Abstract
Functionally competent and self-tolerant T cell repertoire is shaped through positive and negative selection in the cortical and medullary microenvironments of the thymus. The thymoproteasome specifically expressed in the cortical thymic epithelium is essential for the optimal generation of CD8+ T cells. Although how the thymoproteasome governs the generation of CD8+ T cells is not fully understood, accumulating evidence suggests that the thymoproteasome optimizes CD8+ T cell production through the processing of self-peptides associated with MHC class I molecules expressed by cortical thymic epithelial cells. In this review, we describe recent advances in the mechanism of thymoproteasome-dependent generation of CD8+ T cells, focusing on the process of cortical positive selection independent of apoptosis-mediated negative selection.
1. Introduction
T cells play a central role in the immune system by eliciting and regulating antigen-specific immune responses. The ability of T cells to specifically recognize antigens is acquired during their development in the thymus. Within the thymus, lymphoid progenitor cells are induced to express T cell antigen receptors (TCRs) to become T cells in the cortical microenvironment, where cortical thymic epithelial cells (cTECs) provide signals, including DLL4 and IL7, for the specification to generate TCRαβ+ CD4+CD8+ (double positive, DP) thymocytes. cTECs additionally play an essential role in the production of a functionally competent repertoire of TCR recognition specificities, the process referred to as positive selection (Anderson, Jenkinson, Moore, & Owen, 1993; Benoist & Mathis, 1989).
Newly generated DP thymocytes that express individual TCR specificities are selected for survival or death in accordance with their TCR recognition specificities through the interaction with self-peptide-MHC complexes expressed by cTECs (Anderson, Owen, Moore, & Jenkinson, 1994; Laufer, DeKoningt, Markowitz, Lo, & Glimcher, 1996). Only thymocytes that are signaled with low-affinity TCR engagement are thought to receive survival signals, thereby being positively selected, whereas cortical thymocytes that are signaled with high-affinity TCR engagement or that fail to interact with self-peptide-MHC complexes are unable to go through further development, for example, by undergoing apoptosis-mediated negative selection or by being arrested in the thymic cortex (McCaughtry, Baldwin, Wilken, & Hogquist, 2008; Stritesky et al., 2013; Szondy, Garabuczi, Tóth, Kiss, & Köröskényi, 2012). DP thymocytes that are positively selected with self-peptides associated with MHC class I molecules differentiate into CD4−CD8+ (CD8 single positive, CD8SP) T cells, whereas interactions with self-peptides associated with MHC class II molecules promote their differentiation into CD4+CD8− (CD4 single positive, CD4SP) T cells. TCR signals in cortical DP thymocytes induce the expression of chemokine receptor CCR7, so that positively selected cortical thymocytes migrate to the thymic medulla where a subpopulation of medullary thymic epithelial cells (mTECs) express CCR7-ligand chemokine CCL21 and other subpopulations of mTECs express self-antigens to establish self-tolerance in newly arrived thymocytes by inducing the apoptosis-mediated negative selection of self-reactive T cells and by promoting the development of regulatory T cells (Gallegos & Bevan, 2004; Kozai et al., 2017; Kyewski & Derbinski, 2004; Lkhagvasuren, Sakata, Ohigashi, & Takahama, 2013; Takahama, Ohigashi, Baik, & Anderson, 2017; Ueno et al., 2004).
cTECs generate MHC class I-associated and MHC class II-associated self-peptides by degrading endogenous proteins. Proteins in the cytoplasm and the nucleus are degraded by the ubiquitin-proteasome system to generate the peptides, which are transported to the endoplasmic reticulum through the TAP transporter, trimmed by ER aminopeptidases, and loaded onto MHC class I molecules. cTECs express the cTEC-specific form of proteasome, termed the thymoproteasome (Murata et al., 2007; Ohigashi et al., 2013; Ripen, Nitta, Murata, Tanaka, & Takahama, 2011). Accumulating evidence from studies of thymoproteasome-deficient mice has indicated that the thymoproteasome is essential for optimal CD8+ T cell production in the thymus. On the other hand, proteins in endosomal lumens in cTECs are degraded by endosomal-lysosomal endopeptidases, including cathepsin L (Honey, Nakagawa, Peters, & Rudensky, 2002; Nakagawa et al., 1998) and thymus-specific serine protease (TSSP) (Bowlus, Ahn, Chu, & Gruen, 1999; Gommeaux et al., 2009; Viret et al., 2011), and these self-peptides are loaded onto MHC class II molecules in cTECs and are important for the production of CD4+ T cells in the thymus.
Here we would like to discuss the function of the thymoproteasome specifically expressed by cTECs, focusing on recent advances in the role of apoptosis-mediated negative selection in the thymoproteasome-dependent generation of CD8+ T cells.
2. Overview of proteasomes
2.1. Proteasomes, immunoproteasomes, and thymoproteasomes
The proteasome is a large protease complex that is highly conserved in all eukaryotes from yeasts to mammals and responsible for the degradation of ubiquitinated proteins in the cytoplasm and the nucleus (Coux, Tanaka, & Goldberg, 1996). Proteasome-dependent protein degradation is important for not only maintaining the quality of proteins but also mediating various biological processes, including cell cycle, transcription, signal transduction, and immune response (Baumeister, Walz, Zühl, & Seemüller, 1998; Collins & Goldberg, 2017). The proteasome consists of the 20S core particle responsible for catalytic activities and of the regulatory particle that binds to the two ends of the cylindrical 20S core particle. The most common form of the proteasome is the 26S proteasome, which contains two 19S regulatory particles that preprocess protein substrates for degradation by the 20S core particle. The 20S core particle is composed of 28 subunits, which are 2 sets of 7 different α subunits (α1–α7) and 7 different β subunits (β1–β7). Among the 20S core particle subunits, β1, β2, and β5 subunits exhibit proteolytic activities, each with distinct cleavage specificities, namely, caspase-like activity, trypsin-like activity, and chymotrypsin-like activity, respectively (Dick et al., 1998; Kisselev, Akopian, Castillo, & Goldberg, 1999). The proteasome that contains β1, β2, and β5 catalytic subunits is termed the constitutive proteasome and is expressed in a majority of somatic cells in mammalian body. Two other forms of proteasomes, namely, immunoproteasomes and thymoproteasomes, have been documented in vertebrates, which carry the adaptive immune system. The immunoproteasome contains β1i, β2i, and β5i catalytic subunits in place of β1, β2, and β5 subunits, and is expressed in hematopoietic cells and interferon-stimulated cells. In the thymus, the immunoproteasome is widely detected in various cells, including thymocytes, dendritic cells (DCs), and mTECs (Nil, Firat, Sobek, & Eichmann, 2004). In contrast, the thymoproteasome contains β1i, β2i, and β5t catalytic subunits, and is exclusively detectable in cTECs (Murata et al., 2007). The cTEC-specific thymoproteasome is important for optimizing the production of CD8+ T cells in the thymus.
2.2. Substrate specificity of proteasomes affects MHC class I-associated peptides
The substrate specificity of catalytic subunits in proteasomes is mainly determined by the amino acids that compose the S1 substrate-binding pocket within the subunits. The S1 pocket of β5 and β5i is enriched with hydrophobic amino acid residues, thereby being responsible for their chymotrypsin-like activity to generate peptides with hydrophobic C-terminal residues (Huber et al., 2012). The proteasome-degraded peptides bind to MHC class I molecules via anchor residues, including the C-terminus anchor residues, which are enriched with hydrophobic amino acids (Hörig, Young, Papadopoulos, DiLorenzo, & Nathenson, 1999). The β5i-containing immunoproteasome possesses efficient endopeptidase activity to generate C-terminal hydrophobic amino acid residues (Gaczynska, Rock, & Goldberg, 1993), suggesting that the substrate specificity of the immunoproteasome is optimized for the generation of peptides associated with MHC class I molecules. On the contrary, the S1 pocket of β5t is enriched with hydrophilic residues that specifically reduce the chymotrypsin-like activity of the thymoproteasome without affecting other proteolytic activities, including trypsin-like and caspase-like activities (Florea et al., 2010; Murata et al., 2007; Sutoh et al., 2012). A recent study further showed that human thymoproteasomes and immunoproteasomes differ in cleavage preference and peptide products quantitatively and qualitatively (Kuckelkorn et al., 2019). Thus, the thymoproteasome generates peptides that are distinct from those generated by the immunoproteasome.
Indeed, a mass spectrometry analysis of MHC class I-associated peptides generated in fibroblasts ectopically expressing either thymoproteasomes or immunoproteasomes showed a substantial difference in the repertoire of thymoproteasome-dependent and immunoproteasome-dependent peptides (Sasaki et al., 2015). The C-terminal residues of thymoproteasome-dependent peptides remained mostly hydrophobic similar to the immunoproteasome-dependent peptides. However, within 9-mer peptides associated with H-2Db MHC class I molecules, the enrichment of basic and hydrophobic residues at the third position from C-terminus (P3) and of proline residue at the fourth position from C-terminus (P4) was detected in thymoproteasome-expressing cells, in contrast to the enrichment of acidic residues at P3 and basic residues at P4 in immunoproteasome-dependent peptides (Sasaki et al., 2015). Within 8-mer peptides associated with H-2Kb MHC class I molecules, acidic residues and proline were enriched at P2 and P3, respectively, in thymoproteasome-dependent peptides, whereas these amino acid residues were less enriched at P2 and P3 in immunoproteasome-dependent peptides (Sasaki et al., 2015). Therefore, the thymoproteasome is involved in the enrichment of unique P2–P4 amino acid residues in MHC class I-associated peptides. These amino acid residues are found on the TCR-interacting surface of peptide-MHC class I complexes, so that the difference in these amino acid residues between the thymoproteasome- and immunoproteasome-dependent peptides probably causes the difference in their properties for the interactions with TCRs.
3. Thymoproteasome for CD8 T cell generation
3.1. Discovery of β5t and thymoproteasome
The β5t subunit was initially found in a previously unrecognized gene that has high homology to the genes that encode the β5 and β5i subunits of the 20S proteasome, in a search of genome database for proteasome-related genes (Murata et al., 2007). The expression of β5t is specifically detected in cTECs but not in other cells, including other thymic cells such as mTECs, non-TEC thymic stromal cells, and thymocytes (Murata et al., 2007: Ripen et al., 2011; Ohigashi et al., 2013). The cTEC-specific expression of β5t is directly regulated by Foxn1, a transcription factor that plays a crucial role in the development of the thymic epithelium (Uddin et al., 2017; Zuklys et al., 2016). As Foxn1 is expressed in both cTECs and mTECs, additional transcriptional and/or epigenetic mechanisms are suggested to promote the cTEC-specific expression of β5t (Uddin et al., 2017).
The role of the thymoproteasome in the generation of CD8+ T cells was established by the analyses of β5t-deficient mice. The thymus of β5t-deficient mice exhibited normal organization of the cortex and the medulla, with unaltered numbers of cTECs and mTECs. Most strikingly, however, was that the number of CD8+ T cells in the thymus and the peripheral lymphoid organs was specifically reduced in β5t-deficient mice to approximately 20%–30% of the number in control mice, without the alteration of the numbers of CD4+ T cells, γδ T cells, NKT cells, intraepithelial T cells, B cells, DCs, and any other cells in β5t-deficient mice (Murata et al., 2007; Nitta et al., 2010). The β5t deficiency altered the usage of TCR-Vβ and TCR-Vα in CD8+ T cells but not CD4+ T cells (Nitta et al., 2010; Ohigashi et al., 2021; Xing, Jameson, & Hogquist, 2013).Analysis of TCR-transgenic mice showed that CD8+ T cells expressing some TCRs, including HY-TCR and P14-TCR, are more susceptible to the loss of β5t than CD8+ T cells expressing other TCRs, including OT-I-TCR and 2C-TCR (Nitta et al., 2010). Additionally, the thymoproteasome affects the functional competency of CD8+ T cells. The TCR responsiveness in OT-I-TCR-expressing CD8+ T cells generated in the thymus of β5t-deficient mice is impaired, even though β5t deficiency does not significantly reduce the generation of OT-I-TCR T cells (Takada et al., 2015). OT-I-TCR+ CD8+ T cells generated in β5t-deficient thymus preferentially give rise to short-lived effector T cells upon infection with OVA-expressing Listeria monocytogenes, unlike CD8+ T cells generated in control thymus, which predominantly differentiate into the long-term memory T cells upon the infection (Takada et al., 2015). Influenza virus-infected β5t-deficient mice exhibit higher mortality than control mice (Nitta et al., 2010). Thus, the thymoproteasome is important for the generation of functionally competent CD8+ T cell repertoire.
3.2. Impact of thymoproteasome on transcriptome and proteome in cTECs
A recent study of RNA sequencing analysis reported that the expression of cTEC-associated functional genes, such as Ctsl, Prss16, and Cxcl12, was downregulated and the expression of mTEC-associated genes, such as Cldn3, Ctss, and Tnfrsf11a, was upregulated in β5t-deficient cTECs relative to control cTECs, demonstrating that β5t-deficient cTECs acquire mTEC features (Apavaloaei et al., 2019). The authors also showed that the expression of genes encoding cell adhesion molecules and extracellular matrix molecules was elevated in β5t-deficient cTECs and that oxidative stress genes were upregulated in thymocytes and the number of mature CD4SP thymocytes was increased in β5t-deficient mice (Apavaloaei et al., 2019). On the contrary, we reported that none of those alterations in cTECs or thymocytes are reproduced in our β5t-deficient mice of B6 background, and concluded that the loss of β5t does not pervasively affect gene expression in cTECs (Ohigashi et al., 2019). Instead, combined analysis of multiple datasets from the two studies along with quantitative RT-PCR analysis revealed that no genes were consistently elevated in cTECs by the loss of β5t, whereas only 1 gene, Dync1i1, in addition to β5t-encoding Psmb11, was reduced by the loss of β5t. Dync1i1 encodes cytoplasmic dynein 1 intermediate chain 1, although the proteomic analysis did not show that the abundance of this protein was not affected in cTECs in the presence or absence of β5t (Ohigashi et al., 2019). The contradictory results obtained by these two studies may be due to the difference in the genetic background of mice and/or the purity of isolated cTECs for transcriptomic analysis. It should be emphasized that it is important to verify the alterations in gene expression by other methods such as quantitative RT-PCR analysis, especially when the alterations detected in RNA sequencing analysis are modest and not all-or-none.
In addition to transcriptomic analysis, comprehensive proteomic analysis was carried out by using TECs isolated from keratin 5 promoter-driven cyclin D1 (K5D1) transgenic mice (Ohigashi et al., 2019), which showed severe thymic hyperplasia due to enhanced proliferation of immature TECs (Bolner, 2015; Klug et al., 2000; Robles et al., 1996). Similar to transcriptomic profiles, proteomic profiles from tandem mass tag (TMT)-based mass spectrometry analysis showed high similarity between β5t-deficient cTECs and control cTECs in K5D1 background (Ohigashi et al., 2019). Among 5753 proteins quantified by TMT-based mass spectrometry analysis, 264 were significantly more abundant in β5t-deficient cTECs than in control cTECs, and the integration with transcriptomic profiles revealed that 91 of those 264 molecules showed significantly higher mRNA expression in β5t-deficient cTECs than in control cTECs (Fig. 1A). Similarly, 306 proteins were significantly less abundant in β5t-deficient cTECs than in control cTECs, and 121 of those 306 molecules showed significantly lower mRNA expression in β5t-deficient cTECs than in control cTECs (Fig. 1A). It should be noted that molecules significantly different in both mRNA and protein abundance between β5t-deficient and control cTECs were much smaller in number and fold change than those different between cTECs and mTECs (Fig. 1A and B). Nonetheless, other than β5t, which was remarkably reduced in β5t-deficient cTECs, many proteasome components were modestly reduced and β5i and β5 were elevated in protein abundance in β5t-deficient cTECs (Ohigashi et al., 2019). The increase in β5i and β5 appeared to reflect the compensatory expression of these subunits by the loss of β5t, to maintain the proteasomes in β5t-deficient cTECs (Ohigashi et al., 2019). These results indicate that β5t specifically impacts proteasomal composition in cTECs.
Fig. 1.
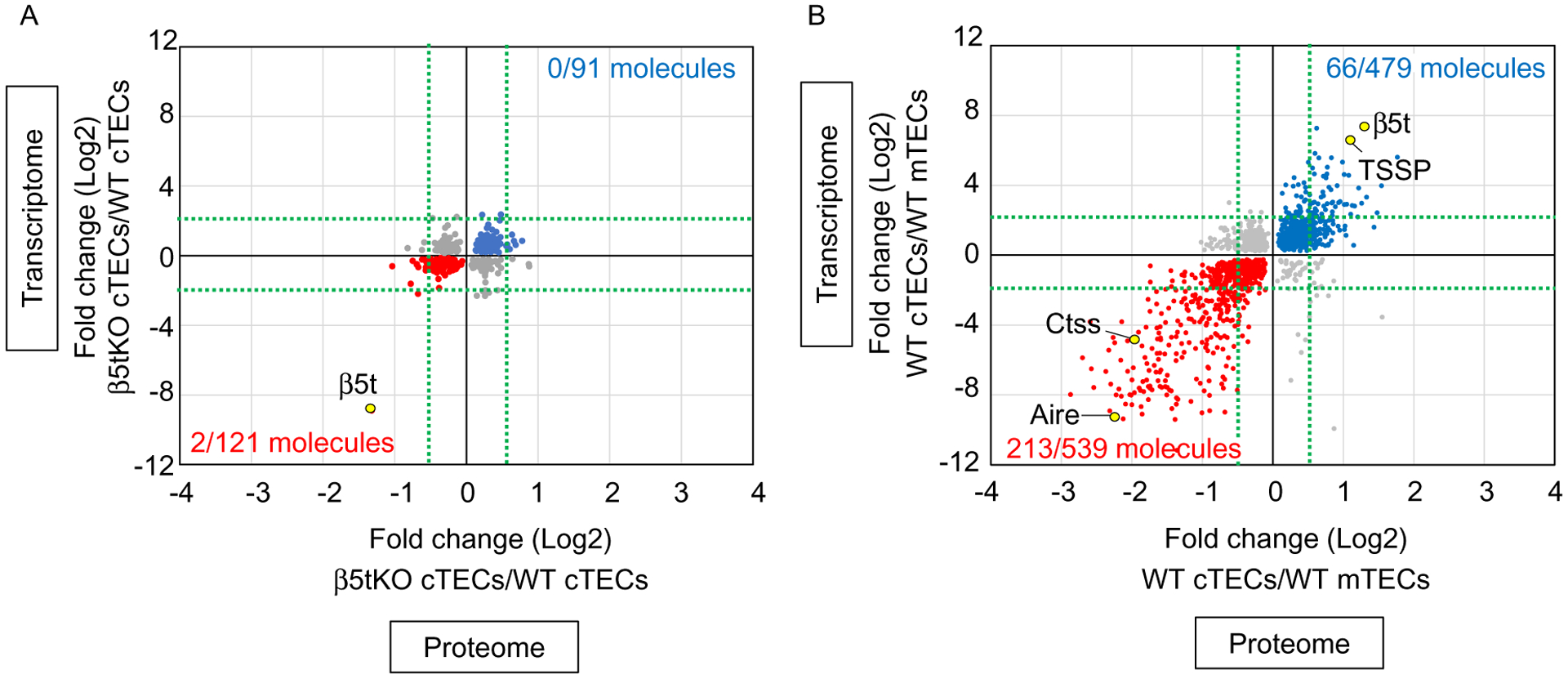
Trans-omics analysis of TECs. Correlation plot analysis of trans-omic (proteomic and transcriptomic) profiles for (A) β5t-deficient cTECs and wildtype (WT) cTECs and (B) WT cTECs and WT mTECs. (A) Among 91 molecules (blue symbols) that were more highly detected (P<0.05) in β5t-deficient cTECs than in WT cTECs in both proteomic and transcriptomic data, none of the molecules showed the fold changes in proteomic data >0.5 and transcriptomic data >2 (indicated by green dot lines). Among 121 molecules (red symbols) that were more highly detected (P<0.05) in WT cTECs than in β5t-deficient cTECs in both proteomic and transcriptomic data, 2 molecules showed the fold changes in proteomic data <−0.5 and transcriptomic data <−2. Among these 2 molecules, only one molecule β5t showed a reproducible reduction in β5t-deficient cTECs in individual quantitative analysis. (B) In the parallel analysis using the same threshold setting, among 479 molecules (blue symbols) that were more highly detected in cTECs than in mTECs, 66 molecules showed the fold changes in proteomic data >0.5 and transcriptomic data >2 (indicated by green dot lines). Among 539 molecules (red symbols) that were more highly detected in mTECs than in cTECs, 213 molecules showed the fold changes in proteomic data <−0.5 and transcriptomic data <−2.
3.3. Impact of β5t deficiency on TCR repertoire in CD8 T cells
To better understand the mechanism of the thymoproteasome-mediated optimization of CD8+ T cell production, we recently examined the impact of the thymoproteasome on the TCR repertoire in CD8+ T cells. The highly diverse TCR repertoire is formed by the somatic recombination of variable (V) and joining (J) segments of TCRα gene and V, diversity (D), and J segments of TCRβ gene (Davis & Bjorkman, 1988). The TCR interacts with self-peptide on MHC molecule through the CDR3 region that is encoded by the rearrangement of V and J segments for TCRα chain and V, D, and J segments for TCRβ chain (Garboczi et al., 1996; Garcia et al., 1996). It was previously estimated that the size of the TCRαβ repertoire is approximately 2×106, with approximately 10 cells each, in the spleen of one young adult wild-type mouse (Casrouge et al., 2000). Accordingly, our deep sequencing analysis showed that approximately 2.5×105 different nucleotide sequences of TCRα and TCRβ genes were equivalently detected in 1×106 CD8+ T cells isolated from β5t-deficient mice and control mice. Importantly, this size of TCR diversity was comparable between CD8+ T cells from β5t-deficient mice and control mice, indicating that the diversity of the TCR repertoire is not reduced in the same number of CD8+ T cells generated in the absence of the thymoproteasome (Ohigashi et al., 2021). More importantly, nucleotide sequences of TCRα and TCRβ genes detected in CD8+ T cells were largely non-overlapped between β5t-deficient and control mice, and the unique sequences of full-length TCRα and TCRβ genes, including CDR3 sequences, were detectable in either β5t-deficient mice or control mice. For example, a TCRα sequence that consists of TRAV12–3 (V region), CDR3 with amino acid sequence CALMGYKLTF, and TRAJ9 (J region), hereinafter referred to as TCRα#1, was preferentially detected in CD8+ T cells from control mice than β5t-deficient mice, whereas another TCRα sequence referred to as TCRα#2, which consists of TRAV12N-3, CDR3 with amino acid sequence CALSDRYNQGKLIF, and TRAJ23, was predominantly detected in CD8+ T cells from β5t-deficient mice (Ohigashi et al., 2021). Consistent with the preferential detection of TCRα#1 in control mice and TCRα#2 in β5t-deficient mice, the thymus reconstitution with bone marrow cells isolated from TCRα#1 or TCRα#2 transgenic and endogenous TCRα-deficient mice showed the preferential generation of TCRα#1-expressing and TCRα#2-expressing CD8+ T cells in control and β5t-deficient recipients, respectively (Ohigashi et al., 2021). Interestingly, the preferential generation of TCRα#1- and TCRα#2-expressing T cells in control and β5t-deficient recipient mice, respectively, was detectable as early as the DP CD69+ CCR7− thymocyte stage, exhibiting the molecular signature of TCR-signaled DP thymocytes still localized in the thymic cortex (Ohigashi et al., 2021). Thus, the thymoproteasome impacts the TCR repertoire of positively selected cortical thymocytes prior to the migration to the thymic medulla.
3.4. Thymic selection in the absence of negative selection
3.4.1. Thymic selection in the absence of medulla
Because it was assumed that the thymoproteasome causes cTECs to express a unique set of MHC class I-associated self-peptides, it was possible for the thymoproteasome to optimize CD8+ T cell generation by avoiding the positive selection of thymocytes that will be subsequently negatively selected by identical MHC class I-associated self-peptides. In this regard, it was interesting to examine whether negative selection might be involved in the thymoproteasome-dependent optimization of CD8+ T cell generation. The thymic medulla is a microenvironment dedicated to the establishment of self-tolerance in T cells. The development of the thymic medulla is basically dependent on the development of mTECs. RelB is a transcription factor activated by the non-canonical NF-κB signaling, and the deficiency in relB severely impairs mTEC development and thymic medulla formation (Burkly et al., 1995; Weih et al., 1995). In relB-deficient mice, the proportions of DN, DP, CD4SP, and CD8SP thymocyte subpopulations are not altered. However, the cellularity of thymocytes is reduced due to severe multiorgan inflammation, which is obvious as early as 2 weeks of age (Cowan et al., 2013; Ohigashi et al., 2021; Weih et al., 1995). It was found that β5t deficiency selectively reduced the number of CD8SP thymocytes in relB-deficient mice even at a young age before the reduction of thymocyte numbers due to the autoimmune inflammation (Ohigashi et al., 2021). Importantly, CD8SP thymocytes were comparably reduced by β5t deficiency in the absence and presence of relB (Ohigashi et al., 2021). Thus, mTEC-dependent negative selection has little contribution to the thymoproteasome-dependent thymic selection of CD8+ T cells.
Accordingly, it was shown that the number of CD8SP thymocytes decreased in β5t-deficient mice in a similar extent even in the absence of CCR7 (Nitta et al., 2010). The CCR7-mediated chemotaxis of positively selected thymocytes to the CCR7-ligand CCL21, which is produced by a subpopulation of mTECs, is essential for positively selected thymocytes to migrate to and accumulate in the thymic medulla (Kozai et al., 2017; Ueno et al., 2004). Thus, the thymoproteasome-dependent optimization of CD8+ T cells is independent of the migration of positively selected thymocytes to the thymic medulla and their accumulation therein.
3.4.2. Thymic selection in the microenvironment where only cTECs present antigens
In the thymic medulla, antigen presentation by hematopoietic cells, such as DCs and B cells, is important for the establishment of self-tolerance in T cells, in addition to the antigen presentation by mTECs (Bonasio et al., 2006; Gallegos & Bevan, 2004; Yamano et al., 2015). Although the formation of medulla is severely impaired in relB-deficient mice, their thymuses contain DCs and B cells (O’Sullivan et al., 2018). Furthermore, it was reported that thymocytes themselves could also induce the negative selection of neighboring thymocytes (Melichar, Ross, Taylor, & Robey, 2015; Schönrich et al., 1993). β2-Microglobulin (β2m) is a component of MHC class I molecules, and β2m deficiency impairs cell surface expression of peptide-MHC class I complexes (Koller, Marrack, Kappler, & Smithies, 1990). Therefore, the reconstitution of hematopoietic cell-depleted relB-deficient thymus with hematopoietic cells derived from β2m-deficient mice enables the generation of a thymus microenvironment where mTECs, DC, B cells, and thymocytes are unable to present MHC class I-associated peptides and cTECs are the only antigen-presenting cells that present self-peptide-MHC class I complexes. Other non-hematopoietic cells, including fibroblasts in the thymus, express limited numbers of MHC class I and class II molecules, and are unlikely to serve as antigen-presenting cells for MHC-associated peptides. It was found that the reconstitution of relB-deficient fetal thymus with β2m-deficient hematopoietic cells resulted in the development of CD8SP TCRβhigh thymocytes, which was still impaired by the β5t deficiency in relB-deficient TECs (Ohigashi et al., 2021). Thus, the thymoproteasome optimizes the development of CD8+ T cells even in the absence of MHC class I-associated intrathymic antigen-presenting cells other than cTECs.
3.4.3. Thymic selection independent of apoptosis-mediated negative selection
It was reported that a substantial number of DP thymocytes underwent negative selection in the thymic cortex, where cortical DCs in addition to cTECs were found to induce the apoptosis of DP thymocytes (Daley, Hu, & Goodnow, 2013; McCaughtry et al., 2008; Stritesky et al., 2013). On the other hand, the transgenic expression of anti-apoptotic molecule Bcl2 in developing thymocytes prevents cortical thymocytes from apoptosis-mediated cell death (Pobezinsky et al., 2012; Punt, Havran, Abe, Sarin, & Singer, 1997; Sentman, Shutter, Hockenbery, Kanagawa, & Korsmeyer, 1991). In I-Aβ-deficient mice, in which cortical DP thymocytes can be engaged to develop into CD8+ lineage T cells by MHC class I-peptide complexes but not into CD4+ lineage T cells by MHC class II-peptide complexes, the generation of DP CD69+CCR7− cortical thymocytes and downstream CD8SP TCRβhigh medullary thymocytes is reduced in β5t-deficient mice compared with control mice (Ohigashi et al., 2021). Importantly, Bcl2 rescues these thymocyte subpopulations in a similar manner in both β5t-deficient mice and control mice, and thus the population sizes of DP CD69+CCR7− cortical thymocytes and CD8SP TCRβhigh medullary thymocytes remain smaller in β5t-deficient mice than control mice regardless of the transgenic expression of Bcl2 (Ohigashi et al., 2021). Thus, the apoptosis-mediated negative selection does not contribute to the β5t-dependent optimization of CD8+ T cells, and the thymoproteasome optimizes the generation of CD8+ T cells independent of apoptosis-mediated negative selection, which can occur in either the thymic cortex or the thymic medulla.
3.5. Thymoproteasome and peptide switch hypothesis
3.5.1. Peptide switch hypothesis
It was previously predicted that the thymus must provide the machinery to present a unique set of MHC-associated peptides for positive selection, to enable successful production of T cells by avoiding the negative selection of positively selected thymocytes through the overlap between positive selection-inducing self-peptide-MHC complexes and negative selection-inducing self-peptide-MHC complexes (Kourilsky & Claverie, 1989; Marrack & Kappler, 1987). More recently, this prediction was described as the “peptide switch hypothesis,” in which the switch of MHC-associated peptides for positive selection in the thymic cortex and for negative selection in the thymus, including the cortex and the medulla, is important for the escape of positively selected thymocytes from negative selection, thereby resulting in the thymic production of T cells (Fig. 2). This model was examined by engineering mice deficient in β1i, β2i, β5i, and β5t subunits (Kincaid, Murata, Tanaka, & Rock, 2016). These mice, called 4KO mice, lack both the thymoproteasome and immunoproteasome anywhere in their bodies, including the thymus. Instead, these mice only express the constitutive proteasome in their bodies. The 4KO mice exhibited more than 90% reduction of CD8+ T cells in the thymus and the spleen relative to control mice, and this reduction was more severe than that detected in β5t-deficient mice (Kincaid et al., 2016; Murata et al., 2007). On the contrary, despite the remarkable reduction of CD8+ T cells, the cellularity of CD4+CD8low CD103+ CD69+CCR7intermediate TCRβintermediate thymocytes, presumably post-positive selection CD8+ lineage cells still localized in the cortex, was comparable between 4KO mice and control mice (Kincaid et al., 2016). However, the number of DP CD69+CCR7high TCRβhigh thymocytes, which were subject to the subsequent negative selection, was strikingly reduced in 4KO mice (Kincaid et al., 2016). Furthermore, the thymus reconstitution with bone marrow cells from mice deficient in Bim, which is a proapoptotic protein important for the negative selection of T cells, showed that the increase of CD8SP thymocyte cellularity by Bim deficiency was more than 4-fold greater in 4KO hosts than wild-type hosts (Kincaid et al., 2016). Based on these findings, the authors concluded that CD8SP thymocytes are lost after positive selection and during negative selection in the 4KO thymuses, where the switch of positive selection-inducing peptides in the cortex and negative selection-inducing peptides in the medulla is absent.
Fig. 2.
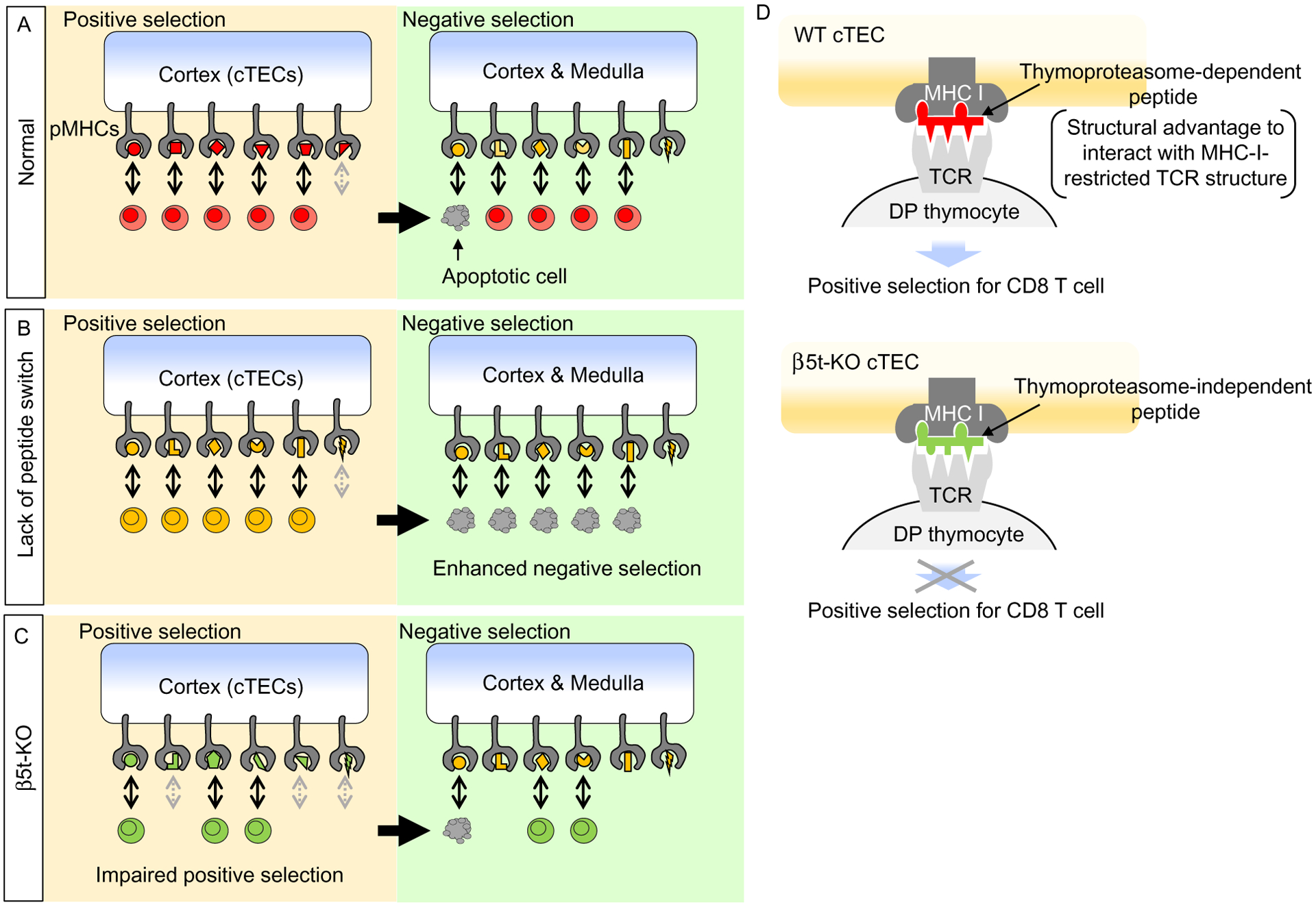
Thymoproteasome and thymic selection. (A) Thymic selection in normal thymus. cTEC-specific thymoproteasome produces a unique set of MHC-I-associated peptides (red symbols) that induce positive selection in the thymic cortex. Positively selected thymocytes (red cells) are screened for negative selection in the cortex and medulla, and a fraction of thymocytes are negatively selected to establish self-tolerance. (B) According to the peptide switch hypothesis, positive selection-inducing MHC-associated peptides must be different from negative selection-inducing MHC-associated peptides. In the conditions that lack the machinery to produce a unique set of MHC-I-associated peptides, a set of MHC-I-associated peptides identical to negative selection-inducing peptides (yellow symbols) are displayed by cTECs and positively select a repertoire of thymocytes in the thymic cortex; however, these thymocytes are subsequently negatively selected in the thymus by the interaction with identical set of MHC-I-associated peptides. Thus, the lack of the peptide switch may not reduce the number of positively selected thymocytes in the cortex (yellow cells) but may enhance the number of negatively selected thymocytes. (C) However, it was shown that in the thymus of β5t-knockout (KO) mice, the number of positively selected thymocytes in the cortex are reduced (green cells), whereas the number of negatively selected thymocytes are not elevated. Thus, the thymoproteasome optimizes CD8+ T cell development by supporting positive selection in the cortex independent of negative selection. The impaired generation of CD8+ T cells in β5t-deficient thymus is primarily due to impaired positive selection in the cortex. (D) How does the thymoproteasome work? The thymoproteasome expressed in WT cTECs produces MHC-I-associated peptides (red) that may possess structural advantage to interact with MHC-I-restricted TCR structures (for example, TCRα#1-containing TCRs as described in Section 3.3) that are preferentially used in efficiently induce positive selection of CD8+ T cells. However, MHC-I-associated peptides (green) displayed by β5t-knockout (KO) cTECs, which are produced independent of the thymoproteasomes, may lack such structural advantage and fail to induce to positive selection of CD8+ T cells.
A more recent study examined T cell development in the thymus that lacked β5i but instead expressed β5t transgene ubiquitously (β5i-KO β5t-Tg). In these mice, antigen-presenting cells localized in the thymic medulla, including mTECs and DCs, expressed the thymoproteasome instead of the immunoproteasome. Similar to β5t-deficient mice, β5i-KO β5t-Tg mice exhibited the reduction in the generation of CD8SP thymocytes (Tomaru et al., 2019). However, the number of DP CD103+ CD69+ TCRβlow thymocytes, which are presumably CD8+ lineage cells after receiving positive selection signals, was unaffected in β5i-KO β5t-Tg mice, suggesting that positive selection is not impaired in these mice. In contrast, DP CD103+ CD69+ TCRβhigh thymocytes, which expressed elevated levels of Bim and Nur77 and were supposed to be generated subsequently, was reduced in number in β5i-KO β5t-Tg mice (Tomaru et al., 2019). Based on these results, it was concluded that cortical DP thymocytes undergo positive selection without obvious abnormality, but negative selection of CD8+ lineage cells is enhanced in β5i-KO β5t-Tg mice, suggesting that the ectopic expression of the thymoproteasome in the thymic medulla causes the loss of the peptide switch between the thymic cortex and medulla, which is important for positively selected CD8+ lineage cells to escape from medullary negative selection.
3.5.2. Thymoproteasome-dependent T cell generation independent of peptide switch
Unimpaired positive selection detected at the DP CD103+ CD69+ thymocyte stage and enhanced negative selection detected at the subsequent CD8SP TCRβhigh thymocyte stage in 4KO mice and β5i-KO β5t-Tg mice were the main basis to support the peptide switch hypothesis for the thymic selection of T cells (Kincaid et al., 2016; Tomaru et al., 2019). On the contrary, our analysis of β5t-deficient MHC class II-deficient mice demonstrated that the loss of β5t caused the impaired positive selection detected as early as the DP CD69+CCR7− thymocyte stage and that the contribution of negative selection detected by Bcl2 transgene was comparable throughout thymocyte development in the presence or absence of β5t, indicating that the thymoproteasome optimizes the development of CD8+ T cells during the cortical positive selection independent of negative selection (Ohigashi et al., 2021; Sections 3.3 and 3.4). That the thymoproteasome optimizes CD8+ T cell development independent of the switch in proteasomes in the thymus was previously described in mice in that β5i-encoding cDNA was knocked into the β5t gene locus and crossed with β5i-deficient mice (Xing et al., 2013). In these β5tβ5i/β5i β5i-KO mice, cTECs expressed β5i instead of β5t, whereas other antigen-presenting cells lacked β5i. Thus, self-peptides displayed in the cortex and the medulla were expected to switch from β5i-dependent peptides to β5-dependent peptides in these mice. However, it was found that the number of CD8SP thymocytes in β5tβ5i/β5i β5i-KO mice was reduced similarly to that in β5t-deficient mice, indicating that CD8SP T cell generation in the absence of β5t is still impaired even in the presence of the switch in proteasomes from β5i to β5 and therefore the probable presence of the switch in self-peptides (Xing et al., 2013). How can we reconcile these results in terms of the contribution of negative selection and the peptide switch mechanism in thymocyte development and selection, including the thymoproteasome-dependent T cell optimization? We consider several issues, as follows.
First of all, the experiments using 4KO mice and β5i-KO β5t-Tg mice relied on the loss of β5i and immunoproteasomes (Kincaid et al., 2016; Tomaru et al., 2019), whereas β5i and immunoproteasomes were not manipulated in the experiments using β5t-deficient MHC class II-deficient mice (Ohigashi et al., 2021). The enhanced negative selection detected in 4KO mice and β5i-KO β5t-Tg mice may be caused by the lack of immunoproteasomes, which are widely distributed in hematopoietic and non-hematopoietic cells in the thymus, including mTECs, DCs, and thymocytes. Immunoproteasome-dependent defects, including the reduced expression of surface MHC-I molecules in the thymus of β5i-deficient mice (Fehling et al., 1994; Kincaid et al., 2012), which are independent of the thymoproteasome, may be responsible for the aberrant thymocyte development in 4KO mice and β5i-KO β5t-Tg mice.
Secondly, the experiments using 4KO mice and β5i-KO β5t-Tg mice examined MHC class I-dependent positive selection in cortical thymocytes by the detection of thymocytes that expressed CD103, integrin αE, on the cell surface (Kincaid et al., 2016; Tomaru et al., 2019), whereas the experiments using β5t-deficient MHC class II-deficient mice examined MHC class I-dependent positive selection in cortical thymocytes by using the genetic deficiency of MHC class II (Ohigashi et al., 2021). It was shown that CD103 is expressed by approximately 80% of mature CD8SP TCRβhigh thymocytes and approximately 3%–4% of mature CD4SP TCRβhigh thymocytes (Grueter et al., 2005), indicating that CD103 expression does not absolutely reflect, although it is associated with, the CD8 lineage of developing thymocytes. Thus, the unimpaired positive selection detected in DP CD103+ thymocytes may be better examined in MHC class I-dependent CD8-lineage DP thymocytes by implementing a technique independent of CD103 expression, for example, by the use of MHC class II-deficient mice.
Thirdly, the impact of negative selection in 4KO mice was examined by crossing with mice deficient in pro-apoptotic protein Bim (Kincaid et al., 2016), whereas the lack of contribution of negative selection in β5t-deficient MHC class II-deficient mice was analyzed by the use of anti-apoptotic Bcl2-transgenic mice (Ohigashi et al., 2021). Transgenic Bcl2 expression was shown to inhibit thymocyte negative selection mediated by pro-apoptotic BH3-only family proteins, which are not limited to Bim but include Puma (Pobezinsky et al., 2012). Thus, the difference between Bim deficiency and Bcl2 transgene may contribute in part to the different interpretations of the impact of negative selection between the results from 4KO mice and those from β5t-deficient MHC class II-deficient mice. Nonetheless, it was shown that Bim deficiency equivalently increased post-positively selected DP CD69+ TCRβ+ thymocytes in β5t-deficient MHC class II-deficient thymus and β5t-sufficient MHC class II-deficient thymus (Xing et al., 2013), suggesting that the different impact of negative selection between 4KO mice and β5t-deficient MHC class II-deficient mice may not only be due to the difference between Bim deficiency and Bcl2 transgene.
Finally, the experiments using β5i-KO β5t-Tg mice relied on β5t-Tg mice, which exhibited impaired degradation of ubiquitinated proteins, pre-mature aging phenotypes, shortened longevity, and metabolic abnormalities (Tomaru et al., 2012). Thus, the aberrant thymocyte development in β5i-KO β5t-Tg mice may result from the combination of multiple abnormalities, including systemic aberrancy in the metabolism, in those transgenic mice.
Thus, the negative selection-dependent peptide switch mechanism is detected in immunoproteasome-deficient mice that either lack or overexpress the thymoproteasome. The thymoproteasome-dependent optimization of CD8+ T cell production occurs during the cortical positive selection and is independent of apoptosis-mediated negative selection, thereby being independent of the “peptide switch.”
3.6. How does thymoproteasome work?
As described above, the thymoproteasome optimizes CD8+ T cell development by supporting positive selection in the cortex independent of negative selection or the peptide switch mechanism. Then, what is the mechanism for the thymoproteasome-dependent optimization of CD8+ T cell production during positive selection? A previous study suggested that the thymoproteasome produces MHC class I-associated peptides that contain TCR-facing amino acid sequences that are advantageous to inducing the positive selection of CD8+ T cells (Sasaki et al., 2015). Those thymoproteasome-dependent peptide sequences may possess structural advantages to interact with TCR structures that are positively selected into CD8+ T cells (Fig. 2). Indeed, thymocytes expressing TCRαβ complexes that contain the TCRα#1 V-J sequence (shown in Section 3.3) are preferentially positively selected by thymoproteasome-expressing cTECs than thymoproteasome-deficient cTECs (Ohigashi et al., 2021). Interestingly, those TCRα#1-expressing thymocytes prefer the development into CD8+ T cells rather than CD4+ T cells (Ohigashi et al., 2021), suggesting that TCR antigen-recognition structures in CD8+ T cells are restrained to those selected by the thymoproteasome-dependent positive selection.
It was shown that the TCR repertoire in pre-selected DP thymocytes is not restricted to MHC class I or MHC class II and that MHC-restricted TCR recognition specificity is imposed by CD4 and CD8 coreceptors during thymic selection (Van Laethem et al., 2007, 2013). TCRα#1-expressing thymocytes are selected preferentially by the thymoproteasome and preferentially develop into CD8+ T cells. Thus, a set of TCR specificities, including TCRα#1-containing TCRs, generated among the pre-selected TCR repertoire may be predestined to recognize the thymoproteasome-dependent MHC class I-associated self-peptides expressed by cTECs and to optimally generate CD8+ T cells. The TCR repertoire in CD8+ T cells may not be random but be restrained to the recognition specificities for thymoproteasome-dependent peptides in the thymic cortex.
4. Concluding remarks
In this review, we described recent advances in the mechanism of thymoproteasome-dependent generation of CD8+ T cells, focusing on the process of cortical positive selection independent of apoptosis-mediated negative selection. It is interesting to note that the thymoproteasome dependency of CD8+ T cell generation is independent of negative selection or the peptide switch mechanism. Obviously, the mechanism of the thymoproteasome-dependent CD8+ T cell development is still unclear. It should be important in the future to identify self-peptide-MHC-I complexes displayed by wild-type and thymoproteasome-deficient mouse cTECs. Recent analysis of thymic epithelial cells in genetically engineered mice that carry an enlarged thymus (Ohigashi et al., 2019) suggested the feasibility of bio-chemically identifying thymoproteasome-dependent MHC-I-associated peptide sequences freshly isolated from mouse cTECs. These analyses will possibly advance our understanding of the fundamental basis for the thymus-dependent positive selection of T cells. We also think that better understanding of thymoproteasome-dependent CD8+ T cell optimization will be useful for future improvement of cytotoxic T cell responses in the clinical setting, including the immunotherapy for cancer.
Acknowledgments
This work was supported by grants from MEXT-JSPS (I.O.) and the Intramural Research Program of the US National Institutes of Health, the National Cancer Institute, and the Center for Cancer Research (Y.T.).
References
- Anderson G, Jenkinson EJ, Moore NC, & Owen JJ (1993). MHC class II-positive epithelium and mesenchyme cells are both required for T-cell development in the thymus. Nature, 362, 70–73. [DOI] [PubMed] [Google Scholar]
- Anderson G, Owen JJ, Moore NC, & Jenkinson EJ (1994). Thymic epithelial cells provide unique signals for positive selection of CD4+CD8+ thymocytes in vitro. Journal of Experimental Medicine, 179, 2027–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apavaloaei A, Brochu S, Dong M, Rouette A, Hardy MP, Villafano G, et al. (2019). PSMB11 orchestrates the development of CD4 and CD8 thymocytes via regulation of gene expression in cortical thymic epithelial cells. The Journal of Immunology, 202, 966–978. [DOI] [PubMed] [Google Scholar]
- Baumeister W, Walz J, Zühl F, & Seemüller E (1998). The proteasome: Paradigm of a self-compartmentalizing protease. Cell, 92, 367–380. [DOI] [PubMed] [Google Scholar]
- Benoist C, & Mathis D (1989). Positive selection of the T cell repertoire: Where and when does it occur? Cell, 58, 1027–1033. [DOI] [PubMed] [Google Scholar]
- Bolner ML (2015). Preventing thymus involution in K5.Cyclin D1 transgenic mice sustains the naive T cell compartment with age. UT GSBS Dissertations and Theses; (p. 636). [Google Scholar]
- Bonasio R, Scimone ML, Schaerli P, Grabie N, Lichtman AH, & von Andrian UH (2006). Clonal deletion of thymocytes by circulating dendritic cells homing to the thymus. Nature Immunology, 7, 1092–1100. [DOI] [PubMed] [Google Scholar]
- Bowlus CL, Ahn J, Chu T, & Gruen JR (1999). Cloning of novel MHC-encoded serine peptidase highly expressed by cortical epithelial cells of the thymus. Cellular Immunology, 196, 80–86. [DOI] [PubMed] [Google Scholar]
- Burkly L, Hession C, Ogata L, Reilly C, Marconi LA, Olson D, et al. (1995). Expression of relB is required for the development of thymic medulla and dendritic cells. Nature, 373, 531–536. [DOI] [PubMed] [Google Scholar]
- Casrouge A, Beaudoing E, Dalle S, Pannetier C, Kanellopoulos J, & Kourilsky P (2000). Size estimate of the alpha beta TCR repertoire of naive mouse splenocytes. The Journal of Immunology, 164, 5782–5787. [DOI] [PubMed] [Google Scholar]
- Collins GA, & Goldberg AL (2017). The logic of the 26S proteasome. Cell, 169, 792–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coux O, Tanaka K, & Goldberg AL (1996). Structure and functions of the 20S and 26S proteasome. Annual Review of Biochemistry, 65, 801–847. [DOI] [PubMed] [Google Scholar]
- Cowan JE, Parnell SM, Nakamura K, Caamano JH, Lane PJL, Jenkinson E, et al. (2013). The thymic medulla is required for Foxp3+ regulatory but not conventional CD4+ thymocyte development. Journal of Experimental Medicine, 210, 675–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley SR, Hu DY, & Goodnow CC (2013). Helios marks strongly autoreactive CD4+ T cells in two major waves of thymic deletion distinguished by induction of PD-1 or NF-κB. Journal of Experimental Medicine, 210, 269–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MM, & Bjorkman PJ (1988). T-cell antigen receptor genes and T-cell recognition. Nature, 334, 395–402. [DOI] [PubMed] [Google Scholar]
- Dick TP, Nussbaum AK, Deeg M, Heinemeyer W, Groll M, Schirle M, et al. (1998). Contribution of proteasomal beta-subunits to the cleavage of peptide substrates analyzed with yeast mutants. Journal of Biological Chemistry, 273, 25637–25646. [DOI] [PubMed] [Google Scholar]
- Fehling H, Swat W, Laplace C, Kühn R, Raiewsky K, Müller U, et al. (1994). MHC class I expression in mice lacking the proteasome subunit LMP-7. Science, 265, 1234–1237. [DOI] [PubMed] [Google Scholar]
- Florea BI, Verdoes M, Li N, van der Linden WA, Geurink PP, van den Elst H, et al. (2010). Activity-based profiling reveals reactivity of the murine thymoproteasome-specific subunit beta5t. Chemistry & Biology, 17, 795–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaczynska M, Rock KL, & Goldberg AL (1993). γ-Interferon and expression of MHC genes regulate peptide hydrolysis by proteasomes. Nature, 365, 264–267. [DOI] [PubMed] [Google Scholar]
- Gallegos AM, & Bevan MJ (2004). Central tolerance to tissue-specific antigens mediated by direct and indirect antigen presentation. Journal of Experimental Medicine, 200, 1039–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, & Wiley DC (1996). Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature, 384, 134–141. [DOI] [PubMed] [Google Scholar]
- Garcia KC, Degano M, Stanfield RL, Brunmark A, Jackson MR, Peterson PA, et al. (1996). An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science, 274, 209–219. [DOI] [PubMed] [Google Scholar]
- Gommeaux J, Grégoire C, Nguessan P, Richelme M, Malissen M, Guerder S, et al. (2009). Thymus-specific serine protease regulates positive selection of a subset of CD4+ thymocytes. European Journal of Immunology, 39, 956–964. [DOI] [PubMed] [Google Scholar]
- Grueter B, Petter M, Egawa T, Laule-Kilian K, Aldrian CJ, Wuerch A, et al. (2005). Runx3 regulates integrin alpha E/CD103 and CD4 expression during development of CD4−/CD8+ T cells. The Journal of Immunology, 175, 1694–1705. [DOI] [PubMed] [Google Scholar]
- Honey K, Nakagawa T, Peters C, & Rudensky A (2002). Cathepsin L regulates CD4+ T cell selection independently of its effect on invariant chain: A role in the generation of positively selecting peptide ligands. Journal of Experimental Medicine, 195, 1349–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörig H, Young ACM, Papadopoulos NJ, DiLorenzo TP, & Nathenson SG (1999). Binding of longer peptides to the H-2Kb heterodimer is restricted to peptides extended at their C terminus: Refinement of the inherent MHC class I peptide binding criteria. The Journal of Immunology, 163, 4434–4441. [PubMed] [Google Scholar]
- Huber EM, Basler M, Schwab R, Heinemeyer W, Kirk CJ, Groettrup M, et al. (2012). Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell, 148, 727–738. [DOI] [PubMed] [Google Scholar]
- Kincaid E, Che J, York I, Escobar H, Reyes-Vargas E, Delgado J, et al. (2012). Mice completely lacking immunoproteasomes show major changes in antigen presentation. Nature Immunology, 13, 129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kincaid EZ, Murata S, Tanaka K, & Rock KL (2016). Specialized proteasome subunits have an essential role in the thymic selection of CD8+ T cells. Nature Immunology, 17, 938–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisselev AF, Akopian TN, Castillo V, & Goldberg AL (1999). Proteasome active sites allosterically regulate each other, suggesting a cyclical bite-chew mechanism for protein breakdown. Molecular Cell, 4, 395–402. [DOI] [PubMed] [Google Scholar]
- Klug DB, Crouch E, Carter C, Coghlan L, Conti CJ, & Richie ER (2000). Transgenic expression of cyclin D1 in thymic epithelial precursors promotes epithelial and T cell development. The Journal of Immunology, 164, 1881–1888. [DOI] [PubMed] [Google Scholar]
- Koller BH, Marrack P, Kappler JW, & Smithies O (1990). Normal development of mice deficient in β2M, MHC class I proteins, and CD8+ T cells. Science, 248, 1227–1230. [DOI] [PubMed] [Google Scholar]
- Kourilsky P, & Claverie JM (1989). MHC restriction, alloreactivity, and thymic education: A common link? Cell, 56, 327–329. [DOI] [PubMed] [Google Scholar]
- Kozai M, Kubo Y, Katakai T, Kondo H, Kiyonari H, Schaeuble K, et al. (2017). Essential role of CCL21 in establishment of central self-tolerance in T cells. Journal of Experimental Medicine, 214, 1925–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuckelkorn U, Stübler S, Textoris-Taube K, Kilian C, Niewienda A, Henklein P, et al. (2019). Proteolytic dynamics of human 20S thymoproteasome. Journal of Biological Chemistry, 294, 7740–7754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyewski B, & Derbinski J (2004). Self-representation in the thymus: An extended view. Nature Review Immunology, 4, 688–698. [DOI] [PubMed] [Google Scholar]
- Laufer TM, DeKoningt J, Markowitz JS, Lo D, & Glimcher LH (1996). Unopposed positive selection and autoreactivity in mice expressing class II MHC only on thymic cortex. Nature, 383, 81–85. [DOI] [PubMed] [Google Scholar]
- Lkhagvasuren E, Sakata M, Ohigashi I, & Takahama Y (2013). Lymphotoxin-β receptor regulates the development of CCL21-expressing subset of postnatal medullary thymic epithelial cells. The Journal of Immunology, 190, 5110–5117. [DOI] [PubMed] [Google Scholar]
- Marrack P, & Kappler J (1987). The T cell receptor. Science, 238, 1073–1079. [DOI] [PubMed] [Google Scholar]
- McCaughtry TM, Baldwin TA, Wilken MS, & Hogquist KA (2008). Clonal deletion of thymocytes can occur in the cortex with no involvement of the medulla. Journal of Experimental Medicine, 205, 2575–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melichar HJ, Ross JO, Taylor KT, & Robey EA (2015). Stable interactions and sustained TCR signaling characterize thymocyte-thymocyte interactions that support negative selection. The Journal of Immunology, 194, 1057–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata S, Sasaki K, Kishimoto T, Niwa S, Hayashi H, Takahama Y, et al. (2007). Regulation of CD8+ T cell development by thymus-specific proteasomes. Science, 316, 1337–1353. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Roth W, Wong P, Nelson A, Farr A, Deussing J, et al. (1998). Cathepsin L: Critical role in Ii degradation and CD4 T cell selection in the thymus. Science, 280, 450–453. [DOI] [PubMed] [Google Scholar]
- Nil A, Firat E, Sobek V, & Eichmann K (2004). Expression of housekeeping and immunoproteasome subunit genes is differentially regulated in positively and negatively selecting thymic stroma subsets. European Journal of Immunology, 34, 2681–2689. [DOI] [PubMed] [Google Scholar]
- Nitta T, Murata S, Sasaki K, Fujii H, Mat Ripen A, Ishimaru N, et al. (2010). Thymoproteasome shapes immunocompetent repertoire of CD8+ T cells. Immunity, 32, 29–40. [DOI] [PubMed] [Google Scholar]
- Ohigashi I, Frantzeskakis M, Jacques A, Fujimori S, Ushio A, Yamashita F, et al. (2021). Thymoproteasome hardwires TCR repertoire of CD8+ T cells with cortical positive selection independent of negative selection. Journal of Experimental Medicine, 218, e20201904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohigashi I, Tanaka Y, Kondo K, Fujimori S, Kondo H, Palin AC, et al. (2019). Trans-omics impact of thymoproteasome in cortical thymic epithelial cells. Cell Reports, 29, 2901–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohigashi I, Zuklys S, Sakata M, Mayer CE, Zhanybekova S, Murata S, et al. (2013). Aire expressing thymic medullary epithelial cells originate from β5t-expressing progenitors. Proceedings of the National Academy of Sciences of the United States of America, 110, 9885–9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan BJ, Yekollu S, Ruscher R, Mehdi AM, Maradana MR, Chidgey AP, et al. (2018). Autoimmune-mediated thymic atrophy is accelerated but reversible in RelB-deficient mice. Frontiers in Immunology, 9, 1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobezinsky LA, Angelov GS, Tai X, Jeurling S, Van Laethem F, Feigenbaum L, et al. (2012). A clonal deletion and the fate of autoreactive thymocytes that survive negative selection. Nature Immunology, 13, 569–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punt JA, Havran W, Abe R, Sarin A, & Singer A (1997). T cell receptor (TCR)-induced death of immature CD4+CD8+ thymocytes by two distinct mechanisms differing in their requirement for CD28 costimulation: Implications for negative selection in the thymus. Journal of Experimental Medicine, 186, 1911–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripen AM, Nitta T, Murata S, Tanaka K, & Takahama Y (2011). Ontogeny of thymic cortical epithelial cells expressing the thymoproteasome subunit beta5t. European Journal of Immunology, 41, 1278–1287. [DOI] [PubMed] [Google Scholar]
- Robles AI, Larcher F, Whalin RB, Murillas R, Richie E, Gimenez-Conti IB, et al. (1996). Expression of cyclin D1 in epithelial tissues of transgenic mice results in epidermal hyperproliferation and severe thymic hyperplasia. Proceedings of the National Academy of Sciences of the United States of America, 93, 7634–7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Takada K, Ohte Y, Kondo H, Sorimachi H, Tanaka K, et al. (2015). Thymoproteasomes produce unique peptide motifs for positive selection of CD8+ T cells. Nature Communications, 6, 7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönrich G, Strauss G, Müller KP, Dustin L, Loh DY, Auphan N, et al. (1993). Distinct requirements of positive and negative selection for selecting cell type and CD8 interaction. The Journal of Immunology, 151, 4098–4105. [PubMed] [Google Scholar]
- Sentman CL, Shutter JR, Hockenbery D, Kanagawa O, & Korsmeyer SJ (1991). bcl-2 inhibits multiple forms of apoptosis but not negative selection in thymocytes. Cell, 67, 879–888. [DOI] [PubMed] [Google Scholar]
- Stritesky GL, Xing Y, Erickson JR, Kalekar LA, Wang X, Mueller DL, et al. (2013). Murine thymic selection quantified using a unique method to capture deleted T cells. Proceedings of the National Academy of Sciences of the United States of America, 110, 4679–4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutoh Y, Konodo M, Ohta Y, Ota T, Tomaru U, Flajnik MF, et al. (2012). Comparative genomic analysis of the proteasome β5t subunit gene: Implications for the origin and evolution of thymoproteasomes. Immunogenetics, 64, 49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szondy Z, Garabuczi E, Tóth K, Kiss B, & Köröskényi K (2012). Thymocyte death by neglect: Contribution of engulfing macrophages. European Journal of Immunology, 42, 1662–1667. [DOI] [PubMed] [Google Scholar]
- Takada K, Van Laethem F, Xing Y, Akane K, Suzuki H, Murata S, et al. (2015). TCR affinity for thymoproteasome-dependent positively selecting peptides conditions antigen responsiveness in CD8+ T cells. Nature Immunology, 16, 1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahama Y, Ohigashi I, Baik S, & Anderson G (2017). Generation of diversity in thymic epithelial cells. Nature Reviews Immunology, 17, 295–305. [DOI] [PubMed] [Google Scholar]
- Tomaru U, Konno S, Miyajima S, Kimoto R, Onodera M, Kiuchi S, et al. (2019). Restricted expression of the thymoproteasome is required for thymic selection and peripheral homeostasis of CD8+ T cells. Cell Reports, 26, 639–651. [DOI] [PubMed] [Google Scholar]
- Tomaru U, Takahashi S, Ishizu A, Miyatake Y, Gohda A, Suzuki S, et al. (2012). Decreased proteasomal activity causes age-related phenotypes and promotes the development of metabolic abnormalities. The American Journal of Pathology, 180, 963–972. [DOI] [PubMed] [Google Scholar]
- Uddin M, Ohigashi I, Motosugi R, Nakayama T, Sakata M, Hamazaki J, et al. (2017). Foxn1-β5t transcriptional axis controls CD8+ T-cell production in the thymus. Nature Communications, 8, 14419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T, Saito F, Gray DHD, Kuse S, Hieshima K, Nakano H, et al. (2004). CCR7 signals are essential for cortex–medulla migration of developing thymocytes. Journal of Experimental Medicine, 200, 493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Laethem F, Sarafova SD, Park JH, Tai X, Pobezinsky L, Guinter TI, et al. (2007). Deletion of CD4 and CD8 coreceptors permits generation of alphabetaT cells that recognize antigens independently of the MHC. Immunity, 27, 735–750. [DOI] [PubMed] [Google Scholar]
- Van Laethem F, Tikhonova AN, Pobezinsky LA, Tai X, Kimura MY, Le Saout C, et al. (2013). Lck availability during thymic selection determines the recognition specificity of the T cell repertoire. Cell, 154, 1326–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viret C, Lamare C, Guiraud M, Fazilleau N, Bour A, Malissen B, et al. (2011). Thymus-specific serine protease contributes to the diversification of the functional endogenous CD4 T cell receptor repertoire. Journal of Experimental Medicine, 208, 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weih F, Carrasco D, Durham SK, Barton DS, Rizzo CA, Ryseck RP, et al. (1995). Multiorgan inflammation and hematopoietic abnormalities in mice with a targeted disruption of RelB, a member of the NF-kappa B/Rel family. Cell, 80, 331–340. [DOI] [PubMed] [Google Scholar]
- Xing Y, Jameson S, & Hogquist K (2013). Thymoproteasome subunit-β5t generates peptide-MHC complexes specialized for positive selection. Proceedings of the National Academy of Sciences of the United States of America, 110, 6979–6984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T, Nedjic J, Hinterberger M, Steinert M, Koser S, Pinto S, et al. (2015). Thymic B cells are licensed to present self antigens for central T cell tolerance induction. Immunity, 42, 1048–1061. [DOI] [PubMed] [Google Scholar]
- Zuklys S, Handel A, Zhanybekova S, Govani F, Keller M, Maio S, et al. (2016). Foxn1 regulates key target genes essential for T cell development in postnatal thymic epithelial cells. Nature Immunology, 17, 1206–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]


