Abstract
Measurements of multiple biomolecules within the same biological sample are important for many clinical applications to enable accurate disease diagnosis or classification. These disease-related biomarkers often exist at very low levels in biological fluids, necessitating ultrasensitive measurement methods. Single-molecule arrays (Simoa), a bead-based digital enzyme-linked immunosorbent assay, is the current state of the art for ultrasensitive protein detection and can detect sub-femtomolar protein concentrations, but its ability to achieve high-order multiplexing without cross-reactivity remains a challenge. Here, a sequential protein capture approach for multiplex Simoa assays is implemented to eliminate cross-reactivity between binding reagents by sequentially capturing each protein analyte and then incubating each capture bead with only its corresponding detection antibody. This strategy not only reduces cross-reactivity to background levels and significantly improves measurement accuracies, but also enables higher-order multiplexing. As a proof of concept, the sequential multiplex Simoa assay is used to measure five different cytokines in plasma samples from Coronavirus Disease 2019 (COVID-19) patients. The ultrasensitive sequential multiplex Simoa assays will enable the simultaneous measurements of multiple low-abundance analytes in a time- and cost-effective manner and will prove especially critical in many cases where sample volumes are limited.
Keywords: cross-reactivity, ELISA, multiplex assay, Simoa, ultrasensitive protein detection
1. Introduction
Proteins are important biomarkers for disease diagnostics and therapeutic monitoring, and are often present at low concentrations in biological fluids, such as blood, saliva, and urine. Accurate quantification of protein biomarkers in these biological fluids, as well as the discovery of new biomarkers, requires ultrasensitive analytical techniques. The current gold standard method for ultrasensitive protein detection is the bead-based digital enzyme-linked immunosorbent assay (ELISA) termed single-molecule arrays (Simoa), which is up to 1000 times more sensitive than conventional ELISA.[1–3] In Simoa, samples are incubated with an excess number of antibody-coated paramagnetic beads compared to the number of target protein molecules, such that each bead binds either zero or one protein molecule.[3] The protein is then labeled with a biotinylated detection antibody and streptavidin-conjugated enzyme, followed by bead loading into femtoliter-sized microwells, where each microwell can fit only one bead. Microwells are sealed with oil and wells containing beads with an enzyme-labeled immunocomplex generate a locally high concentration of fluorescent signal. Single molecule detection is achieved by counting active microwells. The development of Simoa has enabled the discovery of many candidate biomarkers for neurodegenerative, inflammatory and autoimmune, and infectious diseases, as well as for cancer.[4–9]
While many studies frequently report a single specific protein as a candidate biomarker for diagnosis of a certain disease, typically the measurement of one protein biomarker is insufficient for accurate disease diagnosis or classification.[10] Instead, a panel of multiple protein biomarkers is necessary to accurately assess a disease state or monitor the efficacy of a therapeutic.[11–17] One solution to measuring multiple analytes in a biological sample is to divide the sample into subsamples and perform multiple singleplex measurements, where each subsample is used to measure one analyte. Multiple singleplex measurements are ideal when there is sufficient sample volume such that the sample can be divided into several subsamples, provided that each subsample contains the minimum volume required for a specific assay. However, in situations where the sample volume is limited, the number of biomarkers analyzed may be constrained by the number of subsamples that can be obtained. This challenge sometimes can be addressed by diluting the sample to increase the total sample volume. Partitioning and diluting the sample does not always work because some proteins are in such low abundance that they become undetectable.
To address this need, multiplex methods capable of simultaneously detecting multiple proteins in a single sample have been developed.[10,18–21] In Simoa, multiplexing is achieved by using multiple types of beads encoded with dyes of different colors or color intensities.[22] Each unique bead type is coupled to a capture antibody for a specific target protein (Figure 1A). The remaining steps of the Simoa assay follow the same process as described above. Currently, up to a six-plex assay format has been reported using the Simoa technology that measures six cytokines (IL-6, TNF-𝛼, GM-CSF, IL-10, IL1𝛽, and IL-1𝛼) simultaneously in plasma and serum.[23] Multiplex measurements are additionally advantageous for reducing assay times and costs and enabling high-throughput analysis of samples.
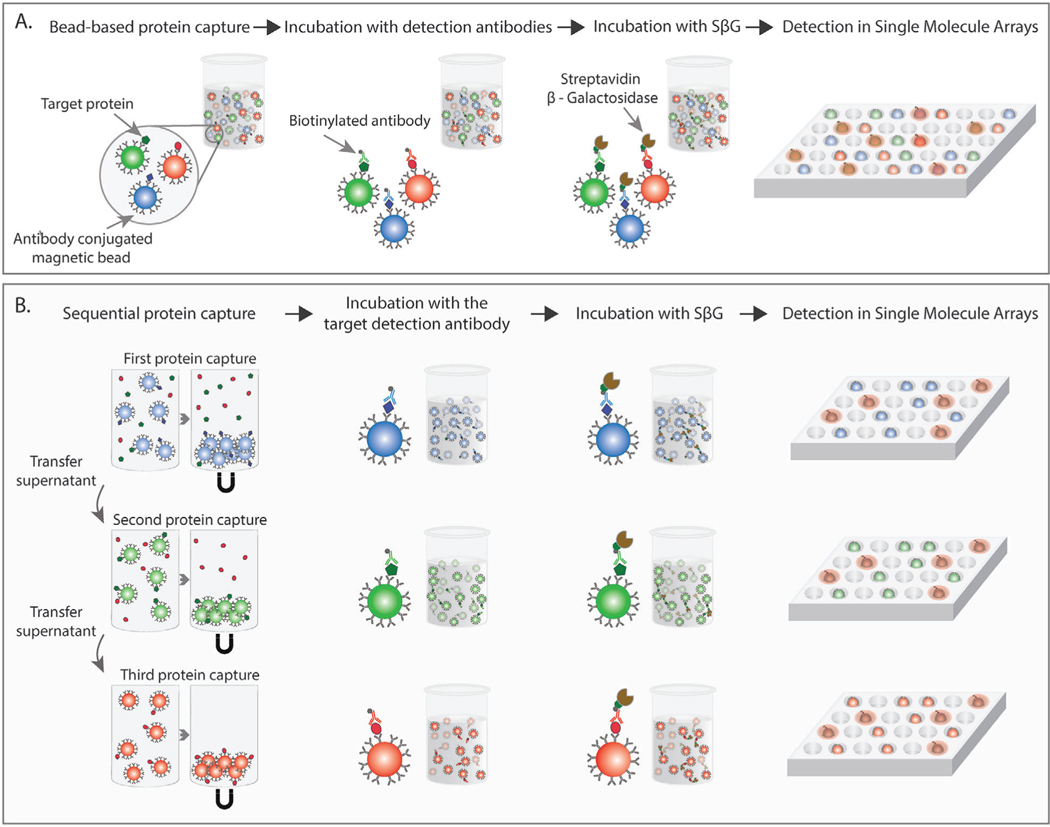
Figure 1.
A) In a standard multiplex Simoa assay, a mixture of magnetic beads coated with different capture antibodies are incubated with a biological sample. Target protein molecules bind to the antibody-coupled beads. In the second step, the beads are introduced to a mixture of detection antibodies, and biotinylated detection antibodies bind to the corresponding target protein. In the third step, the enzyme S𝛽G binds to the biotinylated detection antibody, completing the immune complex. Finally, beads are resuspended in a fluorogenic substrate, loaded into microwell arrays, and sealed with oil. Single-molecule counting is achieved by counting active wells. B) In the sequential multiplex Simoa assay, biological samples are incubated with antibody-coated beads in a sequential fashion (first type of antibody-coupled beads followed by second type of antibody-coupled beads, followed by third type of antibody-coupled beads). Between each bead incubation, the beads are pelleted to the bottom of the well with a magnet and the sample supernatant is transferred to be incubated with the next set of beads. After each protein capture step, each bead type is incubated only with the corresponding detection antibody. The remaining steps follow the steps of a standard Simoa assay.
A major disadvantage of multiplexing that has limited its wider utility in conventional and digital ELISA is cross-reactivity between a binding antibody and an off-target protein.[11,24] In a sandwich ELISA assay, two antibodies (denoted as capture and detection antibodies) with high specificities and affinities to a target protein are required to form the sandwich immunocomplex. In a singleplex assay, the capture antibody or the detection antibody can cross-reactively bind an off-target protein. However, the probability of both a capture and detection antibody binding non-specifically to the same individual molecule tends to zero. In contrast, a multiplex assay contains two or more different capture and detection antibodies in the reaction solution and therefore only one cross-reactive binding event can result in a false signal. Unless the cross-reactivity is sufficiently reduced or quantitatively addressed during analysis, the resulting immunoassays can be unreliable.
Proximity detection approaches have been used to reduce cross-reactivity in multiplex ELISA assays.[18,25,26] In these assays, there is a detectable signal only when both the correct capture and detection antibodies bind to the target protein. An alternative approach is to use spatial or temporal separation methods such that each capture antibody is incubated only with its paired detection antibody.[27–29] In the spatial separation approach, capture antibodies for each target are confined to a specific region or spot, and the corresponding detection antibody is delivered to each correct spot. Accurate alignment of the capture and detection antibodies results in negligible cross reactivity between the different markers.[27,28] In the temporal separation approach, each target is captured sequentially and then incubated with only the correct detection antibody.[29,30] Those methods are beneficial since they allow for quantification of two or more biomarkers simultaneously in one sample without additional dilutions and with negligible cross-reactivity. However, the challenge of adapting such approaches for ultrasensitive multiplex assays with minimal cross reactivity still remains.
Here, we present a multiplex Simoa assay that utilizes sequential protein capture to eliminate assay cross-reactivity and enables higher order multiplexing (Figure 1B). In this method, the entire sample volume is incubated with each capture bead separately in a sequential fashion. This format allows us to eliminate assay cross-reactivity by incubating each detection antibody only with the corresponding capture antibody-coupled beads. We used this sequential multiplexing approach to measure five different cytokines in plasma samples from healthy and Coronavirus Disease 2019 (COVID-19) positive individuals. The combination of this sequential multiplex assay format with ultrasensitive Simoa technology will allow for more accurate quantification of multiple low abundance protein biomarkers in limited volumes of biological fluids.
2. Results
2.1. Validation of the Sequential Multiplex Simoa Assay
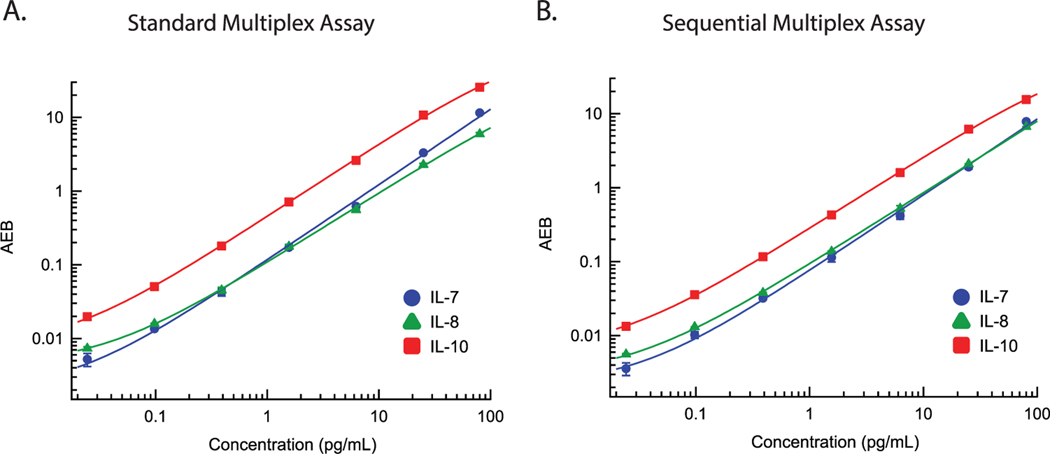
To test whether a sequential multiplex Simoa assay can successfully measure multiple protein analytes with high sensitivities, we first generated calibration curves with a sequential capture method using interleukins 7, 8, and 10 (IL-7, IL-8, and IL-10, respectively). Serial dilutions of standards containing mixtures of IL-7, IL-8, and IL-10 recombinant protein were incubated sequentially with each type of capture bead, as shown in Figure 1B. In the first step, standards were incubated with IL-7 beads in a 96-well plate. After the incubation, IL-7 capture beads were pelleted with a magnet and the standard solutions (supernatants) were transferred to a second 96-well plate. In the second step, the supernatant was incubated with IL-8 beads. After the incubation, IL-8 capture beads were pelleted with a magnet and the standard solutions were transferred to a third 96-well plate. In the third step, the supernatant was incubated with IL-10 beads. An automatic plate washer was used to wash each type of capture bead separately to remove any unbound or nonspecifically adsorbed molecules. Beads were then loaded onto the automated Simoa instrument (HD-X Analyzer), where each bead was incubated only with the corresponding detection antibody, i.e., IL-7 detection antibody with IL-7 capture beads, IL-8 detection antibody with IL-8 capture beads, and IL-10 detection antibody with IL-10 capture beads. Additional details of the sequential multiplex assay are described in the Experimental Section. We also generated calibration curves for IL-7, IL-8, and IL-10 using the standard Simoa multiplex approach (Figure 1A) in which all three types of capture beads were incubated simultaneously with the calibrator solutions, followed by incubating the bead mixtures with a mixture of all three detection antibodies. Each calibration curve was fitted by four-parameter logistic (4PL) regression, which was also used to estimate the limit of detection (LOD) (Table S1, Supporting Information). As shown in Figure 2, sequential protein capture yielded a high dynamic range, with similar sensitivities as the standard multiplex assay for all three proteins. The average enzyme per bead (AEB) values were also similar between the two assays throughout the range of the calibration curve, indicating that the sequential protein capture resulted in negligible losses in signal. The sequential multiplex Simoa assay resulted in slightly higher LODs for IL-7 and IL-8 (Table S1, Supporting Information) compared to the standard multiplex Simoa assay, whereas both assay formats had similar LODs for IL-10. Both assays result in sub-picogram per milliliter LODs for all three markers, indicating there is negligible loss of sensitivity using the sequential multiplex Simoa assay format. To validate the ability of the sequential multiplex assay to detect multiple cytokines in biological fluids, we measured endogenous dilution linearity and recombinant protein spike and recovery in plasma samples for the three proteins (Figure S1 and Table S2, Supporting Information). All three markers diluted linearly between 4× and 32× dilution factors (R2 for linear regression fit > 0.97). Recoveries of spiked recombinant protein in plasma samples were between 75% and 100% for three spiked protein concentrations across all three markers (standard acceptable recoveries are 70–130%).
Figure 2.
Calibrations curves for IL-7 (blue circles), IL-8 (green triangles), and IL-10 (red squares) in the A) standard multiplex assay and B) the sequential multiplex assay. Each curve is fit by 4PL regression. Each marker represents the average of duplicate measurements and the corresponding error bars represent the standard deviation of duplicate measurements.
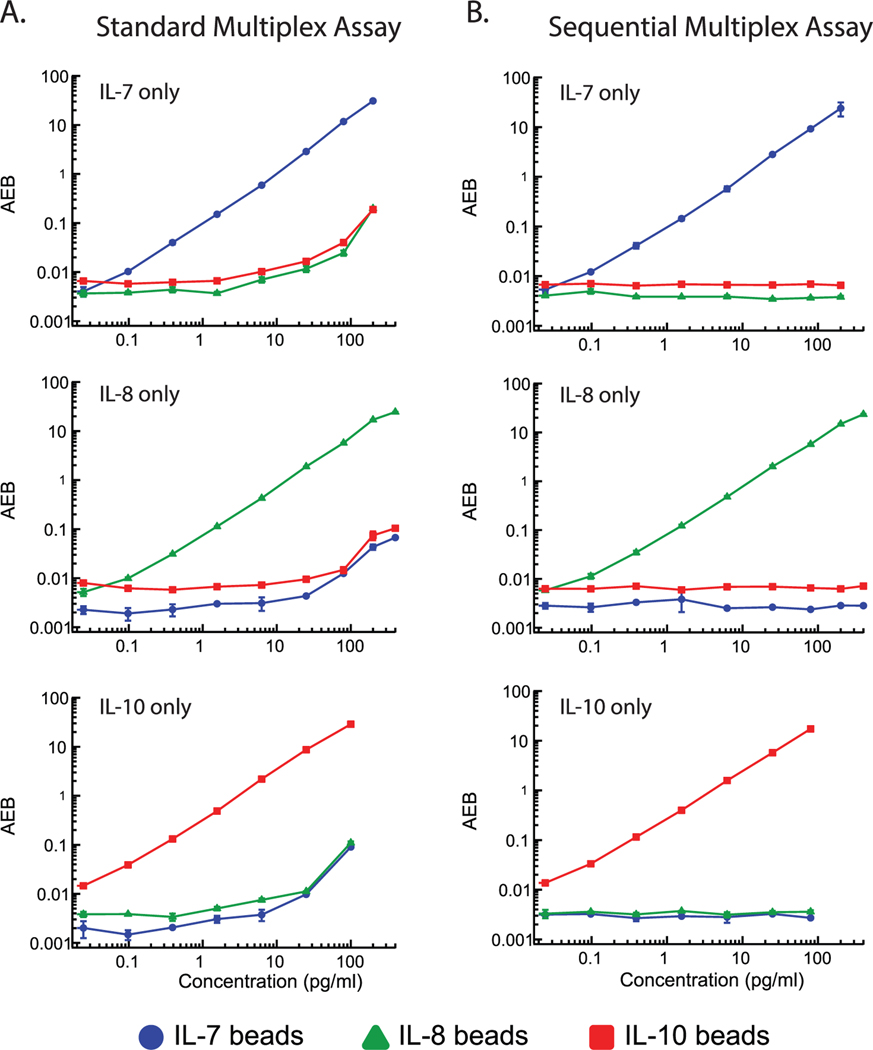
We next assessed whether the sequential multiplex assay reduces cross-reactivity by performing protein dropout experiments for each target protein using both the standard and sequential multiplex assays. In sequential multiplex dropout experiments, serial dilutions of one protein (either IL-7, IL-8, or IL-10) were sequentially incubated with the three bead types coated with antibodies against IL-7, IL-8, and IL-10. In standard multiplex dropout experiments, serial dilutions of one protein were simultaneously incubated with all three bead types coated with antibodies against IL-7, IL-8, and IL-10. The specific binding and cross-reactivity for each analyte concentration were measured and quantified by assessing the AEB response of each protein binding to each bead and calculating the signal-to-background ratios (Figure 3; Table S3, Supporting Information). For example, the IL-7 plots in Figure 3 show measured AEBs for serial dilutions of IL-7 protein binding to IL-7 beads (blue circles), IL-8 beads (green triangles), and IL-10 beads (red squares) in both the standard and sequential multiplex assay formats. The standard multiplex assay demonstrated significant cross-reactivity for all three protein dropout assays, with observable false positive signals on off-target beads at protein concentrations above 10 pg mL−1 (Figure 3A). Above 100 pg mL−1, the false positive signal reached over 50× above background. We attribute this false positive signal to proteins cross-reactively binding to the incorrect capture bead. Because the standard multiplex format contains a mixture of all three detection antibodies, a protein that is cross-reactively bound to an off-target capture bead can be bound by its corresponding detection antibody, producing a false positive signal. An example is IL-7 protein binding to IL-8 capture beads, followed by binding of the IL-7 detection antibody to IL-7 protein. False positive signals can also arise from nonspecific binding of one detector antibody to the wrong capture bead. In contrast, the sequential multiplex assay completely eliminates the cross-reactivities, with no detectable false signals arising on off-target beads across all three dropout assays (Figure 3B). Notably, even at the highest protein concentrations we tested, the measured signals from the off-target beads remained at background levels. Thus, by incubating each detection antibody only with its corresponding capture bead, the number of false positive binding events is vastly reduced, as two cross-reactive binding events are required to produce a false positive signal.
Figure 3.
Protein dropout experiments to assess the cross-reactivities of the A) standard multiplex Simoa assay and B) sequential multiplex Simoa assay. Each plot shows a different experiment wherein only one of the three target proteins is measured at a time to assess the binding to both on-target and off-target capture beads. Each marker represents the average of duplicate measurements and the corresponding error bars represent the standard deviation of duplicate measurements.
2.2. Accuracy of the Sequential Multiplex Simoa Assay
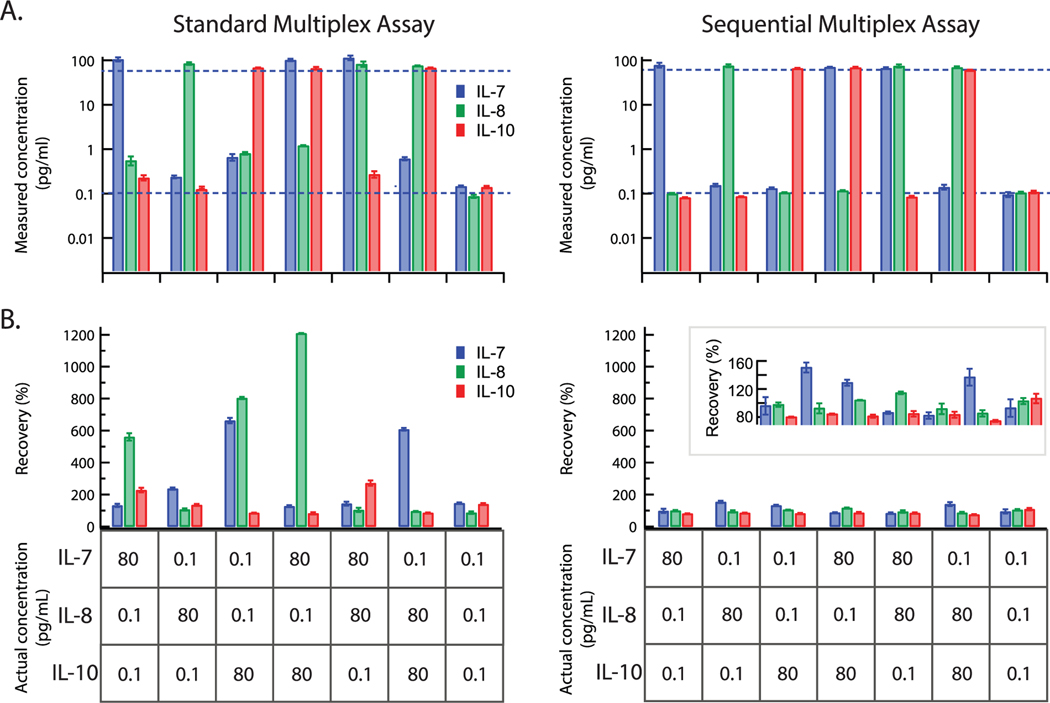
To evaluate the quantification accuracy of the sequential multiplex assay, we measured various combinations of protein mixtures with high (80 pg mL−1) and low (0.1 pg mL−1) concentrations using both the standard and sequential multiplex assay formats. As shown in Figure 4A, for both methods, the high protein concentration was recovered accurately. However, for low protein concentrations, the standard multiplex assay yielded strong inconsistencies between the actual and measured protein concentrations. In order to assess the cross-reactivity for each method, we quantified the recovery of each of the three proteins according to the actual and measured protein concentrations (Figure 4B). Notably, the recoveries in the standard multiplex assay varied significantly between the three proteins in each protein mixture. In some cases, the high levels of cross-reactivity resulted in recoveries of greater than 400%, even reaching up to 1000%. Importantly, the sequential multiplex assay showed high accuracy, with recoveries primarily within 80–120%. The vastly reduced cross-reactivity in the sequential multiplex assay significantly improves measurement accuracy, as high concentrations of one protein do not interfere with measurements of other proteins at low concentrations within the same sample.
Figure 4.
Quantification accuracy of the Simoa multiplex assays. A) Samples consisting of mixtures of IL-7, IL-8, and IL-10 were prepared and measured in both the standard multiplex assay and sequential multiplex assay. The measured concentrations are plotted for both assays. Dotted lines represent the actual concentrations of each protein at either 80 or 0.1 pg mL−1. B) Percent recoveries for each protein in each sample, calculated by dividing the measured concentration by the actual concentration. Actual protein concentrations are displayed in the tables. Error bars represent the standard deviation of duplicate measurements.
2.3. Sequential Five-Plex Simoa Assay in Plasma Samples
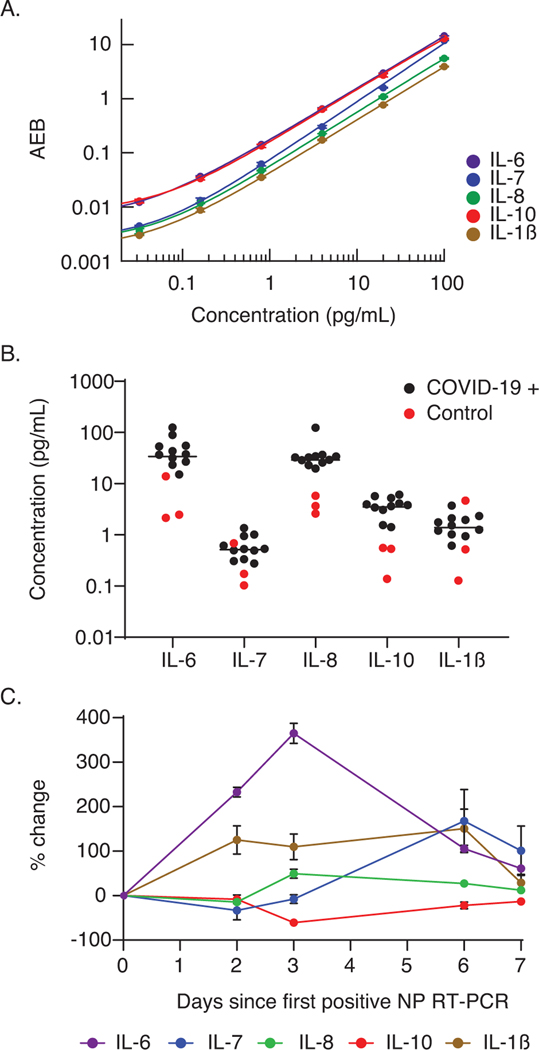
In addition to reducing cross-reactivity to negligible levels, another advantage of the sequential multiplex Simoa assay is that the multiplexing capability does not depend on spectral separation between color coded beads. As a proof of concept, we tested the ability to quantify IL-6, IL-7, IL-8, IL-10, and IL-1𝛽 in clinically relevant plasma samples using a five-plex sequential multiplex Simoa assay. New calibration curves for the five-plex assay were measured, and curves were fitted with 4PL regression (Figure 5A). We measured the five cytokines in 11 plasma samples collected from COVID-19-positive patients, as well as three plasma samples from healthy individuals collected before October 2019 (Figure 5B). Plasma samples were diluted fourfold, and only a total of 50 μL of each sample was required for the five-plex sequential assay (measurements were made in duplicate), which is five times lower than the volume required for measuring the five cytokines using standard singleplex Simoa assays. Elevated cytokine levels were detected in the majority of the COVID-19-positive samples compared to the healthy controls. Furthermore, the quantitative capabilities and ultrasensitivity of the Simoa assay enable detection of small changes in cytokine levels during the first week following a positive reverse transcription polymerase chain reaction test from a nasopharyngeal swab (NP RT-PCR) (Figure 5B). Monitoring multiple cytokine levels with high accuracy is especially important in the case of Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, during which a cytokine storm can occur and potentially result in death.[31]
Figure 5.
Serial monitoring of cytokine levels in plasma using sequential multiplex Simoa assays. A) Calibration curves for the five-plex sequential multiplex Simoa assay; cytokines IL-6 (purple), IL-7 (blue), IL-8 (green), IL-10 (red), and IL-1𝛽 (brown) were measured in this assay. Each curve is fit by 4PL regression. Error bars represent the standard deviation of duplicate measurements. B) Measured concentrations of five cytokines from three healthy (red circles) and 11 COVID-19-positive (black circles) individuals. Each marker represents the average of duplicate measurements. C) Percentage changes in cytokine levels in five serial samples collected from one individual who tested positive for COVID-19. The percentage change was calculated relative to the day of the first positive NP RT-PCR. Error bars represent the standard deviation of duplicate measurements.
3. Discussion
Ultrasensitive multiplex assays are critical for analyzing and quantifying multiple biomarkers in low-volume samples. However, standard multiplex Simoa assays can exhibit cross-reactive binding, limiting the accuracy of multiple protein quantification. We show that cross-reactivity exhibits an especially significant problem when some target proteins in the sample are present at high concentrations, while other proteins are present at low concentrations. Combining the ultrasensitivity of the Simoa assay with sequential protein capture has allowed us to overcome this limitation. We show that the sequential multiplex Simoa assay reduces cross-reactivity to background levels by incubating each capture bead with the corresponding detection antibody. Although nonspecific binding of particular proteins to the wrong beads in prior incubations can still occur, the nonspecifically bound protein will not lead to a false positive signal for other analytes, unlike in the traditional multiplex Simoa assay. As different analytes may have different amounts of nonspecific binding to certain antibody-coated beads types, optimization of the bead incubation order for each set of analytes will minimize effects of nonspecific binding on signal losses for latter analytes.
In the sequential multiplex assay, because the capture beads are not mixed, the degree of multiplexing is no longer limited by the number of different colors of dye-encoded beads that can be simultaneously imaged without spectral overlap. Therefore, additional assays can simply be added to the sequential multiplex assay by adding another incubation step for each new target. We demonstrate this ability to achieve higher order multiplexing by developing a five-plex sequential Simoa assay and measuring five cytokines in clinical plasma samples. The ability to perform high-level multiplexing, i.e., five or more targets, is essential in applications where sample volume is limited.
Although the sequential multiplex Simoa assay shows high accuracy and sensitivity in quantifying multiple proteins simultaneously, this method also has limitations. One limitation is that a fraction of the sample can be lost when the sample is transferred between consecutive incubation steps. This small sample loss can be a major concern when a high plex is required. Additionally, a small amount of bead carryover between sequential steps can reduce the quantification accuracy. Furthermore, protein denaturation may occur and result in further sample loss as a consequence of longer incubations required for each target. However, optimization and automation of the sequential capture process can be used to overcome those limitations. For the five-plex sequential assay presented in this work, we did not observe any effects of sample loss on measured concentrations.
An additional limitation is that since each capture bead incubation step is performed sequentially, the total assay time is increased by approximately 30 min for each additional target, depending on the desired incubation time for each analyte - i.e., each additional marker measured adds additional protein capture, transfer, and wash steps. This also increases the complexity of the assay. The increase in total assay time and need for trained personnel are primarily of concern for point-of-care applications where a result is needed within approximately one hour (from time of sample collection to result). Alternative approaches are possible to separate each detection antibody from off-target antibody-coated beads in multiplex Simoa assays. For instance, spatial separation strategies can be used, in which all the capture beads are simultaneously incubated with the sample and then sorted into separate bead types for individual incubations with the corresponding detection antibodies. As the capture steps for all proteins are performed simultaneously in one mixture and because the subsequent incubations of each bead type with the corresponding detection antibody can be performed in parallel, such strategies can potentially decrease total assay times compared to the sequential multiplex assay.
In summary, we have developed an approach towards high-level multiplexing of ultrasensitive Simoa assays that eliminates cross-reactivity, opening the door for simultaneous measurements of many more analytes than currently possible with the existing Simoa technology. As existing multiplex Simoa and ELISA assays are limited by cross-reactivity and the number of targets that can be simultaneously measured, this sequential multiplex Simoa platform will enable significantly more biological information to be obtained from small volume samples. The capability to achieve higher order multiplexing, combined with the high accuracy and ultrasensitivity of the sequential multiplex Simoa assay, will prove especially important in cases where sample volumes are limited, such as neonatal blood or saliva and single cells,and can potentially improve diagnostic sensitivity and specificity. Furthermore, the vastly improved accuracy of the sequential multiplex Simoa assay can enable multiplex protein detection in samples where proteins exist at concentrations spanning many orders of magnitude. Typically, concentrations of one analyte can interfere with measurements of a low-concentration analyte in standard multiplex Simoa assays, and the dynamic range of the assay is restricted to low concentrations with limited cross-reactivity. The sequential multiplex Simoa assay eliminates cross-reactivity and will thus enable a broad dynamic range across multiple analytes.
4. Experimental Section
Preparation of Antibody-Coated Capture Beads:
Capture antibodies for IL-7 (BioLegend 501302), IL-8 (BD Biosciences 554716), and IL-10 (BioLegend 506801) were received and stored according to the manufacturer’s instructions. Each antibody was buffer exchanged to remove storage buffer; ≈0.13 mg of antibody (in solution) was added to a Millipore Sigma Amicon Ultra-0.5 mL Centrifugal Filter (50 K). Bead conjugation buffer (Quanterix Corp.) was added to the filter for a total volume of 500 μL. The filter device was centrifuged at 14 000 × g for 5 min. After centrifugation, the effluent was discarded and additional bead conjugation buffer was added to the filter (total volume of 500 μL). The centrifugation process was repeated twice for a total of three washes. The filter was then inverted into a new tube and centrifuged at 1000 × g for 2 min. The concentration of the collected antibody was measured using a NanoDrop One (Thermo Fisher). The buffer-exchanged antibodies were diluted to 0.5 mg mL−1 using bead conjugation buffer and stored on ice until ready for use. To prepare beads for conjugation, 2.8 × 108 Quanterix dye-encoded carboxylated paramagnetic beads (2.7 μm) were washed three times with 200 μL of bead wash buffer (Quanterix), three times with 200 μL of bead conjugation buffer, and then resuspended in 190 μL of bead conjugation buffer. Immediately prior to use, 1 mg of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) was dissolved in 100 μL of bead conjugation buffer. Then, 10 μL of EDC was added to the bead suspension and the beads were agitated on a rotator for 30 min. After bead activation with EDC, beads were washed once with 200 μL of bead conjugation buffer, and then resuspended in 200 μL of capture antibody solution. The beads were vortexed for 10 s and agitated on the rotator for 2 h. After conjugation, the beads were washed two times with 200 μL of bead wash buffer. The antibody-conjugated beads were then blocked with BSA for 30 min in 200 μL of bead blocking buffer (Quanterix) under agitation on the rotator. Finally, the antibody-conjugated beads were washed with 200 μL of bead wash buffer, 200 μL of bead diluent (Quanterix), and resuspended in 200 μL of bead diluent. IL-7, IL-8, and IL-10 capture antibodies were conjugated to 488, 700, and 647 nm dye-encoded beads, respectively. Beads were counted using a Beckman Coulter Z1 Particle Counter and stored at 4 °C.
Setup of Standard Multiplex Simoa Assay:
Human recombinant protein standards for IL-7 (207-IL-005), IL-8 (208-IL-010), and IL-10 (217-IL-005) were purchased from R&D Systems. Proteins were received lyophilized, reconstituted in 1× phosphate buffered saline (PBS) with 1% BSA, and stored in aliquots at −80 °C until ready for use. Protein standards were serially diluted in homebrew sample/detector diluent (Quanterix) to the desired concentrations. In a three-step assay configuration, the first step was performed offline the HD-X Analyzer (Quanterix), whereas the second and third steps were performed onboard the HD-X Analyzer, as described below. This assay format was used for standard multiplex Simoa to allow for direct comparison to the sequential multiplex Simoa assay, as described below. Capture beads were diluted in homebrew sample/detector diluent to a concentration of 75 000 beads μL−1 (with 25 000 beads μL−1 per target). For the protein capture step, 100 μL of sample (protein) and 10 μL of capture beads were added to a 96-well plate (Greiner Bio-One 655096) and incubated at room temperature with shaking for 30 min. The beads were washed three times with System Wash Buffer 1 (Quanterix) on a Tecan plate washer. Beads were resuspended in 1× sodium chloride-sodium phosphate buffer with 1.6% dextran sulfate and then transferred to a new 96-well plate (Quanterix) and loaded onto the HD-X Analyzer for analysis. Biotinylated detection antibodies for IL-7 (BioLegend 506601), IL-8 (BD Biosciences 554718), and IL-10 (BioLegend 501501) were combined and diluted into one solution in homebrew sample/detector diluent at a total concentration of 230 ng mL−1. The diluted concentrations for the IL-7, IL-8, and IL-10 biotinylated detection antibodies were 150, 5, and 75 ng mL−1, respectively. Streptavidin-𝛽-galactosidase (S𝛽G) concentrate (Quanterix) was diluted to 100 × 10−12 m in S𝛽G diluent (Quanterix). Biotinylated detection antibody and S𝛽G solutions were placed in plastic bottles (Quanterix) and loaded onto the HD-X Analyzer prior to analysis. Resorufin 𝛽-D-galactopyranoside (RGP), System Wash Buffer 1, System Wash Buffer 2, and Simoa sealing oil were purchased from Quanterix and loaded on the HD-X Analyzer as per the manufacturer’s instructions. Onboard the HD-X Analyzer, all incubations and washes were performed automatically as follows: beads with captured protein were washed, resuspended in 100 μL of biotinylated detection antibody solution, and incubated at room temperature for 5.25 min (7 cadences); washed, resuspended in 100 μL of S𝛽G solution, and incubated at room temperature for 5.25 min (7 cadences); washed; and finally resuspended in RGP solution before loading into microwell arrays for analysis. Following bead loading, the microwell array was sealed with oil and imaged. AEB values were calculated by the software in the HD-X Analyzer.
Setup of Sequential Multiplex Simoa Assay:
IL-7, IL-8, and IL-10 protein standards were serially diluted in homebrew sample/detector diluent to the desired concentrations. In a three-step assay configuration, the first step was performed offline from the HD-X Analyzer, whereas the second and third steps were performed onboard the HD-X Analyzer, as described below. Capture beads were diluted in homebrew sample/detector diluent to a concentration of 50 000 beads μL−1. For the protein capture step, 100 μL of sample (protein) and 10 μL of IL-7 capture beads were added to a 96-well plate (Greiner Bio-One 655096), denoted as plate 1, and incubated at room temperature with shaking for 30 min. After incubation, the 96-well plate was placed on a DynaMag-96 Side Skirted magnet (Thermo Fisher Scientific) for 1 min to allow the beads to pellet at the bottom of each well. The entire sample volume was transferred from plate 1 to a new 96-well plate (plate 2). The beads in plate 1 were resuspended in 100 μL of homebrew sample/detector diluent and washed three times with System Wash Buffer 1 on a Tecan plate washer. This sample incubation, transfer, and washing process was repeated in plate 2 with IL-8 beads and plate 3 with IL-10 beads. Beads from each plate were resuspended in 1× sodium chloride-sodium phosphate buffer with 1.6% dextran sulfate and then transferred into a new 96-well plate (Quanterix) and loaded onto the HD-X Analyzer for analysis. For each sample, IL-7, IL-8, and IL-10 capture beads were transferred into three separate wells in the 96-well plate. Biotinylated detection antibodies for IL-7, IL-8, and IL-10 were diluted into three separate solutions in homebrew sample/detector diluent at final concentrations of 300, 10, and 150 ng mL−1, respectively, and placed in three separate plastic reagent bottles. S𝛽G concentrate was diluted to 100 × 10−12 m in S𝛽G diluent. Biotinylated detection antibody and S𝛽G solutions were placed in plastic bottles and loaded onto the HD-X Analyzer prior to analysis. RGP, System Wash Buffer 1, System Wash Buffer 2, and Simoa sealing oil were loaded on the HD-X Analyzer as per the manufacturer’s instructions. Onboard the HD-X Analyzer, all incubations and washes were performed automatically as follows: beads with captured protein were washed, resuspended in 100 μL of the corresponding biotinylated detection antibody solution (i.e., IL-7 beads were resuspended in IL-7 biotinylated detection antibody solution, IL-8 beads were resuspended in IL-8 biotinylated detection antibody solution, and IL-10 beads were resuspended in IL-10 biotinylated detection antibody solution), and incubated at room temperature for 5.25 min (7 cadences); washed, resuspended in 100 μL of S𝛽G solution, and incubated at room temperature for 5.25 min (7 cadences); washed; and finally resuspended in RGP solution before loading into microwell arrays for analysis. Following bead loading, the microwell array was sealed with oil and imaged. Beads for each marker were analyzed in separate microwell arrays. AEB values were calculated by the software in the HD-X Analyzer.
Protein Detection in Plasma:
Pooled plasma samples were purchased from BioIVT and stored at −80 °C until ready for use. Plasma samples were thawed at room temperature and then centrifuged at 4 °C for 10 min at 2000 × g. The supernatant was collected and centrifuged a second time through a 0.45 × 10−6 m filter at 4 °C for 10 min at 2000 × g. The filtrate was serially diluted in homebrew sample/detector diluent 4-to 32-fold for dilution linearity studies and diluted eightfold for spike and recovery studies. For spike and recovery experiments, different amounts of recombinant IL-7, IL-8, and IL-10 proteins (50, 5 and 0.5 pg mL−1) were spiked into the plasma samples. Clinical samples were obtained from patients presenting to Brigham and Women’s Hospital with viral respiratory symptoms who tested positive for SARS-CoV-2. Pre-pandemic samples, collected from healthy individuals before October 1, 2019, were obtained from the Partner’s Biobank. Clinical samples were collected with prior informed written consent and Institutional Review Board approval (protocol no. 2020P001204). The plasma samples were thawed at room temperature and then centrifuged at 4 °C for 10 min at 2000 × g. The supernatant was collected and diluted fourfold in homebrew sample/detector diluent. IL-6, IL-7, IL-8, IL-10, and IL-1𝛽 were measured in the COVID-19-positive and healthy control samples using the sequential multiplex Simoa assay as described above.
Data Analysis:
For all the data presented in the article, duplicate measurements per sample were obtained and the mean value was plotted. Error bars represent the standard deviation of the two measurements. The calibration curves were fitted using GraphPad Prism version 8.3. All figures were plotted in GraphPad Prism, Igor Pro version 6.3, and Adobe Illustrator version 2015.
Supplementary Material
Acknowledgements
T.G., A.M.M., A.F.O., and C.W. contributed equally to this work. The authors thank Limor Cohen for helpful discussions regarding the experimental design. Partial funding for this work was provided by a gift from the Good Ventures Foundation and a 1F32EB029777-01 grant from the NIBIB at NIH.
D.R.W. has a financial interest in Quanterix Corporation, a company that develops an ultrasensitive digital immunoassay platform. He is an inventor of the Simoa technology, a founder of the company, and also serves on its board of directors. Interests of D.R.W. were reviewed and are managed by BWH and Partners HealthCare in accordance with their conflict of interest policies.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Conflict of Interest
All other authors declare no conflict of interest.
Contributor Information
Tal Gilboa, Department of Pathology, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115, USA; Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston MA 02115, USA.
Adam M. Maley, Department of Pathology, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115, USA Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston MA 02115, USA.
Alana F. Ogata, Department of Pathology, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115, USA Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston MA 02115, USA.
Connie Wu, Department of Pathology, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115, USA; Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston MA 02115, USA.
David R. Walt, Department of Pathology, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115, USA Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston MA 02115, USA.
References
- [1].Rissin DM, Walt DR, Nano Lett. 2006, 6, 520. [DOI] [PubMed] [Google Scholar]
- [2].Rissin DM, Walt DR, J. Am. Chem. Soc 2006, 128, 6286. [DOI] [PubMed] [Google Scholar]
- [3].Rissin DM, Kan CW, Campbell TG, Howes SC, Fournier DR, Song L, Piech T, Patel PP, Chang L, Rivnak AJ, Ferrell EP, Randall JD, Provuncher GK, Walt DR, Duffy DC, Nat. Biotechnol 2010, 28, 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wu C, Maley AM, Walt DR, Crit. Rev. Clin. Lab. Sci 2020, 57, 270. [DOI] [PubMed] [Google Scholar]
- [5].Song L, Hanlon DW, Chang L, Provuncher GK, Kan CW, Campbell TG, Fournier DR, Ferrell EP, Rivnak AJ, Pink BA, Minnehan KA, Patel PP, Wilson DH, Till MA, Faubion WA, Duffy DC, J. Immunol. Methods 2011, 372, 177. [DOI] [PubMed] [Google Scholar]
- [6].Jarolim P, Patel PP, Conrad MJ, Chang L, Melenovsky V, Wilson DH, Clin. Chem 2015, 61, 1283. [DOI] [PubMed] [Google Scholar]
- [7].Olsen DA, Kjaer IM, Brandslund I, J. Immunol. Methods 2018, 459, 63. [DOI] [PubMed] [Google Scholar]
- [8].Startin CM, Ashton NJ, Hamburg S, Hithersay R, Wiseman FK, Mok KY, Hardy J, Lleó A, Lovestone S, Parnetti L, Zetterberg H, Hye A, Strydom A, Alzheimer’s Res. Ther 2019, 11, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ng ASL, Tan YIJ, Lu Z, Ng EYL, Ng SYE, Chia NSY, Setiawan F, Xu Z, Tay KY, Prakash KM, Au WL, Tan E-K, Tan LCS, Ann. Clin. Transl. Neurol 2019, 6, 615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tighe PJ, Ryder RR, Todd I, Fairclough LC, Proteomics: Clin. Appl 2015, 9, 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Juncker D, Bergeron S, Laforte V, Li H, Curr. Opin. Chem. Biol 2014, 18, 29. [DOI] [PubMed] [Google Scholar]
- [12].Bigbee WL, Gopalakrishnan V, Weissfeld JL, Wilson DO, Dacic S, Lokshin AE, Siegfried JM, J. Thorac. Oncol 2012, 7, 698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Suzuki M, Wiers K, Brooks EB, Greis KD, Haines K, Klein-Gitelman MS, Olson J, Onel K, O’neil KM, Silverman ED, Tucker L, Ying J, Devarajan P, Brunner HI, Pediatr. Res 2009, 65, 530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chung L, Moore K, Phillips L, Boyle FM, Marsh DJ, Baxter RC, Breast Cancer Res. 2014, 16, R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Severi G, Fitzgerald LM, Muller DC, Pedersen J, Longano A, Southey MC, Hopper JL, English DR, Giles GG, Mills J, Cancer Med. 2014, 3, 1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ahmad R, Xie L, Pyle M, Suarez MF, Broger T, Steinberg D, Ame SM, Lucero MG, Szucs MJ, Macmullan M, Berven FS, Dutta A, Sanvictores DM, Tallo VL, Bencher R, Eisinger DP, Dhingra U, Deb S, Ali SM, Mehta S, Fawzi WW, Riley ID, Sazawal S, Premji Z, Black R, Murray CJL, Rodriguez B, Carr SA, Walt DR, Gillette MA, Sci. Transl. Med 2019, 11, eaaw8287. [DOI] [PubMed] [Google Scholar]
- [17].Zaidi AH, Gopalakrishnan V, Kasi PM, Zeng X, Malhotra U, Balasubramanian J, Visweswaran S, Sun M, Flint MS, Davison JM, Hood BL, Conrads TP, Bergman JJ, Bigbee WL, Jobe BA, Cancer 2014, 120, 3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cohen L, Walt DR, Chem. Rev 2019, 119, 293. [DOI] [PubMed] [Google Scholar]
- [19].Houser B, Arch. Physiol. Biochem 2012, 118, 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Abhishek J, Manuel M, Circulation 2018, 138, 2482.30524136 [Google Scholar]
- [21].Dupont NC, Wang K, Wadhwa PD, Culhane JF, Nelson EL, J. Reprod. Immunol 2005, 66, 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rissin DM, Kan CW, Song L, Rivnak AJ, Fishburn MW, Shao Q, Piech T, Ferrell EP, Meyer RE, Campbell TG, Fournier DR, Duffy DC, Lab Chip 2013, 13, 2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Rivnak AJ, Rissin DM, Kan CW, Song L, Fishburn MW, Piech T, Campbell TG, Dupont DR, Gardel M, Sullivan S, Pink BA, Cabrera CG, Fournier DR, Duffy DC, J. Immunol. Methods 2015, 424, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ellington AA, Kullo IJ, Bailey KR, Klee GG, Clin. Chem 2009, 55, 1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fredriksson S, Dixon W, Ji H, Koong AC, Mindrinos M, Davis RW, Nat. Methods 2007, 4, 327. [DOI] [PubMed] [Google Scholar]
- [26].Ebai T, Souza De Oliveira FM, Löf L, Wik L, Schweiger C, Larsson A, Keilholtz U, Haybaeck J, Landegren U, Kamali-Moghaddam M, Clin. Chem 2017, 63, 1497. [DOI] [PubMed] [Google Scholar]
- [27].Pla-Roca M, Leulmi RF, Tourekhanova S, Bergeron S, Laforte V, Moreau E, Gosline SJC, Bertos N, Hallett M, Park M, Juncker D, Mol. Cell. Proteomics 2012, 11, M111.011460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Frampton JP, White JB, Simon AB, Tsuei M, Paczesny S, Takayama S, Sci. Rep 2014, 4, 4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Fiema B, Harris AC, Gomez A, Pongtornpipat P, Lamiman K, Vander Lugt MT, Paczesny S, J. Visualized Exp 2012, e4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Poetz O, Henzler T, Hartmann M, Kazmaier C, Templin MF, Herget T, Joos TO, Mol. Cell. Proteomics 2010, 9, 2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chen C, Zhang XR, Ju ZY, He WF, Zhonghua Shaoshang Zazhi 2020, 36, E005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.