Abstract
Purpose:
Phospholipids are key constituents of cell membranes and serve vital functions in the regulation of cellular processes; thus, a method for in vivo detection and characterization could be valuable for detecting changes in cell membranes that are consequences of either normal or pathological processes. Here, we describe a new method to map the distribution of partially restricted phospholipids in tissues.
Methods:
The phospholipids were measured by signal changes caused by relayed nuclear Overhauser enhancement-mediated CEST between the phospholipid Cho headgroup methyl protons and water at around −1.6 ppm from the water resonance. The biophysical basis of this effect was examined by controlled manipulation of head group, chain length, temperature, degree of saturation, and presence of cholesterol. Additional experiments were performed on animal tumor models to evaluate potential applications of this novel signal while correcting for confounding contributions.
Results:
Negative relayed nuclear Overhauser dips in Z-spectra were measured from reconstituted Cho phospholipids with cholesterol but not for other Cho-containing metabolites or proteins. Significant contrast was found between tumor and contralateral normal tissue signals in animals when comparing both the measured saturation transfer signal and a more specific imaging metric.
Conclusion:
We demonstrated specific relayed nuclear Overhauser effects in partially restricted phospholipid phantoms and similar effects in solid brain tumors after correcting for confounding signal contributions, suggesting possible translational applications of this novel molecular imaging method, which we name restricted phospholipid transfer.
Keywords: chemical exchange rotation transfer, chemical exchange saturation transfer, magnetization transfer, nuclear Overhauser enhancement, phospholipids
1 |. INTRODUCTION
Membrane phospholipids play crucial roles in cell development, proliferation, and other vital processes, and changes in their characteristics occur with various pathologies.1–3 However, noninvasive studies of phospholipids in vivo have been hampered by a number of methodological difficulties, including short MR relaxation times and minimal contrast in CT,4 severely limiting imaging of lipid distributions and properties. For example, whereas phosphorus-31 MRS has been used to detect phospholipid metabolites at low spatial resolution,5–11 the proton MRSs resonances of phospholipids themselves are very broad and typically not visible in vivo using conventional methods12 due to the signal’s rapid decay, reflecting the limited motion of cell membranes. Specialized MRS methods have been used ex vivo to study extracts of samples in organic solvent systems,8 but such techniques are not translatable for use in vivo. Thus, a method for in vivo detection and mapping of membrane phospholipids could be highly valuable, especially as the structural and biological significance of phospholipids continues to become clearer. In this paper, we describe a new MRI approach, named restricted phospholipid transfer (RPT) imaging, for in vivo noninvasive measurements of the distribution of ordered phospholipids. The technique exploits a highly specific relayed nuclear Overhauser enhancement (rNOE)-mediated transfer of magnetization between water and specific Cho methyl groups within phospholipids and is sensitive to the composition of phospholipids, as will be shown below.
Transfer of magnetization between water protons and specific macromolecular protons in biological tissues has been investigated by a variety of experimental approaches. For example, labile amide,13,14 amine,15,16 and carbohydrate hydroxyls protons17 (shifted 3.5, 2 to 3, and 1 ppm downfield from the water resonance, respectively) have been studied by CEST, which detects water signal variations caused by chemical exchange between water and solute protons. The atypical signal at −1.6 ppm has been observed in gels18 and with in vivo water exchange filter spectroscopy19–23 and ex vivo nuclear Overhauser effect spectroscopy,24 which is presumably contributed from Cho methyl protons in phosphatidylcholine (PtdCho). Here, we show that a well-resolved magnetization transfer (MT) effect on water can be observed in vivo in rat brain at −1.6 ppm, and extend our previous work by developing a pulse sequence and metric with increased biophysical specificity. We hypothesize that this signal originates from an rNOE between the Cho phospholipid methyl headgroups and water protons, and we here evaluate the factors affecting the magnitude of RPT using model phospholipid systems. The results suggest the signal depends on lipid membrane composition and fluidity. Finally, we explore qualitative and quantitative imaging metrics on rats bearing C6 and 9L brain tumors, revealing significant decreases in the Overhauser effect at −1.6 ppm in tumors compared to normal brain. This signal change can be quantified to produce images with a novel form of contrast, suggesting practical applications of the proposed method.
2 |. METHODS
2.1 |. rNOE-mediated saturation transfer technique
Membrane phospholipids, because of their relatively low concentration and short T2 times, cannot be detected by conventional MRS (proton MRS or phosphorous-31 MRS). However, phospholipids can be detected by measurements of water signal variations caused by MT effects at specific resonance frequencies. MT techniques provide enhanced sensitivity for measuring small proton pools through saturation transfer with water via CEST or rNOE effects. rNOE effects between water protons and Cho phospholipid head groups have previously been observed,19,23,25 but here we clarify the nature of these interactions and exploit them to image Cho phospholipids indirectly by their effect on water signals. Conventionally, MT is measured using continuous wave irradiation. Specifically, a saturation pulse, which usually lasts for several seconds, is applied on the resonance frequency of target molecules. Unaffected water protons then transfer magnetization to these saturated protons via dipolar coupling, resulting in a decrease in the water magnetization. We applied 2 conventional variations of this MT technique, acquiring 1) the water signal as a function of the frequency offset of the irradiation pulse (the Z-spectrum), and 2) the difference of the water signals after irradiation at the resonance frequency of the target molecule and at the corresponding frequency on the opposite side of the water resonance (MTRasym). These are established methods for water-metabolite MT,26 and we include them here in order to establish the sensitivities of the rNOE peak at −1.6 ppm to Cho phospholipids and as a point of comparison for our new quantitative restricted phospholipid transfer (qRPT) imaging method, discussed below.
2.2 |. Quantitative restricted phospholipid transfer imaging
Although conventional MT techniques provide enhanced sensitivity for measuring small pools, they are usually not specific to particular target molecules because the MT signals depend on multiple other tissue parameters, including water relaxation rates and MT with spectrally broad solidlike components in tissues. MTRasym partially addresses these issues and has been extensively applied to amide studies. However, it only partially removes the direct saturation or solid-like MT effects and also has contributions from rNOE effects and solid pool asymmetry about the water resonance. Therefore, the quantification and interpretation of the MT contrast is challenging. Here, we propose a quantitative MT imaging method that is more specific to the target molecules than conventional asymmetric analysis and apply it to the imaging of ordered Cho phospholipids. This quantitative MT method in phospholipid imaging was named qRPT imaging to distinguish it from the more qualitative RPT signal apparent in the dip in the Z-spectrum at −1.6 ppm.
For the purpose of our qRPT study in biological tissue, we separate tissue protons into 3 coupled pools: water, phospholipid, and other solid-like protons. During irradiation at the frequency offset of the phospholipid Cho methyl protons, the steady-state z component of water proton magnetization (Mz) is influenced by rNOE cross-relaxation from the phospholipid pool (fnknw), water proton longitudinal relaxation (rate R1 = 1/T1), direct saturation (coefficient η, which is a function of B1, RF offset, and the sample parameters, most notably T2), and a solid component MT effect (fmkmw). In an approximation of the steady state, assuming complete saturation of the phospholipid pool and the solid component MT pool, and incorporating all B1 effects on the water magnetization into the η term,
| (1) |
| (2) |
Here, fn and fm are the concentrations of the phospholipid pool and the solid component MT pool, respectively. knw is the rNOE cross-relaxation rate between the phospholipid pool and the water pool. kmw is the cross-relaxation rate between the solid component MT pool and the water pool. M0 is the equilibrium magnetization. Equation (2) illustrates that MT signals depend not only on the parameters related to phospholipids (fn, knw) but also multiple other tissue parameters related to water and the solid component MT pool (R1, fm, kmw). Asymmetric analysis (MTRasym) with subtraction of the signals acquired with an irradiation pulse on solute (label) and the symmetrically opposite side of the water peak (reference) has been used to quantify CEST effects. However, Equation (3) shows that this approach cannot fully remove the influence of water relaxation and solid component MT effects.
| (3) |
Here, Sref and Slab are the signals acquired in the reference and label scan, respectively. S0 is the signal acquired with no irradiation. Mz_ref and Mz_lab are the z components of water proton magnetization in the reference and label scan, respectively. This result is problematic because the MTRasym result lacks solute specificity. For example, it is sensitive to changes in the water T1 (≡ 1/R1) and T2 via the nontrivial functional dependence of Equation (3) and η, respectively. (Note that we are ignoring the further complicating effects of asymmetric macromolecular contributions because we will ultimately settle on a metric that has little sensitivity to this phenomenon due to its use of a single frequency for label and reference scans.)
A recently derived27 inverse subtraction approach (MTRRex) can remove the influence of water relaxation and the solid component MT effect. Equation (4) illustrates how the inverse subtraction approach operates. Equation (4) is not fully accurate because of inherent approximations, but it is enough to show the advantages of the inverse approach in removing water relaxation and solid component MT.
| (4) |
However, to quantify the rNOE, this method also requires a reference acquired on the other side of water, which has influences from CEST effects (eg, hydroxyl and amine-water exchange effects at around 1 and 2 ppm, respectively) and asymmetry in the solid proton pool relative to the water resonant frequency. To solve this problem, we describe a new method that combines the inverse approach with our previously developed28 chemical exchange rotation transfer (CERT) metric MTRdouble.
We have used CERT to quantify amide proton transfer without using asymmetric analysis. Supporting Information Figure S1 shows the sequence diagram of CERT, which contains 2 pulsed-MT sequences with the same average irradiation power (Bavg power) but different irradiation flip angles (π or 2π). This can be achieved by varying the pulse duration and delay time.29 For example, a pulsed-MT sequence with an average power of 1 microtesla μT) tesla (T) has a π pulse duration of 10.8 ms, a duty cycle of 50%, a pulse duration of 21.6 ms, and a duty cycle of 50% for 2π pulses.28,30 The signals acquired with π pulses (Sπ and 2π pulses (S2π) are similar to the label scan and reference scan, respectively, in MTRasym, but with the significant advantage of being acquired at a single frequency offset. Hence, MTRdouble avoids several artifacts that originate in the signal dependence on the irradiation frequency offset. Each pulsed-MT sequence contains a series of Gaussian irradiation pulses followed by data acquisition. After each pulse, a crusher gradient was applied to spoil residual transverse magnetization. MTRdouble is calculated using,28,30
| (5) |
Here, we modify the MTRdouble metric by taking the difference of the inverses of the signals. Hence, we get both the benefit of using a single frequency (eg, MTRdouble) and canceling T1 and MT effects (eg, MTRRex), which potentially provides a more specific phospholipid signal. Equation (6) describes the qRPT imaging analysis.
| (6) |
2.3 |. Numerical simulations
To evaluate the influence of multiple tissue parameters on qRPT contrast, simulations were performed with a 3-pool model, which contains a phospholipid pool (the solute pool), a background solid component, and water. Z-spectrum, RPT spectrum and contrast, and MTRasym spectrum and contrast were numerically calculated for a range of sample parameters. We varied the cross-relaxation rate between phospholipid Cho methyl protons and water (knw) (5, 10, 15, 20, 25 Hz), Cho phospholipid fractional population (fs) (0.01, 0.02, 0.03, 0.04, 0.05), water T1 (0.5, 1.0, 1.5, 2.0, 2.5 s), water T2 (30, 60, 90, 120, 150 ms), solid component concentration (fm) (0.03, 0.06, 0.09, 0.12, 0.15), Cho phospholipid Cho methyl proton longitudinal relaxation (T1n) (0.5, 1.0, 1.5, 2.0, 2.5 s), Cho phospholipid Cho methyl proton T2n (5, 10, 15, 20, 25 ms), and solid component-water exchange rate (kmw) (10, 15, 20, 25, 30 s−1). Each parameter was varied individually, with all other parameters remaining at the value in bold. Other simulation parameters included solid component longitudinal and transverse relaxation (1.0 s and 15 μs), phospholipid Cho methyl proton offset of −1.6 ppm, and solid component offset of 0 ppm. Proton T1n and proton T2n are often unknown because they are hard to measure directly given their exchange with water. We choose the phospholipid Cho relaxation values in our simulations to roughly match typical values of metabolites.15,31 The relaxations of solid component were close to values measured previously.32
Simulations used Bloch equations modified with exchange or cross-relaxation terms, which can be written as , where A is a 7 × 7 matrix. The water and phospholipid pool each have 3 equations representing their x, y, and z components. NOE cross-relaxations occur only between the z components of water and phospholipid pools. The solid component MT pool has a single coupled equation representing the z component, with an additional term for saturation effects.32 A super-Lorentzian absorption lineshape (which better fits biological tissue33–35) was used for the solid component MT pool. All numerical simulations of qRPT signal integrated the differential equations through the pulse sequence using the ordinary differential equation solver in MatLab R2014 (MathWorks, Natick, MA).29 Spoiling was modeled by nulling the transverse components of the magnetization before and after each irradiation pulse. Bavg power and duty cycle were set to be 1 μT and 0.5, respectively.
2.4 |. Sample preparation
In order to assess the distinct contributions to the signal, samples with Cho-containing metabolites and other mobile metabolites were prepared using concentrations of 100 mM (except glutamate at 50 mM) in deionized water. Immobile metabolite samples were prepared with 20% (weight/weight) cross-linked bovine serum albumin (BSA) and 100 mM metabolites (except glutamate of 50 mM). Cross-linking was achieved by adding 250 μL of 25% glutaraldehyde to an ice bath-cooled solution of 10 mL aqueous BSA. The BSA sample was prepared by adding 20% (weight/weight) BSA to deionized water. Reconstituted phospholipids (egg PtdCho, DOPC, DSPC, SM, or phosphatidylethanolamine) were dissolved with or without cholesterol in chloroform. Chloroform was then removed under air stream and vacuum. The lipid was resuspended in water with the ratio of 1:3 (lipid:water) by weight. To ensure the lipid was fully dispersed, the samples were placed in a sonication bath for 30 min before 5 freeze/thaw cycles. Detailed lipid composition information is listed in Supporting Information Table S1. Ghost membranes were also prepared from human red blood cells.
2.5 |. Animal preparation
Eight Fischer rats bearing 9L tumors and one Wistar rat bearing a C6 tumor were immobilized and anesthetized with a 2%/98% isoflurane/oxygen mixture. Respiration was monitored to be stable, and a constant rectal temperature of 37°C was maintained throughout the experiments using a warmair feedback system (SA Instruments, Stony Brook, NY). All procedures were approved by the Animal Care and Use Committee at Vanderbilt University (Nashville, TN).
To develop intracranial tumors, rat glioma cells 9L and C6 were used with rats weighing (200-300) g. Briefly, general anesthesia was induced by isoflurane followed by intraperitonial injection of a ketamine (91 mg kg−1) and acepromazine (9.1 mg kg−1) mixture. A 10 μL suspension of 50 thousand 9L or C6 cells in phosphate-buffered saline was injected into the cortex at a depth of 2 mm with a Hamilton syringe and a 30-gauge needle using a stereotactic apparatus (3 mm lateral and 3 mm posterior to the bregma). These rats were subjected to MRI2 to 3 weeks after implantation of tumor cells.
2.6 |. MRI
For experiments on samples of reconstituted phospholipids, we acquired free induction decay signals without imaging. For Z-spectra on 8 rats bearing 9L tumors to study the repeatability of the MT signals at −1.6 ppm, a 2-shot spin-echo echo-planar imaging readout was used for acquisition. The FOV was 32 × 32 mm2, with a slice thickness of 2 mm, matrix of 96 × 96, number of acquisitions 2, continuous wave irradiation power (B1) of 1 μT, and receiver bandwidth 500 kHz. For high-resolution multiparametric MRI imaging on rats bearing C6 tumor, the FOV was 35 × 35 mm2, with a slice thickness of 2 mm, matrix of 128 × 128, number of acquisitions 10, and receiver bandwidth 250 kHz. Irradiation was achieved using a train of pulses with a Bavg power of 1.6 μT; duty cycle of 50 %; pulse repetition time and pulse duration of 13.6 ms and 6.8 ms for radians π, and 27.2 ms and 13.6 ms for radians 2π, respectively. The pulse number for radians π and 2π is 600 and 300, respectively.
In 9L rats, images were acquired with a 2-shot EPI, matrix of 96 × 96, average of 2 acquisitions, and FOV of 32 × 32 mm. RPT imaging was performed with a continuous wave-MT sequence with a B1 of 1.0 μT. In C6 rats, amide proton transfer images were calculated with asymmetric analysis (MTRasym26) using a 4-shot EPI, matrix of 128 × 128, and FOV of 32 × 32 mm. Both qRPT (see Appendix) and amide proton transfer imaging were performed with a Bavg power °f 1.6 μT and duty cycle of 50%.
1H MRS was obtained using the PRESS sequence with the following parameters: voxel size = 7 × 5 × 4 mm3 (which cover the phantom), spectral width = 4 kHz, number of points = 2048, number of acquisitions = 512, TE1 = 8 ms, TE2 = 7 ms, and TR = 3 s. Water suppression was achieved using the variable pulse power and optimized relaxation (VAPOR) delays method. Localized shimming was performed to obtain localized water line width values less than 0.05 ppm.
All measurements were performed on a Varian DirectDrive horizontal 9.4 T magnet with a 38-mm Litz RF coil (Doty Scientific Inc., Columbia, SC).
3 |. RESULTS
3.1 |. Numerical tests of qRPT sensitivity and specificity
Specificity of the quantitative qRPT contrast to Cho phospholipids has been validated through multimodel (water, Cho phospholipid, and solid component MT pools) numerical simulations of the Bloch equations. Figure 1 shows the simulated MT Z-spectra, quantitative qRPT spectra, and conventional MTRasym spectra. Supporting Information Table S2 lists the parameters used in the simulations. Figure 2 shows the plot of simulated quantitative qRPT contrast and conventional MTRasym contrast at 650 Hz (corresponding to −1.6 ppm at 9.4 T) with variations of fn, knw, T1, water transverse relaxation time (T2), fm, solute longitudinal relaxation time (T1s), solute transverse relaxation time (T2s), and semisolid-water coupling rate (kmw). It was found that our proposed quantitative qRPT contrast is proportional to the Cho phospholipid content fn and exchange rate knw and is roughly independent of other parameters, except the T2s, for example, doubling fn (0.02 to 0.04) versus T1 (1 to 2 s) changes qRPT by 98.6 % and −4.3 %, respectively. These relative dependencies are intuitively reasonable given that solute rotation is the essential phenomenon underlying qRPT. Both kmw and T2s influence the rotation effect of solutes. Although qRPT decreases with T1, it is very small. In contrast, the conventional MTRasym depends on all the above parameters. This simulation shows that qRPT contrast is a more specific method to detect ordered Cho phospholipids.
FIGURE 1.
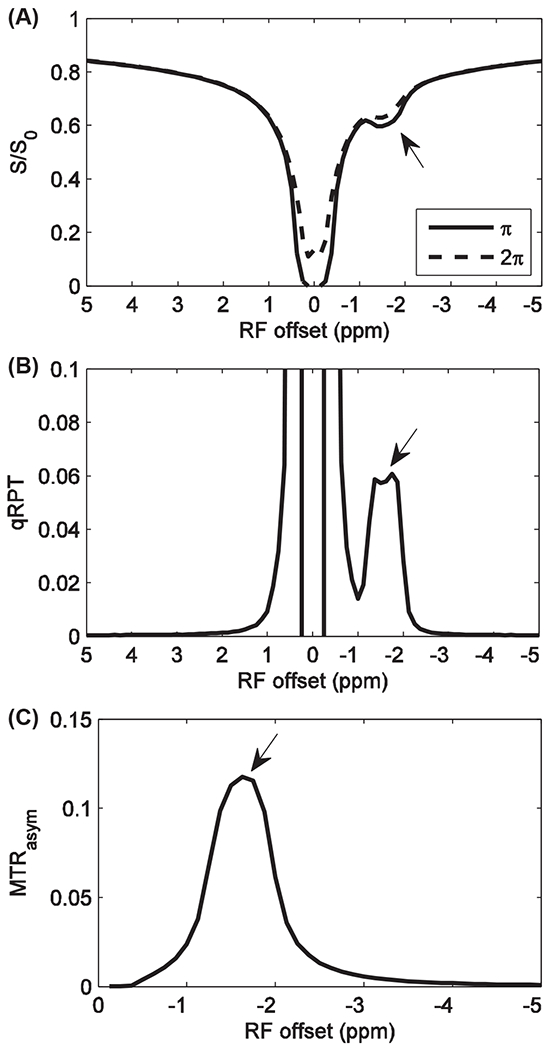
Simulated Z-spectra with irradiation flip angle of π and 2π (A), quantitative qRPT spectra (B), and conventional MTRasym spectra with irradiation flip angle of π (C). Bavg power = 1.0 μT, duty cycle is 50%. Bavg power, average irradiation power; MTRasym, asymmetric magnetization transfer ratio; qRPT, quantitative restricted phospholipid transfer; S/S0, signal/control signal; T, tesla
FIGURE 2.
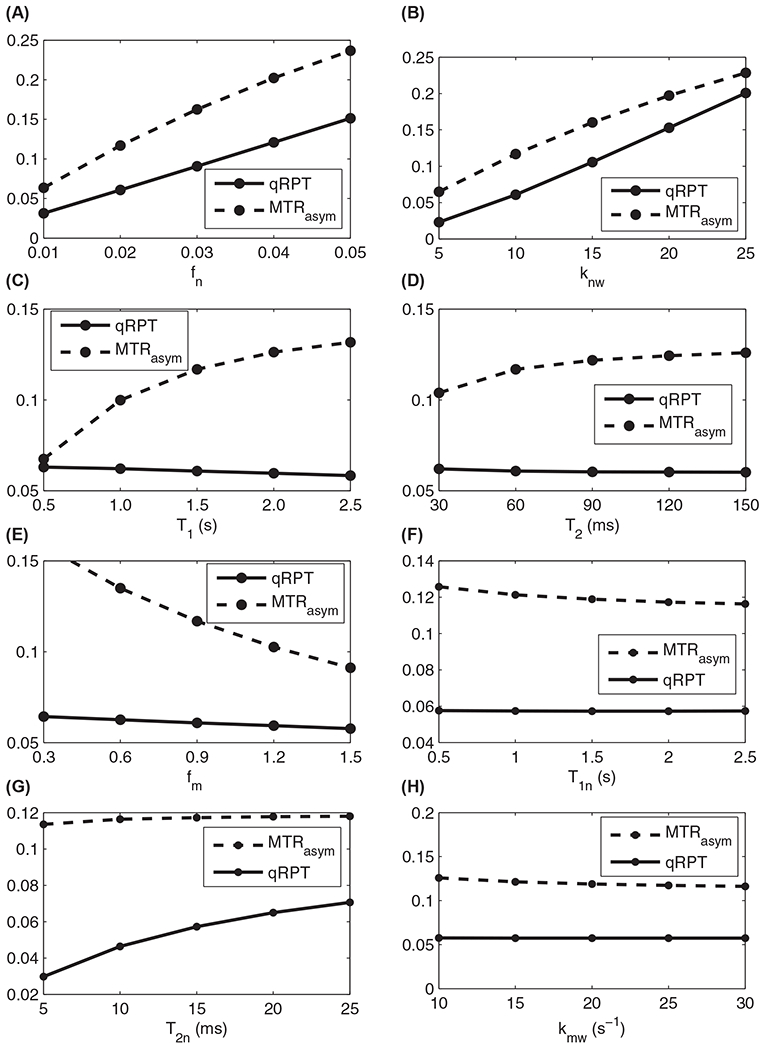
Simulated quantitative qRPT contrast and conventional MTRasym with variations of fn (A), knw (B), T1 (C), T2 (D), fm (E), T1n (F), T2n (G), and kmw (H)
3.2 |. Animal studies
Figure 3 shows the MT Z-spectra and quantitative qRPT spectra on a normal rat brain; it also shows how a mild dip can be seen in the Z-spectra from π pulses near −1.6 ppm and how the π and 2π spectra can be combined to calculate a corresponding qRPT spectra.
FIGURE 3.

Pulsed MT Z-spectra with irradiation flip angle of π and 2π (A) and qRPT spectra (B) on a normal rat brain tissue. Note that the peak at around −1.6 ppm is the quantitative qRPT contrast. The peaks observed at 3.5 and 2 ppm in (B) are from amide and amine in rat brain. Irradiation power was 1.0 μT, and duty cycle is 50%. MT, magnetization transfer
For tumor studies, we explored both qualitative RPT and more specific qRPT measures. For the continuous wave RPT studies, we acquired MT Z-spectra in vivo from 8 rat brains bearing 9L tumors (Figure 4). The MT Z-spectrum of normal tissue shows a distinct decrease of water proton magnetization near −1.6 ppm, whereas tumors show a much less pronounced effect.
FIGURE 4.
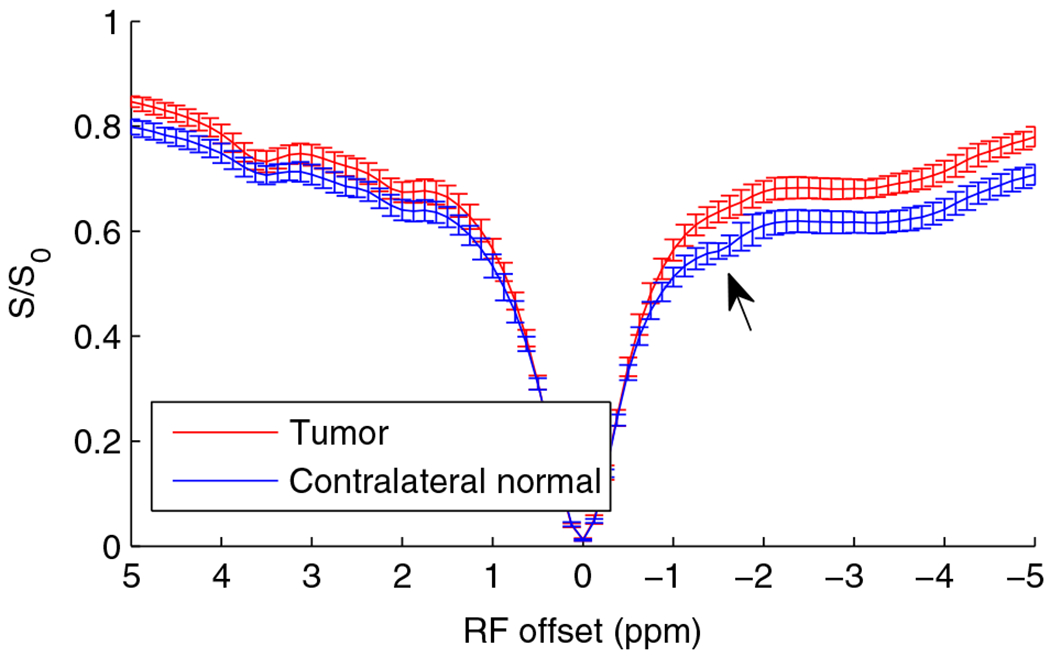
Z-spectra from experiments on 8 rat brains bearing 9L brain tumors (right hemisphere). Note the dip at around −1.6 ppm in the MT Z-spectra in normal tissue but the lack of a clear dip at −1.6 ppm in the tumor. Error bar is across subjects
The continuous wave Z-spectrum in Figure 4 indicates a sensitivity to chemical species that resonates near −1.6 ppm. Although the amplitude of this RPT decrease or dip in normal tissue can be measured (0.031 ± 0.011 in normal tissues and 0.014 ± 0.005 in tumors, giving a P value of 0.001), the resulting metric depends on the details of how the signal at the dip is compared to signals at other frequency offsets. (In our case, we took the maximum difference between the data and a spline interpolation. The spline fit excluded a gap of width 0.75 ppm and constrained location between −0.75 and −2.25 ppm offset from water, chosen by maximizing the spline difference with the signal dip). Furthermore, this approach is inaccurate because it is also affected by several confounding factors, such as direct water saturation, effects of water longitudinal relaxation rate 1/T1, and the presence of solid-like (ie, broad spectrum) coupled macromolecules. Our proposed approach, qRPT imaging, is a related but more specific measure of Cho phospholipid rNOE effects derived from pulsed Z-spectra, which avoids many of the confounds inherent to continuous wave Z-spectra. Figure 5 compares results of different high-resolution multiparametric MRI acquisitions from one C6 tumor-bearing rat brain. There is significant contrast between tumor and contralateral normal tissue for each MRI parameter examined, but each shares a different biophysical basis of varying specificity and sensitivity. Below, we report studies that illustrate the molecular origins of the RPT signals in tissues. (We also examine the experimental power dependence in Supporting Information Figure S2).
FIGURE 5.

Multiparametric MRI maps of a representative rat bearing C6 tumor model. (A) T2-weighted image, (B) T1 map, (C) T2 map, (D) ADC map, (E) CEST imaging at 3.5 ppm (aka APT imaging), and (F) qRPT imaging. Note the significant contrast between tumor and normal tissue in (F), which stems from the Cho phospholipids and may reflect the variation of phospholipid conformation and composition and membrane fluidity. (Phospholipids cause a peak in qRPT spectra but a dip in continuous wave spectra, as in Figure 1). APT, amide proton transfer
3.3 |. Samples showing sensitivity to Cho phospholipids
To explore how different chemical structures and dynamics affect the Cho MT signal, we acquired MT Z-spectra from samples containing Cho and water at 9.4 T (Figure 6A). A sign of NOE depends on the correlation time that characterizes the dipolar interaction between the interacting protons. Small positive rNOE-mediated saturation transfer effects at −1.6 ppm (arrow) were found in simple solutions of Cho, indicating that Cho is substantially unrestricted and correlation time is short. We interpret negative rNOE effects as found in rat brain as arising from molecules in a much more restricted state. (The use and meaning of “restricted” is addressed in the Discussion section, which follows, because it connects to the underlying molecular mechanisms. The present use does not imply a particular correlation time of Cho phospholipid protons but instead indicates that effective dipole–dipole cross-relaxation of the carbon-bound protons is possible as part of a chain of events allowing spin exchange. Possible constituents of this spin exchange chain are discussed below). Figure 6B shows MT Z-spectra from reconstituted phospholipids egg PtdCho and SM with cholesterol. Negative rNOE-mediated saturation transfer effects were found. The MT Z-spectrum for a ghost red blood cell, shown in Figure 6C, has a deviation near −1.6 ppm, which is consistent with assigning the dip to cell membranes, although the size of the observed dip is significantly smaller than that seen in simple reconstituted phospholipids.
FIGURE 6.

MT Z-spectra of sample containing 100 mM Cho and water at 37°C (A), egg PC, SM with presence of cholesterol (B), and ghost membrane (C). Note the positive rNOE in (A) indicating that Cho is in a less restricted state, whereas negative rNOE in (B and C) indicate that the Cho head group of the phospholipids is in a restricted state. Experiments were performed on 9.4T with CW-MT sequence with irradiation power (B1) of 0.25 μT in (A), 0.2 μT in (B), and 0.5 μT in (C). CW, continuous wave; egg PC, egg PtdCho; PtdCho, phosphatidylcholine; rNOE, relayed nuclear Overhauser enhancement; SM, sphingomyelin
We also studied some other potential contributions to the RPT signals. Taurine (Tau), other water-soluble Cho-containing metabolites including phosphocholine (Pcho), glycerylphosphorylcholine, acetylcholine, and the ethanolamine head group of phosphatidylethanolamine also have MRS signals with frequency offsets from water at around −1.6 ppm. Figure 7 shows the MT Z-spectra for those molecules. It was found that there are no signal contributions near −1.6 ppm from taurine, acetylcholine, and glycerylphosphorylcholine. Pcho contributes a small positive rNOE signal. There are negative rNOE signals at around −1.6 ppm from PtdEtn, which indicates possible contributions of phospholipids with non-Cho head groups. However, the signal is weak compared with the MT signals from the Cho phospholipids PtdCho and sphingomyelin. We also measured rNOE effects within a sample of BSA protein. A dip at around −1.9 ppm is found in Figure 7F, which could possibly contribute to the broad dip seen in vivo, although the in vivo effect is centered near the −1.6 ppm Cho phospholipid resonance. Hence, we believe contributions from peptides and proteins can be largely, but not totally,36,37 disregarded.
FIGURE 7.
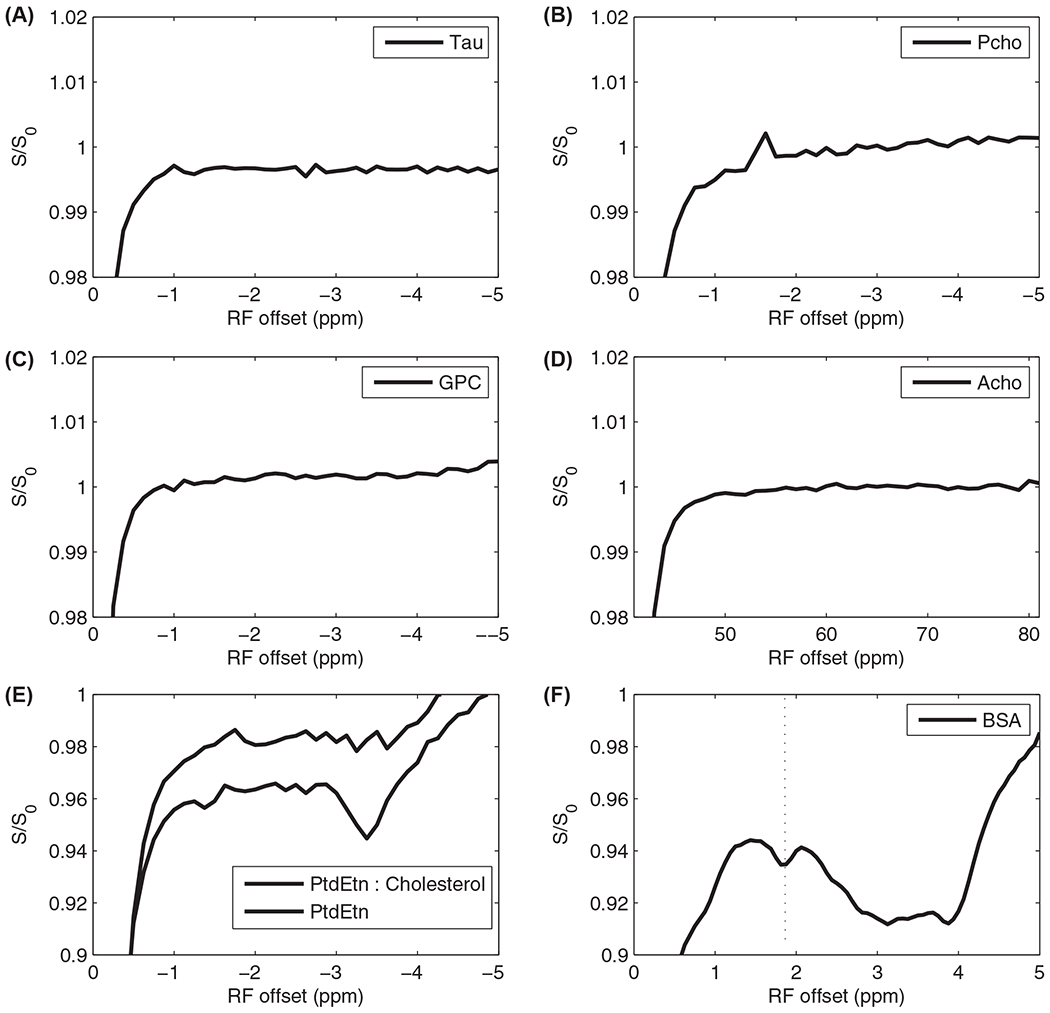
MT Z-spectra from samples containing taurine (A), Pcho (B), GPC (C), Acho (D), PtdEtn with presence and absence of cholesterol (E), and BSA (F). Note the lack of signals from taurine, GPC, and Acho. Pcho contributes positive NOE signal. There are MT signals at around −1.6 ppm from PtdEtn. However, the signal is weak and can be neglected compared with the MT signals from PtdCho and SM. There is an MT signal at around −1.9 ppm from BSA, which could possibly contribute to the broad dip seen in vivo, although the in vivo effect is centered near the −1.6 ppm Cho phospholipid resonance. Experiments were performed at 9.4 T with CW-MT sequence with B1 of 0.25 μT. All samples were measured at 37°C. Acho, acetylcholine; BSA, bovine serum albumin; CW, continuous wave; GPC, glycerylphosphorylcholine; PCho, pholphocholine; PtdEtn, phosphatidylethanolamine
3.4 |. Effects of cholesterol, saturation, chain length, and temperature on the RPT signal
Membranes in vivo are highly complicated systems with widely variable compositions of phospholipids, sterols, and proteins. Phospholipids themselves have varying degrees of unsaturated bonds and different aliphatic chain lengths. In addition, the cholesterol composition in membranes, a key component in membrane MT, is different in different cell types. To begin to explore in a systematic fashion how these different types of lipid compositions affect RPT, a number of different model systems were constructed.
Figure 8 shows the RPT dependence on lipid characteristics. Figure 8A shows the MT Z-spectra of DOPC:Cholesterol and DEPC:Cholesterol, which have the same degree of saturation but varying chain length. (DOPC has 18 carbons, and DEPC has 22 carbons.) Figure 8C compares DSPC: cholesterol and DOPC cholesterol, which have the same chain lengths but varying degrees of saturation. Figure 8E shows results from a collection of phospholipids (with cholesterol) at 25°C and 37°C. Figure 8G shows data from phospholipids (at 37°C) with and without cholesterol. Figure 8B,D,F and G plot the corresponding differences in the Z-spectra, revealing the underlying dependencies of the dip near −1.6 ppm. Note the small effects from varying chain length, larger effects from varying the degree of saturation and temperature, and very large effects from the inclusion of cholesterol. Note that RPT and its dependence on cholesterol is distinct from previous NOE measures of lateral organization in lipid species.38,39 Also note that these dependencies are likely relevant in complex mixture phospholipids. Appendix (see Appendix Figure A1) plots the temperature and cholesterol dependence of DOPC and eggPC, which shows a strong rNOE dependence on the presence of cholesterol and is composed of roughly 45% saturated PtdCho and 55% unsaturated PtdCho, and the acyl chain lengths are 35% and 60% for 16 and 18 carbons, respectively.
FIGURE 8.
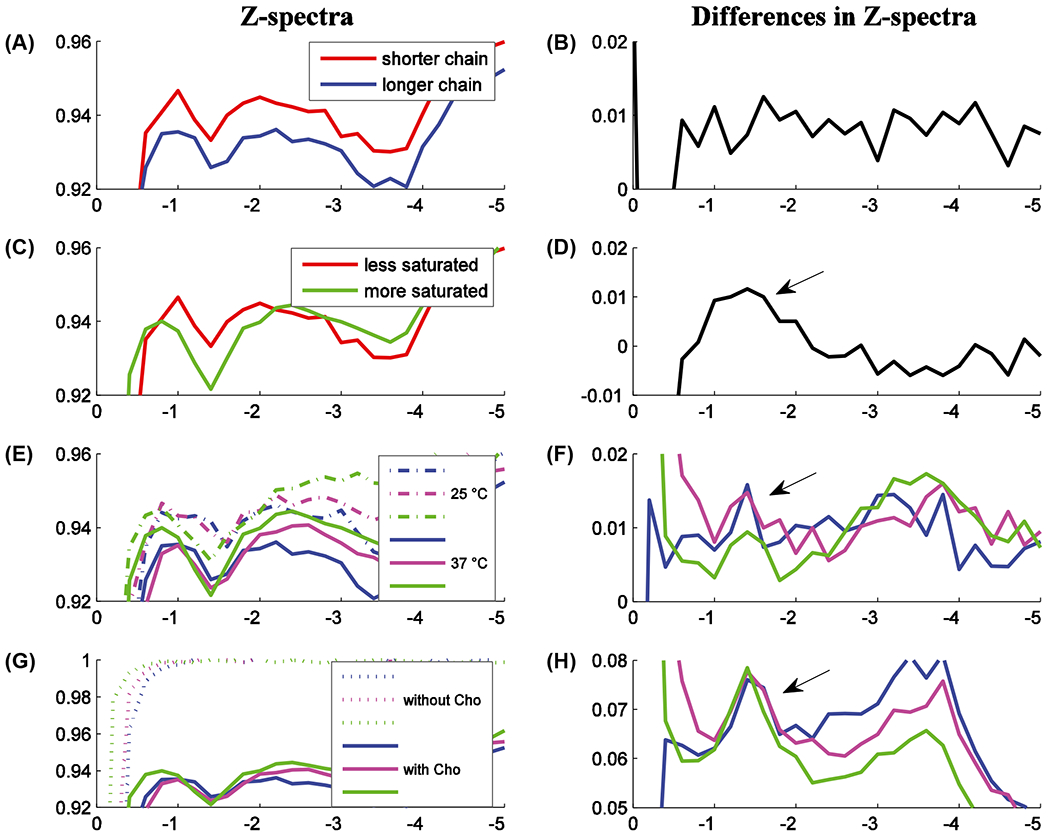
Phospholipid Z-spectra and Z-spectra differences. The samples were chosen to vary (A) chain length (DOPC 18 carbons, DEPC 22 carbons), (C) degree of saturation (less is DOPC, more is DSPC), (E) temperature (blue is DEPC, magenta is POPC, green is DSPC), and (G) inclusion of cholesterol (ditto). All measurements were at 37°C (except in [E]), and all samples include Cho (except in [G]). Figures (B), (D), (F), and (H) give corresponding Z-spectra differences, which reveal the Z-spectra sensitivity. Note the steadily increasing effect size from variations in chain length, temperature, degree of saturation, and (with very large effects) presence of Cho. The Appendix (see Appendix Figure Al) indicates similar dependencies on temperature and Cho for eggPC and DOPC, which were removed for visual clarity
4 |. DISCUSSION
The results above demonstrate that detecting and quantifying the rNOE-based signal dip near −1.6 ppm in RPT imaging is a novel approach for imaging distributions of restricted phospholipids in vivo caused by differences in biophysical characteristics related to rigidity, and hence it may vary with pathology and physiological state.40 Our studies of reconstituted lipid samples verify that these rNOE signals are sensitive to Cho phospholipids and depend on phospholipid composition, although they do not eliminate the possibility of other contributing factors, such as proteins, in vivo. This is consistent with previous magnetization transfer magic-angle spinning measures on reconstituted phospholipids.25 Likewise, previous MT studies on lactate and ethanol in cross-linked BSA have shown that NOEs from these metabolites occur concomitantly with BSA immobilization.41,42 PtdCho and sphingomyelin are the major phospholipid components of eukaryotic cells, accounting for approximately 60% of total phospholipids, and tend to be restrained in relatively rigid membrane structures. Therefore, it is plausible that the observed in vivo MT signals at around −1.6 ppm stem from the head groups of PtdCho and SM. A highly disordered membrane may not be rigid enough to contribute to the rNOE-mediated saturation transfer signals, whereas a more stabilized membrane behaves conversely. Thus, RPT potentially provides insights into biophysical variations not obtainable by other methods.
The proposed RPT imaging method likely has similarities to other measures of mobility, such as lateral diffusion, macromolecular order, and intralipid NOE effects. For example, Figure 8G and Appendix Figure 1C,D indicate a large RPT sensitivity to cholesterol, which reduces the lateral diffusion rate of phospholipids in lipid bilayers,43–48 increases the correlation time accordingly,39,49,50 and has been shown to be the major contribution to lipid NOE cross-relaxation.49 Note that the interplay between these measures is not trivial,51 and one distinct characteristic of RPT is the indirect measure of NOE effects via magnetization exchange with water. No signals show up in the Z-spectra without contacts between water and phospholipids. Membrane structure is crucial to this aspect and determines the accessibility of water.47
NMR relaxation studies combined with molecular modeling find a water translational diffusion constant of 70 ps at the membrane surface.52 This value is too short to create the negative NOE between water and lipids observed here. Therefore, additional mechanisms must be present that act to hinder or reduce the correlation time into the slow-motion regime. There are several possibilities for the mechanism by which the water couples to the −1.6 ppm resonance, and the process may involve multiple steps, similar to previous explanations of the coupling of water with solid-like macromolecules such as proteins and polysaccharides.53 Dipolar through-space interactions must play a role given the absence of chemical exchange with the carbon bound protons, but the exact mechanism and timing is unclear. One possible path is irradiation of restricted Cho protons, followed by through-space dipolar interaction with exchangeable protons on hydroxyl groups. Another route could be irradiation of motionally unrestricted Cho protons followed by bonding and spin exchange with rigid membrane protons, which then transfer magnetization to attached hydroxyl groups, similar to a proposed mechanism involving caffeine.54 Next, chemical exchange between water and hydroxyl protons in Cho would lead to the negative rNOE observed in our studies. This multi-step mechanism would be consistent with our observed dependence on membrane rigidity, and is similar to the mechanism behind a previous observation of an rNOE effect between methylene protons with short T2 (< 100 μs) and water protons.55 In that previous study, a relatively rigid membrane was needed to allow the methylene protons to serve as a spin reservoir in a magnetization transfer process with water. In the current study, the lipid spin reservoir involvement is mediated by the Cho headgroup.
Several studies have shown that membranes isolated from tumor cells present significant alterations in their composition and structural organization, such as cholesterol/phospholipid ratios, fatty acid composition of the phospholipids, and molecular order of the lipids.56,57 In addition, it previously has been reported that a main difference between normal and tumor cell plasma membranes is the status of the membrane fluidity.58 The RPT contrast in tumors shown here is consistent with our previous reports,59–61 and we here additionally avoided confounding factors by using the qRPT metric. The resulting contrast may tentatively be attributed to the variation of membrane fluidity regulated by phospholipid conformation and composition, along with decreased cell density. This change in fluidity likely overwhelms any contributions from changes in phospholipid Cho concentration changes,62 which typically increases in tumors.
Our rNOE results are consistent with and complementary to previous MRS results. Esclassan et al63 has detected a significant MRS signal (near −1.6 ppm from the water resonance) from membrane phospholipids in rat pancreas after stimulation by caerulein. This MRS signal was attributed to a change in conformation (but not content) of PtdCho under hormonal stimulation, allowing enhanced fluidity of membranes and higher mobility of the Cho head group. Several authors64,65 have also reported that phospholipids in rigid structures cannot be detected by MRS but are detectable only in membranes of low viscosity. However, our proposed approach preferentially detects Cho phospholipids in rigid structures, and hence provides complementary information by detecting MRS-invisible signals.66 This complementary relationship between conventional spectroscopy peaks and rNOE effects is not limited to Cho phospholipids but is a general consequence of the opposite effects that molecular motion has on these 2 detection methods. For example, cross-linked bovine serum albumin (BSA) provides a restricting environment for metabolites and water protons, and Figure 9 shows the opposite effects produced in MT Z-spectra and MRS. As an example of no change in solute restriction, in myo-inosito the immobilization does not create rNOE dips in the Z-spectrum and correspondingly has no effect on peaks in the MRS. However, in contrast, for glutamate immobilization it does create rNOE dips and correspondingly diminishes the MRS peaks.
FIGURE 9.

MT Z-spectra and 1H-MRS of immobile and mobile metabolites. Blue and red lines indicate mobile or immobile metabolites, respectively, in all figures. Immobilization does not always induce rNOE dips (as in Ins), but when it does (as in Glu), conventional MRS peaks at the corresponding frequency diminish, showing their complementary nature. Experiments for MT Z-spectra were performed with CW-MT sequence with B1 of 0.25 μT Experiments for 1H-MRS spectra were performed with PRESS on the same voxel. Glu, glutamate
The qRPT imaging analysis method is based on a previously developed apparent exchange-dependent relaxation (AREX) metric, which performs inverse subtraction of label and reference signals normalized by T1w.27,67,68 The goal is to increase the specificity of CEST imaging and is motivated by the analytic form of the steady state signal.27 However, the appropriate T1 normalization of metrics (and the resulting change in image contrast) is still an active area of research.69 Further, using this metric does not fully eliminate confounding and nonspecific residual signal from water, as shown in Figure 1. This residual signal grows with saturation power. Future work may optimize the power to maximize phospholipid specific signal while minimizing this contamination. Finally, although qRPT is designed to maximize specificity, its magnitude is roughly half that of MTRasym (see Figure 2), making sensitivity a key limitation. However, because the metric requires only 3 acquisition points (label, reference, and control), applying multiple averages is a viable approach to increasing sensitivity for clinical translation.
5 |. CONCLUSION
Phospholipids are major tissue components that play important role in regulating multiple disorders. However, phospholipids cannot be measured in vivo by using current imaging techniques. Here, we describe a novel nuclear Overhauser enhancement-mediated MRI signal with sensitivity to Cho phospholipids, which further depends on fluidity and composition (most notably cholesterol) and may prove useful as a probe of phospholipid conformation and composition. This sensitivity has been demonstrated on phantoms. In vivo results, although exhibiting a similar response near −1.6 ppm, may have additional contributions to the signal.
Supplementary Material
Funding information
NIH, Grant Sponsor: R21 EB017873, R01 EB017767, R01 CA184693, T32 EB001628, R01 CA109106, K25 CA168936, R01 EB001744
APPENDIX
ADDITIONAL RESULTS AND DISCUSSIONS
Appendix Figures A1 plots the Z-spectra and Z-spectra differences of eggPC (blue) and DOPC (red). These results are very similar to the results in manuscript Figures 10E-H and were moved to the appendix only to avoid visually confusing line overlaps.
Among our experiments, rat-to-rat variations in the size of the rNOE dip are much larger than the noise in any one measurement, and the dips from some rats are weak and can not be clearly found on the Z-spectra. This might be caused by the membrane composition and order that vary depending on their physiological stages.40
Supporting Information Figure S2 shows that the optimized saturation power is 1 μT.
FIGURE A1.

The Z-spectra and Z-spectra difference of EggPC (blue) and DOPC (red). These results are very similar to the results in manuscript Figures 8E–H and were moved to the appendix only to avoid visually confusing line overlaps. The difference plots (B,D) reveal the dependency of the peak near −1.6 ppm on temperature (small) and Cho (large). (Figure (A) samples included Cho and figure (C) measurements were performed at 37°C)
Footnotes
SUPPORTING INFORMATION
Additional Supporting Information may be found online in the Supporting Information section.
REFERENCES
- 1.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–150. [DOI] [PubMed] [Google Scholar]
- 2.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008:9:112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wymann MP, Schneiter R. Lipid signalling in disease. Nat Rev Mol Cell Biol. 2008;9:162–176. [DOI] [PubMed] [Google Scholar]
- 4.Horwitz AF, Horsley WJ, Klein MP. Magnetic-resonance studies on membrane and model membrane systems—proton magnetic-relaxation rates in sonicated lecithin dispersions. Proc Natl Acad Sci USA. 1972:69:590–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meneses P, Navarro JN, Glonek T. Algal phospholipids by 31P NMR: comparing isopropanol pretreatment with simple chloroform/methanol extraction. Int J Biochem. 1993;25:903–910. [DOI] [PubMed] [Google Scholar]
- 6.Estrada R, Stolowich N, Yappert MC. Influence of temperature on 3 IP NMR chemical shifts of phospholipids and their metabolites I. In chloroform-methanol-water. Anal Biochem. 2008;380:41–50. [DOI] [PubMed] [Google Scholar]
- 7.Komoroski RA, Pearce JM, Griffin WST, Mrak RE, Omori M, Karson CN. Phospholipid abnormalities in postmortem schizophrenic brains detected by P-31 nuclear magnetic resonance spectroscopy: a preliminary study. Psychiatry Res Neuroimaging. 2001;106:171–180. [DOI] [PubMed] [Google Scholar]
- 8.Sotirhos N, Herslof B, Kenne L. Quantitative analysis of phospho-lipids by P-31 NMR. J Lipid Res. 1986;27:386–392. [PubMed] [Google Scholar]
- 9.Merchant TE, Glonek T. P-31 NMR of phospholipid glycerol phosphodiester residues. J Lipid Res. 1990:31:479–486. [PubMed] [Google Scholar]
- 10.Meneses P, Glonek T. High-resolution P-31 NMR of extracted phospholipids. J Lipid Res. 1988;29:679–689. [PubMed] [Google Scholar]
- 11.Metz KR, Dunphy LK. Absolute quantitation of tissue phospholipids using P-31 NMR spectroscopy. J Lipid Res. 1996;37:2251–2265. [PubMed] [Google Scholar]
- 12.Wilhelm MJ, Ong HH, Wehrli SL, et al. Direct magnetic resonance detection of myelin and prospects for quantitative imaging of myelin density. Proc Natl Acad Sci USA. 2012;109:9605–9610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou JY, Payen JF, Wilson DA, Traystman RJ, van Zijl PCM. Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI. Nat Med. 2003;9:1085–1090. [DOI] [PubMed] [Google Scholar]
- 14.Zhou JY, Tryggestad E, Wen ZB, et al. Differentiation between glioma and radiation necrosis using molecular magnetic resonance imaging of endogenous proteins and peptides. Nat Med. 2011;17:130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai KJ, Haris M, Singh A, et al. Magnetic resonance imaging of glutamate. Nat Med. 2012;18:302–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haris M, Singh A, Cai KJ, et al. A technique for in vivo mapping of myocardial creatine kinase metabolism. Nat Med. 2014;20:209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haris M, Cai KJ, Singh A, Hariharan H, Reddy R. In vivo mapping of brain myo-inositol. Neuroimage. 2011;54:2079–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zu Z, Li H, Xie J, Xu J, Gore JC, Gochberg DF. MR imaging of membrane choline phospholipids. Poster Presentation at Chemical Exchange Saturation Transfer (CEST) 2014 Workshop, Colleretto Giacosa, Italy, 2014. [Google Scholar]
- 19.Chen JH, Sambol EB, DeCarolis P, et al. High-resolution MAS NMR spectroscopy detection of the spin magnetization exchange by cross-relaxation and chemical exchange in intact cell lines and human tissue specimens. Magn Reson Med. 2006:55: 1246–1256. [DOI] [PubMed] [Google Scholar]
- 20.Mori S, Abeygunawardana C, van Zijl PCM, Berg JM. Water exchange filter with improved sensitivity (WEX II) to study solvent-exchangeable protons. Application to the consensus zinc finger peptide CP-I. J Magn Reson. 1996:110:96–101. [DOI] [PubMed] [Google Scholar]
- 21.Mori S, Abeygunawardana C, Berg JM, van Zijl PCM. NMR study of rapidly exchanging backbone amide protons in staphylococcal nuclease and the correlation with structural and dynamic properties. J Am Chem Soc. 1997;119:6844–6852. [Google Scholar]
- 22.Zhou J, Wilson DA, Sun PZ, Klaus JA, van Zijl PCM. Quantitative description of proton exchange processes between water and endogenous and exogenous agents for WEX, CEST, and APT experiments. Magn Reson Med. 2004;51:945–952. [DOI] [PubMed] [Google Scholar]
- 23.van Zijl PCM, Zhou J, Mori N, Payen JF, Wilson D, Mori S. Mechanism of magnetization transfer during on-resonance water saturation. A new approach to detect mobile proteins, peptides, and lipids. Magn Reson Med. 2003;49:440–449. [DOI] [PubMed] [Google Scholar]
- 24.Chen JH, Sambol EB, Kennealey PT, et al. Water suppression with-out signal loss in HR-MAS H-1 NMR of cells and tissues. J Magn Reson. 2004;171:143–150. [DOI] [PubMed] [Google Scholar]
- 25.Avni R, Mangoubi O, Bhattacharyya R, Degani H, Frydman L. Magnetization transfer magic-angle-spinning z-spectroscopy of excised tissues. J Magn Reson. 2009;199:1–9. [DOI] [PubMed] [Google Scholar]
- 26.Zhou JY, van Zijl PCM. Chemical exchange saturation transfer imaging and spectroscopy. Prog NMR Spectrosc. 2006;48:109–136. [Google Scholar]
- 27.Zaiss M, Bachert P. Exchange-dependent relaxation in the rotating frame for slow and intermediate exchange: modeling off-resonant spin-lock and chemical exchange saturation transfer. NMR Biomed. 2013;26:507–518. [DOI] [PubMed] [Google Scholar]
- 28.Zu ZL, Janve VA, Xu JZ, Does MD, Gore JC, Gochberg DF. A new method for detecting exchanging amide protons using chemical exchange rotation transfer. Magn Reson Med. 2013;69:637–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zu ZL, Li K, Janve VA, Does MD, Gochberg DF. Optimizing pulsed-chemical exchange saturation transfer imaging sequences. Magn Reson Med. 2011;66:1100–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zu ZL, Janve VA, Li K, Does MD, Gore JC, Gochberg DF. Multi-angle ratiometric approach to measure chemical exchange in amide proton transfer imaging. Magn Reson Med. 2012;68:711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun PZ, Wang EF, Cheung JS, Zhang XA, Benner T, Sorensen AG. Simulation and optimization of pulsed radio frequency irradiation scheme for chemical exchange saturation transfer (CEST) MRI-demonstration of pH-weighted pulsed-amide proton CEST MRI in an animal model of acute cerebral ischemia. Magn Reson Med. 2011;66:1042–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li K, Zu ZL, Xu JZ, et al. Optimized inversion recovery sequences for quantitative T(1) and magnetization transfer imaging. Magn Reson Med. 2010;64:491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sled JG, Pike GB. Quantitative interpretation of magnetization transfer in spoiled gradient echo MRI sequences. J Magn Reson. 2000;145:24–36. [DOI] [PubMed] [Google Scholar]
- 34.Tozer D, Ramani A, Barker GJ, Davies GR, Miller DH, Tofts PS. Quantitative magnetization transfer mapping of bound protons in multiple sclerosis. Magn Reson Med. 2003;50:83–91. [DOI] [PubMed] [Google Scholar]
- 35.Morrison C, Stanisz G, Henkelman RM. Modeling magnetization-transfer for biological-like systems using a semisolid pool with a super-lorentzian lineshape and dipolar reservoir. J Magn Reson B. 1995;108:103–113. [DOI] [PubMed] [Google Scholar]
- 36.Morrison C, Henkelman RM. A model for magnetization-transfer in tissues. Magn Reson Med. 1995;33:475–482. [DOI] [PubMed] [Google Scholar]
- 37.Desmond KL, Moosvi F, Stanisz GJ. Mapping of amide, amine, and aliphatic peaks in the CEST spectra of murine xenografts at 7T. Magn Reson Med. 2014;71:1841–1853. [DOI] [PubMed] [Google Scholar]
- 38.Ulrich EL, Akutsu H, Doreleijers JF, et al. BioMagResBank. Nucleic Acids Res. 2008;36(suppl 1):D402–D408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huster D, Gawrisch K. NOESY NMR crosspeaks between lipid headgroups and hydrocarbon chains: spin diffusion or molecular disorder? J Am Chem Soc. 1999;121:1992–1993. [Google Scholar]
- 40.Huster D, Arnold K, Gawrisch K. Investigation of lipid organization in biological membranes by two-dimensional nuclear Overhauser enhancement spectroscopy. J Phys Chem B. 1999;103:243–251. [Google Scholar]
- 41.Delaat SW, Vandersaag PT, Shinitzky M. Microviscosity modulation during cell-cycle of neuroblastoma-cells. Proc Natl Acad Sci USA. 1977;74:4458–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swanson SD. Protein mediated magnetic coupling between lactate and water protons. J Magn Reson. 1998;135:248–255. [DOI] [PubMed] [Google Scholar]
- 43.Estilaei MR, Matson GB, Meyerhoff DJ. Indirect imaging of ethanol via magnetization transfer at high and low magnetic fields. Magn Reson Med. 2003;49:755–759. [DOI] [PubMed] [Google Scholar]
- 44.Almeida PFF, Vaz WLC, Thompson TE. Lateral diffusion in the liquid phases of dimyristoylphosphatidylcholine/cholesterol lipid bilayers: a free volume analysis. Biochemistry. 1992;31:6739–6747. [DOI] [PubMed] [Google Scholar]
- 45.Filippov A, Orädd G, Lindblom G. The effect of cholesterol on the lateral diffusion of phospholipids in oriented bilayers. Biophys J. 2003;84:3079–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Filippov A, Orädd G, Lindblom G. Influence of cholesterol and water content on phospholipid lateral diffusion in bilayers. Langmuir. 2003;19:6397–6400. [Google Scholar]
- 47.Orädd G, Lindblom G. Lateral diffusion studied by pulsed field gradient NMR on oriented lipid membranes. Magn Reson Chem. 2004;42:123–131. [DOI] [PubMed] [Google Scholar]
- 48.Falck E, Patra M, Karttunen M, Hyvönen MT, Vattulainen I. Lessons of slicing membranes: interplay of packing, free area, and lateral diffusion in phospholipid/cholesterol bilayers. Biophys J. 2004;87:1076–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Korlach J, Schwille P, Webb WW, Feigenson GW. Characterization of lipid bilayer phases by confocal microscopy and fluorescence correlation spectroscopy. Proc Natl Acad Sci USA. 1999;96:8461–8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yau W-M, Gawrisch K. Lateral lipid diffusion dominates NOESY cross-relaxation in membranes. J Am Chem Soc. 2000;122:3971–3972. [Google Scholar]
- 51.Sułkowski WW, Pentak D, Nowak K, Sułkowska A. The influence of temperature, cholesterol content and pH on liposome stability. J Mol Struct. 2005;744:737–747. [Google Scholar]
- 52.Jacobs RE, Oldfield E. NMR of membranes. Prog NMR Spectrosc. 1980;14:113–136. [Google Scholar]
- 53.Victort KG, Korb JP, Bryant RG. Translational dynamics of water at the phospholipid interface. J Phys Chem B. 2013;117:12475–12478. [DOI] [PubMed] [Google Scholar]
- 54.Edzes HT, Samulski ET. The measurement of cross-relaxation effects in the proton NMR spin-lattice relaxation of water in biological systems: hydrated collagen and muscle. J Magn Reson. 1978;31:207–229. [Google Scholar]
- 55.Yadav N, Yang X, Li YG, Li WB, Liu GS, van Zijl PC. Detection of dynamic substrate binding using MRI. In Proceedings of the 25th Annual Meeting of ISMRM, Honolulu, HI, 2017. p. 3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horch RA, Gore JC, Does MD. Origins of the ultrashort-T-2 H-1 NMR signals in myelinated nerve: a direct measure of myelin content? Magn Reson Med. 2011;66:24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sherbet GV. Membrane fluidity and cancer metastasis. Exp Cell Biol. 1989;57:198–205. [DOI] [PubMed] [Google Scholar]
- 58.Podo F Tumour phospholipid metabolism. NMR Biomed. 1999;12:413–439. [DOI] [PubMed] [Google Scholar]
- 59.Hendrich AB, Michalak K. Lipids as a target for drugs modulating multidrug resistance of cancer cells. Curr Drug Targets. 2003;4:23–30. [DOI] [PubMed] [Google Scholar]
- 60.Zu ZL, Xu JZ, Li H, et al. Imaging amide proton transfer and nuclear Overhauser enhancement using chemical exchange rotation transfer (CERT). Magn Reson Med. 2014;72:471–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang XY, Wang F, Jin T, et al. MR imaging of a novel NOE-mediated magnetization transfer with water in rat brain at 9.4T. Magn Reson Med. 2017;78:588–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu JZ, Zaiss M, Zu ZL, et al. On the origins of chemical exchange saturation transfer (CEST) contrast in tumors at 9.4T. NMR Biomed. 2014;27:406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Doblas S, He T, Saunders D, et al. In vivo characterization of several rodent glioma models by 1H MRS. NMR Biomed. 2012;25: 685–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cheminthomas C, Esclassan J, Palevody C, Hollande E. Characterization of a specific signal from human pancreatic tumors heterotransplanted into nude-mice: study by high-resolution H-1-NMR and HPLC. Int J Pancreatol. 1993;13:175–185. [DOI] [PubMed] [Google Scholar]
- 65.Mountford CE, Grossman G, Gatenby PA, Fox RM. High-resolution proton nuclear magnetic-resonance: application to the study of leukemic lymphocytes. Br J Cancer. 1980;41:1000–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meml AH, Nichols JW. Techniques for studying phospholipid membranes. In: Kuo JF, ed. Phospholipids and Cellular Regulation. Vol. 1. Boca Raton, FL: CRC Press; 1985. [Google Scholar]
- 67.Leibfritz D, Dreher W. Magnetization transfer MRS. NMR Biomed. 2001;14:65–76. [DOI] [PubMed] [Google Scholar]
- 68.Zaiss M, Bachert P. Chemical exchange saturation transfer (CEST) and MR Z-spectroscopy in vivo: a review of theoretical approaches and methods. Phys Med Biol. 2013;58:R221–R269. [DOI] [PubMed] [Google Scholar]
- 69.Zaiss M, Xu JZ, Goerke S, et al. Inverse Z-spectrum analysis for spillover-, MT-, and T-1-corrected steady-state pulsed CEST-MRI: application to pH-weighted MRI of acute stroke. NMR Biomed. 2014;27:240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


