7]. One of challenges in the rational design of man-made antibody diversity lies in the complementarity determining region (CDR) H3 loop (CDR-H3) of antibody heavy chain variable region. Traditionally, CDR-H3 loop was randomly diversified by degenerate codons or parsimonious degenerate codons [8], which limited the therapeutic potential of antibody hits. Although TRInucleotide technology and Slonomics technology helped to reduce dysfunctional motifs and to precisely control the codon biases, the CDR-H3 still could not fully mimic that of natural diversity generated by V(D)J recombination and somatic hypermutation process [9, 10]. This limits the translational success of therapeutic antibody candidates derived from in vitro selection of phage or yeast libraries [11]. Recently, massive datasets of human immune repertoires by Briney, Soto and others made it possible for us to survey billions of antibody heavy chain sequences for a better understanding of CDR-H3 natural diversity at immune repertoire level [12–14].
In this study, we aim to understand natural immune diversity of the CDR-H3 loop in a precisely controlled manner. From antibody engineers’ perspective, we observed CDR-H3 length-dependent usage of human IGHJ4 and IGHJ6 gene segments in the natural human immune repertoire. We also observed the biased usage of DH3-JH6 rearrangements in antibodies with long CDR-H3 loops. Inspired by our observations, we also conducted spatial analysis of the VH3–23 antibody paratopes and defined the parameters influencing an antibody’s solvent accessible surface area (SASA). This knowledge sheds light on the design of next-generation synthetic diversity.
MATERIALS AND METHODS
Data source
All antibody sequences were obtained from previous studies, including the IGHV3–23*01 dataset (European Molecular Biology Laboratory accession nos. AM076988–AM083316) [15, 16] and the Dengue virus (DENV) acutely infected patients dataset (the BioProject accession number PRJNA205206) [17]. The DENV dataset was processed as what is described in the publication [17].
Antibody homology modeling and germline usage analysis
The Fv models were generated using Rosetta following the authors’ instructions [18, 19]. In brief, a template was selected. Template CDRs were grafted onto the template frameworks, the frameworks were then assembled according to a template VH-VL orientation. The CDR-H3 of the top ranked model was then de novo modeled and the relative VH-VL orientation was refined via local docking. Kabat numbering was used to number the CDR loops and the SASA was measured using Rosetta. Germline usage analysis was performed by aligning the variable domain sequences to the IMGT reference germline genes using IgBlast [20], the output was parsed using Change-O [21].
Statistical analysis
Statistical analysis was performed using OriginPro 2021. If the dataset conforms to normal distribution, one-way analysis of variance was used [22]. Significance level was set to 0.05. Turkey test was selected for the mean comparison, and the Levene test was selected for the homogeneity of variance test. P-value was obtained by means comparison and overall analysis of variance. If the dataset did not conform to the normal distribution, then non-parametric test method was used. The significance level was set to 0.05. Since the data were not repeated measures, the Kruskal–Wallis ANOVA multi-sample independent non-parametric test was selected [23]. P-value was obtained at the significance level of 0.05. If P > 0.05, there is no significant difference between the two groups of data. If 0.05 > P > 0.01, there is a significant difference between the two sets of data, marked as *. If 0.01 > P > 0.001, there is a significant difference between the two sets of data, marked as **. If P < 0.001, there is a significant difference between the two sets of data, marked as ***.
RESULTS
Antibody heavy chain CDR3-dependent usage of human IGHJ4 and IGHJ6
Antibody CDR H3 loop (CDR-H3) is highly variable. The diversity of CDR-H3 is mainly generated by V(D)J recombination via different mechanisms [24]. Skewed usage of human germline genes such as IGHJ4 and IGHJ6 was previously reported in CDR-H3 [25, 26]. Briney et al. [26] discovered that a skewed small subset of IGHD and IGHJ gene segments were particularly used to encode long CDR-H3s in human peripheral blood antibodies. It, however, remains a still poorly understood correlation. To further understand the exact correlation between CDR-H3 length and skewed human germline gene usage, we analyzed the same set of VH3–23*01 sequences (European Molecular Biology Laboratory accession # AM076988–AM083316) that were extensively validated and studied in the field [15, 16, 27, 28]. This enabled us to elucidate the correlation on a pre-defined antibody framework and genetic background.
Analysis of the same set of unique, productively rearranged human VH3–23*01 gene sequences revealed strong correlation between CDR-H3 length and IGHJ germline gene usage (Fig. 1A). All antibody sequences analyzed in this study encode unique CDR-H3s to eliminate bias introduced by redundancy in the human immune repertoire. As shown in Fig. 1A, when CDR-H3 length increases from 7aa to 23aa, the germline usage of IGHJ4 decreased gradually from 67.57% at 7aa length to 6.11% at 23aa length, while the usage of human IGHJ6 germline gene increased proportionally from 27.03% at 7aa length to 94.90% at 23aa length. At a shorter CDR-H3 length such as 7aa, IGHJ4 was preferentially used (67.57%) over other IGHJ germline genes. At a relatively longer CDR-H3 length such as 23aa, IGHJ6 (94.90%) was predominantly used, while other IGHJ germline genes were very rarely used. CDR-H3 length below 7aa or longer than 23aa was not analyzed in this study due to inadequate number of IGHV3–23*01 sequences available.
Figure 1.
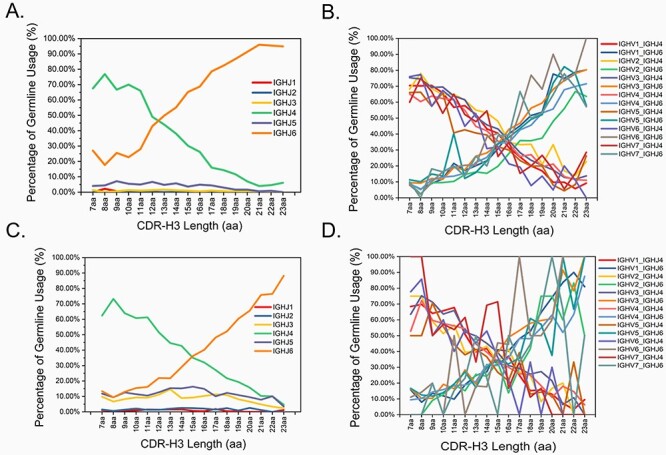
Antibody heavy chain CDR-H3 length-dependent usage of human IGHJ4 and IGHJ6 germline genes in antibody sequences derived from (A) healthy and viral-infection naive cohort of Danish origin; (B) and (C) Nicaraguan individuals acutely exposed to Dengue virus; (D) the accompanying healthy cohort of the DENV study.
The dataset used in Fig. 1A analysis contains antibody sequences isolated from a cohort of healthy and viral infection-naive adults of Danish background. These IGHV3–23*01 sequences were productively rearranged predominantly to IGHJ4 or IGHJ6. To avoid biases incurred from data source, we asked ourselves: (i) is genetic background or health conditions playing a role? And (ii) is this observation reproducible for other human IGHV germline genes? To properly address above concerns, we analyzed a dataset (the BioProject accession number PRJNA205206) containing antibody sequences sampled from 44 Nicaraguan individuals acutely exposed to Dengue virus (DENV) during acute symptomatic dengue at 2–5 days post-symptom onset [17]. Regardless of the germline usage of IGHJ1, IGHJ2, IGHJ3 and IGHJ5, the same germline usage patterns of IGHJ4 and IGHJ6 were observed with antibody sequences derived from a large cohort of patients acutely infected with DENV (Fig. 1C). This suggests that, as CDR-H3 loops go longer, the increased germline usage of IGHJ6 is an intrinsic feature of V(D)J recombination and it is independent of health conditions or ethnic backgrounds. To answer the question if this observation is unique to human IGHV3–23*01 germline gene, we dissected the antibody sequences based on IGHV germline usage from the acute DENV cohort (n = 44) and the accompanying healthy control cohort (n = 8). Figure 1B and D shows the same exact pattern of IGHJ4 and IGHJ6 germline usage in 68 067 and 8539 antibody sequences derived from the acute DENV cohort and the accompanying healthy cohort, respectively.
Above data suggest CDR-H3 length-dependent usage of human IGHJ4 and IGHJ6 germline. Such skewed usage is an intrinsic feature of human heavy chain CDR-H3 diversity generation. It is not impacted by ethnic background, health conditions or the usage of human IGHV germline genes.
We then combined the two datasets and dissected antibody sequences based on human IGHJ germline gene usage. Total 68 023 antibody sequences were analyzed, regardless of health conditions, ethnic background and human IGHV germline gene usage. Figure 2 showed the CDR-H3 length distribution of each IGHJ group, combined (Fig. 2A) or individualized (Fig. 2B). Of the 68 023 antibody sequences, 29 873 (43.92%) belong to the IGHJ4 germline group, while 21 283 (31.29%) belong to the IGHJ6 germline group. Even though only 889 and 991 antibody sequences were obtained in the IGHJ1 and IGHJ2 group, respectively, the Gaussian distribution pattern of the CDR-H3 length distribution of all IGHJ groups suggests that the combined dataset is diverse and scientifically sound for further analysis. Figure 2B showed that the average length of CDR-H3 in natural human antibodies rearranged to human IGHJ6 germline gene is 16.40 amino acids (lower right, n = 21 283). The IGHJ6 antibodies CDR-H3 length distribution peaked at a length of 17 amino acids. The average length of CDR-H3 in natural human antibodies rearranged to human IGHJ4 germline gene is 12.43 amino acids (lower left, n = 29 873). The IGHJ4 antibodies CDR-H3 length distribution peaked at a length of 12 amino acids. This observation is consistent with what's been described in Fig. 1, that is human antibodies with shorter CDR-H3 tend to be preferentially rearranged to human IGHJ4 germline genes, while antibodies with longer CDR-H3 tend to be preferentially rearranged to human IGHJ6 germline genes. CDR-H3 amino acid usage frequency was further determined in each JH antibody group. No statistically significant difference was observed on the overall amino acid usage patterns across all JH antibody groups (Pearson’s Chi-squared test, P > 0.05), albeit some amino acid residues such as tyrosine were differentially used across JH antibody groups (Supplementary Fig. S1, see Supplementary Data available at ABT online).
Figure 2.
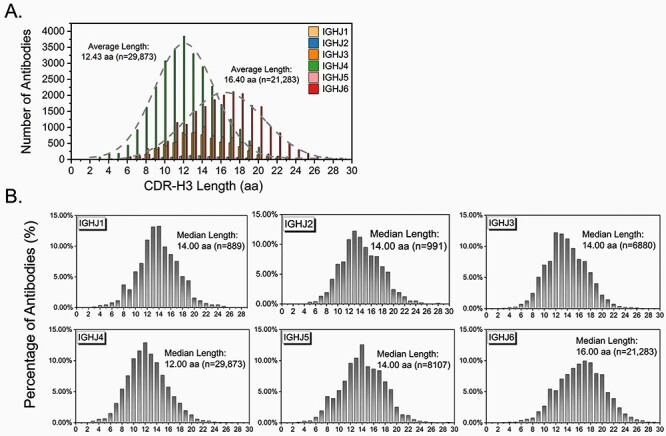
The CDR-H3 length distribution of different human IGHJ antibodies. (A) Combined; (B) individualized.
CDR-H3 length-dependent preference in DH-JH recombination
Recent study by Sankar et al. [29] showed that in natural human antibody repertoire, the antibody CDR-H3 diversity was dynamically shaped by VH, VL and JH germline segment use. Previously, we discovered CDR-H3 length-dependent usage of human IGHD germline genes [28]. Such CDR-H3 length-dependent usage of non-canonical cysteines was extensively characterized in human VH repertoires [30] and in other animals such as chicken [31], bovine [32], shark and camelid [33]. Intrigued by the roles of human IGHD gene segments in shaping human antibody CDR-H3 diversity, we analyzed the human IGHD germline gene usage of natural antibodies derived from 8 healthy donors and 44 DENV acutely infected patients—the same dataset we used in section Antibody heavy chain CDR3-dependent usage of human IGHJ4 and IGHJ6. Not surprisingly, we observed CDR-H3 length-dependent usage of human IGHD3 and IGHD2 germline genes. As CDR-H3 goes longer, the usage of IGHD3 and IGHD2 increased from 24.87 and 14.60% at 7 amino acids length to 57.52 and 30.70% at 23 amino acids length, respectively (Fig. 3A). Such CDR-H3 length-dependent usage of human IGHD2 and IGHD3 is independent of health conditions, as we observed the same germline usage patterns in both healthy donors (Fig. 3A, upper panel, n = 8359) and DENV acutely infected patients (Fig. 3A, lower panel, n = 68 067).
Figure 3.
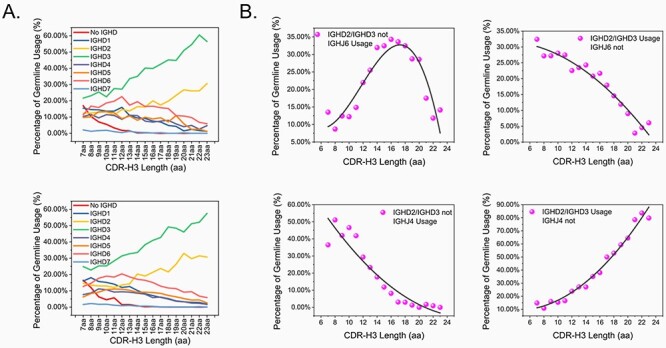
(A) The CDR-H3 length-dependent usage of human IGHD2 and IGHD3 germline genes; (B) IGHD3/IGHD2 were preferentially rearranged to human IGHJ4/IGHJ6 germline genes as CDR-H3 length goes longer.
When CDR-H3 is of 7 amino acids length, in ~17% of the antibodies in the immune repertoire, the CDR-H3 diversity is likely generated by direct rearrangement of IGHV to IGHJ. We could not rule out the possibility that IgBlast might fail to identify the IGHD region, especially when the IGHD is short and extensively mutated. As CDR-H3 loop length increases, the percentage of direct VH-JH recombined sequences gradually decreased to 0%. At 14 amino acids CDR-H3 length or longer, direct rearrangement of IGHV to IGHJ was very rarely observed. This indicates that in antibodies with longer CDR-H3 loops, IGHD gene segments are critical components of CDR-H3 diversity. More intriguingly, human IGHD7 germline gene was rarely used in the human immune repertoire (Fig. 3A).
Given the previous observation of CDR-H3 length-dependent usage of IGHJ4 and IGHJ6 germline genes, we were wondering about potential biases in human IGHD rearrangements to human IGHJ. To answer this question, we turned to the IGHV3–23*01 dataset of Danish origin. As the human IGHV germline usage is fixed to human IGHV3–23*01, the DH-JH recombination analysis is not impacted by the choices of IGHV. We observed CDR-H3 length-dependent preferential rearrangement of IGHD3/IGHD2 gene segments to human IGHJ6 germline gene (Fig. 3B). Of 2125 VH3–23-DH3/DH2 antibody sequences that do not use IGHJ4, a strong correlation was observed between CDR-H3 length and DH3/DH2 germline usage (Fig. 3B, lower right). In contrast, of 1116 VH3–23-DH3/DH2 antibody sequences that do not use IGHJ6, a strong negative correlation was observed between CDR-H3 length and DH3/DH2 germline usage (Fig. 3B, upper right). These data suggest that DH3/DH2 was preferentially rearranged to IGHJ6 as CDR-H3 goes longer. Such a biased preference was not observed with other human IGHD germline genes. Our observation is consistent with a previous study by Briney et al. [26] in the analysis of a B-cell receptor repertoire of a HIV-infected patient cohort. This suggests that the preferential rearrangement of DH3/DH2 to JH6 is universal in CDR-H3 diversity generation. Such a preference is likely required by the intrinsic need to stabilize the long CDR-H3 loops.
Choice of JH only partially contributes to the SASA of an antibody’s paratope
We then moved on to determine if preferentially rearranged DH/JH combinations can be used to guide the design of rationally designed antibody libraries. An essential question to answer is what parameters we can control to finely tune the size of the surface area and the physicochemical property of an antibody’s paratope. The IGHV3–23*01 dataset was again used in this analysis. To avoid bias introduced by irregular framework, all antibody sequences used in this analysis were manually validated to be free of framework insertions or deletions. Similarly, to avoid bias introduced by skewed CDR-H3 amino acid usage, all IGHV3–23*01 sequences were manually validated on the diversity to remove redundant sequences or sequences with irregular CDR loops, framework deletions or insertions. First, we determined the relationship between CDR-H3 length and the SASA of the antibody paratope. Simulated models were built by pairing human IGHV3–23*01 variable region, of various CDR-H3 length ranging from 7aa to 23aa, with human IGKV1–39*01/IGKJ2 variable region. IGKV1–39 was chosen because the VH3–23/VK1–39 pairing was considered a very productive VH/VL pairing with good drug like properties [34, 35]. The SASA values were calculated based on Kabat numbering of the antibody CDR loops. In this analysis, the only variables are the CDR-H3 length and the choices of various JH. As shown in Fig. 4A, a positive correlation was observed. As CDR-H3 went longer, the SASA of antibody paratope grew bigger from 3008.88 ± 116.10 A2 of 7aa length to 3666.42 ± 224.78 A2 of 23aa length. Interestingly, when CDR-H3 is longer than 22 amino acids, the SASA remained stagnant. This observation needs to be further validated with more IGHV3–23*01 sequences of extra CDR-H3 length. In above VH3–23/VK1–39 pairings, the VK1–39 framework was fixed to JK2. We were wondering if the choice of JK would change the SASA of VH3–23/VK1–39 paired antibodies. In this analysis, we chose a dataset with a fixed 10aa length of CDR-H3 and IGKV1–39*01 rearranged to various IGKJ germline genes. Figure 4B showed the statistical analysis of VH3–23/VK1–39 antibody paratopes with various human JKs. No statistically significant difference was observed among the five JK groups (P > 0.05). This indicates that choices of JK minimally impacted the SASA of an antibody’s paratope. We then paired a set of VH3–23*01 of 14aa length with various kappa light chain germline genes rearranged to JK2. Figure 4C showed that VK pairing statistically significantly impacted the SASA of antibody paratopes (P < 0.001). Choices of distal or proximal VK germline also statistically significantly impacted the SASA of antibody paratopes (P < 0.001). In a pairwise analysis, only statistically significant difference was observed between VK3–11, VK3–20 and their distal counterparts (Fig. 4D, P < 0.001). To determine the impact of JH on the SASA of antibody paratope, we paired VH3–23*01 of 14aa length, rearranged to various JH, with VK1–39*01/JK2. As shown in Fig. 4E, in a pairwise analysis, statistically significant difference was observed between JH4/JH5 and JH6 (Fig. 4E, P < 0.001). This indicates that JH6 significantly contributed to the SASA of antibody paratope.
Figure 4.
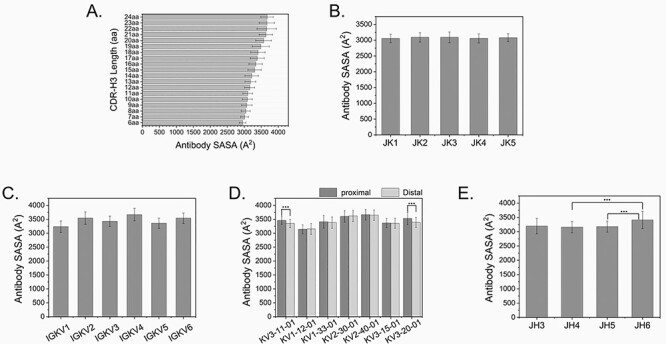
(A) The SASA of an antibody paratope is determined by many factors, including CDR-H3 length; (C and D) VH/VL pairing; (B and E) rearrangement to JK or JH only partially impact the size of SASA.
In this analysis, using a set of precisely controlled data with minimal variables, we confirmed the commonly accepted knowledge in the field that antibody paratope is mainly impacted by the CDR-H3 length and VH/VL germline pairings. We also observed that JK, but not JH, minimally impacted the overall SASA of antibody paratope.
DISCUSSION
The CDR-H3 assembly mechanism, regulation and influence on antibody diversity were extensively reviewed in the field [24, 36, 37]. Recent advances in next-generation sequencing and immune repertoire mining helped us to understand the antibody diversity to a degree to enable man-made variable genes for antibody discovery [7]. In this study, our goal is to understand the components of CDR-H3 diversity in a precisely controlled manner, hoping that the knowledge gained can help rational design of next-generation antibody libraries. Unlike many other studies mining immune repertoire, we utilized a well validated IGHV3–23*01 dataset [15, 16, 27, 28] to keep variables minimal in our study.
We observed CDR-H3 length-dependent usage of IGHJ4 and IGHJ6 germline genes in the immune repertoire. The increased usage of IGHJ6 in antibodies with long CDR-H3 loops validated a well-recognized feature of tyrosine enrichment at the junction of DH-JH recombination [38]. Among all human JH gene segments, JH6 is the longest [39], which explained the increased usage of IGHJ6 germline genes in antibodies with long CDR-H3. Likely, tyrosine residues were introduced repeatedly for the continued diversification at the DH-JH junction as CDR-H3 loops become longer. Tyrosine, on one side, is one of the dominant CDR-H3 residues critical for antibody binding specificity [28, 38]. On the other side, tyrosine-rich motifs were frequently observed in amyloidogenic proteins such as β2-microglobulin [40, 41], suggesting an important role of tyrosine in controlling the integral structure of proteins. Tyrosine motifs at the DH-JH junction likely are not involved in antigen–antibody interactions. In antibodies with long CDR-H3 loops, we hypothesize that tyrosine residues at the DH-JH junction serve a structural role to help assembling and stabilizing the CDR-H3 loop. Such role is currently under-appreciated in the field and it needs to be tested in the future.
A highly skewed DH3-JH6 rearrangement was observed in this study. It correlates well with CDR-H3 loop length but it is independent of ethnic background and health conditions. As both DH3 and JH6 are the longest in their respective families, it is not surprising to observe the biased DH3-JH6 rearrangements in antibodies with long CDR-H3 loops. Interestingly, biased DH3-JH6 rearrangements were also observed in the comparative immune repertoire analysis of a humanized rodent model [39], even though the humanized rodents tend to prefer short DH genes. These suggest that DH3, as well as DH2 [28, 30], serves to stabilize the rigid and protruding CDR-H3 loops by rearrangement with JH6 in antibodies with long CDR-H3 loops. Antibodies with non-canonical cysteine residues in the CDR-H3 were frequently observed in human broadly neutralizing antibodies against HIV-1, human hepatitis C virus, human cytomegalovirus and human influenza virus [42–45]. This suggests that the stabilized CDR-H3 loop may contribute to unique binding attributes such as high affinity or broader binding specificity toward difficult to reach epitopes on the virions [28]. Such unique structural and binding properties in antibodies with longer CDR-H3 loops could be of interest in the next-generation synthetic human antibody libraries to help fight emerging pathogens such as COVID-19.
Above findings inspired us to perform the spatial analysis of the antibody paratope. By computational simulations, in a precisely controlled manner, we defined the spatial patterns of antibody paratope and the influencing parameters such as CDR-H3 length, pairing light chain germline genes, choices of JK and JH, etc. The conformations of CDR-H3 loops are pivotal elements in CDR-H3 diversity design. In this study, the CDR-H3 conformation was not considered a key parameter due to the lack of well-recognized predicative tools. We will keep improving the analysis when it is technically possible and will expand the analysis to all human VH germline genes. The knowledge obtained will help to guide the rational design of next-generation antibody libraries. In the new library design, IGHJ gene fragments will be mixed together proportionally to faithfully reflect the CDR-H3 length-dependent germline usage. CDR-H3 diversity synthesis can be achieved via TRInucleotide technology so that the amino acid distribution pattern follows that of natural immune repertoire. We hope that this will properly address the poor antibody developability issue that the synthetic diversity field is facing. Targeting a pre-defined antigenic epitope, a high-quality antibody library of minimal size and diversity can be designed to facilitate antibody discovery on demand.
FUNDING
This research received no external funding.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
AUTHORS’ CONTRIBUTIONS
Conceptualization, L.C., C.Y. and Y.S.; methodology, L.C. and Y.Y.; validation, K.Y. and H.W.; formal analysis, L.C., K.Y. and H.W.; data curation, K.Y. and H.W.; writing—original draft preparation, L.C., R.W., H.W. and Y.K; writing—review and editing, L.C., C.Y., Y.Y. and Y.S.; visualization, H.W.; supervision, R.W., Y.Y. and L.C.; all authors have read and agreed to the published version of the manuscript.
DATA AVAILABILITY STATEMENT
The following data were used in this study: the set of VH3–23*01 sequences (European Molecular Biology Laboratory accession nos. AM076988–AM083316) and DENV cohort sequences (the Bi-oProject accession number PRJNA205206).
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to acknowledge Ms Mengya Chai on her technical support and Dr Andrew Bradbury for critical reading of the manuscript. The authors would also greatly appreciate Mr Bangqi Zheng’s and Ms Wanmian Huang’s help with the Pearson’s Chi-squared test.
REFERENCES
- 1. Sormanni, P, Aprile, FA, Vendruscolo, M. Third generation antibody discovery methods: in silico rational design. Chem Soc Rev 2018; 47: 9137–57. [DOI] [PubMed] [Google Scholar]
- 2. Laustsen, AH, Greiff, V, Karatt-Vellatt, A et al. Animal immunization, in vitro display technologies, and machine learning for antibody discovery. Trends Biotechnol 2021; 21: 00061–5. [DOI] [PubMed] [Google Scholar]
- 3. Lu, R, Hwang, Y, Liu, I et al. Development of therapeutic antibodies for the treatment of diseases. J Biomed Sci 2020; 27: 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. den Broeder, A, van de Putte, L, Rau, R et al. A single dose, placebo controlled study of the fully human anti-tumor necrosis factor-alpha antibody adalimumab (D2E7) in patients with rheumatoid arthritis. J Rheumatol 2002; 29: 2288–98. [PubMed] [Google Scholar]
- 5. Patnaik, A, Kang, SP, Rasco, D et al. Phase I study of pembrolizumab (MK-3475; anti-PD-1 monoclonal antibody) in patients with advanced solid tumors. Clin Cancer Res 2015; 21: 4286–93. [DOI] [PubMed] [Google Scholar]
- 6. Bradbury, ARM, Sidhu, SS, Dubel, S et al. Beyond natural antibodies: the power of in vitro display technologies. Nat Biotechnol 2011; 29: 245–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Finlay, WJJ, Almagro, JC. Natural and man-made V-gene repertoires for antibody discovery. Front Immunol 2012; 3: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sidhu, SS, Li, B, Chen, Y et al. Phage-displayed antibody libraries of synthetic heavy chain complementarity determining regions. J Mol Biol 2004; 338: 299–310. [DOI] [PubMed] [Google Scholar]
- 9. Knappik, A, Ge, L, Honegger, A et al. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J Mol Biol 2000; 296: 57–86. [DOI] [PubMed] [Google Scholar]
- 10. Zhai, W, Glanville, J, Fuhrmann, M et al. Synthetic antibodies designed on natural sequence landscapes. J Mol Biol 2011; 412: 55–71. [DOI] [PubMed] [Google Scholar]
- 11. Jain, T, Sun, T, Durand, S et al. Biophysical properties of the clinical-stage antibody landscape. PNAS 2017; 114: 944–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Briney, B, Inderbitzin, A, Joyce, C et al. Commonality despite exceptional diversity in the baseline human antibody repertoire. Nature 2019; 566: 393–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Soto, C, Bombardi, RG, Branchizio, A et al. High frequency of shared clonotypes in human B cell receptor repertoires. Nature 2019; 566: 398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kovaltsuk, A, Leem, J, Kelm, S et al. Observed antibody space: a resource for data mining next-generation sequencing of antibody repertoires. J Immunol 2018; 201: 2502–9. [DOI] [PubMed] [Google Scholar]
- 15. Ohm-Laursen, L, Nielsen, M, Larsen, SR et al. No evidence for the use of DIR, D-D fusions, chromosome 15 open reading frames or VH replacement in the peripheral repertoire was found on application of an improved algorithm, JointML, to 6329 human immunoglobulin H rearrangements. Immunology 2006; 119: 265–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ohm-Laursen, L, Barington, T. Analysis of 6912 unselected somatic hypermutations in human VDJ rearrangements reveals lack of strand specificity and correlation between phase II substitution rates and distance to the nearest 3′ activation-induced cytidine deaminase target. J Immunol 2007; 178: 4322–34. [DOI] [PubMed] [Google Scholar]
- 17. Parameswaran, P, Liu, Y, Roskin, KM et al. Convergent antibody signatures in human dengue. Cell Host Microbe 2013; 13: 691–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weitzner, B, Jeliazkov, J, Lyskov, S. Modeling and docking of antibody structures with Rosetta. Nat Protoc 2017; 12: 401–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weitzner, B, Kuroda, D, Marze, M. Blind prediction performance of RosettaAntibody 3.0: grafting, relaxation, kinematic loop modeling, and full CDR optimization. Proteins-structure Function & Bioinformatics 2014; 82: 1611–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jian, Y, Ning, M, Madden, TL. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res 2013; 41: W34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gupta, NT, Heiden, JAV, Uduman, M et al. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics 2015; 31: 3356–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Armstrong, RA, Hilton, AC. Statistical Analysis in Microbiology: Statnotes. Wiley, Hoboken, New Jersey, USA. 2010, 33–7 [Google Scholar]
- 23. Theodorsson-Norheim, E. Kruskal-Wallis test: BASIC computer program to perform nonparametric one-way analysis of variance and multiple comparisons on ranks of several independent samples. Comput Methods Programs Biomed 1986; 23: 57–62. [DOI] [PubMed] [Google Scholar]
- 24. VanDyk, L, Meek, K. Assembly of IgH CDR3: mechanism, regulation, and influence on antibody diversity. Int Rev Immunol 1992; 8: 123–33. [DOI] [PubMed] [Google Scholar]
- 25. Brezinschek, HP, Foster, SJ, Brezinschek, RI et al. Analysis of the human VH gene repertoire. Differential effects of selection and somatic hypermutation on human peripheral CD5(+)/IgM+ and CD5(−)/IgM+ B cells. J Clin Invest 1997; 99: 2488–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Briney, BS, Willis, JR, Crowe, JE Jr. Human peripheral blood antibodies with long HCDR3s are established primarily at original recombination using a limited subset of germline genes. PLoS One 2012; 7: e36750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wei, L, Chahwan, R, Wang, S et al. Overlapping hotspots in CDRs are critical sites for V region diversification. Proc Natl Acad Sci U S A 2015; 112: E728–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen, L, Duan, Y, Benatuil, L et al. Analysis of 5518 unique, productively rearranged human VH3-23*01 gene sequences reveals CDR-H3 length-dependent usage of the IGHD2 gene family. Protein Eng Des Sel 2017; 30: 603–9. [DOI] [PubMed] [Google Scholar]
- 29. Sankar, K, Hoi, KH, Hotzel, I. Dynamics of heavy chain junctional length biases in antibody repertoires. Commun Biol 2020; 3: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Prabakaran, P, Chowdhury, PS. Landscape of non-canonical cysteines in human VH repertoire revealed by immunogenetic analysis. Cell Rep 2020; 31: 107831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu, L, Oficjalska, K, Lambert, M et al. Fundamental characteristics of the immunoglobulin VH repertoire of chickens in comparison with those of humans, mice, and camelids. J Immunol 2012; 188: 322–33. [DOI] [PubMed] [Google Scholar]
- 32. Haakenson, JK, Deiss, TC, Warner, GF et al. A broad role for cysteines in bovine antibody diversity. Immunohorizons 2019; 3: 478–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goldman, ER, Liu, JL, Zabetakis, D et al. Enhancing stability of camelid and shark single domain antibodies: an overview. Front Immunol 2017; 8: 865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tiller, T, Schuster, I, Deppe, D et al. A fully synthetic human fab antibody library based on fixed VH/VL framework pairings with favorable biophysical properties. MAbs 2013; 5: 445–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Teplyakov, A, Obmolova, G, Malia, TJ et al. Structural diversity in a human antibody germline library. MAbs 2016; 8: 1045–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. W., J.D.F. Unraveling V(D)J recombination. Cell 2004; 116: 299–311. [DOI] [PubMed] [Google Scholar]
- 37. Briney, BS, Crowe, JE Jr. Secondary mechanisms of diversification in the human antibody repertoire. Front Immunol 2013; 4: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Birtalan, S, Zhang, Y, Fellouse, FA et al. The intrinsic contributions of tyrosine, serine, glycine and arginine to the affinity and specificity of antibodies. J Mol Biol 2008; 377: 1518–28. [DOI] [PubMed] [Google Scholar]
- 39. Joyce, C, Burton, DR, Briney, BS. Comparisons of the antibody repertoires of a humanized rodent and humans by high throughput sequencing. Sci Rep 2020; 10: 1120–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Anjana, R, Vaishnavi, MK, Sherlin, D et al. Aromatic-aromatic interactions in structures of proteins and protein-DNA complexes: a study based on orientation and distance. Bioinformation 2012; 8: 1220–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee, J, Ju, M, Cho, OH et al. Tyrosine-rich peptides as a platform for assembly and material synthesis. Adv Sci (Weinh) 2019; 6: 1801255–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu, X, Zhou, T, Zhu, J et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 2011; 333: 1593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kong, L, Giang, E, Nieusma, T et al. Hepatitis C virus E2 envelope glycoprotein core structure. Science 2013; 342: 1090–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thomson, CA, Bryson, S, McLean, GR et al. Germline V-genes sculpt the binding site of a family of antibodies neutralizing human cytomegalovirus. EMBO J 2008; 27: 2592–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xiong, X, Corti, D, Liu, J et al. Structures of complexes formed by H5 influenza hemagglutinin with a potent broadly neutralizing human monoclonal antibody. Proc Natl Acad Sci U S A 2015; 112: 9430–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following data were used in this study: the set of VH3–23*01 sequences (European Molecular Biology Laboratory accession nos. AM076988–AM083316) and DENV cohort sequences (the Bi-oProject accession number PRJNA205206).


