ABSTRACT
Both theory and experimental data from pathogens suggest that the production of transmission stages should be strongly associated with virulence, but the genetic bases of parasite transmission/virulence traits are poorly understood. The blood fluke Schistosoma mansoni shows extensive variation in numbers of cercariae larvae shed and in their virulence to infected snail hosts, consistent with expected trade-offs between parasite transmission and virulence. We crossed schistosomes from two populations that differ 8-fold in cercarial shedding and in their virulence to Biomphalaria glabrata snail hosts, and determined four-week cercarial shedding profiles in F0 parents, F1 parents and 376 F2 progeny from two independent crosses in inbred snails. Sequencing and linkage analysis revealed that cercarial production is polygenic and controlled by five QTLs (i.e. Quantitative Trait Loci). These QTLs act additively, explaining 28.56% of the phenotypic variation. These results demonstrate that the genetic architecture of key traits relevant to schistosome ecology can be dissected using classical linkage mapping approaches.
KEYWORDS: Schistosoma parasite, biomphalaria snail host, virulence, transmission, life history traits, genetic crosses, quantitative trait loci (qtl)
INTRODUCTION:
Parasites face a central problem: how to maximize transmission to the next host. This has driven the evolution of a wide variety of lifecycle features to facilitate parasite transmission [1,2]. However, perhaps the most common transmission strategy is to produce vast numbers of infective stages [3, p. 20]. This brute force approach exploits host resources for parasite growth and reproduction, while infective stages may also cause damage as they exit the host to reach the environment [4, p. 200; 5]. Hence, there is a strong expectation that transmission stage production will result in collateral damage to the host, but also that high levels of host mortality will constrain evolution of very high levels of parasite virulence. There is a large body of theoretical work examining this relationship between transmission stage production and virulence evolution [6–8] and empirical studies provide compelling, but not universal support, for this model [9,10]. However, there is limited understanding of the genes and genetic architecture underlying transmission stage production and virulence on which selection can act.
Schistosoma mansoni parasites provide a useful system for examining the genetic basis of transmission/virulence related traits. These parasitic organisms are well suited for genetic studies because (i) the complete lifecycle can be maintained in the laboratory using rodent definitive hosts and freshwater snail intermediate hosts, (ii) parasites have separate sexes which simplifies staging of efficient genetic crosses in the laboratory, (iii) thousands of progeny are produced which provides good statistical power, and (iv) experimental work over the last 75 years has revealed heritable genetic variation in multiple biomedically important traits [11] such as drug resistance [12–16], chronobiology [17–19], host specificity [20–24], and virulence [25–27]. Furthermore, the Schistosoma mansoni genome is fully sequenced and assembled [28,29], and a growing molecular toolkit including molecular sexing tools [30,31], RNAi [32], transfection [33,34], CRISPR [35–37] and a suite of cell biology tools [38–41] improves our ability to link phenotype with genotype. Furthermore, we can also control the genetics of the snail host by generating inbred snail lines. B. glabrata is hermaphrodite, so inbred snail lines can be developed by isolating individual snails and serial inbreeding [42].
In addition of being a tractable model for genetic studies, schistosomes are also important human parasites. Three major schistosome parasite species (i.e. S. mansoni, S. haematobium and S. japonicum) infect over 200 million people in 78 countries [43,44]. These parasites have a complex lifecycle, involving a freshwater snail (intermediate host) and a mammal (definitive host). When parasite eggs are expelled with mammal feces or urine in freshwater, miracidia larvae hatch and actively search for its snail vector. Larvae penetrate the snail head-foot, differentiate into sporocysts that then asexually proliferate to generate daughter sporocysts. These actively consume snail tissue (hepatopancreas and ovotestis), castrating the host snail, and parasite sporocysts can comprise up to 60% of the tissue within infected B. glabrata snails [5]. The daughter sporocysts release cercariae, the mammal-infective larval stage of the parasite. Hundreds to thousands of these motile cercariae exit through the snail body wall and are released into freshwater. Exit through the body wall results in leakage of hemolymph and damage to the snail. This quantitative transmission and virulence related trait – numbers of cercariae produced from the intermediate snail host – is the focus of this paper.
There is strong evidence from laboratory selection experiments that the amount of cercariae produced is heritable: in just three generations, parasites selected for low or high numbers of cercariae showed rapid divergence in this phenotype [25,26]. We have described distinctive life-history strategies in laboratory schistosome populations. In two parasite populations originating from Brazil we observed: i) a “boom-bust” strategy characterized by high transmission measured in term of cercarial production (the total number of larvae released from the intermediate snail host, starting at 4 weeks post-infection) and high virulence to the snail intermediate host resulting in short duration infections, compared with ii) a “slow and steady” strategy characterized by low transmission (few cercariae released), resulting in low virulence to the snail host and a long duration of infection [5]. The high shedding genotype develops larger sporocyst stages than the low shedding genotype, although this is insufficient to explain the difference in cercarial shedding from these two parasites. These patterns are observed when using the same inbred snail line, ruling out an impact of host genetics. Our central goal was to decipher the genetic architecture of a transmission/virulence-related trait in a biomedically important helminth parasite (S. mansoni), to determine whether cercarial shedding is under monogenic or polygenic control, and ultimately to understand the cellular pathways on which selection acts to modulate this trait.
Linkage mapping has previously been used to examine a monogenic drug resistance trait (oxamniquine resistance) in S. mansoni [16,45]. Here we extend this approach to examine a life-history trait: we conducted reciprocal genetic crosses between S. mansoni individuals from two laboratory parasite populations originating from Brazil that show dramatic differences in numbers of cercariae shed (Figure 1). Using classical linkage mapping, we showed that cercariae production in S. mansoni parasite is polygenic, with additive variation at 5 different QTLs (i.e. Quantitative Trait Loci) explaining 28.56% of the variation in cercarial production.
Figure 1.
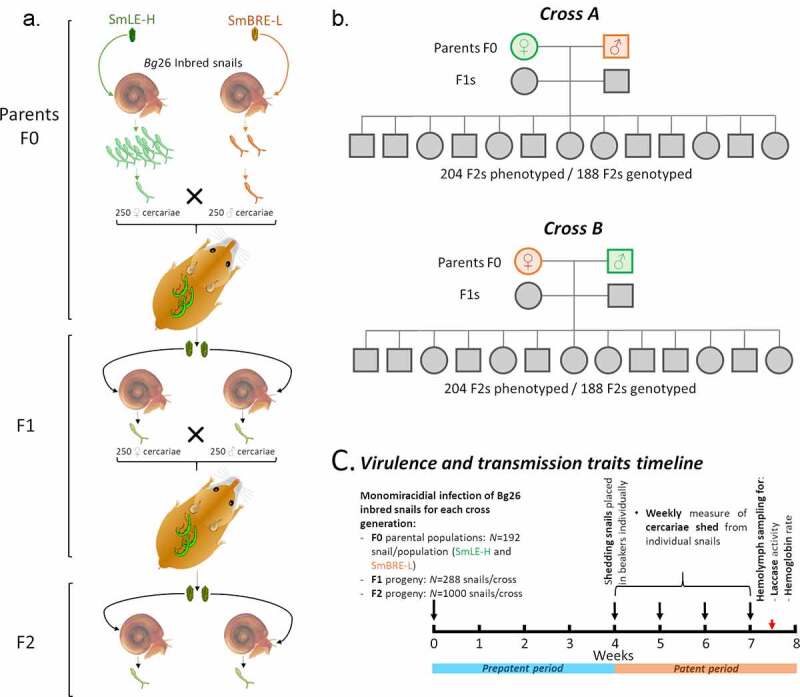
Outline of the genetic cross experimental design. (a-b) We performed two independent reciprocal genetic crosses (crosses A and B, to account for potential sex specific effect(s)) between single genotypes of SmLE-H and SmBRE-L Schistosoma mansoni parasite populations. These two populations exhibit striking differences in terms of transmission stage production (number of cercariae produced). (c) For each parasite generation (F0 parental populations, F1 and F2 progeny), we exposed individual Biomphalaria glabrata Bg26 inbred snail to single miracidium from either the SmLE-H or SmBRE-L populations for F0 (N = 192/population), or the F1 progeny (N = 288/cross), or the F2 progeny (N = 1000/cross). For each infected snail and each generation of S. mansoni parasites, we measured transmission stage production during 4 weeks of the patent period (week 4 to 7 post-infection). We also evaluated the virulence of these generations of crossed parasites by measuring the total laccase-like activity as well as the hemoglobin rate in infected snail hemolymph samples
MATERIALS & METHODS
Ethics statement
This study was performed in accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee of Texas Biomedical Research Institute (permit number: 1419-MA).
Biomphalaria glabrata snails and schistosoma mansoni parasites
Uninfected inbred albino Biomphalaria glabrata snails [line Bg26 derived from 13–16-R1 line; 42] were reared in 10-gallon aquaria containing aerated freshwater at 26–28°C on a 12 L-12D photocycle and fed ad libitum on green leaf lettuce. All snails used in this study had a shell diameter between 8 and 10 mm, as snail size can influence cercarial production [46,47]. For all the experiments we used inbred snails to minimize the impact of snail host genetic background on the parasite life history traits [5].
The SmLE-H S. mansoni population (high shedder, H) was originally obtained from an infected patient in Belo Horizonte (Minas Gerais, Brazil) in 1965 and has since been maintained in laboratory [48], using B. glabrata NMRI and Bg26 populations as intermediate hosts and Syrian golden hamsters (Mesocricetus auratus) as definitive hosts. The SmBRE-L S. mansoni population (low shedder, L) was sampled in 1975 from Recife (East Brazil) [24] and has been maintained in the laboratory in its sympatric albino Brazilian snail host (B. glabrata BgBRE) using hamsters or mice as the definitive host.
Genetic crosses between high shedder and low shedder S. mansoni parasites
Schistosomes have separate sexes which simplifies crosses [16]. To identify QTL(s) controlling the number of cercariae produced by schistosome parasites, we conducted crosses between parasites from our high shedder (SmLE-H) and low shedder (SmBRE-L) populations [5]. We performed reciprocal genetic crosses (cross A: male SmBRE-L x female SmLE-H; cross B: female SmBRE-L x male SmLE-H). Conducting two independent crosses allowed us i) to replicate our cross experiment and ii) to test for the potential influence of sex chromosomes on the transmission phenotype (Figure 1).
The design of the crosses is summarized in Figure 1. To obtain the parental (F0) parasite generation, we exposed individual snails (192 per parasite populations) to single miracidia in 24 well-plates overnight. Exposed snails were then maintained in trays (48 per tray) for 4 weeks as described in 5, in a 26–28°C temperature controlled room with 12 L-12 D photocycle. At four weeks post-exposure, we placed each snail in a well of a 24 well-plate in 1 mL freshwater under artificial light for 2 h to induce cercarial shedding. Shedding was conducted late morning to early afternoon every week from week 4 to week 7 post-infection. All the snail infection experiments and cercarial shedding experiments were conducted in a temperature controlled room (26–28°C). To track cercarial production of individual snails over the 4 weeks of the patent period (4–7 weeks post-infection (PI)), we isolated each infected snail in a uniquely labeled 100 mL glass beaker filled with ~50 mL freshwater at the first shedding, fed them ad libitum with fresh lettuce and kept them in the dark in the 26–28°C temperature controlled room. We determine parasite gender by PCR [30] of clonal cercariae collected from each infected snail in week 4 for the two schistosome populations (i.e. SmLE-H and SmBRE-L). One female and one male genotype, each derived from one infected snail, were randomly selected for producing the next generation. At week 5, cercarial shedding was induced and 2 female golden Syrian hamsters per cross were exposed to 250 cercariae of the female genotype and 250 cercariae of the male genotype. Cercariae from both genders were counted and sampled under a microscope, then transferred into a glass jar containing freshwater. Finally, a hamster is place into the jar (2 h) and is infected though contact with inoculated water. For cross A, the male genotype was SmBRE-L and the female genotype was SmLE-H. For cross B, the male genotype was SmLE-H and the female genotype was SmBRE-L (Figure 1). After 45 days, we euthanized and perfused the hamsters to recover the F0 adult male and female worms for DNA extraction and sequencing. We also collected the livers containing the F1 eggs.
We applied the same procedures for the F1 generation for both crosses (i.e. A and B). We hatched F1 miracidia from eggs recovered from hamster livers following 5. For each cross, we exposed individual Bg26 snails (288 per cross) to single F1 miracidia. At week 4, we isolated infected snails in individual beakers for tracking individual shedding. We collected cercariae of the first shedding for parasite gender determination. At week 5, we infected 4 female hamsters per cross with 250 cercariae of the females and the male genotypes of the selected F1s. We euthanized and perfused hamsters 45 days post infection to recover F1 adult worms and collected livers containing the F2 eggs.
After hatching F2 miracidia from each cross (A and B) from eggs recovered from hamster livers, we exposed 2,000 individual Bg26 snails (1,000 per cross) to single miracidia. After 4 weeks, we isolated the infected snails in individual beakers as above to track individual shedding. For each snail, we collected all the cercariae produced from the first or the second shedding in individual microtubes for further DNA extractions.
Measurement of S. mansoni cercarial production and snail response to infection
We measured larval output (i.e. cercarial production) of F0 (N = 46 SmLE-H and N = 48 SmBRE-L), F1 (N = 102 F1A and N = 110 F1B) and F2 (N = 204 F2A and N = 204 F2B) parasite populations. We used the same inbred snail population for all infections to minimize the impact of host snail genetic variation (Figure 1). All snails were infected with single miracidia to allow examination of cercarial shedding from single parasite genotypes. We also measured the impact of these parasitic infections on the snail host by quantifying snail physiological responses (laccase-like activity and hemoglobin rate in the hemolymph) at end point (Figure 1). The phenotypic datasets are available on Zenodo (DOI: 10.5281/zenodo.4383248).
Snail physiological response to parasitic infection
We placed infected snails in 1 mL freshwater in a 24 well plate under artificial light (as described above) every week for 4 weeks (week 4 to 7 post-exposure). After replacing the snail in its beaker, we sampled three 10 (for the high shedder parasites) or 100 µL (for the low shedder parasites) aliquots from each well, added 20 µL of 20X normal saline and counted the immobilized cercariae under a microscope. We multiplied the mean of the triplicated measurement by the dilution factor (100 or 10) to determine the number of cercariae produced by each infected snail. We assessed cercarial production every week from week 4 to 7 post-exposure in snails infected with F0 (SmLE-H and SmBRE-L), F1 and F2 parasites for each cross (A and B). We use the average number of cercariae produced over this 4 week period for each F2 progeny as the phenotype for our linkage analysis.
Snail physiological response to parasitic infection
During week 7, 3 days after the last cercarial shedding, we collected hemolymph [49] from all surviving snails infected with F0 parental populations (i.e. SmLE-H and SmBRE-L), F1s and F2s. We measured both laccase-like activity [49] and the hemoglobin rates in the hemolymph of each infected snail [5].
Estimation of the minimum number of loci influencing transmission stage production
Castle and Wright proposed an estimate ne of the minimum effective number of genetic factors explaining trait segregation in crosses between two lines based on the phenotypic mean and variance [50,51], where P is the phenotypic mean of the parents (P1 for SmLE-H and P2 for SmBRE-L), Var(F2) is the phenotypic variance of the F2 population progeny and Var(F1) is the phenotypic variance of the F1 population. We calculated ne for each cross independently.
Whole genome and exome sequencing of S. mansoni
We sequenced genomes from F0 parent worms, and exomes from F1 worms used to generate the F2 and from 188 F2s cercariae for both cross A and cross B. We sequenced exomes from F1 and F2 progeny, to reduce costs, and because sparse genotyping data is sufficient to determine the segregation of different genome regions in the progeny and identify QTLs. We sequenced complete genomes from the parents, because this allows us to determine sequence variation within QTL regions, and to identify putative causal SNPs and copy number variants.
gDNA extraction
We extracted gDNA from F0 (For cross A: 73 SmBRE-L males and 70 SmLE-H females; For cross B: 136 SmLE-H males and 104 SmBRE-L females) and F1 (F1A: 228 males and 238 females; F1B: 207 males and 198 females) worms and from F2 cercariae (204 per cross) using the Blood and Tissue kit (Qiagen), following the manufacturer’s protocol, with minor modifications. We homogenized worms in DNA extraction kit lysis buffer using sterile micro pestles. For F2 cercariae, frozen samples were thaw at 4°C, centrifuged 5 minutes at x300 g to pellet the cercariae. We removed the water supernatant, added lysis buffer to the pellet, and vortexed to homogenize. We incubated at 56°C worm and cercariae samples for 2 and 1 hour, respectively. We recovered gDNA in 200 µL of elution buffer. We quantified the worm samples gDNA using the Qubit dsDNA HS Assay Kit (Invitrogen) while F2 cercarial DNA was directly whole genome amplified to provide sufficient DNA for exome capture.
Whole genome amplification (WGA) of F2 cercarial samples
We performed WGA on each F2 gDNA cercarial sample using the Illustra GenomiPhi V2 DNA Amplification kit (GE Healthcare Life Sciences, USA) following Le Clec'h et al. [52]. We quantified the WGA DNA using the Qubit dsDNA HS Assay Kit (Invitrogen).
Whole genome and exome library preparation and sequencing
We prepared whole genome libraries of F0s using the KAPA HyperPlus kit (KAPA Biosystem) according to the manufacturer’s protocol. For each F0 library, we sheared 500 ng of gDNA by adaptive focused acoustics (Duty factor: 10%; Peak Incident Power: 175; Cycles per Burst: 200; Duration: 180 seconds) in AFA tubes, using a Covaris S220 instrument with SonoLab software version 7 (Covaris, Inc., USA), to recover fragmented DNA between 150–200 bp. We used 6 PCR cycles for post-ligation library amplification.
We captured F1 and F2 (188 per cross) S. mansoni exomes using the SureSelectXT2 Target Enrichment System (Agilent). The design of the custom baits used to capture the S. mansoni exome (SureSelect design ID: S0398493) is described in Chevalier et al. [45], and exome capture methodology follows Le Clec'h et al. [52].
We sequenced the libraries on a HiSeq 2500 sequencer (Illumina) using 100 bp pair-end reads. On each sequencing lane, we either pooled 32 exome capture libraries or two whole genome libraries. Raw sequence data are available at the NCBI Sequence Read Archive under accession numbers PRJNA667697.
Bioinformatic analysis
Jupyter notebook and scripts used for processing the sequencing data and mapping QTLs are available on Zenodo (DOI: 10.5281/zenodo.4798591).
Sequence analysis and variant calling
We aligned the sequencing data against the S. mansoni reference genome (schistosoma_mansoni.PRJEA36577.WBPS14) using BWA (v0.7.17) [53] and SAMtools (v1.10) [54]. We used GATK (v4.1.8) [55,56] to mark PCR duplicates and recalibrate base scores. We used the HaplotypeCaller module of GATK to call variants (SNP/indel) and the GenotypeGVCFs module to perform a joint genotyping on each chromosome or unassambled scaffolds. We merged VCF files using the MergeVcfs module. We annotated the variants using snpEff (v.4.3.1 t) [57]. All these steps were automated using Snakemake (v5.14.0) [58].
Linkage analysis
Sexual determination in schistosomes relies on Z and W chromosomes: females are ZW while males are ZZ. In parasite stages exhibiting no sexual dimorphism (i.e. miracidium and cercaria), we can determine sex by PCR using sex specific markers [30]. We took advantage of the sequencing data to perform in silico sexing of F2 cercarial progeny by comparing the read depth along the Z chromosome. The Z chromosome carries a Z-linked region (located between 11 and 44 Mb) which does not recombine with the W chromosome unlike the rest of the Z chromosome (pseudo-autosomal region). While the Z-linked region in males (ZZ) has the same read depth as the pseudo-autosomal region, the Z-linked region in females (ZW) will have half of the read depth of the pseudo-autosomal region (Supplementary Figure 1). We determined the read depth ratio between the Z-linked and pseudo-autosomal regions: a ratio around 1 indicates a male (ZZ) while a ratio of 0.5 corresponds to a female (ZW). We computed the read depth at each base of chromosome Z using SAMtools and BEDtools (v2.29.0) [59]. We used R to first smooth the read depth data which may show local high variation using the runmed function on 101 contiguous sites, then we kept only site showing a read depth of more than 5 to finally compute the ratio on Z-linked over Z pseudo-autosomal read depth. We validated this approach by comparing molecular and in silico sexing of F2s cercariae.
Linkage analysis
We reduced the VCF file using the SelectVariants module of GATK to include sites called in at least half of the samples. The QTL analysis was performed with R (v3.5.1) [60]. We loaded the VCF file using vcfR (v1.10.0) [61], filtered out sites with genotype quality (GQ) less than 30, read depth (DP) less than 10 and with more than 20% of missing data. We used F0 genome sequences to obtain the full genetic information from the parents of the crosses. We selected alternatively fixed variants between parents (i.e., fully informative variants) and retained Mendelian inherited variants only using F1 sequences and converted the genotype data into an R/qtl compatible format. We then ran the scanone function of R/qtl (v1.46.2) [62] on all the recombinant F2s to map QTL in each cross or in the combined crosses (to increase the power of our statistical analysis and linkage mapping) using the expectation–maximization (EM) algorithm. We performed 1,000 permutations for each test.
Having identified QTLs, we selected a subset of around 400 markers total (number of markers per chromosome being proportional to the chromosome size) from the combined crosses and tested QTL interaction using the scantwo function from the same package with 1,000 permutations. We used a subset of markers because this function is computationally intensive. We tested the allelic dominance on the combined crossed using a custom R code.
Candidate gene prioritization
To identify candidate genes involved in cercarial production, we first reduced the VCF file to sites within QTL regions identified previously. We loaded the VCF file in R using vcfR and refined the QTL regions by determining the chromosomal positions corresponding to a drop of 1.8 LOD from the highest LOD score of each QTL. We then retained only genotypes from the cross parents and filtered out sites with a GQ lower than 30 and a DP lower than 6. We used a custom score (genoscore) to objectively prioritize candidate genes in each QTL region:
The genoscore is computed either by gene or gene’s coding sequence (CDS). For each site i of a gene or CDS, we first computed a genotype score (GS) for each cross A and B by associating a score of 20 if genotypes were alternatively fixed in each cross parent or a score of 5 if fixed in one parent and unknown in the other (any other genotype combination received 0) and an impact score (IS) from the snpEff annotation (low impact or modifier mutation = 1, moderate impact = 5, high impact = 10). We summed the results for the n sites present in the gene or CDS. We weighed this sum using the length l of the gene or the CDS, and the square root of the distance d between the position of the highest LOD score (peak) and the start of the gene or the CDS. This was finally modulated by an expression factor e of 1 if the gene was expressed in mature sporocysts from shedding snails [63] or in cercariae [29] or −1 otherwise. Jupyter notebook and scripts used to obtain gene expression are available on Zenodo (DOI: 10.5281/zenodo.4741274).
Gene annotations
To complete the gene annotation of the GFF file, we ran the HHsearch tool from the HH-suite (v3.3.0–0) [64] on each predicted protein sequences. This method relies on generating hidden Markov models (HMM) for a given sequence and compares HMM-HMM alignments. We compared our sequences to three databases (pdb70, scop70 and pfam) and selected the best match if the probability generated by HHsearch was at least of 50%. Gene identifications from the GFF file and the HHsearch analysis were combined in a table and used during the analysis of gene candidates. Jupyter notebook and scripts used for this analysis are available on Zenodo (DOI: 10.5281/zenodo.4741265).
Statistical analysis
All statistical analyzes and graphs were performed using R software (v3.5.1) [60]. When data were not normally distributed (Shapiro test, p < 0.05), we compared results with non-parametric Kruskal-Wallis test followed by pairwise Wilcoxon-Mann-Whitney post-hoc test or a simple pairwise comparison Wilcoxon-Mann-Whitney test. When data followed a normal distribution, we used one–way ANOVA or a pairwise comparison Welsh t-test. We performed correlation analysis using Pearson’s correlation test. The confidence interval of significance was set to 95% and p-values less than 0.05 were considered significant.
Results
Variation in transmission stage production in S. mansoni crosses
We conducted two three-generation (F0 to F2) genetic crosses between high (H) shedding parasite genotypes SmLE-H and low (L) shedding genotypes SmBRE-L. We paired a male genotype SmBRE-L with a female genotype SmLE-H (cross A) and a female genotype SmBRE-L with a male genotype SmLE-H (cross B) to allow examination of sex specific inheritance (Figure 1). The two S. mansoni populations, SmLE-H and SmBRE-L, are both originated from Brazil, and show striking difference in cercarial production [5]. We used the same inbred B. glabrata population (Bg26) in these experiments to minimize the impact of the host genetics, because cercarial shedding can be influenced by snail genotype [65]. Furthermore, we infected snails with single S. mansoni miracidia, so quantitative measure of cercarial production by infected snails can be related to a single schistosome genotype.
Using weekly measures of cercarial shedding over 4 weeks (i.e from week 4 to 7 post-exposure), we observed that SmLE-H population shed 8-fold more cercariae than SmBRE-L (mean (±se) cercariae per shedding: SmBRE-L: 284 ± 19 vs SmLE-H 2352 ± 113; Wilcoxon test, p < 2.2 × 10−16, Figure 2a F0 (average cercariae produced) and 2b F0 (Number of cercariae produced/week); 5). However, the infectivity of these two parasite populations in Bg26 snails was identical. We observed that 26% (46/177) of snails exposed with one SmLE-H miracidia were infected 4 weeks post-exposure (PE), while 26% (48/180) of snails exposed to one SmBRE-L miracidia were infected 4 weeks PE.
Figure 2.
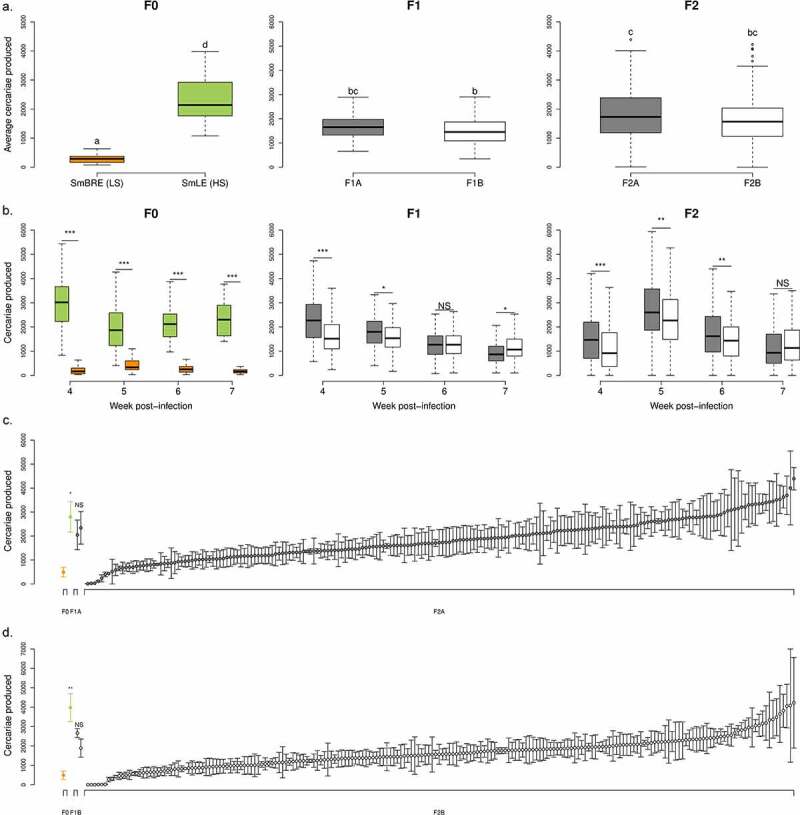
Transmission stage production of two Schistosoma mansoni parental populations (SmLE-H and SmBRE-L) and their progeny (F1s and F2s). (a) Difference in the average number of cercariae produced by SmLE-H and SmBRE-L S. mansoni populations during 4 weeks of the patent period (week 4 to 7 post infection). The SmLE-H population (N = 46) shed more cercariae than the SmBRE-L population (N = 48) [5]. For both crosses A and B, F1 populations (F1A: N = 102; F1B: N = 110) exhibited an intermediate phenotype in terms of cercarial production, compared to F0, while the average production of cercariae by F2s (F2A: N = 204; F2B: N = 204) encompassed both parental and F1s distributions. Cercarial productions from parasite populations or generations not connected by the same letter are significantly different (post-hoc test). (b) Difference in the number of cercariae produced by S. mansoni parental populations (SmLE-H and SmBRE-L) and progeny (F1 and F2) measured by week (week 4 to 7 post infection). (c) Distribution of the cercarial production (mean ± SE) over the 4 weeks of the patent period for the parents (F0) of the cross A, the F1 parents (F1A) and the 204 F2A progeny in rank order. (d) Distribution plot of the cercarial production for the parents (F0), the F1 parents (F1B) and all the 204 F2B progeny for the cross B in rank order. For both crosses (c-d), SmBRE-L and SmLE-H F0 parents exhibited striking differences in cercarial production (cross A: Welsh t-test, p = 0.031; cross B: Welsh t-test, p = 0.013) while F1 showed an intermediate phenotype and F2 encompassed both parental distribution with a gradation from L to H phenotype. NS: No significant difference in cercarial production between the two considered groups, * p < 0.05, ** p ≤ 0.02, *** p ≤ 0.002
We measured the weekly cercarial production of the two S. mansoni F1 populations (i.e. F1A for cross A and F1B for cross B) over 4 weeks (i.e from week 4 to 7 post-exposure). For both crosses, F1 parasite populations shed intermediate numbers of cercariae compared to F0 (i.e. SmLE-H and SmBRE-L parental populations) (Kruskal-Wallis test followed by pairwise Wilcoxon Mann-Whitney post-hoc test, p < 2.2 × 10−16) and showed limited variation in shedding number (Figure 2a – average cercariae produced). However, F1 parasites from cross A produced significantly more cercariae than those from cross B (Wilcoxon test, p = 0.0048, Figure 2a and 2b – Number of cercariae produced/week).
Schistosome gender can also impact the average number of cercariae produced. In the F1A population, male genotypes produced significantly less cercariae than female genotypes (one-way ANOVA, p = 0.0038, Supplementary Figure 2a). We did not see any effect of parasite gender in the F1B progeny (Supplementary Figure 2b).
The F2 parasite progeny from both crosses showed extensive variation in numbers of cercariae shed that encompassed the range seen in the two parental distributions (Kruskal-Wallis test followed by pairwise Wilcoxon Mann-Whitney post-hoc test between F0 populations (i.e. SmLE-H and SmBRE-L) and F2s (i.e. F2A and F2B), p < 2.2 × 10−16, Figure 2a F2 and Figure 2c and 2d). F2 progeny from cross A shed more larvae than cross B (Wilcoxon test, p = 0.0089, Figure 2a-b F2). The F2 progeny have an average cercarial shedding profile across weeks that mimic the SmBRE-L parental profile with a peak in cercarial production at the second shedding week (i.e. week 5 post-infection). However, the average intensity of cercarial production is closer to the SmLE-H parent (Figure 2a-b).
We also observed a significant impact of schistosome gender for the F2 progeny. In the F2B parasite population, male genotypes produced significantly less cercariae than female ones (Kruskal-Wallis test, p = 0.041, Supplementary Figure 2c). We did not find any effect of parasite gender in F2A progeny (Kruskal-Wallis test, p = 0.719, Supplementary Figure 2d).
Linkage analysis of transmission stage production in S. mansoni parasites
We sequenced the whole genome (363 Mb) from parents and the exome (15 Mb) of F1 and F2 progeny from each cross (Figure 1). We found 10,543 (cross A) and 8,779 (cross B) SNPs fixed for alternative alleles (i.e. fully informative markers) in the exome. A classical linkage analysis of the average quantities of cercariae per snail (i.e. schistosome genotype) revealed on the combined crosses three major quantitative trait loci (QTL) involved in transmission stage production: on chr. 1 (LOD = 5.63), chr. 3 (LOD = 8.16) and chr. 5 (LOD = 6.37) (Figure 3a; Supplementary Figure 3a and b). This finding is consistent with the minimum number of loci calculated using the Castle-Wright Estimator [50,51,66–68], which estimates the minimum number of loci determining trait variation from patterns of segregating phenotypic variation. This estimated a minimum of 3.03 loci involved for cross A and 1.85 for cross B. Cercarial gender did not impact the result of the linkage analysis.
Figure 3.
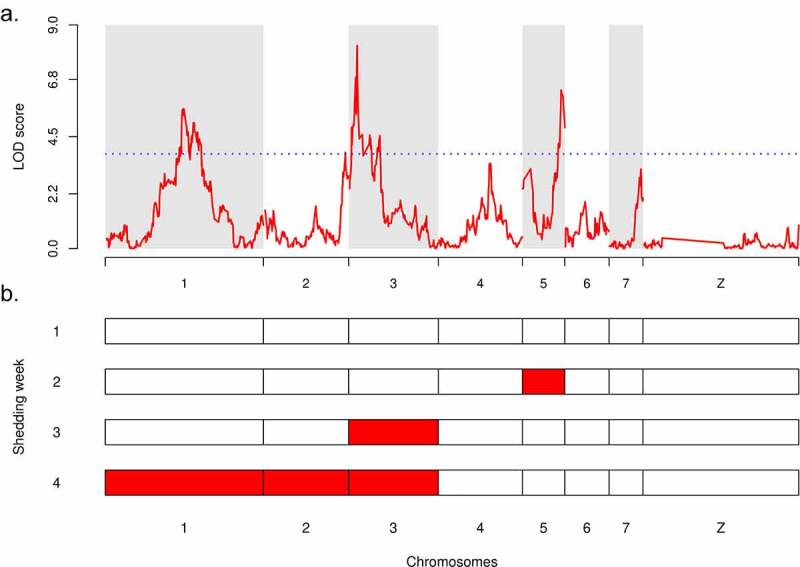
Linkage analysis of the transmission stage production in S. mansoni parasites and QTLs interactions. (a) Linkage analysis of average cercarial production phenotype in combined crosses A and B demonstrated that transmission stage production is a polygenic trait controlled by three major QTLs with statistically significant LOD: on chr. 1 (LOD = 5.63), on chr. 2 (LOD = 8.16) and on chr. 5 (LOD = 6.37). The blue dotted line represents the 1,000 permutation threshold. (b) Genetic architecture of cercarial production is influenced by the shedding week (first shedding week corresponds to week 4 post-infection), demonstrating a sequential pattern of QTL emergence involved in transmission stage production (see also Supplementary Figure 4)
We used a two-dimensional genome scan to search for additional contributing QTLs on the combined crosses and to identify interactions among loci. We found two minor QTLs on chr. 2 (LOD = 3.87) and chr. 4 (LOD = 3.32) which contributed significantly to explain the phenotype variance. The two-QTL scan demonstrated that all five QTLs act additively: we found no evidence for epistasis between the loci (Table 1). Together, these 5 QTLs explained 28.56% of the variation in cercarial production, with a full LOD score of 29.79 (Table 2). The three major QTLs explained most of the phenotype: chr. 1 = 8.10% (p = 5.49 x 10−10), chr. 3 = 6.10% (p = 8.49 x 10−8), chr. 5 = 5.57% (p = 3.31 x 10−7). The two minor QTLs explained less phenotypic variation: chr. 2 = 3.80% (p = 3.38 x 10−5) and chr. 4 = 3.52% (p = 7.06 x 10−5) but these contributions are highly significant.
Table 1.
Summary table of the two-dimensional, two-QTL genome scan identifying QTL interactions (additive or epistatic effects) between the QTL involved in cercarial production in Schistosoma mansoni parasite. LOD values are statistically significant when they are above the 5% threshold LOD for epistatic (interaction) and/or additive effect. We have demonstrated here additive interaction of various loci involved in transmission stage production but no epistatic interaction
| Pairs of loci | Position locus 1 | Position locus 2 | LOD interaction (epistatis) | LOD additive |
|---|---|---|---|---|
| 5% threshold | - | - | 5.68 | 5.84 |
| chr. 1:chr. 2 | 43.84 | 45.94 | 0.611 | 10.48 |
| chr. 1:chr. 3 | 43.84 | 5.44 | 0.186 | 12.40 |
| chr. 1:chr. 4 | 43.84 | 28.95 | 1.020 | 9.05 |
| chr. 1:chr. 5 | 43.84 | 21.56 | 1.367 | 11.88 |
| chr. 2:chr. 4 | 45.94 | 28.90 | 0.738 | 7.48 |
| chr. 2:chr. 5 | 45.94 | 21.56 | 1.244 | 9.60 |
| chr. 3:chr. 5 | 5.44 | 21.56 | 2.587 | 10.96 |
| chr. 4:chr. 5 | 28.90 | 21.56 | 0.622 | 9.20 |
Table 2.
QTLs interactions modeling (additive model) and estimation of the proportion of the cercarial production variance (%var) explained by each of the loci involved in the phenotype. Together, the 5 QTLs explained 28.56% of variation in transmission stage production in S. mansoni parasites
| Loci |
LOD |
%var |
P value |
|---|---|---|---|
| Full additive model (all 5 QTLs) |
29.8 | 28.56 | 0 |
| chr. 1 | 9.52 | 8.10 | 5.49x10−10 |
| chr. 2 | 4.6 | 3.80 | 3.38x10−5 |
| chr. 3 | 7.26 | 6.10 | 8.49x10−8 |
| chr. 4 | 4.26 | 3.52 | 7.06x10−5 |
| chr. 5 | 6.66 | 5.57 | 3.31x10−7 |
Genetic control of shedding varies across time
When we conducted the linkage analysis on our combined crosses using the cercarial quantities produced each week rather than the average across all four weeks (i.e. from week 4 to week 7 post-exposure, see Figure 1), we observed a sequential pattern of QTL emergence (Figure 3b, Supplementary Figure 3). On shedding week 1, none of the identified QTLs passed the permutation threshold (Figure 4b, Supplementary Figure 4a), while on shedding week 2, the QTL on chr. 5 predominated and a QTL on chr. 3 had arisen (Figure 3b, Supplementary Figure 4b). On shedding week 3, the main QTL was on chr. 3 (Figure 3b, Supplementary Figure 4c). Finally, on shedding week 4, the QTL on chr. 1 predominated, along with QTLs on chr. 2 and 3 (Figure 3b, Supplementary Figure 4d). Hence, different parasite genes determine cercarial production across time during the snail infection.
Figure 4.
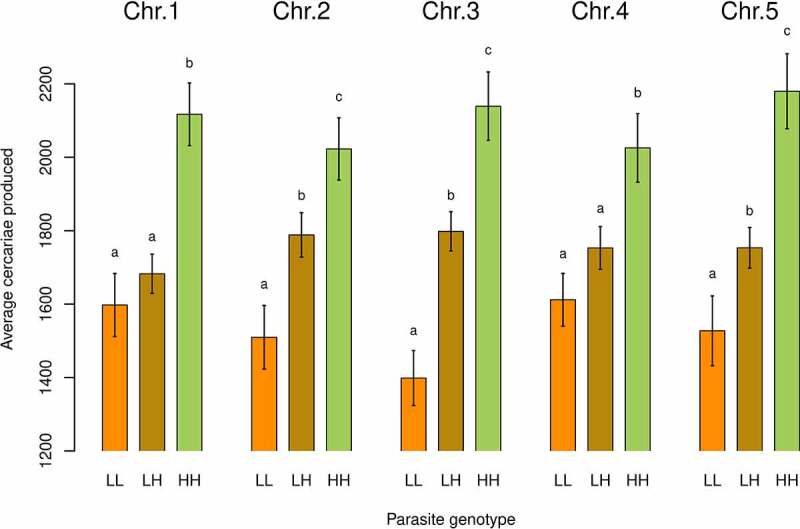
Inheritance of transmission stage production. Impact of the parasite genotype on the average number of cercariae produced for each major and minor QTL linked to transmission stage production. On chr. 1 and chr. 4, cercarial production is not significantly different when parasites are homozygous for the “low shedding” allele (LL) or heterozygous (LH) but only when parasites are homozygous for the “high shedding” allele (i.e. HH). High shedding allele (H) is recessive and low shedding allele (L) is dominant. On chr. 2, chr. 3 and chr.5, cercarial production is significantly different for all the three parasite genotypes encountered (LL, LH and HH). high and low shedding alleles act co-dominantly. Cercarial productions from parasite genotypes (LL, LH, HH) not connected by the same letter for a given locus are significantly different (post-hoc test)
Allelic inheritance and impact on cercarial production in S. mansoni parasites
For each QTL, we determined the interactions between alleles from L and H shedding parents by analyzing the average production of cercariae per genotype (i.e. LL: homozygous for the low shedding allele, LH: heterozygous and HH: homozygous for the high shedding allele; Figure 4) for the combined F2 progeny (i.e. cross A and B analyzed together). Our analysis showed codominance for the chr. 2, 3 and 5 QTLs: F2 progeny with LH genotype produced more cercariae than LL genotype but less than the HH genotype (Figure 4). However, for the chr. 1 and 4 QTLs showed patterns consistent with recessive inheritance. At these loci, F2 progeny with HH genotype produced more cercariae (pairwise comparison using Wilcoxon rank sum test; chr. 1: HH vs. LH: p = 4.2 x 10−5 and HH vs. LL: p = 8.2 x 10−6; chr. 4: HH vs. LH:, p = 0.0109 and HH vs. LL: p = 0.0034) than the other genotypes (i.e. LL and LH; pairwise comparison using Wilcoxon rank sum test; chr. 1: LL vs. LH, p = 0.22; chr. 4: LL vs. LH, p = 0.56; Figure 4).
Candidate genes controlling transmission stage production in S. mansoni parasite
We listed the genes located within the 1.8 LOD-support interval of each QTL and expressed either in daughter sporocysts or in cercariae. We identified 314 genes on chr. 1, 87 on chr. 2, 31 on chr. 3, 76 on chr. 4 and 86 on chr. 5 (Table 3 and Supplementary Table 1). We then prioritized genes using an objective index (“genoscore”, see Materials & Methods section), that accounts for the genotypes of the parents (fixed alternative alleles), the presence of non-synonymous mutations and their impact on the protein predicted by snpEff (see Materials & Methods section), the CDS length and the distance from the QTL peak, and the expression of the gene at the parasite stages of interest (sporocyst or cercariae). Using this objective approach, we identified the three most likely candidate genes within each QTL that may be involved in transmission stage production (Table 4).
Table 3.
Summary table of the total number of genes located within the 1.8 LOD-support interval of each QTL peak, and expressed into the different parasite stages: daughter sporocysts only, cercariae only, both stages or not expressed at all
| Loci | QTL peak absolute coordinates in S. mansoni genome (bp) | Total number of genes | Genes expressed in daughter sporocysts only | Genes expressed in cercariae only | Genes expressed in both stages | Genes not expressed |
|---|---|---|---|---|---|---|
| chr. 1 | 43,844,038 | 329 | 297 | 301 | 284 | 15 |
| chr. 2 | 45,941,343 | 94 | 87 | 83 | 78 | 2 |
| chr. 3 | 4,722,053 | 32 | 28 | 28 | 25 | 1 |
| chr. 4 | 28,695,306 | 79 | 70 | 72 | 66 | 3 |
| chr. 5 | 21,695,876 | 94 | 78 | 82 | 74 | 8 |
Table 4.
Summary table of the 3 top candidate genes for each locus involved in cercarial production in the parasite S. mansoni based on the computed genoscore CDS (see Materials & Methods) and their relative positions to the maximum peak. NA: gene not captured by the S. mansoni baits (exome capture). In bold: selected best candidate genes based on their annotations (GFF or HHsearch)
| Chromosome | Gene ID | GFF annotation | HHsearch annotation | Max. LOD | Genoscore |
|---|---|---|---|---|---|
| chr. 1 | Smp_152850 | Tetratricopeptide-like helical domain superfamily | Putative 70 kda peptidylprolyl isomerase PFL2275c | 5.63 | 1.646 |
| Smp_057230 | Aminoacyl-tRNA synthetase, class II | Seryl-tRNA synthetase (SerRS) | NA | 1.286 | |
| Smp_246820 | No annotation available | hypothetical protein TTHA1254 | 4.05 | 1.114 | |
| chr. 2 | Smp_347740 | Coiled-coil domain-containing protein 39 | Nucleoporin_FG2 | 3.86 | 7.657 |
| Smp_328700 | No annotation available | Engineered transmembrane domain variant | NA | 2.153 | |
| Smp_170430 | FAD dependent oxidoreductase | D-aminoacid oxidase, N-terminal domain | 2.81 | 1.491 | |
| chr. 3 | Smp_343200 | Pleckstrin homology domain | G-protein coupled receptor kinase 2 (beta-adrenergic receptor kinase 1) | 8.16 | 1.653 |
| Smp_190770 | Lanthionine synthetase C-like | Nisin biosynthesis protein NisC | 5.41 | 1.426 | |
| Smp_138060 | No annotation available | Cationic trypsin (e.c.3.4.21.4) | 7.66 | 1.227 | |
| chr. 4 | Smp_313330 | No annotation available | No annotation available | 3.35 | 1.732 |
| Smp_150840 | Zinc finger, C2H2, LYAR-type | 60S ribosomal protein L8, 60S | 2.47 | 1.502 | |
| Smp_334250 | Epithelial sodium channel | Acid-sensing ion channel 1 | 3.42 | 1.257 | |
| chr. 5 | Smp_330680 | No annotation available | No annotation available | NA | 6.997 |
| Smp_330700 | No annotation available | No annotation available | NA | 3.784 | |
| Smp_330760 | IQ motif, EF-hand binding site | Calmodulin, IQ domain-containing protein G | 4.50 | 3.631 |
The genes prioritization on each of the three major QTLs revealed three compelling candidates: (i) peptidylprolyl isomerase (chr. 1) which regulates many biological processes, including intracellular signaling, transcription, inflammation, immunomodulation and apoptosis [69,70], (ii) G-protein coupled receptor kinase 2 (chr. 3) which is involved in cell migration [71] and (iii) a protein with unknown function (chr. 5) which suggests schistosome specific factor yet to be characterized. On chr. 5, another candidate, the Calmodulin IQ domain protein, is of interest: this protein is a calcium sensor and can stimulate changes in the actin cytoskeleton mediated by proteins such as myosin [72].
In the two minor QTLs, the top candidate genes encode a nucleoporin (chr. 2) and a protein of unknown function (chr. 4). The second-best candidate gene on chr. 4 is also of interest: it encodes a LYAR, a cell growth regulating nucleolar protein.
Linkage analysis of snail physiological trait associated with transmission stage production
Laccase-like activity and hemoglobin rate in the Biomphalaria snail hemolymph were correlated (Supplementary Figure 5c and 5f). These metrics provide proxies to evaluate snail health and the impact of schistosome infection [5,49]. Both parameters were negatively correlated with F2 cercarial production (laccase-like activity: Pearson’s correlation test, p = 4.31 × 10−15, correlation coefficient = −0.56; Hemoglobin rate: Pearson’s correlation test, p = 8.014 × 10−12, correlation coefficient = −0.50; Supplementary Figure 5d and 5e). There was no correlation with the F1 cercarial production (laccase-like activity: Pearson’s correlation test, p = 0.426, correlation coefficient = −0.09; Hemoglobin rate: Pearson’s correlation test, p = 0.661, correlation coefficient = −0.05; Supplementary Figure 5a and 5b). This was expected because F1s were heterozygous for markers from each parent at all five QTLs and showed limited variation in cercarial production.
We performed a linkage analysis in each cross and in the combined crosses to investigate parasite genes linked to laccase-like activity and the hemoglobin rate in the snail host hemolymph. We did not find significant QTLs across S. mansoni genome linked to these snail hemolymph phenotypes.
Discussion
How many loci are involved in transmission/virulence?
We determined the genetic architecture of transmission stage production trait in schistosome parasites using classical linkage mapping. We showed that transmission/virulence traits in schistosome are polygenic, involving five loci from which three have major effect. These results are consistent with the idea that relatively small numbers of genes are involved in strongly selected traits such as virulence/transmission. The obvious caveat here is that loci of small effect may be difficult to identify even in well powered linkage analyses. The five loci identified explained 28.56% of the phenotypic variation observed: it is unclear what proportion of the remaining variation is explained by additional undetected QTLs, or reflects the impact of environmental variation or measurement error [73–75].
There is an extensive literature on compatibility between S. mansoni and B. glabrata [22], which is important to distinguish from virulence. We define virulence as parasite induced host mortality and have previously shown that SmLE-H produces more cercariae and causes higher snail mortality than SmBRE-L [5]. Compatibility determines the ability of particular parasites to develop in particular snails. In incompatible host-parasite pairs miracidia are unable to successfully establish: miracidia die soon after snail penetration [22]. In our experiments, both parental parasite genotypes are compatible with Bg26 snails. Twenty-six percent of snails Infected with single miracidia develop infection in for both SmBRE-L and SmLE-H, but the number of cercariae released, and snail mortality differs between these two parasite genotypes. Hence, this work examines difference in virulence rather than compatibility.
Three other studies have used linkage mapping to dissect the genetic basis of parasite transmission/virulence trait (Table 5). In the protozoan parasite Toxoplasma gondii, four sets of genetic crosses were used to identify the genetic basis of acute virulence between different clonal lineages from North America [76]. These crosses identified several QTLs linked to acute virulence and resulted in the identification of a set of polymorphic genes in the GTPase serine/threonine kinase family in secretory organelles called rhoptries [77]. In another protozoan model, the rodent malaria parasite (Plasmodium yoelii yoelii), Pattaradilokrat et al. [7], and Otsuki et al. [79], showed that just one locus on chr. 13 was linked to parasite growth rate and host virulence. Plant parasite nematode system provides a directly comparable macroparasite system in which virulence has been examined using linkage mapping. In the root-knot nematode Meloidogyne hapla, crosses between parasites that differ in ability to produce galls on the roots of the ornamental nightshade Solanum bulbocastanum identified three QTLs linked to nematode transmission success [8].
Table 5.
Do transmission/virulence traits tend to be oligogenic (i.e. controlled by few genes)? Summary table of the number of loci and genes involved in transmission/virulence or growth life-history traits for various eukaryotic parasite systems (reviewed from the literature)
| Parasite system | Phenotypes | Number of loci involved (chr. #) |
Number of gene(s) involved (gene name) |
Percentage of phenotypic variations explained by the loci | References |
|---|---|---|---|---|---|
| Toxoplasma gondii | Acute virulence | 1 (XII) | 1 (ROP5) | - | 97 |
| Acute virulence | 3 (VIIa/VIIb/XII) | 3 (ROP18/ROP16/ROP5) | - | 98, 99 | |
| Acute virulence | 2 (VIIa/Ia) | 1 (ROP18) | - | 100, 101 | |
| Growth | 4 (VIIa/XI/XII/Ia) | - | - | 102 | |
| Migration | 1 (VIIa) | - | - | 101 | |
| Acute virulence | 1 (XII) | 1 (ROP5) | - | 103 | |
| Plasmodium yoelii yoelii (rodent malaria parasite] | Growth rate | 1 (XIII) | 1 (pyebl) | - | 78, 79 |
|
Meloidogyne hapla (plant pathogen nematode) |
Egg mass number | 2 (VII/XIII) | - | - | 80 |
| Total eggs among F2 lines | 2 (IV/VII) | - | - | 80 | |
| Schistosoma mansoni | Transmission stage production/Virulence | 5 (3 major: I/III/V and 2 minor: II/IV) |
5 potential best candidate genes (peptidylprolyl isomerase/G-protein coupled receptor kinase/Calmodulin and Nucleoporin_FG2/LYAR cell growth regulating nucleolar protein) | 28.56% | Present study |
In all these three cases, relatively few loci control transmission/virulence traits, as we see in schistosome parasites (Table 5). This question of “few loci or genes” versus “many loci or genes” controlling key life-history traits is fundamental to our understanding of adaptation in nature. As demonstrated by an increasing number of studies, few large-effect loci (and subsequently genes) have been shown to drive rapid adaptation (reviewed in Messer et al. [81]).
This paper focuses on the genetic basis of transmission/virulence traits in the intermediate snail host. However, schistosome adults infect vertebrate hosts and the complete lifecycle involves six different body plan (eggs, miracidia, sporocysts, cercariae, schistosomulae, adults) [41], so pleiotropic effects of genes on fitness of different lifecycle stages may be important in schistosome evolution. A previous study suggested that high virulence in the snail was inversely correlated with low virulence during the adult stage [25,26]. They suggested that trade-offs between fitness traits in different lifecycle stages should be considered. We have not investigated fitness in other lifecycle stages in this study, but this will be a focus of future work.
How do parasite genes impact numbers of cercariae shed?
Genes may impact numbers of cercariae shed from infected snails by (i) influencing growth and differentiation of sporocysts, (ii) changing the balance of investment between cercarial production and daughter sporocyst production, (iii) influencing the ability of cercariae to escape from the snail host (Figure 5). Two lines of evidence strongly suggest that the genes determining cercarial shedding may act on different processes.
Figure 5.
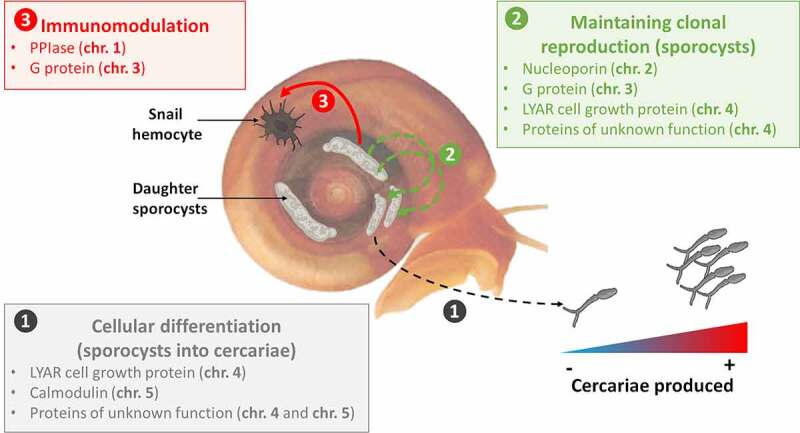
Potential mechanisms involved in production of schistosome transmission stage. our genetic analysis of transmission stage production and virulence in S. mansoni parasites revealed that this complex trait is controlled by five loci with potentially different mechanisms involved to induce and maintain cercarial production over time. this could involve (i) the differentiation of sporocysts cells into cercariae through the calmodulin calcium sensor (chr. 5) (ii) the maintenance of the clonal reproduction of sporocysts and the continuous cercarial production through the G protein (chr. 3) and the nucleoporin (chr. 2), and (iii) the modulation of the snail immune response to protect parasitic cells through the pPIase (chr. 1) and the G protein (chr. 3). the LYAR cell growth protein (chr. 4) could also be involved all along the patent period to regulate the growth and multiplication of sporocysts cells and the development of cercariae. the proteins of unknown function are also of high interest: they could reveal new mechanisms involved in transmission/virulence
First, we previously compared sporocyst size and growth kinetics in the parental populations using qPCR. SmLE-H sporocysts comprise on average 47% of cells within infected snails, while SmBRE-L sporocysts comprise 25% of cells within infected snails [5]. However, this is not sufficient to fully explain the difference in cercariae produced by these two populations, because the SmBRE-L infected snails shed significantly fewer cercariae than predicted from qPCR measures of sporocysts cells in infected snails [5]. These results are consistent with independent action of genes to determine sporocyst size and cercarial production.
Second, when we dissected the genetic architecture of cercarial production over the 4-week patent period of the infection, we observed sequential emergence of QTLs. This is also consistent with the idea that genes influencing cercarial shedding may act on different processes to determine this cercarial production. This sequential emergence of loci could also explain the evolution of sporocysts kinetics trajectories observed in parental population [5]. Moreover, we highlighted that the high shedding alleles acted co-dominantly with the low shedding alleles at QTLs on chr. 2, chr. 3 and chr. 5 but were recessive at QTLs on chr. 1 and chr. 4. These differences in allelic inheritance could also support the existence of different mechanisms involved in the modulation of transmission stage production and virulence: co-dominant loci would involve alleles with dose effect while recessive loci would involve loss-of-function alleles. In the next section we evaluate potential candidate genes and speculate at which stage of the parasite these may play a role.
Candidate genes underlying transmission/virulence traits
The first QTL linked to cercarial production appears on the second shedding week and is located on chr. 5. The three leading candidate genes under this QTL include two genes encoding proteins of unknown function and a gene encoding a calmodulin (CAM) IQ domain protein. The proteins of unknown function are of high interest as they could reveal new mechanisms involved in transmission/virulence and will deserve further attention. CAM IQ domain proteins belong to calcium sensors and can stimulate changes in the actin cytoskeleton mediated by proteins such as myosin [82]. In schistosome, the calcium sensor (CAM IQ domain protein) could modulate the differentiation of daughter sporocysts cells into cercariae and induce the cercarial shedding early in the patent period. Calcium binding proteins are known to play an important role in amoebic pathogenesis and are essential for the Entamoeba histolytica parasite growth [10], while in Toxoplasma gondii parasite, CAM-like proteins are essential and contribute to regulate parasite motility and host cell invasion [83].
During the third shedding week, the major QTL is on chr. 3, where the leading candidate gene is a G-protein coupled receptor kinase 2. In Entamoeba histolytica G-protein are involved in pathogenesis-related cellular processes, such as migration, invasion, phagocytosis and evasion of the host immune response by surface receptor capping [84]. In S. mansoni, this G-protein coupled receptor kinase 2 may stimulate sporocyst expansion or renewal into snail host tissues (i.e. new generation of daughter sporocysts) while escaping host immune defenses, to maintain the parasitic infection and eventually, cercarial production.
During the fourth shedding week, QTLs on chr. 1, chr. 2 and chr. 3 are involved in transmission stage production. The leading candidate gene on chr. 1 is a peptidylprolyl isomerase (PPIase). Most PPIases characterized in parasites belong to the cyclophilin family [85] and are strong immunomodulatory proteins [86]. In the apicomplexan Theileria, a secreted prolyl isomerase modulates host leukocyte transformation [87]; similar immunomodulatory peptidylprolyl isomerases have been demonstrated in two other apicomplexan parasites, Toxoplasma gondii and Neospora caninum [88]. PPIases have been shown to be primary actors of host-parasite interactions and are certainly essential for the development and differentiation of parasitic protozoans (e.g Leishmania parasites), which show a high degree of plasticity in their cellular organization and metabolic status during their infection cycles [89]. In our schistosome model, PPIase may modulate snail host immune response to infection, while maintaining proliferation and viability of sporocysts cells. The best candidate gene under the chr. 2 QTL is a nucleoporin, protein that is essential to microtubule organization and dynamics during mitosis. In the parasite Plasmodium falciparum, nucleoporins are essential for parasite proliferation in human erythrocytes [90]. Nucleoporins are also essential in the transport of mRNA from the nucleus to the cytoplasm after transcription and are involved in cell migration [91]. In schistosome, these proteins might trigger the sporocysts clonal proliferation and cercarial production.
The QTL on chr. 4, revealed by the two-dimensional genome scan, did not pass the permutation threshold at any of the 4-week patent period of the infection. Therefore, this QTL could have a minor but continuous effect throughout the patent period. Interestingly, the leading candidate genes under this QTL encode protein of unknown function followed by a LYAR cell growth regulating nucleolar protein. Proteins of unknown function are again of high interest and will deserve further attention. In addition, the LYAR protein, required for cell proliferation [92] and highly present in early embryos of mammals [93], could also be involved all along the patent period to regulate the growth and multiplication of schistosome sporocysts cells and the development of cercariae.
Prospects for functional analysis
Figure 5 summarizes potential involvement of candidate genes in cercarial shedding in S. mansoni. Functional validation is required to investigate involvement of these candidate genes. While RNAi methods are available for gene knockdown in adult worms [32], delivery of RNA to the sporocysts within the snail is challenging. We have tried several approaches to induce RNAi in daughter sporocysts including injection of ds or siRNA into the snail hemolymph/tissues, but have so far not been able to knockdown genes in this stage. CRISPR provides a possible alternative approach but is still at an early stage of development for S. mansoni [35–37]. Once effective gene manipulation approaches are available for late sporocysts, we will be able to directly examine the involvement of our leading candidate genes.
Stem cells play a central role in schistosome development and reproduction. Wang et al. [16],have demonstrated that primary sporocysts containing different cellular types including stem cells. In our model, we suspect that SmLE-H and SmBRE-L sporocysts may exhibit different cellular trajectories, with differences in development of cell that differentiate to generate cercariae and those that give rise to the next generation of daughter sporocysts [17]. Advances in our understanding of stem-cell differentiation of S. mansoni within the molluskan host coupled with single-cell transcriptomics of daughter sporocysts from our two populations provides and alternative approach to investigate the molecular basis of these transmission-related developmental differences at the cellular and molecular levels [94–96]. Functional and cell biology characterization of this key life history trait underlying transmission and virulence will be the focus of future work on this system.
Supplementary Material
Acknowledgments
We thank Michael S. Blouin (Oregon State University) for the Bg26 snail line, Guillaume Mitta and Benjamin Gourbal (University of Perpignan France) for SmBRE S. mansoni population and Philip LoVerde (UT Health San Antonio) for the SmLE S. mansoni population. We thank the Vivarium of the Southwest National Primate Research Center (SNPRC) for providing rodent care. This research was supported by a Cowles fellowship (WL) from Texas Biomedical Research Institute (13-1328.021), and NIH R01AI133749 (TJCA) and was conducted in facilities constructed with support from Research Facilities Improvement Program grant C06 RR013556 from the National Center for Research Resources. SNPRC research at Texas Biomedical Research Institute is supported by grant P51 OD011133 from the Office of Research Infrastructure Programs, NIH.
WL, FDC and TJCA designed the experiments. WL, FDC, MMW, VM and GAA performed the experiments. WL and FDC performed the data analyses. WL and TJCA drafted the manuscript. All authors read and approved the final manuscript.
Funding Statement
This work was supported by the Foundation for the National Institutes of Health [R01AI133749]; Foundation for the National Institutes of Health (Office of Resaerch Infrastructure Programs) [P51 OD011133]; Foundation for the National Institutes of Health (Research Facilities Improvement Program) [C06 RR013556]; Texas Biomedical Research Institute (US) (Cowles fellowship) [13-1328.021].
Disclosure of potential conflicts of interest
The authors declare that they have no competing interests.
Supplementary material
Supplemental data for this article can be accessed here
References
- [1].Criscione CD, Van Paridon BJ, Gilleard JS, et al. 2020. Clonemate cotransmission supports a role for kin selection in a puppeteer parasite. Proceedings of the National Academy of Sciences 117:5970–5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Vasudevan A, Kumar V, Chiang YN, et al. α2u-globulins mediate manipulation of host attractiveness in Toxoplasma gondii-Rattus novergicus association. ISME J. 2015;9(9):2112–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Loker ES, Hofkin BV.. Parasitology: a conceptual approach. 1st ed. Garland Science; 2015. [Google Scholar]
- [4].Jensen KH, Little TJ, Little T, et al. Empirical support for optimal virulence in a castrating parasite. PLoS Biol. 2006;4(7):e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Le Clec’h W, Diaz R, Chevalier FD, et al. Striking differences in virulence, transmission and sporocyst growth dynamics between two schistosome populations. Parasit Vectors. 2019;12(1):485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Alizon S, Hurford A, Mideo N, et al. Virulence evolution and the trade-off hypothesis: history, current state of affairs and the future. J Evol Biol. 2009;22(2):245–259. [DOI] [PubMed] [Google Scholar]
- [7].Anderson RMMRM. Coevolution of hosts and parasites. Parasitology. 1982;85(2):411–426. [DOI] [PubMed] [Google Scholar]
- [8].Frank SA. Models of parasite virulence. Q Rev Biol. 1996;71(1):37–78. [DOI] [PubMed] [Google Scholar]
- [9].Acevedo MA, Dillemuth FP, Flick AJ, et al. Virulence-driven trade-offs in disease transmission: a meta-analysis. Evolution. 2019;73(4):636–647. [DOI] [PubMed] [Google Scholar]
- [10].Alizon S, Michalakis Y. Adaptive virulence evolution: the good old fitness-based approach. Trends Ecol Evol. 2015;30(5):248–254. [DOI] [PubMed] [Google Scholar]
- [11].Anderson TJC, LoVerde PT, Le Clec’h W, et al. Genetic crosses and linkage mapping in schistosome parasites. Trends Parasitol. 2018;34(11):982–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cioli D, Pica-Mattoccia L, Moroni R. Schistosoma mansoni: hycanthone/oxamniquine resistance is controlled by a single autosomal recessive gene. Exp Parasitol. 1992;75(4):425–432. [DOI] [PubMed] [Google Scholar]
- [13].Greenberg RM. New approaches for understanding mechanisms of drug resistance in schistosomes. Parasitology. 2013;140(12):1534–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Melman SD, Steinauer ML, Cunningham C, et al. Reduced susceptibility to praziquantel among naturally occurring Kenyan isolates of Schistosoma mansoni. PLoS Negl Trop Dis. 2009;3(8):e504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mwangi IN, Sanchez MC, Mkoji GM, et al. Praziquantel sensitivity of Kenyan Schistosoma mansoni isolates and the generation of a laboratory strain with reduced susceptibility to the drug. International journal for parasitology Drugs and drug resistance. 2014;4(3):296–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Valentim CLL, Cioli D, Chevalier FD, et al. Genetic and molecular basis of drug resistance and species-specific drug action in Schistosome parasites. Science. 2013;342(6164):1385–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Théron A. Chronobiology of trematode cercarial emergence: from data recovery to epidemiological, ecological and evolutionary implications. Adv Parasitol. 2015;88:123–164. [DOI] [PubMed] [Google Scholar]
- [18].Théron A, Combes C. Genetic analysis of cercarial emergence rhythms of Schistosoma mansoni. Behav Genet. 1988;18(2):201–209. [DOI] [PubMed] [Google Scholar]
- [19].Théron A, Combes C. [Genetic analysis of the shedding pattern of Schistosoma mansoni cercariae by hybridization of species with early and late emission peaks] Comptes rendus des seances de l’Academie des sciences Serie III. Sciences de la vie. 1983;297(12):571–574. [PubMed] [Google Scholar]
- [20].Files VS, Cram EB. A study on the comparative susceptibility of snail vectors to strains of Schistosoma mansoni . J Parasitol. 1949;35(6):555–560. [PubMed] [Google Scholar]
- [21].Kalbe M, Haberl B, Hertel J, et al. Heredity of specific host-finding behaviour in Schistosoma mansoni miracidia. Parasitology. 2004;128(6):635–643. [DOI] [PubMed] [Google Scholar]
- [22].Mitta G, Gourbal B, Grunau C, et al. The compatibility between biomphalaria glabrata snails and schistosoma mansoni: an increasingly complex puzzle. Adv Parasitol. 2017;97:111–145. [DOI] [PubMed] [Google Scholar]
- [23].Rollinson D, Stothard JR, Southgate VR. Interactions between intermediate snail hosts of the genus Bulinus and schistosomes of the Schistosoma haematobium group. Parasitology. 2001;123(7):S245–S260. [DOI] [PubMed] [Google Scholar]
- [24].Theron A, Rognon A, Gourbal B, et al. Multi-parasite host susceptibility and multi-host parasite infectivity: a new approach of the Biomphalaria glabrata/Schistosoma mansoni compatibility polymorphism. Infect Genet Evol. 2014;26:80–88. [DOI] [PubMed] [Google Scholar]
- [25].Davies CM, Webster JP, Woolhous ME. Trade-offs in the evolution of virulence in an indirectly transmitted macroparasite. Proc Biol Sci. 2001;268(1464):251–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gower CM, Webster JP. Fitness of indirectly transmitted pathogens: restraint and constraint. Evolution. 2004;58(6):1178–1184. [DOI] [PubMed] [Google Scholar]
- [27].Webster JP, Gower CM, Blair L. Do hosts and parasites coevolve? Empirical support from the schistosoma system. Am Nat. 2004;164(Suppl S5):S33–S51. [DOI] [PubMed] [Google Scholar]
- [28].Berriman M, Haas BJ, LoVerde PT, et al. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460(7253):352–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Protasio AV, Tsai IJ, Babbage A, et al. A systematically improved high quality genome and transcriptome of the human blood fluke Schistosoma mansoni. PLoS NeglTrop Dis. 2012;6(1):e1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Chevalier FD, Le Clec’h W, Alves de Mattos AC, et al. Real-time PCR for sexing Schistosoma mansoni cercariae. Mol Biochem Parasitol. 2016;205(1–2):35–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gasser RB, Morahan G, Mitchell GF. Sexing single larval stages of Schistosoma mansoni by polymerase chain reaction. Mol Biochem Parasitol. 1991;47(2):255–258. [DOI] [PubMed] [Google Scholar]
- [32].Krautz-Peterson G, Bhardwaj R, Faghiri Z, et al. RNA interference in schistosomes: machinery and methodology. Parasitology. 2010;137(3):485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mann VH, Suttiprapa S, Skinner DE, et al. Pseudotyped murine leukemia virus for schistosome transgenesis: approaches, methods and perspectives. Transgenic Res. 2014;23(3):539–556. [DOI] [PubMed] [Google Scholar]
- [34].Rinaldi G, Eckert SE, Tsai IJ, et al. Germline transgenesis and insertional mutagenesis in Schistosoma mansoni mediated by murine leukemia virus. PLoS Pathog. 2012;75(7):e1002820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ittiprasert W, Mann VH, Karinshak SE, et al. Programmed genome editing of the omega-1 ribonuclease of the blood fluke, javax.xml.bind.JAXBElement@36467c54. eLife. 2019;8. DOI: 10.7554/eLife.41337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Sankaranarayanan G, Coghlan A, Driguez P, et al. Large CRISPR-Cas-induced deletions in the oxamniquine resistance locus of the human parasite, javax.xml.bind.JAXBElement@4957cc90. Wellcome Open Res. 2020;5:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].You H, Mayer JU, Johnston RL, et al. CRISPR/Cas9-mediated genome editing of Schistosoma mansoni acetylcholinesterase. FASEB J. 2021;35(1):e21205. [DOI] [PubMed] [Google Scholar]
- [38].Collins JNR, Collins JJ. Methods for studying the germline of the human parasite schistosoma mansoni. Methods Mol Biol. 2017;1463:35–47. [DOI] [PubMed] [Google Scholar]
- [39].Wang J, Paz C, Padalino G, et al. Large-scale RNAi screening uncovers therapeutic targets in the parasite Schistosoma mansoni. Science (New York, NY). 2020;369(6511):1649–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wendt G, Zhao L, Chen R, et al. A single-cell RNA-seq atlas of Schistosoma mansoni identifies a key regulator of blood feeding. Science (New York, NY). 2020;369(6511):1644–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wendt GR, Collins JJ. Schistosomiasis as a disease of stem cells. Curr Opin Genet Dev. 2016;40:95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bonner KM, Bayne CJ, Larson MK, et al. Effects of Cu/Zn superoxide dismutase (sod1) genotype and genetic background on growth, reproduction and defense in Biomphalaria glabrata. PLoS NeglTrop Dis. 2012;6(6):e1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Verjee MA. Schistosomiasis: still a cause of significant morbidity and mortality. Res Rep Trop Med. 2019;10:153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].World Health Organization . 2019. Schistosomiasis. https://www.who.int/news-room/fact-sheets/detail/schistosomiasis
- [45].Chevalier FD, Valentim CLL, LoVerde PT, et al. Efficient linkage mapping using exome capture and extreme QTL in schistosome parasites. BMC Genomics. 2014;15(1):617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Gérard C, Ta MH. Schistosoma mansoni–Biomphalaria glabrata : dynamics of the sporocyst population in relation to the miracidial dose and the host size. Can J Zool. 1993;71(9):1880–1885. [Google Scholar]
- [47].Tavalire HF, Blouin MS, Steinauer ML. Genotypic variation in host response to infection affects parasite reproductive rate. Int J Parasitol. 2016;46(2):123–131. [DOI] [PubMed] [Google Scholar]
- [48].Lewis FA, Stirewalt MA, Souza CP, et al. Large-scale laboratory maintenance of Schistosoma mansoni, with observations on three schistosome/snail host combinations. J Parasitol. 1986;72(6):813–829. [PubMed] [Google Scholar]
- [49].Le Clec’h W, Anderson TJC, Chevalier FD. Characterization of hemolymph phenoloxidase activity in two biomphalaria snail species and impact of Schistosoma mansoni infection. Parasit Vectors. 2016;9(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Castle WE. AN IMPROVED METHOD OF ESTIMATING THE NUMBER OF GENETIC FACTORS CONCERNED IN CASES OF BLENDING INHERITANCE. Science (New York, NY). 1921;6(1393):223. [DOI] [PubMed] [Google Scholar]
- [51].Wright SW, Morton NE. Genetic studies on cystic fibrosis in Hawaii. Am J Hum Genet. 1968;20(2):157–169. [PMC free article] [PubMed] [Google Scholar]
- [52].Le Clec’h W, Chevalier FD, McDew-White M, et al. Whole genome amplification and exome sequencing of archived schistosome miracidia. Parasitology. 2018;1–9. DOI: 10.1017/S0031182018000811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Heng L, Handsaker B, Wysoker A, et al. 1000 Genome Project Data Processing Subgroup . The sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Cingolani P, Platts A, Wang LL, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: sNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6(2):80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Köster J, Rahmann S. Snakemake-a scalable bioinformatics workflow engine. Bioinformatics. 2018;34(20):3600. [DOI] [PubMed] [Google Scholar]
- [59].Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].R Core Team . R: a language and environment for statistical computing. In: R Foundation for Statistical Computing - Vienna. Austria: 2018. [Google Scholar]
- [61].Knaus BJ, Grünwald NJ. vcfr : a package to manipulate and visualize variant call format data in R. Mol Ecol Resour. 2017;17(1):44–53. [DOI] [PubMed] [Google Scholar]
- [62].Broman KW, Wu H, Sen S, et al. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19(7):889–890. [DOI] [PubMed] [Google Scholar]
- [63].Buddenborg SK, Kamel B, Hanelt B, et al. The in vivo transcriptome of Schistosoma mansoni in the prominent vector species Biomphalaria pfeifferi with supporting observations from Biomphalaria glabrata. PLoS Negl Trop Dis. 2019;13(9):e0007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Steinegger M, Meier M, Mirdita M, et al. HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinformatics. 2019;20(1):473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Jones-Nelson O, Thiele EA, Minchella DJ. Transmission dynamics of two strains of Schistosoma mansoni utilizing novel intermediate and definitive hosts. Parasitol Res. 2011;109(3):675–687. [DOI] [PubMed] [Google Scholar]
- [66].Cockerham CC. Modifications in estimating the number of genes for a quantitative character. Genetics. 1986;114(2):659–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Hedrick PW. Genetics of populations, Series of books in biology. Science Books International; 1983. [Google Scholar]
- [68].Lande R. The minimum number of genes contributing to quantitative variation between and within populations. Genetics. 1981;99(3–4):541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Ramachandran S, Venugopal A, Kutty VR, et al. Plasma level of cyclophilin A is increased in patients with type 2 diabetes mellitus and suggests presence of vascular disease. Cardiovasc Diabetol. 2014;13(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Wei Y, Jinchuan Y, Yi L, et al. Antiapoptotic and proapoptotic signaling of cyclophilin A in endothelial cells. Inflammation. 2013;36(3):567–572. [DOI] [PubMed] [Google Scholar]
- [71].Randazzo PA, Andrade J, Miura K, et al. 2000. The Arf GTPase-activating protein ASAP1 regulates the actin cytoskeleton. Proceedings of the National Academy of Sciences of the United States of America 97:4011–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Houdusse A, Cohen C. Structure of the regulatory domain of scallop myosin at 2 A resolution: implications for regulation. Structure. 1996;4(1):21–32. [DOI] [PubMed] [Google Scholar]
- [73].Mackay TFC. The genetic architecture of quantitative traits: lessons from Drosophila. Curr Opin Genet Dev. 2004;14(3):253–257. [DOI] [PubMed] [Google Scholar]
- [74].Remington DL, Purugganan MD. Candidate genes, quantitative trait loci, and functional trait evolution in plants. International Journal of Plant Sciences. 2003;164(S3):S7–S20. [Google Scholar]
- [75].Roff DA. A centennial celebration for quantitative genetics. Evolution. 2007;61(5):1017–1032. [DOI] [PubMed] [Google Scholar]
- [76].Behnke MS, Zhang TP, Dubey JP, et al. Toxoplasma gondii merozoite gene expression analysis with comparison to the life cycle discloses a unique expression state during enteric development. BMC Genomics. 2014;30(1):350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Behnke MS, Dubey JP, Sibley LD. Genetic mapping of pathogenesis determinants inToxoplasma gondii. Annu Rev Microbiol. 2016;70(1):63–81. [DOI] [PubMed] [Google Scholar]
- [78].Pattaradilokrat S, Culleton RL, Cheesman SJ, et al. 2009. Gene encoding erythrocyte binding ligand linked to blood stage multiplication rate phenotype in Plasmodium yoelii yoelii. Proceedings of the National Academy of Sciences 106:7161–7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Otsuki H, Kaneko O, Thongkukiatkul A, et al. 2009. Single amino acid substitution in Plasmodium yoelii erythrocyte ligand determines its localization and controls parasite virulence. Proceedings of the National Academy of Sciences of the United States of America 106:7167–7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Thomas VP, Williamson VM. Segregation and mapping in the root-knot nematode meloidogyne hapla of quantitatively inherited traits affecting parasitism. Phytopathology. 2013;103(9):935–940. [DOI] [PubMed] [Google Scholar]
- [81].Messer PW, Ellner SP, Hairston NG. Can population genetics adapt to rapid evolution? Trends Genet. 2016;32(7):408–418. [DOI] [PubMed] [Google Scholar]
- [82].Sahoo N, Labruyère E, Bhattacharya S, et al. Calcium binding protein 1 of the protozoan parasite Entamoeba histolytica interacts with actin and is involved in cytoskeleton dynamics. J Cell Sci. 2004;117(16):3625–3634. [DOI] [PubMed] [Google Scholar]
- [83].Long S, Brown KM, Drewry LL, et al. Calmodulin-like proteins localized to the conoid regulate motility and cell invasion by Toxoplasma gondii. PLoS Pathog. 2017;13(5):e1006379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Bosch DE, Siderovski DP. G protein signaling in the parasite Entamoeba histolytica. Exp Mol Med. 2013;45(3):e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Ünal CM, Steinert M. Microbial peptidyl-prolyl cis/trans isomerases (PPIases): virulence factors and potential alternative drug targets. Microbiol Mol Biol Rev. 2014;78:544–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Wang P, Heitman J. The cyclophilins. Genome Biol. 2005;6(7):226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Marsolier J, Perichon M, DeBarry JD, et al. Theileria parasites secrete a prolyl isomerase to maintain host leukocyte transformation. Nature. 2015;520(7547):378–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Tuo W, Fetterer R, Jenkins M, et al. Identification and characterization of Neospora caninum cyclophilin that elicits gamma interferon production. Infect Immun. 2005;73(8):5093–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Yau W-L, Blisnick T, Taly J-F, et al. Cyclosporin A treatment of Leishmania donovani reveals stage-specific functions of cyclophilins in parasite proliferation and viability. PLoS Negl Trop Dis. 2010;4(6):e729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Dahan-Pasternak N, Nasereddin A, Kolevzon N, et al. PfSec13 is an unusual chromatin-associated nucleoporin of Plasmodium falciparum that is essential for parasite proliferation in human erythrocytes. J Cell Sci. 2013;126(Pt 14):3055–3069. [DOI] [PubMed] [Google Scholar]
- [91].Chatel G, Fahrenkrog B. Dynamics and diverse functions of nuclear pore complex proteins. Nucleus (Austin, Tex). 2012;3(2):162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Hui L, Wang B, Yang A, et al. Ly-1 antibody reactive clone is an important nucleolar protein for control of self-renewal and differentiation in embryonic stem cells. Stem Cells. 2009;27(6):1244–1254. [DOI] [PubMed] [Google Scholar]
- [93].Su L, Hershberger RJ, Weissman IL. LYAR, a novel nucleolar protein with zinc finger DNA-binding motifs, is involved in cell growth regulation. Genes Dev. 1993;7(5):735–748. [DOI] [PubMed] [Google Scholar]
- [94].Wang B, Lee J, Li P, et al. Stem cell heterogeneity drives the parasitic life cycle of Schistosoma mansoni. eLife. 2018;7. DOI: 10.7554/eLife.35449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Collins JJ 3rd, King RS, Cogswell A, et al. An atlas for Schistosoma mansoni organs and life-cycle stages using cell type-specific markers and confocal microscopy. PLoS Negl Trop Dis. 2011;5(3):e1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Wang B, Collins JJ, Newmark PA. Functional genomic characterization of neoblast-like stem cells in larval Schistosoma mansoni. eLife. 2013;2:e00768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Behnke MS, Khan A, Wootton JC, et al. 2011. Virulence differences in Toxoplasma mediated by amplification of a family of polymorphic pseudokinases. Proceedings of the National Academy of Sciences of the United States of America 108:9631–9636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Reese ML, Zeiner GM, Saeij JPJ, et al. 2011. Polymorphic family of injected pseudokinases is paramount in Toxoplasma virulence. Proceedings of the National Academy of Sciences of the United States of America 108:9625–9630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Saeij JPJ, Boyle JP, Coller S, et al. Polymorphic secreted kinases are key virulence factors in toxoplasmosis. Science (New York, NY). 2006;314(5806):1780–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Fentress SJ, Behnke MS, Dunay IR, et al. Phosphorylation of immunity-related GTPases by a Toxoplasma gondii-secreted kinase promotes macrophage survival and virulence. Cell Host Microbe. 2010;8(6):484–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Taylor S, Barragan A, Su C, et al. A secreted serine-threonine kinase determines virulence in the eukaryotic pathogen Toxoplasma gondii. Science (New York, NY). 2006;314(5806):1776–1780. [DOI] [PubMed] [Google Scholar]
- [102].Yamamoto M, Ma JS, Mueller C, et al. ATF6beta is a host cellular target of the Toxoplasma gondii virulence factor ROP18. J Exp Med. 2011;208(7):1533–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Behnke MS, Khan A, Lauron EJ, et al. Rhoptry proteins ROP5 and ROP18 are major murine virulence factors in genetically divergent south american strains of toxoplasma gondii. PLoS Genet. 2015;22(8):e1005434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


