Abstract
Background:
Differential DNA methylation associated with allergy might provide novel insights into shared or unique etiology of asthma, rhinitis and eczema.
Objective:
We sought to identify DNA methylation profiles associated with childhood allergy.
Methods:
Within the European Mechanisms of the Development of ALLergy (MeDALL) consortium, we performed an epigenome-wide association study of whole blood DNA methylation using a cross-sectional design. Allergy is defined as having symptoms from at least one allergic disease (asthma/rhinitis/eczema) and positive serum specific IgE to common aeroallergens. The discovery study included 219 cases and 417 controls at age 4 and 228 cases and 593 controls at age 8 from three birth cohorts, with replication analyses in 325 cases and 1,111 controls. We performed additional analyses on 21 replicated sites in 785 cases and 2,124 controls by allergic symptoms only from eight cohorts, three not previously included in analyses.
Results:
We identified 80 differentially methylated CpG sites (CpGs) which showed 1–3% methylation difference in the discovery phase, of which 21, including five novel CpGs, passed genome-wide significance after meta-analysis. All 21 CpGs were also significantly differentially methylated with allergic symptoms, and shared between asthma, rhinitis and eczema. The 21 CpGs mapped to relevant genes, including ACOT7, LMAN3 and CLDN23. All 21 CpGs were differently methylated in asthma in isolated eosinophils, and ten were replicated in respiratory epithelium.
Conclusion:
Reduced whole blood DNA methylation at 21 CpGs was significantly associated with childhood allergy. The findings provide novel insights into the shared molecular mechanisms underlying asthma, rhinitis and eczema.
Keywords: Epigenetics, DNA methylation, allergy, IgE, children
Capsule Summary
This large-scale epigenome-wide meta-analysis identified 21 CpG sites in whole blood to be consistently associated with childhood allergic disease and implicates shared epigenetic mechanisms of asthma, rhinitis and eczema.
Introduction
Allergic diseases, such as asthma, rhinitis and eczema are of great public health concern and constitute the most prevalent childhood illnesses worldwide.1 Many allergic diseases have a strong hereditary component, yet large-scale genome-wide association studies2–4 (GWAS) indicate that genetic polymorphisms explain only a limited proportion of the susceptibility to allergy. Moreover, the dramatic rise in prevalence of IgE-mediated allergic diseases in Western European countries over the last decades cannot be explained by genetic factors, but likely by environmental factors and life style changes.5 The analysis of DNA methylation in relation to allergic diseases has therefore attracted much interest because epigenetic modification may mediate environmental effects on the development of allergic diseases6,7. So far, many studies have linked the DNA methylome to one single trait, such as asthma8,9 rhinitis10, eczema,11 total12,13 and allergen-specific IgE levels,14–16 as well as food allergy.14,17 However, allergic diseases often coexist in the same individual,18 an overlap that is partly explained by IgE sensitization. A recent large-scale GWAS2 indicated that the majority of genetic risk factors for allergy are shared between asthma, eczema and rhinitis, suggesting many shared causal mechanisms18,19. So far, no study has assessed shared epigenetic factors related to these three allergic diseases.
To address this question, we implemented a large-scale epigenome-wide association study (EWAS) of allergy and whole-blood DNA methylation in three birth cohorts participating in the Mechanisms of the Development of ALLergy (MeDALL) consortium. This is a follow-up of our previous report on childhood asthma.8 In the current study, we investigated DNA methylation associated with IgE-mediated allergic diseases, asthma, rhinitis or eczema in children at ages 4 and 8 years followed by replication and meta-analysis of children aged up to 16 years. We further tested if our differently methylated CpGs were associated with allergic symptoms only without including specific IgE in our case definition, were shared or different between children with asthma, rhinitis or eczema only and also investigated DNA methylation of replicated CpGs in isolated eosinophils and nasal epithelial cells. Finally, we functionally annotated these CpGs.
Methods
The methods section in this article’s Online Repository provides details on the methods used and cohorts included in this study. Study design and structure of this paper is presented in Fig 1.
Fig 1.
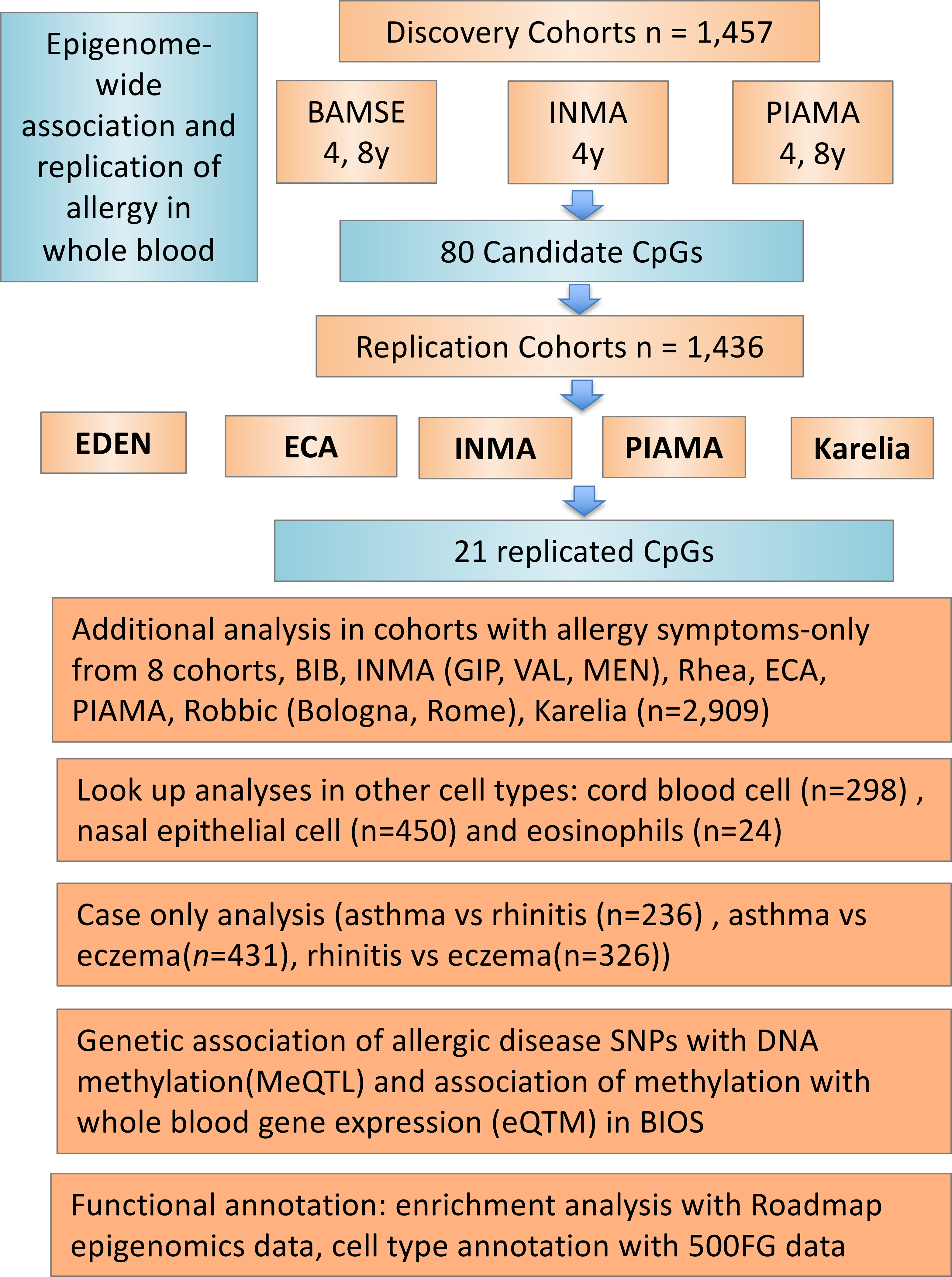
Overview of the study design. The present EWAS study consisted of a discovery and replication phase, followed by a meta-analysis. The top significant CpGs from the discovery phase (n=1,457) were selected for replication from five European cohorts (n=1,436). Subsequently, results from the discovery and replication phases were combined in meta-analysis. The 21 replicated CpGs were further investigated in allergy symptoms-only analysis based on eight cohorts (n=2,909), the look-up analysis in the cord blood and nasal epithelia cell, case only analysis, as well as functional analyses, including MeQTL, eQTM, functional enrichment and cell type annotation.
Allergy definition
The primary outcome of the study is allergy, defined as agreed upon by experts from the MeDALL consortium.18 A child was considered to have allergy if he/she had at least one of three diseases: asthma/rhinitis/eczema, as well as specific IgE against any of the tested allergen (>0.35 lU/mL).
Allergy symptoms-only is defined when a child had at least one of the three diseases: asthma/rhinitis/eczema, without considering specific IgE results information. This definition was also used for comparisons between single diseases.
Detailed definitions of allergy, asthma, rhinitis, eczema and specific IgE positivity in each cohort are provided in the Online Repository.
Study population and design
The present study was conducted within the framework of the MeDALL consortium, and included cohorts with DNA methylation data measured at birth and/or during childhood. The study consisted of two phases, discovery and replication. In discovery and replication, allergic disease endpoint/IgE were collected at the same time as DNA methylation analyses.
In the discovery phase, whole blood DNA methylation was measured in participants of three cohorts, namely, BAMSE (Sweden), INMA (Sabadel [SAB], Spain) and PIAMA (the Netherlands) (Table I and E1). As DNA methylation is age-specific,20 we specified beforehand that the discovery analysis would be performed in two age-groups separately, 4 and 8 years. The discovery study was enriched with cases with allergic disease, and consisted of 219 allergy cases and 417 controls at age 4 years, and 228 allergy cases and 593 controls at age 8 years. After identification of differentially methylated CpGs, we performed replication analysis in 325 allergy cases and 1,111 controls from five cohorts, namely EDEN (France), ECA (Norway), PIAMA (the Netherlands), INMA (MEN, Spain) and Karelia (Finland), and meta-analyzed the results (Table I and E1). Any individuals whose samples have been used in the discovery studies have been excluded in the replication.
Table I.
Study characteristics of discovery and replication cohorts in whole blood DNA methylation analyses
| Cohorta | Country | Age group | Number | Age, years (mean(SD)) | N(%) Male | N(%) allergy (questionnaire + aeroallergen sIgE) | Design |
|---|---|---|---|---|---|---|---|
| BAMSE | Sweden | Pre-school age | 247 | 4.26(0.20) | 134(54.3%) | 117(47.4%) | Discovery |
| INMA-SAB | Spain | 166 | 4.35(0.23) | 84(50.6%) | 38(22.9%) | ||
| PIAMA | The Netherlands | 223 | 4.08(0.21) | 119(53.4%) | 64(28.7%) | ||
| BAMSE | Sweden | School age | 235 | 8.32(0.41) | 125(53.2%) | 96(40.9%) | |
| BAMSE-epi | Sweden | 383 | 8.30(0.52) | 207(54.0%) | 75(19.6%) | ||
| PIAMA | The Netherlands | 203 | 8.06(0.28) | 101(49.8%) | 57(28.1%) | ||
| EDEN | France | Pre-school age | 143 | 5.65(0.10) | 87(60.8%) | 22(15.4%) | Replication |
| BIB | United Kingdom | 514 | 4.53(0.36) | 278(54.1%) | NA | ||
| INMA-GIP | Spain | 74 | 4 | 36(48.6%) | NA | ||
| Rhea | Greece | 490 | 4.26(0.27) | 254(51.8%) | NA | ||
| ECA | Norway | School age | 213 | 10.89(0.78) | 112(52.6%) | 35(16.4%) | |
| INMA-VAL | Spain | 58 | 7.50(0.20) | 31(53.4%) | NA | ||
| PIAMA | The Netherlands | 361 | 8.30(0.46) | 193(53.5%) | 46 (12.7%) | ||
| Robbic-Bologna | Italy | 123 | 7.82(0.51) | 63(51.2%) | NA | ||
| Robbic-Rome | Italy | 214 | 8.73(0.27) | 108(50.5%) | NA | ||
| Karelia | Finland | Adolescent age | 98 | 15.74(1.59) | 38 (38.8%) | 35(35.7%) | |
| INMA-MEN | Spain | 75 | 14 | 35(46.7%) | 14(18.7%) | ||
| PIAMA | The Netherlands | 546 | 16.35(0.22) | 271(49.6%) | 115(21.1%) |
We conducted an allergy symptoms-only additional analysis to test the association of allergy replicated CpGs in 785 cases and 2,124 controls from eight cohorts, including BIB (UK), EDEN (France), ECA (Norway), Karelia (Finland), PIAMA, RHEA (Greece), Robbic (Italy; subsets Bologna and Rome), and INMA (Spain; subsets Gipuzkoa [GIP], Menorca [MEN] and Valencia [VAL]) (Table I and E1).
Furthermore, we investigated potential shared or disease specific DNA methylation markers by comparing three non-overlapping groups of cases that had a single allergic disease: only asthma or only rhinitis or only eczema. Thus, the following groups were compared: asthma vs rhinitis (118 vs 118), asthma vs eczema (252 vs 179) and rhinitis vs eczema (156 vs 170). Information on IgE sensitization status was not considered in the case-only analyses.
We investigated whether cord blood DNA methylation levels at replicated CpGs could predict allergy at age 4–5 years in 298 children from the EDEN, and INMA-SAB cohorts (Table E2). We subsequently studied if the allergy replicated CpGs in whole blood were also differentially methylated in nasal brushed samples from 16-year-old children in the PIAMA cohort and isolated eosinophils (16 asthma cases, 8 controls, age 2–56 years) from families from the Saguenay-Lac-Saint-Jean (SLSJ) region in Canada21 (Table E2). Detailed information of these cohorts is provided in supplementary documents.
Medical Ethical Committees of all institutions approved this study. Written informed consent was obtained from parents or legal guardians of all participating children.
DNA methylation measurements
In the discovery analysis, DNA methylation was measured by Illumina Infinium HumanMethylation450 BeadChips (450K), whereas in the cohorts contributing to the replication, it was measured by either 450K or iPlex (Agena Biosciences). We applied the same quality control (QC) of DNA methylation data and covariates as published previously.8
Functional follow-up of significant DNA methylation findings
We searched for the effects of genetic variants on DNA methylation variation (cis and trans- methylation quantitative trait loci (meQTLs)) by using the public Biobank-Based Integrative Omics Studies (BIOS) Consortium database (the BIOS QTL browser, http://genenetwork.nl/biosqtlbrowser/). We associated blood CpG methylation to whole genome gene expression by RNA-Seq in the BIOS consortium dataset.22 CpGs were annotated by Genomic Regions Enrichment of Annotations Tool (GREAT).23 Functional enrichment analysis was done by overlapping our replicated CpGs with histone marks and chromatin states of 27 blood cell types in the Roadmap epigenome project.24 Gene expression sets associated with replicated CpGs were interrogated for cell type specificity using the 500 functional genomics (500FG) dataset.25,26
Statistical analyses
Allergy-associated differentially methylated CpGs were identified by robust linear regression to account for potential outliers and heteroscedasticity in the data,27 adjusted for sex, cohort and technical co-variables. We used models without adjustment for blood cell type as main model for discovery and replication, and report cell type corrected models by inclusion of estimated cell type proportions28 as covariates in sensitivity analyses. For regional analysis, comb-p29 was used to identify differentially methylated regions (DMRs) by using the summary statistics from models of single CpG site analysis.
We defined genome-wide significance using Bonferroni correction (P <1.14×10−7, 439,306 tests). In each age group, genome-wide significant CpGs were taken further for replication. If less than 10 CpGs met the genome-wide significance level, the top 10 CpGs were selected for replication. We performed inverse variance-weighted fixed-effects meta-analyses with METAL,30 or random effects meta-analysis31, the latter in case of a heterogeneity χ2 test P < 0.05. Our replicated CpGs comprised those that were significantly associated with allergy in the meta-analysis of the replication samples (Bonferroni correction, P < 0.0006, 80 tests) and passed the epigenome-wide significance threshold using Bonferroni correction (P <1.14×10−7) in the meta-analysis of all studies (discovery and replication). To avoid unstable effect estimates due to small numbers, we set a minimum of 10 cases per participating cohort to be included in the analyses. Detailed description of the functional follow-up is provided in the Online Supplemental Methods.
Results
Discovery analyses
The baseline characteristics of the study populations are presented in Table I and E1. In the discovery population, allergy prevalence ranged from 22.9% to 47.4% at the age of 4 years, and 19.6% to 40.9% at age 8 years. In the replication population with IgE data available, 12.7% to 16.4% of children fulfilled the definition of allergy at preschool and school age, respectively, and 18.7% to 35.7% during adolescence.
At age 4 years, the discovery EWAS analysis identified one genome-wide significant CpG-site, namely cg15344640, annotated to Max dimerization protein 3 (MXD3) and Lectin, Mannose Binding 2 (LMAN2) gene (P=2.97 × 10−8; Table E3, Fig 2a, Fig E1a, genomic inflation factor λ=1.15). Based on our pre-specified plan, we selected the top 10 CpGs for replication at age 4 years (Table E3). At age 8 years, 74 CpGs passed genome-wide significance threshold with the strongest association (P=1.76 × 10−10) found for cg03695871 annotated to Adaptor Related Protein Complex 5 Subunit Beta 1 (AP5B1) and Ribonuclease H2 Subunit C (RNASEH2C) genes (Table E4, Fig 2b, Fig E1b, λ=1.09). White blood cell composition was estimated in all discovery cohorts, and we did not observe significant, consistent cell composition differences between allergy and control group in seven cell types in our discovery cohorts at age 4 and 8 years (Fig E2, Table E5). However, the association of CpG methylation with allergy became less significant after eosinophil and neutrophil correction: p values after correction ranged from 1.71 × 10−2 to 1.12 × 10−5 vs 3.58 × 10−7 to 8.27 × 10−10 before correction (Table E3–4), which did suggest that at least part of the results was driven by cell type. Between the top 10 CpGs at age 4 years and the 74 genome-wide significant CpGs at age 8, four CpGs (cg13628444, cg20885063, cg11456013 and cg03131767) were overlapping, thus 80 unique CpGs were selected for replication. DMR analyses identified regions near genes that were previously implicated in the development of allergy such as RUNX3, IL1B, IL13 and AP5B1 (Table E6–7).
Fig 2.
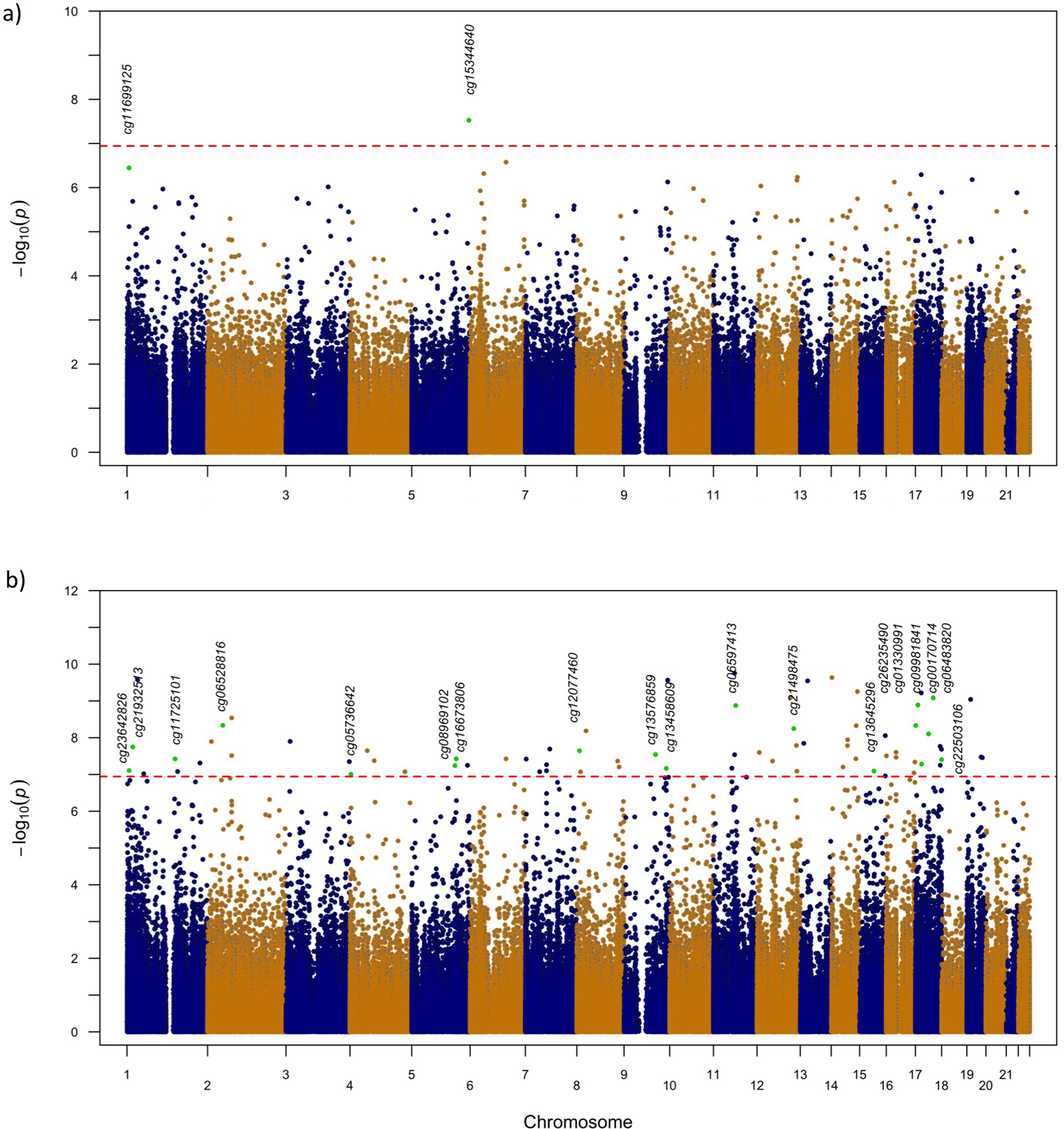
Manhattan plots from the epigenome-wide association studies performed in three European birth cohorts of childhood allergic disease (discovery). A total of 439,306 CpGs were tested for association. The red dotted horizontal line represents the Bonferroni-corrected threshold (p<1.14 × 10–7) of genome-wide significance. All 21 replicated CpGs are marked as green circles and annotated with CpGs name. (A) Results for the EWAS analysis in children aged 4 years (n=636) from the three European cohorts BAMSE, INMA and PIAMA. (B) Results for the EWAS meta-analysis in children aged 8 years (n=821) from BAMSE and PIAMA.
Replication and meta-analysis
Iplex assays for 70 of these 80 CpGs were available using the Iplex design and passed Quality Control (QC). Two CpGs out of 10 selected at age 4 and 19 out of 74 significant CpGs at age 8 years were significantly associated with allergy across childhood in the replication setting (Table II). Of all 21 replicated CpGs, allergic subjects had lower methylation levels than controls. Genes annotated to these 21 CpGs were investigated via literature review (Table E8). In the meta-analysis of the discovery and replication results (Table II), the most significant association (P=9.84 ×10−21) was observed for cg06483820 annotated to A-Kinase Anchoring Protein 1 (AKAP1) and Musashi RNA Binding Protein 2 (MSI2). Forest plots showing cohort-specific and combined estimates and 95% CIs of all replicated and not replicated CpGs are shown in Fig E3 –E6 in this article’s Online Repository.
Table II.
Description of 21 unique replicated allergic disease-related CpG sites across childhood
| CpGs | Age group | Chra | Base pair positionb | Gene namec | Discovery coefd | Discovery P valuee | Houseman corrected P valuef |
Eosinophils corrected P valueg |
Replication Coefh | Replication P valuei | Meta- analysisCoefj | Meta- analysis P valuek |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cg11699125 | 4 years | 1 | 6341327 | GPR153, ACOT7 | −0.0190 | 3.58 × 10−7 | 1.02× 10−5 | 1.66× 10−4 | −0.0306 | 2.87× 10−14 | −0.0271 | 5.84× 10−19 |
| cg15344640 | 5 | 176774729 | MXD3, LMAN2 | −0.0100 | 2.97× 10−8 | 9.46× 10−7 | 1.12× 10−5 | −0.0104 | 1.62× 10−7 | −0.0100 | 2.53× 10−14 | |
| cg23642826 | 8 years | 1 | 6645463 | ZBTB48, KLHL21 | −0.0178 | 7.77× 10−8 | 7.87× 10−5 | 1.71× 10−2 | −0.0127 | 2.88× 10−8 | −0.0140 | 2.60× 10−14 |
| cg21932513 | 1 | 17751974 | RCC2, PADI4 | −0.0161 | 1.79× 10−8 | 2.74× 10−5 | 2.81× 10−3 | −0.0140 | 3.31× 10−7 | −0.0150 | 5.34× 10−14 | |
| cg11725101 | 1 | 148739457 | NBPF16 | −0.0125 | 3.77× 10−8 | 9.03× 10−7 | 1.50× 10−3 | −0.0077 | 5.22× 10−4 | −0.0100 | 3.58× 10−10 | |
| cg06528816 | 2 | 47242277 | TTC7A, CALM2 | −0.0209 | 4.61× 10−9 | 1.81× 10−6 | 3.50× 10−4 | −0.0169 | 1.57× 10−10 | −0.0183 | 8.47× 10−18 | |
| cg05736642 | 4 | 3123132 | RGS12, HTT | −0.0128 | 9.90× 10−8 | 4.67× 10−6 | 2.46× 10−4 | −0.0129 | 2.90× 10−6 | −0.0124 | 1.37× 10−12 | |
| cg16673806 | 5 | 137771872 | REEP2 | −0.0160 | 3.76× 10−8 | 4.94× 10−6 | 1.51× 10−3 | −0.0098 | 6.56× 10−5 | −0.0116 | 5.74× 10−11 | |
| cg08969102 | 5 | 133563532 | PPP2CA | −0.0168 | 5.73× 10−8 | 1.09× 10−5 | 1.25× 10−3 | −0.0103 | 3.74× 10−5 | −0.0129 | 3.81× 10−11 | |
| cg12077460 | 8 | 8702053 | ERI1,CLDN23 | −0.0156 | 2.27× 10−8 | 1.03× 10−6 | 6.39× 10−5 | −0.0144 | 2.98× 10−6 | −0.0150 | 3.73× 10−13 | |
| cg13576859 | 9 | 97403129 | FBP1 | −0.0196 | 2.88× 10−8 | 5.63× 10−7 | 1.64× 10−4 | −0.0128 | 1.05× 10−7 | −0.0147 | 4.91× 10−14 | |
| cg13458609 | 9 | 130608923 | ENG, FPGS | −0.0188 | 6.84× 10−8 | 1.52× 10−6 | 8.92× 10−4 | −0.0164 | 1.53× 10−8 | −0.0174 | 7.08× 10−15 | |
| cg06597413 | 11 | 69251883 | CCND1, TPCN2 | −0.0172 | 1.33× 10−9 | 3.91× 10−6 | 3.98× 10−4 | −0.0123 | 2.25× 10−7 | −0.0137 | 4.20× 10−15 | |
| cg21498475 | 12 | 113737469 | SLC24A6, TPCN1 | −0.0155 | 5.64× 10−9 | 2.20× 10−6 | 1.69× 10−3 | −0.0121 | 2.20× 10−9 | −0.0134 | 1.78× 10−16 | |
| cg13645296 | 15 | 64275810 | HERC1, DAPK2 | −0.0208 | 8.06× 10−8 | 1.41× 10−7 | 4.61× 10−5 | −0.0220 | 3.17× 10−11 | −0.0215 | 1.70× 10−17 | |
| cg26235490 | 17 | 1047567 | ABR, TIMM22 | −0.0132 | 4.64× 10−9 | 9.61× 10−7 | 2.20× 10−3 | −0.0153 | 1.92× 10−10 | −0.0142 | 1.07× 10−17 | |
| cg01330991 | 17 | 7648108 | KDM6B, DNAH2 | −0.0166 | 1.29× 10−9 | 1.58× 10−6 | 4.23× 10−4 | −0.0098 | 9.32× 10−6 | −0.0122 | 2.42× 10−13 | |
| cg09981841 | 17 | 19177387 | EPN2, B9D1 | −0.0171 | 5.21× 10−8 | 1.89× 10−4 | 2.50× 10−2 | −0.0090 | 4.25× 10−4 | −0.0122 | 4.63× 10−10 | |
| cg00170714 | 17 | 40724562 | PSMC3IP, MLX | −0.0147 | 7.87× 10−9 | 1.23× 10−5 | 1.57× 10−3 | −0.0113 | 6.92× 10−7 | −0.0129 | 2.13× 10−14 | |
| cg06483820 | 17 | 55167149 | MSI2, AKAP1 | −0.0159 | 8.27× 10−10 | 7.96× 10−7 | 4.21× 10−4 | −0.0140 | 1.41× 10−12 | −0.0146 | 9.84× 10−21 | |
| cg22503106 | 17 | 81040906 | METRNL | −0.0198 | 3.95× 10−8 | 3.07× 10−6 | 3.32× 10−3 | −0.0143 | 2.28× 10−5 | −0.0174 | 5.68× 10−12 |
Chromosome
Basepair position according to Genome build 37
the CpGs were annotated by GREAT version 3.0.0 (Genomic Regions of Annotations Tool, http://bejerano.stanford.edu/great/)
Discovery - coef: Regression coefficient from discovery study
Discovery- P value: P value from discovery study
Houseman P value: P-value from discovery study after correction for estimated cell counts with default Houseman setting (T cells (CD8T), T cells (CD4T), Natural Killer (NK) cells, B cell, Monocytes, Granulocytes)
Eosinophils P value: P value from discovery study after correction for estimated cell counts (T cells (CD8T), T cells (CD4T), Natural Killer (NK) cells, B cell, Monocytes, Eosinophils, neutrophils)
Replication Coef: Regression coefficient from replication study
Replication P value: P value from meta-analysis of replication study
Meta-analysis coef: regression coefficient from meta-analysis of discovery and replication
Meta analysis P value: P value from meta-analysis of discovery and replication.
Additional replication and functional analyses
In an additional analysis, all 21 abovementioned CpGs were significantly associated with allergy defined by symptoms only (Table III, Fig E7). In a comparison of cases with only asthma, only rhinitis, or only eczema while not taking specific IgE into account, none of the 21 CpGs was differentially methylated (i.e. disease-specific) in relation to any of these three individual diseases (Table E9).
Table III.
Independent meta-analysis of the association of DNA methylation at 21 replicated CpG sites with allergic disease defined on the basis of symptoms without considering IgE levels in 8 replication cohorts
| CpG site | Gene namec | Meta-analysis coef | Meta-analysis P value | Direction |
|---|---|---|---|---|
| cg11699125 | GPR153, ACOT7 | −0.0189 | 1.78E-09 | ----+----+-+ |
| cg15344640 | MXD3, LMAN2 | −0.0064 | 2.95E-04 | --+-++---+-- |
| cg23642826 | ZBTB48, KLHL21 | −0.0079 | 8.06E-07 | ----++------ |
| cg21932513 | RCC2, PADI4 | −0.0093 | 8.26E-07 | ----+++--+-- |
| cg11725101 | NBPF16 | −0.0074 | 5.28E-05 | ----?+-+-+-+ |
| cg06528816 | TTC7A, CALM2 | −0.0111 | 3.51E-08 | ----+----?-- |
| cg05736642 | RGS12, HTT | −0.0069 | 4.73E-05 | ----++------ |
| cg16673806 | REEP2 | −0.0064 | 3.43E-05 | ----++---+-- |
| cg08969102 | PPP2CA | −0.0089 | 2.73E-06 | ----?+------ |
| cg12077460 | ERI1,CLDN23 | −0.0117 | 1.90E-06 | ----++---+-- |
| cg13576859 | FBP1 | −0.0083 | 1.88E-08 | ----++---+-- |
| cg13458609 | ENG, FPGS | −0.0122 | 5.63E-08 | --+-?+------ |
| cg06597413 | CCND1, TPCN2 | −0.0084 | 1.71E-09 | ----+----+-- |
| cg21498475 | SLC24A6, TPCN1 | −0.0088 | 1.48E-07 | --+------?-+ |
| cg13645296 | HERC1, DAPK2 | −0.0172 | 2.78E-12 | ---------?-- |
| cg26235490 | ABR, TIMM22 | −0.0097 | 3.17E-09 | -----+---+-- |
| cg01330991 | KDM6B, DNAH2 | −0.008 | 3.28E-08 | ----++---+-- |
| cg09981841 | EPN2, B9D1 | −0.0075 | 7.01E-05 | --+-++------ |
| cg00170714 | PSMC3IP, MLX | −0.007 | 1.74E-04 | ----?+-----+ |
| cg06483820 | MSI2, AKAP1 | −0.0086 | 1.01E-09 | ----+------- |
| cg22503106 | METRNL | −0.0108 | 8.34E-07 | ----++---+-- |
Coef: Regression coefficient from meta-analysis; P value:
P value from meta-analysis of the replication cohorts only, without considering the discovery population.
For each cohort participating in the analysis, “+” indicates a positive direction of effect, “-” indicates a negative direction of effect, and “?” indicates missing information for that CpG in a given cohort. Cohort order is as follows: BIB, Rhea, INMA (GIP), EDEN, INMA (VAL), PIAMA (age 8), Robbic (Rome), Robbic (Bologna), ECA, INMA (MEN), PIAMA (age 16), Karelia.
Methylation of the 21 allergy-associated CpGs in 298 cord blood DNA samples at birth was not associated with allergy development during early childhood (Table E10), while 10 out of 21 tested CpGs were associated with allergic disease with nominal significance (P <0.05) in nasal respiratory epithelial cells of 16-year-old children of the PIAMA cohort, with matching direction of effect (Table E11).
In the SLSJ cohort, we found all 21 CpG sites to be significantly associated with asthma within eosinophils: Participants with asthma had on average 19% lower DNA methylation of these 21 CpG sites compared to those without asthma. (Table E12).
Functional analyses using Roadmap epigenomics data showed that 17 out of 21 allergy-associated CpGs were significantly enriched for enhancer markers in whole blood Roadmap and Encode data (Tables E13 and E14).
We next investigated whether DNA methylation of the 21 allergy-associated CpGs is driven by allergy-associated genetic variants. Looking up MeQTL from the BIOS QTL browser (https://genenetwork.nl/biosqtlbrowser/), we found one out of 21 CpGs, cg06528816, annotated to tetratricopeptide repeat domain 7A (TTC7A), and calmodulin 2 (CALM2) to be significantly associated in trans to the asthma-associated polymorphism rs2381416 in the Interleukin-33 gene (FDR p value =0.03) (Table E15).32 Interestingly, rs2381416-A has been associated with higher methylation and lower eosinophil levels and higher neutrophil percentage levels (Table E15 and E16).33 Expression quantitative trait methylation (eQTM) of the 21 CpGs with gene expression in cis in 2,353 adults identified 15 CpGs to be associated with 38 blood gene transcripts (Table E17). All CpGs that showed significant trans-eQTM with whole-blood gene expression are shown in Table E18. In the trans-eQTM analysis, we identified four gene expression patterns in whole blood by hierarchical clustering (Fig E8), with cluster 1 and 3 showing distinct patterns. In Cluster 1, CpG methylation was correlated with gene expression of C-C chemokine receptor type 7 (CCR7), Charged Multivesicular Body Protein 7 (CHMP7) and Lymphoid Enhancer Binding Factor 1 (LEF1). In cluster 3, CpG methylation was correlated with gene expression of Sialic Acid Binding Ig Like Lectin 8 (SIGLEC8), Oligodendrocyte Transcription Factor 2 (OLIG2), Adenosine A3 Receptor (ADORA3) and Interleukin 5 Receptor Subunit Alpha (IL5RA), all genes related to blood eosinophils. An RNA-Seq dataset of 89 individuals with 75 measured immune cell (sub)-populations25 from 500 functional genomics project (500FG) showed clustered RNA-transcripts associated with allergy CpGs to be strongly associated with effect/memory CD8 and NK cells and naïve CD8+ CD4+ cells. (Fig E9).
Discussion
This large consortium-based meta-analysis identified 21 CpGs in whole blood to be associated with childhood allergic disease. Consistently lower methylation levels at two CpGs were observed in children with allergy at the age of 4 years and 19 CpGs at the age of 8 years, which were all replicated in independent cohorts up to adolescence. These CpGs associated with allergy defined by asthma, rhinitis or eczema in combination with specific IgE, but also when allergy was defined by disease symptoms only. We found no evidence for single disease-specific CpG methylation signatures, suggesting shared DNA methylation signatures of allergy in blood. DNA methylation at these 21 CpGs in cord blood was not predictive of allergy during early childhood, indicating that postnatal changes in DNA methylation may play an important role in the development of allergy.
Five out of 21 differentially methylated CpGs represent novel findings in the context of EWAS on allergy. One of them is cg21932513, annotated to Regulator of chromosome condensation 2 (RCC2) and Peptidyl arginine deiminase 4 (PADI4). PADI4, being a well-established genetic risk factor for rheumatoid arthritis,34 has recently been implicated in acute peanut allergic reactions.35 Other novel findings include cg11725101, mapping to Neuroblastoma Breakpoint Family Member 16 (NBPF16); cg16673806 - receptor accessory protein 2 (REEP2); cg26235490 - ABR activator of RhoGEF and GTPase (ABR) and translocase of inner mitochondrial membrane 22 (TIMM22), as well as cg01330991 - lysine demethylase 6B (KDM6B) and dynein axonemal heavy chain 2 (DNAH2). These genes have not been previously implicated in allergic disease.
Some of the differentially methylated CpGs found in this study were previously reported. We identified two overlapping CpGs, cg15344640 (MXD3, LMAN2), and cg06483820 (MSI2, AKAP1) that were found in our previous MeDALL EWAS on asthma,18 as well as six overlapping CpGs (cg11699125, cg21498475, cg00170714, cg13576859, cg08969102, cg13458609) with the PACE consortium study on asthma.8 Further, two of 21 CpGs were previously reported in relation to total or specific IgE levels: cg11699125 belonging to G Protein-Coupled Receptor 153 (GPR153) and Acyl-CoA Thioesterase 7 (ACOT7) was found in the Project Viva birth cohort in USA,36 as well as in two studies of Hispanic children.11 Cg11699125 (GPR153, ACOT7), and cg12077460 mapping to Exoribonuclease 1 (ERI1), and Claudin 23 (CLDN23) were previously reported in the Isle of Wight cohort.16 Taken together, these nine CpGs represent replicated DNA methylation markers of allergic disease across multiple populations.
Our top significant CpG, cg06483820, mapped to AKAP1 and MSI2. A-kinase anchoring proteins have been identified in T-cells and contribute to the maintenance of T-cell homeostasis.37 The second most significant finding was cg11699125 in ACOT7, known to play a role in inflammation responses through the production of arachidonic acid and is involved in increased prostaglandin production.38 ACOT7 is particularly interesting, since differential methylation in ACOT7 was previously identified in an EWAS of blood in relation to both total and allergen-specific IgE,13,39 asthma,8,40,41 as well as in an EWAS of nasal epithelium to asthma.42 In this study, we also observed differential methylation of cg11699125 in nasal respiratory epithelial cells associated with allergy. Thus, ACOT7 methylation may relate to asthma, IgE sensitization in both blood and epithelial cells, and provide a cross-tissue allergy associated methylation site. In fact, we observed replication of 10 of the 21 CpGs in respiratory nasal epithelial cells that further support our findings in whole blood. This suggests that many allergy associated DNA methylation markers may be replicable across different tissues. To further investigate this possibility, we did a look-up of published nasal methylation sites that were significantly associated with allergic asthma43 or IgE sensitization15 in our whole blood DNA methylation data at ages 4 and 8 years, and found many CpGs to be significantly associated in blood (Table E19–20).
A recent large genome-wide association study on allergic disease defined by having either asthma, rhinitis or eczema identified 136 SNPs to be significantly associated with allergic disease 2, with case only analyses strongly suggesting that 130 of these 136 SNPs are associated with multimorbidity rather than with single disease phenotypes. Our epigenetic study is the first to confirm this finding for DNA methylation in blood: all 21 CpGs were similarly associated with either allergic disease and no disease specificity could be shown. This could be due to shared epigenetic signatures of IgE sensitization in blood cells, as disease specificity may be more visible in the end organ. We acknowledge that a limitation of this analysis is the lack of data on IgE sensitization, so we suggest that further studies that compare epigenetic patterns in lung, nose and skin in combination with IgE sensitization are needed to address this important question.
Integration of genetic and DNA methylation data revealed two genes that were implicated both by genetic and epigenetic studies. First, LMAN2, which encodes a type I transmembrane lectin that shuttles between the endoplasmic reticulum, the Golgi apparatus, and the plasma membrane. This gene has previously been reported in allergy GWAS 2 and EWAS of childhood asthma.8 Thus, both genetic and epigenetic markers in LMAN2 are associated with allergy, which prompted us to investigate if allergy associated SNPs could be related to our top CpGs. However, no significant association between SNPs and CpGs in LMAN2 was found, suggesting that genetic and epigenetic factors may independently relate to or co-interact in allergy. In contrast, we did identify a strong association between the allergy associated SNP rs2381416 in IL33 at chromosome 9 and CpG methylation in cg06528816 at chromosome 2. The latter finding may be driven by a direct effect of rs238141 on cg06528816 methylation in this gene, or an indirect effect of this SNP regulating blood eosinophils, which is then reflected by the eosinophil associated CpG site cg06528816.
Correlation of our 21CpGs with genome-wide gene expression revealed clusters of associated genes. One cluster included genes such as CCR7, playing a role in allergic airway inflammation, and TCF7, involved in the pathogenesis of lung diseases. Another cluster was inversely associated with cg11699125 and contained genes such as IDO1, earlier implicated in the development of immune responses, HRASLS5, SPNS3, CCL23, ASB2, OLIG2 and SEMA7A, known to be correlated with blood eosinophil counts, CEBPE, involved in activation of granulocyte production47 and PTGDR2, linked to asthma.
Through clustering of the 21 allergy-associated CpGs with immune-cell-specific gene expression signatures, we were also able to annotate the allergy-associated CpGs to activated immune cell subsets. We observed one cluster consisting of genes related to eosinophil function, such as SIGLEC8, OLIG2, ADORA3, IL5RA. The direction of effect suggests that the lower DNA methylation in allergy found in this study relates to a higher eosinophil level. The second cluster is in line with previous findings from the MeDALL EWAS asthma study that whole-blood DNA from children with allergic diseases carries CpG methylation marks associated with lower activity of naïve T-cells and higher activity of effector and memory CD8 T-cells and natural killer cells.18 This allergy-associated shift in cell state might be due to past exposure, such as childhood infections or microbial stimulation. Thus, we interpret the DNA methylation signatures as markers of activated cell subsets in peripheral blood, reflecting current or past cell activation or suppression events in allergy development.
Our study has some strengths and limitations. This is the largest DNA methylation study in childhood allergy, using independent laboratory methods and cohorts for replication. The study uses detailed allergy phenotype definition, as well as availability of samples at multiple ages and nationalities. We further replicated our findings in five cohorts that defined allergy by symptoms and IgE sensitization, but also found that all 21 sites were replicated in eight cohorts where allergy was defined by symptoms only. This suggests that our allergy-associated CpGs represent robust findings in the context of childhood allergy from the age of 4 years onwards.
Previous studies have shown that differences in methylation patterns can be attributed to variability in cell type composition when whole blood is used in DNA methylation studies.26 Thus, DNA methylation may reflect allergy associated changes in cell type composition in whole blood, cell specific changes within the DNA methylome, or a combination of the two, as was shown for asthma in the MeDALL study.18 Since no blood cell-specific DNA methylation data in allergic disease are available, we could only replicate our findings in purified eosinophils for asthma. Here, we found that all CpG sites were significantly associated with asthma, indicating that our results are at least partly driven by DNA methylation changes within eosinophils. We also replicated 10 out of the 21 CpGs in nasal epithelial brushed cells, further underlining the robustness of our findings. Another limitation is that the Illumina 450K platform covered only about 1.6% of all methylation sites of the genome. One would expect to find more allergy-associated CpGs using whole-methylome sequencing in the future. Due to the small numbers of cases with single diseases in the case only analysis, we have limited power to detect unique signatures of single diseases. In our analysis of allergy prediction at birth, we investigated cord blood samples, and not peripheral blood as was done later in life. Recent analyses showed that 70 % of DNA methylation sites are in good agreement between cord and peripheral blood49, which suggests that analyzing cord blood may have introduced an additional source of variation in our analysis.
Our analyses were based on Caucasian populations, and it remains to be investigated whether the findings can be extrapolated to other ethnic groups.
In conclusion, our study identified consistently lower DNA methylation levels in 21 CpGs to be associated with allergy in childhood from ages 4 to 16 years, showing that these methylation sites are shared between asthma, rhinitis and eczema and implicated eosinophils. Our allergy associated DNA methylation profiles are indicative of lower presence or activity of naïve T cells and higher activity or presence of eosinophils and effector and memory CD8 T cells and natural killer cells in childhood allergy.
Supplementary Material
Acknowledgements
We are grateful to all children and families that participated in this study. We especially thank Professor Dirkje Postma for helpful comments and support during the course of this study. MGN was supported by an ERC Advanced Grant (833247) and a Spinoza Grant of the Netherlands Organization for Scientific Research. Catherine Laprise is part of the Quebec Respiratory Health Network (RHN; https://rsr-qc.ca/en/), the investigator of CHILD Study, the director of the Centre intersectoriel en santé durable de l’UQAC and the chairholder of the Canada Research Chair in the Environment and Genetics of Respiratory Disorders and Allergy (http://www.chairs.gc.ca).
Funding
The Mechanisms of the Development of ALLergy (MeDALL) EU project was supported by the seventh Framework programme (grant agreement number 261357). The Biobank-Based Integrative Omics Studies (BIOS) Consortium was funded by BBMRI-NL, a research infrastructure financed by the Dutch government (NWO 184.021.007). Cohort specific funding is described in the Online Supplement.
The funders of the study had no role in the design of the study, data gathering, analysis, interpretation, writing of the report, or in the decision to submit the report for publication.
Abbreviations used:
- 450K
Illumina Infinium HumanMethylation450 BeadChip
- 500FG
500 Functional Genomics project
- BAMSE
Barn/Children, Allergy, Milieu, Stockholm, Epidemiology
- BIB
Born in Bradford
- BIOS
Biobank-based Integrative Omics Studies
- CpGs
CpG sites
- DMR
Differentially Methylated Regions
- ECA
Environment and Childhood Asthma study in Oslo
- EDEN
Déterminants pré et post-natals du développement de la santé de l’enfant
- eQTM
expression quantitative trait methylation
- EWAS
Epigenome-wide association study
- FDR
False discovery rate
- GREAT
Genomic Regions Enrichment of Annotations Tool
- GWAS
Genome-wide association study
- IgE
Immunoglobulin E
- INMA-SAB
Infancia y Medio Ambiente–Sabadel
- MeDALL
Mechanisms of the Development of ALLergy
- meQTLs
methylation quantitative trait loci
- PIAMA
Prevention and Incidence of Asthma and Mite Allergy
- QC
Quality control
- RHEA
Mother-Child Cohort in Crete
- SNP
Single nucleotide polymorphism
Footnotes
Disclosure of potential conflict of interest:
Dr. van Hage reports personal fees from Thermo Fisher Scientific and ALK, personal fees from Biomay AG, Vienna, Austria and Hycor Biomedical LLC, CA, US., outside the submitted work. Dr. Koppelman participated in advisory board meetings for GSK and Pure-IMS, outside the submitted work. All other authors declare no competing interests.
References
- 1.Asher MI, Montefort S, Björkstén B, Lai CK, Strachan DP, Weiland SK, et al. Worldwide time trends in the prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and eczema in childhood: ISAAC Phases One and Three repeat multicountry cross-sectional surveys. The Lancet. 2006;368:733–43. [DOI] [PubMed] [Google Scholar]
- 2.Ferreira MA, Vonk JM, Baurecht H, Marenholz I, Tian C, Hoffman JD, et al. Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology. Nat Genet. 2017;49:1752–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Demenais F, Margaritte-Jeannin P, Barnes KC, Cookson WOC, Altmüller J, Ang W, et al. Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat Genet. 2018;50:42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paternoster L, Standl M, Waage J, Baurecht H, Hotze M, Strachan DP, et al. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat Genet. 2015;47:1449–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linneberg A The increase in allergy and extended challenges. Allergy. 2011;66:1–3. [DOI] [PubMed] [Google Scholar]
- 6.Tost J A translational perspective on epigenetics in allergic diseases. J Allergy Clin Immunol. 2018;142:715–26. [DOI] [PubMed] [Google Scholar]
- 7.Qi C, Xu C-J, Koppelman GH. The role of epigenetics in the development of childhood asthma. Expert Rev Clin Immunol. 2019;15:1287–302. [DOI] [PubMed] [Google Scholar]
- 8.Xu C-J, Söderhäll C, Bustamante M, Baïz N, Gruzieva O, Gehring U, et al. DNA methylation in childhood asthma: an epigenome-wide meta-analysis. Lancet Respir Med. 2018;6:379–88. [DOI] [PubMed] [Google Scholar]
- 9.Reese SE, Xu C-J, den Dekker HT, Lee MK, Sikdar S, Ruiz-Arenas C, et al. Epigenome-wide meta-analysis of DNA methylation and childhood asthma. J Allergy Clin Immunol. 2019;143:2062–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qi C, Jiang Y, Yang IV, Forno E, Wang T, Vonk JM, et al. Nasal DNA methylation profiling of asthma and rhinitis. J Allergy Clin Immunol [Internet]. 2020; Available from: http://www.sciencedirect.com/science/article/pii/S0091674920300348 [DOI] [PMC free article] [PubMed]
- 11.Quraishi BM, Zhang H, Everson TM, Ray M, Lockett GA, Holloway JW, et al. Identifying CpG sites associated with eczema via random forest screening of epigenome-scale DNA methylation. Clin Epigenetics. 2015;7:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang L, Willis-Owen SAG, Laprise C, Wong KCC, Davies GA, Hudson TJ, et al. An epigenome-wide association study of total serum immunoglobulin E concentration. Nature. 2015;520:670–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen W, Wang T, Pino-Yanes M, Forno E, Liang L, Yan Q, et al. An epigenome-wide association study of total serum IgE in Hispanic children. J Allergy Clin Immunol. 2017;140:571–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng C, Van Meel ER, Cardenas A, Rifas-Shiman SL, Sonawane AR, Glass KR, et al. Epigenome-wide association study reveals methylation pathways associated with childhood allergic sensitization. Epigenetics. 2019;1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forno E, Wang T, Qi C, Yan Q, Xu C-J, Boutaoui N, et al. DNA methylation in nasal epithelium, atopy, and atopic asthma in children: a genome-wide study. Lancet Respir Med. 2019;7:336–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, Kaushal A, Merid SK, Melén E, Pershagen G, Rezwan FI, et al. DNA methylation and allergic sensitizations: A genome-scale longitudinal study during adolescence. Allergy. 2019;74:1166–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paparo L, Nocerino R, Bruno C, Di Scala C, Cosenza L, Bedogni G, et al. Randomized controlled trial on the influence of dietary intervention on epigenetic mechanisms in children with cow’s milk allergy: the EPICMA study. Sci Rep. 2019;9:2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinart M, Benet M, Annesi-Maesano I, Berg A von, Berdel D, Carlsen KCL, et al. Comorbidity of eczema, rhinitis, and asthma in IgE-sensitised and non-IgE-sensitised children in MeDALL: a population-based cohort study. Lancet Respir Med. 2014;2:131–40. [DOI] [PubMed] [Google Scholar]
- 19.Lemonnier N, Melén E, Jiang Y, Joly S, Ménard C, Aguilar D, et al. A novel whole blood gene expression signature for asthma, dermatitis and rhinitis multimorbidity in children and adolescents. Allergy [Internet]. 2020. [cited 2020 Apr 13];n/a. Available from: 10.1111/all.14314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu C-J, Bonder MJ, Söderhäll C, Bustamante M, Baïz N, Gehring U, et al. The emerging landscape of dynamic DNA methylation in early childhood. BMC Genomics. 2017;18:25–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laprise C The Saguenay-Lac-Saint-Jean asthma familial collection: the genetics of asthma in a young founder population. Genes Immun. 2014;15:247–55. [DOI] [PubMed] [Google Scholar]
- 22.Bonder MJ, Luijk R, Zhernakova DV, Moed M, Deelen P, Vermaat M, et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat Genet. 2017;49:131–8. [DOI] [PubMed] [Google Scholar]
- 23.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol [Internet]. 2010;28. Available from: 10.1038/nbt.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aguirre-Gamboa R, Joosten I, Urbano PCM, van der Molen RG, van Rijssen E, van Cranenbroek B, et al. Differential Effects of Environmental and Genetic Factors on T and B Cell Immune Traits. Cell Rep. 2016;17:2474–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Oosting M, Smeekens SP, Jaeger M, Aguirre-Gamboa R, Le KTT, et al. A Functional Genomics Approach to Understand Variation in Cytokine Production in Humans. Cell. 2016;167:1099–1110.e14. [DOI] [PubMed] [Google Scholar]
- 27.Fox J, Weisberg S. An R Companion to Applied Regression. SAGE; 2011. 473 p. [Google Scholar]
- 28.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics [Internet]. 2012;13. Available from: 10.1186/1471-2105-13-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pedersen BS, Schwartz DA, Yang IV, Kechris KJ. Comb-p: software for combining, analyzing, grouping and correcting spatially correlated P-values. Bioinformatics. 2012;28:2986–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viechtbauer W Conducting Meta-Analyses in R with the metafor Package. J Stat Softw [Internet]. 2010;36. Available from: http://www.jstatsoft.org/v36/i03/
- 32.Grotenboer NS, Ketelaar ME, Koppelman GH, Nawijn MC. Decoding asthma: Translating genetic variation in IL33 and IL1RL1 into disease pathophysiology. J Allergy Clin Immunol. 2013;131:856–865.e9. [DOI] [PubMed] [Google Scholar]
- 33.Astle WJ, Elding H, Jiang T, Allen D, Ruklisa D, Mann AL, et al. The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell. 2016;167:1415–1429.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki A, Yamada R, Chang X, Tokuhiro S, Sawada T, Suzuki M, et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet. 2003;34:395–402. [DOI] [PubMed] [Google Scholar]
- 35.Watson CT, Cohain AT, Griffin RS, Chun Y, Grishin A, Hacyznska H, et al. Integrative transcriptomic analysis reveals key drivers of acute peanut allergic reactions. Nat Commun. 2017;8:1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng C, Cardenas A, Rifas-Shiman SL, Hivert M-F, Gold DR, Platts-Mills TA, et al. Epigenome-wide association study of total serum immunoglobulin E in children: a life course approach. Clin Epigenetics. 2018;10:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wehbi VL, Taskén K. Molecular Mechanisms for cAMP-Mediated Immunoregulation in T cells – Role of Anchored Protein Kinase A Signaling Units. Front Immunol. 2016;7:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakuma S, Usa K, Fujimoto Y. The regulation of formation of prostaglandins and arachidonoyl-CoA from arachidonic acid in rabbit kidney medulla microsomes by linoleic acid hydroperoxide. Prostaglandins Other Lipid Mediat. 2006;79:271–7. [DOI] [PubMed] [Google Scholar]
- 39.Everson TM, Lyons G, Zhang H, Soto-Ramírez N, Lockett GA, Patil VK, et al. DNA methylation loci associated with atopy and high serum IgE: a genome-wide application of recursive Random Forest feature selection. Genome Med. 2015;7:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang IV, Pedersen BS, Liu A, O’Connor GT, Teach SJ, Kattan M, et al. DNA methylation and childhood asthma in the inner city. J Allergy Clin Immunol. 2015;136:69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arathimos R, Suderman M, Sharp GC, Burrows K, Granell R, Tilling K, et al. Epigenome-wide association study of asthma and wheeze in childhood and adolescence. Clin Epigenetics. 2017;9:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang IV, Pedersen BS, Liu AH, O’Connor GT, Pillai D, Kattan M, et al. The nasal methylome and childhood atopic asthma. J Allergy Clin Immunol. 2017;139:1478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cardenas A, Sordillo JE, Rifas-Shiman SL, Chung W, Liang L, Coull BA, et al. The nasal methylome as a biomarker of asthma and airway inflammation in children. Nat Commun. 2019;10:3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Afshar R, Strassner JP, Seung E, Causton B, Cho JL, Harris RS, et al. Compartmentalized chemokine-dependent regulatory T-cell inhibition of allergic pulmonary inflammation. J Allergy Clin Immunol. 2013;131:1644–1652.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu Y, Wang W, Wang X. Roles of transcriptional factor 7 in production of inflammatory factors for lung diseases. J Transl Med. 2015;13:273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salazar F, Hall L, Negm OH, Awuah D, Tighe PJ, Shakib F, et al. The mannose receptor negatively modulates the Toll-like receptor 4–aryl hydrocarbon receptor–indoleamine 2,3-dioxygenase axis in dendritic cells affecting T helper cell polarization. J Allergy Clin Immunol. 2016;137:1841–1851.e2. [DOI] [PubMed] [Google Scholar]
- 47.Bigler J, Boedigheimer M, Schofield JPR, Skipp PJ, Corfield J, Rowe A, et al. A Severe Asthma Disease Signature from Gene Expression Profiling of Peripheral Blood from U-BIOPRED Cohorts. Am J Respir Crit Care Med. 2016;195:1311–20. [DOI] [PubMed] [Google Scholar]
- 48.Huang T, Hazen M, Shang Y, Zhou M, Wu X, Yan D, et al. Depletion of major pathogenic cells in asthma by targeting CRTh2. JCI Insight [Internet]. 2016. [cited 2020 Mar 26];1. Available from: https://insight.jci.org/articles/view/86689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang Y, Wei J, Zhang H, Ewart S, Rezwan FI, Holloway JW, et al. Epigenome wide comparison of DNA methylation profile between paired umbilical cord blood and neonatal blood on Guthrie cards. Epigenetics. 2020;15:454–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


