Keywords: dysbiosis, gut microbiome, necrotizing enterocolitis, preterm, sepsis
Abstract
Advances in metagenomics have allowed a detailed study of the gut microbiome, and its role in human health and disease. Infants born prematurely possess a fragile gut microbial ecosystem that is vulnerable to perturbation. Alterations in the developing gut microbiome in preterm infants are linked to life-threatening diseases such as necrotizing enterocolitis (NEC) and late-onset sepsis; and may impact future risk of asthma, atopy, obesity, and psychosocial disease. In this mini-review, we summarize recent literature on the origins and patterns of development of the preterm gut microbiome in the perinatal period. The host-microbiome-environmental factors that portend development of dysbiotic intestinal microbial patterns associated with NEC and sepsis are reviewed. Strategies to manipulate the microbiome and mitigate dysbiosis, including the use of probiotics and prebiotics will also be discussed. Finally, we explore the challenges and future directions of gut microbiome research in preterm infants.
INTRODUCTION
The human gastrointestinal (GI) tract harbors a complex and diverse microbial community which is dominated by bacteria, but also includes viruses, archaea, fungi, and other eukaryotes. This complex ecosystem with 1012−14 cells from 100 to 1,000 species is considered to be the most dense microbial habitat on earth (1). Compared with ∼23,000 genes of human genome, the gut microbiome has been referred to as a “hidden” metabolic organ and encodes over 3 million genes producing thousands of metabolites (2). Briefly, 90% of the GI microbiota is composed of bacteria from two major phyla namely Bacteroidetes and Firmicutes. Other phyla consistently found in human gut include Proteobacteria, Actinobacteria, Fusobacteria, and Verrucomicrobia (3). At lower taxonomic levels however, individuals differ considerably in composition of fecal microbiota and each individual may have his or her own distinctive pattern of microbial profile (4).
Studies suggest that the early pattern of infant gut microbial colonization is critical for proper development of the human GI tract. The neonatal gut microbiota plays an essential role in the acquisition of postnatal intestinal endotoxin tolerance (5) and specifically regulates maturation of regulatory T-cells (CD4+, Foxp3+), natural killer cell, and gamma delta T-cells (6). Dysbiosis or imbalance in gut microbial communities during this critical period is linked to several diseases including inflammatory, metabolic, neurologic, cardiovascular, and GI illnesses (7). Compared with term neonates, infants born prematurely are at greater risk for disruptions to the gut microbiota. Herein, we briefly review commonly used metagenomic methods for microbiome analysis, compare the challenges of preterm versus term gut microbiome, delineate antecedents and consequences of preterm gut dysbiosis, and discuss advances in microbiome modulation therapy in preterm infants. We focus predominantly on the bacterial communities as most research has been done in this area.
METHODS OF MICROBIAL PROFILING WITH 16S AND SHOTGUN SEQUENCING
Currently available high-throughput sequencing technologies used to profile the genomic composition of a microbial community in a culture-independent manner can be subdivided into two different approaches, namely 16S rRNA gene sequencing and shotgun metagenomics. In 16S rRNA sequencing, the 16S rRNA gene is used as a genetic fingerprint to decipher bacterial phylogeny and taxonomy. A component of the 30S subunit of prokaryotic ribosome, the 16S rRNA gene is 1,500 bp long and comprises nine variable regions (V1–V9) interspersed among the conserved regions of its sequence. These hypervariable regions are unique to each bacterial taxon, whereas the conserved regions allow for the development of universal primers that bind to known sequences shared among most bacteria. During data analysis, amplified 16S rRNA sequences are assigned to operational taxonomic units (OTUs) and OTU clustering methods using a moderately stringent sequence identity threshold allowing for basic taxonomic assignments (family level or higher) (8). Amplicon sequencing targeting the 16S rRNA gene is limited by differences arising from the different variable region chosen for PCR amplification, sequencing errors, poor discriminatory power for some genera, and severely limited resolution at the species level due to short-read amplicon sequencing, potential for contamination, and sequencing platform used (9–11).
Unlike 16S sequencing, which only targets 16S rRNA sequences, shotgun metagenomics generates whole genome sequencing data that can be compared with existing reference databases for accurate taxonomic identification. By sequencing all genetic information within a sample, shotgun metagenomic studies can extract both taxonomic and functional information from complex microbial communities and therefore guide phenotypic studies to understand their potential roles in health and disease. Although shotgun metagenomics can be used to extract species- and even strain-level information through computational approaches, it can still be challenging to differentiate between closely related or coexisting bacterial taxa (12). This has mandated the need for extensive and well-characterized collections of reference genomes including those from the Human Microbiome Project and the Human Gastrointestinal Bacteria Genome Collection (13, 14).
CHALLENGES OF THE PRETERM INFANT THAT SHAPE THE GUT MICROBIOME IN COMPARISON WITH THE TERM INFANT
Preterm infants face several unique environmental and host conditions that negatively impact the development of their gut microbiome. Even before birth, ∼25%–30% of preterm infants are exposed to microbes in the context of preterm premature rupture of membranes and intra-amniotic infection (15). Cesarean section, with subsequent gut colonization by skin microbiota rather than vaginal and rectal microbiota from vaginal delivery, is also more common in preterm than term infants (16). After delivery, most preterm infants need stabilization and subsequent care in the neonatal intensive care unit which, while necessary, is also invasive and results in exposure to the hospital microbial environment. Consequently, as risk of serious nosocomial infection is high, preterm infants are often exposed to powerful broad-spectrum antibiotics throughout their hospital stay. Preterm nutrition and feeding practices—such as delayed introduction of enteral feedings, frequent withholding of feeds, use of acid-suppressive medications, and provision of exclusive human milk diet or formula—are also major modifiers of gut microbiota composition (17). Together, these environmental factors interact with the intrinsically immature GI tract and immune system of the preterm infant in a highly complex but poorly defined process that significantly impact the developing preterm gut microbiome (18, 19).
In general, the intestinal microbiota in the preterm infant differs from the term infant in having delayed colonization, fewer bacterial species, and less diversity and abundance (18, 20, 21). There is also a predilection to being colonized by potentially pathogenic facultative anaerobes (e.g., Enterobacter, Escherichia, and Klebsiella) and decreased levels of commensal strictly anaerobic organisms (e.g., Bifidobacterium, Bacteroides, and Clostridium) (Table 1) (27). A shared patterned progression of microbial communities from Bacilli to Gammaproteobacteria to Clostridia has also been described in preterm infants (28). The pace of this patterned progression is predominantly influenced by gestational age, whereas the mode of delivery, antibiotic exposure, and type of feeding appear to have less impact (28).
Table 1.
Major differences in bacterial gut colonization of preterm infants compared with term infants
| Bacterial Groups | Important Characteristics and Function |
|---|---|
| Decreased in preterm infants | |
| Bacteroidetes (e.g., Bacteroides fragilis) | Anti-inflammatory function through surface component polysaccharide A (22) |
| Actinobacteria (e.g., Bifidobacterium infantis, Bifidobacterium breve) | Anti-inflammatory function through secretion of short chain fatty acids (23), and acceleration of maturation of intestinal innate immune response genes (24) |
| Increased in preterm infants | |
| Firmicutes (e.g., Enterococcus faecalis, Staphylococcus) | Gram-positive cocci that can act as opportunistic pathogens, especially in patients with prolonged hospitalization or has received multiple antibiotic therapy (25) |
| Proteobacteria (e.g., Escherichia coli, Klebsiella pneumoniae) | Gram-negative bacteria commonly associated with opportunistic nosocomial infections; have an outer membrane that contains the endotoxin lipopolysaccharide (26) |
Whether the differences in gut microbiota of preterm versus term infant constitute true dysbiosis or merely represent the common gut microbiome signature in very-low-birth weight infants is debatable. Current studies do not clarify whether a distinct pattern of dysbiosis that portends disease susceptibility exists, apart from a modest increase in Enterobacteriaceae in infants who develop necrotizing enterocolitis (NEC), as further discussed in consequences of dysbiosis in preterm infants.
ANTECEDENTS OF DYSBIOSIS IN PRETERM INFANTS
Prenatal Factors
The development of intestinal microbiome in the preterm neonate and the proclivity to a dysbiotic signature are influenced by several prenatal factors (Fig. 1). Maternal gestational diet appears to alter the composition of the first stool after birth (meconium), with depletion of Bacteroides observed with high-fat diet (29). Other studies have described an interactive effect between maternal diet and mode of delivery on the neonatal gut microbiome, with fruit intake increasing intestinal Streptococcus/Clostridium in infants delivered vaginally, whereas increased dairy intake was associated with higher abundance of Clostridium among infants delivered by cesarean section (30). In pregnancies complicated by intrauterine amniotic infection or chorioamnionitis, a higher relative abundance of Mycoplasmataceae and phylum Bacteroidetes has been identified in stool of infants postnatally (31). Although previous studies had suggested the presence of microbial communities in the placenta and amniotic fluid of healthy pregnancies potentially colonizing the gut prenatally, more vigorous recent studies have disproved the existence of a distinct placental microbiome (32, 33).
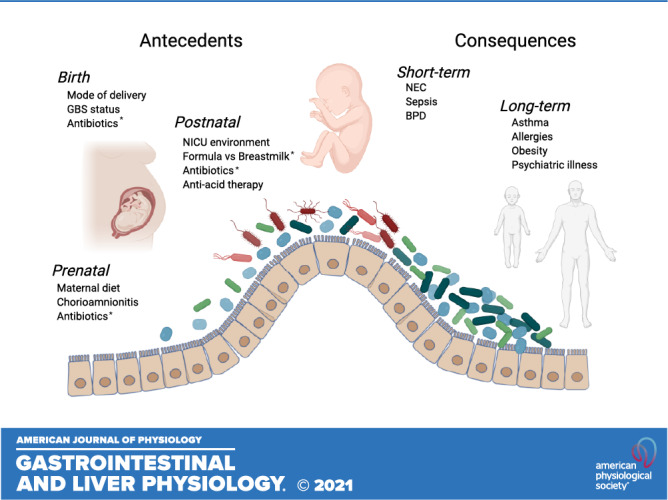
Figure 1.
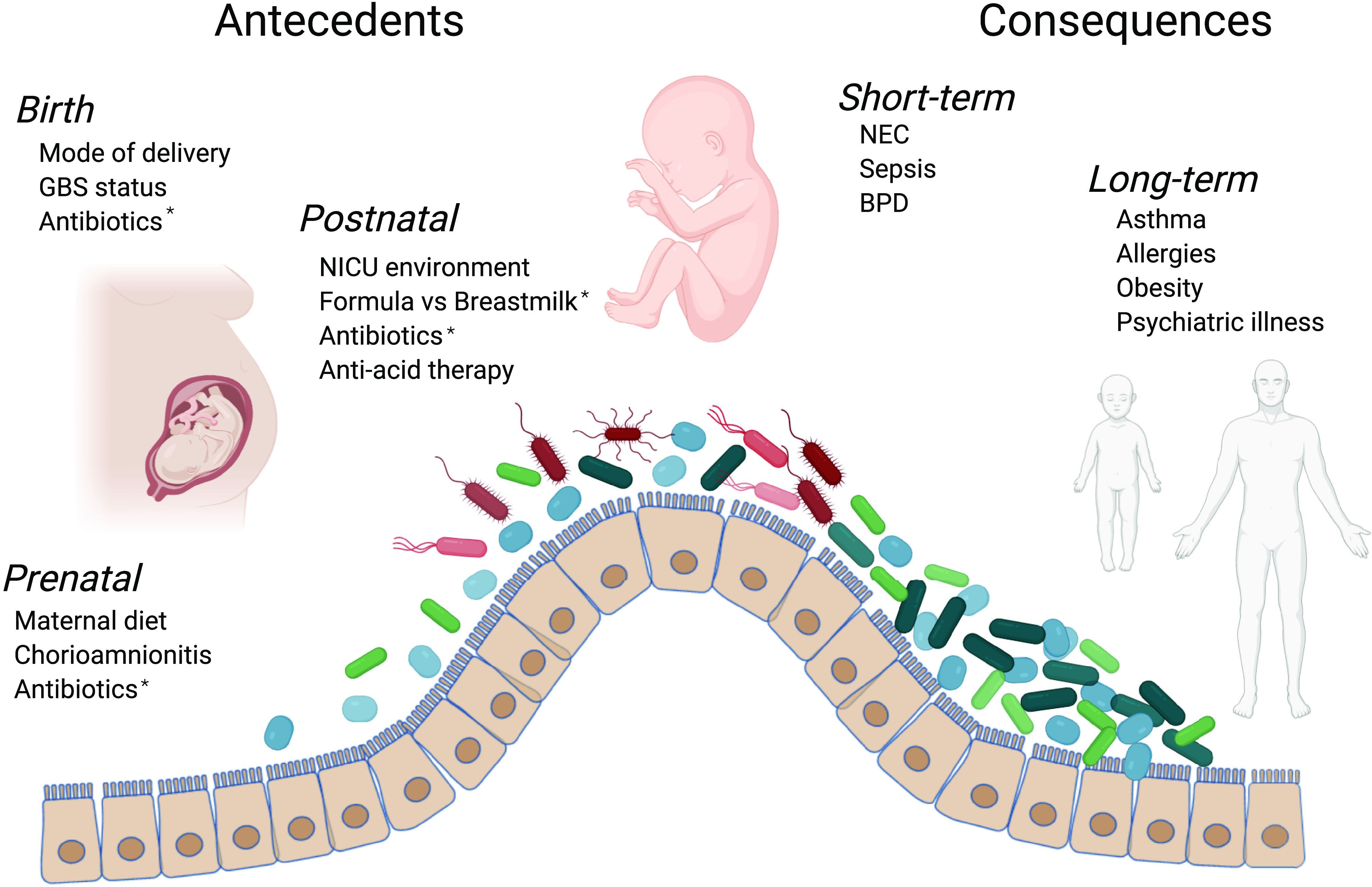
Summary of antecedents and consequences of gut dysbiosis in preterm infants. Several prenatal, birth, and postnatal factors negatively impact the developing gut microbiome of the preterm infant. The resulting dysbiotic gut microbial communities during this critical period of development have been linked to both short-term and long-term morbidities. *Highlights factors considered to have relatively greater impact than others. Figure 1 created using www.biorender.com.
Intrapartum antibiotics (IAP) administered to prevent Group B streptococcal transmission has also been shown to have significant early effects on the composition of gut microbiome in neonates that persist through the first year of life (34–36). Nogacka et al. (36) showed a persistent increase in Proteobacteria and Firmicutes with decreases in Acinetobacter and Bacteroides in association with higher occurrence of some bacterial genes that code for β-lactamase resistance. Others have shown that the use of IAP was associated with decreases in Bifidobacteria spp. at 7 days of life, whereas Lactobacillus spp. remains unchanged (35). Furthermore, the effects of IAP on the infant microbiome may be selective with penicillin suppressing Bacteroides spp., and cephalosporin delaying the increase in Bifidobacteria (34). These studies suggest that maternal gestational diet, chorioamnionitis, and IAP influence the composition of the neonatal microbiome and these effects can last well beyond the neonatal period.
Postnatal Factors
Several postnatal factors influence the development of gut microbiome and can program intestinal dysbiosis (Fig. 1). The mode of birth, i.e., vaginal versus cesarean, has been shown to influence the neonatal microbiome (16, 37, 38). Infants born by cesarean section have less intestinal microbial diversity, decreased colonization with Bifidobacteria, Bacteroides, and Lactobacilli, and tended to have increased skin microbiota such as Streptococcus and Staphylococcus in the first weeks after birth (16, 38). Interestingly, differences related to mode of delivery are less evident with maturity, likely because other factors like postnatal diet impact the gut microbiota (37).
Early exposure to parenteral antibiotics is also a risk factor for gut dysbiosis (39, 40). Fouhy et al. (41) demonstrated that early (<48 h) antibiotic treatment with ampicillin and gentamicin was associated with increases in Proteobacteria and decreased abundance of Actinobacteria, Bifidobacteria, and Lactobacillus. Gibson et al. (42) demonstrated that broad spectrum antibiotics such as meropenem, cephalosporins, and ticarcillin-clavulanate decreased species richness whereas vancomycin and gentamicin had nonuniform results. They also noted emergence of antibiotics resistant genes that was antibiotic- and species-specific. Zwittink et al. (43) noted that the use of short- or long-term amoxicillin/ceftazidime treatment was associated with significant higher abundance of Enterococcus during the first two postnatal weeks at the expense of Bifidobacterium and Streptococcus.
Nutrition is also a strong modifier gut microbiome. Preterm infants fed with formula milk exhibit a less diverse gut microbiome with less enrichment of Clostridiales and Bifidobacteria compared with preterm infants fed mothers’ own milk (44). In summary, major postnatal factors linked to a dysbiotic gut signature in preterm infants include mode of delivery, exposure to antibiotics and other medications, and type of milk feeding.
CONSEQUENCES OF DYSBIOSIS IN PRETERM INFANTS
Necrotizing Enterocolitis
NEC is the most widely recognized consequence of gut dysbiosis in preterm infants (Fig. 1). The role of gut bacteria in NEC is demonstrated from mouse models showing absence of disease in a germ-free environment, direct studies of human stools showing dysbiotic signatures, and demonstration of bacterial invasion by fluorescent techniques in surgically resected human tissues (45–47). Clusters of NEC outbreaks related to specific organisms that have varied by institution have also been reported, providing additional evidence of the role of microbes in NEC pathogenesis. With advances in metagenomics, several studies have demonstrated that preterm infants who develop NEC have altered gut microbiota signatures compared with preterm infants who do not develop NEC, though these signatures have varied between studies (48).
In a small study, Heida et al. (49) identified enrichment of Clostridrium perfringens in meconium of preterm infants who subsequently developed NEC, suggesting that a pathogenic microbial signature for NEC may be present in the very first day of life. However, meconial or very early stool signatures for NEC have not been demonstrated in subsequent studies (21, 50). In one of the largest longitudinal studies in preterm infants, Warner et al. (50) studied >2,400 stool samples from 122 infants born with a birthweight <1,500 g to demonstrate that NEC was associated with Gammaproteobacteria abundance, and relative depletion of Negativicutes, an anaerobe. Interestingly, these patterns emerged typically after the first 4 weeks of life. Other interesting observations were that antibiotic use increased proportions of Bacilli, whereas vaginal birth was associated with decreased proportions of Bacilli, and the volume of human milk feeds did not impact bacterial community architecture.
Similar to the landmark study by Warner et al. (50), several investigators have reported an enrichment of potentially pathogenic Gram-negative bacteria in the intestine in addition to a relative decrease in the abundance of Firmicutes and Bacteroides before the onset of NEC (21, 50–53). Furthermore, although preterm infants in general have low gut microbial diversity compared with term infants, several studies have noted further reduced levels of diversity in preterm infants who develop NEC compared with preterm infants without NEC (21, 48, 54, 55). A meta-analysis of published studies confirmed a relative abundance of Gammaproteobacteria, with decreased relative proportions of Firmicutes and Bacteroides preceding NEC, but did not confirm reduced microbial diversity before NEC onset (21). Gammaproteobacteria, which include several well-known pathogenic species that have the endotoxin lipopolysaccharide, can induce excessive intestinal and systemic inflammation through stimulation of Toll-like receptors (45); whereas Firmicutes and Bacteroides can enhance gut development by facilitating intestinal epithelial cell differentiation and preserving mucin and tight junction integrity (56).
Although these studies have primarily focused on bacterial signatures, a potential role for viruses such as norovirus, rotavirus, and cytomegalovirus (CMV) in NEC causation is also recognized (48, 57). For example, case reports (58) have described infants with CMV-associated NEC, and retrospective studies using surgical specimens (59, 60) have detected CMV in intestinal tissue of preterm infants with NEC or spontaneous intestinal perforation. Infants with CMV-associated NEC tend to have lower gestational age and thrombocytopenia than non-CMV associated cases. The relative impact of viruses on NEC remains poorly characterized as reported rates of detection of CMV and other viruses can vary depending on the sample (blood, stool, and intestinal tissue), technique (immunohistochemistry, polymerase chain reaction, enzyme-linked immunosorbent assay), and sample size of NEC cohort (61–63). The potential role of fungi and the mycobiome in NEC also remain largely unexplored (64).
Although metagenomic studies provide a snapshot of microbial patterns associated with NEC, more rigorous studies are needed to elucidate their mechanistic role and establish their contribution in NEC. More recently, multi-omic studies combining metagenomics, metatranscriptomics, and metabolomics are beginning to shed new light on bacterial replication rates, quorum sensing patterns, and metabolic profiles associated with intestinal dysbiosis preceding NEC (21, 52, 55). For example, Morrow et al. (51) evaluated the urinary metabolome and gut microbiome of 11 preterm infants with NEC and 21 matched controls. Using this two complementary “-omic” approaches, the authors found that the urinary metabolite alanine was directly correlated with Firmicutes and indirectly correlated with Proteobacteria in the gut, suggesting that specific intestinal bacteria can influence production and utilization of specific metabolites which may be useful as surrogate biomarkers for early diagnosis of NEC. Although still at its infancy, these novel multi-omic approaches are anticipated to further advance our understanding and provide a full mechanistic picture of NEC pathogenesis (65).
Sepsis and Other Diseases
The consequences of dysbiosis in the preterm infants extend beyond the predisposition to develop NEC (Fig. 1). Stewart et al. (40) noted that the bacteria isolated from a blood culture during sepsis corresponded to the dominant bacterial genera in the gut microbiome, and that Bifidobacteria abundance was observed in infants who did not develop sepsis or NEC. Other studies have also noted decreased diversity and lower abundance of Bifidobacteria among infants who developed sepsis (66). Potential mechanisms by which Bifidobacteria can protect against sepsis include reduced bacterial translocation through enhanced intestinal barrier function and increased production of beneficial short-chain fatty acids like acetate (67, 68). Sepsis related to antibiotic-resistant strains of bacteria originating from the gut is also increased in preterm infants treated with broad spectrum antibiotics that increase dysbiosis (40, 42, 50). Although not the focus here, dysbiosis in preterm infants has also been linked to bronchopulmonary dysplasia (a chronic debilitating lung condition of infants) and growth failure (69, 70).
Potential Long-Term Consequences
Epidemiological studies suggest a link between early disturbances to the gut microbiome from antibiotics or cesarean delivery with later childhood diseases including allergy (71–73), obesity (74, 75), and attention-deficit hyperactivity disorder (76). A recent population-based cohort study using the Rochester Epidemiological Project found similar associations between early-life antibiotic exposure and increased risk for several childhood immunological, metabolic, and neurobehavioral health conditions (77). Although these studies did not differentiate term from preterm infants, the aforementioned stressors that preterm infants experience in the hospital environment puts them at greatest risk for early gut microbial dysbiosis (78). Additional studies in mice provide supporting evidence for the hypothesis that early-life perturbations during key developmental periods can have long-term consequences. For example, in a mouse model of early gut microbial disruption with low-dose penicillin, long-lasting metabolic effects of increased fat mass were observed even with limited antibiotic administration where microbial communities recovered after cessation of antibiotics (79).
MICROBIOME MODULATION THERAPY
With accumulating evidence of how gut microbiome perturbations contribute to preterm diseases, novel strategies that reverse or alleviate dysbiosis are being explored.
Probiotics
Probiotics are defined as living microorganisms that confer health benefits when administered in adequate amounts. In addition to favorable alteration of gut microbiome, probiotics are now recognized to confer benefit through other mechanisms such as colonization resistance against pathogenic bacteria, enhanced gut barrier function, and immunomodulation (80). Numerous clinical trials have demonstrated the safety and efficacy of prophylactic administration of probiotics in preterm infants for the prevention of NEC. A recent network meta-analysis (63 trials, 15,712 preterm infants) identified that a multispecies combination that contains Lactobacillus and Bifidobacterium provided superior benefits for NEC prevention compared with single species or other multispecies probiotic combinations for reducing mortality and NEC in preterm infants (81).
Prebiotics
Prebiotics are nutrient substrates that selectively enrich beneficial microbiota in the gut. The most important source of prebiotics for preterm infants is human milk oligosaccharides (HMO)—complex sugars not otherwise digestible by the human gut but utilized by specific microbes such as Bifidobacteria and Bacteroides to promote their growth and activity. In addition to stimulating probiotic growth, prebiotics also help improve intestinal mobility and gut barrier function. Artificial or nonhuman milk oligosaccharides designed to function in a similar manner to HMOs have also been tested in small clinical trials with promising results (82). Other active areas of investigation include synbiotics which are combination products that contain both prebiotics and probiotics (83).
Postbiotics
Postbiotics are metabolic byproducts of probiotic bacteria that can exert positive biological activity in the host. In a recent study, Zheng et al. (23) demonstrated that short-chain fatty acids, such as butyrate and acetate produced by Bifidobacterium infantis from metabolism of complex carbohydrates expressed in breastmilk, have anti-inflammatory effects in immature enterocytes. Other studies have shown how butyrate can also enhance intestinal barrier function and regulate mucosal immunity (84). By using metabolites instead of bacteria, postbiotics potentially provide an effective, simpler, and safer alternative to ingestion of live microorganisms. A related strategy is the use of para-probiotics, which are inactivated microbial cells or cell fractions typically produced by heat killing of probiotics (85).
Fecal Microbiota Transplantation
In fecal microbiota transplantation (FMT), fecal material from a healthy donor is transplanted to the recipient’s GI tract to restore microbial homeostasis. Compared with probiotics, FMT allows more robust and longer-lasting community of diverse microbes with a single dose. Safety concerns include inadvertent introduction or promotion of pathogenic species and transference of antibiotic resistance. FMT in preterm infants remains largely unexplored except in experimental animal research (86). A similar thematic approach is vaginal seeding, wherein bacteria from the mother’s vaginal tract is transferred to the face and body of infants born via cesarean section (87).
CONCLUSIONS
The concept of the holobiont, consisting of the human host and the resident microbial communities, is important in considering perturbations of intestinal microbial assembly in preterm infants (7, 88). The development of a symbiotic intestinal ecosystem is a central event for successful adaptation to postnatal life. Maternal disease, intrauterine infection, and perinatal use of antibiotics can disrupt establishment of coevolved microbial communities, which are integral for the optimal function and development of the intestine, the immune system, the brain, and other physiological processes (7, 88). Restoration to a normal microbial assembly is further impeded in the preterm neonate fed formula milk, exposed to long-term antibiotics and deviant microbial dispersal patterns in a pathogen-rich milieu, and microbial community priority effects. These adverse influences result in the establishment of a dysbiotic microbial signature characterized by a relative abundance in Gammaproteobacteria, decreased diversity, and a modest decrease in Bifidobacteria. Several studies link these dysbiotic signatures to increased vulnerability to life-threatening neonatal illness such as NEC and sepsis, and increasingly to childhood asthma, allergic disease, disorders of mood, and obesity (7, 88). Efforts at restoring microbial community structure using probiotics have shown promise in alleviating NEC and sepsis risk. Other approaches based on prebiotics or enteral immune therapy, however, warrant careful trials. Understanding the role of host genetics in programming the neonatal gut microbial assembly, combining metabolomic and transcriptomic signatures concurrent with dysbiotic microbiota signatures, and personalized approaches in reconstituting the deviant microbial assembly in preterm neonates to prevent disease and ensure healthy outcomes remain highly significant areas for translational research (55, 89, 90).
GRANTS
This study was supported by institutional funds at Children’s Mercy Hospital (to V. Sampath and A. Cuna), R01DK117296 (to V. Sampath), and K08DK125735 (to A. Cuna).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
A.C. and V.S. conceived and designed research; A.C., M.J.M., and V.S. prepared figures; A.C., M.J.M., S.U., and V.S. drafted manuscript; A.C., M.J.M., I.A., S.U., and V.S. edited and revised manuscript; A.C., M.J.M., I.A., S.U., and V.S. approved final version of manuscript.
ACKNOWLEDGMENTS
Figure 1 was created using www.biorender.com.
REFERENCES
- 1.Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J 474: 1823–1836, 2017. doi: 10.1042/BCJ20160510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valdes AM, Walter J, Segal E, Spector TD. Role of the gut microbiota in nutrition and health. BMJ 361: k2179, 2018. doi: 10.1136/bmj.k2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, MetaHIT Consortium, et al. Enterotypes of the human gut microbiome. Nature 473: 174–180, 2011. [Erratum in Nature 474: 666, 2011]. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science 308: 1635–1638, 2005. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lotz M, Gutle D, Walther S, Menard S, Bogdan C, Hornef MW. Postnatal acquisition of endotoxin tolerance in intestinal epithelial cells. J Exp Med 203: 973–984, 2006. doi: 10.1084/jem.20050625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanidad KZ, Zeng MY. Neonatal gut microbiome and immunity. Curr Opin Microbiol 56: 30–37, 2020. doi: 10.1016/j.mib.2020.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamburini S, Shen N, Wu HC, Clemente JC. The microbiome in early life: implications for health outcomes. Nat Med 22: 713–722, 2016. doi: 10.1038/nm.4142. [DOI] [PubMed] [Google Scholar]
- 8.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10: 996–998, 2013. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 9.Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12: 38, 2011. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tremblay J, Singh K, Fern A, Kirton ES, He S, Woyke T, Lee J, Chen F, Dangl JL, Tringe SG. Primer and platform effects on 16S rRNA tag sequencing. Front Microbiol 6: 771, 2015. doi: 10.3389/fmicb.2015.00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Youssef N, Sheik CS, Krumholz LR, Najar FZ, Roe BA, Elshahed MS. Comparison of species richness estimates obtained using nearly complete fragments and simulated pyrosequencing-generated fragments in 16S rRNA gene-based environmental surveys. Appl Environ Microbiol 75: 5227–5236, 2009. doi: 10.1128/AEM.00592-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quince C, Walker AW, Simpson JT, Loman NJ, Segata N. Shotgun metagenomics, from sampling to analysis. Nat Biotechnol 35: 833–844, 2017. doi: 10.1038/nbt.3935. [DOI] [PubMed] [Google Scholar]
- 13.Forster SC, Kumar N, Anonye BO, Almeida A, Viciani E, Stares MD, Dunn M, Mkandawire TT, Zhu A, Shao Y, Pike LJ, Louie T, Browne HP, Mitchell AL, Neville BA, Finn RD, Lawley TD. A human gut bacterial genome and culture collection for improved metagenomic analyses. Nat Biotechnol 37: 186–192, 2019. doi: 10.1038/s41587-018-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, , et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59–65, 2010. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet 371: 75–84, 2008. doi: 10.1016/S0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rutayisire E, Huang K, Liu Y, Tao F. The mode of delivery affects the diversity and colonization pattern of the gut microbiota during the first year of infants' life: a systematic review. BMC Gastroenterol 16: 86, 2016. doi: 10.1186/s12876-016-0498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collado MC, Cernada M, Neu J, Perez-Martinez G, Gormaz M, Vento M. Factors influencing gastrointestinal tract and microbiota immune interaction in preterm infants. Pediatr Res 77: 726–731, 2015. doi: 10.1038/pr.2015.54. [DOI] [PubMed] [Google Scholar]
- 18.Groer MW, Luciano AA, Dishaw LJ, Ashmeade TL, Miller E, Gilbert JA. Development of the preterm infant gut microbiome: a research priority. Microbiome 2: 38, 2014. doi: 10.1186/2049-2618-2-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yasmin F, Tun HM, Konya TB, Guttman DS, Chari RS, Field CJ, Becker AB, Mandhane PJ, Turvey SE, Subbarao P, Sears MR, Investigators CS, Scott JA, Dinu I, Kozyrskyj AL; CHILD Study Investigators. Cesarean section, formula feeding, and infant antibiotic exposure: separate and combined impacts on gut microbial changes in later infancy. Front Pediatr 5: 200, 2017. doi: 10.3389/fped.2017.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arboleya S, Sanchez B, Milani C, Duranti S, Solis G, Fernandez N, de los Reyes-Gavilan CG, Ventura M, Margolles A, Gueimonde M. Intestinal microbiota development in preterm neonates and effect of perinatal antibiotics. J Pediatr 166: 538–544, 2015. doi: 10.1016/j.jpeds.2014.09.041. [DOI] [PubMed] [Google Scholar]
- 21.Pammi M, Cope J, Tarr PI, Warner BB, Morrow AL, Mai V, Gregory KE, Kroll JS, McMurtry V, Ferris MJ, Engstrand L, Lilja HE, Hollister EB, Versalovic J, Neu J. Intestinal dysbiosis in preterm infants preceding necrotizing enterocolitis: a systematic review and meta-analysis. Microbiome 5: 31, 2017. doi: 10.1186/s40168-017-0248-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorreja F, Rush ST, Kasper DL, Meng D, Walker WA. The developmentally regulated fetal enterocyte gene, ZP4, mediates anti-inflammation by the symbiotic bacterial surface factor polysaccharide A on Bacteroides fragilis. Am J Physiol Gastrointest Liver Physiol 317: G398–G407, 2019. doi: 10.1152/ajpgi.00046.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng N, Gao Y, Zhu W, Meng D, Walker WA. Short chain fatty acids produced by colonizing intestinal commensal bacterial interaction with expressed breast milk are anti-inflammatory in human immature enterocytes. PLoS One 15: e0229283, 2020. doi: 10.1371/journal.pone.0229283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong CB, Iwabuchi N, Xiao JZ. Exploring the science behind Bifidobacterium breve M-16V in infant health. Nutrients 11: 1724, 2019. doi: 10.3390/nu11081724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dahl C, Stigum H, Valeur J, Iszatt N, Lenters V, Peddada S, Bjornholt JV, Midtvedt T, Mandal S, Eggesbo M. Preterm infants have distinct microbiomes not explained by mode of delivery, breastfeeding duration or antibiotic exposure. Int J Epidemiol 47: 1658–1669, 2018. doi: 10.1093/ije/dyy064. [DOI] [PubMed] [Google Scholar]
- 26.Wandro S, Osborne S, Enriquez C, Bixby C, Arrieta A, Whiteson K. The microbiome and metabolome of preterm infant stool are personalized and not driven by health outcomes including necrotizing enterocolitis and late-onset sepsis. mSphere 3: e00104–e00118, 2018. doi: 10.1128/mSphere.00104-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gritz EC, Bhandari V. The human neonatal gut microbiome: a brief review. Front Pediatr 3: 17, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.La Rosa PS, Warner BB, Zhou Y, Weinstock GM, Sodergren E, Hall-Moore CM, Stevens HJ, Bennett WE Jr, Shaikh N, Linneman LA, Hoffmann JA, Hamvas A, Deych E, Shands BA, Shannon WD, Tarr PI. Patterned progression of bacterial populations in the premature infant gut. Proc Natl Acad Sci USA 111: 12522–12527, 2014. [Erratum in Proc Natl Acad Sci USA 111: 17336, 2014]. doi: 10.1073/pnas.1409497111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu DM, Antony KM, Ma J, Prince AL, Showalter L, Moller M, Aagaard KM. The early infant gut microbiome varies in association with a maternal high-fat diet. Genome Med 8: 77, 2016. doi: 10.1186/s13073-016-0330-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lundgren SN, Madan JC, Emond JA, Morrison HG, Christensen BC, Karagas MR, Hoen AG. Maternal diet during pregnancy is related with the infant stool microbiome in a delivery mode-dependent manner. Microbiome 6: 109, 2018. doi: 10.1186/s40168-018-0490-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puri K, Taft DH, Ambalavanan N, Schibler KR, Morrow AL, Kallapur SG. Association of chorioamnionitis with aberrant neonatal gut colonization and adverse clinical outcomes. PLoS One 11: e0162734, 2016. doi: 10.1371/journal.pone.0162734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep 6: 23129, 2016. doi: 10.1038/srep23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Goffau MC, Lager S, Sovio U, Gaccioli F, Cook E, Peacock SJ, Parkhill J, Charnock-Jones DS, Smith GCS. Human placenta has no microbiome but can contain potential pathogens. Nature 572: 329–334, 2019. doi: 10.1038/s41586-019-1451-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coker MO, Hoen AG, Dade E, Lundgren S, Li Z, Wong AD, Zens MS, Palys TJ, Morrison HG, Sogin ML, Baker ER, Karagas MR, Madan JC. Specific class of intrapartum antibiotics relates to maturation of the infant gut microbiota: a prospective cohort study. BJOG 127: 217–227, 2020. doi: 10.1111/1471-0528.15799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corvaglia L, Tonti G, Martini S, Aceti A, Mazzola G, Aloisio I, Di GD, Faldella G. Influence of intrapartum antibiotic prophylaxis for group B streptococcus on gut microbiota in the first month of life. J Pediatric Gastroenterol Nutr 62: 304–308, 2016. doi: 10.1097/MPG.0000000000000928. [DOI] [PubMed] [Google Scholar]
- 36.Nogacka A, Salazar N, Suarez M, Milani C, Arboleya S, Solis G, Fernandez N, Alaez L, Hernandez-Barranco AM, de Los Reyes-Gavilan CG, Ventura M, Gueimonde M. Impact of intrapartum antimicrobial prophylaxis upon the intestinal microbiota and the prevalence of antibiotic resistance genes in vaginally delivered full-term neonates. Microbiome 5: 93, 2017. doi: 10.1186/s40168-017-0313-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chu DM, Ma J, Prince AL, Antony KM, Seferovic MD, Aagaard KM. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med 23: 314–326, 2017. doi: 10.1038/nm.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA 107: 11971–11975, 2010. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Esaiassen E, Fjalstad JW, Juvet LK, van den Anker JN, Klingenberg C. Antibiotic exposure in neonates and early adverse outcomes: a systematic review and meta-analysis. J Antimicrob Chemother 72: 1858–1870, 2017. doi: 10.1093/jac/dkx088. [DOI] [PubMed] [Google Scholar]
- 40.Stewart CJ, Embleton ND, Marrs ECL, Smith DP, Fofanova T, Nelson A, Skeath T, Perry JD, Petrosino JF, Berrington JE, Cummings SP. Longitudinal development of the gut microbiome and metabolome in preterm neonates with late onset sepsis and healthy controls. Microbiome 5: 75, 2017. doi: 10.1186/s40168-017-0295-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fouhy F, Guinane CM, Hussey S, Wall R, Ryan CA, Dempsey EM, Murphy B, Ross RP, Fitzgerald GF, Stanton C, Cotter PD. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob Agents Chemother 56: 5811–5820, 2012. doi: 10.1128/AAC.00789-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gibson MK, Wang B, Ahmadi S, Burnham CA, Tarr PI, Warner BB, Dantas G. Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nat Microbiol 1: 16024, 2016. doi: 10.1038/nmicrobiol.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zwittink RD, van Zoeren-Grobben D, Renes IB, van Lingen RA, Norbruis OF, Martin R, Groot Jebbink LJ, Knol J, Belzer C. Dynamics of the bacterial gut microbiota in preterm and term infants after intravenous amoxicillin/ceftazidime treatment. BMC Pediatr 20: 195, 2020. doi: 10.1186/s12887-020-02067-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu W, Judge MP, Maas K, Hussain N, McGrath JM, Henderson WA, Cong X. Systematic review of the effect of enteral feeding on gut microbiota in preterm infants. J Obstet Gynecol Neonatal Nurs 47: 451–463, 2018. doi: 10.1016/j.jogn.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jilling T, Simon D, Lu J, Meng FJ, Li D, Schy R, Thomson RB, Soliman A, Arditi M, Caplan MS. The roles of bacteria and TLR4 in rat and murine models of necrotizing enterocolitis. J Immunol 177: 3273–3282, 2006. doi: 10.4049/jimmunol.177.5.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Remon JI, Amin SC, Mehendale SR, Rao R, Luciano AA, Garzon SA, Maheshwari A. Depth of bacterial invasion in resected intestinal tissue predicts mortality in surgical necrotizing enterocolitis. J Perinatol 35: 755–762, 2015. doi: 10.1038/jp.2015.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waligora-Dupriet AJ, Dugay A, Auzeil N, Huerre M, Butel MJ. Evidence for clostridial implication in necrotizing enterocolitis through bacterial fermentation in a gnotobiotic quail model. Pediatr Res 58: 629–635, 2005. doi: 10.1203/01.PDR.0000180538.13142.84. [DOI] [PubMed] [Google Scholar]
- 48.Morowitz MJ, Poroyko V, Caplan M, Alverdy J, Liu DC. Redefining the role of intestinal microbes in the pathogenesis of necrotizing enterocolitis. Pediatrics 125: 777–785, 2010. doi: 10.1542/peds.2009-3149. [DOI] [PubMed] [Google Scholar]
- 49.Heida FH, van Zoonen A, Hulscher JBF, Te KB, Wessels R, Kooi EMW, Bos AF, Harmsen HJM, de Goffau MC. A necrotizing enterocolitis-associated gut microbiota is present in the meconium: results of a prospective study. Clin Infect Dis 62: 863–870, 2016. doi: 10.1093/cid/ciw016. [DOI] [PubMed] [Google Scholar]
- 50.Warner BB, Deych E, Zhou Y, Hall-Moore C, Weinstock GM, Sodergren E, Shaikh N, Hoffmann JA, Linneman LA, Hamvas A, Khanna G, Rouggly-Nickless LC, Ndao IM, Shands BA, Escobedo M, Sullivan JE, Radmacher PG, Shannon WD, Tarr PI. Gut bacteria dysbiosis and necrotising enterocolitis in very low birthweight infants: a prospective case-control study. Lancet 387: 1928–1936, 2016. doi: 10.1016/S0140-6736(16)00081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morrow AL, Lagomarcino AJ, Schibler KR, Taft DH, Yu Z, Wang B, Altaye M, Wagner M, Gevers D, Ward DV, Kennedy MA, Huttenhower C, Newburg DS. Early microbial and metabolomic signatures predict later onset of necrotizing enterocolitis in preterm infants. Microbiome 1: 13, 2013. doi: 10.1186/2049-2618-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olm MR, Bhattacharya N, Crits-Christoph A, Firek BA, Baker R, Song YS, Morowitz MJ, Banfield JF. Necrotizing enterocolitis is preceded by increased gut bacterial replication, Klebsiella, and fimbriae-encoding bacteria. Sci Adv 5: eaax5727, 2019. doi: 10.1126/sciadv.aax5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, Hoenig JD, Malin KJ, Qamar S, Petrof EO, Sun J, Antonopoulos DA, Chang EB, Claud EC. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J 3: 944–954, 2009. doi: 10.1038/ismej.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dobbler PT, Procianoy RS, Mai V, Silveira RC, Corso AL, Rojas BS, Roesch LFW. Low microbial diversity and abnormal microbial succession is associated with necrotizing enterocolitis in preterm infants. Front Microbiol 8: 2243, 2017. doi: 10.3389/fmicb.2017.02243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sher Y, Olm MR, Raveh-Sadka T, Brown CT, Sher R, Firek B, Baker R, Morowitz MJ, Banfield JF. Combined analysis of microbial metagenomic and metatranscriptomic sequencing data to assess in situ physiological conditions in the premature infant gut. PLoS One 15: e0229537, 2020. doi: 10.1371/journal.pone.0229537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu Y, Lu J, Oliphant K, Gupta N, Claud K, Lu L. Maternal administration of probiotics promotes gut development in mouse offsprings. PLoS One 15: e0237182, 2020. doi: 10.1371/journal.pone.0237182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Coggins SA, Wynn JL, Weitkamp JH. Infectious causes of necrotizing enterocolitis. Clin Perinatol 42: 133–154, 2015. doi: 10.1016/j.clp.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tran L, Ferris M, Norori J, Stark M, Craver R, Dowd S, Penn D. Necrotizing enterocolitis and cytomegalovirus infection in a premature infant. Pediatrics 131: e318–e322, 2013. doi: 10.1542/peds.2011-1971. [DOI] [PubMed] [Google Scholar]
- 59.Omarsdottir S, Agnarsdottir M, Casper C, Orrego A, Vanpee M, Rahbar A, Soderberg-Naucler C. High prevalence of cytomegalovirus infection in surgical intestinal specimens from infants with necrotizing enterocolitis and spontaneous intestinal perforation: a retrospective observational study. J Clin Virol 93: 57–64, 2017. doi: 10.1016/j.jcv.2017.05.022. [DOI] [PubMed] [Google Scholar]
- 60.Panesso-Gomez S, Shimamura M, Conces M, Talavera MM, Moallem M, Sanchez PJ, Malleske DT. Detection of cytomegalovirus in intestinal tissue of infants with necrotizing enterocolitis or spontaneous intestinal perforation. J Pediatr 214: 34–40, 2019. doi: 10.1016/j.jpeds.2019.07.038. [DOI] [PubMed] [Google Scholar]
- 61.Cheng C, He Y, Xiao S, Ai Q, Yu J. The association between enteric viruses and necrotizing enterocolitis. Eur J Pediatr 180: 225–232, 2021. doi: 10.1007/s00431-020-03746-w. [DOI] [PubMed] [Google Scholar]
- 62.Skeath T, Stewart C, Waugh S, Embleton N, Cummings S, Berrington J. Cytomegalovirus and other common enteric viruses are not commonly associated with NEC. Acta Paediatr 105: 50–52, 2016. doi: 10.1111/apa.13110. [DOI] [PubMed] [Google Scholar]
- 63.Ullrich T, Tang YW, Correa H, Garzon SA, Maheshwari A, Hill M, Matta P, Krishnan MK, Weitkamp JH. Absence of gastrointestinal pathogens in ileum tissue resected for necrotizing enterocolitis. Pediatr Infect Dis J 31: 413–414, 2012. doi: 10.1097/INF.0b013e318242534a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.James SA, Phillips S, Telatin A, Baker D, Ansorge R, Clarke P, Hall LJ, Carding SR. Preterm infants harbour a rapidly changing mycobiota that includes Candida pathobionts. J Fungi (Basel) 6: 273, 2020. doi: 10.3390/jof6040273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Neu J. Necrotizing enterocolitis: a multi-omic approach and the role of the microbiome. Dig Dis Sci 65: 789–796, 2020. doi: 10.1007/s10620-020-06104-w. [DOI] [PubMed] [Google Scholar]
- 66.Mai V, Torrazza RM, Ukhanova M, Wang X, Sun Y, Li N, Shuster J, Sharma R, Hudak ML, Neu J. Distortions in development of intestinal microbiota associated with late onset sepsis in preterm infants. PLoS One 8: e52876, 2013. doi: 10.1371/journal.pone.0052876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, Taylor TD, Itoh K, Kikuchi J, Morita H, Hattori M, Ohno H. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469: 543–547, 2011. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 68.Ling X, Linglong P, Weixia D, Hong W. Protective effects of bifidobacterium on intestinal barrier function in LPS-induced enterocyte barrier injury of Caco-2 monolayers and in a rat NEC model. PLoS One 11: e0161635, 2016. doi: 10.1371/journal.pone.0161635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ryan FJ, Drew DP, Douglas C, Leong LEX, Moldovan M, Lynn M, Fink N, Sribnaia A, Penttila I, McPhee AJ, Collins CT, Makrides M, Gibson RA, Rogers GB, Lynn DJ. Changes in the composition of the gut microbiota and the blood transcriptome in preterm infants at less than 29 weeks gestation diagnosed with bronchopulmonary dysplasia. mSystems 4: e00484–e00519, 2019. doi: 10.1128/mSystems.00484-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Younge NE, Newgard CB, Cotten CM, Goldberg RN, Muehlbauer MJ, Bain JR, Stevens RD, O'Connell TM, Rawls JF, Seed PC, Ashley PL. Disrupted maturation of the microbiota and metabolome among extremely preterm infants with postnatal growth failure. Sci Rep 9: 8167, 2019. doi: 10.1038/s41598-019-44547-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, Kuzeljevic B, Gold MJ, Britton HM, Lefebvre DL, Subbarao P, Mandhane P, Becker A, McNagny KM, Sears MR, Kollmann T, Investigators CS, Mohn WW, Turvey SE, Finlay BB; CHILD Study Investigators. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med 7: 307ra152, 2015. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 72.Mitre E, Susi A, Kropp LE, Schwartz DJ, Gorman GH, Nylund CM. Association between use of acid-suppressive medications and antibiotics during infancy and allergic diseases in early childhood. JAMA Pediatr 172: e180315, 2018. doi: 10.1001/jamapediatrics.2018.0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ni J, Friedman H, Boyd BC, McGurn A, Babinski P, Markossian T, Dugas LR. Early antibiotic exposure and development of asthma and allergic rhinitis in childhood. BMC Pediatr 19: 225, 2019. doi: 10.1186/s12887-019-1594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ajslev TA, Andersen CS, Gamborg M, Sorensen TI, Jess T. Childhood overweight after establishment of the gut microbiota: the role of delivery mode, pre-pregnancy weight and early administration of antibiotics. Int J Obes (Lond) 35: 522–529, 2011. doi: 10.1038/ijo.2011.27. [DOI] [PubMed] [Google Scholar]
- 75.Leong KSW, McLay J, Derraik JGB, Gibb S, Shackleton N, Taylor RW, Glover M, Audas R, Taylor B, Milne BJ, Cutfield WS. Associations of prenatal and childhood antibiotic exposure with obesity at age 4 years. JAMA Netw Open 3: e1919681, 2020. doi: 10.1001/jamanetworkopen.2019.19681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Axelsson PB, Clausen TD, Petersen AH, Hageman I, Pinborg A, Kessing LV, Bergholt T, Rasmussen SC, Keiding N, Lokkegaard ECL. Investigating the effects of cesarean delivery and antibiotic use in early childhood on risk of later attention deficit hyperactivity disorder. J Child Psychol Psychiatry 60: 151–159, 2019. doi: 10.1111/jcpp.12961. [DOI] [PubMed] [Google Scholar]
- 77.Aversa Z, Atkinson EJ, Schafer MJ, Theiler RN, Rocca WA, Blaser MJ, LeBrasseur NK. Association of infant antibiotic exposure with childhood health outcomes. Mayo Clin Proc 96: 66–77, 2020. [33208243] doi: 10.1016/j.mayocp.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Groer MW, Gregory KE, Louis-Jacques A, Thibeau S, Walker WA. The very low birth weight infant microbiome and childhood health. Birth Defects Res C Embryo Today 105: 252–264, 2015. doi: 10.1002/bdrc.21115. [DOI] [PubMed] [Google Scholar]
- 79.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, Kim SG, Li H, Gao Z, Mahana D, Zarate RJ, Rogers AB, Robine N, Loke P, Blaser MJ. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158: 705–721, 2014. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cuna A, Yu W, Menden HL, Feng L, Srinivasan P, Chavez-Bueno S, Ahmed I, Umar S, Sampath V. NEC-like intestinal injury is ameliorated by Lactobacillus rhamnosus GG in parallel with SIGIRR and A20 induction in neonatal mice. Pediatr Res 88: 546–555, 2020. doi: 10.1038/s41390-020-0797-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morgan RL, Preidis GA, Kashyap PC, Weizman AV, Sadeghirad B, McMaster Probiotic P, Synbiotic W; McMaster Probiotic, Prebiotic, and Synbiotic Work Group. Probiotics reduce mortality and morbidity in preterm, low-birth-weight infants: a systematic review and network meta-analysis of randomized trials. Gastroenterology 159: 467–480, 2020. doi: 10.1053/j.gastro.2020.05.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chi C, Buys N, Li C, Sun J, Yin C. Effects of prebiotics on sepsis, necrotizing enterocolitis, mortality, feeding intolerance, time to full enteral feeding, length of hospital stay, and stool frequency in preterm infants: a meta-analysis. Eur J Clin Nutr 73: 657–670, 2019. doi: 10.1038/s41430-018-0377-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nandhini LP, Biswal N, Adhisivam B, Mandal J, Bhat BV, Mathai B. Synbiotics for decreasing incidence of necrotizing enterocolitis among preterm neonates—a randomized controlled trial. J Matern Fetal Neonatal Med 29: 821–825, 2016. doi: 10.3109/14767058.2015.1019854. [DOI] [PubMed] [Google Scholar]
- 84.Liu J, Zhu H, Li B, Lee C, Alganabi M, Zheng S, Pierro A. Beneficial effects of butyrate in intestinal injury. J Pediatr Surg 55: 1088–1093, 2020. doi: 10.1016/j.jpedsurg.2020.02.036. [DOI] [PubMed] [Google Scholar]
- 85.Deshpande G, Athalye-Jape G, Patole S. Para-probiotics for preterm neonates-the next frontier. Nutrients 10: 871, 2018. doi: 10.3390/nu10070871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Prado C, Michels M, Avila P, Burger H, Milioli MVM, Dal-Pizzol F. The protective effects of fecal microbiota transplantation in an experimental model of necrotizing enterocolitis. J Pediatr Surg 54: 1578–1583, 2019. doi: 10.1016/j.jpedsurg.2018.10.045. [DOI] [PubMed] [Google Scholar]
- 87.Mueller NT, Dominguez-Bello MG, Appel LJ, Hourigan SK. 'Vaginal seeding' after a caesarean section provides benefits to newborn children: FOR: does exposing caesarean-delivered newborns to the vaginal microbiome affect their chronic disease risk? The critical need for trials of 'vaginal seeding' during caesarean section. BJOG 127: 301, 2020. doi: 10.1111/1471-0528.15979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dominguez-Bello MG, Godoy-Vitorino F, Knight R, Blaser MJ. Role of the microbiome in human development. Gut 68: 1108–1114, 2019. doi: 10.1136/gutjnl-2018-317503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cuna A, George L, Sampath V. Genetic predisposition to necrotizing enterocolitis in premature infants: current knowledge, challenges, and future directions. Semin Fetal Neonatal Med 23: 387–393, 2018. doi: 10.1016/j.siny.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kurilshikov A, Wijmenga C, Fu J, Zhernakova A. Host genetics and gut microbiome: challenges and perspectives. Trends Immunol 38: 633–647, 2017. doi: 10.1016/j.it.2017.06.003. [DOI] [PubMed] [Google Scholar]


