Abstract
Bats are the second most diverse order of mammals and key species for ecosystem functioning, providing a wide range of ecosystem services, from pest control to seed dispersal. Chiropterans are known for hosting a large diversity of viruses, in some cases with little or no effect to their health. Here, we report on the results of a screening for DNA (Herpesviridae) and RNA viruses (Rhabdovirus and Pneumovirus), finding a high prevalence and wide diversity of both Beta- and Gamma-Herpesvirus in insectivorous and hematophagous bats of the southern cone of South America. Our findings suggest that bats in the southern neotropics harbor a high diversity of herpesviruses and, at least in some cases, the viral community in the bat species is more strongly associated with ecological traits of the hosts, rather than their taxonomy. The presence of a separate clade into the Gammaherpesvirinae subfamily in the common vampire bat suggests the independent circulation of herpesviruses in hematophagous and insectivorous bats and highlights the properness of these viruses to track vampire bats’ population structure for rabies studies. Hence, we suggest that as other pathogens viruses may be used to track the population dynamics of their hosts, including movement and demographics.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10393-021-01530-2.
Keywords: Herpesvirus, Chiroptera, Uruguay, Rabies virus, Pneumovirus
Introduction
Bats (Order Chiroptera) are known as the second (after rodents) most diverse and widely disperse mammal order in the world, comprising over 1300 species (Teeling et al. 2018). They represent about 20% of all known mammal species and are grouped in two suborders: Yinpterochiroptera and Yangochiroptera, consisting in seven and 14 families, respectively (Teeling et al. 2018). Bats possess a unique set of features among mammals, including the potential to fly long distances, migratory and gregarious habits, and longevity. On top of these attributes, bats provide key ecosystem services, ranging from insect population control to seed dispersal and forest regeneration (Teeling et al. 2018). In recent years, interest in bats as possible reservoir hosts for emerging viral pathogens to humans has increased in line with increased reporting on serological evidences, viral isolation and detection of viruses from bat tissues (Calisher et al. 2006; Brook and Dobson 2015). However, the fact that bats can harbor a wide variety of viruses is not enough to trigger the emergence or re-emergence of pathogens that threat human health. This relationship must be explored in studies that involve not only bats’ features but also natural changes and anthropogenic modifications of the environment (López-Baucells et al. 2017; Plowright et al. 2017; Botto Nuñez et al. 2019a).
At least 24 viral families of viruses have been known to infect bat families from both suborders (Hayman 2016). Some highly infectious ARN viruses such as Henipavirus (Paramyxoviridae) (Chua et al. 2000), severe acute respiratory syndrome coronavirus (SARS-CoV) (Coronaviridae) (Li et al. 2005), filoviruses (Filoviridae) (Leroy et al. 2005), or rabies virus (Rhabdoviridae), have been demonstrated to be related with the zoonotic emergence and human infections. In the case of Pneumoviridae, a clade of viruses related with mouse pneumovirus has been reported from bats of the Yinpterochiroptera suborder (Drexler et al. 2012).
Among DNA viruses, herpesviruses in bats have been reported in an early study by Tandler where cytomegalovirus-like particles were detected in bats by electron microscopy images (Tandler 1996; Wibbelt et al. 2007). The order Herpesvirales is a group of DNA viruses, which consists of three families: Alloherperviridae, Malacoherpesviridae and Herpesviridae (Davison et al. 2009). The latter is divided in three subfamilies Alpha-, Beta- and Gammaherpesvirinae (Alpha-HV, Beta-HV, Gamma-HV, respectively) according to their viral structure, biological properties, sequences similarities, replicative cycles and cytopathogenicity. Herpesviridae family accounts for the most diversity within the herpesviruses, with ten out of the thirteen genera grouped in the family (Sasaki et al. 2014).
Despite that numerous studies of viral discovery have been carried out in bats around the world, little is known about Uruguayan bats and the viruses they harbor. There are 22 bat species recorded in Uruguay, belonging to Vespertilionidae, Molossidae and Phyllostomidae families, with frugivorous, insectivore and hematophagous diets (Botto Nuñez et al. 2019b). The most recent viral study performed in Uruguayan bats focused on the antigenic and genetic characterization of rabies viruses (RABV) detected in bats and livestock, during the first outbreak of sylvatic rabies in the country, occurred in 2007 (Guarino et al. 2013). Hence, the aim of this study is to carry on the first molecular detection and genetic characterization of DNA and RNA viruses from different species of bats from Uruguay, focusing on herpesvirus (HV), (RABV) and pneumovirus (PNE).
Material and Methods
Ethic Statements
The capture and manipulation of the wild animals was made by trained and authorized personal under two scientific capture permits issued by the National Fauna Office at the Secretary of Livestock, Agriculture and Fisheries (MGAP): case files: 176/2012 (Aug. 4th 2012) and 161/14 (Apr. 4th 2014).
Area of Study
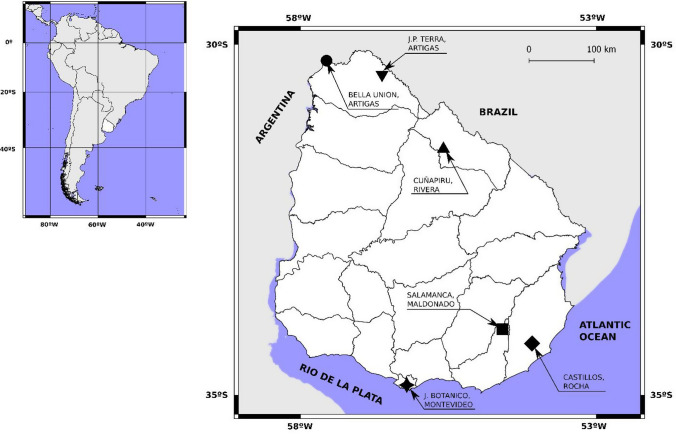
Uruguay is located at 30°–35° (South latitude), and 53°–58° (West longitude) between Argentina and Brazil in America Latina. It is divided administratively in 19 departments (Fig. 1) and has a temperate climate with four clearly delimitated seasons, with an annual average temperature of 17.5º C.
Figure 1.
Continental position of Uruguay and geographic distribution of the samples within the country, showing the 19 s-order administrative units (Departments). Reference: inverted triangle Artigas (Juan P. Terra), circle Artigas (Bella Unión), triangle Rivera (Cuñapirú), square Maldonado (Salamanca), diamond Rocha (Castillos), four pointed star Montevideo.
Sample Collection
A total of 77 oral swabs from nine different species of bats: Desmodus rotundus, Eumops bonariensis, Molossops temminckii, Molossus molossus, Molossus rufus, Tadarida brasiliensis, Eptesicus diminutus, Eptesicus furinalis and Myotis spp. (Table 1), were collected between year 2013 and 2015 at five departments (Artigas, Rivera, Maldonado, Rocha and Montevideo) of Uruguay (Fig. 1). The oral swabs were picked up from bats and placed in cryotubes with 200 µL of viral buffer (AVL® buffer from a Kit of Qiagen, Hilden, Germany), stored at room temperature during transport to the laboratory, where they were processed immediately. Sampled bats were captured using mist nets at the entry of identified roost sites, at a rural zone at an abandoned electric power station, in a forest and also at the botanic garden of Montevideo city. When bats were removed from the net, they were placed in a smooth cloth bag. Sex, reproductive condition, date of capture, taxonomic identity and external measurements were recorded. Species were identified using the last available national field guide, based on external characteristics (González and Martínez-Lanfranco 2010). The bats from the Rivera colony were wing banded as part of a long-term study in the colony. When collected, voucher specimens were deposited at the mammal scientific collection of the National Museum of Natural History in Montevideo (MNHN). Collected voucher specimens were: sample 19, catalog number EEM2327 and sample 24, catalog number EEM2326 (see supplementary Table 3).
Table 1.
Species of Bats, Number of Samples, Georeferenced Place and Year of Capture.
| Family | Species | Samples | Locality | Department | Georeference | Year |
|---|---|---|---|---|---|---|
| Molossidae | Eumops bonariensis | 1 | Colonia J.P. Terra | Artigas | 30.44 S, 56.63 W | 2013 |
| Molossops temminckii | 2 | Colonia J.P. Terra | Artigas | 30.44 S, 56.63 W | 2013 | |
| Tadarida brasiliensis | 28 | Usina Cuñapirú | Rivera | 31.52 S, 55.59 W | 2015 | |
| Molossus molossus | 3 | Castillos | Rocha | 34.29 S, 54.06 W | 2013 | |
| Molossus molossus | 1 | Colonia J. P. Terra | Artigas | 30.44 S, 56.63 W | 2013 | |
| Molossus rufus | 1 | Bella Unión | Artigas | 30.23 S, 57.57 W | 2015 | |
| Phyllostomidae | Desmodus rotundus | 19 | Grutas de Salamanca | Maldonado | 34.48 S, 54.61 W | 2013 |
| Vespertilionidae | Eptesicus furinalis | 2 | Castillos | Rocha | 34.29 S, 54.06 W | 2013 |
| Eptesicus furinalis | 2 | Colonia J.P. Terra | Artigas | 30.44 S, 56.63 W | 2013 | |
| Eptesicus furinalis | 1 | Jardín botánico | Montevideo | 34.86 S, 56.20 W | 2015 | |
| Eptesicus diminutus | 2 | Colonia J. P. Terra | Artigas | 30.44 S, 56.63 W | 2013 | |
| Myotis spp. | 2 | Colonia J. P. Terra | Artigas | 30.44 S, 56.63 W | 2013 | |
| Myotis spp. | 13 | Usina Cuñapirú | Rivera | 31.52 S, 55.59 W | 2015 |
Extraction of Viral Nucleic Acids and Controls
Total viral nucleic acids of all oral swab samples were extracted using a commercial kit QIAmp® Viral RNA (Qiagen, Hilden, Germany) according with manufacturer’s instructions. Five hundred microliters of the same AVL ® buffer was added to the cryotubes with the swab to complete a volume easy to handle. Also 5.6 µL of RNA carrier (Qiagen) was added and incubated 10 min at room temperature to continue with the manufacturer’s instructions. Nucleic acid extractions were aliquoted and frozen at − 80 °C until the molecular analysis.
A positive control for RABV assay was kindly supplied by Dr. H. Guarino from Veterinary Faculty. Total RNA from a brain sample of a field-strain-infected mouse was extracted with TRIZOL® according to the manufacturer´s instructions.
For PNE assays control, a prototype (Long) strain of Respiratory Syncytial Virus (RSV) was propagated in HEp-2 cell cultures. The total RNA extraction from the infected culture was prepared using TRIZOL® protocol with minor modification.
For HV assay, a human Herpes Simplex type 1 strain (HSV) available at the Virology laboratory was propagated in Vero cells. The protocol followed was similar that the one employed to produce the PNE control.
PCR Screening
RABV Assays
A 250 bp fragment of the nucleoprotein gene was amplified by reverse transcription (RT) using specific primers (Supplementary Table 1) and a nested polymerase chain reaction (nested PCR) assay according to Bordignon et al. and Beltran et al. with minor modification (Bordignon et al. 2005; Beltran et al. 2014).
PNE Assays
A 250 bp fragment of the RNA polymerase gene was amplified in a RT-nested PCR using degenerated primers (Supplementary Table 1) according to Tong et al. with minor modifications (Tong et al. 2008).
HV Assays
A fragment between 215 and 315 bp of the DNA polymerase gene was amplified in a multiplex-nested PCR according to Vandevanter et al., with five degenerated oligonucleotides (Supplementary Table 1) (Vandevanter et al. 1996). RT-PCR assays are described in detail in the Supplementary information.
Sequencing and Phylogenetic Analysis
Purification of PCR products of positive samples and nucleotide sequencing (plus and minus strands) were carried out at Macrogen Inc., Korea, with the same primers used in the nested PCR assays.
Sequences were reviewed and edited with BioEdit v7.2.5 software (Hall 1999) and compared with GenBank database sequences, using the nucleotide Basic Local Alignment Search Tool (BLASTn) (Camacho et al. 2009) (Supplementary Table 2). Alignments were constructed using ClustalW (Thompson et al. 1994), and Muscle (Edgar 2004). To estimate the most suitable model of nucleotide substitution, ModelGenerator v0.85 software was used (Keane et al. 2006). Phylogenetic reconstruction was done under the maximum likelihood (ML) criterion, using PhyML software (Guindon et al. 2010). Statistical supports of the tree nodes were calculated by approximate likelihood ratio test (aLRT).
Data Analysis
Total HV prevalence (i.e., Beta-HV and Gamma-HV together) and prevalence of each subfamily (i.e., Beta-HV and Gamma-HV) were analyzed using logistic regressions. We fitted six univariate regressions for each analysis (i.e., Beta-HV + Gamma-HV, Beta-HV, Gamma-HV). For the univariate analyses we used as predictors the host species identity (both as multinomic and comparing T. brasiliensis [the one with the highest prevalence] with all the others); the locality of capture (both as multinomic and comparing Cuñapirú [the one with the highest prevalence] with all the others), sex of the host and age class (adult vs juvenile). For those variables with a significant effect on the viral infection, we calculated the odds ratios (OR) as a measure of strength of the association (Supplementary Tables 4–6). We used Akaike’s information criterion and Akaike’s weights to compare the resulting models. All analyses were performed in R v3.6.2.(R Core Team 2019).
Results
A total of 77 samples from nine autochthonous bats species, collected in 2013 and 2015, at five departments of Uruguay were screened for both DNA and RNA viruses (Table 2 and Supplementary Table 3). Tadarida brasiliensis (n = 28), was the most frequent bat species, while D. rotundus (n = 19) and Myotis spp. (n = 15) accounted for the second and third more frequently sampled bat species. The other bat species were less represented: E. furinalis (n = 5), M. molossus (n = 4), E. diminutus (n = 2) together with M. temminckii (n = 2), M. rufus (n = 1) and E. bonariensis (n = 1).
Table 2.
Rhabdovirus, Pneumovirus and Herpesvirus results: Positive Samples/Samples Analyzed (% [91% Exact Binomial Confidence Interval]).
| Family | Bat species | Rhabdoviridae | Herpesviridae | ||
|---|---|---|---|---|---|
| Pneumoviridae | Beta | Gamma | |||
| Phyllostomidae | D. rotundus | 0/19 (0% [0.0–17.6]) | 0/19 (0% [0.0–17.6]) | 0/14 (0% [0.0–23.2]) | 6/14 (43% [17.7–71.1]) |
| Molossidae | E. bonariensis | 0/1 (0% [0.0–97.5]) | 0/1 (0% [0.0–97.5]) | 0/1 (0% [0.0–97.5]) | 0/1 (0% [0.0–97.5]) |
| M. temminckii | 0/2 (0% [0.0–84.2]) | 0/2 (0% [0.0–84.2]) | 1/2 (50% [12.6–98.7]) | 0/2 (0% [0.0–84.2]) | |
| M. molossus | 0/4 (0% [0.0–60.2]) | 0/4 (0% [0.0–60.2]) | 2/4 (0.5% [6.7–93.2]) | *1/4 (25% [0.6–80.6]) | |
| M. rufus | 0/1 (0% [0.0–97.5]) | 0/1 (0% [0.0–97.5]) | 0/1 (0% [0.0–97.5]) | 1/1 (100% [2.5–100.0]) | |
| T. brasiliensis | 0/28 (0% [0.0–12.3]) | 0/28 (0% [0.0–12.3]) | 16/28 (57% [37.2, 75.5]) | 9/28 (32% [15.9–52.4]) | |
| Vespertilionidae | E. diminutus | 0/2 (0% [0.0–84.2]) | 0/2 (0% [0.0–84.2]) | 1/1 (100% [2.5–100.0]) | 0/1 (0% [0.0–97.5]) |
| E. furinalis | 0/5 (0% [0.0–52.2]) | 0/5 (0% [0.0–52.2]) | 1/5 (20% [0.5–71.6]) | 1/5 (20% [0.5–71.6]) | |
| Myotis spp. | 0/15 (0% [0.0–21.8]) | 0/15 (0% [0.0–21.8]) | 6/15 (40% [16.3–67.7]) | 2/15 (13% [1.7–40.5]) | |
| Total | 0/77 (0% [0.0–4.7]) | 0/77 (0% [0.0–4.7]) | 27/71 (38% [26.8–50.3]) | 20/71 (28% [18.1–40.1]) | |
(*) One of the Samples of M. molossus Could Not be Assigned to Beta- or Gammaherpesvirus.
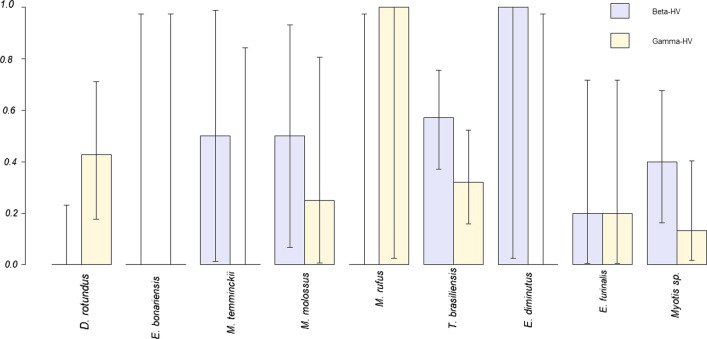
None of the analyzed samples was positive for RABV or PNE amplification (Table 2 and Supplementary Table 3). On the other hand, 48 of the 71 analyzed samples for HV (67.6%) resulted positive. The distribution of positive samples by species was as follows; T. brasiliensis (n = 25/28, 89.3%), D. rotundus (n = 6/14, 42.8%), Myotis spp. (n = 8/15, 53.3%), E. furinalis (n = 2/5, 40%), M. molossus (n = 4/4, 100%), E. diminutus (n = 1/1, 100%), M. temminckii (n = 1/2, 50%), M. rufus (n = 1/1, 100%), Eumops bonariensis (n = 0/1, 0%) (Table 2, Fig. 3).
Figure 3.
Observed Beta- and Gamma-HV prevalence across species. Error bars show exact binomial 95% confidence intervals. Except for D. rotundus and M. rufus, there is a general pattern of more frequent Beta-HV infection. Given sample sizes, T. brasiliensis and Myotis sp. are the species providing more informative prevalence values for both viral subfamilies.
Phylogenetic Analysis
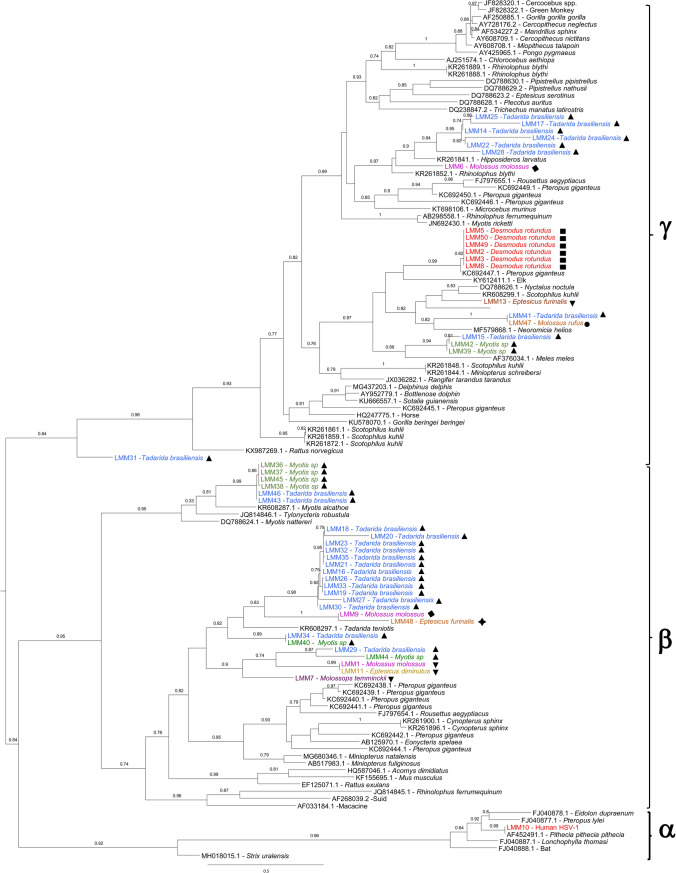
From these positive samples (laboratory code LMM*), high-quality sequences were obtained in 47 amplicons and subsequently included in the phylogenetic analysis. One sample (LMM4) was not included in the analysis, despite been amplified, due to poor sequence quality. Partial polymerase sequences were edited and aligned with 72 sequences downloaded from GenBank, representative of the three subfamilies of Herpesviridae and isolated from diverse bat species and other vertebrates. Maximum likelihood phylogeny showed that viruses from the Alpha-, Beta- and Gamma-HV subfamilies consistently grouped together with significant aLRT support. The samples obtained in this study grouped with sequences from Beta-HV, and Gamma-HV. No Alpha -HV were found in these samples. Twenty-seven samples (56.3%) grouped with sequences from Beta-HV, whereas 20 (41.6%) corresponded to Gamma-HV. Sample codes and species are indicated on the phylogenetic tree, together with accession numbers and hosts for the sequences downloaded from GenBank (Fig. 2). Among Gamma-HV, D. rotundus viruses clustered in a monophyletic group (aLRT = 0.99) together with KC692447.1, an HV collected in a Pteropus giganteus from Bangladesh (Anthony et al. 2013). The whole group displayed a nucleotide identity of 98% (available upon request). Gamma-HV from insectivorous bats grouped in different clusters, and displayed nucleotide identities between 55–76% when compared with other HV. A group of samples from T. brasiliensis from Rivera clustered in a monophyletic group (aLRT = 0.94), and together with an HV from M. molossus captured in Rocha grouped in a subclade of Gamma-HV (aLRT = 0.99) which included diverse HV from Asian and European bats and other vertebrates. A second group of Gamma-HV clustered in another subclade (aLRT = 0.97) together with Gamma-HV from other vertebrates, including bats. These HV were found in several bat species and localities: two from T. brasiliensis, and two Myotis spp., all from Rivera; one E. furinalis and one M. rufus, both captured in Artigas. Although not directly related, these HV grouped in the same subclade as the D. rotundus Gamma-HV. One T. brasiliensis (LMM31) captured in Rivera was the most divergent of the group (40 to 52% identity), displaying a basal position in the Gamma-HV clade (aLRT = 0.84) (Fig. 2).
Figure 2.
Phylogenetic tree of partial Herpesviridae DNA polymerase gene. Positive samples are coded as LMM* and species and localities are shown. The final alignment was 495 nucleotide length. Phylogeny was inferred under the maximum likelihood (ML) criterion and the GTR model of nucleotide substitution. Statistical supports of the nodes were calculated by aLRT and values > to 0.75 are shown. Downloaded sequences are identified by the GenBank accession number. Sequences were submitted to GenBank and DDBJ. The accession numbers are as follows: MT036389-036394, MT036631-036634, MT066061, MT074099-074101, MT906861-906862, MT903351-903357, MT876198, MT936425-936434, MW029377-029384, LC579821-579822, LC578846, LC583830-583031, LC589954.
Among Beta-HV, samples obtained in this study grouped in two main branches. Most HV from T. brasiliensis included in the study captured in Rivera, together with an HV from M. molossus from Rocha, one HV from Myotis spp. (Rivera) and the only HV from Montevideo (in E. furinalis) formed a group with moderate support (aLRT = 0.82); the only sequence from the GenBank database in the group was KR608297.1 (a Beta-HV from Tadarida teniotis, Spain) (Pozo et al. 2016). Twelve of the 16 Beta-HV from T. brasiliensis collected in Rivera displayed a nucleotide identity of 90–100% and formed a monophyletic group with aLRT = 0.98 (Table 2 and Fig. 2).
In the same branch, a subclade (aLRT = 0.90) included viruses from different bat species and two localities: T. brasiliensis, and Myotis spp., both from Rivera and a group of HV from M. molossus, E. diminutus and M. temminckii all captured in Artigas.
The second main branch in the Beta-HV displayed an aLRT = 0.95 and included viruses from four Myotis spp. and two T. brasiliensis all from Rivera, and clustered together with HV from bats from Germany, Spain and China. As well as for the Gamma-HV from insectivorous bats reported in this work, the Beta-HV shared between 57 and 72% nucleotide identity with the previously reported sequences retrieved from GenBank.
For most species, except for M. rufus (just one individual) and D. rotundus, infection with Beta-HV infection was more frequent than Gamma-HV (Fig. 3). Only D. rotundus showed infection exclusively with Gamma-HV, while M. temminckii and E diminutus were only infected by Beta-HV. However, sample sizes in these species were small.
Logistic Regression
In the logistic regression models, T. brasiliensis showed an increased risk for HV infection, both when combining Beta- and Gamma-HV (OR = 6.88, p value < 0.01) and when considering only Beta-HV (OR = 3.74, p value = 0.01). The bats captured at Cuñapirú also showed higher risk of being infected both when combining Beta- and Gamma-HV (OR = 3.73, p value = 0.01) and when considering only Beta-HV (OR = 5.87, p value < 0.01). Finally, juvenile bats showed reduced risk of infection, but only when considering together Beta- and Gamma-HV (OR 0.10, p value = 0.05) (Supplementary Tables 4–6). In the multivariate models, the significant effects of the host species identity and the locality of capture were lost, suggesting an interaction between both variables (T. brasiliensis is the most frequent species captured in Cuñapirú).
Discussion
In this study, 77 samples from nine different species of autochthonous bats from Uruguay were analyzed in order to gain knowledge about the viruses they can harbor. In our country, the first record of viruses associated with bats was the report of RABV from both hematophagous and insectivorous bat species during the 2007 livestock rabies outbreak (Guarino et al. 2013). Despite this, the absence of positive samples for RABV in this work (Table 2) may be explained by the low prevalence observed in wild bat populations. Furthermore, when samples are obtained from free-caught individuals, and not following rabies cases or outbreaks, prevalences have been shown to be significantly lower than those calculated from submitted individuals (Sheeler-Gordon and Smith 2001; Escobar et al. 2013, 2015; Astorga et al. 2015). In our study, only 77 individuals were sampled, and assuming a prevalence of 0.1%, as reported for a large study in Mexico and Paraguay not including submissions of suspected animals) (Sheeler-Gordon and Smith 2001), the probability of detecting a positive sample is as low as 7%. Hence, a negative result should not be interpreted as absence of infection. In accordance with this, our confidence interval for the prevalence (0.0–4.7%) includes the expected prevalence in the individuals analyzed (Sheeler-Gordon and Smith 2001). While the correlation among prevalence estimated from brain samples and from saliva swabs is not completely established (Almeida et al. 2005; Aguilar-Setien et al. 2005), our approach has the advantage of allowing long-term monitoring of the populations, because individuals can be returned to the wild wing banded for later recapture and individual identification, enabling stronger inferences on disease dynamics (Gamble et al. 2020).
For PNE assays, we used a set of degenerate oligonucleotides potentially capable to detect the diversity into the Pneumoviridae family; however, our study did not find PNE positive bats. To our knowledge, the only report of these viruses corresponds to a mouse MPV-related Pneumovirus, detected in Pteropodidae bats (Drexler et al. 2012). Although our bats are not related to the Pteropodidae family, there are reports of similar viruses associated with phylogenetically distant bat families (Wray et al. 2016).
Despite reporting negative results for Rhabdoviridae and Pneumoviridae, the present study contributes to set a baseline for viral circulation in bats in Uruguay and allows for longitudinal studies through wing-banding surveyed individuals.
The distribution of bat sequences across the Herpesviridae tree shows that the sequences analyzed belong to the subfamilies Beta- and Gamma-HV, showing high genetic diversity among them and compared with the HV sequences from other vertebrate species. In order to avoid possible sample contamination, a human strain of HV (LMM10) was included as a control. The position of this sequence in the tree as well as the other HV sequences included in the analysis clusters consistently into the Alpha- Beta- and Gamma-HV clades as reported in previous studies. Regarding the bat samples analyzed, several independent clades in both Gamma- and Beta-HV subfamilies were found, with variable divergence from other HV that cluster together. The comparison of sequence divergences among HV from insectivorous bats and with previously reported HV suggests the finding of new HV species; however, this should be confirmed with further sequencing (i.e., gB glycoprotein gene) (Wibbelt et al. 2007).
HV genetic material was detected in eight of nine bat species and at all capture sites. However, due to the latent phase in HV infections, prevalence may be underestimated since oral swabs were used for detection. Higher numbers of positive samples for Beta-HV (n = 27) in comparison with Gamma-HV (n = 20) were detected, and this bias is even more evident when analyzing only insectivorous bats (27 and 14 positive samples, respectively (Table 2, Fig. 2 and Supplementary Tables 4–6). Overall, given the sample sizes, the most informative prevalences are those for T. brasiliensis and Myotis sp. both for Beta- and Gamma-HV (Fig. 3). Further studies are required in these two species, together with D. rotundus that showed a distinctive infection pattern dominated by Gamma-HV.
The samples from Rivera department, T. brasiliensis and Myotis spp., represent together the largest number of analyzed samples (43/71) corresponding to a mixed colony which roost in the dwellings of an abandoned dam and mineral processing plant, sampled in the same year (2015) (Genta et al. 2017; Botto Nuñez et al. 2018).
In this shelter, both species are found in close contact, to the extreme of pups of both species clustering together, suggesting a common nursery (Botto Nuñez et al. 2018, 2019b). This close association could be favorable for exchange of viruses, especially those present in the saliva, and this can be observed in the phylogenetic tree, where samples from both species cluster together in several clades in Beta-HV as well as in Gamma-HV subfamilies (Fig. 3). The high identity of sequences found in some viruses detected from T. brasiliensis and Myotis spp., with 96% identity at Gamma-HV (LMM15, 42 and 39), and between 74% (LMM44 and LMM29) to 99% (LMM36, 37, 45, 38, 46, 43, and LMM 34 and LMM40) in Beta-HV could strengthen the idea of viral exchange. The phylogenetic structure of the detected viruses suggests that the association is more likely driven by ecological similarities, rather than phylogenetic closeness of the hosts.
The primer sets used in this study to amplify the polymerase gene have been extensively used for HV detection in bats, although lately they have been reported to be biased toward Gamma-HV (Anthony et al. 2015). Considering this bias, and the larger abundance of Beta-HV in that mixed colony, the distribution of positive individuals for both Beta-HV and Gamma-HV could be reflecting a pattern of coinfections. Under this assumption, it is possible that Beta-HV are widely present and that Gamma-HV positive bats might be also Beta-HV positive, with larger sharing of viruses in that colony. Further work to test for coinfections and to characterize their patterns (if present) will be needed to provide a more comprehensive ecological explanation.
Insectivorous bats show a high diversity of viruses and no phylogenetic, temporal, or geographic signal is evident when analyzing the topology of the phylogenetic tree of the HV (Fig. 2).
The viruses found in D. rotundus, sampled from one colony within a two years period, form an independent, strongly supported clade, in exclusion of insectivorous species (Fig. 2). This structure suggests the ecological isolation of this species, even when it can share shelters with insectivorous species in the country (Botto Nuñez et al. 2019b), something that is reinforced by the proved circulation of different rabies virus variants in insectivorous and vampire bats (Guarino et al. 2013; Botto Nuñez et al. 2019a).
It is important also to note that the migratory behavior of T. brasiliensis recorded in the colony (Botto Nuñez et al. 2018) can allow those bats to mix the viral communities of insectivorous bats across their migration areas. Currently available data suggest that only females of T. brasiliensis migrate north during winter, potentially exposing them to contact with different colonies in their wintering areas (Botto Nuñez et al. 2018). Upon return, and during the birth season, colonies of T. brasiliensis and Myotis spp. have been shown to share the roosts, even sharing nursery areas (Unpublished data). Myotis spp. are assumed to have in the country only local short-range movements (Unpublished data). The combination of these two species sharing roosts and the differences in migratory behavior may result in a fission/fusion dynamic for the groups that would allow for easier exchange of viruses across colonies (Gilbert et al. 2006; Olsen et al. 2006; Hill et al. 2012; Tian et al. 2015; Boulinier et al. 2016). Furthermore, these highly prevalent viruses can be used as markers to track migration routes and hence can be used to inform conservation policies for a species protected by international treaties (Hutson et al. 2001; Mickleburgh et al. 2002; Botto Nuñez et al. 2019b).
While we cannot rule out a sampling artifact, given the disbalance in species frequencies, T. brasiliensis appeared with significant higher prevalences, both when analyzing all HV together and when analyzing only Beta-HV infection (Supplementary Tables 4–6). The also disbalanced distribution of the species across capture sites does not allow to completely separate the effect of the host identity and the locality of capture, as most T. brasiliensis were sampled in Cuñapirú. While the infection and transmission dynamics of Beta- and Gamma-HV might be different, the significant effect of the age class on the total prevalence is consistent with a long-lasting infection.
These associations highlight the need to provide a comprehensive ecological background to virologic findings. Even in a country with limited knowledge of its chiropteran fauna (González and Lessa 2014; Botto Nuñez et al. 2019b), the ecology and behavior of the bat species can contribute to the interpretation of virologic findings, and in turn, these findings can be used as tools to more in-depth studies of movement and association. This work reveals the need for more detailed studies on the virome of bats from the South American southern cone and for larger collaborative studies among disciplines.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
GBN was funded by a Fulbright Graduate Scholarship and Bat Conservation International (Student Research Scholarship for Global Bat Conservation Priorities). We want to thank the collaboration of the members of the Program for Conservation of Bats from Uruguay (PCMU), and especially to Ana Laura Rodales and Enrique González for their help with the field work and the collection of samples for this study.
Footnotes
Lucía Moreira Marrero and Germán Botto Nuñez have contributed equally to this work.
Contributor Information
Adriana Delfraro, Email: adriana@fcien.edu.uy.
Sandra Frabasile, Email: sfrabasile@fcien.edu.uy.
References
- Aguilar-Setien A, Loza-Rubio E, Salas-Rojas M, Brisseau N, Cliquet F, Pastoret PP, Rojas-Dotor S, Tesoro E, Kretschmer R. Salivary excretion of rabies virus by healthy vampire bats. Epidemiol Infect. 2005;133:517–522. doi: 10.1017/S0950268805003705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida MF, Martorelli LFA, Aires CC, Sallum PC, Durigon EL, Massad E. Experimental rabies infection in haematophagous bats Desmodus rotundus. Epidemiol Infect. 2005;133:523–527. doi: 10.1017/S0950268804003656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony SJ, Epstein JH, Murray KA, Navarrete-Macias I, Zambrana-Torrelio CM, Solovyov A, Ojeda-Flores R, Arrigo NC, Islam A, Ali Khan S, Hosseini P, Bogich TL, Olival KJ, Sanchez-Leon MD, Karesh WB, Goldstein T, Luby SP, Morse SS, Mazet JAK, Daszak P, Lipkin WI. A Strategy To Estimate Unknown Viral Diversity in Mammals. Mbio. 2013 doi: 10.1128/mbio.00598-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony SJ, Islam A, Johnson C, Navarrete-Macias I, Liang E, Jain K, Hitchens PL, Che X, Soloyvov A, Hicks AL, Ojeda-Flores R, Zambrana-Torrelio C, Ulrich W, Rostal MK, Petrosov A, Garcia J, Haider N, Wolfe N, Goldstein T, Morse SS, Rahman M, Epstein JH, Mazet JK, Daszak P, Lipkin WI. Non-random patterns in viral diversity. Nat Commun. 2015;6:8147. doi: 10.1038/ncomms9147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astorga F, Escobar LE, Poo-Muñoz DA, Medina-Vogel G. Dog ownership, abundance and potential for bat-borne rabies spillover in Chile. Prev Vet Med. 2015;118:397–405. doi: 10.1016/j.prevetmed.2015.01.002. [DOI] [PubMed] [Google Scholar]
- Beltran FJ, Dohmen FG, Del Pietro H, Cisterna DM. Diagnosis and molecular typing of rabies virus in samples stored in inadequate conditions. J Infect Dev Ctries. 2014;8:1016–1021. doi: 10.3855/jidc.4136. [DOI] [PubMed] [Google Scholar]
- Bordignon J, Brasil-dos-Anjos G, Bueno CR, Salvatiera-Oporto J, Davila AMR, Grisard EC, Zanetti CR. Detection and characterization of rabies virus in Southern Brazil by PCR amplification and sequencing of the nucleoprotein gene. Arch Virol. 2005;150:695–708. doi: 10.1007/s00705-004-0448-y. [DOI] [PubMed] [Google Scholar]
- Botto Nuñez G, Becker DJ, Plowright RK. The emergence of vampire bat rabies in Uruguay within a historical context. Epidemiol Infect. 2019;147:1–8. doi: 10.1017/S0950268819000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto Nuñez G, Genta M, Díaz M, Rodales AL, González EM. Circannual sex distribution of the Brazilian Free-tailed Bat, Tadarida brasiliensis (Chiroptera:Molossidae), suggests migration in colonies from Uruguay. Mastozool Neotrop. 2018;25:213–219. doi: 10.31687/saremMN.18.25.1.0.17. [DOI] [Google Scholar]
- Botto Nuñez G, González EM, Rodales AL. Conservación De Los Murciélagos (Mammalia: Chiroptera) De Uruguay: Estado Actual Y Perspectivas. Mastozoología Neotrop. 2019;26:1–33. doi: 10.31687/saremmn.19.26.1.0.05. [DOI] [Google Scholar]
- Boulinier T, Kada S, Ponchon A, Dupraz M, Dietrich M, Gamble A, Bourret V, Duriez O, Bazire R, Tornos J, Tveraa T, Chambert T, Garnier R, McCoy KD (2016) Migration, Prospecting, Dispersal? What Host Movement Matters for Infectious Agent Circulation? In: Integrative and Comparative Biology. Oxford University Press, pp 330–342 [DOI] [PubMed]
- Brook CE, Dobson AP. Bats as “special” reservoirs for emerging zoonotic pathogens. Trends Microbiol. 2015;23:172–180. doi: 10.1016/j.tim.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. Bats: Important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006;19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua KB, Bellini WJ, Rota PA, Harcourt BH, Tamin A, Lam SK, Ksiazek TG, Rollin PE, Zaki SR, Shieh W, Goldsmith CS, Gubler DJ, Roehrig JT, Eaton B, Gould AR, Olson J, Field H, Daniels P, Ling AE, Peters CJ, Anderson LJ, Mahy BW. Nipah virus: a recently emergent deadly paramyxovirus. Science. 2000;288:1432–1435. doi: 10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- Davison AJ, Eberle R, Ehlers B, Hayward GS, McGeoch DJ, Minson AC, Pellett PE, Roizman B, Studdert MJ, Thiry E. The order Herpesvirales. Arch Virol. 2009;154:171–177. doi: 10.1007/s00705-008-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler JF, Corman VM, Müller MA, Maganga GD, Vallo P, Binger T, Gloza-Rausch F, Rasche A, Yordanov S, Seebens A, Oppong S, Sarkodie YA, Pongombo C, Lukashev AN, Schmidt-Chanasit J, Stöcker A, Carneiro AJB, Erbar S, Maisner A, Fronhoffs F, Buettner R, Kalko EKV, Kruppa T, Franke CR, Kallies R, Yandoko ERN, Herrler G, Reusken C, Hassanin A, Krüger DH, Matthee S, Ulrich RG, Leroy EM, Drosten C. Bats host major mammalian paramyxoviruses. Nat Commun. 2012 doi: 10.1038/ncomms1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar LE, Peterson AT, Favi M, Yung V, Pons DJ, Medina-Vogel G. Ecology and Geography of Transmission of Two Bat-Borne Rabies Lineages in Chile. PLoS Negl Trop Dis. 2013 doi: 10.1371/journal.pntd.0002577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar LE, Restif O, Yung V, Favi M, Pons DJ, Medina-Vogel G. Spatial and temporal trends of bat-borne rabies in Chile. Epidemiol Infect. 2015;143:1486–1494. doi: 10.1017/S095026881400226X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble A, Garnier R, Chambert T, Gimenez O, Boulinier T. Next-generation serology: integrating cross-sectional and capture–recapture approaches to infer disease dynamics. Ecology. 2020 doi: 10.1002/ecy.2923. [DOI] [PubMed] [Google Scholar]
- Genta M, Díaz M, Botto G (2017) Propuesta de Sitio de Importancia para la Conservación de los Murciélagos (SICOM), Usina de Cuñapirú (Dpto. Rivera, Uruguay). 1–11
- Gilbert M, Xiao X, Domenech J, Lubroth J, Martin V, Slingenbergh J. Anatidae migration in the western Palearctic and spread of highly pathogenic avian influenza H5N1 virus. Emerg. Infect. Dis. 2006;12:1650–1656. doi: 10.3201/eid1211.060223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González EM, Lessa EP (2014) Historia de la mastozoología en Uruguay. In: Ortega J, Martínez JL, Tirira DG (eds) Historia de la mastozología en Latinoamérica, las Guyanas y el Caribe. Editorial Murciélago Blanco y Asociación Ecuatoriana de Mastozoología, Quito y México DF, pp 381–404
- González EM, Martínez-Lanfranco JA (2010) Mamíferos de Uruguay. Guía de campo e introducción a su estudio y conservación. Vida Silvestre – Museo Nacional de Historia Natural, Ediciones de la Banda Oriental, Montevideo
- Guarino H, Castilho JG, Souto J, Oliveira RN, Carrieri ML, Kotait I. Antigenic and genetic characterization of rabies virus isolates from Uruguay. Virus Res. 2013;173:415–420. doi: 10.1016/j.virusres.2012.12.013. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascue O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0.". Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hayman DTS. Bats as Viral Reservoirs. Annu Rev Virol. 2016;3:77–99. doi: 10.1146/annurev-virology-110615-042203. [DOI] [PubMed] [Google Scholar]
- Hill NJ, Takekawa JY, Ackerman JT, Hobson KA, Herring G, Cardona CJ, Runstadler JA, Boyce WM. Migration strategy affects avian influenza dynamics in mallards (Anas platyrhynchos) Mol Ecol. 2012;21:5986–5999. doi: 10.1111/j.1365-294X.2012.05735.x. [DOI] [PubMed] [Google Scholar]
- Hutson AM, Mickleburgh SP, Racey PA (eds) (2001) Microchiropteran bats. Global survey and conservation action plan. IUCN/SSC Chiroptera Specialist Group, Gland
- Keane TM, Creevey CJ, Pentony MM, J NT, McInerney JO, Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol. 2006;6:29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, Délicat A, Paweska JT, Gonzalez J-P, Swanepoel R. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, Zhang J, McEachern J, Field H, Daszak P, Eaton BT, Zhang S, Wang LF (2005) Bats are natural reservoirs of SARS-like coronaviruses. Science (80- ) 310:676–679 [DOI] [PubMed]
- López-Baucells A, Rocha R, Fernández-Llamazares Á. When bats go viral: Negative framings in virological research imperil bat conservation. Mamm Rev. 2017 doi: 10.1111/mam.12110. [DOI] [Google Scholar]
- Mickleburgh SP, Hutson AM, Racey PA. A review of the global conservation status of bats. Oryx. 2002;36:18–34. doi: 10.1017/S0030605301000011. [DOI] [Google Scholar]
- Olsen B, Munster VJ, Wallensten A, Waldenström J, Osterhaus ADME, Fouchier RAM (2006) Global patterns of influenza A virus in wild birds. Science (80-. ). 312:384–388 [DOI] [PubMed]
- Plowright RK, Parrish CR, Mccallum H, Hudson PJ, Ko AI, Graham L, Lloyd-smith JO. Pathways to zoonotic spillover. Nat Rev Microbiol. 2017;15:502–510. doi: 10.1038/nrmicro.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo F, Juste J, Vázquez-Morón S, Aznar-López C, Ibáñez C, Garin I, Aihartza J, Casas I, Tenorio A, Echevarría JE. Identification of novel betaherpesviruses in iberian bats reveals parallel evolution. PLoS One. 2016 doi: 10.1371/journal.pone.0169153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2019) R Core Team (2019) R A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. - References - Scientific Research Publishing
- Sasaki M, Setiyono A, Handharyani E, Kobayashi S, Rahmadani I, Taha S, Adiani S, Subangkit M, Nakamura I, Sawa H, Kimura T. Isolation and Characterization of a Novel Alphaherpesvirus in Fruit Bats. J Virol. 2014;88:9819–9829. doi: 10.1128/JVI.01277-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeler-Gordon LL, Smith JS. Survey of Bat Populations From Mexico and Paraguay for Rabies. J Wildl Dis. 2001;37:582–593. doi: 10.7589/0090-3558-37.3.582. [DOI] [PubMed] [Google Scholar]
- Tandler B. Cytomegalovirus in the principal submandibular gland of the little brown bat, Myotis lucifugus. J Comp Pathol. 1996;114:1–9. doi: 10.1016/S0021-9975(96)80057-0. [DOI] [PubMed] [Google Scholar]
- Teeling E, Vernes S, Davalos LM, Ray DA, Gilbert MTP, Myers E, Bat1K Consortium (2018) Bat Biology, Genomes, and the Bat1K Project: To Generate Chromosome-Level Genomes for all Living Bat Species. Annu Rev Anim Biosci 6:annurev–animal–022516–022811. 10.1146/annurev-animal-022516-022811 [DOI] [PubMed]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Zhou S, Dong L, Van Boeckel TP, Cui Y, Wu Y, Cazelles B, Huang S, Yang R, Grenfell BT, Xu B. Avian influenza H5N1 viral and bird migration networks in Asia. Proc Natl Acad Sci U S A. 2015;112:172–177. doi: 10.1073/pnas.1405216112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Chern SWW, Li Y, Pallansch MA, Anderson LJ. Sensitive and broadly reactive reverse transcription-PCR assays to detect novel paramyxoviruses. J Clin Microbiol. 2008;46:2652–2658. doi: 10.1128/JCM.00192-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandevanter DR, Warrener P, Bennett L, Schultz ER, Coulter S, Garber RL, Rose TM. Detection and analysis of diverse herpesviral species by consensus primer PCR. J Clin Microbiol. 1996;34:1666–1671. doi: 10.1128/jcm.34.7.1666-1671.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibbelt G, Kurth A, Yasmum N, Bannert M, Nagel S, Nitsche A, Ehlers B. Discovery of herpesviruses in bats. J Gen Virol. 2007;88:2651–2655. doi: 10.1099/vir.0.83045-0. [DOI] [PubMed] [Google Scholar]
- Wray AK, Olival KJ, Morán D, Lopez MR, Alvarez D, Navarrete-Macias I, Liang E, Simmons NB, Lipkin WI, Daszak P, Anthony SJ. Viral Diversity, Prey Preference, and Bartonella Prevalence in Desmodus rotundus in Guatemala. Ecohealth. 2016;13:761–774. doi: 10.1007/s10393-016-1183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.