Abstract
Parent-child similarity is a function both of genetic and environmental transmission. In addition, genetic effects not transmitted to offspring may drive parental behavior, thereby affecting the rearing environment of the child. Measuring genetic proclivity directly, through polygenic risk scores (PRSs), provides a way to test for the effect of non-transmitted parental genotype, on offspring outcome, termed a “genetic nurture” effect. In other words, if and how parental genomes might affect their children through the environment. The current study used polygenic risk scores for smoking initiation, cigarettes per day, and drinks per week to predict substance use in a sample of 3,008 twins, assessed prospectively from age 17 to age 29, and their parents, from the Minnesota Center for Twin and Family Research. Mixed effects models were used to test for a genetic nurture effect whereby parental PRSs predict offspring tobacco and alcohol use after statistically adjusting for offspring’s own PRS. Parental smoking initiation PRS predicted offspring cigarettes per day at age 24 (β=0.103; 95% confidence interval (CI) [0.03, 0.17]) and alcohol use at age 17 (β=0.091; 95% CI [0.04, 0.14]) independent of shared genetics. There was also a suggestive independent association between the parent PRS and offspring smoking at age 17 (β=0.096; 95% CI [0.02, 0.17]). Mediation analyses provided some evidence for environmental effects of parental smoking, alcohol use, and family socioeconomic status. These findings, and more broadly the molecular genetic method used, have implications on the identification of environmental effects on developmental outcomes such as substance use.
Keywords: nicotine, alcohol, genetic nurture, polygenic risk score, gene-environment correlation
It is widely accepted that nearly all complex traits and behaviors follow both genetic and environmental transmission (Polderman et al., 2015). Because of their potential to be modified, a primary goal of research is in determining whether, and to what extent, environmental factors causally influence developmental outcomes. A problem in identifying causal exposure effects, however, is the fact that genetic and environmental influences are not independent of each other. The idea that environmental exposures are associated with genetic risk has been termed gene-environment correlation (rGE). There are three types of rGE: passive, evocative, and reactive (Jaffee & Price, 2008; Scarr & McCartney, 1983). We focus only on passive rGE here, which occurs when parents pass on genes to their offspring that influence behaviors while also fostering an environment that influences risk.
Detecting the presence of passive rGE is important for understanding the potential causal effect of environmental exposures on offspring outcomes (Jaffee & Price, 2012). Passive rGE can imply that shared genes confound the relationship between a rearing environmental exposure and offspring behaviors, inducing a spurious correlation between the two. In this case the environmental exposure is non-causal and modifying it will likely have no impact on outcomes. Another possibility is that while there might be some confounding by genotype, there is also evidence of an environmental exposure effect that is independent of the genes transmitted by parents. This would be consistent with a causal effect of the environmental exposure on offspring behaviors and would suggest that modifying this exposure may be a worthwhile goal in affecting outcomes. To date, much of the evidence for passive rGE has been provided by twin and adoption studies, by showing that environmental exposures are themselves heritable or through correlations between adoptive parents and their adopted children despite not transmitting any genes (Jaffee & Price, 2007; Kendler & Baker, 2007).
These methods, however, have several limitations. Adoption studies can be difficult to conduct with large samples while twin studies can only investigate passive rGE with exposures that differ within a twin pair. The effect of family-level exposures (i.e., rearing socioeconomic status or parental educational attainment) on offspring developmental outcomes would be missed using a twin design (Plomin, 2014). More recently, molecular genetic approaches have been used to test for passive rGE. These designs measure the genotype directly, testing for environmental effects on offspring outcomes through the parent’s genotype that is not passed to their offspring, termed a “genetic nurture” effect (Kong et al., 2018). For example, parents have genes that influence their risk of substance use, some of which will be randomly transmitted to their offspring. Other (non-transmitted) genes may influence some parental phenotype (environmental exposure) that in turn affects offspring substance use, independent of the genetic risk transmitted from parent to offspring.
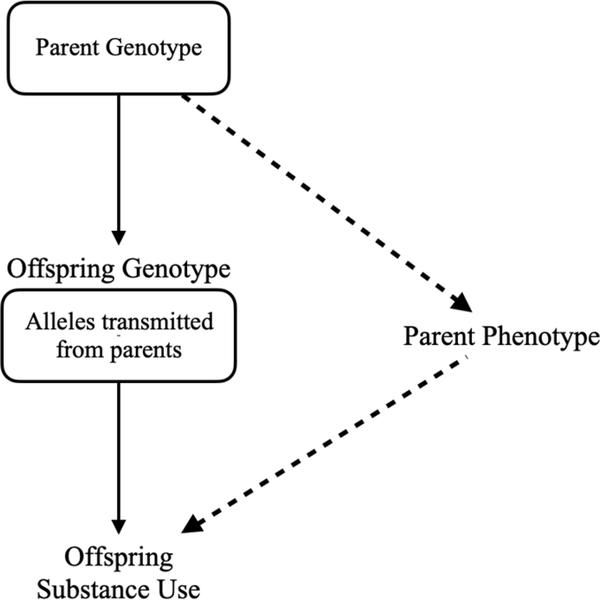
While some studies have used single genes to test for passive rGE (O’Brien et al., 2013), polygenic risk scores may provide greater power in detecting genetic nurture effects. A polygenic risk score (PRS) is a weighted sum of risk alleles associated with a phenotype. Scores are computed for each individual based on single-nucleotide polymorphisms (SNPs) associated with phenotypes of interest, like tobacco and alcohol use, in previous large-scale genome wide association studies (GWAS). The current study makes use of polygenic risk scores for tobacco and alcohol behaviors to test for the presence of a genetic nurture effect in a large parent-offspring sample. We first derived polygenic scores for smoking initiation, cigarettes per day, and drinks per week and assessed their predictive power for all offspring outcomes. We then examined whether the influence of parental substance use on offspring substance use outcomes is due only to the genetic risk they pass (measured here by polygenic scores), consistent with the presence of passive rGE and a non-causal environmental exposure effect on offspring outcomes, or if there was an independent effect of the parental polygenic scores, as a measure of genetic risk, through some form of parental phenotype, on offspring substance use (Figure 1). The logic follows that parents have a genetic risk, or proclivity, for alcohol and tobacco use, as measured here by polygenic scores, that affects the substance use of their children through risk genes passed directly from parents to offspring. In the simplest case the effect of parental polygenic scores on offspring substance use would be completely accounted for by the offspring’s own polygenic score. However, if there exists a unique contribution of parental polygenic scores to offspring alcohol and tobacco use, this suggests that the parent genotype influences some part of the environment that in turn influences offspring substance use; there is a direct and indirect path from parent polygenic score to offspring outcomes. The presence of a parent PRS effect on offspring outcome, independent of offspring’s own PRS, provides evidence for a genetic nurture effect and is equivalent in interpretation to finding an effect of non-transmitted alleles on offspring outcomes (Bates et al., 2018; Kong et al., 2018). The supplemental material of Willoughby et al. (2019) includes a detailed comparison of the current method and that of partitioning transmitted and non-transmitted alleles as in Kong et al. (2018), showing the equivalence of the genetic nurture effects across both methods.
Figure 1.
Example of passive gene-environment correlation where parental genotype can confound the relationship between a rearing environmental exposure and offspring substance use.
Note. Solid lines represent substance use risk genes that are passed directly from parent to offspring. Dashed lines represent an indirect effect of the parental risk genes on offspring substance use, through a parent phenotype, that is independent of shared genetics.
If parent polygenic scores are associated with offspring alcohol and tobacco use independent of shared genes, it may be possible to identify rearing environmental effects that are associated with offspring outcomes. Because offspring may model parental behavior, parental smoking and alcohol use are potential environmental effects to evaluate (Gilman et al., 2009; Rossow et al., 2016; Scherrer et al., 2012). Another environmental effect of interest is parent, or rearing, socioeconomic status (SES). Previous research has shown robust associations between rearing SES and both alcohol and tobacco use, though these relationships may be particularly complex (Calling et al., 2019; Gilman et al., 2003; Gilman et al., 2008; Wellman et al., 2016). There are also significant genetic correlations between education attainment, a component of SES, and measures of tobacco use, though the genetic covariance with alcohol use appears to be weaker (Liu et al., 2019). While rearing SES may not directly increase or decrease substance use risk, growing up in a higher SES home may influence peer group choice and access to material advantages and activities out of school that in turn effect smoking and alcohol use behaviors (Bradley & Corwyn, 2002). Thus, we explored whether parental smoking, alcohol use, and socioeconomic status mediated potential genetic nurture effects.
Method
Sample
The current sample included parents and offspring from the Minnesota Center for Twin and Family Research (MCTFR), a longitudinal study of risk for substance misuse (Iacono et al., 1999; Keyes et al., 2009). MCTFR participants were ascertained through Minnesota state birth records from the birth years 1972 to 1994 and are largely representative of families in Minnesota at the time of recruitment (e.g., approximately 98% of the sample is Caucasian). The current sample is drawn from three cohorts of same-sex twin pairs assessed up to six times between the ages of 11 and 29 (see Supplementary Materials for a more detailed description of each cohort).
For validating the derived PRSs, all individuals with genotype and phenotypic outcome data are included, resulting in samples sizes ranging from N=756–3,008 for offspring and N=2,614–2,876 for parents. Table 1 provides approximate sample sizes for each outcome. Of note, the sample size for cigarettes per day at age 29 is far smaller than at earlier ages because data on smoking was only collected from one cohort due to funding limitations at the time. In testing our core hypothesis of a genetic nurture effect, all offspring with genotype and phenotypic outcome data, as well as genotype data from at least one parent, are included. This results in sample sizes ranging from N=693–2,880 from N=434–1,491 families, which are slightly smaller than those shown in Table 1 due to the availability of parental genotype data (18% of the offspring sample had only maternal genotype data, 5% had paternal genotype data, and 72% of the sample had genotype data for both mothers and fathers). The effects of sample attrition are minimal and are further described in the Supplementary Material. All studies were approved by the University of Minnesota Internal Review Board through the Minnesota Center for Twin and Family Research under Study Numbers 00001992, 8704M00020, 1305M32821, and 9109M04330, and all parents and offspring gave written informed consent or assent, as appropriate.
Table 1.
Descriptive statistics for each outcome, split by age and generation. Sample sizes include all participants with phenotypic outcome and genotype data.
| N | Female (%) | M | SD | |
|---|---|---|---|---|
| Cigarettes per day (among smokers) | ||||
| Offspring (at age 17) | 1,423 | 46% | 6.95 | 9.69 |
| Offspring (at age 24) | 1,671 | 41% | 8.38 | 10.31 |
| Offspring (at age 29) | 756 | 38% | 8.62 | 10.80 |
| Parents (maximum lifetime) | 2,614 | 53% | 15.49 | 15.46 |
| Drinking index | ||||
| Offspring (at age 17) | 3,008 | 53% | 1.08 | 1.00 |
| Offspring (at age 24) | 2,249 | 54% | 2.02 | 0.75 |
| Offspring (at age 29) | 2,309 | 53% | 1.72 | 0.73 |
| Parents (at twin intake; mean age 42.8 years) | 2,876 | 55% | 1.76 | 0.70 |
Note: M = mean; SD = standard deviation; mean values for cigarettes per day are computed by assigning the midpoint of each level as we do not have a continuous measure of cigarettes per day available at each age.
Phenotypic Measures
Outcome measures were collected in offspring at the age 17, 24, and 29 assessments using either the Substance Abuse Module (Robins et al., 1987) of the Composite International Diagnostic Interview (Robins et al., 1988) or the Computerized Substance Use Questionnaire, depending on the age of the participant. Parental substance use outcomes were collected at their offspring’s intake assessment. The tobacco outcome variable is a binned measure of cigarettes per day (CPD) reported in the previous 12 months, including all forms of tobacco. The CPD measure is coded only for those who have reported ever smoking and ranges from 1 (less than one cigarette/one cigar or pipe/one pinch per day) to 6 (2 or more packs/20 or more cigars or pipes/2 or more tins per day). In this way, we are interested in smoking progression among those who have ever been exposed. The current analysis used offspring CPD at ages 17, 24, and 29, while the parent measure of CPD is their lifetime maximum.
The alcohol outcome variable is a composite measure of alcohol consumption (ALC) at ages 17, 24, and 29 in offspring and at the twin intake in parents (mean age of 42.8 years old). The drinking index incorporates measures of quantity, frequency, number of intoxications, and maximum number of drinks reported in the previous 12 months. Specifically, it consists of four self-report alcohol use items: frequency of alcohol use (scored from 0=never to 5=at least once per day), average number of drinks per drinking event (scored from 0=never drank to 6=30 or more), maximum number of drinks in a 24-hour period (scored from 0=never drank to 6=30 or more), and number of times intoxicated (scored from 0=never to 6=50 or more). This composite index provides a more comprehensive view of overall alcohol exposure than a single component measure. These measures were initially summed before using integrative data analysis and item response theory methods to estimate individual scores on a common latent trait. Additional detail on the creation of the drinking index can be found in McGue et al. (2014).
Genotyping and polygenic scoring
Participants were genotyped on 527,829 single nucleotide polymorphism (SNP) markers using Illumina’s Human660W-Quad array (Miller et al., 2012). European genotypes were then imputed to Haplotype Reference Consortium (HRC) version 1 using the Michigan imputation server. Basic quality control steps removed variants with a minor allele frequency < 1%, missing genotype call rate > 5%, individuals with missingness > 1%, and then finally subset to approximately 1.2 million HapMap3 variants.
Polygenic risk scores were created for parents and offspring for cigarettes per day among ever smokers (CigDay-PRS), smoking initiation (SmkInit-PRS), and drinks per week (DrnkWk-PRS). Because we made no specific hypotheses regarding maternal and paternal genetic nurture effects we use the mean of mother and father polygenic scores as the parental PRS measure. Only participants of European ancestry, the vast majority of the overall sample, were included as this was the population used in deriving the weights from the GSCAN consortium (Martin et al., 2017). Weights for computing PRSs in the current study come from the GWAS & Sequencing Consortium of Alcohol and Nicotine use (GSCAN), with the MCTFR sample removed (Liu et al., 2019). Each PRS was computed using LDpred software, which accounts for the effects of linkage disequilibrium (Vilhjálmsson et al., 2015). More information about the calculation of polygenic scores is included in the Supplementary Material.
Covariate Measures
Base covariates include sex, age at reported use (to account for the variation in actual age at each target assessment), and year of birth (up to a quadratic term, to account for potential cohort effects). We also adjusted for the first ten genetic principal components to better account for possible spurious associations due to population stratification (Price et al., 2006). In follow-up mediation analyses we added parent cigarettes per day, drinking index scores, and rearing socioeconomic status (SES). Parental cigarettes per day and drinking index scores are measured in the same way as in the offspring, while rearing SES was defined as the mean of maximum education attained by either parent on a 5-point scale (1= <High School, 2=High School, 3=Some College, 4= College, 5=Professional Degree) and maximum occupational status reported by either parent (coded on a 1–7 Hollingshead scale only for those in a full-time job and reversed so that higher scores reflect higher occupational status).
Statistical Analysis
Data were analyzed in R using mixed effect models with family ID as a random effect (intercept) to account for the correlated nature of the data. Linear mixed effects models were used to estimate beta coefficients (βs) for both tobacco and alcohol outcomes (Bates et al., 2015; see the Supplemental Material for exact model specification). Because our measure of smoking is a binned variable, we re-ran the main CPD analyses using ordinal mixed effects models (Archer et al., 2018). Interpretation of results remained unchanged and are reported in Supplemental Table 3. Here we treat CPD as a continuous measure using the midpoint of each CPD level to facilitate better comparison of effects across tobacco and alcohol use. All estimates are standardized, are accompanied by 95% confidence intervals (CIs). Given the multiple, correlated tests, we use a correction based on the effective number of tests as estimated using the eigenvalues of a correlation matrix of related variables (Derringer, 2018; Nyholt, 2004). The per-test α (0.05) is then divided by the number of effective tests, resulting in a significance threshold of p < 0.0058. For ease of interpretation, we report cross-sectional results by outcome and offspring age (age 17, 24, and 29) in the main text though more parsimonious repeated measures models were also fit and are reported in the Supplemental Tables 6 and 7. General conclusions remain unchanged across both methods of statistical analysis. Lastly, because the base covariates used in the current analysis differ from those in the discovery GWAS, we report the main results using only the same covariates as the discovery GWAS in Supplemental Tables 4 and 5. The results are again unchanged. For follow-up mediation analyses, we use a bootstrap method to estimate confidence intervals of indirect effects based on 500 replications.
Results
Descriptive Statistics
Descriptive statistics for phenotypic outcomes are given in Table 1. As expected due to secular changes in tobacco use, offspring smokers at all ages report fewer cigarettes per day than smoking parents (because we do not have a continuous measure of cigarettes per day for all offspring, we used the midpoint of each CPD level when computing the means and standard deviations). Mean drinking index scores increase from age 17 to age 24 before falling slightly at age 29, reaching a level comparable with what parents report. Correlations among polygenic scores and outcomes in parents and offspring are included in the Supplemental Figure 1. We additionally note that there is no evidence of genetic assortative mating based on the polygenic scores as derived here (Supplemental Figure 2).
Associations between PRSs and outcomes
Beginning with the tobacco outcome of cigarettes per day, we tested the predictive accuracy of all three polygenic risk scores at offspring ages 17, 24, and 29 (Table 2). As expected, CigDay-PRS predicts offspring CPD at all ages, though interestingly, SmkInit-PRS has the strongest predictive effect for offspring CPD across age. There is evidence of a small, but positive, predictive effect of DrnkWk-PRS on the CPD outcomes at age 17. The effect of DrnkWk-PRS at age 24 and age 29 is negligible as evidenced by a confidence interval that crosses zero. Supplemental Table 1 additionally includes p-values and variance explained by each PRS across age for both parents and offspring.
Table 2.
Association between own and parental polygenic scores and offspring cigarettes per day.
| Parental PRS model |
|||
|---|---|---|---|
| Offspring PRS Alone | Offspring PRS | Parent PRS | |
| β (95% CI) | β (95% CI) | β (95% CI) | |
| CigDay polygenic scores | |||
| Age 17 | 0.154* (0.098, 0.210) |
0.134* (0.060, 0.207) |
0.033 (−0.046, 0.112) |
| Age 24 | 0.169* (0.118, 0.220) |
0.143* (0.077, 0.209) |
0.047 (−0.022, 0.116) |
| Age 29 | 0.141* (0.062, 0.220) |
0.101 (−0.005, 0.207) |
0.097 (−0.014, 0.207) |
| SmkInit polygenic scores | |||
| Age 17 | 0.169* (0.112, 0.225) |
0.118 (0.046, 0.190) |
0.095 (0.019, 0.170) |
| Age 24 | 0.181* (0.130, 0.231) |
0.129* (0.063, 0.196) |
0.103* (0.033, 0.172) |
| Age 29 | 0.187* (0.111, 0.264) |
0.177* (0.078, 0.276) |
0.029 (−0.077, 0.136) |
| DrnkWk polygenic scores | |||
| Age 17 | 0.071 (0.015, 0.127) |
0.093 (0.020, 0.165) |
−0.032 (−0.110, 0.045) |
| Age 24 | 0.034 (−0.018, 0.085) |
0.043 (−0.025, 0.111) |
−0.021 (−0.092, 0.050) |
| Age 29 | 0.063 (−0.016, 0.142) |
0.110 (0.008, 0.212) |
−0.088 (−0.197, 0.021) |
Note. All models adjust for the covariates of sex, age at assessment, year of birth (up to a quadratic term), and the first 10 genetic principal components. All polygenic scores were scaled to have a mean of 0 and standard deviation of 1 prior to analysis. PRS = polygenic risk score; β = standardized beta coefficient; CI = confidence interval. Parent PRS is defined as the mean of mother and father polygenic scores. Parental PRS model indicates that both offspring and parent PRS are included (i.e., the parent PRS coefficient is the association with offspring outcome with inclusion of offspring’s own PRS). Asterisks denote p-values surviving correction for the number of effective tests (p < 0.0058).
Similar analysis for the alcohol outcome of drinking index scores is shown in Table 3. As expected, DrnkWk polygenic scores predict drinking index scores at all ages. Similar to tobacco, SmkInit polygenic scores have the strongest predictive effect for offspring drinking, though this effect declines across age. There is a near zero effect of CigDay-PRS on alcohol outcomes at all ages as evidenced by confidence intervals that include zero. Supplemental Table 2 additionally includes p-values and variance explained by each PRS across age for both parents and offspring.
Table 3.
Association between own and parental polygenic scores and offspring drinking index scores.
| Parental PRS model |
|||
|---|---|---|---|
| Offspring PRS Alone | Offspring PRS | Parent PRS | |
| β (95% CI) | β (95% CI) | β (95% CI) | |
| CigDay polygenic scores | |||
| Age 17 | 0.033 (−0.006, 0.072) |
0.008 (−0.040, 0.057) |
0.043 (−0.012, 0.097) |
| Age 24 | 0.034 (−0.011, 0.079) |
0.027 (−0.032, 0.086) |
0.015 (−0.049, 0.079) |
| Age 29 | 0.053 (0.008, 0.097) |
0.071 (0.012, 0.130) |
−0.023 (−0.086, 0.039) |
| SmkInit polygenic scores | |||
| Age 17 | 0.117* (0.079, 0.155) |
0.075* (0.028, 0.123) |
0.091* (0.037, 0.144) |
| Age 24 | 0.125* (0.081, 0.170) |
0.109* (0.052, 0.166) |
0.047 (−0.014, 0.108) |
| Age 29 | 0.113* (0.069, 0.157) |
0.100* (0.043, 0.156) |
0.051 (−0.009, 0.110) |
| DrnkWk polygenic scores | |||
| Age 17 | 0.088* (0.050, 0.126) |
0.089* (0.039, 0.135) |
0.004 (−0.050, 0.058) |
| Age 24 | 0.101* (0.057, 0.145) |
0.100* (0.043, 0.158) |
0.009 (−0.053, 0.071) |
| Age 29 | 0.127* (0.083, 0.171) |
0.140* (0.083, 0.198) |
−0.014 (−0.074, 0.047) |
Note. All models adjust for the covariates of sex, age at assessment, year of birth (up to a quadratic term), and the first 10 genetic principal components. All polygenic scores were scaled to have a mean of 0 and standard deviation of 1 prior to analysis. PRS = polygenic risk score; β = standardized beta coefficient; CI = confidence interval. Parent PRS is defined as the mean of mother and father polygenic scores. Parental PRS model indicates that both offspring and parent PRS are included (i.e., the parent PRS coefficient is the association with offspring outcome with inclusion of offspring’s own PRS). Asterisks denote p-values surviving correction for the number of effective tests (p < 0.0058).
Genetic nurture effects through parental PRSs
Parental polygenic risk scores alone predict offspring outcomes following similar patterns of own PRS prediction. Parent CigDay and SmkInit-PRSs predict offspring smoking at all ages, while parent DrnkWk and SmkInit-PRSs predict alcohol use. The effect of parental SmkInit-PRS on outcomes is stronger than for the other parent polygenic scores. After controlling for offspring’s own polygenic scores, parental SmkInit-PRS remains a significant predictor of offspring’s CPD outcomes at age 24 (Table 2; β=0.103, 95% CI [0.03, 0.17], p=.004). There is some evidence for an effect at age 17 (β=0.095, 95% CI [0.02, 0.17], p=.013) but it is not significant based on our more stringent p-value cut-off of 0.0058 to account for multiple testing.
Parental SmkInit-PRS no longer has an effect at age 29 (β=0.029, 95% CI [−0.08, 0.14], p=.58). There is no evidence of an independent predictive effect of parental DrnkWk scores on offspring CPD after controlling for offspring’s own PRS. While the parental CigDay-PRS effect increases in magnitude over development from β=0.033 at age 17 to β=0.097 at age 29, the small offspring sample size at age 29 results in wide confidence intervals overlapping zero. Full results for CPD are shown in Table 2. Results from similar analyses treating CPD as a binned (ordinal) outcome variable are shown in Supplemental Table 3.
For offspring alcohol outcomes, after controlling for offspring’s own polygenic scores, parental SmkInit polygenic scores predict offspring’s alcohol use at age 17 (β=0.091, 95% CI [0.04, 0.14], p=.001) but no longer have an effect at ages 24 and 29 (β=0.047, 95% CI [−0.01, 0.11], p=.13, and β=0.051, 95% CI [−0.01, 0.11], p=.09, respectively). There is no evidence of an independent predictive effect of parental DrnkWk or CigDay scores after controlling for offspring’s own PRS. Full results for alcohol use are shown in Table 3.
Results from repeated measures models are shown in Supplemental Tables 6 and 7. The pattern of the results is generally consistent with cross-sectional models suggesting an effect of parental SmkInit-PRS, after controlling for offspring’s own PRS, for offspring cigarettes per day (β=0.084, 95% CI [0.04, 0.13], p=.002) and alcohol use (β=0.065, 95% CI [0.03, 0.10], p=.0004). There is no evidence, however, that the strength of the identified genetic nurture effects changes significantly across offspring age, though we may be underpowered to detect such an interaction effect should one exist.
We then tested whether parental substance use or rearing SES mediate the relationship between parental SmkInit polygenic scores and offspring outcomes. Mediation analysis is often hindered by low power, so we run these analyses only in the repeated measures models for each offspring outcome. Parental SmkInit-PRSs significantly predicted their own outcomes (potential mediators) of cigarettes per day (β=0.216, 95% CI [0.18, 0.25]), drinking index scores (β=0.092, 95% CI [0.06, 0.13]), and socioeconomic status (β=−0.143, 95% CI [−0.18, −0.10]). All potential mediators were associated with both offspring smoking and drinking outcomes, with the exception of rearing SES and offspring drinking index scores, and parental alcohol use and offspring cigarettes per day. All standardized coefficients and mediation plots are shown in Supplemental Figure 3. We tested the significance for each indirect effect using bootstrapping with 500 replications (95% confidence intervals were computed by determining the indirect effects at the 2.5th and 97.5th percentiles), shown in Supplemental Table 8. While indirect effects were relatively small in magnitude there was some evidence for parental alcohol use mediating the relationship between parental SmkInit-PRS and offspring drinking index scores (indirect effect = 0.018 95% CI [0.016, 0.019]), parental (rearing) SES mediating the relationship between parental SmkInit-PRS and offspring smoking (indirect effect = 0.023 95% CI [0.019, 0.026]), and parental tobacco use mediating the relationship between parental SmkInit-PRS and both offspring tobacco and alcohol use outcomes (indirect effect = 0.040 95% CI [0.034, 0.045] and indirect effect = 0.023 95% CI [0.019, 0.027], respectively).
Discussion
Using molecular genetic methods, we have provided evidence of a genetic nurture effect for offspring alcohol use in late adolescence and tobacco use in early adulthood. This methodology can separate environmental influences on outcomes from shared genetic transmission, allowing us to disentangle the potentially confounded relationship between rearing environmental exposures and offspring outcome. Our results show that there is an association between parental genotype and offspring smoking and alcohol use in early development independent of the genetic risk that is transmitted. This unique effect of parental genes must operate through the environment, through form of parental phenotype, to influence offspring behaviors. We found that the relationship between parental genotype and offspring substance use is potentially mediated by rearing socioeconomic status, as well as parent’s own substance use.
Overall, a common pattern that appears is the strong predictive effect of SmkInit polygenic scores for both tobacco and alcohol outcomes across age and generation, relative to other polygenic scores. In most cases, the SmkInit-PRS effect on drinking index scores is larger than that of the DrnkWk-PRS. We believe there are two possible explanations for this that are not mutually exclusive. One, the effect of SmkInit-PRSs is strongest because of greater precision, owing to a much larger sample size and number of detected variants in the original GSCAN GWAS (Liu et al., 2019). The GWAS of smoking initiation (SmkInit) included N=1,232,091 participants and identified 378 associated variants compared to N=337,334 participants and 55 variants for CigDay and N=941,280 participants and 99 associated variants for DrnkWk. Consistent with this idea, in the current sample we find that the variance in offspring CPD explained ranged from 2.3%−3.0% for CigDay-PRS, 0.1%−0.5% for DrnkWk-PRS, and 2.9%−3.7% for SmkInit-PRS (Nakagawa et al., 2017). We find a similar pattern for variance explained in offspring drinking index scores by CigDay-PRS, R2=0.1%−0.3%, DrnkWk-PRS, R2=0.8%−1.6%, and SmkInit-PRS, R2=1.4%−1.7%. It may be the case that as sample sizes continue to increase and a greater number of genetic variants are identified, the polygenic scores for CigDay and DrnkWk will reach a predictive accuracy closer to that of smoking initiation. A second possible explanation is that what the SmkInit-PRSs are capturing is simply a better predictor of both tobacco and alcohol use, and that this is unique to smoking initiation. The genetic correlations between SmkInit and CigDay is r=0.33 and between SmkInit and DrnkWk is r=0.34 (Liu et al., 2019), so it may be that each polygenic score is indexing somewhat different genetic risk. In this case, even with increasing sample sizes and identified variants, SmkInit-PRSs will continue to have the strongest predictive accuracy for tobacco and alcohol outcomes.
Parental polygenic risk scores predict offspring outcomes in a similar way as offspring’s own PRS (i.e., parental CigDay and SmkInit-PRS predict offspring cigarettes per day and parental SmkInit and DrnkWk-PRS predict offspring alcohol outcomes). While the effect size of parent polygenic scores vary by offspring age (e.g., a declining effect of parental SmkInit-PRS and increasing effect of parental CigDay and DrinkWk-PRSs on offspring CPD from ages 17 to 29), all confidence intervals are overlapping suggesting the effect of parental PRS on offspring substance use remains relatively stable over offspring development, following a similar pattern of the effect of offspring’s own PRS.
Finally, our main hypothesis of genetic nurture effects was partially supported. We found evidence for a genetic nurture effect of parental SmkInit-PRS on offspring CPD at age 24 and offspring drinking index scores at age 17 suggesting that some environmental factor (e.g. parent phenotype) associated with parental SmkInit polygenic scores influences offspring outcomes in late adolescence and early adulthood independent of shared genes. The size of the parental SmkInit-PRS on offspring CPD, after controlling for offspring PRS, effect is about 57% the size of offspring own SmkInit-PRS at age 24. The effect of parental SmkInit-PRS on offspring drinking at age 17 is approximately 78% of the size of offspring’s own SmkInit-PRS effect. These findings support the presence of passive rGE and an effect of some parenting phenotype, influenced by the genetic risk variants associated with smoking initiation, on offspring substance use and are consistent with prior work in adopted families that provides evidence for environmental mediation of parental smoking on adopted offspring use (Keyes et al., 2008; Samek et al., 2014). There was also a trend for an independent effect of parental CigDay-PRS and DrnkWk-PRS (in the opposite direction) on offspring CPD as evidenced by an increasing effect size over offspring age after accounting for offspring’s own scores. The reduced offspring CPD sample size at age 29, however, results in wide confidence intervals making it difficult to determine whether these are real effects. Results were generally consistent across cross-sectional and longitudinal models, and were robust to the inclusion of different base covariates.
While longitudinal models did not show a significant interaction between parental SmkInit-PRS and offspring age for either outcome, we do see a general decline in the magnitude of the independent prediction of parental SmkInit-PRS as offspring reach adulthood (age 29) in cross sectional models. Presumably, the influence of the rearing, parental environment on substance use would be strongest while offspring live in the home and would diminish once they leave. In other words, at younger ages when offspring either live in the home with parents or are frequently present in the home as is common during college years, it may be that the offspring are more influenced by the environment that parents model or create. After this age, when offspring are more likely to live on their own, this environmental effect would diminish such that offspring substance use is driven in larger part by shared genetic factors or environmental influences they themselves control. This would be largely consistent with prior research that shows that genetic factors explain an increasing amount of variance in tobacco and alcohol use over adolescence and young adulthood, almost mirroring a decline in the variance explained by shared environmental factors (Kendler et al., 2008).
Given the evidence supporting some genetic nurture effects for both substances, we explored whether rearing SES and/or parental substance use explained, at least in part, the relationship between parental SmkInit-PRS and offspring outcomes. While the size of the indirect effects was relatively small, we found some evidence for potential environmental mediation of genetic nurture effects through parental SES and substance use. This is generally consistent with the literature that has attempted to identify causal influences on substance use (Gilman et al., 2009; Slutske et al., 2008). We note that parental SES is negatively correlated with both alcohol and tobacco use (r=−0.06 and r=−0.13, respectively), while parental alcohol and tobacco use are positively correlated with each other (r=0.22) in the current sample. Given the relatively small mediation effects, combined with our sample size, we did not further investigate the possible joint and independent effects among the mediators. Replication of our main results as well as more in-depth mediation analysis is a prime avenue for further research.
Why we find a genetic nurture effect for parental SmkInit-PRS on both tobacco and alcohol use, and not for CigDay and DrnkWk scores, may be for similar reasons as to why own SmkInit-PRSs have the strongest predictive effect for all outcomes. As mentioned above, one reason may be greater precision in parental SmkInit polygenic scores due to the larger discovery sample size. A second possibility is that there is something unique about parental SmkInit-PRSs not captured by the other scores. Smoking initiation PRSs differ from the CigDay and DrnkWk scores in that they index the genetic risk for smoking exposure rather than progression or consumption measures of substance use. It may be the case that the genetic variants associated with smoking initiation influence parental phenotypes, like attained educational and occupational status and own substance use, to a greater extent than those associated with progression, like cigarettes per day and drinks per week. Indeed, the correlations between parental PRSs and their own educational attainment are r=−0.13 for SmkInitPRS, r=−0.06 for CigDayPRS, and r=0.03 for DrnkWkPRS. These correlations are consistent with prior research showing an inverse relationship between smoking initiation and educational attainment (r=−0.27) but no relationship between smoking progression and educational outcomes in those who have already initiated (r=−0.05; McCaffery et al., 2008). Taken together, this suggests that the genetic variants that influence smoking initiation influence own substance use and attained SES to a greater extent than the variants influencing substance use/progression (i.e., CigDay and DrnkWk) which may explain, in part, why we found a genetic nurture effect of parental SmkInit-PRS and not for CigDay or DrnkWk scores. This does not necessarily rule out the possibility that the effect only for smoking initiation is also due to the imprecision of DrnkWk and CigDay polygenic scores.
While the current study uses weights from the largest GWAS of alcohol and tobacco phenotypes to derive the polygenic scores, these scores only explain a small portion of variance in each outcome. The variance explained in tobacco and alcohol use by each polygenic score is somewhat smaller than what was reported in the original GWAS (Liu et al., 2019), in part likely due to our using somewhat different outcome phenotypes. As we continue to increase GWAS sample sizes and are able to explain more of the variance in tobacco and alcohol use behaviors we will have an increased ability to identify genetic nurture effects. This will also help to clarify whether the small, but highly statistically significant effect of smoking initiation polygenic scores in all models is due to enhanced precision, or something unique about these scores.
A second limitation in the interpretation of the current results is that the causal mechanisms by which environmental factors influence offspring early tobacco and alcohol use are difficult to fully explain. We attempted to explore three possible environmental mediators, parental alcohol and tobacco use and rearing SES, though we caution that these results may be underpowered and require independent replication. We also did not test the joint and independent effects of each mediator (i.e., does parental smoking have an effect independent of parental alcohol use?). Lastly, though importantly, the mediating effects identified were of small magnitude, implying that these environmental factors may not have large effects on offspring substance use after controlling for shared genetics. Nevertheless, they remain potential targets for further study.
Other limitations include the lack of diverse sample. The current sample was restricted to those of European ancestry, which is the same population from which the polygenic score weights were derived, so these results may not generalize to non-European samples. The ancestrally homogenous sample along with statistical adjustment for the first 10 genetic principal components in all models, however, provides for (possibly incomplete) control of confounding due to population structure. In other words, it is possible that lingering population structure, not entirely controlled for by the genetic homogeneity and principal component correction, could confound the relationship between parental genotype and offspring outcome. Additionally, we used only one measure each for tobacco and alcohol outcomes. It would be of interest whether the genetic nurture effects seen for cigarettes per day and drinking index scores remain for other measures of use and abuse. It may be that there is evidence for genetic nurture effects only for certain types, or severities, of substance use behavior rather than a broader tobacco and alcohol effect. It would be of interest whether the genetic nurture effects are substance specific or if they index externalizing behaviors more broadly. Lastly, the vastly reduced CPD sample size at age 29 limits our ability to identify the possible genetic nurture effect of parental CigDay polygenic scores. While there is trend for an increasing effect over offspring age, the confidence intervals at age 29 preclude us from identifying an independent genetic effect.
To conclude, molecular genetic methods are becoming increasingly useful tools for testing a variety of genetic and environmental effects. These may include exploring how the strength of genetic effects change across time, environmental mediation of genetic effects over the lifespan, gene-by-environment interactions, and reciprocal sibling effects. Here we showed how summary statistics from large genome-wide associations, coupled with a genetically informative sample, provide insights on the relationship between shared environmental factors (socioeconomic status and parent substance use) and offspring alcohol and tobacco use. Better understanding causal relationships in observational data, as we have attempted to do here, provides the foundation for identifying, selecting, and characterizing interventions to reduce negative developmental outcomes, like substance use. As we continue to increase GWAS sample sizes and better understand the effect of genetic variation, the better we will be able to elucidate the causal structure between genes, environment, and outcomes.
Supplementary Material
Footnotes
We have no known conflict of interest to disclose.
References
- Bates D, Mächler M, Bolker B, & Walker S (2015). Fitting Linear Mixed-Effects Models Using lme4. Journal of Statistical Software, 67(1), 1–48. doi: 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- Bates TC, Maher BS, Medland SE, McAloney K, Wright MJ, Hansell NK, Kendler KS, Martin NG, & Gillespie NA (2018). The Nature of Nurture: Using a Virtual-Parent Design to Test Parenting Effects on Children’s Educational Attainment in Genotyped Families. Twin Research and Human Genetics, 21(2), 73–83. 10.1017/thg.2018.11 [DOI] [PubMed] [Google Scholar]
- Bradley RH, & Corwyn RF (2002). Socioeconomic Status and Child Development. Annual Review of Psychology, 53(1), 371–399. 10.1146/annurev.psych.53.100901.135233 [DOI] [PubMed] [Google Scholar]
- Calling S, Ohlsson H, Sundquist J, Sundquist K, & Kendler KS (2019). Socioeconomic status and alcohol use disorders across the lifespan: A co-relative control study. PLOS ONE, 14(10), e0224127. 10.1371/journal.pone.0224127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derringer J (2018). A simple correction for non-independent tests. PsyArXiv. 10.31234/osf.io/f2tyw [DOI] [Google Scholar]
- Gilman SE, Abrams DB, & Buka SL (2003). Socioeconomic status over the life course and stages of cigarette use: Initiation, regular use, and cessation. Journal of Epidemiology & Community Health, 57(10), 802–808. 10.1136/jech.57.10.802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman Stephen E., Martin LT, Abrams DB, Kawachi I, Kubzansky L, Loucks EB, Rende R, Rudd R, & Buka SL (2008). Educational attainment and cigarette smoking: A causal association? International Journal of Epidemiology, 37(3), 615–624. 10.1093/ije/dym250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman Stephen E., Rende R, Boergers J, Abrams DB, Buka SL, Clark MA, Colby SM, Hitsman B, Kazura AN, Lipsitt LP, Lloyd-Richardson EE, Rogers ML, Stanton CA, Stroud LR, & Niaura RS (2009). Parental Smoking and Adolescent Smoking Initiation: An Intergenerational Perspective on Tobacco Control. Pediatrics, 123(2), e274–e281. 10.1542/peds.2008-2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG, Carlson SR, Taylor J, Elkins IJ, & Mcgue M (1999). Behavioral disinhibition and the development of substance-use disorders: Findings from the Minnesota Twin Family Study. Development and Psychopathology, 11(04), 869–900. https://doi.org/null [DOI] [PubMed] [Google Scholar]
- Jaffee SR, & Price TS (2007). Gene–environment correlations: A review of the evidence and implications for prevention of mental illness. Molecular Psychiatry, 12(5), 432–442. 10.1038/sj.mp.4001950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffee Sara R, & Price TS (2008). Genotype–environment correlations: Implications for determining the relationship between environmental exposures and psychiatric illness. Psychiatry, 7(12), 496–499. 10.1016/j.mppsy.2008.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffee Sara R., & Price TS (2012). The implications of genotype–environment correlation for establishing causal processes in psychopathology. Development and Psychopathology, 24(4), 1253–1264. 10.1017/S0954579412000685 [DOI] [PubMed] [Google Scholar]
- Archer Kellie J., Hedeker Donald, Nordgren Rachel and Gibbons Robert D. (2018). mixor: Mixed-Effects Ordinal Regression Analysis. R package version 1.0.4. https://CRAN.R-project.org/package=mixor
- Kendler KS, & Baker JH (2007). Genetic influences on measures of the environment: A systematic review. Psychological Medicine, 37(5), 615–626. 10.1017/S0033291706009524 [DOI] [PubMed] [Google Scholar]
- Kendler KS, Schmitt E, Aggen SH, & Prescott CA (2008). Genetic and Environmental Influences on Alcohol, Caffeine, Cannabis, and Nicotine Use From Early Adolescence to Middle Adulthood. Archives of General Psychiatry, 65(6), 674–682. 10.1001/archpsyc.65.6.674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes MA, Malone SM, Elkins IJ, Legrand LN, McGue M, & Iacono WG (2009). The Enrichment Study of the Minnesota Twin Family Study: Increasing the Yield of Twin Families at High Risk for Externalizing Psychopathology. Twin Research and Human Genetics : The Official Journal of the International Society for Twin Studies, 12(5), 489–501. 10.1375/twin.12.5.489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes M, Legrand LN, Iacono WG, & McGue M (2008). Parental Smoking and Adolescent Problem Behavior: An Adoption Study of General and Specific Effects. American Journal of Psychiatry, 165(10), 1338–1344. 10.1176/appi.ajp.2008.08010125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Frigge ML, Vilhjalmsson BJ, Young AI, Thorgeirsson TE, Benonisdottir S, Oddsson A, Halldorsson BV, Masson G, Gudbjartsson DF, Helgason A, Bjornsdottir G, Thorsteinsdottir U, & Stefansson K (2018). The nature of nurture: Effects of parental genotypes. Science, 359(6374), 424–428. 10.1126/science.aan6877 [DOI] [PubMed] [Google Scholar]
- Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, Datta G, Davila-Velderrain J, McGuire D, Tian C, Zhan X, Choquet H, Docherty AR, Faul JD, Foerster JR, Fritsche LG, Gabrielsen ME, Gordon SD, Haessler J, … Vrieze S (2019). Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nature Genetics, 51, 237–244. 10.1038/s41588-018-0307-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, Daly MJ, Bustamante CD, & Kenny EE (2017). Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. American Journal of Human Genetics, 100(4), 635–649. 10.1016/j.ajhg.2017.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffery JM, Papandonatos GD, Lyons MJ, Koenen KC, Tsuang MT, & Niaura R (2008). Educational attainment, smoking initiation and lifetime nicotine dependence among male Vietnam-era Twins. Psychological Medicine, 38(9), 1287–1297. 10.1017/S0033291707001882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGue M, Malone S, Keyes M, & Iacono WG (2014). Parent–Offspring Similarity for Drinking: A Longitudinal Adoption Study. Behavior Genetics, 44(6), 620–628. 10.1007/s10519-014-9672-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MB, Basu S, Cunningham J, Eskin E, Malone SM, Oetting WS, Schork N, Sul JH, Iacono WG, & McGue M (2012). The Minnesota Center for Twin and Family Research Genome-Wide Association Study. Twin Research and Human Genetics, 15(6), 767–774. 10.1017/thg.2012.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Johnson PC, & Schielzeth H (2017). The coefficient of determination R 2 and intra-class correlation coefficient from generalized linear mixed-effects models revisited and expanded. Journal of the Royal Society Interface, 14(134), 20170213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholt DR (2004). A Simple Correction for Multiple Testing for Single-Nucleotide Polymorphisms in Linkage Disequilibrium with Each Other. American Journal of Human Genetics, 74(4), 765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien TC, Mustanski BS, Skol A, Cook EH, & Wakschlag LS (2013). Do dopamine gene variants and prenatal smoking interactively predict youth externalizing behavior? Neurotoxicology and Teratology, 40, 67–73. 10.1016/j.ntt.2013.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomin R (2014). Genotype-Environment Correlation in the Era of DNA. Behavior Genetics, 44(6), 629–638. 10.1007/s10519-014-9673-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polderman TJC, Benyamin B, de Leeuw CA, Sullivan PF, van Bochoven A, Visscher PM, & Posthuma D (2015). Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nature Genetics, 47(7), 702–709. 10.1038/ng.3285 [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, & Reich D (2006). Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics, 38(8), 904–909. 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- Robins L, Babor T, & Cottler L (1987). Composite International Diagnostic Interview: Expanded Substance Abuse Module. Washington University. [Google Scholar]
- Robins LN, Wing J, Wittchen H, & et al. (1988). The composite international diagnostic interview: An epidemiologic instrument suitable for use in conjunction with different diagnostic systems and in different cultures. Archives of General Psychiatry, 45(12), 1069–1077. 10.1001/archpsyc.1988.01800360017003 [DOI] [PubMed] [Google Scholar]
- Rossow I, Keating P, Felix L, & McCambridge J (2016). Does parental drinking influence children’s drinking? A systematic review of prospective cohort studies. Addiction, 111(2), 204–217. 10.1111/add.13097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samek DR, Keyes MA, Hicks BM, Bailey J, McGue M, & Iacono WG (2014). General and Specific Predictors of Nicotine and Alcohol Dependence in Early Adulthood: Genetic and Environmental Influences. Journal of Studies on Alcohol and Drugs, 75(4), 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarr S, & McCartney K (1983). How people make their own environments: A theory of genotype greater than environment effects. Child Development, 54(2), 424–435. [DOI] [PubMed] [Google Scholar]
- Scherrer JF, Xian H, Pan H, Pergadia ML, Madden PAF, Grant JD, Sartor CE, Haber JR, Jacob T, & Bucholz KK (2012). Parent, sibling and peer influences on smoking initiation, regular smoking and nicotine dependence. Results from a genetically informative design. Addictive Behaviors, 37(3), 240–247. 10.1016/j.addbeh.2011.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slutske WS, D’Onofrio BM, Turkheimer E, Emery RE, Harden KP, Heath AC, & Martin NG (2008). Searching for an environmental effect of parental alcoholism on offspring alcohol use disorder: A genetically informed study of children of alcoholics. Journal of Abnormal Psychology, 117(3), 534–551. 10.1037/a0012907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilhjálmsson BJ, Yang J, Finucane HK, Gusev A, Lindström S, Ripke S, Genovese G, Loh P-R, Bhatia G, Do R, Hayeck T, Won H-H, Ripke S, Neale BM, Corvin A, Walters JTR, Farh K-H, Holmans PA, Lee P, … Price AL (2015). Modeling Linkage Disequilibrium Increases Accuracy of Polygenic Risk Scores. The American Journal of Human Genetics, 97(4), 576–592. 10.1016/j.ajhg.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellman RJ, Dugas EN, Dutczak H, O’Loughlin EK, Datta GD, Lauzon B, & O’Loughlin J (2016). Predictors of the Onset of Cigarette Smoking: A Systematic Review of Longitudinal Population-Based Studies in Youth. American Journal of Preventive Medicine, 51(5), 767–778. 10.1016/j.amepre.2016.04.003 [DOI] [PubMed] [Google Scholar]
- Willoughby EA, McGue M, Iacono WG, Rustichini A, & Lee JJ (2019). The role of parental genotype in predicting offspring years of education: Evidence for genetic nurture. Molecular Psychiatry, 1–9. 10.1038/s41380-019-0494-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.