Abstract
Plant hormones, produced in response to environmental stimuli, regulate almost all aspects of plant growth and development. Ethylene is a gaseous plant hormone that plays pleotropic roles in plant growth, plant development, fruit ripening, stress responses, and pathogen defenses. After decades of research, the key components of ethylene signaling have been identified and characterized. Although the molecular mechanisms of the sensing of ethylene signal and the transduction of ethylene signaling have been studied extensively, how chromatin influences ethylene signaling and ethylene response is a new area of research. This review describes the current understanding of how chromatin modifications, specifically histone acetylation, regulate ethylene signaling and the ethylene response.
Keywords: chromatin, ethylene response, ethylene signaling, histone acetylation, transcription
1. Introduction to Ethylene Signaling
Ethylene, a gaseous plant hormone, is important for a myriad of physiological and developmental processes including seed germination, plant growth, fruit ripening, organ abscission, and senescence. It is also involved in the responses to stresses such as drought, cold, flooding, and infection.[1,2] The common aquatic ancestor of plants, which existed about 450 million years ago, possessed an ethylene signaling pathway that was similar to that of modern Arabidopsis.[3] The typical “triple response phenotype” of Arabidopsis seedlings grown in the dark and treated with ethylene has enabled scientists to identify genes that when mutated cause hyper- and hyposensitive responses to ethylene.[4–6]
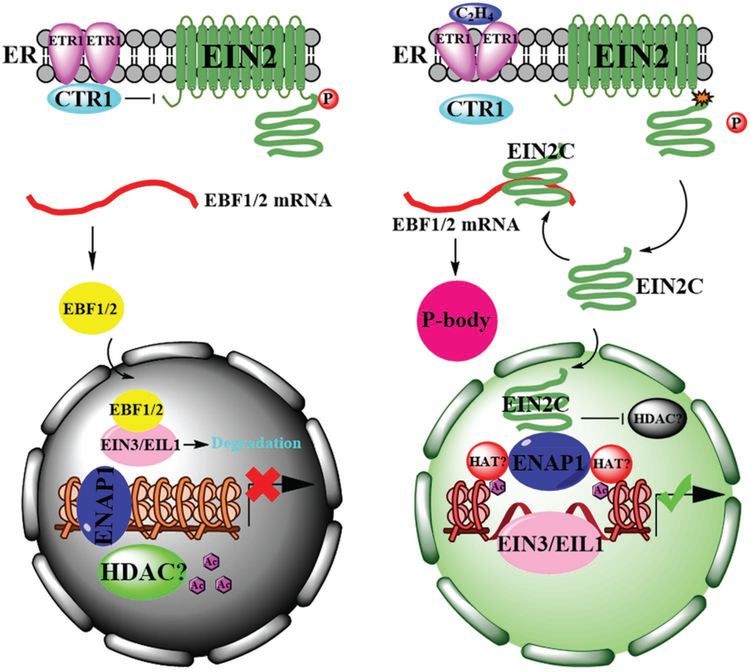
A model of ethylene signaling pathway has been established based on decades of genetic and molecular biology research (Figure 1). In brief, ethylene is perceived by five receptors anchored to the endoplasmic reticulum (ER): ethylene response1 (ETR1), ETR2, ethylene response sensor1 (ERS1), ERS2, and ethylene insensitive4 (EIN4).[7–10] A copper transporter responsive-to-antagonist1 (RAN1) is required for both ethylene binding and the function of receptors.[11] reversion-to-ethylene sensitivity1 (RTE1) colocalizes with ETR1 to promote its function.[12] Upon the perception of ethylene, the functions of receptors and of the downstream factor constitutive triple response1 (CTR1) are repressed,[13] activating EIN2, which is another ER anchored protein.[14] The activated EIN2 transduces the ethylene signal to two main downstream transcription factors EIN3 and ein3-like1 (EIL1).[15] The activated EIN3 and EIL1 bind to the promoter regions of genes encoding downstream factors to regulate their expression.[16] In the absence of ethylene, EIN2 protein and EIN3 protein levels are regulated by F-box proteins.[17] Also, upon perception of ethylene, the cleaved EIN2 carboxy-terminal (C-terminal) domain translocates to the cytosol and binds to mRNAs that encode ein3-binding f box protein1 (EBF1) and EBF2. This complex moves to the processing body (P-body) where EBF1 and EBF2 are degraded by ethylene insensitive5 (EIN5), which is a 5′→3′ exoribonuclease that degrades these mRNAs in the presence of ethylene.[6,18–20] As EBF1 and EBF2 target EIN3 and EIL1 for degradation in the nucleus, inhibition of translation of EBF1 and EBF2 stimulate the ethylene response.
Figure 1.
A proposed model of how EIN2 controls signal transduction in response to ethylene. In the absence of ethylene (left panel), EIN2 is localized on the ER membrane and is constitutively phosphorylated by CTR1.[78] EBF1 and EBF2 proteins target EIN3 and EIL1 for degradation in the nucleus.[18,19] ENAP1 docks on chromatin and histone proteins are deacetylated potentially by HDACs in the nucleus, resulting in suppression of ethylene responses.[75] Upon the perception of ethylene (right panel), the activities of the receptor ETR1 and of CTR1 are suppressed, resulting in dephosphorylation of EIN2,[13,78] which leads to proteolytic cleavage and release of the EIN2 C-terminal domain (EIN2-C), which rapidly translocates to the nucleus and to the cytosol.[6,20,22] In the nucleus, EIN2-C interacts with ENAP1 and this complex likely recruits an unknown HAT to acetylate specific histone residues.[75] Simultaneously, the activities of HDACs are inhibited, leading to a relax chromatin status at ethylene-responsive loci, resulting in EIN3/EIL1-dependent transcription regulation.[70] In the cytosol, EIN2-C binds to the 3′ untranslated regions of EBF1 and EBF2 and targets these mRNAs to the P-body inhibiting the translation of EBF1 and EBF2.[6,20] This in turn promotes the accumulation of EIN3 and EIL1. Abbreviations: P, phosphorylation; Ac, acetyl group; C2H4, ethylene.
Among the many important factors that are involved in ethylene signaling, EIN2 is the key mediator of the signal from the ER to the nucleus. The Arabidopsis knockout ein2 mutant shows complete insensitivity in all examined ethylene-regulated responses.[14] The EIN2 gene encodes a 1294-amino acid protein, and EIN2 is localized to the ER membrane in the absence of ethylene. Its amino terminus (N-terminus) has a predicted 12-fold hydrophobic transmembrane domain with sequence similarity to the conserved NRAMP family of metal ion transporters,[14] although as of now, no metal transport activity has been shown for EIN2.[21] The C-terminus of EIN2 has a hydrophilic domain, and overexpression of the C-terminal domain activates the ethylene responses in both etiolated seedlings and light-grown Arabidopsis plants.[14,22]
Both genetic and molecular studies have demonstrated that EIN3 and EIL1 are the positive regulators that are necessary and sufficient for the ethylene response.[15,17,23] In the absence of nuclear-localized EIN3, plants are insensitive to ethylene both at the morphological and molecular levels.[15,17] The EIN3 binding motif was identified after analysis of the genes such as ERF1 and EDF2 that are highly upregulated by ethylene followed by validation using an electrophoresis mobility shift assay (EMSA).[16,24–29] Using the EMSA assay, EIN3 was shown to form a homodimer in the presence of DNA in vitro.[16] A number of transcription factors form homodimers or heterodimers, which increases specificity and affinity for certain DNA motifs. Whether dimerization is necessary for function of EIN3 in vivo is currently unknown.
2. Histone Acetylation in the Ethylene Response
In eukaryotes, transcription factor binding is mainly determined by the state of the genome packaging with specific structural proteins, mainly histones. A histone modification is a covalent post-translational modification (PTM) to histone proteins which includes methylation, phosphorylation, acetylation, ubiquitylation, and sumoylation. The PTMs made to histones can impact gene expression by altering chromatin structure or recruiting histone modifiers. The histone modifications involved in plant hormone signaling have been reviewed recently by Yamamuro et al.[30] The studies of chromatin regulation in ethylene signaling have focused primarily on histone acetylation regulation. In Arabidopsis, there are 12 histone acetyl transferases (HATs) classified into four families.[31] The HATs that are involved in particular plant hormone responses are listed in Table 1.
Table 1.
HATs that are involved in plant hormone responses.
| HAT family | Protein name | Species | Physical interacting partners | Plant hormone responsea) | Associated histone acetylation | Refs. |
|---|---|---|---|---|---|---|
| GNAT | GCN5 | Arabidopsis | N/A | Represses the ABA response | N/A | [32] |
| PRZ1 | Positively regulate gene expression in response to Auxin | H3K9, H3K14, H3K27, H4K8, and H4K12 | [33–37] | |||
| N/Ab) | together with CLV1 to suppress the ethylene response | H3K9 and H3K14 | [34] | |||
| ELP3 | Arabidopsis | ELP1, ELP2, ELP4, and ELP6 | Represses the ABA response | N/A | [38–40] | |
| N/A | Represses the Auxin response | H3K14 | [38] | |||
| N/A | Represses the ethylene response | N/A | [38] | |||
| N/A | Represses the JA response | N/A | [38] | |||
| OsHAG702 | Rice | N/A | OsHAG702 expression is elevated by ABA | N/A | [41] | |
| OsHAG703 | Rice | N/A | OsHAG703 expression is elevated by ABA | N/A | [41] | |
| HvGCN5 | Barley | N/A | HvGCN5 expression is elevated by ABA | N/A | [42] | |
| HvELP3 | Barley | N/A | HvELP3 expression is elevated by ABA | N/A | [42] | |
| MYST | OsHAM701 | Rice | N/A | OsHAM701 expression is elevated by ABA | N/A | [41] |
| HvMYST | Barley | N/A | OHvMYST expression is elevated by ABA | N/A | [42] | |
| HAC | HAC1 | Arabidopsis | N/A | Represses the GA response | N/A | [43] |
| N/A | Represses the ethylene response | N/A | [44] | |||
| HAC4 | Arabidopsis | N/A | Represses the ethylene response | N/A | [44] | |
| HAC5 | Arabidopsis | N/A | Represses the ethylene response | N/A | [44] | |
| OsHAC701 | Rice | N/A | OsHAC701 expression is elevated by ABA | N/A | [41] | |
| OsHAC703 | Rice | N/A | OsHAC703 expression is elevated by ABA | N/A | [41] | |
| N/A | OsHAC703 expression is reduced by SA | N/A | [41] | |||
| OsHAC704 | Rice | N/A | OsHAC704 expression is reduced by SA | N/A | [41] |
ABA, abscisic acid; JA, jasmonic acid; GA, gibberellic acid; SA, salicylic acid
N/A, not available.
A few studies have focused specifically on HATs that are involved in the ethylene response. For example, in Arabidopsis, GCN5 and CLV1 act together to repress the ethylene-induced genes expression.[34] The gcn5clv1 mutant displays a hypersensitive ethylene response phenotype.[34] The acetylation of H3K9 and H3K14 in the promoter regions of ethylene responsive genes are elevated in the gcn5clv1 mutant, and the elevation is associated with the upregulation of expression of these genes.[34] Thus, there is an anticorrelation between histone acetylation levels as well as expression of ethylene-regulated genes and the function of histone acetyltransferase GCN5 and CLV1. HAC1 and HAC5, from the HAC family, which are homologs of CBP/p300 from mammals,[31,71] also repress ethylene-regulated genes. The hac1hac5 double mutant has a constitutive triple response phenotype: root and hypocotyl elongation, an exaggerated apical hook, and a thickening of the hypocotyls.[44] It was expected that gene expression would be downregulated in the hac1hac5 double mutant due to the reduction of histone acetylation levels; however, the downstream ethylene responsive genes are elevated in hac1hac5 double mutant,[44] suggesting an indirect regulation of ethylene responsive genes by HAC1 and HAC5.
In addition to HATs, histone deacetylases (HDACs) are involved in responses of plants to hormones as summarized in Table 2. Several HDACs are implicated in the ethylene response. For example, levels of HDA19 are specifically elevated by ethylene treatment.[45] Interestingly, the expression of ethylene responsive gene ERF1 is increased in 35S:HDA19 transgenic plants, however, the levels of histone H3 acetylation in ERF1 gene are decreased in 35S:HDA19 transgenic plants, suggesting that HDA19 indirectly influences ERF1 gene expression. In general, acetylation neutralizes the positive charges of lysine residues and decreases the interaction between histone and DNA, leading to a more relaxed chromatin structure, which is associated with transcriptional activation. In contrast, deacetylation induces a compact chromatin structure, which is associated with transcriptional repression.[72–74] Notably, the studies mentioned above revealed anticorrelations between HAT or HDAC activity and histone acetylation levels or expression of the genes regulated by ethylene. This anticorrelation strongly suggests that either the effects of HATs and HDACs on expression of ethylene-regulated genes are indirect or that other regulatory mechanisms are involved.
Table 2.
HDACs that are involved in plant hormone responses.
| HDAC family | Protein name | Species | Physical interacting partners | Plant hormone responsea) | Histone acetylation sites | Refs. |
|---|---|---|---|---|---|---|
| RPD3/HDA1 | HDA6 | Arabidopsis | JAZ1, JAZ3, and JAZ9 | Represses the JA response | N/Ab) | [45–47] |
| EIN3/EIL1 | Represses the ethylene response | N/A | [47] | |||
| HDC1 | Repress the ABA response | H3K9 and H3K14 | [48,49] | |||
| HD2C | Represses the ABA response | N/A | [50] | |||
| BIN2 | Promotes the BR response | N/A | [51] | |||
| HDA9 | Arabidopsis | N/A | Auxin signaling is influenced by HDA9 | N/A | [52] | |
| HDA19 | Arabidopsis | N/A | JA | N/A | [45] | |
| SNL1 and SNL2 | Represses the ethylene response | H3K9, H3K14, and H3K18 | [45,53,54] | |||
| SAP18, ERF3, and ERF4 | AtSAP18, HDA19, and ERF3 act together to repress transcription of genes involved in ethylene response | N/A | [55] | |||
| SNL1 and SNL2 | Represses ethylene signaling to establish seed dormancy | H3K9 and H3K18 | [56] | |||
| WRKY38 and WRKY62 | Promotes SA-mediated disease resistance | H3K9 | [57,58] | |||
| SIN3 | Represses the ABA response | N/A | [59,60] | |||
| N/A | Represses the ABA response | N/A | [61] | |||
| HDC1 | Represses the ABA response | H3K9 and H3K14 | [49] | |||
| MSI1 | Represses the ABA response | H3K9 | [54] | |||
| SNL1 and SNL2 | Responses to ABA | H3K9, H3K14 and H3K18 | [53,54] | |||
| SNL1 and SNL2 | Promotes ABA signaling to establish seed dormancy | H3K9 and H3K18 | [56] | |||
| SNL1 and SNL2 | Represses Auxin levels during seed germination | H3K9 and H3K18 | [56] | |||
| BZR1 | Promotes BZR1- targeted genes expression | N/A | [56,62] | |||
| OsHDA705 | Rice | N/A | JA induces the accumulation of HDA705 | N/A | [63] | |
| HvHDAC1s | Barley | N/A | JA regulates HvHDAC1s genes expression | N/A | [64,65] | |
| HD2 | HD2A | Arabidopsis | N/A | Represses the ABA response | N/A | [50,66] |
| HD2B | Arabidopsis | N/A | HD2B expression is repressed by ABA | N/A | [50,66] | |
| HD2C | Arabidopsis | HDA6 | Represses the ABA response | H3K9 and H3K14 | [50,66,67] | |
| HD2D | Arabidopsis | N/A | HD2D expression is repressed by ABA | N/A | [50,66] | |
| OsHDT701 | Rice | N/A | Represses the ABA response | N/A | [63,68] | |
| N/A | Represses the GA response | N/A | [63,68] | |||
| OsHDT702 | Rice | N/A | OsHDT702 expression is induced by SA | N/A | [63,68] | |
| N/A | OsHDT702 expression is induced by JA | N/A | [63] | |||
| N/A | OsHDT702 expression is repressed by ABA | N/A | [63] | |||
| HvHDAC2–1 | Barley | N/A | HvHDAC2–1 expression is induced by JA | N/A | [65] | |
| N/A | HvHDAC2–1 expression is induced by ABA | N/A | [65] | |||
| N/A | HvHDAC2–1 expression is induced by SA | N/A | [65] | |||
| HvHDAC2–2 | Barley | N/A | HvHDAC2–2 expression is induced by JA | N/A | [65] | |
| N/A | HvHDAC2–2 expression is repressed by ABA | N/A | [65] | |||
| SIR2 | SRT2 | Arabidopsis | N/A | Suppresses SA biosynthesis | N/A | [69] |
| SRT2 | Arabidopsis | ENAP1 | Promotes the ethylene response | H3K9 | [70] |
JA, jasmonic acid; ABA, abscisic acid; BR, brassinosteroid; SA, salicylic acid; GA, gibberellic acid
N/A, not available.
Recent studies from our lab provide evidence that levels of H3K14Ac and H3K23Ac, but not the classical histone acetylation marks H3K9Ac, H3K18Ac, and H3K27Ac, in the promoters of ethylene-regulated genes are positively correlated with gene expression.[75,76] Interestingly, even though the levels of H3K9Ac are not regulated by ethylene, the levels of H3K9Ac in the promoters of ethylene upregulated genes are higher than in promoters of ethylene downregulated genes both with and without ethylene treatment.[75,76] Presumably, H3K9Ac is a pre-existing mark that labels genes regulated by ethylene, whereas the elevation of H3K14Ac and H3K23Ac is required for gene activation. In a recent study from our lab demonstrated that a low level of H3K9Ac over ethylene-repressed genes leads to a downregulation of gene expression. Two HDACs, SRT1 and SRT2, partially mediate the transcriptional repression by regulating the levels of H3K9 acetylation during ethylene signaling, and evidence suggests that these are direct regulators of the ethylene response.[70]
It is now well established that many of the effects exerted by transcription factors in eukaryotes are mediated through interactions with coregulators that modify the chromatin state, resulting in a more open (in case of activation) or closed conformation (in case of repression). These coactivators typically consist of (or recruit) chromatin modifier complexes that either displace or evict nucleosomes or covalently modify histones to loosen their interactions with DNA. The discovery of the regulation of histone acetylation in ethylene response is presumably the tip of an iceberg of chromatin regulation necessary for the responses to hormones such as ethylene.
3. EIN2 Mediates the Interplay between Ethylene Signaling and Histone Acetylation
The work of Zhang et al. revealed for the first time that H3K14Ac and H3K23Ac are positively associated with gene expression in response to ethylene and that the ethylene-induced change of histone acetylation is EIN2 dependent.[75] By ectopically expressing the EIN2 C-terminus in an ein2–5 mutant using the CRISPR/dCas9 system, Zhang et al. demonstrated that the EIN2 C-terminal domain directly regulates histone acetylation.[77] However, the EIN2 C-terminal domain has no previously characterized histone or a DNA binding domain. Furthermore, no binding of EIN2 to DNA was detected by standard chromatin immunoprecipitation coupled with quantitative PCR (ChIP-qPCR) or ChIP-seq protocols (unpublished data), suggesting that EIN2 does not bind DNA or histones directly. The question of how the EIN2 C-terminal domain regulates histone acetylation in response to ethylene was partially answered by the identification of ein2 nuclear associated protein1 (ENAP1), a histone binding protein that interacts with the EIN2 C-terminus as shown using ChIP–reChIP experiments.[77] In the presence of ethylene, the EIN2 C-terminal fragment is translocated into the nucleus where it interacts with ENAP1. This complex associates with chromatin over ethylene-responsive genes to regulate acetylation of H3K14 and H3K23, leading to an EIN3-dependent transcriptional regulation (Figure 1).
At the molecular level, the mechanisms involving EIN2, ENAP1, and EIN3 in the regulation of histone acetylation during the ethylene response remain unclear. Specifically, how the target loci for histone acetylation are determined, how acetylation at different lysines is coordinated to regulate gene expression in the ethylene response, and whether other histone modifications are involved are unknown. Moreover, the biochemical functions of the full-length EIN2 and the EIN2 C-terminus are unknown. One possibility is that the EIN2 C-terminal domain is itself a HAT; however, no HAT activity of EIN2 C-terminus has been detected (unpublished results from the Qiao lab). Alternatively, the C-terminal fragment of EIN2 may act as a scaffolding protein to recruit and assemble histone modification complexes in the presence of ethylene. If it is the case, identification of HAT or HDAC that functions in cooperation with the EIN2 C-terminal domain with a positive correlation would validate this assumption.
4. Conclusions
Ethylene is important for plant development and stress responses, and understanding this response is relevant due to the function of this hormone in agricultural crops during unfavorable environmental conditions. As a key component in the ethylene signaling pathway, EIN2 mediates the connection between upstream receptors bound to the ER membrane to the downstream factors in the nucleus and P-body that are involved in translational regulation of ethylene-responsive genes. The discovery of the importance of the C-terminal domain of EIN2 in the ethylene response revealed for the first time a direct connection between ethylene signaling and chromatin regulation. Although it is clear that EIN2 integrates into EIN3-dependent transcriptional regulation, the detailed molecular mechanism of how EIN2 transduces ethylene signaling to mediate histone acetylation is still largely unknown. Characterization of the biochemical function of the EIN2 C-terminus and identification of the HAT or HDAC involved are priorities. Many levels of regulation are involved in establishment of chromatin structure; histone acetylation is just one of these. Further study to establish whether other chromatin modifications are involved in mediating ethylene signaling and whether the higher order structure of chromatin is involved are of interest.
Acknowledgements
L.W. and F.Z. contributed equally to this work. The authors thank all the members from Qiao lab for the support with research and advice on this manuscript. This work was supported by grant from the National Institutes of Health (R01GM115879-01) to H.Q.
Biography

Likai Wang received his Ph.D. degree in cell biology from Huazhong Agricultural University in December 2014, China. He is currently working as a postdoctoral research fellow in Prof. Hong Qiao’s group at The University of Texas at Austin. His research interest is to uncover the gene expression regulation in response to ethylene using both experimental and computational methods.

Fan Zhang received his Ph.D. degree in biology from The Institute of Botany of the Chinese Academy of Sciences in 2014. He did postdoctoral training in Prof. Hong Qiao’s lab in The University of Texas at Austin during 2014–2018. He is currently a professor of the Key Laboratory of Horticultural Plant Biology of Ministry of Education, College of Horticulture and Forestry Sciences, Huazhong Agricultural University. His research focuses on the molecular mechanisms of plant flower senescence.

Hong Qiao received her Ph.D. degree in developmental and cell biology from The Institute of Genetics and Developmental Biology of Chinese Academy of Sciences. She did her postdoctoral training at the Salk Institute for Biology Studies. She is currently an assistant professor in the Department of Molecular biosciences at The University of Texas at Austin. Her research focuses on chromatin regulation in plant hormone ethylene and environmental stress responses.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Likai Wang, Institute for Cellular and Molecular Biology, The University of Texas at Austin, Austin, TX 78712, USA; Department of Molecular Biosciences, The University of Texas at Austin, Austin, TX 78712, USA.
Fan Zhang, Institute for Cellular and Molecular Biology, The University of Texas at Austin, Austin, TX 78712, USA; Department of Molecular Biosciences, The University of Texas at Austin, Austin, TX 78712, USA.
Hong Qiao, Institute for Cellular and Molecular Biology, The University of Texas at Austin, Austin, TX 78712, USA; Department of Molecular Biosciences, The University of Texas at Austin, Austin, TX 78712, USA.
References
- [1].Lund ST, Stall RE, Klee HJ, Plant Cell 1998, 10, 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ju C, Chang C, Plant Physiol 2015, 169, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ju C, Van de Poel B, Cooper ED, Thierer JH, Gibbons TR, Delwiche CF, Chang C, Nat. Plants 2015, 1, 14004. [DOI] [PubMed] [Google Scholar]
- [4].Bleecker AB, Estelle MA, Somerville C, Kende H, Science 1988, 241, 1086. [DOI] [PubMed] [Google Scholar]
- [5].Ecker JR, Science 1995, 268, 667. [DOI] [PubMed] [Google Scholar]
- [6].Li W, Ma M, Feng Y, Li H, Wang Y, Ma Y, Li M, An F, Guo H, Cell 2015, 163, 670. [DOI] [PubMed] [Google Scholar]
- [7].Chang C, Kwok SF, Bleecker AB, Meyerowitz EM, Science 1993, 262, 539. [DOI] [PubMed] [Google Scholar]
- [8].Sakai H, Hua J, Chen QG, Chang C, Medrano LJ, Bleecker AB, Meyerowitz EM, Proc. Natl. Acad. Sci. USA 1998, 95, 5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hua J, Chang C, Sun Q, Meyerowitz EM, Science 1995, 269, 1712. [DOI] [PubMed] [Google Scholar]
- [10].Hua J, Sakai H, Nourizadeh S, Chen QG, Bleecker AB, Ecker JR, Meyerowitz EM, Plant Cell 1998, 10, 1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hirayama T, Kieber JJ, Hirayama N, Kogan M, Guzman P, Nourizadeh S, Alonso JM, Dailey WP, Dancis A, Ecker JR, Cell 1999, 97, 383. [DOI] [PubMed] [Google Scholar]
- [12].Resnick JS, Rivarola M, Chang C, Plant J 2008, 56, 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR, Cell 1993, 72, 427. [DOI] [PubMed] [Google Scholar]
- [14].Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR, Science 1999, 284, 2148. [DOI] [PubMed] [Google Scholar]
- [15].Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR, Cell 1997, 89, 1133. [DOI] [PubMed] [Google Scholar]
- [16].Solano R, Stepanova A, Chao Q, Ecker JR, Genes Dev 1998, 12, 3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Guo H, Ecker JR, Cell 2003, 115, 667. [DOI] [PubMed] [Google Scholar]
- [18].Olmedo G, Guo H, Gregory BD, Nourizadeh SD, Aguilar-Henonin L, Li H, An F, Guzman P, Ecker JR, Proc. Natl. Acad. Sci. USA 2006, 103, 13286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Potuschak T, Vansiri A, Binder BM, Lechner E, Vierstra RD, Genschik P, Plant Cell 2006, 18, 3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Merchante C, Brumos J, Yun J, Hu Q, Spencer KR, Enriquez P, Binder BM, Heber S, Stepanova AN, Alonso JM, Cell 2015, 163, 684. [DOI] [PubMed] [Google Scholar]
- [21].Cho YH, Yoo SD, Front. Plant Sci. 2014, 5, 733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Qiao H, Shen Z, Huang SS, Schmitz RJ, Urich MA, Briggs SP, Ecker JR, Science 2012, 338, 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chang KN, Zhong S, Weirauch MT, Hon G, Pelizzola M, Li H, Huang SS, Schmitz RJ, Urich MA, Kuo D, Nery JR, Qiao H, Yang A, Jamali A, Chen H, Ideker T, Ren B, Bar-Joseph Z, Hughes TR, Ecker JR, eLife 2013, 2, e00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Eyal Y, Meller Y, Lev-Yadun S, Fluhr R, Plant J 1993, 4, 225. [DOI] [PubMed] [Google Scholar]
- [25].Meller Y, Sessa G, Eyal Y, Fluhr R, Plant Mol. Biol 1993, 23, 453. [DOI] [PubMed] [Google Scholar]
- [26].Ohme-Takagi M, Shinshi H, Plant Mol. Biol 1990, 15, 941. [DOI] [PubMed] [Google Scholar]
- [27].Sessa G, Meller Y, Fluhr R, Plant Mol. Biol 1995, 28, 145. [DOI] [PubMed] [Google Scholar]
- [28].Shinshi H, Usami S, Ohme-Takagi M, Plant Mol. Biol 1995, 27, 923. [DOI] [PubMed] [Google Scholar]
- [29].Sato F, Kitajima S, Koyama T, Yamada Y, Plant Cell Physiol 1996, 37, 249. [DOI] [PubMed] [Google Scholar]
- [30].Yamamuro C, Zhu JK, Yang Z, Mol. Plant 2016, 9, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Pandey R, Muller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA, Nucleic Acids Res 2002, 30, 5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hark AT, Vlachonasios KE, Pavangadkar KA, Rao S, Gordon H, Adamakis ID, Kaldis A, Thomashow MF, Triezenberg SJ, Biochim. Biophys. Acta, Gene Regul. Mech 2009, 1789, 117. [DOI] [PubMed] [Google Scholar]
- [33].Anzola JM, Sieberer T, Ortbauer M, Butt H, Korbei B, Weinhofer I, Mullner AE, Luschnig C, Proc. Natl. Acad. Sci. USA 2010, 107, 10308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Poulios S, Vlachonasios KE, J. Exp. Bot 2016, 67, 905. [DOI] [PubMed] [Google Scholar]
- [35].Benhamed M, Bertrand C, Servet C, Zhou DX, Plant Cell 2006, 18, 2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mao Y, Pavangadkar KA, Thomashow MF, Triezenberg SJ, Biochim. Biophys. Acta, Gene Struct. Expression 2006, 1759, 69. [DOI] [PubMed] [Google Scholar]
- [37].Stockinger EJ, Mao Y, Regier MK, Triezenberg SJ, Thomashow MF, Nucleic Acids Res 2001, 29, 1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Nelissen H, De Groeve S, Fleury D, Neyt P, Bruno L, Bitonti MB, Vandenbussche F, Van Der Straeten D, Yamaguchi T, Tsukaya H, Witters E, De Jaeger G, Houben A, Van Lijsebettens M, Proc. Natl. Acad. Sci. USA 2010, 107, 1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chen ZZ, Zhang HR, Jablonowski D, Zhou XF, Ren XZ, Hong XH, Schaffrath R, Zhu JK, Gong ZH, Mol. Cell. Biol 2006, 26, 6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhou XF, Hua DP, Chen ZZ, Zhou ZJ, Gong ZZ, Plant J 2009, 60, 79. [DOI] [PubMed] [Google Scholar]
- [41].Liu X, Luo M, Zhang W, Zhao JH, Zhang JX, Wu KQ, Tian LN, Duan J, BMC Plant Biol 2012, 12, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Papaefthimiou D, Likotrafiti E, Kapazoglou A, Bladenopoulos K, Tsaftaris A, Plant Physiol. Biochem 2010, 48, 98. [DOI] [PubMed] [Google Scholar]
- [43].Heisel TJ, Li CY, Grey KM, Gibson SI, Front. Plant Sci 2013, 4, 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Li C, Xu J, Li J, Li QY, Yang HC, Plant Cell Physiol 2014, 55, 426. [DOI] [PubMed] [Google Scholar]
- [45].Zhou CH, Zhang L, Duan J, Miki B, Wu KQ, Plant Cell 2005, 17, 1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wu K, Zhang L, Zhou C, Yu CW, Chaikam V, J. Exp. Bot 2008, 59, 225. [DOI] [PubMed] [Google Scholar]
- [47].Zhu ZQ, An FY, Feng Y, Li PP, Xue L, Mu A, Jiang ZQ, Kim JM, To TK, Li W, Zhang XY, Yu Q, Dong Z, Chen WQ, Seki M, Zhou JM, Guo HW, Proc. Natl. Acad. Sci. USA 2011, 108, 12539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Chen LT, Luo M, Wang YY, Wu KQ, J. Exp. Bot 2010, 61, 3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Perrella G, Lopez-Vernaza MA, Carr C, Sani E, Gossele V, Verduyn C, Kellermeier F, Hannah MA, Amtmann A, Plant Cell 2013, 25, 3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Luo M, Wang YY, Liu XC, Yang SG, Lu Q, Cui YH, Wu KQ, J. Exp. Bot 2012, 63, 3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hao YH, Wang HJ, Qiao SL, Leng LN, Wang XL, Proc. Natl. Acad. Sci. USA 2016, 113, 10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lee K, Park OS, Jung SJ, Seo PJ, J. Plant Physiol 2016, 191, 95. [DOI] [PubMed] [Google Scholar]
- [53].Wang Z, Cao H, Sun YZ, Li XY, Chen FY, Carles A, Li Y, Ding M, Zhang C, Deng X, Soppe WJJ, Liu YX, Plant Cell 2013, 25, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Mehdi S, Derkacheva M, Ramstrom M, Kralemann L, Bergquist J, Hennig L, Plant Cell 2016, 28, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Song CP, Galbraith DW, Plant Mol. Biol 2006, 60, 241. [DOI] [PubMed] [Google Scholar]
- [56].Wang Z, Chen FY, Li XY, Cao H, Ding M, Zhang C, Zuo JH, Xu CN, Xu JM, Deng X, Xiang Y, Soppe WJJ, Liu YX, Nat. Commun 2016, 7, 13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Choi SM, Song HR, Han SK, Han M, Kim CY, Park J, Lee YH, Jeon JS, Noh YS, Noh B, Plant J 2012, 71, 135. [DOI] [PubMed] [Google Scholar]
- [58].Kim KC, Lai ZB, Fan BF, Chen ZX, Plant Cell 2008, 20, 2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Chen LT, Wu K, Plant Signaling Behav 2010, 5, 1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang PC, Zhu JK, Plant Cell 2005, 17, 2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Ryu H, Cho H, Bae W, Hwang I, Nat. Commun 2014, 5, 4138. [DOI] [PubMed] [Google Scholar]
- [62].Wang CM, Shang JX, Chen QX, Oses-Prieto JA, Bai MY, Yang YH, Yuan M, Zhang YL, Mu CC, Deng ZP, Wei CQ, Burlingame AL, Wang ZY, Sun Y, Mol. Cell. Proteomics 2013, 12, 3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Fu WQ, Wu KQ, Duan J, Biochem. Biophys. Res. Commun 2007, 356, 843. [DOI] [PubMed] [Google Scholar]
- [64].Demetriou K, Kapazoglou A, Bladenopoulos K, Tsaftaris AS, Plant Mol. Biol. Rep 2010, 28, 9. [Google Scholar]
- [65].Demetriou K, Kapazoglou A, Tondelli A, Francia E, Stanca MA, Bladenopoulos K, Tsaftaris AS, Physiol. Plant 2009, 136, 358. [DOI] [PubMed] [Google Scholar]
- [66].Colville A, Alhattab R, Hu M, Labbe H, Xing T, Miki B, Plant Cell Rep 2011, 30, 1969. [DOI] [PubMed] [Google Scholar]
- [67].Sridha S, Wu KQ, Plant J 2006, 46, 124. [DOI] [PubMed] [Google Scholar]
- [68].Zhao JH, Zhang JX, Zhang W, Wu KL, Zheng F, Tian LN, Liu XC, Duan J, Front. Plant Sci 2015, 5, 764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Wang CZ, Gao F, Wu JG, Dai JL, Wei CH, Li Y, Plant Cell Physiol 2010, 51, 1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Zhang F, Wang L, Ko EE, Shao K, Qiao H, Plant Cell 2018, 30, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Li C, Xu J, Li J, Li Q, Yang H, Plant Signaling Behav 2014, 9, e28173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Fletcher TM, Hansen JC, Crit. Rev. Eukaryotic Gene Expression 1996, 6, 149. [DOI] [PubMed] [Google Scholar]
- [73].Steger DJ, Workman JL, BioEssays 1996, 18, 875. [DOI] [PubMed] [Google Scholar]
- [74].Luger K, Richmond TJ, Curr. Opin. Genet. Dev 1998, 8, 140. [DOI] [PubMed] [Google Scholar]
- [75].Zhang F, Qi B, Wang L, Zhao B, Rode S, Riggan ND, Ecker JR, Qiao H, Nat. Commun 2016, 7, 13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Wang L, Zhang F, Rode S, Chin KK, Ko EE, Kim J, Iyer VR, Qiao H, BMC Genomics 2017, 18, 538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Zhang F, Wang L, Qi B, Zhao B, Ko EE, Riggan ND, Chin K, Qiao H, Proc. Natl. Acad. Sci. USA 2017, 114, 10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Ju C, Yoon GM, Shemansky JM, Lin DY, Ying ZI, Chang J, Garrett WM, Kessenbrock M, Groth G, Tucker ML, Cooper B, Kieber JJ, Chang C, Proc. Natl. Acad. Sci. USA 2012, 109, 19486. [DOI] [PMC free article] [PubMed] [Google Scholar]