Abstract
PURPOSE
Breast cancer risks for CHEK2 and ATM pathogenic variant (PV) carriers are modified by an 86-single nucleotide polymorphism polygenic risk score (PRS) and individual clinical factors. Here, we describe comprehensive risk prediction models for women of European ancestry combining PV status, PRS, and individual clinical variables.
MATERIALS AND METHODS
This study included deidentified clinical records from 358,095 women of European ancestry who received testing with a multigene panel (September 2013 to November 2019). Model development included CHEK2 PV carriers (n = 4,286), ATM PV carriers (n = 2,666), and women negative for other breast cancer risk gene PVs (n = 351,143). Odds ratios (ORs) were calculated using multivariable logistic regression with adjustment for familial cancer history. Risk estimates incorporating PV status, PRS, and Tyrer-Cuzick v7.02 were calculated using a Fixed-Stratified method that accounts for correlations between risk factors. Stratification of PV carriers into risk categories on the basis of remaining lifetime risk (RLR) was assessed in independent cohorts of PV carriers.
RESULTS
ORs for association of PV status with breast cancer were 2.01 (95% CI, 1.88 to 2.16) and 1.83 (95% CI, 1.68 to 2.00) for CHEK2 and ATM PV carriers, respectively. ORs for PRS per one standard deviation were 1.51 (95% CI, 1.37 to 1.66) and 1.45 (95% CI, 1.30 to 1.64) in CHEK2 and ATM PV carriers, respectively. Using the combined model (PRS plus Tyrer-Cuzick plus PV status), RLR was low (≤ 20%) for 24.2% of CHEK2 PV carriers, medium (20%-50%) for 63.8%, and high (> 50%) for 12.0%. Among ATM PV carriers, RLR was low for 31.5% of patients, medium for 58.5%, and high for 9.7%.
CONCLUSION
In CHEK2 and ATM PV carriers, risk assessment including PRS, Tyrer-Cuzick, and PV status has the potential for more precise direction of screening and prevention strategies.
INTRODUCTION
Genetic testing for inherited pathogenic variants (PVs) in breast cancer risk genes is an established tool for identifying women at increased risk for breast cancer. Women with PVs in moderate-risk genes, such as ATM and CHEK2, have an approximately two-fold higher risk for breast cancer compared with women in the general population and are candidates for screening at younger age, with consideration of breast magnetic resonance imaging (MRI) in addition to mammography.1,2 PVs in BRCA1, BRCA2, PALB2, and several other genes confer a higher risk for breast cancer, and guidelines recommend that carriers be offered the option of risk-reducing mastectomy in addition to intensified screening incorporating breast MRI.1,2
CONTEXT
Key Objective
Can a model combining polygenic risk, pathogenic variant (PV) status, and clinical risk factors further stratify breast cancer risk for carriers of CHEK2 or ATM PVs?
Knowledge Generated
The combined model categorized CHEK2 and ATM PV carriers into multiple risk categories on the basis of remaining lifetime risk. This included shifting risk up or down compared with PV status and clinical risk factors alone.
Relevance
Combining polygenic risk and other risk factors for PV carriers can provide more personalized breast cancer risk estimation, which can be used to inform appropriate screening and prevention decisions.
There is considerable evidence demonstrating that breast cancer risk in women carrying PVs in both moderate- and high-risk genes can be modified by many of the same clinical and family history factors that influence breast cancer risk in women without such PVs. For example, hormonal and reproductive factors may affect breast cancer risk in women with PVs in BRCA1 and BRCA2.3-6 It has also been shown that a stronger family history of breast cancer correlates with higher risks for women with PVs in CHEK2 and PALB2.7-9 Incorporating these clinical factors into a comprehensive risk assessment tool for women with PVs in breast cancer risk genes may allow for more precise individualized risk estimation.
In addition to the genetic risk associated with PVs in known breast cancer risk genes, there is a growing body of evidence highlighting the contribution of common, low-risk genetic variants (single nucleotide polymorphisms [SNPs]) to inherited breast cancer risk. Individually, these variants contribute small incremental risks. However, the contributions of multiple low-risk variants can be pooled to create a polygenic risk score (PRS) capable of stratifying unaffected women into risk categories ranging from below general population risk to risks equal to, or higher than, that seen in carriers of PVs in moderate-risk breast cancer genes.10-12 It has been shown that a PRS can also accurately modify the risks associated with PVs in moderate- and high-risk genes,12-14 including a recent study demonstrating that an 86-SNP PRS significantly modifies breast cancer lifetime risk for BRCA1, BRCA2, ATM, CHEK2, and PALB2 PV carriers.15 In this study, we developed and validated a breast cancer risk model for unaffected women carrying PVs in ATM and CHEK2, using a previously described 86-SNP PRS in combination with the clinical and family history factors captured by the Tyrer-Cuzick model v7.02.16
MATERIALS AND METHODS
Patient Population
The data set included patients who underwent clinical testing for hereditary cancer risk with a multigene panel. Women were eligible if they were 18-84 years old and of European ancestry (Ashkenazi and non-Ashkenazi), as the PRS was developed and validated in women of European ancestry. Two nonoverlapping patient sets including both PV carriers and noncarriers, separated by time of testing, were used for model development and evaluation of risk stratification by the final models (Data Supplement). PV carriers were patients who tested positive for a PV in CHEK2 or ATM, whereas noncarriers included those who tested negative for a PV in known breast cancer predisposition genes (BRCA1, BRCA2, PTEN, ATM, PALB2, CHEK2, NBN, TP53, CDH1, BARD1, and STK11).
At the time of this analysis, the testing laboratory did not classify CHEK2 I157T or S428F as pathogenic, and thus, women carrying these variants were not included as carriers in this analysis. Women with ATM c.7271T>G were excluded because of higher penetrance compared with other ATM PVs. Women were excluded if they were homozygous for a PV, were compound heterozygous, had a PV in > 1 breast cancer risk gene, or had ductal carcinoma in situ, lobular carcinoma in situ, or atypical hyperplasia without a subsequent breast cancer diagnosis. Women were also excluded if they were submitted from states that disallow the research use of samples after completion of genetic testing. This work was performed with a waiver of informed consent and oversight from an institutional review board (Advarra Institutional Review Board previously Quorum, #33893/1).
Tyrer-Cuzick variable information was collected using provider-completed test request forms starting in May 2017. For analyses involving Tyrer-Cuzick and its composite variables, only women tested after this date were included in the analysis (Data Supplement).
Genetic Testing
Testing was performed using a multigene panel in a Clinical Laboratory Improvement Amendments–approved and College of American Pathology–approved laboratory (Myriad Genetic Laboratories Inc, Salt Lake City, UT) by next-generation sequencing. Genes were included in the multigene panel on the basis of evidence of association with one or more hereditary cancers as described previously.17 The panel consisted of at least 25 genes, with additional genes being added in July 2016 and February 2019 for a total panel size of 36 genes by the end of the eligibility timeframe (Data Supplement). All relevant breast cancer risk genes listed above were included on the panel for the duration of the study period. Hybridization probes for 86 SNP markers were also included in the sequencing panel. Details regarding the composition of the 86-SNP panel have been previously published.16 Residual samples or test materials were not stored for later use per state regulations.
Statistical Methods
Associations and interactions between variables
Associations between PV status (CHEK2 or ATM) and Tyrer-Cuzick variables were tested using logistic regression, adjusting for age and Ashkenazi ancestry. Similarly, associations between PRS and Tyrer-Cuzick variables were analyzed using a linear regression, adjusting for age and ancestry. Other multivariable logistic regression analyses were adjusted for personal and familial cancer history as previously described.16 Unless stated otherwise, all regressions were adjusted for age (binned; Data Supplement). Logistic regression analyses were used to determine if there was a significant interaction between Tyrer-Cuzick variables and PV status or PRS as a predictor of breast cancer. Women were excluded if their self-reported information (from the test request form) was discrepant or improbable (Data Supplement). P values were reported as two-sided without adjustment for multiple testing. Analyses were completed using R version 3.5.3 or higher.
Combining PV risk with the Tyrer-Cuzick model
PV-associated and Tyrer-Cuzick–estimated breast cancer risks were combined according to the Fixed-Stratified method that prevents double counting of information from correlated risk factors in a manner equivalent to full multivariable coestimation.18 A detailed explanation of the statistical equations used is given in the Data Supplement.
Risk classification
Remaining lifetime risk (RLR) of breast cancer was calculated according to Tyrer-Cuzick v7.02, Tyrer-Cuzick plus PV status, and the final combined model (Tyrer-Cuzick plus PV status plus 86-SNP PRS). No adjustments were made for competing mortality. RLR was classified as low (≤ 20%), medium (20%-50%), or high (> 50%), determined on the basis of guideline-recommended thresholds for consideration of enhanced screening (20%) and risk-reducing mastectomy (50%).2,19
RESULTS
Patient Population
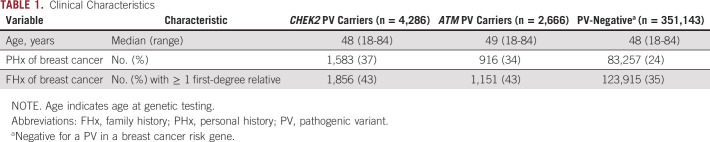
The development data set included 4,286 women with a CHEK2 PV, 2,666 women with an ATM PV, and 351,143 PV-negative women (Table 1). Age at genetic testing was similar between these three groups, with the two PV-carrying data sets containing a greater proportion of women with a personal and/or family history of breast cancer than the PV-negative women. The PRS distribution was similar among PV carriers and noncarriers unaffected by breast cancer (Data Supplement). We observed no association between PRS and PV status (Data Supplement). A summary of Tyrer-Cuzick variables for all patients is presented in the Data Supplement.
TABLE 1.
Clinical Characteristics
Model Development for PV Carriers
A step-wise approach was taken for model development for each PV-carrying population (CHEK2 and ATM) separately. To verify that the common founder mutation CHEK2 1100delC was equivalent to other CHEK2 mutations in relative cancer risk, odds ratios (ORs) were calculated using multivariable logistic regression for 4,286 CHEK2 PV carriers (OR 2.01; 95% CI, 1.88 to 2.16), for 681 CHEK2 missense PV carriers (OR 2.09; 95% CI, 1.76 to 2.48), 2,407 CHEK2 1100delC carriers (OR 2.01; 95% CI, 1.83 to 2.20), and 1,198 carriers of other CHEK2 PVs (OR 1.98; 95% CI, 1.74 to 2.26). Comparable ORs between groups indicated that specific CHEK2 PVs were unlikely to have variable impact on risk, and therefore, CHEK2 PV status could be treated as a binomial factor (carrier v noncarrier). A previous publication indicated that breast cancer risks for CHEK2 PV carriers could be age-dependent.20 Using the larger PV carrier and noncarrier population in this study, no age dependence was observed for breast cancer risk for CHEK2 PV carriers (data not shown). On the basis of these findings, no additional corrections were required to account for specific PV type or age in the model.
Associations between CHEK2 PV carrier status and factors contained in the Tyrer-Cuzick risk model were evaluated to determine what adjustments were required (Table 2). CHEK2 PV status was strongly associated with family history of breast cancer (P < 10–13, Table 2) and was therefore combined according to the Fixed-Stratified method to avoid double counting. CHEK2 PV carriers were more likely to be premenopausal at the time of testing than noncarriers. No other factor in the Tyrer-Cuzick model showed evidence of association with CHEK2 PV status after correcting for multiple comparisons. Interactions were similarly evaluated, and no risk factors contained in the Tyrer-Cuzick model showed evidence of interaction with CHEK2 PV status, indicating that all factors conferred the same risk to carriers and noncarriers alike.
TABLE 2.
Associations Between Mutation Status and Factors in the Tyrer-Cuzick Model, After Adjusting for Age (Bins) and Ashkenazi Ancestry
To develop the model for ATM PV carriers, a similar approach was taken as for CHEK2 PV carriers. The OR for association of ATM PV with breast cancer was 1.83 (95% CI, 1.68 to 2.00). There was no age dependence observed for relative risk of breast cancer for ATM PV carriers. Family history of breast cancer was the only factor contained in the Tyrer-Cuzick model associated with ATM PV status (P < 10–6) and was combined with Tyrer-Cuzick variables according to the Fixed-Stratified method. No Tyrer-Cuzick variables showed evidence of interaction with ATM PV status after correcting for multiple comparisons.
To compare with previous work showing that the effect size for breast cancer risk modification by the PRS was similar in PV carriers and noncarriers,15 standardized ORs were calculated and compared with the previously published values for non-carriers. No difference was observed between noncarriers (OR 1.47; 95% CI, 1.45 to 1.49),15 CHEK2 PV carriers (OR 1.51; 95% CI, 1.37 to 1.66), and ATM PV carriers (OR 1.45; 95% CI, 1.30 to 1.64; Data Supplement). To combine the 86-SNP PRS into models with PV status and Tyrer-Cuzick variables, associations and interactions between the PRS and Tyrer-Cuzick variables were also evaluated. For CHEK2 PV carriers, the 86-SNP PRS was associated with family history of breast cancer (P = 5.9 × 10–6), but not with any other factors included in the Tyrer-Cuzick model (Table 3). No significant association between the 86-SNP PRS and family history of breast cancer was observed for ATM PV carriers (P = .10), although the estimate was in the same direction as CHEK2 PV carriers and noncarriers.21,22 Family history was again combined into the models for CHEK2 and ATM PV carriers using a Fixed-Stratified method to avoid double counting of risk information. For CHEK2 PV carriers, there was a marginal interaction between PRS and family history, although it did not remain significant after correcting for multiple comparisons. No other factors from the Tyrer-Cuzick risk model showed evidence of interaction with the 86-SNP PRS for either PV carrier group, indicating that all factors conferred the same risk to women with high or low 86-SNP scores.
TABLE 3.
Associations Between the 86-SNP PRS and Factors in the Tyrer-Cuzick Model, After Adjusting for Age (Bins) and Ashkenazi Ancestry
Risk Stratification: CHEK2 PV Carriers
Using an independent data set of CHEK2 PV carriers unaffected by breast cancer (n = 459), we evaluated risk stratification by three models: Tyrer-Cuzick alone, Tyrer-Cuzick plus CHEK2 PV risk, and the combined model containing PRS risk, CHEK2, and Tyrer-Cuzick. Using Tyrer-Cuzick alone, 250 women (54.5%) had low RLR (≤ 20%), whereas 209 (45.5%) had medium RLR of breast cancer (20%-50%). No women were categorized as having high RLR (> 50%) by the Tyrer-Cuzick model alone (Fig 1). With the addition of CHEK2 risk to the Tyrer-Cuzick model, average estimated risk increased, with the majority of patients having medium RLR (Fig 1). In total, 82 women (17.9%) had low RLR, 339 (73.9%) had medium RLR, and 38 (8.3%) had high RLR of breast cancer (Fig 1).
FIG 1.
Stratification of remaining lifetime risk for unaffected patients with CHEK2 PV (n = 459) on the basis of (A) the Tyrer-Cuzick model alone, (B) a combination of CHEK2 and Tyrer-Cuzick, and (C) the PRS, CHEK2, and Tyrer-Cuzick combined or for patients with ATM PV (n = 216) on the basis of (D) the Tyrer-Cuzick model alone, (E) a combination of ATM and Tyrer-Cuzick, and (F) the PRS, ATM, and Tyrer-Cuzick combined. PRS, polygenic risk score; PV, pathogenic variant.
As expected, by using the total combined risk model (Tyrer-Cuzick plus CHEK2 risk plus 86-SNP PRS), average estimated risk was not substantially different from that with CHEK2 and Tyrer-Cuzick alone, but the risk range for the same patient population was wider (Fig 1). Using this combined model, 111 women (24.2%) had low RLR, 293 (63.8%) had medium RLR, and 55 (12.0%) had high RLR of breast cancer (Fig 1). The addition of the 86-SNP PRS to the model substantially increased or decreased RLR estimates for individual patients (Fig 2), sometimes shifting patients across the different risk thresholds (Data Supplement). In this cohort of CHEK2 PV carriers, 71 women (15.5%) were categorized as having lower RLR when using the combined model including the PRS compared with the Tyrer-Cuzick plus CHEK2 model. Conversely, 59 women (12.9%) were categorized as having higher RLR.
FIG 2.
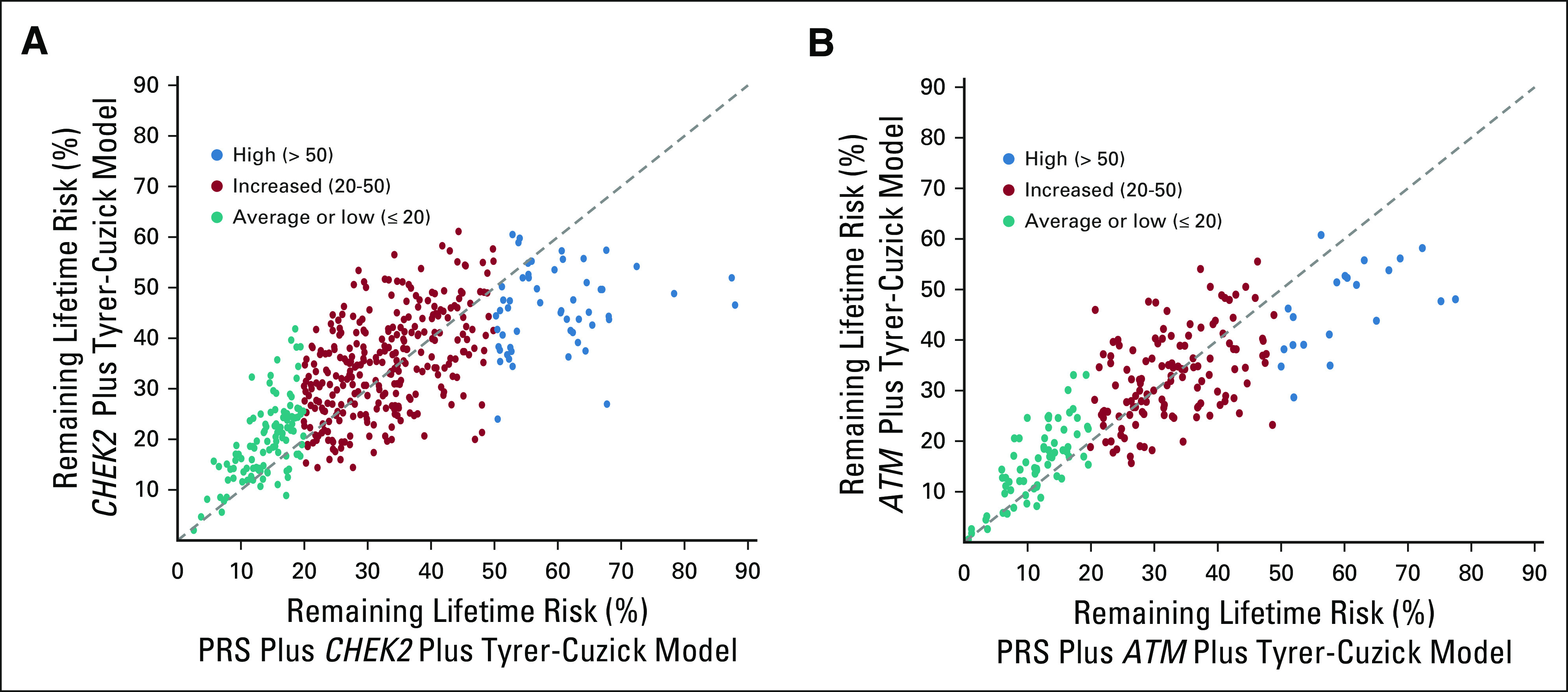
Scatterplot of risk distribution for (A) unaffected CHEK2 PV carriers (n = 459) or (B) unaffected ATM PV carriers (n = 216) on the basis of CHEK2 and Tyrer-Cuzick alone or with the addition of the 86-SNP PRS. PRS, polygenic risk score; PV, pathogenic variant; SNP, polygenic risk score.
Risk Stratification: ATM PV Carriers
Using a population of ATM PV carriers unaffected by breast cancer (n = 216) independent of the model development population, risk stratification was evaluated using the three models. Similar to what was observed for CHEK2, the distribution of risk changed with the addition of each factor (ATM PV status and PRS) to the Tyrer-Cuzick risk model for ATM PV carriers. Using the Tyrer-Cuzick risk model alone, 126 women (58.3%) had low RLR, whereas 90 (41.7%) had medium RLR and none had high RLR of breast cancer (Fig 1). With the addition of ATM risk, 58 women (26.9%) had low RLR, 145 (67.1%) had medium RLR, and 13 (6.0%) had high RLR of breast cancer (Fig 1). The addition of ATM risk to Tyrer-Cuzick shifted risks higher, pushing some women above the 50% threshold for high RLR.
Using the combined model including the 86-SNP–based risk, 68 women (31.5%) had low RLR, 127 (58.5%) had medium RLR, and 21 (9.7%) had high RLR of breast cancer (Fig 1). Adding the 86-SNP PRS to the model results in wider risk distribution. Among ATM PV carriers, 44 women (20.3%) were recategorized when comparing the Tyrer-Cuzick plus ATM model with the combined model including PRS (Fig 2, Data Supplement). This included 21 women (9.7%) whose RLR categorization was lower and 23 women (10.6%) whose categorization was higher.
DISCUSSION
This study presents a novel combination of validated clinical and molecular risks into models that provide more precise individualized breast cancer risk estimates for women of European ancestry carrying CHEK2 or ATM PVs. Using a large clinical testing population with thousands of CHEK2 and ATM carriers, substantial stratification of risk with tight CIs was achieved through combination of the Tyrer-Cuzick risk model (v7.02), PV-associated risk estimates, and an 86-SNP PRS. When risk thresholds of 20% and 50% were used for medium and high risk of breast cancer, respectively, estimated risk categorization changed for a large proportion of patients. This included patients who shifted between medium- and high-risk categories and, possibly more significantly, some whose RLR was no longer higher than average.
Improved risk stratification should lead to more personalized prevention and screening strategies, on the basis of current guidelines. For example, patients with a lifetime risk of breast cancer > 20% are candidates for more intensive screening starting at an earlier age with consideration of MRI in addition to mammography.2,19,23-25 Currently, all women with PVs in CHEK2 and ATM would be considered appropriate for this enhanced screening, which introduces increased expense, patient burden, and the potential for false alarms and overtreatment. The models presented here demonstrate that a substantial proportion of women with ATM and CHEK2 PVs might have personalized risk estimates below the 20% threshold and are therefore less likely to benefit from enhanced screening. Improved risk stratification may have an even greater impact for the fraction of ATM and CHEK2 PV carriers whose risk was shifted above 50%, which, by analogy with recommendations for high-penetrance breast cancer risk genes, is the point at which risk-reducing mastectomy might be considered in place of other high-risk screening or chemoprevention risk management strategies.19
This study has a number of limitations. First, the present study includes potential ascertainment bias because it is based on a clinical testing population cohort. However, it has been previously shown that this potential bias can be avoided by accounting for family history in the logistic regression model.1,16,18,26 Second, this study used only women of European ancestry. Further studies are required to examine polygenic breast cancer risk for women of non-European ancestry, including PV carriers and noncarriers. Additionally, the clinical factors included in this risk assessment include PV carrier status, factors from the Tyrer-Cuzick model, and a SNP-based PRS. Incorporation of additional clinical factors such as breast density may be warranted to further customize risk calculations.27 Finally, this study was performed using an 86-SNP PRS and a prospectively tested clinical patient cohort collected as early as 2013 without storage of residual test materials per state regulations. The 86-SNP PRS was developed using the most impactful SNPs published at the time, although recent studies have reported additional PRSs containing more SNPs, which may be tested prospectively using large cohorts in the future. Recent literature has shown that increasing the number of SNPs in a PRS provides only incrementally more information than a PRS with a smaller SNP composition.21,28 Although future work may expand the SNP profiles for commercially available PRSs, this and other recent work show that these risk models provide important clinical information to inform individual patient cancer risks.15,16,18
This work combined a validated PRS with mutation status and the Tyrer-Cuzick model according to a previously validated methodology.18 Validation in an independent patient cohort would be beneficial but is currently infeasible because of the rarity of PVs in unbiased study populations. By incorporating polygenic variant-conferred risk for moderate penetrance genes such as CHEK2 and ATM into other clinically accepted risk assessment tools, it is possible to refine short-term and lifetime risk stratification. Personalized risk assessment using tools such as presented here can result in more appropriate targeting of screening and risk management strategies for breast cancer prevention. Overall, the precise combination of PRS, Tyrer-Cuzick, and PV status may reduce the overuse of costly screening and prevention methods while ensuring that these resources are appropriately considered and prioritized for patients at the highest risk for breast cancer.
Shannon Gallagher
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Elisha Hughes
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Allison W. Kurian
Research Funding: Myriad Genetics
Other Relationship: Ambry Genetics, Color Genomics, GeneDx/BioReference, InVitae, Genentech
Susan M. Domchek
Honoraria: AstraZeneca, Clovis Oncology, Bristol Myers Squibb
Research Funding: AstraZeneca, Clovis Oncology
Open Payments Link: https://openpaymentsdata.cms.gov/physician/917904
Judy Garber
Consulting or Advisory Role: Novartis, GTx, Helix BioPharma, Konica Minolta, Aleta BioTherapeutics, H3 Biomedicine, Kronos Bio
Research Funding: Novartis, Ambry Genetics, InVitae, Myriad Genetics
Other Relationship: Susan G. Komen for the Cure, AACR, Diana Helis Henry Medical Foundation, James P. Wilmot Foundation, Adrienne Helis Malvin Medical Research Foundation, Breast Cancer Research Foundation, Facing Our Risk of Cancer Empowered
Braden Probst
Employment: Myriad Genetics
Brian Morris
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Placede Tshiaba
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Stephanie Meek
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Eric Rosenthal
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Benjamin Roa
Employment: Myriad Genetics
Leadership: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Research Funding: Myriad Genetics
Patents, Royalties, Other Intellectual Property: Intellectual Property held by employer Myriad Genetics
Travel, Accommodations, Expenses: Myriad Genetics
Thomas P. Slavin
Employment: Myriad Genetics
Leadership: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Susanne Wagner
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Patents, Royalties, Other Intellectual Property: Co-author of patents held by Myriad Genetics, no royalties
Jeffrey Weitzel
Speakers' Bureau: AstraZeneca
Alexander Gutin
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics, Gilead Sciences
Jerry S. Lanchbury
Employment: Myriad Genetics
Leadership: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Patents, Royalties, Other Intellectual Property: I am an inventor on Multiple patents filed by Myriad Genetics
Travel, Accommodations, Expenses: Myriad Genetics
Mark Robson
Consulting or Advisory Role: Change HealthCare
Research Funding: AstraZeneca, Pfizer, Merck
Other Relationship: Research to Practice, Clinical Care Options, Physicans' Education Resource, Invitae, Pfizer
Uncompensated Relationships: Merck, Pfizer, Daiichi Sankyo, Epic Sciences
Open Payments Link: https://openpaymentsdata.cms.gov/physician/612669/summary
No other potential conflicts of interest were reported.
DATA SHARING STATEMENT
The data that support the findings of this study are available on reasonable request from the corresponding author. The individual patient data are not publicly available because of privacy and ethical restrictions.
AUTHOR CONTRIBUTIONS
Conception and design: Shannon Gallagher, Elisha Hughes, Alexander Gutin, Jerry S. Lanchbury, Mark Robson
Administrative support: Jerry S. Lanchbury
Collection and assembly of data: Shannon Gallagher, Elisha Hughes, Braden Probst, Brian Morris, Placede Tshiaba, Benjamin Roa, Jerry S. Lanchbury
Data analysis and interpretation: Shannon Gallagher, Elisha Hughes, Allison W. Kurian, Susan M. Domchek, Judy Garber, Braden Probst, Placede Tshiaba, Stephanie Meek, Eric Rosenthal, Thomas P. Slavin, Susanne Wagner, Jeffrey Weitzel, Jerry S. Lanchbury, Mark Robson
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by the authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Shannon Gallagher
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Elisha Hughes
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Allison W. Kurian
Research Funding: Myriad Genetics
Other Relationship: Ambry Genetics, Color Genomics, GeneDx/BioReference, InVitae, Genentech
Susan M. Domchek
Honoraria: AstraZeneca, Clovis Oncology, Bristol Myers Squibb
Research Funding: AstraZeneca, Clovis Oncology
Open Payments Link: https://openpaymentsdata.cms.gov/physician/917904
Judy Garber
Consulting or Advisory Role: Novartis, GTx, Helix BioPharma, Konica Minolta, Aleta BioTherapeutics, H3 Biomedicine, Kronos Bio
Research Funding: Novartis, Ambry Genetics, InVitae, Myriad Genetics
Other Relationship: Susan G. Komen for the Cure, AACR, Diana Helis Henry Medical Foundation, James P. Wilmot Foundation, Adrienne Helis Malvin Medical Research Foundation, Breast Cancer Research Foundation, Facing Our Risk of Cancer Empowered
Braden Probst
Employment: Myriad Genetics
Brian Morris
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Placede Tshiaba
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Stephanie Meek
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Eric Rosenthal
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Benjamin Roa
Employment: Myriad Genetics
Leadership: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Research Funding: Myriad Genetics
Patents, Royalties, Other Intellectual Property: Intellectual Property held by employer Myriad Genetics
Travel, Accommodations, Expenses: Myriad Genetics
Thomas P. Slavin
Employment: Myriad Genetics
Leadership: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Susanne Wagner
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Patents, Royalties, Other Intellectual Property: Co-author of patents held by Myriad Genetics, no royalties
Jeffrey Weitzel
Speakers' Bureau: AstraZeneca
Alexander Gutin
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics, Gilead Sciences
Jerry S. Lanchbury
Employment: Myriad Genetics
Leadership: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Patents, Royalties, Other Intellectual Property: I am an inventor on Multiple patents filed by Myriad Genetics
Travel, Accommodations, Expenses: Myriad Genetics
Mark Robson
Consulting or Advisory Role: Change HealthCare
Research Funding: AstraZeneca, Pfizer, Merck
Other Relationship: Research to Practice, Clinical Care Options, Physicans' Education Resource, Invitae, Pfizer
Uncompensated Relationships: Merck, Pfizer, Daiichi Sankyo, Epic Sciences
Open Payments Link: https://openpaymentsdata.cms.gov/physician/612669/summary
No other potential conflicts of interest were reported.
REFERENCES
- 1.Kurian AW, Hughes E, Handorf EA, et al. Breast and ovarian cancer penetrance estimates derived from germline multiple-gene sequencing results in women JCO Precis Oncol 11–122017 [DOI] [PubMed] [Google Scholar]
- 2.Daly MB, Pilarski R, Berry M, et al. NCCN Clinical Practice Guidelines in Oncology, Genetic/Familial High-Risk Assessment: Breast and Ovarian (version 3.2019) https://www.nccn.org/professionals/physician_gls/pdf/genetics_bop.pdf [Google Scholar]
- 3.Andrieu N, Goldgar DE, Easton DF, et al. Pregnancies, breast-feeding, and breast cancer risk in the International BRCA1/2 Carrier Cohort Study (IBCCS) J Natl Cancer Inst 98535–5442006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park B, Hopper JL, Win AK, et al. Reproductive factors as risk modifiers of breast cancer in BRCA mutation carriers and high-risk non-carriers Oncotarget 8102110–1021182017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans DG, Harkness EF, Howel S, et al. Young age at first pregnancy does protect against early onset breast cancer in BRCA1 and BRCA2 mutation carriers Breast Cancer Res Treat 167779–7852018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toss A, Grandi G, Cagnacci A, et al. The impact of reproductive life on breast cancer risk in women with family history or BRCA mutation Oncotarget 89144–91542017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang X, Leslie G, Doroszuk A, et al. Cancer risks associated with germline PALB2 pathogenic variants: An International Study of 524 Families J Clin Oncol 38674–6852020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cybulski C, Wokolorczyk D, Jakubowska A, et al. Risk of breast cancer in women with a CHEK2 mutation with and without a family history of breast cancer J Clin Oncol 293747–37522011 [DOI] [PubMed] [Google Scholar]
- 9.Weischer M, Bojesen SE, Ellervik C, et al. CHEK2*1100delC genotyping for clinical assessment of breast cancer risk: Meta-analyses of 26,000 patient cases and 27,000 controls J Clin Oncol 26542–5482008 [DOI] [PubMed] [Google Scholar]
- 10.Mavaddat N, Pharoah PD, Michailidou K, et al. Prediction of breast cancer risk based on profiling with common genetic variants. J Natl Cancer Inst. 2015;107:djv036. doi: 10.1093/jnci/djv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lakeman IMM, Rodríguez-Girondo M, Lee A, et al. Validation of the BOADICEA model and a 313-variant polygenic risk score for breast cancer risk prediction in a Dutch prospective cohort Genet Med 221803–18112020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barnes DR, Rookus MA, McGuffog L, et al. Polygenic risk scores and breast and epithelial ovarian cancer risks for carriers of BRCA1 and BRCA2 pathogenic variants Genet Med 221653–16662020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muranen TA, Greco D, Blomqvist C, et al. Genetic modifiers of CHEK2*1100delC-associated breast cancer risk Genet Med 19599–6032017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuchenbaecker KB, McGuffog L, Barrowdale D, et al. Evaluation of polygenic risk scores for breast and ovarian cancer risk prediction in BRCA1 and BRCA2 mutation carriers. J Natl Cancer Inst. 2017;109:djw302. doi: 10.1093/jnci/djw302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallagher S, Hughes E, Wagner S, et al. Association of a polygenic risk score with breast cancer among women carriers of high- and moderate-risk breast cancer genes. JAMA Netw Open. 2020;3:e208501. doi: 10.1001/jamanetworkopen.2020.8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes E, Tshiaba P, Gallagher S, et al. Development and validation of a clinical polygenic risk score to predict breast cancer risk JCO Precis Oncol 4585–5922020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Judkins T, Leclair B, Bowles K, et al. Development and analytical validation of a 25-gene next generation sequencing panel that includes the BRCA1 and BRCA2 genes to assess hereditary cancer risk. BMC Cancer. 2015;15:215. doi: 10.1186/s12885-015-1224-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes E, Tshiaba P, Wagner S, et al. Integrating clinical and polygenic factors to predict breast cancer risk in women undergoing genetic testing JCO Precis Oncol 5307–3162021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bevers TB, Ward JH, Ahrendt GM, et al. NCCN Clinical Practice Guidelines in Oncology: Breast Cancer Risk Redution (Version 1.2020) https://www.nccn.org/professionals/physician_gls/pdf/breast_risk.pdf [Google Scholar]
- 20.Schmidt MK, Hogervorst F, van Hien R, et al. Age- and tumor subtype–specific breast cancer risk estimates for CHEK2*1100delC carriers J Clin Oncol 342750–27602016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mavaddat N, Michailidou K, Dennis J, et al. Polygenic risk scores for prediction of breast cancer and breast cancer subtypes Am J Hum Genet 10421–342019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuchenbaecker KB, Hopper JL, Barnes DR, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers JAMA 3172402–24162017 [DOI] [PubMed] [Google Scholar]
- 23.The American Society of Breast Surgeons . Consensus Guideline on Diagnostic and Screening Magnetic Resonance Imaging of the Breast. 2017. https://www.breastsurgeons.org/docs/statements/Consensus-Guideline-on-Diagnostic-and-Screening-Magnetic-Resonance-Imaging-of-the-Breast.pdf [Google Scholar]
- 24.The American Society of Breast Surgeons . Position Statement on Screening Mammography. 2019. https://www.breastsurgeons.org/docs/statements/Position-Statement-on-Screening-Mammography.pdf [Google Scholar]
- 25.Saslow D, Boetes C, Burke W, et al. American Cancer Society guidelines for breast screening with MRI as an adjunct to mammography CA Cancer J Clin 5775–892007 [DOI] [PubMed] [Google Scholar]
- 26.Rothman KJ, Greenland S, Lash T. Modern Epidemiology. Philadelphia, PA: Lippincott Williams & Wilkins; 2008. [Google Scholar]
- 27.Lee CI, Chen LE, Elmore JG.Risk-based breast cancer screening: Implications of breast density Med Clin North Am 101725–7412017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shieh Y, Fejerman L, Lott PC, et al. A polygenic risk score for breast cancer in US Latinas and Latin American women J Natl Cancer Inst 112590–5982020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available on reasonable request from the corresponding author. The individual patient data are not publicly available because of privacy and ethical restrictions.