Abstract
Short-chain fatty acids (SCFAs) are mainly produced by intestinal microbiota and play an important role in many host biological processes such as immune system development, glucose and energy homeostasis, and regulation of immune response and inflammation. In addition, they participate in the regulation of anorectic hormones, which have a role in appetite control, tumor suppression, and regulating central and peripheral nervous systems. As such, there is great interest in monitoring levels of SCFAs in various biological samples. Due to the highly hydrophilic and volatile characteristics of SCFAs, optimizing extraction and sample preparation procedures is often a central component to further improve SCFA quantification. Here, we describe a rapid and highly sensitive analytical method for measuring SCFAs in human serum and feces. Briefly, SCFAs are protected by adding sodium hydroxide, followed by a one-step extraction (pH > 7). Then, SCFAs are quantified with gas chromatography coupled to mass spectrometry (GC-MS) after derivatization by N-tert-Butyldimethylsilyl-N-Methyltrifluoroacetamide (MTBSTFA). This method demonstrates excellent sensitivity, linearity, and derivatization efficiency for simultaneous determination of 14 different SCFAs. Further, this validated method can be successfully applied to quantify SCFAs in micro-scale biological samples. In summary, we describe efficient and advanced sample preparation and detection procedures, which are critically needed for monitoring SCFA concentrations in human biological samples.
Basic Protocol 1:
SCFA Extraction and Detection from Fecal and Serum Samples with Gas Chromatography-Mass Spectrometry
Keywords: gas chromatography, mass spectrometry, metabolomics, microbiome, short-chain fatty acids
INTRODUCTION
Short-chain fatty acids (SCFAs) are fatty acids with less than six carbon atoms encompassing both saturated aliphatic SCFAs (including acetic acid, propionic acid, butyric acid, valeric acid, among others) and branched chain SCFAs (including isobutyric acid, 2-methylbutyric acid, isovaleric acid, and others). The former are mainly produced from dietary fibers metabolized by anaerobic intestinal microbiota (Ríos-Covián et al., 2016; Cummings, 1981), while the latter are predominantly derived from the catabolism of branched-chain amino acids (Macfarlane and Macfarlane, 2003).
SCFAs provide a valuable opportunity to study the human gut microbiome, which consists of trillions of microbes (Mallick et al., 2019), is involved in numerous physiological processes (Flint et al., 2012a), and is vital to the host’s health (Qin et al., 2010). For instance, development of a competent immune system is strongly influenced by the production of gut-derived metabolites such as SCFAs and lipopolysaccharides (Daliri et al., 2017), while a number of other critical functions have also been ascribed to the intricate cross-talk between host and microbiota, such as synthesis of SCFAs and vitamins, adipose storage, and regulation of angiogenesis, as well as certain aspects of behavior (Cryan and O’Mahony, 2011; Yamashiro, 2018). Interestingly, results have increasingly shown that metabolome-derived ligands play major roles in disease propagation (Dvořák et al., 2020; Scoville et al., 2019) and, conversely, may also modulate toxic effects of environmental contaminants (Lim et al., 2020; Dempsey et al., 2019; Li et al., 2018). Given the enormous metabolic potential of the intestinal microbiota (Holmes et al., 2012), there is great interest in characterizing metabolic interactions between the host and microbes (Nicholson et al., 2012). Metabolomics, the comprehensive study of small molecular-weight metabolites and their dynamic changes in biological systems (Patti et al., 2012; Chong et al., 2018; Wishart, 2008; Gu et al., 2015; Shi et al., 2019; Gowda et al., 2008; Shi et al., 2020; Jasbi et al., 2019c, 2019b) has been increasingly applied to this end (Holmes et al., 2012; Nicholson et al., 2012). Considering the ubiquitous presence of SCFA-activated G-protein-coupled receptors (GPCRs) present on multiple cell types, including intestinal epithelial cells, macrophages, dendritic cells, and mast cells (Valdes et al., 2018; Cox and Blaser, 2013; Kim et al., 2013), research efforts have centered on characterizing SCFAs such as acetate, propionate, and butyrate produced by gut microbes under anaerobic conditions in the large intestine by fermentation of dietary fibers (Flint et al., 2012b). SCFAs have been shown to be associated with health outcomes such as glucose and energy homeostasis, and regulation of immune response and inflammation, in addition to regulation of anorectic hormones, which have a role in appetite control, tumor suppression, and regulating central and peripheral nervous systems (Cox and Blaser, 2013; Kim et al., 2013; Bienenstock et al., 2015; Hamer et al., 2008).
Generally, SCFAs are produced and absorbed in the large intestine (Sakata, 1987). In addition to providing energy for enterocytes, they play an important role in physiological activity and biological homeostasis. For example, SCFAs can activate 5’ adenosine monophosphate-activated protein kinase to regulate metabolic homeostasis and inhibit lipolysis in adipocytes through GPCRs (Samuel et al., 2008). They are also involved in the interaction between host microbiota and brain function, given their inherent ability to cross the blood-brain barrier (Erny et al., 2017). Recently, numerous studies have indicated SCFAs to be associated with colorectal cancers, diarrheal disorders, inflammatory bowel diseases, autism spectrum disorder (ASD), Rett syndrome, and many metabolic diseases such as obesity, hypertension, and diabetes (Pingitore et al., 2017; Natarajan et al., 2016; Roelofsen et al., 2010; Strati et al., 2016; Zhao et al., 2017; Sun et al., 2017; Roy et al., 2006). Moreover, some SCFAs such as propionic acid and butyric acid show specific functions in many tissues and organs. For example, propionic acid is an energy source of intestinal cells (Schwarz et al., 2017; Wolever et al., 1991). Given the demonstrated importance of SCFAs in physiological and pathological processes, quantitative and qualitative analysis of these volatile fatty acids in biological samples is of increasing interest to scientific researchers of various disciplines (Pluznick, 2016; Miller, 2012; Wong et al., 2006).
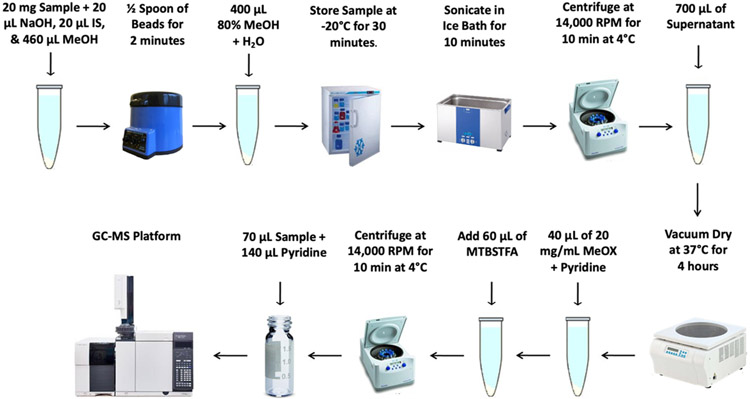
The protocol described here (Basic Protocol 1) expounds a highly sensitive, rapid, accurate, and applicable GC-mass spectrometry (GC-MS) method for quantification of multiple SCFAs in biological samples (such as serum and fecal) in micro-scale (Jasbi et al., 2019a). Due to their production in the large intestine and absorption into the bloodstream, fecal and serum samples are often used as typical specimens for the measurement of SCFAs, and are used here to illustrate the protocol. In this approach, sodium hydroxide is added to the samples at the beginning to reduce the volatility of SCFAs under basic conditions, and then, SCFAs are extracted and derivatized by N-tert-Butyldimethylsilyl-N-Methyltrifluoroacetamide (MTBSTFA) (See Figure 1 for a summary of the fecal sample preparation method). The advantage of using MTBSTFA is that the derivatization reaction can smoothly proceed under 60 °C without any assistance such as ultrasonication (Lotti et al., 2017), and the derivatives can be directly injected into GC-MS without further extraction (Lotti et al., 2017; He et al., 2018; Hoving et al., 2018; Zhao et al., 2006).
Figure 1.
Overview of the fecal sample preparation method described in Basic Protocol 1 to detect SCFAs using GC-MS.
STRATEGIC PLANNING
Fecal and serum samples for use in this protocol can be obtained using previously described human sample extraction methods (Lotti et al., 2017; Zheng et al., 2013). After the subjects fast overnight (12 h), serum and fecal samples should be collected the subsequent morning, and prepared at 4 °C or stored at −20 °C (short-term) or −80 °C (long-term). During preparation, the samples should remain at 4 °C until dried in the vacuum concentrator. Due to drying time lengths, the volume of samples prepared at a given time should not exceed the vacuum concentrator capacity. If the entire protocol cannot be completed on a single run, a suggested stopping point is right before sample derivatization.
All solutions should be prepared daily before sample preparation. Any additional materials should be arranged prior to the protocol or during hands-off time.
The retention time and quantification mass for each SCFA are determined using its chemical standard (Table 1). Users should prepare calibration curves with the 14 SCFAs to determine their concentrations. They can do this by preparing a series of standard solutions across a range of concentrations (after normalization to the Internal Standard (hexanoic acid-6,6,6-d3 (Sigma-Aldrich, cat. no. 498727) and measuring signal response; operators can plot the relationship to interpolate concentrations of the analytes. Standards are commercially available (Acetic acid (Thermo Fisher Scientific, cat. no. MFCD00036152), Propionic acid (Sigma-Aldrich, cat. no. 402907), Isobutyric acid (Sigma-Aldrich, cat. no. I1754), Butyric acid (Sigma-Aldrich, cat. no. B103500), 2-Methylbutyric acid (Sigma-Aldrich, cat. no. W269514), Isovaleric acid (Sigma-Aldrich, cat. no. 129542), Valeric acid (Sigma-Aldrich, cat. no. 240370), 2-Methylpentanoic acid (Sigma-Aldrich, cat. no. W275409), 3-Methylpentanoic acid (Sigma-Aldrich, cat. no. W343706), Isocaproic acid (Sigma-Aldrich, cat. no. 277827), Caproic acid (Sigma-Aldrich, cat. no. 153745), 2-Methylhexanoic acid (Sigma-Aldrich, cat. no. 338273), 4-Methylhexanoic acid (Sigma-Aldrich, cat. no. 559016), Heptanoic acid (Sigma-Aldrich, cat. no. 75190)).
Table 1.
Retention time (RT) and quantification mass (m/z) of 14 SCFA chemical standards.
| Compound | Retention time (min) | Quantitation Ion (m/z) |
|---|---|---|
| Acetic acid | 5.86 | 117 |
| Propionic acid | 7.32 | 131 |
| Isobutyric acid | 7.53 | 145 |
| Butyric acid | 7.92 | 145 |
| Isovaleric acid | 8.03 | 159 |
| 2-Methylbutyric acid | 8.45 | 159 |
| Valeric acid | 9.00 | 159 |
| 2-Methylpentaboic acid | 9.35 | 173 |
| 3-Methylpentanoic acid | 9.67 | 173 |
| Isocaproic acid | 9.72 | 173 |
| Caproic acid | 10.23 | 173 |
| 2-Methylhexanoic acid | 10.55 | 187 |
| 4-Methylhexanoic acid | 11.14 | 187 |
| Heptanoic acid | 11.55 | 187 |
BASIC PROTOCOL 1: SCFA EXTRACTION AND DETECTION FROM FECAL AND SERUM SAMPLES WITH GAS CHROMATOGRAPHY-MASS SPECTROMETRY
The protocol described here, employed in our previous work (Jasbi et al., 2019a), is a rapid and highly sensitive method for quantifying the presence of 14 SCFAs, namely acetic acid, propionic acid, isobutyric acid, butyric acid, isovaleric acid, 2-methylbutyric acid, valeric acid, 2-methylpentanoic acid, 3-methylpentanoic acid, isocaproic acid, caproic acid, 2-methylhexanoic acid, 4-methylhexanoic acid, and heptanoic acid, in either serum or fecal samples through GC-MS. The sample preparation method described below has shown to reliably detect branched chain and saturated aliphatic SCFAs with good intra- and inter-day precision.
We describe sample preparation steps for either fecal (Steps 1a to 16a) or serum (Steps 1b to 12b) samples, before outlining the steps required for GC-MS analysis and data extraction. During preparation, SCFAs are preserved through the use of sodium hydroxide (NaOH), extracted with the addition of methanol (MeOH), and derivatized with MTBSTFA. Optimal conditions include using extracting solutions with a pH = 9, and a derivatization time and temperature of 30 minutes and 60 °C, respectively.
All reagents should be of analytical grade, handled with care, and capped when not in use. All aqueous solutions should be prepared with ultrapure water (filtered deionized water) and stored at room temperature unless otherwise noted. Follow all standard institutional waste disposal protocols.
MATERIALS
Human fecal sample or Human serum sample (see Strategic Planning)
Methanol (MeOH; Sigma-Aldrich, cat. no. 34860)
Pyridine (Sigma-Aldrich, cat. no. 360570)
0.5 M Sodium hydroxide (NaOH; Thermo Fisher Scientific cat. no. 60-014-41) in water
0.1 M Sodium hydroxide (NaOH; Thermo Fisher Scientific cat. no. 60-014-41) in water
Internal Standard solution (200 μM hexanoic acid-6,6,6-d3 in water)
Derivatization solution (see Reagents and Solution)
N-tert-Butyldimethylsilyl-N-methyltrifluoroacetamide (MTBSTFA; Sigma-Aldrich, cat. no. 394882)
1.5 mL Eppendorf tubes
Pipet and pipet tips
2 mL Glass vials
100 mL Graduated cylinder
Vortex mixer (Fisher Scientific, cat. no. 14955151)
Centrifuge with speed, time, and temperature control (Thermo Scientific, cat. no. 75004270)
Vacuum concentrator (CentriVap Concentrator, cat. no. 7810010)
0.5 mm diameter Stainless Steel beads (Next Advance, Troy, NY, cat. no. SSB05)
0.10 g measuring spoon (Next Advance, Troy, NY, cat. no. MSP01)
Tissue homogenizer (Next Advance, Troy, NY, cat. no. BBY5M)
Digital dry bath (Thermo Scientific, cat. no. 88870002)
Scientific balance with mg readability (VWR-64B)
Ultrasonic liquid processor (VWR 97043-996)
Agilent 7820A GC-5977B MSD system (Agilent Technologies, Santa Clara, CA)
Agilent HP-5 ms capillary column (30 m x 250 μm x 0.25 μm)
Agilent MassHunter Workstation Software Quantitative Analysis (B.09.00)
Protocol steps
Fecal Sample Preparation (Figure 1)
-
1a.
Weigh out 20 mg ± 1.5 mg of each biological sample in a 1.5 mL Eppendorf tube.
-
2a.
Add 20 μL of a 0.5 M NaOH solution, 20 μL of Internal Standard solution, and 460 μL of MeOH to each sample.
-
3a.
Add 0.6 g of stainless-steel beads to the tube and homogenize for 2 min in the tissue homogenizer.
-
4a.
Prepare an 80% v/v solution of MeOH in water and add 400 μL of it to the sample.
-
5a.
Vortex mixture for 30 s and store at −20 °C for 30 min.
-
6a.
Sonicate in ultrasonic liquid processor in ice bath for 10 min at 4 °C.
Prepare ice bath for use prior to sonication.
-
7a.
Centrifugate at 14,000 RPM for 10 min at 4 °C.
-
8a.
Transfer 700 μL of supernatant into a new 1.5 mL Eppendorf tube.
The remaining solution may be stored long-term at −80 °C for future analysis of total protein concentration.
-
9a.
Dry the supernatant in the vacuum concentrator at 37 °C for 240 min.
Drying time is dependent upon total sample size and vacuum concentrator capacity. Fecal sample sizes greater than the vacuum concentrator capacity will result in additional drying time. If it cannot be immediately dried, samples should be stored at −20 °C short-term or at −80 °C long-term. This is a preferred stopping point if the protocol cannot be completed in a single run.
-
10a.
Derivatize samples by adding 40 μL of derivatization solution and placing the samples in the digital dry bath for 90 min at 60 °C.
-
11a.
Add 60 μL of MTBSTFA and incubate the mixture at 60 °C for 30 min.
-
12a.
Vortex mixture for 30 s
-
13a.
Centrifuge at 14,000 RPM for 10 min at 4 °C.
-
14a.
Transfer 70 μL of the supernatant into a new 2 mL glass vial and dilute with 140 μL of pyridine.
-
15a.
Proceed to GC-MS Analysis and Data Extraction.
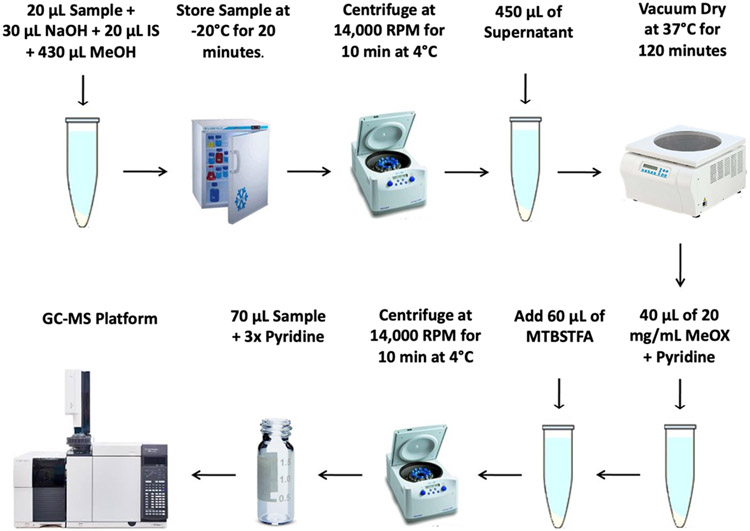
Serum Sample Preparation (Figure 2)
Figure 2.
Overview of the serum sample preparation method described in Basic Protocol 1 to detect SCFAs using GC-MS.
-
1b.
Transfer 20 μL of each serum sample to a 1.5 mL Eppendorf tube
-
2b.
Add 30 μL of 0.1 M aqueous NaOH, 20 μL of Internal Standard solution, and 430 μL of MeOH.
-
3b.
Vortex mixture for 10 s and store at −20 °C for 20 min.
Check pH to confirm the mixture is at ~9.
-
4b.
Centrifuge at 14,000 RPM for 10 min at 4 °C.
-
5b.
Transfer 450 μL of the supernatant into a new 1.5 mL Eppendorf tube.
-
6b.
Dry the supernatant under vacuum at 37 °C for 120 min.
Drying time is dependent upon total sample size and vacuum concentrator capacity. Serum sample sizes greater than the vacuum concentrator capacity will result in additional drying time. If it cannot be immediately dried, samples should be stored at −20 °C short-term or at −80 °C long-term. This is a preferred stopping point if the protocol cannot be completed in a single run.
-
7b.
Derivatize samples with 40 μL of derivatization solution in the digital dry bath for 90 min at 60 °C.
-
8b.
Add 60 μL of MTBSTFA and incubate the mixture at 60 °C for 30 min.
-
9b.
Vortex mixture for 30 s
-
10b.
Centrifuge at 14,000 RPM for 10 min at 4 °C.
-
11b.
Transfer 70 μL of supernatant into a new 2 mL glass vial and dilute with 140 μL of pyridine.
-
12b.
Proceed to GC-MS Analysis and Data Extraction
GC-MS Analysis and Data Extraction
-
16.
Set injection volume to 1 μL and solvent delay time to 5 min for each sample.
-
17.
Hold initial oven temperature at 60 °C for 1 min.
-
18.
Increase temperature to 325 °C at a rate of 10 °C/min and hold at 325 °C for 10 min.
-
19.
Hold helium at a constant flow rate of 20 mL/min through the column.
-
20.
Set the temperatures of the front inlet, transfer line, and electron impact (EI) ion source to 250 °C, 290 °C, and 230 °C, respectively.
-
21.
Set electron energy to −70 eV and collect the mass spectral data in full scan mode (m/z 50-600).
-
22.
Transfer data to the Agilent Quantitative Analysis software.
-
23.
Create a new batch with all of the appropriate samples selected and the desired method applied.
-
24.
After designating the method, select to integrate the data.
The appropriate peaks should be integrated according to their chemical standard retention times (see Table 1).
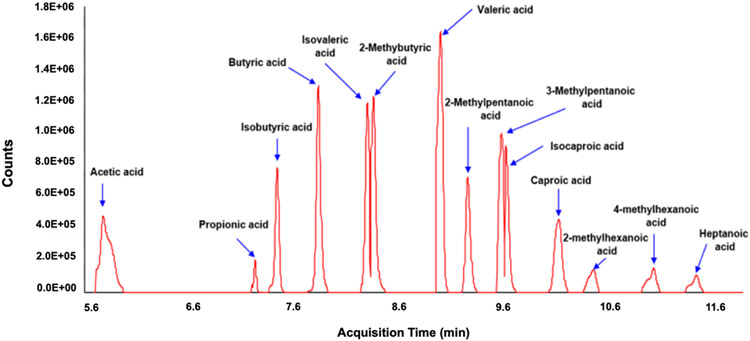
Figure 3 shows the total ion chromatogram of 14 SCFA standards. Agilent MassHunter Workstation Software Quantitative Analysis (B.09.00) can be used to process the GC-MS data for compound identification, peak selection, and quantification. The concentrations of SCFAs in biological samples should be calculated using the calibration curves constructed from the corresponding signals of SCFAs standards (see Strategic Planning).
Figure 3.
Total ion chromatogram of 14 SCFA standards.
REAGENTS AND SOLUTIONS
All aqueous solutions should be prepared using ultrapure water. The reagents should be at least of analytical grade. Abide by all necessary waste disposal protocols as they apply. Inspect all pipettes for readiness prior to use.
Derivatization Solution
MeOX (Sigma-Aldrich, cat. no. 226904)
Pyridine (Sigma-Aldrich, cat. no. 360570)
Add 20 mg of MeOX per 1 mL of pyridine solution. Prepare daily (i.e., prepare fresh each time) and at room temperature. The solution may require minor shaking to dissolve MeOX, and should be prepared in a fume hood. Handle reagents with care and cap tightly when not in use.
COMMENTARY
Background Information
Some techniques such as high-performance liquid chromatography, nuclear magnetic resonance, and capillary electrophoresis have been applied in quantitative and qualitative analyses of SCFAs, although gas chromatography (GC) is still the most common choice for analysis, due to the high volatility of short chain fatty acids (Nusbaum et al., 2018; Primec et al., 2017; Huda-Faujan et al., 2010; Gu et al., 2013). In 1952, A.T. James et al. first used GC to separate volatile fatty acids (James and Martin, 1952), leading to a variety of GC-based methods that have been explored for the analysis of SCFAs in biological samples (Hoving et al., 2018; He et al., 2018; Lotti et al., 2017; Zheng et al., 2013; Zhao et al., 2006; Moreau et al., 2003). Most of the extraction procedures reported in the literature describe using acidic phase separation (Lotti et al., 2017; He et al., 2018; Zhao et al., 2006) and a two-step extraction (Zheng et al., 2013; Moreau et al., 2003) with highly volatile organic solvents, including hexane or diethyl ether. Due to their highly hydrophilic and volatile characteristics, complex sample preparation procedures under acidic conditions could affect method precision during determination of SCFAs, especially for a large batch of samples. Therefore, optimization of extraction and GC derivatization procedures is a key factor to further improve quantification of SCFAs in biological samples.
There are significant advantages conferred by the SCFA detection method described here compared to previous methods, including decreased analytical burden and sample analysis time. We extract SCFAs under basic conditions, which reduces volatility of SCFAs. This extraction approach allows us to dry and concentrate the samples, further improving the SCFA detection sensitivity. The advantage of using MTBSTFA compared to many other derivatization reagents is that the derivatization reaction can smoothly proceed under 60 °C without any assistance such as ultrasonication (Lotti et al., 2017), and the derivatives can be directly injected into GC-MS without further extraction (Lotti et al., 2017; He et al., 2018; Hoving et al., 2018; Zhao et al., 2006). This is the first description of a protocol in which SCFAs are extracted under an alkaloid environment (pH > 7) and derivatized by MTBSTFA to enhance GC-MS detection of SCFAs. The 14 SCFAs monitored in the current protocol are highly preserved in human serum and fecal samples and are of high biological relevance. Notably, these 14 SCFAs just serve as a proof-of-concept; in theory, the method may be expanded to detect other SCFAs of interest.
Critical Parameters
Samples should be prepared upon collection at 4 °C or stored promptly at −20 °C (short-term) or at −80 °C (long-term). In order to preserve the integrity of the samples, freeze-thaw cycles should be minimized (Wang et al., 2019; Stevens et al., 2019; González-Domínguez et al., 2020). In addition, the recovery of SCFAs is decreased with each day the samples remain at 4 °C or higher. It is thus prudent to only thaw the samples being prepared and analyze them with GC-MS in a timely manner. With the objective of reducing the time in which samples are thawed, all solutions should be prepared prior to starting the procedure. Ultrapure water should be used for all solutions. If the samples cannot be immediately dried after transferring the supernatant, samples should be stored at −20 °C short-term or at −80 °C long-term. Typically, it is best to not exceed the capacity of the vacuum concentrator with the volume of samples prepared at a given time.
Robust calibration curves (R2 ≥ 0.95) are essential for calculation of results. Users should prepare a new calibration curve for every batch run.
Troubleshooting
Table 2 describes a few common problems users may face when following the protocol, along with possible causes and recommended solutions.
Table 2.
Troubleshooting guide for SCFA detection with GC-MS.
| Problem | Possible Cause(s) | Solution |
|---|---|---|
| Vacuum pressure is consistently higher than that recommended for flow rate | Air leaks or other vacuum problems | Consult operation manual |
| Spurious chromatogram peaks and poor repeatability | Backflash in response to flooding the injection line | Inject less sample volume |
| Poor recovery of analyte from sample | Ensure that freeze/thaw cycles have been kept to ≤ 2 and that samples are stored at −80 °C for long-term storage | |
| Shifting retention, baseline disturbances | Column bleed, column contamination, column aging | Replace with new column and condition before use |
Understanding Results
This protocol is able to provide good quantitation of 14 SCFAs from biological samples. Figure 3 shows the total ion chromatogram of 14 SCFA standards, and Table 1 shows the retention time and quantification mass (m/z) values of 14 SCFA chemical standards. After the data is extracted from the Quantitative Analysis software, it may be exported for quantitation. Figure 4 shows an excellent calibration curve, using valeric acid as an example. A calibration curve demonstrates concentration (μL/mL) plotted against peak intensity for the SCFA. It may be generated by analyzing different concentrations of a SCFA standard through GC-MS. The previous concentrations are used to generate a curve with their measured intensities. Calibration curves may be used to identify an unknown SCFA concentration (from the test samples) through its peak intensity.
Figure 4.
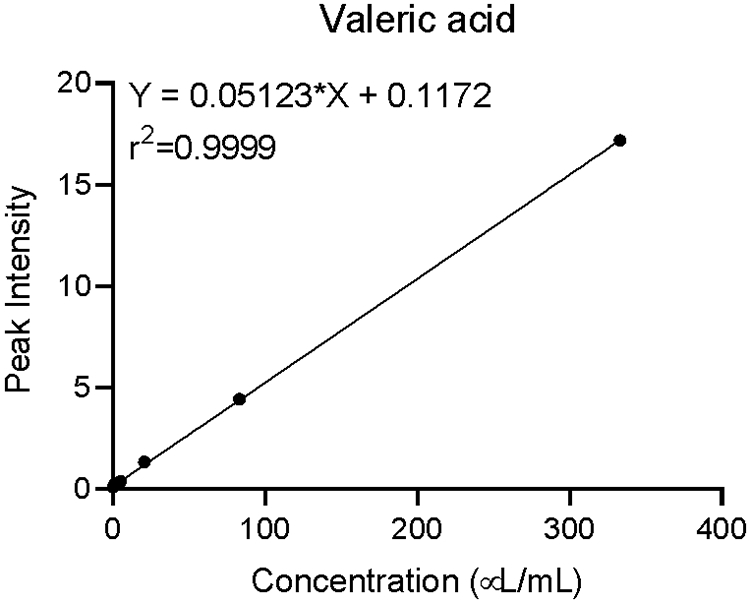
Calibration curve for valeric acid plotted as concentration vs peak intensity. A calibration curve is created by first preparing a set of standard solutions with known concentrations of the analyte; the instrument response is measured for each and plotted vs. concentration of the standard solution.
Typical concentrations of detected short-chain fatty acids may vary significantly between fecal and serum samples and will most likely be indicative of experimental or conditional factors. Furthermore, normal concentrations of individual SCFAs will vary. For example, as can be determined using established metabolomics databases such as the Human Metabolomics Database (HMDB), normal concentrations of acetic acid in human blood (male and female) may range from 26.8 to 64.2 μM (Psychogios et al., 2011); operators should consult relevant literature when interpreting their obtained results. A reduced yield of SCFAs may be attributed to extended periods of sample thawing and/or preparation at or above 4 °C.
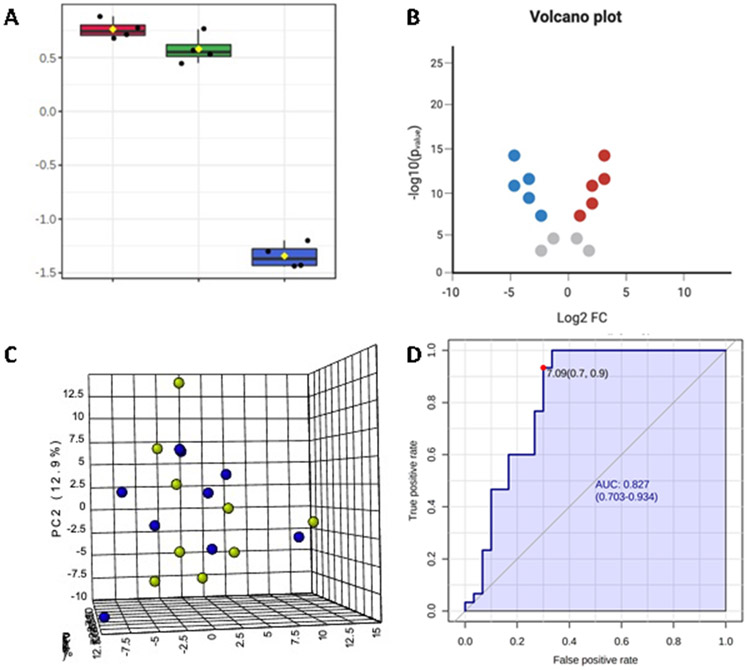
When multiple samples are measured, the data can be grouped and entered into a statistical software. For example, the analyses observed in our unpublished studies in Figure 5, such as box and volcano plots, principal component analysis (PCA), and receiver operating characteristic (ROC) curve can be performed to reveal integral information regarding statistical significance, effect size, variance explained, as well as sensitivity and specificity. In general, it is best practice to run duplicates (i.e., two technical replicates per sample) for data validation.
Figure 5.
Examples of typical data analyses that can be used for the 14 SCFAs from human serum and fecal samples: A) Box plot of one-way ANOVA showing log-transformed concentrations in a conventional three-group study design, B) Volcano plot of significant SCFAs and fold-change magnitude between two experimental groups, C) Principal components analysis (PCA) of two groups (green/blue) between three PC components, and D) Receiver operating characteristic (ROC) curve of multivariate model for discrimination between two experimental groups.
Time Considerations
Quickly performing the extraction and derivatization protocol is paramount to avoid SCFA loss during preparation. Serum sample preparation can be completed in six hours, with two hours of hands-on time for 60 samples. The protocol for fecal samples is longer and may involve up to ten hours, with a hands-on time of three hours for 60 samples. The drying time is dependent upon total sample size and type of sample prepared. With practice and a vacuum concentrator system with the appropriate capacity, 60 fecal samples can be prepared for GC-MS analysis in one day. Due to drying length, it is the preferred stopping point.
Acknowledgements
This work was supported by the NIH (1R01ES030197-01, 1P01HL146369-01A1).
Footnotes
Conflict of Interest Statement
The authors declare no conflict of interest.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
LITERATURE CITED
- Bienenstock J, Kunze W, and Forsythe P 2015. Microbiota and the gut-brain axis. Nutrition Reviews 73:28–31. [DOI] [PubMed] [Google Scholar]
- Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, Wishart DS, and Xia J 2018. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Research 46:W486–W494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LM, and Blaser MJ 2013. Pathways in microbe-induced obesity. Cell Metabolism 17:883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan JF, and O’Mahony SM 2011. The microbiome-gut-brain axis: From bowel to behavior. Neurogastroenterology and Motility 23:187–192. [DOI] [PubMed] [Google Scholar]
- Cummings JH 1981. Short chain fatty acids in the human colon. Gut 22:763–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daliri EBM, Wei S, Oh DH, and Lee BH 2017. The human microbiome and metabolomics: Current concepts and applications. Critical Reviews in Food Science and Nutrition 57:3565–3576. [DOI] [PubMed] [Google Scholar]
- Dempsey JL, Wang D, Siginir G, Fei Q, Raftery D, Gu H, and Yue Cui J 2019. Pharmacological Activation of PXR and CAR Downregulates Distinct Bile Acid-Metabolizing Intestinal Bacteria and Alters Bile Acid Homeostasis. Toxicological Sciences 168:40–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvořák Z, Kopp F, Costello CM, Kemp JS, Li H, Vrzalová A, Štěpánková M, Bartoňková I, Jiskrová E, Poulíková K, et al. 2020. Targeting the pregnane X receptor using microbial metabolite mimicry. EMBO Molecular Medicine 12:e11621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erny D, Hrabě de Angelis AL, and Prinz M 2017. Communicating systems in the body: how microbiota and microglia cooperate. Immunology 150:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint HJ, Scott KP, Duncan SH, Louis P, and Forano E 2012a. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3:289–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint HJ, Scott KP, Louis P, and Duncan SH 2012b. The role of the gut microbiota in nutrition and health. Nature Reviews Gastroenterology and Hepatology 9:577–589. [DOI] [PubMed] [Google Scholar]
- González-Domínguez R, González-Domínguez Á, Sayago A, and Fernández-Recamales Á 2020. Recommendations and best practices for standardizing the pre-analytical processing of blood and urine samples in metabolomics. Metabolites 10:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda GN, Zhang S, Gu H, Asiago V, Shanaiah N, and Raftery D 2008. Metabolomics-based methods for early disease diagnostics. Expert Review of Molecular Diagnostics 8:617–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Gowda GAN, Neto FC, Opp MR, and Raftery D 2013. RAMSY: Ratio Analysis of Mass Spectrometry to Improve Compound Identification. Analytical Chemistry 85:10771–10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Zhang P, Zhu J, and Raftery D 2015. Globally Optimized Targeted Mass Spectrometry: Reliable Metabolomics Analysis with Broad Coverage. Analytical Chemistry 87:12355–12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, and Brummer RJ 2008. Review article: The role of butyrate on colonic function. Alimentary Pharmacology and Therapeutics 27:104–119. [DOI] [PubMed] [Google Scholar]
- He L, Prodhan MAI, Yuan F, Yin X, Lorkiewicz PK, Wei X, Feng W, McClain C, and Zhang X 2018. Simultaneous quantification of straight-chain and branched-chain short chain fatty acids by gas chromatography mass spectrometry. Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences 1092:359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E, Li JV, Marchesi JR, and Nicholson JK 2012. Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metabolism 16:559–564. [DOI] [PubMed] [Google Scholar]
- Hoving LR, Heijink M, van Harmelen V, van Dijk KW, and Giera M 2018. GC-MS analysis of short-chain fatty acids in feces, cecum content, and blood samples. In Methods in Molecular Biology pp. 247–256. Humana Press Inc. [DOI] [PubMed] [Google Scholar]
- Huda-Faujan N, Abdulamir AS, Fatimah AB, Anas OM, Shuhaimi M, Yazid AM, and Loong YY 2010. The Impact of the Level of the Intestinal Short Chain Fatty Acids in Inflammatory Bowel Disease Patients Versus Healthy Subjects. The Open Biochemistry Journal 4:53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, and Martin 1952. Gas-liquid partition chromatography; the separation and micro-estimation of volatile fatty acids from formic acid to dodecanoic acid. The Biochemical Journal 50:679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasbi P, Baker O, Shi X, Gonzalez LA, Wang S, Anderson S, Xi B, Gu H, and Johnston CS 2019a. Daily red wine vinegar ingestion for eight weeks improves glucose homeostasis and affects the metabolome but does not reduce adiposity in adults. Food and Function 10:7343–7355. [DOI] [PubMed] [Google Scholar]
- Jasbi P, Mitchell NM, Shi X, Grys TE, Wei Y, Liu L, Lake DF, and Gu H 2019b. Coccidioidomycosis Detection Using Targeted Plasma and Urine Metabolic Profiling. Journal of Proteome Research 18:2791–2802. [DOI] [PubMed] [Google Scholar]
- Jasbi P, Wang D, Cheng SL, Fei Q, Cui JY, Liu L, Wei Y, Raftery D, and Gu H 2019c. Breast cancer detection using targeted plasma metabolomics. Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences 1105:26–37. [DOI] [PubMed] [Google Scholar]
- Kim MH, Kang SG, Park JH, Yanagisawa M, and Kim CH 2013. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 145:396–406. [DOI] [PubMed] [Google Scholar]
- Li CY, Dempsey JL, Wang D, Lee SW, Weigel KM, Fei Q, Bhatt DK, Prasad B, Raftery D, Gu H, et al. 2018. PBDEs altered gut microbiome and bile acid homeostasis in male C57BL/6 mice. Drug Metabolism and Disposition 46:1226–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JJ, X L, HJ L, D W, H G, and JY C 2020. Gut Microbiome Critically Impacts PCB-induced Changes in Metabolic Fingerprints and the Hepatic Transcriptome in Mice. Toxicological Sciences 177:168–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotti C, Rubert J, Fava F, Tuohy K, Mattivi F, and Vrhovsek U 2017. Development of a fast and cost-effective gas chromatography–mass spectrometry method for the quantification of short-chain and medium-chain fatty acids in human biofluids. Analytical and Bioanalytical Chemistry 409:5555–5567. [DOI] [PubMed] [Google Scholar]
- Macfarlane S, and Macfarlane GT 2003. Regulation of short-chain fatty acid production. Proceedings of the Nutrition Society 62:67–72. [DOI] [PubMed] [Google Scholar]
- Mallick H, Franzosa EA, Mclver LJ, Banerjee S, Sirota-Madi A, Kostic AD, Clish CB, Vlamakis H, Xavier RJ, and Huttenhower C 2019. Predictive metabolomic profiling of microbial communities using amplicon or metagenomic sequences. Nature Communications 10:3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S 2012. Cellular and Physiological Effects of Short-Chain Fatty Acids. Mini-Reviews in Medicinal Chemistry 4:839–845. [DOI] [PubMed] [Google Scholar]
- Moreau NM, Goupry SM, Antignac JP, Monteau FJ, Le Bizec BJ, Champ MM, Martin LJ, and Dumon HJ 2003. Simultaneous measurement of plasma concentrations and 13C-enrichment of short-chain fatty acids, lactic acid and ketone bodies by gas chromatography coupled to mass spectrometry. Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences 784:395–403. [DOI] [PubMed] [Google Scholar]
- Natarajan N, Hori D, Flavahan S, Steppan J, Flavahan NA, Berkowitz DE, and Pluznick JL 2016. Microbial short chain fatty acid metabolites lower blood pressure via endothelial G protein-coupled receptor 41. Physiological Genomics 48:826–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, and Pettersson S 2012. Host-gut microbiota metabolic interactions. Science 336:1262–1267. [DOI] [PubMed] [Google Scholar]
- Nusbaum DJ, Sun F, Ren J, Zhu Z, Ramsy N, Pervolarakis N, Kunde S, England W, Gao B, Fiehn O, et al. 2018. Gut microbial and metabolomic profiles after fecal microbiota transplantation in pediatric ulcerative colitis patients. FEMS microbiology ecology 94:fiy133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti GJ, Yanes O, and Siuzdak G 2012. Metabolomics: the apogee of the omics trilogy. Nature Reviews Molecular Cell Biology 13:263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingitore A, Chambers ES, Hill T, Maldonado IR, Liu B, Bewick G, Morrison DJ, Preston T, Wallis GA, Tedford C, et al. 2017. The diet-derived short chain fatty acid propionate improves beta-cell function in humans and stimulates insulin secretion from human islets in vitro. Diabetes, Obesity and Metabolism 19:257–265. [DOI] [PubMed] [Google Scholar]
- Pluznick JL 2016. Gut microbiota in renal physiology: focus on short-chain fatty acids and their receptors. Kidney International 90:1191–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primec M, Mičetić-Turk D, and Langerholc T 2017. Analysis of short-chain fatty acids in human feces: A scoping review. Analytical Biochemistry 526:9–21. [DOI] [PubMed] [Google Scholar]
- Psychogios N, Hau DD, Peng J, Guo AC, Mandal R, Bouatra S, Sinelnikov I, Krishnamurthy R, Eisner R, Gautam B, et al. 2011. The human serum metabolome. PLoS One 6(2):e16957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ríos-Covián D, Ruas-Madiedo P, Margolles A, Gueimonde M, De los Reyes-Gavilán CG, and Salazar N 2016. Intestinal short chain fatty acids and their link with diet and human health. Frontiers in Microbiology 7:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelofsen H, Priebe MG, and Vonk RJ 2010. The interaction of short-chain fatty acids with adipose tissue: Relevance for prevention of type 2 diabetes. Beneficial Microbes 1:433–437. [DOI] [PubMed] [Google Scholar]
- Roy CC, Kien CL, Bouthillier L, and Levy E 2006. Short-chain fatty acids: ready for prime time? Nutrition in Clinical Practice 21:351–366. [DOI] [PubMed] [Google Scholar]
- Sakata T 1987. Stimulatory effect of short-chain fatty acids on epithelial cell proliferation in the rat intestine: a possible explanation for trophic effects of fermentable fibre, gut microbes and luminal trophic factors. British Journal of Nutrition 58:95–103. [DOI] [PubMed] [Google Scholar]
- Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, Hammer RE, Williams SC, Crowley J, Yanagisawa M, et al. 2008. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proceedings of the National Academy of Sciences of the United States of America 105:16767–16772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz A, Bruhs A, and Schwarz T 2017. The Short-Chain Fatty Acid Sodium Butyrate Functions as a Regulator of the Skin Immune System. Journal of Investigative Dermatology 137:855–864. [DOI] [PubMed] [Google Scholar]
- Scoville DK, Li CY, Wang D, Dempsey JL, Raftery D, Mani S, Gu H, and Cui JY 2019. Polybrominated Diphenyl Ethers and Gut Microbiome Modulate Metabolic Syndrome-Related Aqueous Metabolites in Mice. Drug Metabolism and Disposition 47:928–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Wang S, Jasbi P, Turner C, Hrovat J, Wei Y, Liu J, and Gu H 2019. Database-Assisted Globally Optimized Targeted Mass Spectrometry (dGOT-MS): Broad and Reliable Metabolomics Analysis with Enhanced Identification. Analytical Chemistry 91:13737–13745. [DOI] [PubMed] [Google Scholar]
- Shi X, Xi B, Jasbi P, Turner C, Jin Y, and Gu H 2020. Comprehensive Isotopic Targeted Mass Spectrometry: Reliable Metabolic Flux Analysis with Broad Coverage. Analytical Chemistry 92:11728–11738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens VL, Hoover E, Wang Y, and Zanetti KA 2019. Pre-analytical factors that affect metabolite stability in human urine, plasma, and serum: A review. Metabolites 9:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strati F, Cavalieri D, Albanese D, De Felice C, Donati C, Hayek J, Jousson O, Leoncini S, Pindo M, Renzi D, et al. 2016. Altered gut microbiota in Rett syndrome. Microbiome 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Wu W, Liu Z, and Cong Y 2017. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. Journal of Gastroenterology 52:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes AM, Walter J, Segal E, and Spector TD 2018. Role of the gut microbiota in nutrition and health. BMJ 361:36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Gu H, Palma-Duran SA, Fierro A, Jasbi P, Shi X, Bresette W, and Tasevska N 2019. Influence of storage conditions and preservatives on metabolite fingerprints in urine. Metabolites 9:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS 2008. Metabolomics: applications to food science and nutrition research. Trends in Food Science & Technology 19:482–493. [Google Scholar]
- Wolever TMS, Spadafora P, and Eshuis H 1991. Interaction between colonic acetate and propionate in humans. American Journal of Clinical Nutrition 53:681–687. [DOI] [PubMed] [Google Scholar]
- Wong JMW, De Souza R, Kendall CWC, Emam A, and Jenkins DJA 2006. Colonic health: Fermentation and short chain fatty acids. Journal of Clinical Gastroenterology 40:235–243. [DOI] [PubMed] [Google Scholar]
- Yamashiro Y 2018. Gut Microbiota in Health and Disease. Annals of Nutrition and Metabolism 71:242–246. [DOI] [PubMed] [Google Scholar]
- Zhao G, Nyman M, and Jönsson JÅ 2006. Rapid determination of short-chain fatty acids in colonic contents and faeces of humans and rats by acidified water-extraction and direct-injection gas chromatography. Biomedical Chromatography 20:674–682. [DOI] [PubMed] [Google Scholar]
- Zhao R, Chu L, Wang Y, Song Y, Liu P, Li C, Huang J, and Kang X 2017. Application of packed-fiber solid-phase extraction coupled with GC–MS for the determination of short-chain fatty acids in children’s urine. Clinica Chimica Acta 468:120–125. [DOI] [PubMed] [Google Scholar]
- Zheng X, Qiu Y, Zhong W, Baxter S, Su M, Li Q, Xie G, Ore BM, Qiao S, Spencer MD, et al. 2013. A targeted metabolomic protocol for short-chain fatty acids and branched-chain amino acids. Metabolomics 9:818–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.