Abstract
In a competitive microbial environment nutrient acquisition is a major contributor to the survival of any individual bacterial species, and the ability to access uncommon energy sources can provide a fitness advantage. One set of soluble carbohydrates that have attracted increased attention for use in biotechnology and biomedicine are the α-diglucosides. Maltose is the most well studied member of this class, however the remaining four less common α-diglucosides (trehalose, kojibiose, nigerose, and isomaltose) are increasingly used in processed food and fermented beverages. The consumption of trehalose has recently been shown to be a contributing factor in gut microbiome disease as certain pathogens are using α-diglucosides to outcompete native gut flora. Kojibiose and nigerose have also been examined potential prebiotics and alternative sweeteners for a variety of foods. Compared to the study of maltose metabolism our understanding of the synthesis and degradation of uncommon α-diglucosides is lacking and several fundamental questions that remain unanswered, particularly in regards to the regulation of bacterial metabolism for α-diglucosides. Therefore this minireview attempts to provide a focused analysis of uncommon α-diglucoside metabolism in bacteria, and suggests some future directions for this research area that could potentially accelerate biotechnology and biomedicine developments.
Keywords: α-diglucoside, carbohydrate active enzyme, isomaltose, kojibiose, maltose, nigerose, sakibiose, trehalose
INTRODUCTION
One major determinant of bacterial survival is the ability to acquire and utilize nutrients obtained from their environment. Competition to obtain nutrients is a major driver of bacterially community development and affects both species composition and metabolic potential (Guignard et al. 2017). Bacterial species that exploit diverse nutrient sources gain a competitive advantage over those that are selective or restrictive in nutrient acquisition. For example, within the human gut microbiome alterations in diet greatly altered the composition of gut microbiota (Sanz et al. 2005; Holscher 2017). Of particular biomedical interest, it was recently shown that the ability of some pathogenic bacteria to use rare nutrient sources is a determinant of disease establishment and progression (Sanz et al. 2005; Chen et al. 2007). At the same time, a deeper understanding of how bacteria compete for nutrients has resulted in two major therapeutic approaches, probiotics that attempt to introduce new bacteria to an environment, and prebiotics that aim to alter the composition of bacteria that are already existing in an environment (Holscher 2017; Tsai et al. 2019). A deeper understanding of the nutrient utilization constrains of bacteria is not only important for enhancing human health, but for also to advance biotechnology and industrial applications. Engineering bacterial strains to utilize rare nutrient sources has been shown to increase feedstock conversion efficiency and increased metabolite yields (Keasling 2010; Cairns et al. 2019). As a consequence, characterizing the mechanisms that bacterial species employ to metabolize uncommon nutrient sources provides insight into the physiological functions that are important from both ecological and industrial perspectives (Peralta-Yahya et al. 2012).
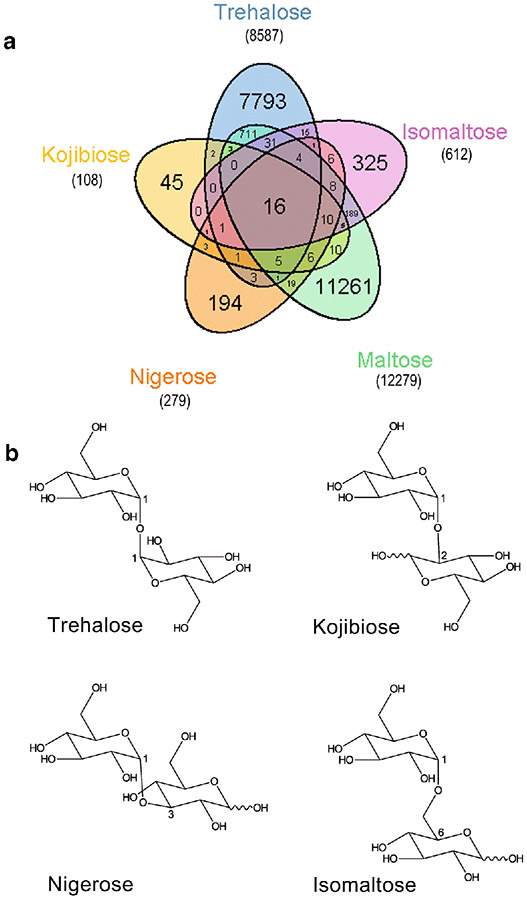
One metabolite that is drawing increased interested due to its biomedical and industrial importance is the α-diglucoside. These disaccharides all share the common feature of a being comprised of two glucose molecules joined by an α-glycosidic bond (Fig. 1a). The five α-diglucosides are trehalose, kojibiose, nigerose, maltose, and isomaltose, and these sugars differ only in the position of their glycosidic bond. These disaccharides can serve as a source of nutrition for humans and are found in a wide variety mushrooms, grains, fruits, vegetables, and more recently processed foods and beverages (Da Costa Leite et al. 2000; Elbein et al. 2003; Gerschenson et al. 2017). For a discussion of human disaccharide metabolism, several recent reviews have been published that explore this topic in detail (Rose et al. 2018; Kluch et al. 2020). Perhaps not surprisingly, not all α-diglucosides have been studied equally, and an analysis of PubMed indexed articles in April 2020 found a disproportionate number of studies (over 11,000) of maltose metabolism (Fig. 1b). Furthermore, maltose metabolism has been summarized from a variety of contexts and there are a number of excellent reviews published (Van Der Maarel et al. 2002; Ajita and Thirupathihalli 2014; Lentze 2018), and therefore we have excluded it from this review. Trehalose is the second most studied α-diglucoside and also has been reviewed (Elbein et al. 2003; Avonce et al. 2006; Ruhal et al. 2013), however we have included it in this review as several reports have been recently published that shed new light on bacterial trehalose degradation. For the remaining three α-diglucosides, there has not been a robust summary of past research concerning bacterial metabolism or discussion of emerging research areas. Consequently, the purpose of this review is to encapsulate the current understanding of the diversity of bacterial α-diglucoside metabolism and highlighting critical structural features that affect enzyme activity when a substrate is one of the uncommon α-diglucosides trehalose, kojibiose, nigerose, or isomaltose. The enzymes included for discussion were chosen based on the availability of physiological (in vivo) data or biochemical (in vitro) data that went beyond standard kinetic characterization. We conclude the review with a roadmap for the fundamental questions that still remain to be answered for bacterial α-diglucoside metabolism, specifically regarding advancing biotechnology and biomedicine.
Fig. 1. Literature analysis and structure of α-diglucosides.
(a) Literature analysis of those articles available on PubMed indicating representation of the different α-diglucosides and their overlapping content. Numbers within the Venn diagram represent unique identifiers for each grouping while numbers within parenthesis represent total articles containing the above term. Data collected in April 2020. Figure generated using R and Adobe Illustrator. (b) Chair structures of trehalose, kojibiose, nigerose, and isomaltose are represented. Numbers indicate carbons involved in the glycosidic bond. Structures were generated using ChemDoodle.
TREHALOSE (α-1,1-α DIGLUCOSIDE)
Biological Functions
Bacteria most commonly use trehalose as a stress protectant (Argüelles 2000; Elbein et al. 2003; Avonce et al. 2006). A variety of studies in E. coli have shown trehalose accumulation under heat, cold, osmotic, and other abiotic stress factors (Newman et al. 1993; Argüelles 2000; Iordachescu and Imai 2008). There are three modes of action believed to confer the protectant activity; chemical stability, water replacement, and sugar glass formation (Iturriaga et al. 2009; Jain and Roy 2009; Luyckx and Baudouin 2011). Trehalose is a non-reducing diglucoside, which makes it incapable of forming reactive aldehyde or ketone groups and its glycosidic linkage is relatively low energy (1 kcal/mol compared to the 27 kcal/mol glycosidic bond of sucrose) (Ohtake and Wang 2011). Furthermore, trehalose lacks internal hydrogen bonds, which facilitates a water replacement mode of stress protection (Brown et al. 1972). Specifically, the ability to form numerous hydrogen bonds with phospholipids and membrane proteins increases their stability during osmotic and thermal stress (Leslie et al. 1995; Leidy et al. 2004; Choi et al. 2006; Albertorio et al. 2007). These stabilized hydrogen-bond interactions are additionally beneficial as trehalose enters its glassy state, allowing an amorphous solid to surround, stabilize, and protect other macromolecules upon desiccation or high temperatures (Roos 1993; Miller et al. 1997; Liu et al. 1997).
In addition to stress mitigation, certain species of bacteria also use trehalose as a structural molecule or for energy storage. The Mycobacterium and Corynebacterium families possess members which incorporate modified trehalose in the form of trehalose dimycolate or trehalose monomycolate into their membrane glycolipids, making it a popular target for drug development in treating tuberculosis or diphtheria (De Smet et al. 2000; Nobre et al. 2014). Trehalose interactions with membrane phospholipids and membrane proteins during freeze-drying allow for the stabilization of these molecules, thereby increasing survival of frozen cells (Leslie et al. 1995). As carbon and energy storage molecule, Streptomyces griseus has been observed to accumulate 25% trehalose by dry weight under glucose abundant conditions, but as low as 1% under glucose limiting conditions, suggesting that glucose is converted to trehalose in carbohydrate abundant conditions (McBride and Ensign 1987). Conversely, S. griseus spores accumulate trehalose when grown in nutrient limiting conditions. The concentration of trehalose varies from 2% to 20% depending upon whether cells were grown in liquid or solid media with previous exposure to nutrient limiting conditions (Kendrick and Ensign 1983; McBride and Ensign 1987). Spores with larger quantities of trehalose had increased heat and desiccation resistance but a reduced germination rate, suggesting a trade-off between spore survival and ease of departure from a dormant state (McBride and Ensign 1987).
Finally, trehalose serves as a carbon and energy source for many bacteria, and exploiting trehalose metabolism has increasingly been of interest for modulating the composition of the human gut microbiome. For example, trehalose has recently been studied as a potential prebiotic, as it selectively promotes the growth of beneficial Bifidobacterium and their production of bacteriocins (Sanz et al. 2005; Chen et al. 2007). Conversely, an epidemiological study indicated that higher trehalose consumption correlated with increased virulence of Clostridium difficile (Collins et al. 2018). Conflicting arguments also present themselves in terms of oral pathogens, with some studies indicating increased growth of the common oral pathogen Streptococcus mutans while others note decreased growth in relation to trehalose consumption (Neta et al. 2000; Hodoniczky et al. 2012a).
Biosynthesis
There are five potential biosynthetic pathways to generate trehalose in bacteria. The first pathway discovered, and arguably the most widely distributed, is direct synthesis of trehalose from monosaccharides (Cabib and Leloir 1958). This reaction involves the combination of glucose 6-phosphate and UDP-glucose to form trehalose 6-phosphate, which is performed by trehalose 6-phosphate synthase (TPS; E.C. 2.4.1.15). Subsequently, Tre-6-P is dephosphorylated by trehalose-phosphatase (TP; E.C. 3.1.3.12) (Elbein et al. 2003). First identified in S. cerevisiae, it has since been characterized in several bacterial species such as E. coli, Mycobacterium, and Xanthomonas, as well as archaea, plants, and fungi (Elbein et al. 2003; Avonce et al. 2006; Carroll et al. 2007; Paul et al. 2008).
The next most prevalent pathway converts maltoligosaccharides into trehalose, which is also the preferred industrial method to generate this diglucoside (Song et al. 2016; Cai et al. 2018). The first step involves maltooligosyltrehalose synthase (E.C. 5.4.99.15) converting the reducing end of a maltooligosaccharide from an α-1,4 bond to an α-1,1 bond. Next, trehalose is cleaved from the oligosaccharide by maltooligosyltrehalose trehalohydrolase (E.C. 3.2.1.141) (Song et al. 2016). As maltooligosaccharides are an integral reactant, organisms possessing this pathway are typically starch producing or starch consuming. Initially identified in Arthrobacter, this two-step generation of trehalose is known as either the MTSase MTHase pathway or the TreY – TreZ pathway (Nakada et al. 1955; Elbein et al. 2003). A pBLAST genome analysis performed in 2006 indicated that approximately 50% of bacteria and 10% of archaea possess this pathway (Avonce et al. 2006).
The remaining three biosynthetic pathways employ single-step conversion mechanisms, however they are less prevalent within trehalose producing organisms and not used for commercial production of trehalose. The TreP (E.C. 2.4.1.64) pathway has been shown to use glucose 1-phosphate and glucose to generate trehalose and inorganic phosphate in vitro, specifically under low phosphate concentrations (Wannet et al. 1998). This pathway was initially identified in fungi, but has since been observed in few bacteria, such as Thermoanaerobacter tengcongensis (Wannet et al. 1998; Elbein et al. 2003; Avonce et al. 2006). The trehalose glycosyltransferring synthase (TreT; E.C. 2.4.1.245) pathway is capable of generating trehalose from ADP-glucose and glucose, and has been observed in both archaea and bacteria (Qu et al. 2004; Avonce et al. 2006; Nobre et al. 2008). The final pathway; trehalose synthase, has only been observed within bacteria. Similar to the TreY-TreZ pathway, it converts α-1,4 bonds to α-1,1 bonds, however, it is highly specific for maltose (Nishimoto et al. 1996; Avonce et al. 2006).
Degradation and Metabolism
When used as a nutrient source by bacteria trehalose can be cleaved during transport into the cell or by the action of extracellular or cytoplasmic enzymes. Trehalose transported through the phosphotransferase system (PTS) is cleaved by trehalose-6-phosphate hydrolases (E.C. 3.2.1.93), and generates glucose and glucose 6-phosphate for entry into metabolism (Fig. 2a) (Elbein et al. 2003; Ruhal et al. 2013). The PTS systems of a diverse range of bacteria have been reviewed elsewhere (Kotrba et al. 2001; Saier 2015), therefore we will focus here on non-membrane associated trehalases.
Fig. 2. Enzyme class reactions with representative members.
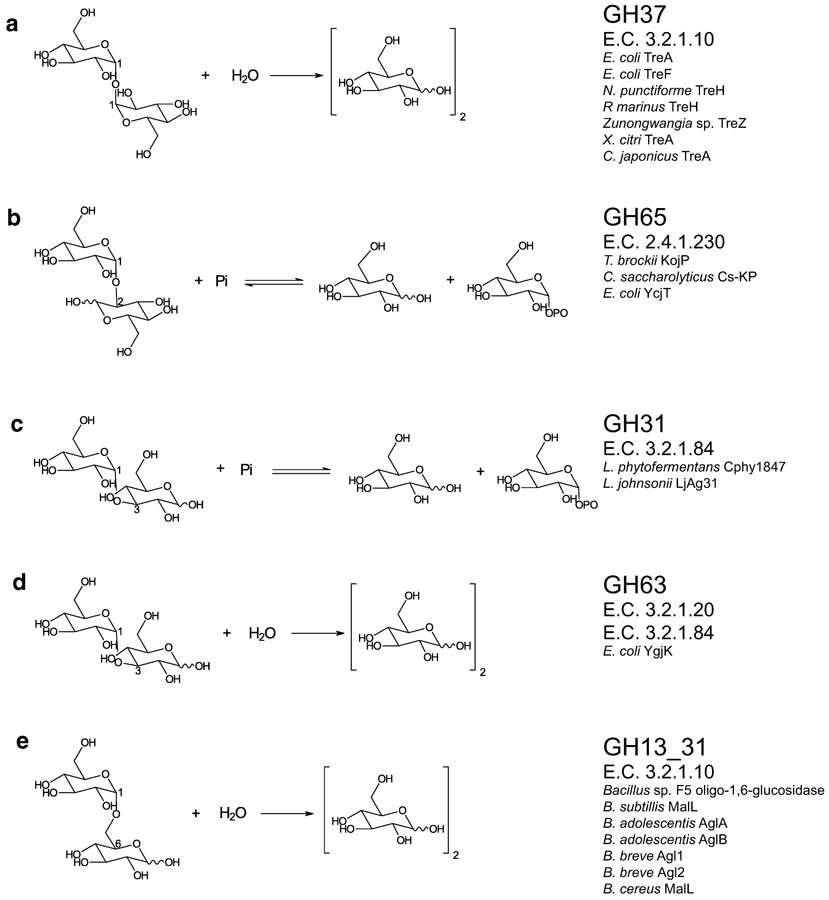
Enzyme reactions for those glycoside hydrolases (GH) and enzyme classes (E.C.) discussed. Enzymes representative of their respective classes are listed for (a) GH37 trehalases, (b) GH65 kojibiose phosphorylases, (c) GH 31 nigerose phosphorylases, (d) GH63 nigerose hydrolases, and (e) GH 13_31 isomaltose hydrolases.
Soluble trehalases (E.C. 3.2.1.28) split trehalose to produce two glucose molecules. As classified by the Carbohydrate Active enZyme (CAZyme) database, trehalases can belong to glycoside hydrolase (GH) families 15, 37, and 65 (Lombard et al. 2014). Enzymes belonging to the GH15 and GH65 families have been characterized as cleaving a diverse range of substrates, including trehalose (Lombard et al. 2014; Sakaguchi 2020). In this review, we will only discuss the GH37 enzymes, as this family has exclusively trehalase activity. Trehalases of the GH37 family have been identified in species of 178 genera (Lombard et al. 2014), and suggests that despite trehalose being relatively low in abundance in the environment it is still a metabolite with the potential to provide a competitive edge for those organisms capable of consuming it.
Biochemical characterization of GH37 trehalases has suggested that these enzymes are largely comparable despite being made by diverse bacteria. Bacterial trehalases from a broad range of species have been physiologically, biochemically, or structurally characterized and representative examples include E. coli TreA and TreF, E. cloacae TreA, N. punctiforme TreH, R. marinus TreH, C. japonicus Tre37A, X. citri sp. citri TreA, and Zunongwangia sp. TreZ (Table 1) (Boos et al. 1987; Horlacher et al. 1996; Uhland et al. 2000; Gibson et al. 2007; Jorge et al. 2007; Yoshida and Sakamoto 2009; Cheng et al. 2016; Adhav et al. 2019). Summarized below are several key points from the characterization of these GH37 enzymes, with a particular emphasis on the structural features important for catalysis.
Table 1.
Biochemical characterization of bacterial GH37 trehalases.
| Enzyme | VMAX (μM min−1) |
KM (mM) |
KCAT (min−1) |
Specific activity (μmol min−1 mg−1) |
pH | Temp (°C) |
Source |
|---|---|---|---|---|---|---|---|
| E. coli TreA | 0.012 | 0.41 | 1.2 x 103 | 6.6 x 101 | 5.5 | 37 | (Gibson et al. 2007) |
| E. coli TreF | 54 | 1.9 | N/A | 6.0 x 101 | 6 | N/A | (Horlacher et al. 1996) |
| E. cloacae TreA | 200 | 1.47 | 6.3 x 103 | 1.4 x 103 | 5 | 55 | (Adhav et al. 2019) |
| N. punctiforme TreH | N/A | 18 | N/A | 2.5 x 10−2 | 7.5 | N/A | (Yoshida and Sakamoto 2009) |
| R. marinus TreH | 81 | 0.16 | N/A | 1.3 x 101 | 6.5 | 88 | (Jorge et al. 2007) |
| Zunongwangia sp. TreZ | N/A | 0.99 | 1.6 x 104 5795 | 2.6 x 102 | 6 | 50 | (Cheng et al. 2016) |
| X. citri sp. citri TreA | 55.038 | 0.077 | N/A | 5.5 x 101 | 8 | 25 | (Alexandrino et al. 2016) |
| C. japonicus Tre37A | N/A | N/A | N/A | 7.3 x 103 | 6 | 20-30 | (Garcia et al. 2020) |
The structure of the E. coli trehalase TreA in complex with competitive inhibitors validoxylamine A and 1-thiatrehazolin provided insight into the GH37 catalytic mechanism (Gibson et al. 2007). This periplasmic enzyme contains a diagnostic (α/α)6 barrel structure characteristic of many α-glycosidases. X-ray crystallography of the enzyme complexed with competitive inhibitor indicated that residues Asp312 and Glu496 act as the catalytic acid and base, respectively. Additionally, the pseudo sugar ring present in validoxylamine A and the glucose moiety present in 1-thiatrehazolin exposed the presence of a large hydrophilic binding platform in the enzyme. The positioning of the +1 and −1 subsites relative to the competitive inhibitors suggested that significant conformational changes are required during substrate biding and cleavage due to the presence of an expansive binding pocket.
Mutational analysis of the cytoplasmic E. coli TreF identified two residues, Thr172 and Ala82, which when mutated increased the catalytic activity of the enzyme six-fold (Uhland et al. 2000). Amino acid sequence analysis by Sakaguchi et al. suggested that these are not conserved residues, but Thr172 appeared to be located within a sequence motif that is likely surrounded by tryptophan residues involved in the hydrophilic binding platform (Gibson et al. 2007; Adhav et al. 2019; Sakaguchi 2020). It was argued that a mutation in this region may affect the surrounding hydrophilic binding platform and can generate a more accessible structure with an increased rate of binding. It was less clear why the second mutation, Ala82Thr, was increased TreF activity, as this residue was not located within a conserved region.
Subsequently, two structures of the periplasmic E. cloacae TreA (PxEcTre) were solved, one in complex with validoxylamine A and one without any substrates (Gibson et al. 2007; Adhav et al. 2019). As observed with the E. coli TreA, the E. cloacae GH37 enzyme had an (α/α)6 barrel structure and the positioning of the acid and base acting residues, Asp312 and Glu496, were also similar. Interestingly, the structure solved without substrate identified a lid loop that helped explain the large conformational changes observed when substrate was bound, as well as a side loop that further stabilized substrate binding. Furthermore, mutational analysis found a critical non-catalytic residue (Trp447) that was required for proper binding pocket formation. A second non-catalytic residue that was found to be important for activity but not enzyme stability was Glu279, which was predicted form a salt bridge. Both of these residues are conserved in several bacterial trehalases.
Studies of Zunongwangia sp. TreZ utilized site-directed mutagenesis and error-prone-PCR to generate mutants with increased catalytic efficiency (Cheng et al. 2016). Two mutations, Tyr227His and Arg442Gly, were observed to act both individually and synergistically to increase the substrate specificity and reaction rate of TreZ. The authors argued that these mutations improved activity via expansion of the substrate binding pocket. The presence of both mutations resulted in a catalytic efficiency three-fold greater than that of the wild-type protein. Interestingly, Tyr227 appears to be conserved amongst characterized trehalases, while Arg442 is not.
The saprophytic soil bacterium Cellvibrio japonicus possesses two periplasmic GH37 trehalases, Tre37A and Tre37B (Garcia et al. 2020). Growth curve analysis indicated that only one of these, Tre37A, is required for trehalose utilization. This agreed with biochemical data, which found that Tre37A was a potent trehalase while Tre37B had no detectable activity (Table 1). Amino acid alignment of the C. japonicus trehalases alongside other characterized trehalases found that they possessed the conserved catalytic domains, signature motifs, and hydrophilic bonding platform identified from E. coli TreA (Gibson et al. 2007; Garcia et al. 2020).
KOJIBIOSE (α-1,2 DIGLUCOSIDE)
Biological Functions
Initially isolated in 1953 from sake, kojibiose is a reducing sugar is also found in beer, honey, caramelized glucose, and as a byproduct of dextran formation (Aso and Shibasaki 1953; Watanabe and Aso 1960; Matsuda et al. 1961). Kojibiose is classified as a rare sugar because it occurs infrequently in nature and in small quantities (Granström et al. 2004; Beerens et al. 2017). For example, honey contains approximately 0.5 g to 1.5 g kojibiose for every 100 g total mass, depending upon the origin of the honey (Schievano et al. 2017). Currently, the only known role for kojibiose in nature is as an energy source used for its glucose moieties. Regardless of its availability, kojibiose has garnered increased interest as a potential prebiotic and for its role as an α-glucosidase inhibitor (Ogawa et al. 1998; Sanz et al. 2005). Single culture studies of human gut microbiome indicate that Bifidobacterium, Lactobacillus, Clostridium, Bacteroides, and Eubacterium can grow using kojibiose as a sole carbon source (Chaen et al. 2001; Nakada et al. 2003; Gibson et al. 2004). Studies with bacterial consortia from fecal material suggested that kojibiose had a high selectivity for Bifidobacterium and resulted in greater acetic acid yields than many other disaccharides tested (Sanz et al. 2005).
Kojibiose can act as an α-glucosidase inhibitor, and is being explored for the treatment for different diseases. For example, drug development to treat HIV infections is testing kojibiose, as the virus is reliant on α-glucosidases to produce the glycoproteins that interact with host CD4 cells during virus reproduction (Kwong et al. 1998; Ogawa et al. 1998). Kojibiose alone behaves as a weak competitive inhibitor for α-glucosidase, but kojibiose pseudodisaccharides, which are saccharides possessing five membered rings where oxygen is present in the 1- and 3- position, have been discovered with increased potency (Ogawa et al. 1998; Barker and Rose 2013). Kojibiose is also being explored as a treatment for diabetes, as inhibiting α-glucosidase activity would reduce carbohydrate metabolism and glucose absorbance in the human gut (Phung et al. 2010).
Synthesis
Early attempts to isolate kojibiose from natural sources returned incredibly low quantities of the desired sugar (Watanabe and Aso 1959; Matsuda et al. 1961). Although honey was observed to contain approximately 0.5% - 1.5% kojibiose by weight, purification from this source had a return rate much less than 0.001% and required numerous concentration and purification steps, likely due in part to the wide variety of carbohydrates present within honey (Watanabe and Aso 1959). The low natural abundance of kojibiose, coupled with the poor recovery from natural sources, has led to increased interest in the chemical production of this α-glucoside. One described method involves the acetolysis of dextrans utilizing a mixture of acetic anhydride and sulfuric acid (Matsuda et al. 1961; Duke et al. 1973). The source of dextrans typically comes from Leuconostoc mesernteroides NRRL B-1299, which forms higher than average proportions of α-1,2 glycosidic linkages than other species. While this method is able to produce kojibiose from dextrans, the products require intensive purification that renders it not industrially viable. An alternative chemical method employed branched glucopyranose and zinc chloride in a modified Koenigs-Knorr reaction. This method generates a mixture of glucose, kojibiose, and the β-diglucoside sophorose, and as with previous attempts to chemically produce kojibiose resulted in low yields (McGrath et al. 1969).
Biosynthesis of kojibiose has also been successful, specifically a four-step reaction can generate kojibiose with a 38% yield and 99% purity (Diez-Munico et al. 2014). A mixture of sucrose and lactose is initially incubated with dextransucrase from L. mesenteroides to form 4’-galactosyl-kojibiose and fructose. A S. cerevisiae strain then metabolizes excess fructose and sucrose from the reaction mixture prior to incubation with β-galactosidase from K. lactis. The β-galactosidase cleaves 4’-galactosyl-kojibiose to release kojibiose and galactose (see Fig. 2 from Diez-Munico et al. 2014). The kojibiose is purified with liquid chromatography and while this method can generate large quantities of purified kojibiose, as outlined above it requires multiple steps and extensive processing to remove microbiological contaminants and undesirable side products.
A more efficient in vitro method of kojibiose production employs a single sucrose phosphorylase obtained from B. adolescentis (Verhaeghe et al. 2016). This method produces maltose and kojibiose by transglycosylation of glucose and sucrose (see Scheme 1 from Verhaeghe et al., 2016). While the wild-type enzyme preferentially forms maltose, introduction of L314I and Q345S mutations into the active site shifted production towards kojibiose. Incubation of the reaction mixture with S. cerevisiae was sufficient for removal of undesirable side products and resulted in a kojibiose purity of 99.5%. Additionally, the modified enzyme appeared to maintain a high rate of activity throughout a one-week incubation period at 55 °C, which would allow it to be used for multiple production cycles.
Degradation and Metabolism
Phosphorylases are transferase enzymes that catalyze the reversible transfer of phosphates, which allows for both synthesis and degradation to be mediated by a single enzyme. Bacterial production and degradation of kojibiose is dependent upon one of two enzyme classes; kojibiose phosphorylase (E.C. 2.4.1.230, GH65) (Fig. 2b) or sucrose phosphorylase (E.C. 2.4.1.7, GH13_18), though sucrose phosphorylase activity on kojibiose is considered an exlcusively in vitro non-specific side reaction. As kojibiose phosphorylases are natively more specific than sucrose phosphorylases for the production and degradation of kojibiose, this review will focus on kojibiose phosphorylases. For a review on sucrose phosphorylases, two reviews provide an excellent background on those enzymes (Puchart 2015; Franceus and Desmet 2020). Here we will limit our discussion to the GH65 enzymes, with particular emphasis on the structural features important for catalysis.
Bacterial kojibiose phosphorylases degrade kojibiose to D-glucose and β-D-glucopyranose 1-phosphate in vivo, but have also been shown to produce kojibiose in vitro when certain equilibrium conditions are favored (Chaen et al. 1999; Chaen et al. 2001; Yamamoto et al. 2006; Okada et al. 2014). The GH65 family, and therefore potential kojibiose phosphorylase activity, is widely distributed amongst Gram-positive bacteria, and examples can be found in Bacillus, Clostridioides, Enterobacter, and Lactobacillus genera (Lombard et al. 2014). However, the GH65 family also has members with α-trehalase, maltose phosphorylase, and nigerose phosphorylase activities, therefore without experimental data a definitive assessment of a GH65 enzyme can be difficult based on sequence similarly alone. Therefore, we will limit our discussion of kojibiose phosphorylases to those that have been biochemically characterized, and include enzymes from Thermoanaerobacter brockii, Calicellulosiruptor saccharolyticus, and Escherichia coli (Chaen et al. 1999; Chaen et al. 2001; Yamamoto et al. 2006; Okada et al. 2014).
The first kojibiose phosphorylase identified belonged to thermophilic spore forming bacterium T. brockii ATCC 35047, and was designated KojP (Chaen et al. 1999). This enzyme was able to both synthesize and degrade kojibiose, though it showed a greater KCAT during kojibiose degradation. Additionally, the enzyme showed greater specificity for kojibiose degradation compared to the degradation of other disaccharides. This is in direct contrast to the synthesis reaction, which produced a wide array of products, including kojibiose but also minor amounts of trehalose, sorbose, nigerose, maltose, and isomaltose when using different acceptor substrates. These minor products suggested that T. brockii KojP may have broad specificity for the formation of α-1,2 glycosidic bonds (Chaen et al. 1999). Site-directed mutagenesis studies of T. brockii KojP found that Asp362Asn, Lys612Gln, or Glu642Gln changes abolished kojibiose degradative capabilities. In contrast, the mutation of Asp459Asn resulted in a near seven-fold increase in KM for kojibiose and glucose substrates. This mutation likely affected the active site in such a way as to eliminate phosphorolysis, while maintaining transglycosylation and even increase specificity for these two saccharides (Yamamoto et al. 2004). These residues are conserved among other characterized kojibiose phosphorylases, with Asp362 acting as part of the hydrophilic substrate-bonding site, and Lys612 and Glu642 being hydrophilic bonding residues. Additionally, the Asp459 residue critical for kojibiose degradation is located near a loop region identified as being critical for kojibiose phosphorylation in C. saccharolyticus (Okada et al. 2014). Mutation of these residues in other kojibiose phosphorylases may produce similar increases in kojibiose formation as well as decreased kojibiose phosphorylation. A separate study generated a T. brockii kojibiose phosphorylase (KojP) and trehalose phosphorylase (TreP) chimera and observed a shifted ratio in favor of kojibiose production, however, the overall rate of the reaction was reduced (Yamamoto et al. 2006). Together, these studies identified the critical elements that alter the regioselectivity of T. brockii KojP, which will be important to ensure a high selectivity for kojibiose formation if used in selective in vitro biotechnology applications.
The study of a kojibiose phosphorylase from the anaerobe Calicellulosiruptor saccharolyticus provided a detailed crystal structure (Yamamoto et al. 2011; Okada et al. 2014). Biochemical characterization of the C. saccharolyticus kojibiose phosphorylase (Cs-KP, locus ID: Csac_0439) indicated that the pH and temperature optima for kojibiose synthesis were distinct from the optima for kojibiose degradation (Table 2) (Yamamoto et al. 2011). Additionally, the specific activity for synthesis is six-fold greater than that of the degradation, further indicating that the production of kojibiose is favored and dependent on the concentration of reactants. Optimization of reaction conditions was not determined, as the syntheses also generated a variety of products and favored the production of kojitriose over kojibiose. Nevertheless, the dependency on pH and temperature to dictate the directionality of Cs-KP might be exploited industrially towards kojibiose synthesis.
Table 2.
Kojibiose phospholyase synthase (S) and phosphorolytic (P) activities.
| Enzyme | VMAX (μM min−1) |
KM (mM) |
KCAT (min−1) |
Specific activity (μmol min−1 mg−1) |
pH | Temp (°C) |
Source | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | P | S | P | S | P | S | P | S | P | S | P | ||
| T. brockii KojP | N/A | N/A | 3.52 | 0.77 | 3.9 x 103 | N/A | N/A | 71.3 | 5.5 | 5.5 | 65 | 65 | (Chaen et al. 1999) |
| C. saccharolyticus Cs-KP | N/A | N/A | N/A | N/A | N/A | N/A | N/A | 7.34 | 5.5 | 6 | 75 | 85 | (Yamamoto et al. 2011) |
| E. coli YcjT | N/A | N/A | 1.7 | 1.05 | 48 | 66 | N/A | N/A | N/A | N/A | N/A | N/A | (Mukherjee et al. 2018) |
Crystal structures of Cs-KP in complex with either glucose and phosphate or kojibiose and sulfate identified an extended loop region critical in forming the active site and several residues involved in substrate interactions (Okada et al. 2014). As is characteristic for many GH65 enzymes, a single acidic catalytic residue (Glu483) was observed in addition to a molecule of phosphate maintained in the correct orientation by three amino acid residues for a nucleophilic attack. The structures indicated that both glucose substrates and kojibiose substrates are complexed in similar confirmations, with several conserved hydrophilic residues stabilizing the substrate at the +1 and −1 subsites. Site-directed mutagenesis identified Glu392 as being critical for kojibiose degradation, though mutation of resides Trp391 and Thr417 also reduced kojibiose catalysis. The Trp391 and Glu392 residues precede the critical loop region and are conserved amongst kojibiose phosphorylases. Interestingly, the less-impactful Thr417 residue is located within the loop region, critical for substrate interaction, and shares identity with the highly active T. brockii KojP but not the lesser active E. coli YcjT. Although the loop structure was identified as being crucial for kojibiose interaction, it shares reduced identity in comparison to other kojibiose phosphorylase regions.
Recently, an E. coli kojibiose phosphorylase (YcjT) was identified and characterized (Mukherjee et al. 2018). A gene cluster of unknown function was observed in E. coli, containing an assortment of putative transporters in addition to two putative hydrolases/phosphorylases that shared homology with sucrose phosphorylases from Bifidobacterium (Sprogøe et al. 2004; Mukherjee et al. 2018). However, YcjT showed only moderate identity with T. brockii KojP (30.4%) and Cs-KP (32.2%). Kojibiose synthesis activity was observed in solutions containing 8.0 mM β-D-glucose-1-phosphate, while degradation was observed in solutions containing 5 mM phosphate (Mukherjee et al. 2018). Examination of the amino acid sequence indicated the presence of a Ser409Thr mutation, deviating from the conserved hydrophilic bonding template and loop region identified from C. saccharolyticus kojibiose phosphorylase (Okada et al. 2014; Mukherjee et al. 2018). Interestingly, while quantifiable enzyme activity was observed, E. coli proved incapable of growth on media containing kojibiose as the sole carbon source. Consequently, it is currently unknown what physiological role YcjT plays in E. coli metabolism.
NIGEROSE (α-1,3 DIGLUCOSIDE)
Biological Functions
Nigerose, also known as sakebiose, is a reducing sugar originally isolated from the degradation of nigeran, a polysaccharide found in Aspergillus niger (Konishi and Shindo 1997; Ma et al. 2019). As the synonym suggests, this α-glucoside can also be found in sake, as well as beer, honey, as a product of glucose caramelization and as a byproduct of dextran degradation (Aso and Shibasaki 1953; Matsuda et al. 1961; Konishi and Shindo 1997; Ma et al. 2019). NMR quantification of the carbohydrates found in honey observed 0.23 g to 0.65 g nigerose in 100 g honey, though the concentration varied depending upon origin (Schievano et al. 2017). Additionally, nigerose has also been found in the red algae Dilsea edulis, albeit in small quantities (Peat et al. 1957). Consequently, nigerose is often considered an environmentally rare sugar, as it occurs in few organisms and in small quantities (Granström et al. 2004; Kraus et al. 2016).
Nigerose has been evaluated for its potential as a prebiotic, alternative sweetener, cryoprotectant, immunoprotectant, and possible mechanism for drug delivery (Murosaki et al. 1999; Sanz et al. 2005; Al-Otaibi et al. 2018). This α-glucoside has high potential for use as a reduced calorie sweetener (Hodoniczky et al. 2012a; Hodoniczky et al. 2012b). It is resistant to degradation by the common human oral pathogen, Streptococcus mutans, which is an advantage over other sweeteners such as glucose, trehalose, and sucrose (Hodoniczky et al. 2012a). In addition to its potential as a sweetener and oral prebiotic, there is some evidence for its potential as a digestive prebiotic. Inoculation of complex fecal cultures with nigerose proved to be somewhat selective for the growth of Bifidobacterium and Bacteroides and reduced the growth of Clostridium, however, these trends were more pronounced when kojibiose and sophorose were evaluated (Sanz et al. 2005). Increased resistance to Lactobacillus plantarum infection was also observed when mice were given oral doses of nigerose and mixed nigerans, further identifying nigerose as a potential therapeutic for infectious disease (Murosaki et al. 1999). The addition of nigerose to human cell lines prior to cryopreservation resulted in increased viability after thawing, which suggests an industrial application if more widely adopted (Al-Otaibi et al. 2018).
Biosynthesis
Nigerose research has primarily concentrated on production of this α-glucoside, with in vitro methods developed to degrade dextrans with nigerose phosphorylases or sucrose phosphorylases (Robyt 2008; Nihira et al. 2014; Franceus et al. 2019). Recent reports have identified and characterized few phosphorylases able to generate nigerose with and without extensive mutagenesis (Nihira et al. 2012; Nihira et al. 2014; Kraus et al. 2016; Franceus et al. 2019). While phosphorylase reactions are typically reversible, rational design of enzymes and careful control of conditions are able to force the nigerose forming reaction (Nihira et al. 2014). In this review, we will summarize the current knowledge concerning bacterial nigerose phosphorylases (GH65; E.C. 2.4.1.297) (Fig. 2c) and a nigerose-active sucrose phosphorylase mutant (CH13_18; E.C. 2.4.1.7) from Bacillus subtillis (Table 3) (Kitao et al. 1994; Stam et al. 2006; Kraus et al. 2016).
Table 3.
Biochemical characterization of nigerose synthase (S) and degradative (D) activities.
| Enzyme | VMAX (μM min−1) |
KM (mM) |
KCAT (min−1) |
Specific activity (μmol min−1 mg−1) |
pH | Temp (°C) |
Source | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | D | S | D | S | D | S | D | S | D | S | D | ||
| L. phytofermentans Cphy1874 | N/A | N/A | 3.3 | 1.7 | 6600 | 4.0 x 103 | 54 | 32 | 7 | 7 | 40 | 40 | (Nihira et al. 2012) |
| B. adolescentis BaSP mutant | N/A | N/A | 305 | N/A | 10.2 | N/A | N/A | 14.5 | N/A | N/A | N/A | N/A | (Franceus et al. 2019) |
| E. coli YgjK | N/A | N/A | 1.7 | N/A | 1.9 x 103 | N/A | N/A | N/A | 5 | N/A | 42 | N/A | (Kurakata et al. 2008) |
| L. johnsonii LjAg31 | N/A | N/A | 1.79 | N/A | 2 x 103 | N/A | N/A | N/A | 6 | N/A | 35 | N/A | (Kang et al. 2009) |
At the time of this review the only characterized bacterial nigerose phosphorylase (locus ID: Cphy1874) that has been characterized is from Lachnoclostridium phytofermentans, previously known as Clostridium phytofermentans (Nihira et al. 2012; Yutin and Galperin 2013). Cphy1874 was observed to weakly degrade kojibiose (<1%), and produce small quantities after long periods of incubation. Conversely, shorter periods of incubation resulted in a high level of specificity for reactions involving nigerose. Nigerose production was observed when utilizing glucose, however, nigerose analogues were produced when a wide variety of substrates such as, galactose, xylose, and 1,5-anhydro-D-glucitol were used as acceptors (Nihira et al. 2012). NMR spectra analysis of products formed by incubation with glucose indicated that α-1,3 glucosides were the major product formed. Some transglycosylation of nigerose into kojibiose was observed after extended periods of incubation, which hinted that the ratio of glucose, nigerose, and kojibiose influences product formation (Nihira et al. 2014). The authors conclude that the specificity for both nigerose degradation and production over other α-glucosides by Cphy1874 might be desirable for industrial nigerose production. A subsequent study using Cphy1874 successfully produced nigerose from sucrose using a one-pot reaction with three additional enzymes (see Figure 3A from Nihira et al., 2014) (Nihira et al. 2014; Franceus and Desmet 2020). Briefly, a sucrose phosphorylase from Bifidobacterium longum JCM1217 (locus ID: BLLJ_0106) used phosphate to cleave sucrose and yielded α-D-glucose-1-phosphate that was subsequently converted into α-D-glucose-6-phosphate by α-phosphoglucomutase from Thermococcus kodakarensis KOD1 (locus ID: TK1108). The α-D-glucose-6-phosphate was then converted to β-D-glucose-1-phosphate by β-phosphoglucomutase from Lactococcus sp. (Sigma-Aldrich; product ID P4109). The final step in the one-pot reaction had Cphy1874 combine β-D-glucose-1-phosphate and D-glucose to yield nigerose and inorganic phosphate. The phosphate would then allow for sucrose phosphorylase to continue the initial reaction. When incubated for 100 hours approximately 67% reaction yield of nigerose was obtained. The reaction schema allowed for utilization of all side products formed and mitigated nigerose degradation as substrates were continually being produced while limiting phosphate to either the sucrose phosphorylase or the nigerose phosphorylase reactions (Nihira et al. 2014).
Broad-range sucrose phosphorylases are another potential source of nigerose production. One such sucrose phosphorylase from Leuconostoc mesenteroides, LmSPase, was observed to form both kojibiose and nigerose upon incubation with sucrose and D-glucose, however, kojibiose production was more than double that of nigerose; 7.5% and 3.2% w:v reaction mixture respectively (Kitao et al. 1994). Increasing the reaction time produced no significant change in product concentration. Similar reactions were observed when the initial reactants consisted of solely sucrose, α-D-glucose-1-phosphate and D-glucose, or α-D-glucose-1-phosphate and sucrose, although the products were greatly reduced in these reactions.
The limited success using LmSPase to generate nigerose was enough that other sucrose phosphorylases were also characterized and included one from Bifidobacterium adolescentis (BaSP) (Franceus et al. 2019; Franceus and Desmet 2020). Initial studies of BaSP activity noted a strong preference for the formation of kojibiose or maltose, but no significant production of nigerose (Kraus et al. 2016). However, a Gln345Phe mutation of the loop region affecting binding of acceptor and donor substrates resulted in production of nigerose and maltose in equal concentrations. Addition of DMSO to 40% concentration increased nigerose production six-fold and further decreases the concentration of maltose produced. Introducing two additional mutations to this loop region, Asp342Gly and Tyr344Gln, resulted in a nine-fold increase in relative activity and five-fold increase in nigerose:maltose production over that of the single mutation. Addition of a fourth mutation to the acceptor site loop, Arg135Tyr, further increased the relative activity by 36-fold and the nigerose:nmaltose production by seven-fold (Franceus et al. 2019; Franceus and Desmet 2020). When the quadruple mutant was tested for nigerose production at a larger scale it was observed to have a 43% reaction yield (Franceus et al. 2019).
Degradation and Metabolism
Due to its low natural abundance and high cost to produce at an industrial scale, nigerose metabolism has focused on synthesis and not degradation. While the degradation of this α-glucoside has received less attention, two enzymes with nigerose hydrolytic activities have been observed, one from E. coli (YgjK) and one from Lactobacillus johnsonii (LjAg31, locus ID: LJ_0569) (Fig. 2d) (Kurakata et al. 2008; Kang et al. 2009; Miyazaki et al. 2013). Interestingly, E. coli YgjK is classified as a GH63 enzyme (E.C. 3.2.1.20 and E.C. 3.2.1.84), and members of the GH63 family are exo-acting α-glucosidases with characterized prokaryotic GH63 enzymes functioning primarily as mannosylglycerate hydrolases (Lombard et al. 2014). The L. johnsonii enzyme LjAg31 is classified as a GH31 enzyme (E.C. 3.2.1.84), a family which possess a broad range of activities, including; α-glucosidase, α-galactosidase, α-mannosidase, α-xylosidase, among others. Other enzymes with the classification of E.C. 3.2.1.84 are capable of hydrolyzing α-1,3 glycosidic linkages, however these enzymes are typically exo-acting and cleave at the reducing end of α-1,3 glucans to produce glucose (Zonneveld 1972; Ait-lahsen et al. 2001). We have summarized below the work done to date on the YgiK and LjAg31 enzymes.
Thus far E. coli possesses the only enzyme currently characterized as a GH63 nigerose hydrolase, YgjK (Lombard et al. 2014). An NCBI BlastP search of the amino acid sequence of this enzyme found homologs within Enterobacter, Vibrionacae, and Alteromonas orders (matrix BLOSUM62, expect threshold 1x10−5). Although the identification of homologous in this manner initially seems promising, it is likely that these sequences will need to be analyzed individually for structural similarities to YgjK. The solved structure of E. coli YgjK showed two distinct domains and a cleaved N-terminus, which indicated secretion into the periplasm or extracellular matrix (Tonozuka et al. 2004; Kurakata et al. 2008). The proposed catalytic residues are Asp501 and Glu727 acting as the acid and base, respectively (Barker and Rose 2013). These residues are not conserved among the other characterized nigerose-acting enzymes. It was also found that YgjK bound glucose, mannose, and galactose (Barker and Rose 2013). This result agrees with biochemical data that YgjK can hydrolyze trehalose, maltooligosaccharides, and kojibiose, however the enzyme displayed a strong preference for α-1,3 glycosidic linkages (Table 3) (Kurakata et al. 2008). The overall low affinity for nigerose suggests that it is likely not the preferred substrate of YgjK. This is corroborated by the inability of E. coli to grow using nigerose as a sole carbon source. Mutation of the catalytic Glu727Ala or the binding pocket residue Asp324Asn converted YgjK into a glucosynthase, producing 2-O-α-D-glucopyranosyl-D-galactopyranose from galactose and β-D-glucofuranose and no longer had any nigerose hydrolase activity (Miyazaki et al. 2013). These mutational studies of YgjK highlight the versatility and promise of GH63 nigerose hydrolases to produce high-value oligosaccharides for biotechnology and biomedicine.
The second enzyme with nigerose hydrolase activity was obtained from L. johnsonii (Kang et al. 2009). The cytoplasmc L. johnsonii LjAg31 enzyme shows activity against a broad range of substrates including maltose, isomaltose, kojibiose, and nigerose. Despite being able to cleave several authentic α-diglucoside substrates, LjGH31 had the greatest affinity for the artificial substrate pNPG. This is interesting as the majority of characterized GH31 enzymes exhibit α-glucosidase activities while this enzyme shows a high affinity for transglucosylation, forming both pNP α-nigeroside and pNP α-isomaltoside when incubated with pNP α-maltoside (Kang et al. 2009; Lombard et al. 2014). The authors proposed that the LjAg31 enzyme warranted classification as an α-1,3 glucosidase due to the KCAT/KM value for nigerose being the largest observed amongst all substrates (Kang et al. 2009). If the substrate specificities and catalytic efficiencies are compared between the two nigerose hydrolase enzymes of E. coli and L. johnsonii, it is observed that LjAg31 has a 128-fold higher affinity, but a similar catalytic rate for nigerose (Kurakata et al. 2008; Kang et al. 2009). An in-depth sequence comparison of these two hydrolytic enzymes may provide insight as to substrate specificity and catalytic efficiency, however this may prove difficult as the two enzymes share low identity (~15%).
ISOMALTOSE (α-1,6 DIGLUCOSIDE)
Biological Functions
Isomaltose was initially described as a reducing, non-fermentable sugar produced by the condensation of glucose, and as its name suggests, has historically been described as an isomer of maltose (Wolfrom et al. 1949; Bacon and Bacon 1954). When isolated from starch, isomaltose was described simply as an anomalous branching unit serving to generate cross-linked chains of maltose (Meyer and Bernfeld 1940; Tsuchiya et al. 1949). Later studies stripped away α-1,4 maltose units from amylopectin and uncovered that cross-linking branch points consisted of α-1,6 isomaltose (Montgomery et al. 1949; Tsuchiya et al. 1949).
Isomaltose has been studied as an alternative sweetener, prebiotic, potential therapeutic for health issues such as constipation and similar digestive problems, a potential marker for macular degeneration, and for increased conversion efficiency of oligosaccharides containing α-1,6 mixed linkages (Sanz et al. 2005; Harrion et al. 2007; Yen et al. 2011; Abe et al. 2017; Luo et al. 2017; Fatoki et al. 2018). The human gut has only slight capacity for isomaltose hydrolysis and absorption, which allows this α-glucoside to have a reduced glycemic index when compared to more readily absorbed carbohydrates (Ang and Linn 2014). These health benefits are enhanced due to the prebiotic properties of isomaltose (Kohmoto et al. 1988). Early studies indicated that various strains of Bifidobacterium commonly found within the human gastrointestinal tract were capable of utilizing isomaltose. Studies using human fecal matter indicated that isomaltose had a small positive selective pressure on Bifidobacterium, Bacteroides, Eubacterium, and Lactobacillus, and a small negative selective pressure on Clostridium (Sanz et al. 2005). Incubation with isomaltose produced large quantities of acetic acid and minute quantities of lactic acid, propionic acid, and butyric acid, which are common metabolic byproducts of Bifidobacterium anaerobic metabolism (Sanz et al. 2005). As isomaltose is commonly found in foods such as sake, honey, and caramel, albeit in small quantities (Watanabe and Aso 1959; Watanabe and Aso 1960; Matsuda et al. 1961). For example, honey was determined to contain between 0.8 g and 4.75 g isomaltose per 100 g starting material, depending upon the origin of the honey, putting its concentration above that of both nigerose and kojibiose (Schievano et al. 2017). In addition to a primary function as a nutrient source, isomaltose has also been examined for its biomedical potential. A study involving a wide range of people in various states of health noticed that the concentration of isomaltose in urine strongly correlated with periods of fasting (Vitek et al. 1972). This may have potential in identifying populations with insufficient or unstable food sources who may need assistance. Furthermore, due to the low glycemic index of isomaltose it has been studied as an alternative sweetener for people with diabetes (Ang and Linn 2014). There is also some evidence that isomaltose has weak antimicrobial properties (Memarzadeh 2009; Fatoki et al. 2018). Specifically, a pharmaceutical involving chlorine mono- and di- halogenated isomaltose has been patented as an oral mucosa cell permeation enhancer and low-grade antimicrobial (US Patent 2008/0171721 A1; 2008). These qualities in conjunction with its ubiquity in a variety of food and beverages potentially make isomaltose a convenient method of self-treatment before more potent antibiotics are required (Watanabe and Aso 1960).
Biosynthesis
Initially, isomaltose generation was an undesirable byproduct of starch degradation using α-amylases (Crabb and Shetty 1999). Common amylose starches are composed of primarily α-1,4 bound glucose, but up to 5% of the glycosidic bonds can be α-1,6 branch points (Pfister and Zeeman 2016). Typically, starch is degraded using α-amylases which cleave α-1,4 glycosidic bonds and release glucose, maltooligosaccharides, and α-1,6 isomaltose (Gopinath et al. 2017). Recently, isomaltose and isomaltooligosaccharides (IMOs) have gained interest as potential prebiotics and alternative sweeteners (Sanz et al. 2005; Harrion et al. 2007; Yen et al. 2011; Abe et al. 2017; Luo et al. 2017; Fatoki et al. 2018). IMO producing enzymes are largely described as glycoside hydrolases which also possess transferase activities (Lombard et al. 2014). Five such GH classes have been observed; GH13 (E.C. 3.2.1.133), GH31 (E.C. 3.2.1.20), GH57 (E.C. 2.4.1.18), GH66 (E.C. 3.2.1.11), and GH70 (E.C. 2.4.1.-) (Casa-Villegas et al. 2018). For a structural analysis of the enzymes involved in IMO synthesis, please refer to the following source (Casa-Villegas et al. 2018).
Commercially, GH31 transglycosylases from Aspergillus sp. are used due their strong preference towards generating isomaltose over other disaccharides, and also for bond synthesis rather than bond hydrolysis (Mangas-Sánchez and Adlercreutz 2015; Casa-Villegas et al. 2017). One transglycosylase from A. nidulans is capable of 50% conversion of maltose to other carbohydrate products (Kato et al. 2002; Goffin et al. 2010). Increasing the concentration of reactants is one proposed method for increasing conversion rates, while a second method involves combining enzymes (Viktor et al. 2013; Niu et al. 2017). Specifically, combining transglycosylating enzymes with amylases, glucoamylases or pullulanases reduces the formation of undesired byproducts and increases conversion to isomaltooligosaccharides (Viktor et al. 2013; Casa-Villegas et al. 2017; Niu et al. 2017).
Bacterial IMO generating enzymes have been found within each of the GH13, GH31, and GH70 classes, though the production of larger IMOs are favored over that of isomaltose (Lombard et al. 2014; Casa-Villegas et al. 2018). For that reason, here we will focus on the mutant Arg325Gln SmuA protein obtained from Protaminobacter rubrum (Lee et al. 2008). P. rubrum SmuA is classified as a GH13 sucrose isomerase, and the wild-type variant preferentially produces isomaltulose from sucrose, although a small shift towards producing isomaltose is observed when the reaction is supplemented with 5% glucose. Introduction of an Arg325Gln mutation within the conserved fructose-binding site present within sucrose isomerases shifted product yield away from isomaltulose and towards isomaltose. Isomaltose production was further increased when variant SmuA was incubated at 35°C and supplemented with 15% (w:v) glucose. Under these conditions, the relative activity for isomaltose production reached 46%. As few bacterial enzymes possess the required transglycosylation abilities, and fewer still preferentially produce isomaltose, future research may benefit from a rational engineering of bacterial sucrose isomerases in a similar manner.
Degradation and Metabolism
Many organisms possess transglycosylases capable of altering the α-1,6 bond present in starch and other dextrans to the more readily digested α-1,4 bond (Souza 2010). However, transglycosylation can generate products that do not have α-1,4 bonds, and several transglycosylases characterized thus far displayed low specific activity (Ajita and Thirupathihalli 2014; Mangas-Sánchez and Adlercreutz 2015). Alternatively, direct and efficient isomaltose hydrolysis can be performed by isomaltases (GH13_31; E.C.3.2.1.10), which are part of the oligo-1,6-glucosidase family (Fig. 2e, Table 4) (Lombard et al. 2014).
Table 4.
Biochemical characterization of bacterial GH13_31 oligo-1,6 glucosidases with high relative isomaltase activity.
| Enzyme | VMAX (μM min−1) |
KM (mM) |
KCAT (min−1) |
Specific activity (μmol min−1 mg−1) |
pH | Temp (°C) |
Source(s) |
|---|---|---|---|---|---|---|---|
| Bacillus sp. F5 oligo-1,6-glucosidase | N/A | N/A | N/A | 35a | 6 – 6.5 | 45 | (Yamamoto and Horikoshi 1990) |
| B. subtillis MalL | 162 | 0.455 | N/A | 2.02 | 7 | 42 | (Schönert et al. 1998; Schönert et al. 1999) |
| B. adolescentis AglA | 228 | 1.05 | N/A | N/A | 6.6a | 37b | (Van Den Broek et al. 2003) |
| B. adolescentis AglB | 113 | 0.47 | N/A | N/A | 6.8a | 47b | (Van Den Broek et al. 2003) |
| B. breve Agl1 | 35.9 | 8.3 | 1848 | 14.1 | 5.5b | 37c | (Pokusaeva et al. 2009) |
| B. breve Agl2 | 42.3 | 9 | 2178 | 20.4 | 5.5b | 30c | (Pokusaeva et al. 2009) |
| B. cereus MalL | N/A | 3.0 | N/A | 229a | 5.2 | 62 | (Suzuki and Tomura 1986) |
Reported in U/mg
Values reported for pnp-glucanpyranoside, noted to be similar for isomaltose
Temperature and pH values reported for palatinose and used for determination of isomaltose
One of the first characterized oligo-1,6 glucosidases from Bacillus sp. F5 (MalL; gene ID: O16G_BACF5) was heterologously expressed in E. coli, purified, and the kinetic parameters determined for several potential substrates (Yamamoto and Horikoshi 1990). The recombinant enzyme had the greatest level of activity on panose, a trisaccharide with mixed α-1,6 and α-1,4 glucosidic linkages, followed isomaltooligosaccharides, and then maltose. The broad specificity observed has been observed in other oligo-1,6 glucosidases, however the high level of activity towards smaller isomaltooligosaccharides is particularly interesting as many enzymes of this classification tend towards hydrolyzing larger oligosaccharides (Yamamoto and Horikoshi 1990; Niu et al. 2017).
Another isomaltose hydrolyzing GH13_31 from Bacillus subtilis (now also called MalL, previously identified as yvdL, locus ID: BSU_34560) was characterized as part of larger study to determine the mechanisms of maltose metabolism (Schönert et al. 1998; Schönert et al. 1999). It was found that the malL gene was highly up-regulated during growth of B. subtillus on maltose, though catabolite repression was noted when cells were grown with both maltose and glucose. Furthermore, malL induction was observed during B. subtillis growth on more complex substrates possessing α-1,4 bonds, such as amylose, starch, and even glycogen (Schönert et al. 1999) Deletion of the malL gene resulted in an inability to utilize maltose (Schönert et al. 1998). Purified MalL was observed to have the greatest affinity for maltose, then isomaltose, and finally sucrose. The VMAX (μmol min−1 mg protein−1) for these substrates is reversed, with MalL cleaving sucrose fastest, then isomaltose and finally maltose. Almost no activity was observed on larger isomaltooligosacharides (Schönert et al. 1999). In addition to these substrates, some activity was observed on p-nitrophenyl-α-D-pyranoside (PNPG) (Schönert et al. 1998).
Later studies involving B. subtillis MalL analyzed 268 single amino acid mutations and identified four that altered activity, Val200Ala, Val200Ser, Val200The, and Gly202Phe. The Val200Ser mutation led to an approximately two-fold reduction in KM and a 1.2-fold increase in KCAT while the Gly202Phe variation experienced a 2.2-fold reduction in KM and a nearly five-fold reduction in KCAT. Structural analysis indicated that the Val200Ser mutation induced additional hydrogen bond formation, thereby reducing conformational states and enzyme activity. Alternatively, the Gly202Phe mutation increased the flexibility of MalL, which resulted in increased energy requirements during the transition stage and a reduction in KCAT.
Biochemical characterization of Bifidobacterium breve Agl1 and Agl2 reported high specific activity towards isomaltose and panose (Pokusaeva et al. 2009). No activity was observed on maltose or other substrates with α-1,4 bonds present. Additionally, activity on isomaltotriose was significantly reduced for both Alg1 and Agl2. Interestingly, growth analysis of B. breve indicates that it can utilize both isomaltose and isomaltotriose as primary carbon sources, with culture grown on isomaltotriose exhibiting increased growth over that of isomaltose. Enzyme kinetics analyses revealed that both enzymes have similar temperature and pH optima, but the Agl1 protein broader ranges before experiencing a loss of activity. The authors report that Southern hybridization with the agl sequences indicated the presence of a homologous agl gene in other Bifidobacterium species, and a Bifidobacterium adolescentis genomic library screen identified the aglA and aglB genes as putative α-glucosidases (Van Den Broek et al. 2003). Later metabolic studies argued that members of Bifidobacterium preferentially hydrolyze larger oligosaccharides with high degrees of polymerization, which may provide a source of α-1,6 linkages for degradation by the agl gene families (Andersen et al. 2013; Hu et al. 2013).
One additional enzyme with high relative isomaltase activity was identified from Bacillus cereus (malL, locus ID: BAA11354.1) (Suzuki and Tomura 1986; Kizaki et al. 1993; Watanabe et al. 2001). While the enzyme was categorized as an oligo-1,6-glucosidase, it was most active on isomaltose with decreasing activity as the length of the oligosaccharide increased (Suzuki and Tomura 1986). Additionally, no notable activity was observed on α-1,4 glycosidic linkages Site-directed mutational analysis allowed for the identification of three catalytic residues; Asp199, Glu255 and Asp329, and two substrate binding residues; His103 and His328 (Watanabe et al. 2001). All of these residues are conserved amongst the characterized isomaltases summarized in this review, and provide the framework for optimization efforts if these or related enzymes were to be used industrially.
FUTURE OUTLOOK
The α-diglucosides are becoming increasingly prevalent in consumer goods as their beneficial properties are being discovered (Ogawa et al. 1998; Sanz et al. 2005; Gerschenson et al. 2017; Mooradian et al. 2017). This has led to an increase in research regarding α-diglucoside production. However, as they become more prevalent in food and consumer goods it will become increasingly important to understand how these sugars influence human health, particularly the gut microbiome. As has been previously observed with increased trehalose consumption, there may be unforeseen consequences regarding pathogenicity and virulence of gut bacteria (Dalwai et al. 2006; Collins et al. 2018). From an industrial standpoint, engineering organisms to generate these metabolites requires a complete understanding of their production, transport, and degradation (Keasling 2010; Abe et al. 2017). Therefore, a complete understanding of both biosynthesis and degradation are integral from an industrial standpoint. Below we propose four major focus questions to direct future work to identify and characterize mechanisms of bacterial α-diglucosides metabolism, with one goal being to accelerate the development of biomedicine and biotechnology applications.
What is the prevalence of bacterial α-diglucoside metabolic enzymes in diverse microbial consortia?
Currently, one substantial challenge that hinders identification of α-diglucoside metabolic enzymes is homology. As outlined in this review the amino acid sequence of trehalases, kojibiose phosphorylases, and isomaltases may only have moderate levels of identity. However, physiological and biochemical activities are not necessarily correlated with sequence identity. For example, C. japonicus Tre37B shares 44% identity with the characterized E. coli TreA but has no detectible trehalase activity (Garcia et al. 2020). Identifying potential targets becomes increasingly difficult if protein identity is inherently small, which is the case for the nigerose active enzymes discussed in this review (< 25% identity). Physiological experiments used to identify additional organisms capable of metabolizing these rare carbohydrates can be used as a basis to predict additional homologous genes. Additionally, those organisms described within this review can be used to identify potential transporters; a critical step for future metabolic engineering.
How are bacterial α-diglucoside metabolic enzymes regulated?
Physiological data are distinctly under-represented in the available literature regarding α-disaccharide metabolism, and this is likely a consequence the difficulty predicting complete metabolic pathways for diglucoside utilization. Furthermore, it would take significant work to generate the mutant strains required to characterize diglucoside metabolism from a variety of bacteria. However, physiological data are going to be critical for identifying potential enzymes and transport mechanisms. For example, the B. subtillis isomaltose metabolic system has been physiologically characterized and through those experiments it was discovered that a putative isomaltose active enzyme was essential for isomaltose metabolism (Mangas-Sánchez and Adlercreutz 2015). A second example comes from the C. japonicus trehalose metabolic system, where phenotypic analysis found that Tre37A is required for trehalose degradation but the predicted trehalase Tre37B was dispensable (Garcia et al. 2020). Alternatively, the presence of detectible enzyme activity does not correlate with physiological relevance. This was observed with the E. coli YgjK enzyme, which was capable of hydrolyzing nigerose, but E. coli is incapable of utilizing nigerose as a sole carbon source (Tonozuka et al. 2004). Finally, three novel C. japonicus strains capable of metabolizing kojibiose, nigerose, and isomaltose have recently been identified, however, predictive genomic analyses have not yet identified genes likely to act on said carbohydrates and phenotypic data will be essential to identify beneficial mutations (Garcia et al. 2019). Therefore, future experiments to generate physiological data will complement in vitro data and facilitate in silico predictions to discover and characterize bacterial diglucoside metabolic enzymes.
How are α-diglucosides transported by bacteria?
Current knowledge indicates that α-diglucosides are degraded and the resulting glucose moieties enter glycolysis (Kanehisa 2000). Amino acid sequence analysis of characterized kojibiose phosphorylases, nigerose phosphorylases, nigerose hydrolases, and isomaltases suggested that these enzymes are largely cytoplasmic, suggesting a transport requirement that may be substrate specific (Almagro Armenteros et al. 2019). Previous work to engineer E. coli to metabolize isomaltose not only an isomaltase obtained from B. subtillis, but also an isomaltose specific phosphotransferase system (PTS) component from K. pneumoniae (Abe et al. 2017). In the absence of a PTS, the transportation systems for kojibiose and nigerose are poorly understood. Operon analysis of the T. brockii genome indicated that there are possible permeases surrounding the kojP gene which may be important for kojibiose transport into the cell (Chaen et al. 1999). Similarly, predicted permeases can be found near the L. phytofermentans cphy1874 gene and the E. coli ycjT genes (Nihira et al. 2012; Mukherjee et al. 2018). In addition to permeases, TonB-dependent transporters may be involved in carbohydrate metabolism. Genome analysis of the C. japonicus genome, which does not have a PTS, found the presence of a TonB-dependent receptor upstream of the tre37A gene (Garcia et al. 2019). While deletion of this TonB-dependent receptor gene produced no notable physiological effect, it is possible that the entire complex would need to be deleted to elicit a phenotype. Future work will not only need to identify α-diglucoside transporters, but also determine their substrate specificity to maximize their utility in biomedical or biotechnology applications.
What are the principles to most efficiently optimize bacterial α-diglucoside metabolic enzymes?
Study of the α-diglucosides kojibiose, nigerose, and isomaltose are primarily limited by their availability. Many of those enzymes involved in biosynthesis are promiscuous and produce a wide variety of products, or are also capable of degradation (Table 2, Table 3). Several studies have focused on generating mutations in these enzymes for more efficient production of the desired α-diglucoside, and include reducing the variety of products formed, increasing substrate KM, VMAX, and KCAT, and minimizing the enzyme’s ability to hydrolyze the desired product.
Closing Remarks
Bacterial species capable of utilizing diverse nutrient sources gain a competitive advantage over those species with more restricted nutrient sources (Guignard et al. 2017). Therapeutics such as prebiotics and probiotics take advantage of this competition to alter the nutrient and bacterial composition, with the goal of improving human health (Holscher 2017; Tsai et al. 2019). Despite being understudied, α-diglucosides show great promise as a prebiotic, in addition to being an alternative sweetener, diagnostic tool, or therapeutic (Murosaki et al. 1999; Sanz et al. 2005; Luo et al. 2017; Al-Otaibi et al. 2018). To capitalize on their potential, future research will need to emphasize not only the discovery of α-diglucoside metabolic enzymes, but characterizing their physiological function in order to engineer them for maximal utility in biomedical and biotechnology applications.
KEY POINTS.
α-diglucosides are increasingly important, but understudied bacterial metabolites.
Kinetically superior α-diglucosides enzymes require few amino acid substitutions.
In vivo studies are required to realize the biotechnology potential of α-diglucosides.
Funding.
This work was supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under Award Number DE-SC0014183 and the Meyerhoff Graduate Fellows Program NIGMS Initiative for Maximizing Student Development Grant T32-GM066706.
Footnotes
Conflicts of interest/Competing interests. The authors declare that they have no conflicts of interest.
Ethics approval. Not applicable.
Consent to participate. Not applicable.
Consent for publication. Not applicable.
Availability of data and material. Not applicable.
Code availability. Not applicable.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
REFERENCES
- Abe K, Kuroda A, Takeshita R (2017) Engineering of Escherichia coli to facilitate efficient utilization of isomaltose and panose in industrial glucose feedstock. Appl Microbiol Biotechnol 101:2057–2066. 10.1007/s00253-016-8037-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhav A, Harne S, Bhide A, Giri A, Gayathri P, Joshi R (2019) Mechanistic insights into enzymatic catalysis by trehalase from the insect gut endosymbiont Enterobacter cloacae. FEBS J 286:1700–1716. 10.1111/febs.14760 [DOI] [PubMed] [Google Scholar]
- Ait-lahsen H, Soler A, Rey M, De La Cruz J, Monte E, Llobell A (2001) An antifungal exo-1,3-glucanase (AGN13.1) from the biocontrol fungus Trichoderma harzianum. Appl Environ Microbiol 67:5833–5839. 10.1128/AEM.67.12.5833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajita S, Thirupathihalli M (2014) α-Amylase production and applications: a review. J Appl Environ Microbiol 2:166–175. 10.12691/jaem-2-4-10 [DOI] [Google Scholar]
- Al-Otaibi NAS, Cassoli JS, Martins-De-Souza D, Slater NKH, Rahmoune H (2018) Human leukemia cells (HL-60) proteomic and biological signatures underpinning cryo-damage are differentially modulated by novel cryo-additives. Gigascience 8:1–13. 10.1093/gigascience/giy155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertorio F, Chapa VA, Chen X, Diaz AJ, Cremer PS (2007) The α,α-(1→1) linkage of trehalose is key to anhydrobiotic preservation. J Am Chem Soc 129:10567–10574. 10.1038/jid.2014.371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrino AV, Goto LS, Novo-Mansur MTM (2016) TreA codifies for a trehalase with involvement in xanthomonas citri subsp. citri Pathogenicity. PLoS One 11:1–13. 10.1371/journal.pone.0162886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H (2019) SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37:420–423. 10.1038/s41587-019-0036-z [DOI] [PubMed] [Google Scholar]
- Andersen JM, Barrangou R, Hachem MA, Lahtinen SJ, Goh YJ, Svensson B, Klaenhammer TR (2013) Transcriptional analysis of oligosaccharide utilization by Bifidobacterium lactis Bl-04. BMC Genomics 14:1. 10.1186/1471-2164-14-312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang M, Linn T (2014) Comparison of the effects of slowly and rapidly absorbed carbohydrates on postprandial glucose metabolism in type 2 diabetes mellitus patients: A randomized trial. Am J Clin Nutr 100:1059–1068. 10.3945/ajcn.113.076638 [DOI] [PubMed] [Google Scholar]
- Argüelles JC (2000) Physiological roles of trehalose in bacteria and yeasts: A comparative analysis. Arch Microbiol 174:217–224. 10.1007/s002030000192 [DOI] [PubMed] [Google Scholar]
- Aso K, Shibasaki K (1953) Studies on the unfermentable sugars I. II. III. Tohoku J Agric Res 3:337–358 [Google Scholar]
- Avonce N, Mendoza-Vargas A, Morett E, Iturriag G (2006) Insights on the evolution of trehalose biosynthesis. BMC Evol Biol 6:1–15. 10.1186/1471-2148-6-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon EE, Bacon JS (1954) The occurrence of isomaltose among the products of heating glucose in dilute mineral acid. Biochem J 58:396–402. 10.1042/bj0580396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MK, Rose DR (2013) Specificity of processing α-glucosidase I is guided by the substrate conformation: Crystallographic and in silico studies. J Biol Chem 288:13563–13574. 10.1074/jbc.M113.460436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerens K, De Winter K, Van De Walle D, Grootaert C, Kamiloglu S, Miclotte L, Van De Wiele T, Van Camp J, Dewettinck K, Desmet T (2017) Biocatalytic synthesis of the rare sugar kojibiose: process scale-up and application testing. J Agric Food Chem 65:6030–6041. 10.1021/acs.jafc.7b02258 [DOI] [PubMed] [Google Scholar]
- Boos W, Ehmann U, Bremer E, Middendorf A, Postma P (1987) Trehalase of Escherichia coli. Mapping and cloning of its structural gene and identification of the enzyme as a periplasmic protein induced under high osmolarity growth conditions. J bioche 262:13212–13218 [PubMed] [Google Scholar]
- Brown GM, Rohrer DC, Berking B, Beevers CA, Gould RO, Simpson R (1972) The crystal structure of α,α-trehalose dihydrate from three independent X-ray determinations. Acta Crystallogr Sect B Struct Crystallogr Cryst Chem 28:3145–3158. 10.1107/s0567740872007654 [DOI] [Google Scholar]
- Cabib E, Leloir LF (1958) The biosynthesis of trehalose phosphate. J Biol Chem 231:259–275 [PubMed] [Google Scholar]
- Cai X, Seitl I, Mu W, Zhang T, Stressler T, Fischer L, Jiang B (2018) Biotechnical production of trehalose through the trehalose synthase pathway: current status and future prospects. Appl Microbiol Biotechnol 102:2965–2976. 10.1007/s00253-018-8814-y [DOI] [PubMed] [Google Scholar]
- Cairns TC, Zheng X, Zheng P, Sun J, Meyer V (2019) Moulding the mould: Understanding and reprogramming filamentous fungal growth and morphogenesis for next generation cell factories. Biotechnol Biofuels 12:1–18. 10.1186/s13068-019-1400-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JD, Pastuszak I, Edavana VK, Pan YT, Elbein AD (2007) A novel trehalase from Mycobacterium smegmatis - Purification, properties, requirements. FEBS J 274:1701–1714. 10.1111/j.1742-4658.2007.05715.x [DOI] [PubMed] [Google Scholar]
- Casa-Villegas M, Marín-Navarro J, Polaina J (2018) Amylases and related glycoside hydrolases with transglycosylation activity used for the production of isomaltooligosaccharides. Amylase 2:17–29. 10.1515/amylase-2018-0003 [DOI] [Google Scholar]
- Casa-Villegas M, Marín-Navarro J, Polaina J (2017) Synthesis of isomaltooligosaccharides by Saccharomyces cerevisiae cells expressing Aspergillus niger α-glucosidase. ACS Omega 2:8062–8068. 10.1021/acsomega.7b01189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaen H, Nishimoto T, Nakada T, Fukuda S, Kurimoto M, Tsujisaka Y (2001) Enzymatic synthesis of kojioligosaccharides using kojibiose phosphorylase. J Biosci Bioeng 92:177–182. 10.1016/s1389-1723(01)80221-8 [DOI] [PubMed] [Google Scholar]
- Chaen H, Yamamoto T, Nishimoto T, Nakada T, Fukuda S, Sugimoto T, Kurimoto M, Tsujisaka Y (1999) Purification and characterization of a novel phosphorylase, kojibiose phosphorylase, from Thermoanaerobium brockii. J Appl Glycosci 46:423–429. 10.5458/jag.46.423 [DOI] [Google Scholar]
- Chen YS, Srionnual S, Onda T, Yanagida F (2007) Effects of prebiotic oligosaccharides and trehalose on growth and production of bacteriocins by lactic acid bacteria. Lett Appl Microbiol 45:190–193. 10.1111/j.1472-765X.2007.02167.x [DOI] [PubMed] [Google Scholar]
- Cheng Q, Gao H, Hu N (2016) A trehalase from Zunongwangia sp.: Characterization and improving catalytic efficiency by directed evolution. BMC Biotechnol 16:6–13. 10.1186/s12896-016-0239-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Cho KW, Jeong K, Jung S (2006) Molecular dynamics simulations of trehalose as a ‘dynamic reducer’ for solvent water molecules in the hydration shell. Carbohydr Res 341:1020–1028. 10.1016/J.CARRES.2006.02.032 [DOI] [PubMed] [Google Scholar]
- Collins J, Robinson C, Danhof H, Knetsch CW, van Leeuwen HC, Lawley TD, Auchtung JM, Britton RA (2018) Dietary trehalose enhances virulence of epidemic Clostridium difficile. Nature 553:291–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb WD, Shetty JK (1999) Commodity scale production of sugars from starches. Curr Opin Microbiol 2:252–256. 10.1016/S1369-5274(99)80044-7 [DOI] [PubMed] [Google Scholar]
- Da Costa Leite JM, Trugo LC, Costa LSM, Quinteiro LMC, Barth OM, Dutra VML, De Maria CAB (2000) Determination of oligosaccharides in Brazilian honeys of different botanical origin. Food Chem 70:93–98. 10.1016/S0956-7135(99)00115-2 [DOI] [Google Scholar]
- Dalwai F, Spratt DA, Pratten J (2006) Modeling shifts in microbial populations associated with health or disease. Appl Environ Microbiol 72:3678–3684. 10.1128/AEM.72.5.3678-3684.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet KAL, Weston A, Brown IN, Young DB, Robertson BD (2000) Three pathways for trehalose biosynthesis in mycobacteria. Microbiology 146:199–208. 10.1099/00221287-146-1-199 [DOI] [PubMed] [Google Scholar]
- Diez-Munico M, Montilla A, Moreno FJ, Herrero M (2014) A sustainable biotechnological process for the efficient synthesis of kojibiose. Green Chem 16:2219–2226 [Google Scholar]
- Duke J, Little N, Goldstein IJ (1973) Preparation of cyrstalline [alpha]-kojibiose octaacetate from dextran B-1299-S: Its conversion into p-nitrophenyl and p-isothiocyanatophenyl [beta]-kojibioside. Carbohydr Res 27:193–198 [DOI] [PubMed] [Google Scholar]
- Elbein AD, Pan YT, Pastuszak I, Carroll D (2003) New insights on trehalose: A multifunctional molecule. Glycobiology 13:17–27. 10.1093/glycob/cwg047 [DOI] [PubMed] [Google Scholar]
- Fatoki T, Sanni D, Momodu D, Ugboko H, Adeseko C, Faleye B (2018) Evaluation of empirical functions and fate of isomaltose. J Appl Life Sci Int 16:1–10. 10.9734/jalsi/2018/39370 [DOI] [Google Scholar]
- Franceus J, Desmet T (2020) Sucrose phosphorylase and related enzymes in Glycoside Hydrolase Family 13: discovery, application and engineering. Int J Mol Sci 21:2526. 10.3390/ijms21072526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceus J, Dhaene S, Decadt H, Vandepitte J, Caroen J, Van Der Eycken J, Beerens K, Desmet T (2019) Rational design of an improved transglucosylase for production of the rare sugar nigerose. Chem Commun 55:4531–4533. 10.1039/c9cc01587f [DOI] [PubMed] [Google Scholar]
- Garcia CA, Narrett JA, Gardner JG (2020) Trehalose degradation in Cellvibrio japonicus exhibits no functional redundancy and is solely dependent on the Tre37A enzyme. Appl Environ Microbiol 1–15. 10.1128/aem.01639-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia CA, Narrett JA, Gardner JG (2019) Complete genome sequences of Cellvibrio japonicus strains with improved growth when using α-diglucosides. Microbiol Resour Announc 8:3–5. 10.1128/mra.01077-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerschenson LN, Rojas AM, Fissore EN (2017) Chapter 3 - Carbohydrates. In: Galanakis CM (ed) Nutraceutical and Functional Food Components; effects of innvative processing techniques. Academic Press, pp 39–101 [Google Scholar]
- Gibson GR, Probert HM, Van Loo J, Rastall RA, Roberfroid MB (2004) Dietary modulation of the human colonic microbiota: updating the concept of prebiotics. Nutr Res Rev 17:259–275. 10.1079/nrr200479 [DOI] [PubMed] [Google Scholar]
- Gibson RP, Gloster TM, Roberts S, Warren RAJ, Storch De Gracia I, García Á, Chiara JL, Davies GJ (2007) Molecular basis for trehalase inhibition revealed by the structure of trehalase in complex with potent inhibitors. Angew Chemie - Int Ed 46:4115–4119. 10.1002/anie.200604825 [DOI] [PubMed] [Google Scholar]
- Goffin D, Wathelet B, Blecker C, Deroanne C, Malmendier Y, Paquot M (2010) Comparison of the glucooligosaccharide profiles produced from maltose by two different transglucosidases from Aspergillus niger. Biotechnol Agron Soc Environ 14:607–616 [Google Scholar]
- Gopinath SCB, Anbu P, Arshad MKM, Lakshmipriya T, Voon CH, Hashim U, Chinni SV. (2017) Biotechnological processes in microbial amylase production. Biomed Res Int 2017:. 10.1155/2017/1272193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granström TB, Takata G, Tokuda M, Izumori K (2004) Izumoring: a novel and complete strategy for bioproduction of rare sugars. J Biosci Bioeng ce Bioeng 97:89–94. 10.1263/jbb.97.89 [DOI] [PubMed] [Google Scholar]
- Guignard MS, Leitch AR, Acquisti C, Eizaguirre C, Elser JJ, Hessen DO, Jeyasingh PD, Neiman M, Richardson AE, Soltis PS, Soltis DE, Stevens CJ, Trimmer M, Weider LJ, Woodward G, Leitch IJ (2017) Impacts of nitrogen and phosphorus: From genomes to natural ecosystems and agriculture. Front Ecol Evol 5:. 10.3389/fevo.2017.00070 [DOI] [Google Scholar]
- Harrion MD, Purdue JC, Hoffman AJ (2007) Food products comprising a slowly digestible or digestion resistant carbohydrate composition. U.S. paten [Google Scholar]
- Hodoniczky J, Morris CA, Rae AL (2012a) A systematic evaluation of oral and intestinal utilization of oligosaccharides as potential sweeteners. Food Chem `132:1951–1958 [Google Scholar]
- Hodoniczky J, Morris CA, Rae AL (2012b) Oral and intestinal digestion of oligosaccharides as potential sweeteners: A systematic evaluation. Food Chem 132:1951–1958. 10.1016/j.foodchem.2011.12.031 [DOI] [Google Scholar]
- Holscher HD (2017) Dietary fiber and prebiotics and the gastrointestinal microbiota. Gut Microbes 8:172–184. 10.1080/19490976.2017.1290756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlacher R, Uhland K, Klein W, Ehrmann M, Boos W (1996) Characterization of a cytoplasmic trehalase of Escherichia coli. J Bacteriol 178:6250–6257. 10.1128/jb.178.21.6250-6257.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Ketabi A, Buchko A, Gänzle MG (2013) Metabolism of isomalto-oligosaccharides by Lactobacillus reuteri and Bifidobacteria. Lett Appl Microbiol 57:108–114. 10.1111/lam.12076 [DOI] [PubMed] [Google Scholar]
- Iordachescu M, Imai R (2008) Trehalose biosynthesis in response to abiotic stresses. J Integr Plant Biol 50:1223–1229. 10.1111/j.1744-7909.2008.00736.x [DOI] [PubMed] [Google Scholar]
- Iturriaga G, Suárez R, Nova-Franco B (2009) Trehalose metabolism: from osmoprtection to signaling. Int J Mol Sci 10:3793–3810. 10.3390/ijms10093793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain NK, Roy I (2009) Effect of trehalose on protein structure. Protein Sci 18:24–36. 10.1002/pro.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorge CD, Sampaio MM, Hreggvidsson GÓ, Kristjánson JK, Santos H (2007) A highly thermostable trehalase from the thermophilic bacterium Rhodothermus marinus. Extremophiles 11:115–122. 10.1007/s00792-006-0021-6 [DOI] [PubMed] [Google Scholar]
- Kanehisa M (2000) Post-genome informatics. Oxford University Press [Google Scholar]
- Kang MS, Okuyama M, Mori H, Kimura A (2009) The first α-1,3-glucosidase from bacterial origin belonging to glycoside hydrolase family 31. Biochimie 91:1434–1442. 10.1016/j.biochi.2009.07.018 [DOI] [PubMed] [Google Scholar]
- Kato N, Suyama S, Shirokane M, Kato M, Kobayashi T, Tsukagoshi N (2002) Novel alpha-glucosidase from Aspergillus nidulans with strong transglycosylation activity. Appl Environ Microbiol 68:1250–1256. 10.1128/AEM.68.3.1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keasling JD (2010) Manufacturing molecules through metabolic engineering. Science (80- ) 330:1355–1358. 10.7312/will90914-008 [DOI] [PubMed] [Google Scholar]
- Kendrick KE, Ensign JC (1983) Sporulation of Streptomyces griseus in submerged culture. J Bacteriol 155:357–366. 10.1128/jb.155.1.357-366.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitao S, Horiuchi T, Yoshida S, Kusakabe I, Sekine H (1994) Formation of kojibiose and nigerose by sucrose phosphorylase. Biosci Biotechnol Biochem 58:790–791. 10.1271/bbb.58.790 [DOI] [PubMed] [Google Scholar]
- Kizaki H, Hata Y, Watanabe K, Katsube Y, Suzuki Y (1993) Polypeptide folding of Bacillus cereus ATCC7064 oligo-1,6-glucosidase revealed by 3.0 a resolution X-ray analysis. J Biochem 113:646–649. 10.1093/oxfordjournals.jbchem.a124097 [DOI] [PubMed] [Google Scholar]
- Kluch M, Socha-Banasiak A, Pacześ K, Borkowska M, Czkwianianc E (2020) The role of disaccharidases in the digestion - diagnosis and significance of their deficiency in children and adults. Pol Merkur Lekarski 49:275–278 [PubMed] [Google Scholar]
- Kohmoto T, Fukui F, Takaku H, Machida Y, Arai M, Mitsuoka T (1988) Effect of isomalto-oligosaccharides on human fecal flora. Bifidobact Microflora 7:61–69. 10.12938/bifidus1982.7.2_61 [DOI] [Google Scholar]
- Konishi YY, Shindo K (1997) Production of nigerose, nigerosyl glucose, and nigerosyl maltose by Acremonium sp. S4G13. Chem Pharm Bull 61:439–442. 10.1271/bbb.61.439 [DOI] [PubMed] [Google Scholar]
- Kotrba P, Inui M, Yukawa H (2001) Bacterial phosphotransferase system (PTS) in carbohydrate uptake and control of carbon metabolism. J Biosci Bioeng 92:502–517 [DOI] [PubMed] [Google Scholar]
- Kraus M, Görl J, Timm M, Seibel J (2016) Synthesis of the rare disaccharide nigerose by structure-based design of a phosphorylase mutant with altered regioselectivity. Chem Commun 52:4625–4627. 10.1039/c6cc00934d [DOI] [PubMed] [Google Scholar]
- Kurakata Y, Uechi A, Yoshida H, Kamitori S, Sakano Y, Nishikawa A, Tonozuka T (2008) Structural insights into the substrate specificity and function of Escherichia coli K12 YgjK, a glucosidase belonging to the Glycoside Hydrolase Family 63. J Mol Biol 381:116–128 [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA (1998) Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659. 10.1016/j.physbeh.2017.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Kim JH, Kim SY, Lee JK (2008) Isomaltose production by modification of the fructose-binding site on the basis of the predicted structure of sucrose isomerase from “Protaminobacter rubrum.” Appl Environ Microbiol 74:5183–5194. 10.1128/AEM.00181-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidy C, Gousset K, Ricker J, Wolkers WF, Tsvetkova NM, Tablin F, Crowe JH (2004) Lipid phase behavior and stabilization of domains in membranes of platelets. Cell Biochem Biophys 40:123–148. 10.1385/CBB:40:2:123. [DOI] [PubMed] [Google Scholar]
- Lentze MJ (2018) The history of maltose-active disaccharidases. J Pediatr Gastroenterol Nutr 66:4–6. 10.1097/MPG.0000000000001960 [DOI] [PubMed] [Google Scholar]
- Leslie SB, Israeli E, Lighthart B, Crowe JH, Crowe LM (1995) Trehalose and sucrose protect both membranes and proteins in intact bacteria during drying. Appl Environ Microbiol 61:3592–3597. 10.1128/aem.61.10.3592-3597.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Schmidt RK, Teo B, Karplus PA, Brady JW (1997) Molecular Dynamics Studies of the Hydration of α,α-Trehalose. J Am Chem Soc 119:7851–7862. 10.1021/ja970798v [DOI] [Google Scholar]
- Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:490–495. 10.1093/nar/gkt1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Deng T, Yuan W, Deng H, Jin M (2017) Plasma metabolomic study in Chinese patients with wet age-related macular degeneration. BMC Ophthalmol 17:1–9. 10.1186/s12886-017-0555-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luyckx J, Baudouin C (2011) Trehalose: An intriguing disaccharide with potential for medical application in ophthalmology. Clin Ophthalmol 5:577–581. 10.2147/OPTH.S18827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Okuyama M, Tagami T, Kikuchi A, Klahan P, Kimura A (2019) Novel α-1,3/α-1,4-glucosidase from Aspergillus niger exhibits unique transglucosylation to generate high levels of nigerose and kojibiose. J Agric Food Chem 67:3380–3388. 10.1021/acs.jafc.8b07087 [DOI] [PubMed] [Google Scholar]
- Mangas-Sánchez J, Adlercreutz P (2015) Enzymatic enzymatic preparation of oligosaccharides by transglycosylation : A comparative study of glucosidases. "Journal Mol Catal B, Enzym 122:51–55. 10.1016/j.molcatb.2015.08.014 [DOI] [Google Scholar]
- Matsuda K, Watanabe H, Fujimoto K, Aso K (1961) Isolation of nigerose and kojibiose from dextrans. Nature 191:278. 10.1038/191278a0 [DOI] [PubMed] [Google Scholar]
- McBride MJ, Ensign JC (1987) Effects of intracellular trehalose content on Streptomyces griseus spores. J Bacteriol 169:4995–5001. 10.1128/jb.169.11.4995-5001.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath D, Lee EE, O’Colla PS (1969) The chemical synthesis of polysaccharides : Part II. (1→2)-, (1→3)-, and (1→4)-linked glucans. Carbohydr Res 11:461–465 [Google Scholar]
- Memarzadeh B (2009) Halogenated alkyl di- and trisaccharides, pharmaceutical formulations, diagnostic kits and methods of treatment. US Patent 2008/0171721 A1; 2008.
- Meyer KH, Bernfeld P (1940) Recherches sur l’amidon V. L’amylopectine. HCA; 23: [DOI] [PubMed] [Google Scholar]
- Miller DP, de Pablo JJ, Corti H (1997) Thermophysical properties of trehalose and its concentrated aqueous solutions. Pharm Res 14:578–590 [DOI] [PubMed] [Google Scholar]
- Miyazaki T, Ichikawa M, Yokoi G, Kitaoka M, Mori H, Kitano Y, Nishikawa A, Tonozuka T (2013) Structure of a bacterial glycoside hydrolase family 63 enzyme in complex with its glycosynthase product, and insights into the substrate specificity. FEBS J 280:4560–4571. 10.1111/febs.12424 [DOI] [PubMed] [Google Scholar]
- Montgomery EM, Weakley FB, Hilbert GE (1949) Isolation of 6-[α-D glucopyranosyl]-D-glucose (isomaltose) from enzymic hydrolyzates of starch. J Am Chem Soc 71:1682–1687. 10.1021/ja01173a038 [DOI] [Google Scholar]
- Mooradian AD, Smith M, Tokuda M (2017) The role of artificial and natural sweeteners in reducing the consumption of table sugar: A narrative review. Clin Nutr ESPEN 18:1–8. 10.1016/j.clnesp.2017.01.004 [DOI] [PubMed] [Google Scholar]
- Mukherjee K, Narindoshvili T, Raushel FM (2018) Discovery of a kojibiose phosphorylase in Escherichia coli K-12. Biochemistry 57:2857–2867. 10.1016/j.physbeh.2017.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murosaki S, Muroyama K, Yamamoto Y, Kusaka H, Liu T, Yoshikai Y (1999) Immunopotentiating activity of nigerooligosaccharides for the t helper 1-like immune response in mice. Biosci Biotechnol Biochem 63:373–378 [DOI] [PubMed] [Google Scholar]
- Nakada T, Maruta K, Mitsuzumi H, Kubota M, Chaen H, Sugimoto T, Kurimoti M, Tsujisaka T (1955) Purification and characterization of a novel enzyme, maltooligosyl trehalose trehalohydrolase, from Arthrobacter sp. Q36. Biosci Biotechnol Biochem 59:2215–2218 [DOI] [PubMed] [Google Scholar]
- Nakada T, Nishimoto T, Chaen H, Fukuda S (2003) Kojioligosaccharides: application of kojibiose phosphorylase on the formation of various kojioligosaccharides. In: Oligosaccharides in Food and Agriculture. American Chemical Society, pp 104–117 SE–9 [Google Scholar]
- Neta T, Takada N, Hirasawa M (2000) Low-cariogenicity of trehalose as a substrate. J Dent 28:571–576 [DOI] [PubMed] [Google Scholar]
- Newman YM, Ring SG, Colaco C (1993) The role of trehalose and other carbohydrates in biopreservation. Biotechnol Genet Eng Rev 11:263–294. 10.1080/02648725.1993.10647903 [DOI] [PubMed] [Google Scholar]
- Nihira T, Miyajima F, Chiku K, Nishimoto M, Kitaoka M, Ohtsubo K, Nakai H (2014) One pot enzymatic production of nigerose from common sugar resources employing nigerose phosphorylase. J Appl Glycosci 61:75–80. 10.5458/jag.jag.jag-2013_012 [DOI] [Google Scholar]
- Nihira T, Nakai H, Chiku K, Kitaoka M (2012) Discovery of nigerose phosphorylase from Clostridium phytofermentans. Appl Microbiol Biotechnol 93:1513–1522. 10.1007/s00253-011-3515-9 [DOI] [PubMed] [Google Scholar]
- Nishimoto T, Nakano M, Nakada T, Chaen H, Fukuda S, Sugimoto T, Kurimoto M, Tsujisaka Y, Fukada S, Sugimoto T, Kurimoto M, Tsujisaka Y (1996) Purification and properties of a novel enzyme, trehalose synthase, from Pimelobacter sp. R48. Biosci Biotechnol Biochem 60:640–644 [DOI] [PubMed] [Google Scholar]
- Niu D, Qiao J, Li P, Tian K, Liu X, Singh S, Lu F (2017) Highly efficient enzymatic preparation of isomalto-oligosaccharides from starch using an enzyme cocktail. Electron J Biotechnol 26:46–51. 10.1016/j.ejbt.2016.12.002 [DOI] [Google Scholar]
- Nobre A, Alarico S, Fernandes C, Empadinhas N, Da Costa MS (2008) A unique combination of genetic systems for the synthesis of trehalose in Rubrobacter xylanophilus: Properties of a rare actinobacterial TreT. J Bacteriol 190:7939–7946. 10.1128/JB.01055-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobre A, Alarico S, Maranha A, Mendes V, Empadinhas N (2014) The molecular biology of mycobacterial trehalose in the quest for advanced tuberculosis therapies. Microbiol (United Kingdom) 160:1547–1570. 10.1099/mic.0.075895-0 [DOI] [PubMed] [Google Scholar]
- Ogawa S, Ashiura M, Uchida C (1998) Synthesis of α-glucosidase inhibitors: Kojibiose-type pseudodisaccharides and a related pseudotrisaccharide. Carbohydr Res 307:83–95. 10.1016/S0008-6215(98)00045-7 [DOI] [PubMed] [Google Scholar]
- Ohtake S, Wang YJ (2011) Trehalose: current use and future applications. J Pharm Sci 101:2020–2053. 10.1002/jps [DOI] [PubMed] [Google Scholar]
- Okada S, Yamamoto T, Watanabe H, Nishimoto T, Chaen H, Fukuda S, Wakagi T, Fushinobu S (2014) Structural and mutational analysis of substrate recognition in kojibiose phosphorylase. FEBS J 281:778–786. 10.1111/febs.12622 [DOI] [PubMed] [Google Scholar]
- Paul MJ, Primavesi LF, Jhurreea D, Zhang Y (2008) Trehalose metabolism and signaling. Annu Rev Plant Biol 59:417–441. 10.1146/annurev.arplant.59.032607.092945 [DOI] [PubMed] [Google Scholar]
- Peat S, Turvey JR, Evans JM (1957) Isolation of nigerose from floridean starch. Nature 179:261. 10.1038/179261a0 [DOI] [PubMed] [Google Scholar]
- Peralta-Yahya PP, Zhang F, Del Cardayre SB, Keasling JD (2012) Microbial engineering for the production of advanced biofuels. Nature 488:320–328. 10.1038/nature11478 [DOI] [PubMed] [Google Scholar]
- Pfister B, Zeeman SC (2016) Formation of starch in plant cells. Cell Mol Life Sci 73:2781–2807. 10.1007/s00018-016-2250-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phung OJ, Scholle JM, Talwar M, Coleman CI (2010) Effect of noninsulin antidiabetic drugs added to metformin therapy on glycemic control, weight gain, and hypoglycemia in type 2 diabetes. JAMA - J Am Med Assoc 303:1410–1418. 10.1001/jama.2010.405 [DOI] [PubMed] [Google Scholar]
- Pokusaeva K, O’Connell-Motherway M, Zomer A, Fitzgerald GF, Van Sinderen D (2009) Characterization of two novel α-glucosidases from Bifidobacterium breve UCC2003. Appl Environ Microbiol 75:1135–1143. 10.1128/AEM.02391-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchart V (2015) Glycoside phosphorylases: Structure, catalytic properties and biotechnological potential. Biotechnol Adv 33:261–276. 10.1016/j.biotechadv.2015.02.002 [DOI] [PubMed] [Google Scholar]
- Qu Q, Lee SJ, Boos W (2004) TreT, a novel trehalose glycosyltransferring synthase of the hyperthermophilic archaeon Thermococcus litoralis. J Biol Chem 279:47890–47897. 10.1074/jbc.M404955200 [DOI] [PubMed] [Google Scholar]
- Robyt JF (2008) Mechanisms involved in the biosynthesis of polysaccharides. Biologia (Bratisl) 63:980–988. 10.2478/s11756-008-0168-y [DOI] [Google Scholar]
- Roos Y (1993) Melting and glass transitions of low molecular weight carbohydrates. Carbohydr Res 238:39–48. 10.1016/0008-6215(93)87004-C [DOI] [Google Scholar]
- Rose DR, Chaudet MM, Jones K (2018) Structural studies of the intestinal α-glucosidases, maltase-glucoamylase and sucrase-isomaltase. J Pediatr Gastroenterol Nutr 66:S11–S13. 10.1097/MPG.0000000000001953 [DOI] [PubMed] [Google Scholar]
- Ruhal R, Kataria R, Choudhury B (2013) Trends in bacterial trehalose metabolism and significant nodes of metabolic pathway in the direction of trehalose accumulation. Microb Biotechnol 6:493–502. 10.1111/1751-7915.12029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MHJ (2015) The bacterial phosphotransferase system: new frontiers 50 years after its discovery. J Mol Microbiol Biotecnnol 25:73–78. 10.1159/000381215.The [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi M (2020) Diverse and common features of trehalases and their contributions to microbial trehalose metabolism. Appl Microbiol Biotechnol 104:1837–1847. 10.1007/s00253-019-10339-7 [DOI] [PubMed] [Google Scholar]
- Sanz ML, Gibson GR, Rastall RA (2005) Influence of disaccharide structure on prebiotic selectivity in vitro. J Agric Food Chem 53:5192–5199. 10.1021/jf050276w [DOI] [PubMed] [Google Scholar]
- Schievano E, Tonoli M, Rastrelli F (2017) NMR quantification of carbohydrates in complex mixtures. A challenge on honey. Anal Chem 89:13405–13414. 10.1021/acs.analchem.7b03656 [DOI] [PubMed] [Google Scholar]
- Schönert S, Buder T, Dahl MK (1998) Identification and enzymatic characterization of the maltose-inducible α-glucosidase MalL (sucrase-isomaltase-maltase) of Bacillus subtilis. J Bacteriol 180:2574–2578. 10.1128/jb.180.9.2574-2578.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönert S, Buder T, Dahl MK (1999) Properties of maltose-inducible α-glucosidase MalL (sucrase-isomaltase-maltase) in Bacillus subtilis: Evidence for its contribution to maltodextrin utilization. Res Microbiol 150:167–177. 10.1016/S0923-2508(99)80033-3 [DOI] [PubMed] [Google Scholar]
- Song X, Tang S, Jiang L, Zhu L, Huang H (2016) Integrated biocatalytic process for trehalose production and separation from maltose. Ind Eng Chem Res 55:10566–10575. 10.1021/acs.iecr.6b02276 [DOI] [Google Scholar]
- de Souza PM (2010) Application of microbial amylase in industry. Brazillian J Microbiol 41:850–861. 10.1590/S1517-83822010000400004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprogøe D, van den Broek LAM, Mirza O, Kastrup JS, Voragen AGJ, Gajhede M, Skov LK (2004) Crystal structure of sucrose phosphorylase from Bifidobacterium adolescentis. Biochemistry 43:1156–1162. 10.1021/bi0356395 [DOI] [PubMed] [Google Scholar]
- Stam MR, Danchin EGJ, Rancurel C, Coutinho PM, Henrissat B (2006) Dividing the large glycoside hydrolase family 13 into subfamilies: Towards improved functional annotations of α-amylase-related proteins. Protein Eng Des Sel 19:555–562. 10.1093/protein/gzl044 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Tomura Y (1986) Purification and characterization of Bacillus coagulans oligo- 1,6- glucosidase. Eur J Biochem 158:77–83. 10.1111/j.1432-1033.1986.tb09723.x [DOI] [PubMed] [Google Scholar]
- Tonozuka T, Uechi A, Mizuno M, Ichikawa K, Nishikawa A, Sakano Y (2004) Crystallization and preliminary X-ray analysis of Escherichia coli K12 YgjK protein, a member of glycosyl hydrolase family 63. Acta Crystallogr Sect D Biol Crystallogr 60:1284–1285. 10.1107/S0907444904009631 [DOI] [PubMed] [Google Scholar]
- Tsai YL, Lin TL, Chang CJ, Wu TR, Lai WF, Lu CC, Lai HC (2019) Probiotics, prebiotics and amelioration of diseases. J Biomed Sci 26:1–8. 10.1186/s12929-018-0493-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya HM, Montgomery EM, Corman J (1949) Hydrolysis of the α-1,6-glucosidic linkage in isomaltose by culture filtrate of Aspergillus Niger NRRL 330. J Am Chem Soc 71:3265. 10.1021/ja01177a537 [DOI] [Google Scholar]
- Uhland K, Mondigler M, Spiess C, Prinz W, Ehrmann M (2000) Determinants of translocation and folding of TreF, a trehalase of Escherichia coli. J Biol Chem 275:23439–23445. 10.1074/jbc.M002793200 [DOI] [PubMed] [Google Scholar]
- Van Den Broek LAM, Struijs K, Verdoes JC, Beldman G, Voragen AGJ (2003) Cloning and characterization of two α-glucosidases from Bifidobacterium adolescentis DSM20083. Appl Microbiol Biotechnol 61:55–60. 10.1007/s00253-002-1179-1 [DOI] [PubMed] [Google Scholar]
- Van Der Maarel MJEC, Van Der Veen B, Uitdehaag JCM, Leemhuis H, Dijkhuizen L (2002) Properties and applications of starch-converting enzymes of the α-amylase family. J Biotechnol 94:137–155. 10.1016/S0168-1656(01)00407-2 [DOI] [PubMed] [Google Scholar]
- Verhaeghe T, De Winter K, Berland M, De Vreese R, D’Hooghe M, Offmann B, Desmet T (2016) Converting bulk sugars into prebiotics: Semi-rational design of a transglucosylase with controlled selectivity. Chem Commun 52:3687–3689. 10.1039/c5cc09940d [DOI] [PubMed] [Google Scholar]
- Viktor MJ, Rose SH, Van Zyl WH, Viljoen-Bloom M (2013) Raw starch conversion by Saccharomyces cerevisiae expressing Aspergillus tubingensis amylases. Biotechnol Biofuels 6:. 10.1186/1754-6834-6-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitek V, Vitek K, Lin H-C, Cowley RA (1972) Isomaltose excretion in health, severe injury, and disease. Metabolism 21:701–712 [DOI] [PubMed] [Google Scholar]
- Wannet WJB, Op den Camp HJM, Wisselink HW, van der Drift C, Van Griensven LJLD, Vogels GD (1998) Purification and characterization of trehalose phosphorylase from the commercial mushroom Agaricus bisporus. Biochem Biophys Acta 1425:177–188 [DOI] [PubMed] [Google Scholar]
- Watanabe K, Miyake K, Suzuki Y (2001) Identification of catalytic and substrate-binding site residues in Bacillus cereus ATCC7064 oligo-1,6-glucosidase. Biosci Biotechnol Biochem 65:2058–2064 [DOI] [PubMed] [Google Scholar]
- Watanabe T, Aso K (1960) Studies on Honey II. isolation of kojibiose, nigerose, maltose, and isomaltose from honey. Tohuku J Agric Res 11:109–115 [Google Scholar]
- Watanabe T, Aso K (1959) Isolation of Kojibiose from Honey. Nature 183:1740. [DOI] [PubMed] [Google Scholar]
- Wolfrom ML, Georges LW, Miller IL (1949) Crystalline Derivatives of Isomaltose. J Am Chem Soc 71:125–127. 10.1021/ja01169a034 [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Horikoshi K (1990) Nucleotide sequence of alkalophilic Bacillus oligo-1,6-glucosidase gene and the properties of the gene product by Escherichia coli HB 101. Denpun Kagaku 37:137–144 [Google Scholar]
- Yamamoto T, Maruta K, Mukai K, Yamashita H, Nishimoto T, Kubota M, Fukuda S, Kurimoto M, Tsujisaka Y (2004) Cloning and sequencing of kojibiose phosphorylase gene from Thermoanaerobacter brockii ATCC35047. J Biosci Bioeng 98:99–106. 10.1263/jbb.98.99 [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Nishio-Kosaka M, Izawa S, Aga H, Nishimoto T, Chaen H, Fukuda S (2011) Enzymatic properties of recombinant kojibiose phosphorylase from Caldicellulosiruptor saccharolyticus ATCC43494. Biosci Biotechnol Biochem 75:1208–1210. 10.1271/bbb.110116 [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Yamashita H, Mukai K, Watanabe H, Kubota M, Chaen H, Fukuda S (2006) Construction and characterization of chimeric enzymes of kojibiose phosphorylase and trehalose phosphorylase from Thermoanaerobacter brockii. Carbohydr Res 341:2350–2359. 10.1016/j.carres.2006.06.024 [DOI] [PubMed] [Google Scholar]
- Yen CH, Tseng YH, Kuo YW, Lee MC, Chen HL (2011) Long-term supplementation of isomalto-oligosaccharides improved colonic microflora profile, bowel function, and blood cholesterol levels in constipated elderly people-A placebo-controlled, diet-controlled trial. Nutrition 27:445–450. 10.1016/j.nut.2010.05.012 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Sakamoto T (2009) Water-stress induced trehalose accumulation and control of trehalase in the cyanobacterium Nostoc punctiforme IAM M-15. J Gen Appl Microbiol 55:135–145. 10.2323/jgam.55.135 [DOI] [PubMed] [Google Scholar]
- Yutin N, Galperin MY (2013) A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ Microbiol 15:2631–2641. 10.1111/1462-2920.12173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonneveld BJM (1972) A new type of enzyme, an exo-splitting α-1,3 glucanase from non-induced cultures of Aspergillus nidulans. Biochem Biophys Acta 258:541–547 [DOI] [PubMed] [Google Scholar]