Abstract
The phosphorylated pathway of serine biosynthesis is initiated with 3-phosphoglycerate dehydrogenase (PGDH). The liverwort Marchantia polymorpha possesses an amino acid-sensitive MpPGDH which is inhibited by l-serine and activated by five proteinogenic amino acids, while the eudicot Arabidopsis thaliana has amino acid-sensitive AtPGDH1 and AtPGDH3 as well as amino acid-insensitive AtPGDH2. In this study, we analyzed PGDH isozymes of the representative land plants: the monocot Oryza sativa (OsPGDH1–3), basal angiosperm Amborella trichopoda (AmtriPGDH1–2), and moss Physcomitrium (Physcomitrella) patens (PpPGDH1–4). We demonstrated that OsPGDH1, AmtriPGDH1, PpPGDH1, and PpPGDH3 were amino acid-sensitive, whereas OsPGDH2, OsPGDH3, AmtriPGDH2, PpPGDH2, and PpPGDH4 were either sensitive to only some of the six effector amino acids or insensitive to all effectors. This indicates that PGDH sensitivity to effectors has been diversified among isozymes and that the land plant species examined, except for M. polymorpha, possess different isozyme types in terms of regulation. Phylogenetic analysis suggested that the different sensitivities convergently evolved in the bryophyte and angiosperm lineages. Site-directed mutagenesis of AtPGDH1 revealed that Asp538 and Asn556 residues in the ACT domain are involved in allosteric regulation by the effectors. These findings provide insight into the evolution of PGDH isozymes, highlighting the functional diversification of allosteric regulation in land plants.
Keywords: allosteric regulation, enzymatic activity, enzyme activation, inhibition, l-amino acids, phosphorylated pathway
Introduction
Serine is an important molecule that acts as a protein building block and precursor for various biomolecules, including nucleic acid bases, phospholipids, sphingolipids, and amino acids, such as tryptophan and cysteine. The phosphorylated pathway of serine biosynthesis is common in bacteria, animals, and plants [1–3] and consists of three reactions catalyzed by 3-phosphoglycerate dehydrogenase (PGDH), phosphoserine aminotransferase, and phosphoserine phosphatase (Figure 1). The first committed enzyme, PGDH, oxidizes 3-phosphoglycerate (3-PGA), one of the products of glycolysis in all organisms and of the Calvin cycle in photosynthetic autotrophs, to form 3-phosphohydroxypyruvate. In some bacteria, such as Mycobacterium tuberculosis and Escherichia coli, the phosphorylated pathway is regulated by negative feedback through allosteric inhibition of PGDH by l-serine [4,5]. A PGDH in M. tuberculosis (MtPGDH) is biochemically well-studied and its crystal structure has been solved [6]. In addition to the catalytic domain, MtPGDH possesses allosteric substrate binding (ASB) and aspartate kinase-chorismate mutase-tyrA (ACT) domains [7,8] at its C-terminal region, both of which are involved in regulating its enzymatic activity. MtPGDH mainly forms a homotetramer, and binding of l-serine to the ACT domain causes a conformational change in the homotetramer that leads to a decrease in the maximum velocity of enzymatic activity. Serine inhibition is enhanced in the presence of the phosphate ion, which binds to the ASB domain [6]. In contrast, the PGDH of E. coli (EcPGDH) possesses only the ACT domain [9].
Figure 1. Schematic of the phosphorylated pathway of serine biosynthesis.
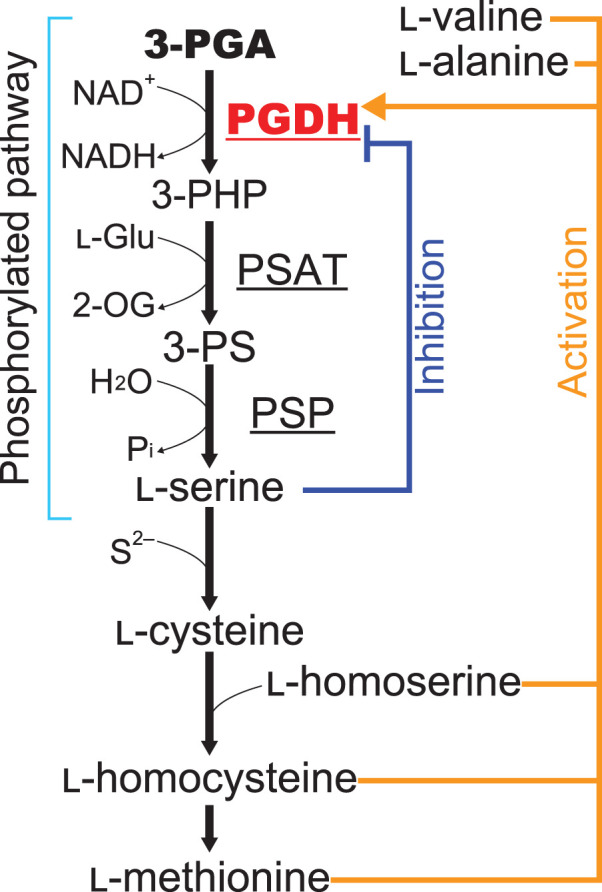
PGDH, 3-phosphoglycerate dehydrogenase; PSAT, phosphoserine aminotransferase; PSP, phosphoserine phosphatase; 3-PHP, 3-phosphohydroxypyruvate; 3-PS, 3-phosphoserine; l-Glu, l-glutamate; 2-OG, 2-oxoglutarate. Regulation of PGDH by amino acids in plants is shown.
In plants, serine is synthesized in two additional pathways: the glycolate and glycerate pathways [2]. The glycolate pathway, which takes place in the mitochondria and is part of the photorespiratory pathway, is the most important source of serine among the three pathways, at least in photosynthetic tissues [2]. The glycerate pathway occurs in the cytosol and may be a major source of serine during dark periods in C3 plants and in non-photosynthetic tissues [10]. On the other hand, the phosphorylated pathway takes place in the plastid and is considered to function mainly in photosynthetic organs during dark periods and in non-photosynthetic tissues [2].
The enzymatic activity of PGDH from Pisum sativum was previously found to be inhibited by serine and activated by methionine at 10 mM [11]. The eudicot (angiosperm) Arabidopsis thaliana possesses three plastid-localized PGDH isozymes encoded by AtPGDH1, AtPGDH2, and AtPGDH3 [12,13]. All three isozymes contain ASB and ACT domains, but AtPGDH2 is not inhibited by l-serine [13,14]. AtPGDH1 is involved in the biosynthesis of tryptophan-derived metabolites including indole-3-acetic acid and in adaptation to high CO2 conditions, under which photorespiration and the serine supply via the glycolate pathway are suppressed [13]. AtPGDH3 plays a crucial role in stromal NADH supply and eventually in photosynthetic performance [15]. Our previous study revealed that AtPGDH1 and AtPGDH3 are not only feedback-inhibited by l-serine but also activated by l-alanine, l-valine, l-methionine, l-homoserine, and l-homocysteine in a cooperative manner [14] (Figure 1). We also revealed that AtPGDH1 and AtPGDH3 predominantly form homotetramers, whereas AtPGDH2 formed an equilibrium of homooctamers and homotetramers. Among the above six effector amino acids, the sulfur-containing l-homocysteine showed the lowest half maximal effective concentration (EC50). Furthermore, inhibition of these AtPGDHs by l-serine and activation by the activator amino acids affected each other, indicating that the serine supply via the phosphorylated pathway involving AtPGDH1 and AtPGDH3 is regulated by the ratio of l-serine and activator amino acids. This, in turn, regulates the balance of various metabolic pathways, including tryptophan biosynthesis and sulfur metabolism [14]. A lack of the phosphorylated pathway perturbs sulfur assimilation and sulfur homeostasis between photosynthetic and non-photosynthetic tissues [16].
Domain swapping between the amino acid-sensitive AtPGDH1 and amino acid-insensitive AtPGDH2 was conducted to construct two chimeric enzymes possessing the N-terminal half (containing the catalytic domain) of AtPGDH2 and C-terminal half (containing the ASB and ACT domains) of AtPGDH1, and vice versa [14]. The results suggested that l-serine inhibits AtPGDH1 enzymatic activity by binding to the ACT domain [14]. Although the binding site of the activator amino acids was not clearly identified, the results suggested that cooperative activation by the effector amino acids requires the formation of higher-order structures of the C- and N-terminal half regions via intra- and/or inter-molecular interactions [14].
We identified a single-copy gene encoding PGDH in the basal land plant (bryophyte) Marchantia polymorpha (MpPGDH) [17]. MpPGDH also possesses the ASB and ACT domains in its C-terminal region. Similar to AtPGDH1 and AtPGDH3, MpPGDH forms a homotetramer in vitro, and is inhibited by l-serine and activated by l-alanine, l-valine, l-methionine, l-homoserine, and l-homocysteine, with the lowest EC50 for l-homocysteine [17]. These findings suggest that PGDH regulation by amino acids is conserved in land plants, regardless of the presence or absence of cooperativity. In addition, the lack of amino acid-insensitive PGDH in M. polymorpha evokes a question when such isozymes as AtPGDH2 emerged during land plant evolution. To address this issue, in this study, we identified PGDH isozymes in three representative land plant species with available genome sequences, namely, the monocot (angiosperm) Oryza sativa, basal angiosperm Amborella trichopoda, and moss (bryophyte) Physcomitrium (Physcomitrella) patens, and examined their regulation by effector amino acids. The findings suggest that PGDH functional diversification in terms of regulation occurred independently in the bryophyte and angiosperm lineages. Furthermore, we conducted site-directed mutagenesis of AtPGDH1 at the amino acid residues corresponding to the key residues for serine binding in MtPGDH, which identified two key residues for regulation and indicated that PGDH regulation occurs in an allosteric manner.
Materials and methods
Multiple alignment of amino acid sequences for construction of protein expression vectors
The amino acid sequences of PGDHs from Arabidopsis thaliana (AtPGDH1, AtPGDH2, and AtPGDH3), Oryza sativa (OsPGDH1, OsPGDH2, and OsPGDH3), Amborella trichopoda (AmtriPGDH1 and AmtriPGDH2), Physcomitrium (Physcomitrella) patents (PpPGDH1, PpPGDH2, PpPGDH3, and PpPGDH4), Marchantia polymorpha (MpPGDH), Aphanothece halophytica (AhPGDH), and Mycobacterium tuberculosis (MtPGDH) were aligned using CLUSTALW in MEGA7 software [18] with the BLOSUM matrix. Alignments were visualized using ESPript (http://espript.ibcp.fr) [19]. Accession numbers were summarized in Supplementary Table S2.
Reagents
NAD disodium salt, d-(–)-3-phosphoglyceric acid disodium salt, and l-homocysteine were purchased from Sigma-Aldrich Co., Ltd. (St. Louis, MO, USA). l-Alanine, l-valine, l-methionine, l-homoserine, l-serine, and N-Tris(hydroxymethyl)methyl-3-aminopropanesulfonic acid (TAPS), DTT, NADH, and isopropyl-β-d-thiogalactopyranoside (IPTG) were purchased from FUJIFILM Wako Pure Chemical Industries, Ltd. (Osaka, Japan).
Genetic complementation of Escherichia coli serine auxotroph mutant
Genetic complementation of the E. coli serine auxotroph mutant by PGDH genes was performed as described previously [17,20]. Complementation vectors were constructed as described as follows. Regions corresponding to mature enzymes were amplified using the primers shown in Supplementary Table S1, which were ligated into the NcoI and KpnI sites of the expression vector pTV118N (Takara Bio, Inc., Shiga, Japan) using the In-Fusion HD cloning kit. cDNA synthesis and cloning into the NcoI and KpnI sites of the pTV118N vector of PpPGDH1 and AmtriPGDH2 were performed by artificial gene synthesis (Eurofins Genomics Inc., Tokyo, Japan). The E. coli l-serine-auxotroph JW 2880 strain (TG1ΔserA::KmFRT) [21] was transformed with the pTV118N expression vectors possessing cDNAs of OsPGDHs, PpPGDHs, and AmtriPGDHs and grown on M9 media with or without 0.2 mM l-serine. The Escherichia coli strain JW2880 was provided by NBRP E. coli Strain at the National Institute of Genetics, Japan.
Preparation of recombinant enzymes
Full-length cDNA clones of OsPGDH1 and OsPGDH2 were obtained from NIAS DNA Bank (accession codes AK120939 and AK243399, respectively). cDNA clones of PpPGDH2, PpPGDH3, and PpPGDH4 were obtained from RIKEN BioResource Center (accession code pdp33831, pdp82465, and pdp12194, respectively). cDNA of OsPGDH3 and AmtriPGDH1 was obtained by artificial gene synthesis (Integrated DNA Technologies, Inc., Coralville, IA, USA). cDNA synthesis and cloning into a heterologous expression vector (as described below) of PpPGDH1 and AmtriPGDH2 were performed by artificial gene synthesis (Eurofins Genomics Inc.).
Transit peptide sequences of OsPGDH1, OsPGDH2, OsPGDH3, PpPGDH1, PpPGDH2, PpPGDH3, PpPGDH4, AmtriPGDH1, and AmtriPGDH2 were predicted by comparing their amino acid sequences with that of MtPGDH. Regions corresponding to mature enzymes were amplified using the primers provided in Supplementary Table S1, which were assembled at the SpeI and NotI sites of the expression vector pPAL7 (Bio-Rad, Hercules, CA, USA) using an In-Fusion HD cloning kit (Takara Bio, Inc.) [22].
All recombinant enzymes without the transit peptide were expressed in E. coli BL21 CodonPlus (DE3)-RIPL cells (Agilent Technologies, Santa Clara, CA, USA). Pre-cultivation was performed in Luria-Bertani (LB) liquid medium containing 100 μg/ml carbenicillin and 30 μg/ml chloramphenicol at 37°C for 12 h. Next, 2% of these cultures was used to inoculate 150 ml LB liquid medium containing 100 μg/ml carbenicillin and 30 μg/ml chloramphenicol and grown at 20°C until the optical density at 600 nm (OD600) reached 0.5. IPTG was added at a final concentration of 0.5 mM, and the cells were further incubated for 12 h at 20°C.
Tag-free recombinant proteins were prepared by affinity purification using Profinity eXact Purification Resin (Bio-Rad). Briefly, cell pellets were obtained by centrifugation of E. coli cultures at 9000×g, and the cell pellets were resuspended in 100 mM sodium phosphate buffer (pH 9.0) and sonicated on ice for 10 min. Crude extracts were then centrifuged for 10 min at 9000×g. The supernatants were applied to the Profinity eXact Purification Resin, followed by in-column incubation at 20°C for 1 h to eliminate the affinity tag eXact Fusion-Tag from the recombinant proteins. The buffer of eluted fractions was immediately exchanged with 100 mM sodium phosphate buffer (pH 9.0) by ultrafiltration using an Amicon Ultra-4 Centrifugal Filter Unit (MWCO 10 000; Merck-Millipore, Billerica, MA, USA). All recombinant proteins were analyzed by SDS-PAGE using a 10% polyacrylamide gel.
Spectrophotometric assays of recombinant PGDH enzymes
A spectrophotometric assay was performed as previously described [14,17]. The optimal pH for AtPGDHs ranged from 9.0 to 10.0 [14,17], whereas that for AhPGDH was 9.0 [23]. The enzyme assay was conducted at pH 9.0 in 100-μl reaction mixtures containing 0.1 M TAPS (pH 9.0), 1 mM DTT, 10 mM 3-PGA, 1 mM NAD+, 0.1 M NaCl, and approximately 3.0–4.0 μg recombinant enzyme. The reaction mixtures were pre-incubated without 3-PGA at 25°C for 10 min, and the reactions were initiated by adding 3-PGA [24,25]. 3-PGA oxidation activities were determined by the increased absorbance of NADH (340 nm), as detected with a UV-2700 spectrophotometer (Shimadzu, Kyoto, Japan). Reaction mixtures without substrates were used as negative controls. Kinetic parameters for apparent Michaelis constants () and apparent maximum velocities () were calculated by fitting specific activities to Michaelis–Menten equations under various concentrations of substrates using the Enzyme Kinetics Module in SigmaPlot 14 (Systat Software, San Jose, CA). Initial velocities were determined from the slopes of the plots of NADH formation versus incubation time within 15 s. The dose response of PGDHs to l-alanine, l-valine, l-methionine, l-homoserine, l-homocysteine, and l-serine were determined by fitting the percentage relative activities at various effector concentrations to the Hill equation [26,27] using Global Curve Fitting in SigmaPlot 14, as described previously [14].
The specific activities of PGDHs were determined at 10 mM 3-PGA and 1 mM NAD+ in the presence of various concentrations of amino acids as follows: for AmtriPGDHs and PpPGDHs, l-serine, l-alanine, l-valine, l-homoserine and l-methionine were added at 0, 0.1, 0.2, 0.5, 0.7, 1, 2, 5, 7, 10, 20, 50 mM and l-homocysteine was added at 0, 0.001, 0.002, 0.005, 0.007, 0.01, 0.02, 0.05, 0.07, 0.1, 0.2, 0.5 mM; for OsPGDHs, l-alanine, l-serine, l-homoserine, were added at 0, 0.1, 0.2, 0.5, 0.7, 1, 2, 5, 7, 10, 20, 50 mM; for OsPGDH1, l-methionine were added at 0, 0.01, 0.02, 0.05, 0.07, 0.1, 0.2, 0.5, 0.7, 1, 2, 5, 7, 10, 20, 50 mM; for OsPGDH2 and OsPGDH3, l-methionine were added at 0, 0.1, 0.2, 0.5, 0.7, 1, 2, 5, 7, 10, 20, 50 mM; for OsPGDH1, l-homocysteine were added at 0, 0.001, 0.002, 0.005, 0.007, 0.01, 0.02, 0.05, 0.07, 0.1, 0.2, 0.5, 0.7, 1 mM; for OsPGDH2 and OsPGDH3, l-homocysteine were added at 0, 0.01, 0.02, 0.05, 0.07, 0.1, 0.2, 0.5, 0.7, 1, 2, 5, 7, 10 mM; for AhPGDH, l-alanine, l-homocysteine, l-homoserine, l-methionine, l-serine and l-valine were added at 0, 0.1, 0.2, 0.5, 0.7, 1, 2, 5, 7, 10 mM.
Site-directed mutagenesis
Using the pPAL7 (Bio-Rad) vector carrying AtPGDH1, site-directed mutagenesis for AtPGDH1-Q536A, AtPGDH1-D538A, and AtPGDH-N556A were performed by inverse PCR using overlapping primers for alanine substitution (Supplementary Table. S1). After amplification, E. coli DH5α competent cells were transformed with the reaction mixtures, and the clones carrying the expected mutation were selected by sequencing. Heterologous expression and purification, determination of kinetics parameters, and dose response analysis were performed as described above.
Phylogenetic analysis
Amino acid sequences of PGDHs were retrieved from the databases Phytozome12.1 (https://phytozome.jgi.doe.gov/pz/portal.html) [28], Gymno PLAZA 1.0 (https://bioinformatics.psb.ugent.be/plaza/versions/gymno-plaza/) [29], CoGe (https://genomevolution.org/coge/) [30,31], Ensemble Plant (http://plants.ensembl.org/index.html) [32], and Genbank (https://www.ncbi.nlm.nih.gov/genbank/) [33]. These amino acid sequences (Supporting Table S2) were aligned with Clustal Omega (1.2.2) [34] implemented in Geneious Prime 2021.0.3 (Biomatters Ltd., Auckland, New Zealand) with automatically adjusted settings on number of sequences. The obtained multiple sequence alignment was manually trimmed for the N-terminal predicted transit peptide by comparing amino acid sequence of MtPGDH, and then any sites containing at least 80% gaps were removed using the Mask Alignment function in Geneious Prime. Bayesian phylogenetic analysis was conducted by MrBayes (ver. 3.2.7a) [35] on CIPRES Science Gateway (http://www.phylo.org/) [36] using MtPGDH as the outgroup with the following parameters: aamodelpr = fixed(mixed); ngen = 20 000 000; nruns = 2: nchains = 4: temp = 0.2: relburnin = yes; burninfrac = 0.25. Phylogenetic tree was rendered with FigTree version 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/). The alignment used to generate the phylogenetic tree is available upon request.
Results
Genetic complementation of an E. coli serine auxotrophic mutant by land plant PGDHs
We found PGDH isozymes in O. sativa (OsPGDH1–3), A. trichopoda (AmtriPGDH1–2), and P. patens (PpPGDH1–4) by homology searching. Their enzymatic activities in vivo and in vitro have not been determined. Therefore, to investigate whether OsPGDHs, AmtriPGDHs, and PpPGDHs are functional in living cells, we performed genetic complementation experiments in a serine auxotrophic mutant of E. coli, in which PGDH was disrupted [21]. The cDNA of each PGDH without its transit peptide sequence was expressed under the lac promoter in the mutant. We observed that all PGDH isozymes, except AmtriPGDH2, complemented serine auxotrophy, indicating that they participate in serine biosynthesis in vivo in E. coli (Supplementary Figure S1).
Kinetic parameters of recombinant PGDH isozymes
Next, we examined the biochemical properties of OsPGDHs, AmtriPGDHs, and PpPGDHs using the respective recombinant proteins expressed in E. coli. AmtriPGDH2 was also examined to determine why it did not complement the E. coli serine auxotroph mutant. SDS-PAGE analysis indicated that E. coli expressed approximately 60 kDa proteins, which was consistent with the theoretical molecular weights of PGDHs without the transit peptide (Supplementary Figure S2).
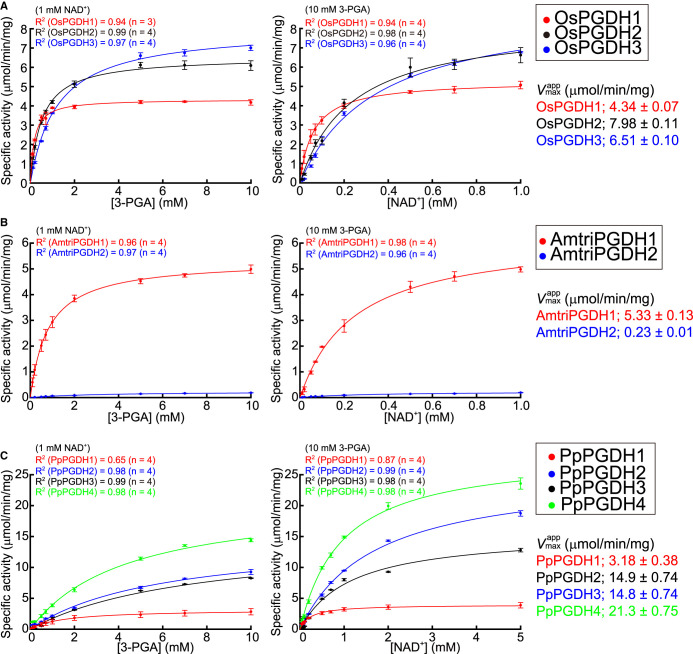
The apparent Michaelis constant () and maximum velocity () for 3-PGA and NAD+ were calculated at pH 9.0 (Figure 2). The values of OsPGDHs and AmtriPGDH1 ranged from 0.18 to 1.21 mM (for 3-PGA) and from 0.062 to 0.245 mM (for NAD+) (Figure 2A,B and Table 1), which were comparable to those of AtPGDHs and MpPGDH [14,17]. The values of OsPGDHs and AmtriPGDH1 ranged from 4.34 to 7.98 µmol min−1·mg−1, which are similar to those of AtPGDHs and MpPGDH (Figure 2A,B). Thus, OsPGDHs and AmtriPGDH1 gave specific constants (kcat/Kmapp) comparable to those of AtPGDHs and MpPGDH. However, those of AmtriPGDH2 were extremely low (Table 1), which may perhaps explain the failure of genetic complementation in the E. coli serine auxotrophic mutant (Supplementary Figure S1). In contrast, the and values of PpPGDH2-4 were higher than those of AtPGDHs, MpPGDH, OsPGDHs, and AmtriPGDHs (Figure 2C and Table 1).
Figure 2. Michaelis–Menten plots of PGDH isozymes.
(A) OsPGDHs, (B) AmtriPGDHs, and (C) PpPGDHs. In each panel, specific activities at various concentrations of 3-PGA (left) and NAD+ (right) are shown. Data are presented as the means and standard errors from two technical replicates, using enzymes purified from two independent batches of cells (basically n = 4).
Table 1. Kinetic parameters of PGDH isozymes.
| Enzyme name | kcat (s−1) | (mM) | kcat/ (M−1·s−1) | ||
|---|---|---|---|---|---|
| 3-PGA | NAD+ | 3-PGA (×104) | NAD+ (×104) | ||
| OsPGDH1 | 16.5 ± 0.2 | 0.175 ± 0.016 | 0.062 ± 0.007 | 9.4 ± 0.8 | 2.67 ± 0.30 |
| OsPGDH2 | 30.7 ± 0.4 | 1.212 ± 0.053 | 0.348 ± 0.025 | 2.53 ± 0.11 | 0.88 ± 0.064 |
| OsPGDH3 | 24.9 ± 0.3 | 0.519 ± 0.033 | 0.234 ± 0.029 | 4.80 ± 0.31 | 1.06 ± 0.13 |
| AmtriPGDH1 | 20.4 ± 0.4 | 0.815 ± 0.070 | 0.245 ± 0.021 | 2.51 ± 0.22 | 0.83 ± 0.07 |
| AmtriPGDH2 | 0.88 ± 0.03 | 3.186 ± 0.327 | 0.313 ± 0.038 | 0.0276 ± 0.0029 | 0.028 ± 0.003 |
| PpPGDH1 | 12.1 ± 1.4 | 1.550 ± 0.566 | 0.237 ± 0.038 | 0.78 ± 0.30 | 0.51 ± 0.10 |
| PpPGDH2 | 57.6 ± 2.8 | 5.971 ± 0.603 | 1.677 ± 0.068 | 0.96 ± 0.10 | 0.34 ± 0.02 |
| PpPGDH3 | 56.4 ± 2.8 | 7.533 ± 0.661 | 1.156 ± 0.098 | 0.74 ± 0.07 | 0.48 ± 0.04 |
| PpPGDH4 | 81.9 ± 2.8 | 4.392 ± 0.350 | 0.952 ± 0.054 | 1.86 ± 0.16 | 0.86 ± 0.05 |
The values were calculated from Figure 2 and are presented with the standard errors (n = 4, except OsPGDH1 (n = 3)).
Regulation of PGDH isozymes by effector amino acids
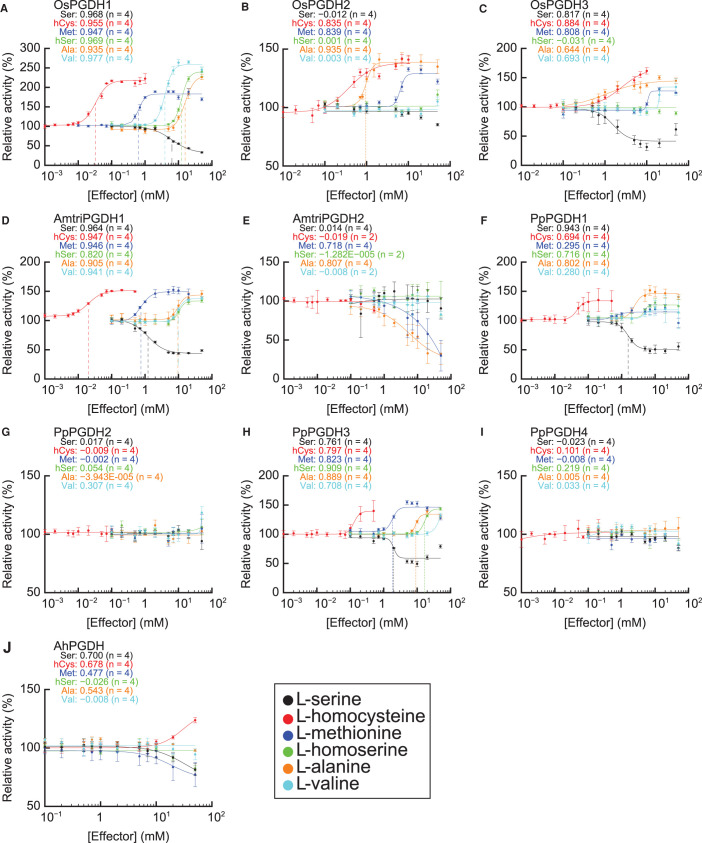
We previously reported that AtPGDH1 and AtPGDH3 from the eudicot A. thaliana and MpPGDH from the liverwort M. polymorpha were inhibited by l-serine and activated by l-alanine, l-valine, l-methionine, l-homoserine, and l-homocysteine, whereas AtPGDH2 was not [14,17]. To clarify whether l-amino acid-mediated PGDH regulation is conserved in land plant lineages, we examined the 3-PGA-oxidation activity of PGDH isozymes from land plants O. sativa (monocot), A. trichopoda (basal angiosperm), and P. patens (moss) in response to different doses of effector amino acids (Figure 3). We also investigated AhPGDH from the cyanobacterium Aphanothece halophytica, whose biochemical properties have already been characterized (Supplementary Figure S3) [23].
Figure 3. Dose responses of PGDH isozymes to effector amino acids.
Specific activities of (A) OsPGDH1, (B) OsPGDH2, (C) OsPGDH3, (D) AmtriPGDH1, (E) AmtriPGDH2, (F) PpPGDH1, (G) PpPGDH2, (H) PpPGDH3, (I) PpPGDH4, and (J) AhPGDH at various concentrations of effectors were measured and are expressed relative to their activities in the absence of effectors. Data are presented as the means and standard errors from two technical replicates using enzymes purified from two independent batches of cells (basically n = 4). Regression curves of Hill equations are shown with coefficients of determination (R2). Three-letter codes of amino acids followed by numerical values in each graph show the values of R2. Ser, l-serine; hCys, l-homocysteine; Met, l-methionine; hSer, l-homoserine; Ara, l-alanine; Val, l-valine.
The results showed that OsPGDH1 and AmtriPGDH1 were inhibited by serine and activated by the activator amino acids, exhibiting sigmoidal dose-response curves fitted to the Hill equation (determination coefficient R2 > 0.9) [26] (Figure 3A,D). The Hill coefficients of OsPGDH1 for all effectors were >1.5 (Table 2), indicating that these amino acids inhibited (l-serine) or activated (l-alanine, l-valine, l-methionine, l-homoserine, and l-homocysteine) enzymatic activity in a cooperative manner. Among the effectors, the EC50 of l-homocysteine was lowest, followed by l-methionine, in OsPGDH1 (Table 2). AmtriPGDH1 was also inhibited by l-serine and activated by the other amino acids (Figure 3D). The Hill coefficients for the effectors other than l-homoserine were >1.5 (Table 2), indicating that AmtriPGDH1 was cooperatively regulated by these amino acids. Again, l-homocysteine showed the lowest EC50 value among all tested amino acids. In contrast, only some of the tested amino acids regulated OsPGDH2 and OsPGDH3 (Figure 3B,C). OsPGDH2 was activated by l-homocysteine, l-alanine, and l-methionine to a lesser extent than OsPGDH1, whereas its activity was not affected by l-valine, l-homoserine, or l-serine at any of the tested concentrations. OsPGDH3 was inhibited by l-serine to a similar degree as OsPGDH1 and activated by l-homocysteine, l-alanine, l-methionine, and l-valine to a lesser extent compared to OsPGDH1. The responses of AmtriPGDH2 to the effector amino acids also differed from those of AmtriPGDH1 and OsPGDH1: the enzyme was inhibited by l-alanine and l-methionine and was not regulated by the other effectors (Figure 3E).
Table 2. EC50 and Hill coefficients of PGDH isozymes.
| Amino acid | OsPGDH1 | OsPGDH2 | AmtriPGDH1 | PpPGDH1 | PpPGDH3 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| EC50 (mM) | Hill coefficient | EC50 (mM) | Hill coefficient | EC50 (mM) | Hill coefficient | EC50 (mM) | Hill coefficient | EC50 (mM) | Hill coefficient | |
| l-Serine | 6.4 ± 0.6 | 1.5 ± 0.2 | — | — | 1.2 ± 0.1 | 1.7 ± 0.2 | 1.6 ± 0.1 | 3.2 ± 0.6 | — | — |
| l-Homocysteine | 0.033 ± 0.003 | 2.4 ± 0.4 | — | — | 0.020 ± 0.002 | 2.1 ± 0.4 | — | — | — | — |
| l-Methionine | 0.64 ± 0.03 | 4.2 ± 0.8 | — | — | 0.75 ± 0.05 | 2.7 ± 0.5 | — | — | — | — |
| l-Homoserine | 12.4 ± 0.6 | 4.0 ± 0.5 | — | — | — | — | — | — | 16.8 ± 1.2 | 5.2 ± 1.5 |
| l-Alanine | 15.8 ± 1.1 | 3.1 ± 0.5 | 0.95 ± 0.04 | 5.3 ± 1.0 | 9.6 ± 0.7 | 4.0 ± 1.0 | — | — | — | — |
| l-Valine | 3.9 ± 0.2 | 3.7 ± 0.4 | — | — | 9.8 ± 0.5 | 4.5 ± 1.0 | — | — | — | — |
The values were calculated from Figure 3 when regression curves showed R2 values > 0.9 and are presented with the standard errors (n = 4).
Among the moss PGDHs, PpPGDH1, and PpPGDH3 were amino acid-sensitive, whereas PpPGDH2 and PpPGDH4 were not affected by the effector amino acids (Figure 3F–I). The EC50 values for l-homocysteine in PpPGDH1 and PpPGDH3 were the lowest among the effectors, which was also the case for AtPGDH1, AtPGDH3, OsPGDH1, and AmtriPGDH1. In contrast, cyanobacterial AhPGDH was neither inhibited by l-serine nor activated by l-homocysteine and the other activator amino acids (Figure 3J).
These results indicate that O. sativa, A. trichopoda, and P. patens contain at least one isozyme such as MpPGDH that is regulated by all effector amino acids and possess other isozymes whose regulation is diversified.
Identification of key amino acid residues for regulation of AtPGDH1
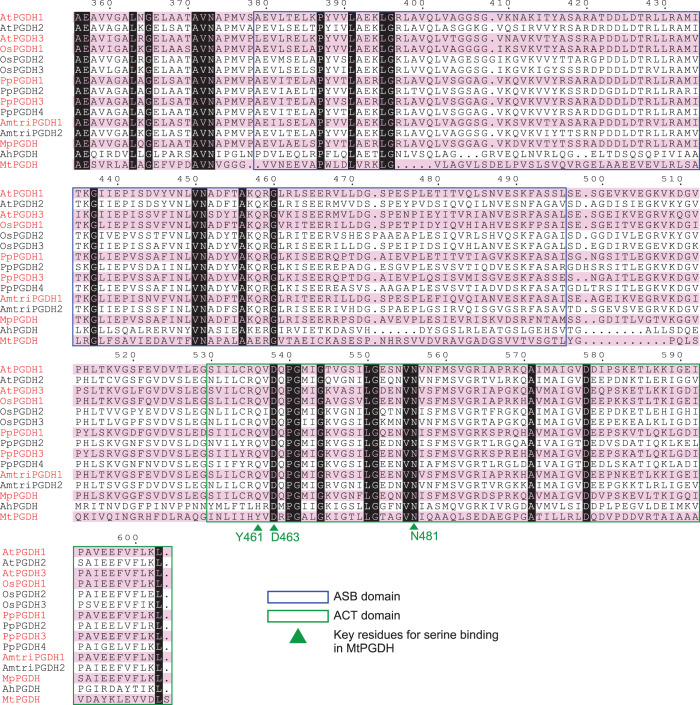
To clarify difference between amino acid-sensitive and -insensitive isozymes, we searched the amino acid sequences of PGDH isozymes and evaluated which protein motif is responsible for the regulation. Our previous domain swapping experiment between amino acid-sensitive AtPGDH1 and amino acid-insensitive AtPGDH2 suggested that l-serine binds the ACT domain of AtPGDH1 to inhibit its enzymatic activity, although the binding site of the activator amino acids has not been clearly identified [14]. In this study, we compared the amino acid sequences of the PGDH isozymes of the land plant species as well as those of A. halophytica and M. tuberculosis (Supplementary Figure S4), focusing on the C-terminal regions containing the ASB and ACT domains (Figure 4).
Figure 4. Multiple sequence alignments of PGDH regulatory domains.
The C-terminal amino acid sequences of PGDHs from A. thaliana (AtPGDHs), O. sativa (OsPGDHs), A. trichopoda (AmtriPGDHs), P. patens (PpPGDHs), M. polymorpha (MpPGDH), A. halophytica (AhPGDH), and M. tuberculosis (MtPGDH) are shown. Isozymes regulated by all six effector amino acids are highlighted by red font and pink shading. The predicted ASB domains and ACT domains are surrounded by blue and green rectangles, respectively. Green triangles represent the key residues for serine binding in MtPGDH.
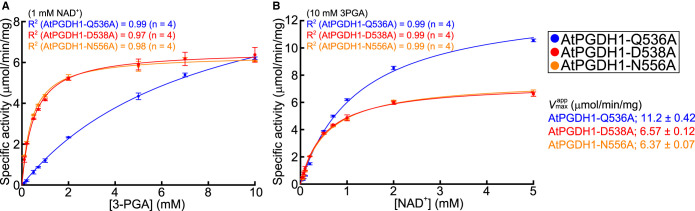
In the case of MtPGDH inhibition by l-serine, Tyr461 (Y461), Asp463 (D463), and Asn481 (N481) residues in the ACT domain (Figure 4, arrowheads) formed hydrogen bond networks with a carboxyl group of l-serine [37]. Multiple sequence alignment (Figure 4) indicated that amino acid residues corresponding to D463 and N481 were completely conserved among all PGDHs examined. In contrast, residues corresponding to Y461 were commonly substituted with Gln (Q) in the land plant PGDHs. Therefore, to determine whether the triad of Gln, Asp, and Asn in the land plant PGDHs is involved in PGDH regulation by the effector amino acids, we conducted site-directed mutagenesis of AtPGDH1 at Q536, D538, and N556 (which correspond to Y461, D463, and N481 in MtPGDH, respectively, Supplementary Figure S4). We constructed three alanine-substituted AtPGDH1 enzymes: AtPGDH1-Q536A, AtPGDH1-D538A, and AtPGDH1-N556A. A spectrophotometric assay using the recombinant enzymes indicated that AtPGDH1-D538A and AtPGDH1-N556A had slightly different enzyme kinetic parameters, whereas AtPGDH1-Q536A showed a lower specific constant for 3-PGA (Figure 5 and Table 3) compared to wild-type AtPGDH1.
Figure 5. Michaelis–Menten plots of alanine-substituted AtPGDH1 enzymes.
Specific activities at various concentrations of 3-PGA (A) and NAD+ (B) are shown. Data are presented as the means and standard errors from two technical replicates by using enzymes purified from two independent batches of cells (n = 4).
Table 3. Kinetic parameters of alanine-substituted AtPGDH1 enzymes.
| Enzyme name | kcat (s−1) | (mM) | kcat/ (M−1·s−1) | ||
|---|---|---|---|---|---|
| 3-PGA | NAD+ | 3-PGA (×104) | NAD+ (×105) | ||
| AtPGDH1-Q536A | 42.9 ± 1.6 | 7.87 ± 0.55 | 1.22 ± 0.05 | 0.54 ± 0.04 | 0.34 ± 0.02 |
| AtPGDH1-D538A | 25.1 ± 0.4 | 0.51 ± 0.03 | 0.51 ± 0.03 | 4.88 ± 0.38 | 0.48 ± 0.02 |
| AtPGDH1-N556A | 24.4 ± 0.2 | 0.42 ± 0.01 | 0.57 ± 0.03 | 5.78 ± 0.26 | 0.42 ± 0.02 |
| AtPGDH11 | 15 | 0.50 ± 0.05 | 0.050 ± 0.002 | 3.0 | 3.0 |
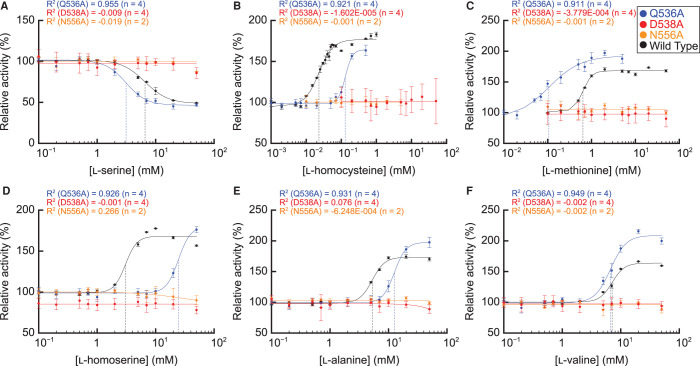
We next analyzed the dose responses of the alanine-substituted AtPGDH1 enzymes to the effector amino acids. AtPGDH1-D538A and AtPGDH1-N556A completely lost their cooperative inhibition by l-serine and activation by the five activator amino acids (Figure 6). In contrast, AtPGDH1-Q536A was inhibited and activated by the effector amino acids with slightly different EC50 values compared to wild-type AtPGDH1 (Table 4). The Hill coefficients were greater than 2 except for that of l-methionine, indicating that Q536 is not directly involved in cooperative inhibition and activation by effector amino acids other than l-methionine. These results suggest that D538 and N556 in the ACT domain of AtPGDH1 are necessary for cooperative inhibition and activation by the effectors (Figure 6), and that the regulation occurs in an allosteric manner. Additionally, Q536 affects AtPGDH1 catalytic activity (Figure 5).
Figure 6. Dose response of alanine-substituted AtPGDH1 enzymes to effector amino acids.
Specific activities of AtPGDH1-Q536A, AtPGDH1-D538A, and AtPGDH1-N556A at various concentrations of (A) l-serine, (B) l-homocysteine, (C) l-methionine, (D) l-homoserine, (E) l-alanine, and (F) l-valine, relative to their activities measured in the absence of effectors, are shown. Data are presented as the means and standard errors from two technical replicates by using enzymes purified from two independent batches of cells (basically n = 4). For comparison, the activities of wild-type AtPGDH1 reported in [14] are also indicated.
Table 4. EC50 and Hill coefficients of alanine-substituted AtPGDH-Q536A.
| Mode | Amino acid | AtPGDH11 | AtPGDH1-Q536A | ||
|---|---|---|---|---|---|
| EC50 (mM) | Hill coefficient | EC50 (mM) | Hill coefficient | ||
| Inhibitor | l-Serine | 6.6 ± 0.3 | 2.2 ± 0.3 | 3.1 ± 0.3 | 2.5 ± 0.5 |
| l-Homocysteine | 0.023 ± 0.001 | 2.1 ± 0.2 | 0.130 ± 0.013 | 4.3 ± 1.1 | |
| l-Methionine | 0.67 ± 0.03 | 4.7 ± 0.6 | 0.106 ± 0.024 | 1.2 ± 0.3 | |
| Activator | l-Homoserine | 3.0 ± 0.3 | 4.4 ± 0.8 | 24.5 ± 5.6 | 4.2 ± 3.4 |
| l-Alanine | 5.3 ± 0.2 | 4.0 ± 0.5 | 12.6 ± 1.1 | 4.4 ± 1.0 | |
| l-Valine | 7.1 ± 0.2 | 4.1 ± 0.5 | 6.7 ± 0.4 | 3.3 ± 0.7 | |
Discussion
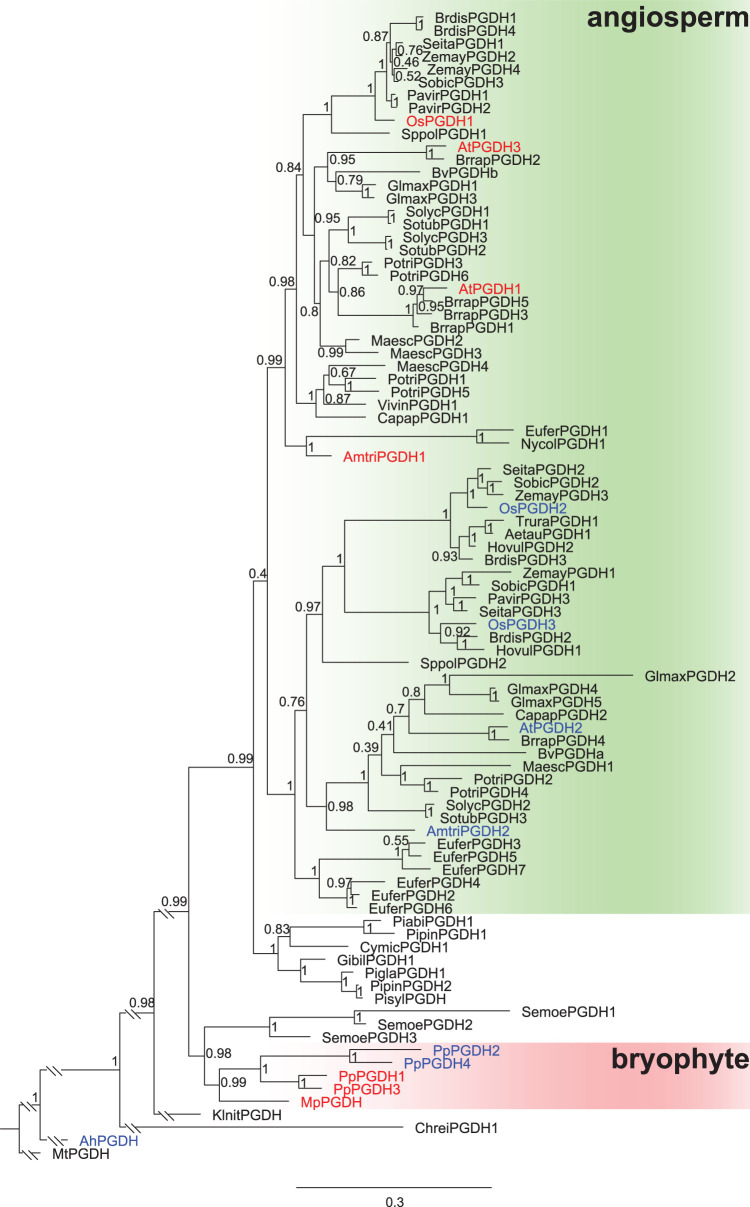
Our biochemical analysis revealed that all land plants examined possess PGDH isozyme(s) which are inhibited by l-serine and activated by l-alanine, l-valine, l-methionine, l-homoserine, and l-homocysteine. Except for M. polymorpha, these plants also contain isozymes with diverse sensitivities to effector amino acids. Phylogenetic analysis of PGDHs from various land plant species, including bryophytes, lycophyte, gymnosperms, and angiosperms, indicated that angiosperm PGDHs were divided into two subclades (Figure 7, Supplementary Table S2). One subclade (hereafter called as sub. I) includes PGDH isozymes that are inhibited by l-serine and activated by five l-amino acids; these were the PGDH isozymes from the eudicot A. thaliana (AtPGDH1 and AtPGDH3), monocot O. sativa (OsPGDH1), and basal angiosperm A. trichopoda (AmtriPGDH1) (Figure 7, shown in red letters). The other subclade (sub. II) includes isozymes that are diverse in terms of amino acid sensitivity. AtPGDH2 is insensitive to all effector amino acids, whereas OsPGDH2 and OsPGDH3 are sensitive to some effectors (Figure 7, shown in blue letters). AmtriPGDH2 was also regulated by some of the effectors (Figure 3), although its specificity constant was extremely low (Figure 2 and Table 1). We found that almost all angiosperm species examined possess both sub. I and sub. II PGDHs (Figure 7). The seven gymnosperm PGDHs examined formed a clade sister to both sub. I and sub. II (Figure 7), suggesting that the two angiosperm subclades were separated after the divergence from the gymnosperm lineage.
Figure 7. Phylogenetic tree of PGDHs across multiple plant lineages.
A phylogenetic tree of PGDHs from eudicots, monocots, basal angiosperms, gymnosperms, lycophyte, bryophytes, charophyte, chlorophyte, cyanobacterium, and actinobacterium were generated and the branch length was modified. The original phylogenetic tree is shown in Supplementary Figure S5. Red and blue letters indicate the isozymes sensitive to six effector amino acids and those insensitive or sensitive to some effectors, respectively. Posterior probabilities are indicated at the nodes. Bar indicates substitutions per site.
In the bryophyte PGDH clade, four PGDH isozymes of the moss P. patens were separated into two groups, each corresponding to the amino acid-sensitive type (PpPGDH1 and PpPGDH3) and amino acid-insensitive type (PpPGDH2 and PpPGDH4). The liverwort M. polymorpha has the single PGDH isozyme MpPGDH belonging to the bryophyte clade. Given that MpPGDH is sensitive to all six effector amino acids [17], the phylogeny suggests that the ancestral PGDH in land plants was likely amino acid-sensitive and that PGDH isozymes at least partially free from regulation by the six effector amino acids were later acquired independently in different land plant lineages via gene duplication events during evolution. In addition, we showed that the cyanobacterium A. halophytica possesses only the amino acid-insensitive type of PGDH (Figures 3 and 7). It is widely accepted that the eukaryotic photosynthetic organelle (plastid) originated from endosymbiosis of cyanobacteria in the plantae ancestor [38]. Therefore, further studies of PGDHs in green plant lineages before the divergence of land plants would reveal the origin of amino acid-mediated PGDH regulation.
PGDH duplication and functional diversification appear to have been necessary for the evolution of land plants to adequately control the serine supply in different tissues at different developmental stages. In A. thaliana, three AtPGDH genes exhibited different tissue-specific expression patterns [12,13]. The loss-of-function mutant of AtPGDH1 exhibited embryonic lethality, whereas those of AtPGDH2 and AtPGDH3 showed no drastic visible phenotype [12,13], demonstrating the functional diversification of PGDH isozymes. Because serine functions as a precursor of stress-related specialized (secondary) metabolites such as glucosinolates in Brassicaceae plants [39] and glycine betaine in Poaceae plants [40], the serine supply may be involved in environmental stress responses in plants. It is likely that some PGDH paralogs are regulated to fulfill this demand. In fact, AtPGDH1 is under the control of MYB34 and MYB51, which are transcription factors that regulate tryptophan-derived glucosinolate biosynthesis [13]. In Beta vulgaris, BvPGDHa was induced whereas BvPGDHb was repressed under salt stress [41]. Similarly, AtPGDH1 and AtPGDH2 were induced and AtPGDH3 was repressed by salt stress, although the metabolic functions of these genes under salt stress are yet to be identified [42]. However, as the serine content increased in A. thaliana after salt treatment [42], regulation of PGDH may also play important roles in regulating the serine supply in response to salt stress. The transcriptional regulatory mechanism by salt does not appear to be related to the diversification of sub. I and sub. II PGDHs, as sub. I contains both the salt-inducible AtPGDH1 and salt-repressible AtPGDH3. The pattern of occurrence of amino acid-sensitive isozymes and salt-regulated homologs in land plant lineages suggests that PGDH represents a point in the serine biosynthesis that is easily controllable by environmental stresses such as salinity. Recent study revealed redox regulation of AtPGDH1 associated with the redox-active Cys pair uniquely found in land plant PGDH [43].
Our previous study using AtPGDH1–AtPGDH2 chimeric enzymes indicated that some features of the AtPGDH1 N-terminal region are necessary for the five activator amino acids to fully activate PGDH [14]. These results suggest that regulation of PGDH enzymatic activity by the effector amino acids requires not only effector binding to the ACT domain but also the transmission of resultant structural changes from the ACT domain at the C-terminal region to the catalytic domain at the N-terminal region or from the effector-bound PGDH monomer to the other monomers [14,17]. In this study, site-directed mutagenesis of AtPGDH1 showed that the D538 and N556 residues in the ACT domain are necessary for cooperative inhibition by l-serine and activation by the activator amino acids (Figure 6). These residues correspond to the serine-binding sites of MtPGDH from a mycobacterium, suggesting that they are involved in binding of the effector amino acids in AtPGDH1, and binding of the effector to the ACT domain regulates catalytic activity via an allosteric effect. However, these aspartic acid (D) and asparagine (N) residues were conserved in all PGDHs we examined, including amino acid-insensitive isozymes (Figure 4). The Q536 residue of AtPGDH1, which is conserved in land plant PGDHs, is not essential for regulation but is involved in enzyme kinetics (Figures 5 and 6). At the primary protein structure level, no amino acid residue is conserved only in amino acid-sensitive isozymes (Figure 4 and Supplementary Figure S4). Protein structure analysis performed in the presence of the effector amino acids would help us understand the molecular mechanism of PGDH regulation by effector amino acids.
In conclusion, functionally diversified PGDH enzymes in terms of amino acid-mediated allosteric regulation convergently evolved in the bryophyte and angiosperm lineages. Although the protein structural differences between amino acid-sensitive and -insensitive isozymes require further analysis, our findings reveal the binding sites of allosteric effectors in land plant PGDHs which has long been unknown [11,44] and provide insight into the biological importance of the phosphorylated pathway of serine biosynthesis in land plants.
Abbreviations
- 3-PGA
3-phosphoglycerate
- ACT
aspartate kinase-chorismate mutase-tyrA
- ASB
allosteric substrate binding
- EC50
half maximal effective concentration
- PGDH
3-phosphoglycerate dehydrogenase
Data Availability
All data are included in the main manuscript and in the Supplementary data file.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the JSPS Grants-in-Aid for Scientific Research on Innovative Areas number JP25113010 to M.Y.H. and for Young Scientists (B) number JP26870855 to E.O.
CRediT Author Contribution
Masami Yokota Hirai: Conceptualization, supervision, funding acquisition, investigation, writing — original draft. Eiji Okamura: Conceptualization, funding acquisition, investigation, writing — original draft. Kinuka Ohtaka: Investigation, writing — original draft. Ryuichi Nishihama: Investigation, writing — original draft. Kai Uchida: Investigation. Ayuko Kuwahara: Investigation. Keiichi Mochida: Investigation.
Supplementary Material
References
- 1.Ros, R., Cascales-Minana, B., Segura, J., Anoman, A.D., Toujani, W., Flores-Tornero, M.et al. (2013) Serine biosynthesis by photorespiratory and non-photorespiratory pathways: an interesting interplay with unknown regulatory networks. Plant Biol. (Stuttg.) 15, 707–712 10.1111/j.1438-8677.2012.00682.x [DOI] [PubMed] [Google Scholar]
- 2.Ros, R., Munoz-Bertomeu, J. and Krueger, S. (2014) Serine in plants: biosynthesis, metabolism, and functions. Trends Plant. Sci. 19, 564–569 10.1016/j.tplants.2014.06.003 [DOI] [PubMed] [Google Scholar]
- 3.Hausler, R.E., Ludewig, F. and Krueger, S. (2014) Amino acids–a life between metabolism and signaling. Plant Sci. 229, 225–237 10.1016/j.plantsci.2014.09.011 [DOI] [PubMed] [Google Scholar]
- 4.Grant, G.A. (2012) Contrasting catalytic and allosteric mechanisms for phosphoglycerate dehydrogenases. Arch. Biochem. Biophys. 519, 175–185 10.1016/j.abb.2011.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pizer, L.I. (1963) The pathway and control of serine biosynthesis in Escherichia coli. J. Biol. Chem. 238, 3934–3944 10.1016/S0021-9258(18)51809-3 [DOI] [PubMed] [Google Scholar]
- 6.Xu, X.L. and Grant, G.A. (2014) Regulation of Mycobacterium tuberculosis D-3-phosphoglycerate dehydrogenase by phosphate-modulated quaternary structure dynamics and a potential role for polyphosphate in enzyme regulation. Biochemistry (Mosc.) 53, 4239–4249 10.1021/bi500469d [DOI] [PubMed] [Google Scholar]
- 7.Chipman, D.M. and Shaanan, B. (2001) The ACT domain family. Curr. Opin. Struct. Biol. 11, 694–700 10.1016/S0959-440X(01)00272-X [DOI] [PubMed] [Google Scholar]
- 8.Grant, G.A. (2006) The ACT domain: a small molecule binding domain and its role as a common regulatory element. J. Biol. Chem. 281, 33825–33829 10.1074/jbc.R600024200 [DOI] [PubMed] [Google Scholar]
- 9.Thompson, J.R., Bell, J.K., Bratt, J., Grant, G.A. and Banaszak, L.J. (2005) Vmax regulation through domain and subunit changes. The active form of phosphoglycerate dehydrogenase. Biochemistry (Mosc.) 44, 5763–5773 10.1021/bi047944b [DOI] [PubMed] [Google Scholar]
- 10.Igamberdiev, A.U. and Kleczkowski, L.A. (2018) The glycerate and phosphorylated pathways of serine synthesis in plants: the branches of plant glycolysis linking carbon and nitrogen metabolism. Front. Plant Sci. 9, 318 10.3389/fpls.2018.00318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slaughter, J.C. and Davies, D.D. (1975) 3-Phosphoglycerate dehydrogenase from seedlings of Pisum sativum. Methods Enzymol. 41, 278–281 10.1016/S0076-6879(75)41063-1 [DOI] [PubMed] [Google Scholar]
- 12.Toujani, W., Munoz-Bertomeu, J., Flores-Tornero, M., Rosa-Tellez, S., Anoman, A.D., Alseekh, S.et al. (2013) Functional characterization of the plastidial 3-phosphoglycerate dehydrogenase family in Arabidopsis. Plant Physiol. 163, 1164–1178 10.1104/pp.113.226720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benstein, R.M., Ludewig, K., Wulfert, S., Wittek, S., Gigolashvili, T., Frerigmann, H.et al. (2013) Arabidopsis phosphoglycerate dehydrogenase1 of the phosphoserine pathway is essential for development and required for ammonium assimilation and tryptophan biosynthesis. Plant Cell 25, 5011–5029 10.1105/tpc.113.118992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okamura, E. and Hirai, M.Y. (2017) Novel regulatory mechanism of serine biosynthesis associated with 3-phosphoglycerate dehydrogenase in Arabidopsis thaliana. Sci. Rep. 7, 3533 10.1038/s41598-017-03807-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Höhner, R., Day, P.M., Zimmermann, S.E., Lopez, L.S., Krämer, M., Giavalisco, P.et al. (2021) Stromal NADH supplied by PHOSPHOGLYCERATE DEHYDROGENASE3 is crucial for photosynthetic performance. Plant Physiol. 185, 142–167 10.1093/plphys/kiaa117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anoman, A.D., Flores-Tornero, M., Benstein, R.M., Blau, S., Rosa-Tellez, S., Brautigam, A.et al. (2019) Deficiency in the phosphorylated pathway of serine biosynthesis perturbs sulfur assimilation. Plant Physiol. 180, 153–170 10.1104/pp.18.01549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akashi, H., Okamura, E., Nishihama, R., Kohchi, T. and Hirai, M.Y. (2018) Identification and biochemical characterization of the serine biosynthetic enzyme 3-phosphoglycerate dehydrogenase in Marchantia polymorpha. Front. Plant Sci. 9, 956 10.3389/fpls.2018.00956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar, S., Stecher, G. and Tamura, K. (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robert, X. and Gouet, P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho, C.L., Noji, M., Saito, M. and Saito, K. (1999) Regulation of serine biosynthesis in Srabidopsis. Crucial role of plastidic 3-phosphoglycerate dehydrogenase in non-photosynthetic tissues. J. Biol. Chem. 274, 397–402 10.1074/jbc.274.1.397 [DOI] [PubMed] [Google Scholar]
- 21.Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M.et al. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the keio collection. Mol. Syst. Biol. 2, 2006.0008 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berrow, N.S., Alderton, D., Sainsbury, S., Nettleship, J., Assenberg, R., Rahman, N.et al. (2007) A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 35, e45 10.1093/nar/gkm047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waditee, R., Bhuiyan, N.H., Hirata, E., Hibino, T., Tanaka, Y., Shikata, M.et al. (2007) Metabolic engineering for betaine accumulation in microbes and plants. J. Biol. Chem. 282, 34185–34193 10.1074/jbc.M704939200 [DOI] [PubMed] [Google Scholar]
- 24.Dey, S., Hu, Z., Xu, X.L., Sacchettini, J.C. and Grant, G.A. (2005) D-3-Phosphoglycerate dehydrogenase from Mycobacterium tuberculosis is a link between the Escherichia coli and mammalian enzymes. J. Biol. Chem. 280, 14884–14891 10.1074/jbc.M414488200 [DOI] [PubMed] [Google Scholar]
- 25.Saski, R. and Pizer, L.I. (1975) Regulatory properties of purified 3-phosphoglycerate dehydrogenase from Bacillus subtilis. Eur. J. Biochem. 51, 415–427 10.1111/j.1432-1033.1975.tb03941.x [DOI] [PubMed] [Google Scholar]
- 26.Goutelle, S., Maurin, M., Rougier, F., Barbaut, X., Bourguignon, L., Ducher, M.et al. (2008) The Hill equation: a review of its capabilities in pharmacological modelling. Fundam. Clin. Pharmacol. 22, 633–648 10.1111/j.1472-8206.2008.00633.x [DOI] [PubMed] [Google Scholar]
- 27.Hill, A.V. (1910) The possible effects of the aggregation of the molecules of hæmoglobin on its dissociation curves. J. Physiol. 40, iv–vii 10.1113/jphysiol.1910.sp001386 [DOI] [Google Scholar]
- 28.Goodstein, D.M., Shu, S., Howson, R., Neupane, R., Hayes, R.D., Fazo, J.et al. (2012) Phytozome: a comparative platform for Green plant genomics. Nucleic Acids Res. 40, D1178–D1186 10.1093/nar/gkr944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Proost, S., Van Bel, M., Vaneechoutte, D., Van de Peer, Y., Inze, D., Mueller-Roeber, B.et al. (2015) PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res. 43, D974–D981 10.1093/nar/gku986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lyons, E. and Freeling, M. (2008) How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 53, 661–673 10.1111/j.1365-313X.2007.03326.x [DOI] [PubMed] [Google Scholar]
- 31.Lyons, E., Pedersen, B., Kane, J., Alam, M., Ming, R., Tang, H.et al. (2008) Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant Physiol. 148, 1772–1781 10.1104/pp.108.124867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howe, K.L., Contreras-Moreira, B., De Silva, N., Maslen, G., Akanni, W., Allen, J.et al. (2020) Ensembl genomes 2020-enabling non-vertebrate genomic research. Nucleic Acids Res. 48, D689–D695 10.1093/nar/gkz890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clark, K., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J. and Sayers, E.W. (2016) Genbank. Nucleic Acids Res. 44, D67–D72 10.1093/nar/gkv1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sievers, F., Wilm, A., Dineen, D., Gibson, T.J., Karplus, K., Li, W.et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D.L., Darling, A., Hohna, S.et al. (2012) Mrbayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, M.A., Pfeiffer, W. and Schwartz, T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. 2010 Gateway Computing Environments Workshop (GCE) 1-8 10.1109/GCE.2010.5676129 [DOI]
- 37.Dey, S., Burton, R.L., Grant, G.A. and Sacchettini, J.C. (2008) Structural analysis of substrate and effector binding in Mycobacterium tuberculosis D-3-phosphoglycerate dehydrogenase. Biochemistry (Mosc.) 47, 8271–8282 10.1021/bi800212b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reyes-Prieto, A., Weber, A.P. and Bhattacharya, D. (2007) The origin and establishment of the plastid in algae and plants. Annu. Rev. Genet. 41, 147–168 10.1146/annurev.genet.41.110306.130134 [DOI] [PubMed] [Google Scholar]
- 39.Halkier, B.A. and Gershenzon, J. (2006) Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57, 303–333 10.1146/annurev.arplant.57.032905.105228 [DOI] [PubMed] [Google Scholar]
- 40.Annunziata, M.G., Ciarmiello, L.F., Woodrow, P., Dell'Aversana, E. and Carillo, P. (2019) Spatial and temporal profile of glycine betaine accumulation in plants under abiotic stresses. Front. Plant Sci. 10, 230 10.3389/fpls.2019.00230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kito, K., Tsutsumi, K., Rai, V., Theerawitaya, C., Cha-Um, S., Yamada-Kato, N.et al. (2017) Isolation and functional characterization of 3-phosphoglycerate dehydrogenase involved in salt responses in sugar beet. Protoplasma 254, 2305–2313 10.1007/s00709-017-1127-7 [DOI] [PubMed] [Google Scholar]
- 42.Rosa-Tellez, S., Anoman, A.D., Alcantara-Enguidanos, A., Garza-Aguirre, R.A., Alseekh, S. and Ros, R. (2020) PGDH family genes differentially affect Arabidopsis tolerance to salt stress. Plant Sci. 290, 110284 10.1016/j.plantsci.2019.110284 [DOI] [PubMed] [Google Scholar]
- 43.Yoshida, K., Ohtaka, K., Hirai, M.Y. and Hisabori, T. (2020) Biochemical insight into redox regulation of plastidial 3-phosphoglycerate dehydrogenase from Arabidopsis thaliana. J. Biol. Chem. 295, 14906–14915 10.1074/jbc.RA120.014263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, Q., Qi, Y., Yin, N. and Lai, L. (2014) Discovery of novel allosteric effectors based on the predicted allosteric sites for Escherichia coli D-3-phosphoglycerate dehydrogenase. PLoS One 9, e94829 10.1371/journal.pone.0094829 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the main manuscript and in the Supplementary data file.