Abstract
We aimed to investigate novel emerging severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) lineages in Japan that harbor variants in the spike protein receptor-binding domain (RBD). The total nucleic acid contents of samples from 159 patients with coronavirus disease 2019 (COVID-19) were subjected to whole genome sequencing. The SARS-CoV-2 genome sequences from these patients were examined for variants in spike protein RBD. In January 2021, three family members (one aged in their 40s and two aged under 10 years old) were found to be infected with SARS-CoV-2 harboring W152L/E484K/G769V mutations. These three patients were living in Japan and had no history of traveling abroad. After identifying these cases, we developed a TaqMan assay to screen for the above hallmark mutations and identified an additional 14 patients with the same mutations. The associated virus strain was classified into the GR clade (Global Initiative on Sharing Avian Influenza Data [GISAID]), 20B clade (Nextstrain), and R.1 lineage (Phylogenetic Assignment of Named Global Outbreak [PANGO] Lineages). As of April 22, 2021, R.1 lineage SARS-CoV-2 has been identified in 2,388 SARS-CoV-2 entries in the GISAID database, many of which were from Japan (38.2%; 913/2,388) and the United States (47.1%; 1,125/2,388). Compared with that in the United States, the percentage of SARS-CoV-2 isolates belonging to the R.1 lineage in Japan increased more rapidly over the period from October 24, 2020 to April 18, 2021. R.1 lineage SARS-CoV-2 has potential escape mutations in the spike protein RBD (E484K) and N-terminal domain (W152L); therefore, it will be necessary to continue to monitor the R.1 lineage as it spreads around the world.
Author summary
A novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), emerged in December 2019 in Wuhan, China. SARS-CoV-2 had evolved and spread around the world, threatening human life. Several mutations, which alter the viral fitness, virulence, and transmissibility, were identified in SARS-CoV-2. Here, we detected R.1 lineage SARS-CoV-2 harboring mutations in spike protein. The R.1 lineage have spread at the beginning of 2021 in Japan. This lineage has potential escape mutations in the spike protein receptor-binding domain (E484K) and N-terminal domain (W152L). We also developed a novel TaqMan assay targeting the hallmark mutations occurring in the spike proteins of R.1 lineage for screening. Our data indicate that emergent variants are dominated by the natural selection and need to be monitored by genomic epidemiological research.
Introduction
Most mutations that occur during viral evolution are neutral and not to influence viral properties. However, some mutations are selectively propagated owing to their positive influence on viral fitness, virulence, and transmissibility [1,2]. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) “Variants of Concern” have emerged within the last few months, largely belonging to three major lineages, B.1.1.7, B.1.351, and P.1 [3–6]. These emerging lineages are all characterized by multiple mutations in the SARS-CoV-2 spike protein, raising concerns that they may escape monoclonal antibody therapy or vaccine-elicited antibodies. The B.1.1.7 lineage is estimated to have emerged in late September 2020 and has become the dominant strain of SARS-CoV-2 in the United Kingdom [3,7,8]. The B.1.351 lineage has become the dominant strain of SARS-CoV-2 in South Africa, where it was first detected in October 2020 [4]. The P.1 lineage was first identified in four travelers from Brazil and has been associated with cases of reinfection [5,6,9].
The hallmark mutation shared by the B.1.1.7, B.1.351, and P.1 lineages is N501Y, located in the receptor-binding domain (RBD) of the spike protein [3]. SARS-CoV-2 variants with this mutation are thought to be more transmissible and possibly more virulent [7,10–13]. The other hallmark mutation of the B.1.351 and P.1 lineages is E484K, which has been shown to reduce the neutralizing activity of antibodies, thus raising concerns about vaccine efficacy [14–19].
In this study, we conducted a genetic surveillance and identified the SARS-CoV-2 R.1 lineage, which harbors an E484K mutation in the RBD, via whole genome sequencing and TaqMan assay. We report the first detailed genetic characterization of the SARS-CoV-2 R.1 lineage, its familial transmission, and its viral evolution as assessed by phylogenetic analysis. The SARS-CoV-2 R.1 lineage has spread worldwide and remains especially prevalent in Japan.
Materials and methods
Ethics statement
The Institutional Review Board of the Clinical Research and Genome Research Committee at Yamanashi Central Hospital approved this study and the use of an opt-out consent method (Approval No. C2019-30). The requirement for written informed consent was waived owing to it being an observational study and the urgent need to collect COVID-19 data.
Patients and sample collection
A total of 192 patients in our hospital were confirmed to have coronavirus disease 2019 (COVID-19), 159 of whom were selected to provide samples for subsequent genome analysis. Nasopharyngeal swab samples were collected by using cotton swabs and placed in 3 ml of viral transport media (VTM) purchased from Copan Diagnostics (Murrieta, CA, United States). We used 200 μl of VTM for nucleic acid extraction, performed within 2 h of sample collection.
Quantitative reverse transcription-polymerase chain reaction (RT-qPCR)
Total nucleic acid was isolated from the nasopharyngeal swab samples using the MagMAX Viral/Pathogen Nucleic Acid Isolation Kit (Thermo Fisher Scientific; Waltham, MA, United States) [20,21]. Following the protocol developed by the National Institute of Infectious Diseases in Japan [22], we performed one-step RT-qPCR to detect SARS-CoV-2 on a StepOnePlus Real-Time PCR System (Thermo Fisher Scientific). The viral load, measured as absolute copy number, was determined using a serially diluted DNA control targeting the nucleocapsid gene of SARS-CoV-2 (Integrated DNA Technologies; Coralville, IA, United States) [23,24].
Whole-genome sequencing
SARS-CoV-2 genomic RNA was reverse transcribed into cDNA and amplified by using the Ion AmpliSeq SARS-CoV-2 Research Panel (Thermo Fisher Scientific) on the Ion Torrent Genexus System in accordance with the manufacturer’s instructions [25]. Sequencing reads were processed, and their quality was assessed by using Genexus Software with SARS-CoV-2 plugins. The sequencing reads were mapped and aligned by using the torrent mapping alignment program. After initial mapping, a variant call was performed by using the Torrent Variant Caller. The COVID19AnnotateSnpEff plugin was used for the annotation of variants. Assembly was performed with the Iterative Refinement Meta-Assembler [26].
Clade and lineage classification
The viral clade and lineage classifications were conducted by using the Global Initiative on Sharing Avian Influenza Data (GISAID) database [27], Nextstrain [28], and Phylogenetic Assignment of Named Global Outbreak (PANGO) Lineages [29]. The sequences of the three initially identified R.1 variants were deposited in the GISAID EpiCoV database (Accession Nos. EPI_ISL_1164927, EPI_ISL_1164928, and EPI_ISL_1164929).
Global sequencing data on the SARS-CoV-2 R.1 variant through April 22, 2021 were exported from the GISAID EpiCoV database. We searched for lineage “R.1” and found 2,388 available metadata entries for the R.1 lineage for the period from October 24, 2020 to April 18, 2021, when excluding two data entries lacking a specific collection date. We also searched for the total number of samples by applying the following parameters: host “human”, location “Asia /Japan” or “North America/USA”, collection date “October 24, 2020 to April 18, 2021”. During this period, a total of 17,432 samples were registered in Japan, and 235,373 samples were registered in the United States.
Rapid detection of R.1 lineage SARS-CoV-2 samples with a novel TaqMan assay
We designed a Custom TaqMan assay for detecting SARS-CoV-2 spike protein with the W152L and G769V mutations (Thermo Fisher Scientific). We also used a TaqMan SARS-CoV-2 Mutation Panel for detecting spike E484K (ID: ANU7GMZ, Thermo Fisher Scientific). TaqPath 1-Step RT-qPCR Master Mix CG was used as the master mix. The TaqMan MGB probes for the wild-type and variant alleles were labelled with VIC dye and FAM dye fluorescence, respectively.
Results
Household transmission of SARS-CoV-2 harboring a spike protein with the E484K mutation
As part of our ongoing genomic surveillance of SARS-CoV-2, we began to investigate SARS-CoV-2 spike protein RBD variants in Kofu city, Japan [25]. We previously identified P.1 lineage SARS-CoV-2, which harbors the K417T/E484K/N501Y mutations, in one patient [25]. A consecutive analysis detected the W152L/E484K/G769V mutations in three patients who provided samples on January 14, 2021. All three patients belonged to the same family: a man in his 40s (the father), a boy under 10 years old, and a girl under 10 years old. This family lived in Japan and had no history of travel to foreign countries. The mother of this family was also infected with SARS-CoV-2 around the same time as the other family members, but we were unable to obtain her samples because she was tested at another hospital. These results suggest that the family members were infected with the same virus lineage and that household transmission occurred.
Genetic characterization defines the SARS-CoV-2 R.1 lineage
Our sequencing analysis of the SARS-CoV-2 isolates from the three family members identified the same 21 mutations in each isolate; these comprised 13 missense, six synonymous, and two intergenic variants. Among the missense mutations, four were in the spike protein (W152L, E484K, D614G, and G769V), four were in ORF1ab (T4692I, N6301S, L6337M, and I6525T), one was in the membrane protein (F28L), and four were in the nucleocapsid protein (S187L, R203K, G204R, and Q418H).
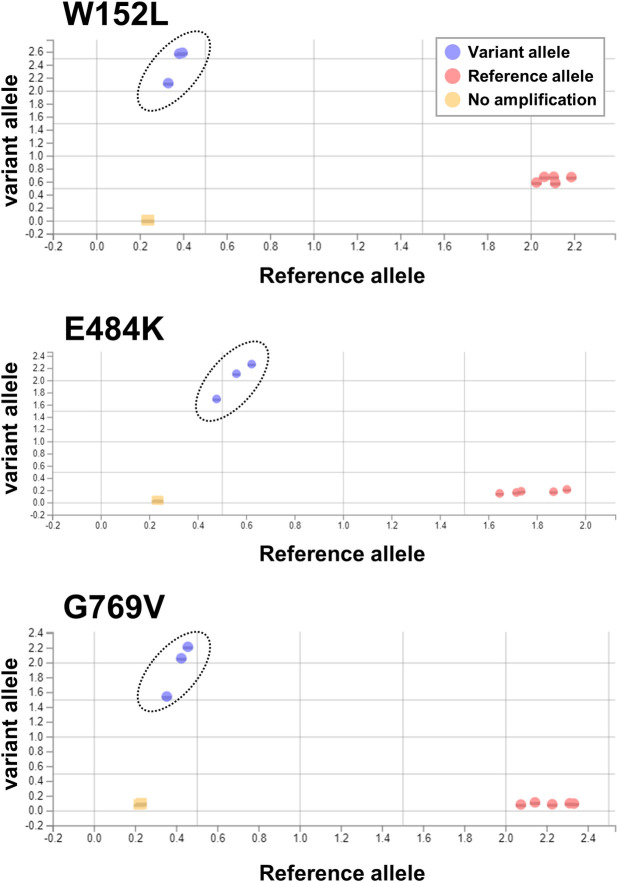
To examine the phylogeny based on the identified genetic variations, we analyzed SARS-CoV-2 genomic data using GISAID, Nextstrain, and Pangolin. The sequences of our SARS-CoV-2 were assigned as GR clade (GISAID), 20B clade (Nextstrain), and R.1 lineage (PANGO lineage). The R.1 lineage is a sublineage of the B.1.1.316 lineage, and the common mutations found in the R.1 lineage are listed in Table 1. For the rapid detection of SARS-CoV-2 R.1 lineage isolates, we designed a TaqMan assay that detects the hallmark spike protein mutations (W152L, E484K, and G769V) (Table 1). This TaqMan assay can successfully discriminate the wild-type and variant alleles in each position (Fig 1). Using the TaqMan assay, we identified an additional 14 patients who were infected with SARS-CoV-2 R.1 lineage. We further confirmed the results via whole genome sequencing. These findings suggest that the novel TaqMan assay targeting SARS-CoV-2 R.1 lineage hallmark mutations is a useful tool for detecting the presence of R.1 lineage isolates in SARS-CoV-2-positive PCR samples.
Table 1. Common mutations in the SARS-CoV-2 R.1 lineage.
| Gene | Mutation |
|---|---|
| ORF1b | P314L |
| ORF1b | G1362R |
| ORF1b | P1936H |
| S | W152L |
| S | E484K |
| S | D614G |
| S | G769V |
| N | S187L |
| N | R203K |
| N | G204R |
| N | Q418H |
ORF, open reading frame; S, spike; N, nucleocapsid
Fig 1. TaqMan assay for discriminating R.1 lineage SARS-CoV-2 by its spike variant allele.
We analyzed samples of R.1 lineage SARS-CoV-2 (n = 3), which harbors a spike variant with the W152L/E484K/G769V mutations and samples of SARS-CoV-2 without these three mutations (n = 5) using a TaqMan assay. An allelic discrimination plot of the results, showing the spike variants with W152L (upper), E484K (middle), and G769V (lower). The dots indicate the mutant alleles (purple), reference allele (pink), or no amplification (orange). The dotted circles indicate the mutant alleles of each variant.
Epidemiological event of SARS-CoV-2 R.1 lineage
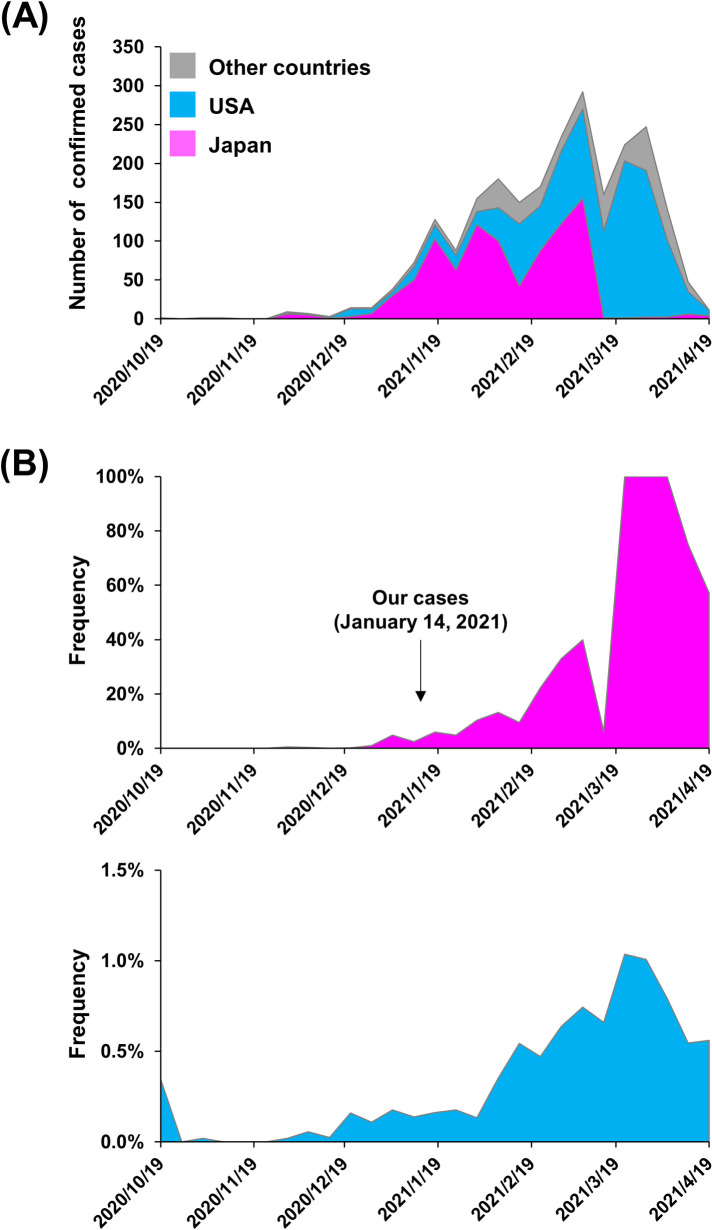
To investigate the global distribution of SARS-CoV-2 R.1 lineage, we next collected registration data from the EpiCoV of GISAID database [27]. As of March 5, 2021, a total of 2,388 SARS-CoV-2 samples with R.1 lineage had been registered from all over the world, with the majority being from Japan (38.2%; 913/2,388) or the United States (47.1%; 1,125/2,388) (Fig 2A and Table 2). R.1 lineage SARS-CoV-2 was first reported at the end of October 2020 in Texas, United States and was first detected in Japan at the end of November 2020. The change in the number of R.1 lineage SARS-CoV-2 samples has followed similar trends in the United States, Japan, and other countries (Fig 2A).
Fig 2. Timeline of SARS-CoV-2 R.1 lineage emergence and country-specific percentages of global cases.
(A) Timeline of the number of confirmed cases of infection with R.1 lineage SARS-CoV-2. The plot, created based on the data in Table 1, shows the case load for Japan (pink), the United States (light blue), and other countries (gray). (B) The percentage of R.1 lineage SARS-CoV-2 strains relative to the total number of registered samples during the period is shown for Japan (upper panel) and the United States (lower panel). The arrow indicates the date of infection for the initial three individuals infected with R.1 lineage SARS-CoV-2 who were identified in this study.
Table 2. Global numbers and percentages of confirmed R.1 lineage SARS-CoV-2 cases.
| Country | Number of confirmed cases (n = 2,388) | Frequency |
|---|---|---|
| USA | 1,125 | 47.1% |
| Japan | 913 | 38.2% |
| Germany | 100 | 4.2% |
| Austria | 69 | 2.9% |
| Belgium | 35 | 1.5% |
| Nether lands | 30 | 1.3% |
| Sweden | 26 | 1.1% |
| United Kingdom | 19 | 0.8% |
| Trinidad and Tobago | 18 | 0.8% |
| Spain | 12 | 0.5% |
| France | 8 | 0.3% |
| Canada | 7 | 0.3% |
| Ghana | 6 | 0.3% |
| Turkey | 3 | 0.1% |
| Australia | 3 | 0.1% |
| Croatia | 2 | 0.1% |
| Switzer land | 2 | 0.1% |
| Nigeria | 1 | 0.04% |
| China | 1 | 0.04% |
| United Arab Emirates | 1 | 0.04% |
| Denmark | 1 | 0.04% |
| Finland | 1 | 0.04% |
| Italy | 1 | 0.04% |
| Portugal | 1 | 0.04% |
| Slovakia | 1 | 0.04% |
| New Zealand | 1 | 0.04% |
| Aruba | 1 | 0.04% |
| Total | 2,388 | 100% |
Regarding the period from October 24, 2020 to April 18, 2021, the number of R.1 lineage SARS-CoV-2 samples deposited in GISAID began an ongoing increase in February 2021 (Fig 2B), with the percentage of these isolates in Japan exceeding 20% since the beginning of March 2021 (Fig 2B). While the percentage of R.1 lineage SARS-CoV-2 remained low in the United States, it grew rapidly in Japan (Fig 2B). We also observed that the numbers and percentage of R.1 lineage SARS-CoV-2 isolates increased in our district of Japan (S1 Fig).
Phylogenetic analysis of SARS-CoV-2 R.1 lineage
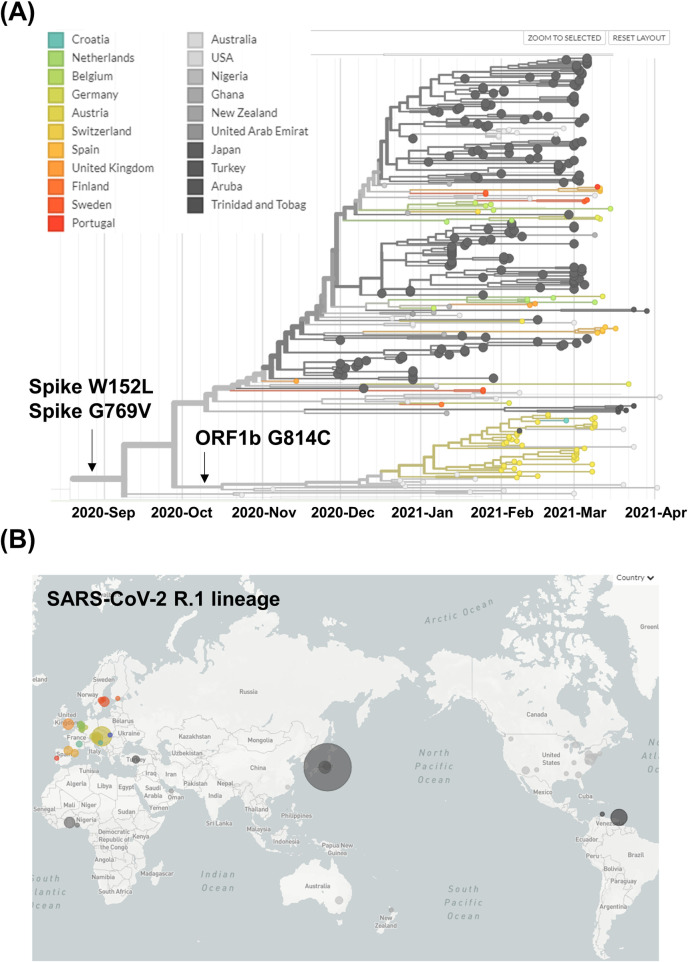
To determine the timing of the emergence of the SARS-CoV-2 R.1 lineage and its acquisition of characteristic mutations, we analyzed a phylogeny of SARS-CoV-2 carrying the E484K mutation, generated from global data. It revealed that the SARS-CoV-2 R.1 lineage forms a monophyletic clade (Figs 3A and S2). The parental lineage harboring the E484K mutation later acquired spike protein W152L and G769V mutations, and the SARS-CoV-2 R.1 lineage was predicted to have emerged around September 9, 2020 (Fig 3A). Subsequently, a sublineage, which has been observed in Austria and the United States, diverged after acquiring the ORF1b G814C mutation around October 19, 2020 (Fig 3A). A parental lineage without the ORF1b G814C mutation has been prevalent mainly in Japan and the United States (Fig 3A). In addition to being detected in Japan, the SARS-CoV-2 R.1 lineage has been observed in 27 additional countries worldwide (Fig 3B and Table 2). Collectively, these results demonstrate that the SARS-CoV-2 R.1 lineage is spreading rapidly, especially in Japan.
Fig 3. Phylogenetic tree of R.1 lineage SARS-CoV-2.
(A) Global data on the SARS-CoV-2 R.1 lineage prevalence over time. The R.1 lineage acquired its spike W152L and G769V mutations at the root. The sublineage harbors an ORF1b G814C mutation. (B) Geographic distribution of the SARS-CoV-2 R.1 lineage. World map showing the geographic distribution of SARS-CoV-2 R.1 lineage as of April 22, 2021. The size of the circle indicates the number of samples. Nextstrain, https://nextstrain.org, CC-BY-4.0 license.
Discussion
COVID-19 vaccines have been approved in many countries within a year of the initial appearance of this disease, which is an unprecedented scientific achievement [30]. However, the emergence of mutations in SARS-CoV-2 spike RBD and N-terminal domain (NTD) are of great concern because of their potential contribution to immune escape [31,32]. In the present study, we observed an expansion in Japan of R.1 lineage SARS-CoV-2 harboring the spike RBD E484K and spike NTD W152L mutations, which have potential significance for immune escape. Previous reports showed that COVID-19 convalescent and mRNA vaccine-elicited sera/plasma have reduced neutralizing activity against SARS-CoV-2 harboring the E484K mutation [17–19]. Worryingly, the E484K mutation has been identified in SARS-CoV-2 “Variants of Concern”, such as B.1.351 (South Africa) and P.1 (Brazil), and in SARS-CoV-2 “Variants of Interest”, such as B.1.526 (New York, NY, USA), B.1.525 (United Kingdom/Nigeria), and P.2 (Brazil) [33–35]. Furthermore, the W152L mutation is located in the N3 loop of the NTD and may be related to decreased antibody neutralization activity [36]. The new emergent lineage B.1.429, which has a substitution in the same codon 152 (W152C), was first identified in California, United States; this lineage is defined as a “Variant of Interest” by the US Centers for Disease Control and Prevention (CDC) [35]. The W152C mutation is located in the NTD antigenic supersite (designated site i) and is also associated with reduced recognition of the NTD by neutralizing monoclonal antibodies [32,37].
There is concern that SARS-CoV-2 with these immune escape mutations may evade the host immune response. In Brazil, a patient was re-infected with SARS-CoV-2 carrying the E484K mutation, indicating that this mutation is related to escape from neutralizing antibodies in recovered patients [38]. Additionally, a patient who received the second shot of mRNA-1273 vaccine (Moderna) was infected with SARS-CoV-2 harboring the E484K mutation, despite having a serum neutralizing antibody titer that is normally sufficient to prevent infection [39]. Recently, R.1 lineage SARS-CoV-2 was detected at a skilled nursing facility in Kentucky, in residents and healthcare personnel who had received BNT162b2 mRNA vaccines (Pfizer-BioNTech) [40]. These results indicate that even after COVID-19 vaccination, the risk of infection is not completely eliminated. Although COVID-19 mRNA vaccines have demonstrated high efficacy for decreasing the risk of SARS-CoV-2 transmission and severe outcomes from COVID-19 [41–43], it is still necessary to monitor vaccinated individuals for new emergent lineages that have acquired novel escape mutations.
There are limitations to this epidemiological report on the SARS-CoV-2 R.1 lineage. The amount of SARS-CoV-2 sequencing data available from different countries does not reflect the actual prevalence of specific variants because the extent of sequencing analysis conducted varies among countries [30]. If the sequences of nearly all SARS-CoV-2-positive PCR specimens is analyzed, the results can represent the whole picture of prevailing virus lineages; unfortunately, this is not the case here. However, although the number of samples in our study is small, we analyzed all the SARS-CoV-2-positive PCR samples in our hospital and found that the percentage of samples with R.1 lineage SARS-CoV-2 increased to about 50% by the end of the study period (S1 Fig). Along with the R.1 lineage prevalence, the B.1.1.7 (United Kingdom) lineage prevalence is also increasing in Japan [44]. Further investigation is needed to determine whether these two lineages of SARS-CoV-2 will circulate dominantly in Japan.
Circulating SARS-CoV-2 acquires approximately one or two mutations per month. This seems to be very slow; however, the more the virus circulates in population, the more opportunity it has to change [45]. Therefore, genomic surveillance in real-time will provide us with important insights into the effectiveness of vaccines, the development of antibody therapy, and public health.
Supporting information
(A) The number of R.1 lineage SARS-CoV-2 samples identified by TaqMan assay and whole genome analysis during the period from January to April 2021. The pink color indicates R.1 lineage strains, and the gray color indicates other strains. (B) The percentage of SARS-CoV-2 strains detected over the period from January to April 2021 that belong to the R.1 lineage.
(TIF)
The dotted pink line shows a monophyletic clade containing the SARS-CoV-2 R.1 lineage.
(TIF)
(PDF)
Acknowledgments
We also thank Masato Kondo, Ryota Tanaka, Kazuo Sakai, Manami Nagano, Takuhito Fukami, and Ryo Kitamura (Thermo Fisher Scientific) for technical help, all of the medical and ancillary hospital staff for their support, and the patients for their participation. We thank all researchers who share genome data on GISAID (http://www.gisaid.org) (S1 File). We thank Katie Oakley, PhD, from Edanz Group (https://jp.edanz.com/ac) for editing a draft of this manuscript.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This study was supported by a Grant-in-Aid for the Genome Research Project from Yamanashi Prefecture (to Y.H. and M.O.), Grant-in-Aid for Early-Career Scientists 18K16292 (to Y.H.) and Grant-in-Aid for Scientific Research (B) 20H03668 (to Y.H.) from the Japan Society for the Promotion of Science (JSPS) KAKENHI, a Research Grant for Young Scholars (to Y.H.) from Satoshi Omura Foundation, the YASUDA Medical Foundation (to Y.H.), the Uehara Memorial Foundation (to Y.H.), and Medical Research Grants from the Takeda Science Foundation (to Y.H.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hou YJ, Chiba S, Halfmann P, Ehre C, Kuroda M, Dinnon KH, et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370:1464–1468. doi: 10.1126/science.abe8499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graham RL, Becker MM, Eckerle LD, Bolles M, Denison MR, Baric RS A live, impaired-fidelity coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease. Nat Med. 2012;18:1820–1826. doi: 10.1038/nm.2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrew R, Nick L, Oliver P, Wendy B, Jeff B, Alesandro C, et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. virological 2020. Available from: https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 [Google Scholar]
- 4.Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv 2020. 10.1101/2020.12.21.20248640 [DOI] [Google Scholar]
- 5.Faria NR, Claro IM, Candido D, Franco LAM, Andrade PS, Coletti TM, et al. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. Virological 2020. Available from: https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586. [Google Scholar]
- 6.Fujino T, Nomoto H, Kutsuna S, Ujiie M, Suzuki T, Sato R, et al. Novel SARS-CoV-2 Variant in Travelers from Brazil to Japan. Emerging Infectious Disease. 2021;27:1243–1245. doi: 10.3201/eid2704.210138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, et al. Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: Insights from linking epidemiological and genetic data. medRxiv 2021. 10.1101/2020.12.30.20249034 [DOI] [Google Scholar]
- 8.European Centre for Disease Prevention and Control: Rapid increase of a SARS-CoV-2 variant with multiple spike protein mutations observed in the United Kingdom. 2020:1–13. Available from: https://www.ecdc.europa.eu/sites/default/files/documents/SARS-CoV-2-variant-multiple-spike-protein-mutations-United-Kingdom.pdf [Google Scholar]
- 9.Felipe N, Valdinete N, Victor S, André C, Fernanda N, George S, et al. Phylogenetic relationship of SARS-CoV-2 sequences from Amazonas with emerging Brazilian variants harboring mutations E484K and N501Y in the Spike protein. Virological 2021. Available from: https://virological.org/t/phylogenetic-relationship-of-sars-cov-2-sequences-from-amazonas-with-emerging-brazilian-variants-harboring-mutations-e484k-and-n501y-in-the-spike-protein/585 [Google Scholar]
- 10.Washington NL, Gangavarapu K, Zeller M, Bolze A, Cirulli ET, Barrett KMS, et al. Genomic epidemiology identifies emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. medRxiv 2021. doi: 10.1101/2021.02.06.21251159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou B, Thao TTN, Hoffmann D, Taddeo A, Ebert N, Labroussaa F, et al. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature 2021;592:122–127. doi: 10.1038/s41586-021-03361-1 [DOI] [PubMed] [Google Scholar]
- 13.Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday J, et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372:eabg3055. doi: 10.1126/science.abg3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wibmer CK, Ayres F, Hermanus T, Madzivhandila M, Kgagudi P, Oosthuysen B, et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med 2021;27:622–625. doi: 10.1038/s41591-021-01285-x [DOI] [PubMed] [Google Scholar]
- 15.Liu Z, VanBlargan LA, Bloyet LM, Rothlauf PW, Chen RE, Stumpf S, et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe 2021;29:477–488. doi: 10.1016/j.chom.2021.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, et al. Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition. Cell Host & Microbe 2021; 29:44–57. doi: 10.1016/j.chom.2020.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Schmidt F, Weisblum Y, Muecksch F, Barnes CO, Finkin S, et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature 2021;592:616–622. doi: 10.1038/s41586-021-03324-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie X, Liu Y, Liu J, Zhang X, Zou J, Fontes-Garfias CR, et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat Med 2021;27:620–621. doi: 10.1038/s41591-021-01270-4 [DOI] [PubMed] [Google Scholar]
- 19.Wu K, Werner AP, Koch M, Choi A, Narayanan E, Stewart-Jones GBE, et al. Serum Neutralizing Activity Elicited by mRNA-1273 Vaccine. New England Journal of Medicine 2021;384:1468–1470. doi: 10.1056/NEJMc2102179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirotsu Y, Maejima M, Shibusawa M, Nagakubo Y, Hosaka K, Amemiya K, et al. Pooling RT-qPCR testing for SARS-CoV-2 in 1000 individuals of healthy and infection-suspected patients. Sci Rep 2020; 10:18899. doi: 10.1038/s41598-020-76043-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirotsu Y, Maejima M, Nakajima M, Mochizuki H, Omata M: Environmental cleaning is effective for the eradication of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in contaminated hospital rooms: A patient from the Diamond Princess cruise ship. Infect Control Hosp Epidemiol. 2020; 419:1105–1106. doi: 10.1017/ice.2020.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shirato K, Nao N, Katano H, Takayama I, Saito S, Kato F, et al. Development of Genetic Diagnostic Methods for Novel Coronavirus 2019 (nCoV-2019) in Japan. Jpn J Infect Dis. 2020; 73:304–307 doi: 10.7883/yoken.JJID.2020.061 [DOI] [PubMed] [Google Scholar]
- 23.Hirotsu Y, Mochizuki H, Omata M: Double-quencher probes improve detection sensitivity toward Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in a reverse-transcription polymerase chain reaction (RT-PCR) assay. J Virol Methods. 2020; 284:113926. doi: 10.1016/j.jviromet.2020.113926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Omata M, Hirotsu Y, Sugiura H, Maejima M, Nagakubo Y, Amemiya K, et al. The dynamic change of antibody index against Covid-19 is a powerful diagnostic tool for the early phase of the infection and salvage PCR assay errors. Journal of Microbiology, Immunology and Infection 2021;S1684–1182. doi: 10.1016/j.jmii.2020.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirotsu Y, Omata M: Discovery of a SARS-CoV-2 variant from the P.1 lineage harboring K417T/E484K/N501Y mutations in Kofu, Japan. J Infect. 2021;S0163–4453. doi: 10.1016/j.jinf.2021.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shepard SS, Meno S, Bahl J, Wilson MM, Barnes J, Neuhaus E: Viral deep sequencing needs an adaptive approach: IRMA, the iterative refinement meta-assembler. BMC Genomics. 2016; 17:708. doi: 10.1186/s12864-016-3030-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shu Y, McCauley J: GISAID: Global initiative on sharing all influenza data—from vision to reality. Euro Surveill 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 2018; 34:4121–4123. doi: 10.1093/bioinformatics/bty407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rambaut A, Holmes EC, O’Toole A, Hill V, McCrone JT, Ruis C, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020; 5:1403–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.COVID-19 vaccines: acting on the evidence. Nature Medicine 2021; 272:183–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen RE, Zhang X, Case JB, Winkler ES, Liu Y, VanBlargan LA, et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nature Medicine 2021;27:717–726. doi: 10.1038/s41591-021-01294-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCallum M, De Marco A, Lempp FA, Tortorici MA, Pinto D, Walls AC, et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 2021;184:2332–2347. doi: 10.1016/j.cell.2021.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lasek-Nesselquist E, Lapierre P, Schneider E, George KS, Pata J: The localized rise of a B.1.526 SARS-CoV-2 variant containing an E484K mutation in New York State. medRxiv 2021. 10.1101/2021.02.26.21251868 [DOI] [Google Scholar]
- 34.Annavajhala MK, Mohri H, Zucker JE, Sheng Z, Wang P, Gomez-Simmonds A, et al. A Novel SARS-CoV-2 Variant of Concern, B.1.526, Identified in New York. medRxiv 2021. doi: 10.1101/2021.02.23.21252259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.CDC: Available from: https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/variant-surveillance/variant-info.html.
- 36.Chi X, Yan R, Zhang J, Zhang G, Zhang Y, Hao M, et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020; 3696504:650–655. doi: 10.1126/science.abc6952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCallum M, Bassi J, Marco AD, Chen A, Walls AC, Iulio JD, et al. SARS-CoV-2 immune evasion by variant B.1.427/B.1.429. bioRxiv. 2021. doi: 10.1101/2021.03.31.437925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nonaka CK, Franco MM, Gräf T, de Lorenzo Barcia CA, de Ávila Mendonça RN, de Sousa KAF, et al. Genomic Evidence of SARS-CoV-2 Reinfection Involving E484K Spike Mutation, Brazil. Emerging Infectious Disease. 2021;27:1522–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hacisuleyman E, Hale C, Saito Y, Blachere NE, Bergh M, Conlon EG, et al. Vaccine Breakthrough Infections with SARS-CoV-2 Variants. New England Journal of Medicine 2021. doi: 10.1056/NEJMoa2105000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alyson MC, Sarah F, Patricia L, Vaneet A, Matt J, Karim G, et al. Outbreak Associated with a SARS-CoV-2 R.1 Lineage Variant in a Skilled Nursing Facility After Vaccination Program—Kentucky, March 2021. Morb Mortal Wkly Rep 2021;70:639–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. New England Journal of Medicine. 2020; 384:403–416. doi: 10.1056/NEJMoa2035389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. New England Journal of Medicine. 2020; 383:2603–2615. doi: 10.1056/NEJMoa2034577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dagan N, Barda N, Kepten E, Miron O, Perchik S, Katz MA, et al. BNT162b2 mRNA Covid-19 Vaccine in a Nationwide Mass Vaccination Setting. New England Journal of Medicine 2021;384:1412–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sekizuka T, Itokawa K, Hashino M, Okubo K, Ohnishi A, Goto K, et al. A Discernable Increase in the Severe Acute Respiratory Syndrome Coronavirus 2 R.1 Lineage Carrying an E484K Spike Protein Mutation in Japan. medRxiv 2021. 10.1101/2021.04.04.21254749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wise J: Covid-19: The E484K mutation and the risks it poses. BMJ 2021, 372:n359. doi: 10.1136/bmj.n359 [DOI] [PubMed] [Google Scholar]