Abstract
In vertebrate animals, motor and sensory efferent neurons carry information from the central nervous system (CNS) to peripheral targets. These two types of efferent systems sometimes bear a close resemblance, sharing common segmental organization, axon pathways, and chemical messengers. Here, we focus on the development of the octavolateral efferent neurons (OENs) and their interactions with the closely-related facial branchiomotor neurons (FBMNs) in zebrafish. Using live-imaging approaches, we investigate the birth, migration, and projection patterns of OENs. We find that OENs are born in two distinct groups: a group of rostral efferent neurons (RENs) that arises in the fourth segment, or rhombomere (r4), of the hindbrain and a group of caudal efferent neurons (CENs) that arises in r5. Both RENs and CENs then migrate posteriorly through the hindbrain between 18 and 48 hours post-fertilization, alongside the r4-derived FBMNs. Like the FBMNs, migration of the r4-derived RENs depends on function of the segmental identity gene hoxb1a; unlike the FBMNs, however, both OEN populations move independently of prickle1b. Further, we investigate whether the previously described ‘pioneer’ neuron that leads FBMN migration through the hindbrain is an r4-derived FBMN/REN or an r5-derived CEN. Our experiments verify that the pioneer is an r4-derived neuron and reaffirm its role in leading FBMN migration across the r4/5 border. In contrast, the r5-derived CENs migrate independently of the pioneer. Together, these results indicate that the mechanisms OENs use to navigate the hindbrain differ significantly from those employed by FBMNs.
Keywords: zebrafish, hindbrain, neurons, lateral line system, facial nerve, cell movement, Hox genes
Graphical Abstract
1. Introduction
Integration of the central nervous system (CNS) with peripheral targets is achieved through the projections of afferent and efferent neurons, which carry information towards and away from the CNS, respectively. Most efferent neurons are motor neurons, although sensory efferents also localize their somas in the CNS, sending their axon termini to the periphery to provide an important level of regulatory control over sensory inputs. Occasionally these two classes of efferents display a close relationship, as is the case with the facial branchiomotor neurons (FBMNs) and octavolateral efferent neurons (OENs). FBMNs form the motor component of the VIIth cranial nerve, innervating structures of the second pharyngeal arch, while OENs contribute sensory efferent innervation to the ear and lateral line as part of the VIIIth cranial nerve. Shared attributes between these two efferent populations, including the position of their somas, close coupling of their projections, and reliance on common neurotransmitters have led some researchers to consider the OENs a subset of the branchiomotor column, albeit one that innervates sensory structures rather than muscle (Roberts & Meredith, 1989).
There has been significant focus on the specification and migration of the FBMNs, aided in the zebrafish by the development of transgenic lines driven by the islet1 (isl1) regulatory sequences (Higashijima et al., 2000; Mapp et al., 2010; Uemura et al., 2005). However, the islet1 transgenes also label the closely associated but comparatively understudied OENs. Here, we set out to describe OEN development and migration, with special attention to the qualities that set OENs apart from FBMNs.
The migration of the FBMNs begins in the fourth segment, or rhombomere (r), of the vertebrate hindbrain. FBMNs are specified in r4 with input from the homeodomain transcription factor Hoxb1, encoded by hoxb1a in zebrafish (Goddard et al., 1996a; McClintock et al., 2002; Rohrschneider et al., 2007; Studer et al., 1996). After their births in r4, FBMNs in zebrafish, mice, and humans migrate posteriorly in two parallel streams, leaving a trailing axon behind them that exits the hindbrain to innervate second pharyngeal arch derivatives (Chandrasekhar et al., 1997). Time-lapse microscopy in zebrafish has shown that FBMNs migrate tangentially along the medial floor plate through r5, after which they undergo a short radial migration before clustering into bilateral nuclei in r6 and r7 (Grant & Moens, 2010; Wanner et al., 2013). Previously, we demonstrated that the initial migration of FBMNs across the r4/r5 border is led by a pioneer neuron, and that ablation of the pioneer results in a partial or complete block to FBMN migration (Wanner & Prince, 2013). Additionally, FBMN migration relies on function of several PCP factors such as Scrib, Vangl2, and Pk1b (Mapp et al., 2011; Walsh et al., 2011; Wanner et al., 2013; Zakaria et al., 2014) and activity of the transcriptional repressor Rest, which is required to keep FBMNs in an immature and migratory state (Kok et al., 2012; Love & Prince, 2015). Furthermore, FBMN migration is a collective process in which cell-cell contacts between neurons are partially mediated by Cadherin-2 (Rebman et al., 2016; Stockinger et al., 2011; Wanner & Prince, 2013).
In contrast, comparatively little is known about the OENs. In zebrafish and most other anamniote vertebrates, OENs project to the lateral line system (LLS), forming part of a sensory circuit that provides information on the surrounding aquatic environment (Blaxter, 1987; Roberts & Meredith, 1989). While the anatomy of the LLS varies between species, its fundamental sensory organ, the neuromast, is conserved. Neuromasts contain mechanosensory hair cells that transmit information to the central nervous system via basally located afferent nerve termini (Flock & Wersgll, 1962). The neuromasts are also supplied by the OENs, which are thought to play both excitatory and inhibitory roles, modulating hair cell sensitivity based on the behavioral state of the animal (Bricaud et al., 2001; Meredith & Roberts, 1987). Single OENs generally innervate multiple neuromasts; however, not every neuromast within an individual animal receives efferent innervation (Blaxter, 1987). Furthermore, in many anamniote species including catfish, goldfish, zebrafish, and Xenopus, single OENs often supply not only the LLS but also the inner ear (Bell, 1981; Bleckmann et al., 1991; Hellniann & Fritzsch, 1996; Metcalfe et al., 1985). As such, they are often considered analogous to the vestibulocochlear efferents of amniotes, which make up the efferent component of the VIIIth cranial nerve.
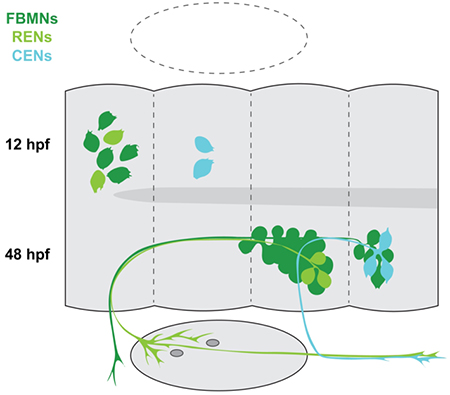
In zebrafish, three octavolateral efferent nuclei have been characterized by backfilling from the periphery at 24 hours post-fertilization (hpf) and beyond (Bricaud et al., 2001; Metcalfe et al., 1985). The most anteriorly-located of the OENs comprise the diencephalic efferents to the lateral line (DELL). In addition, two separate nuclei of rhombencephalic OENs have been described, the rostral and caudal efferent neurons (RENs and CENs, respectively). Our study is focused on the rhombencephalic OENs (schematized in Figure 1a).
Figure 1: Single-cell photoconversions reveal that OENs migrate concurrently with FBMNs.
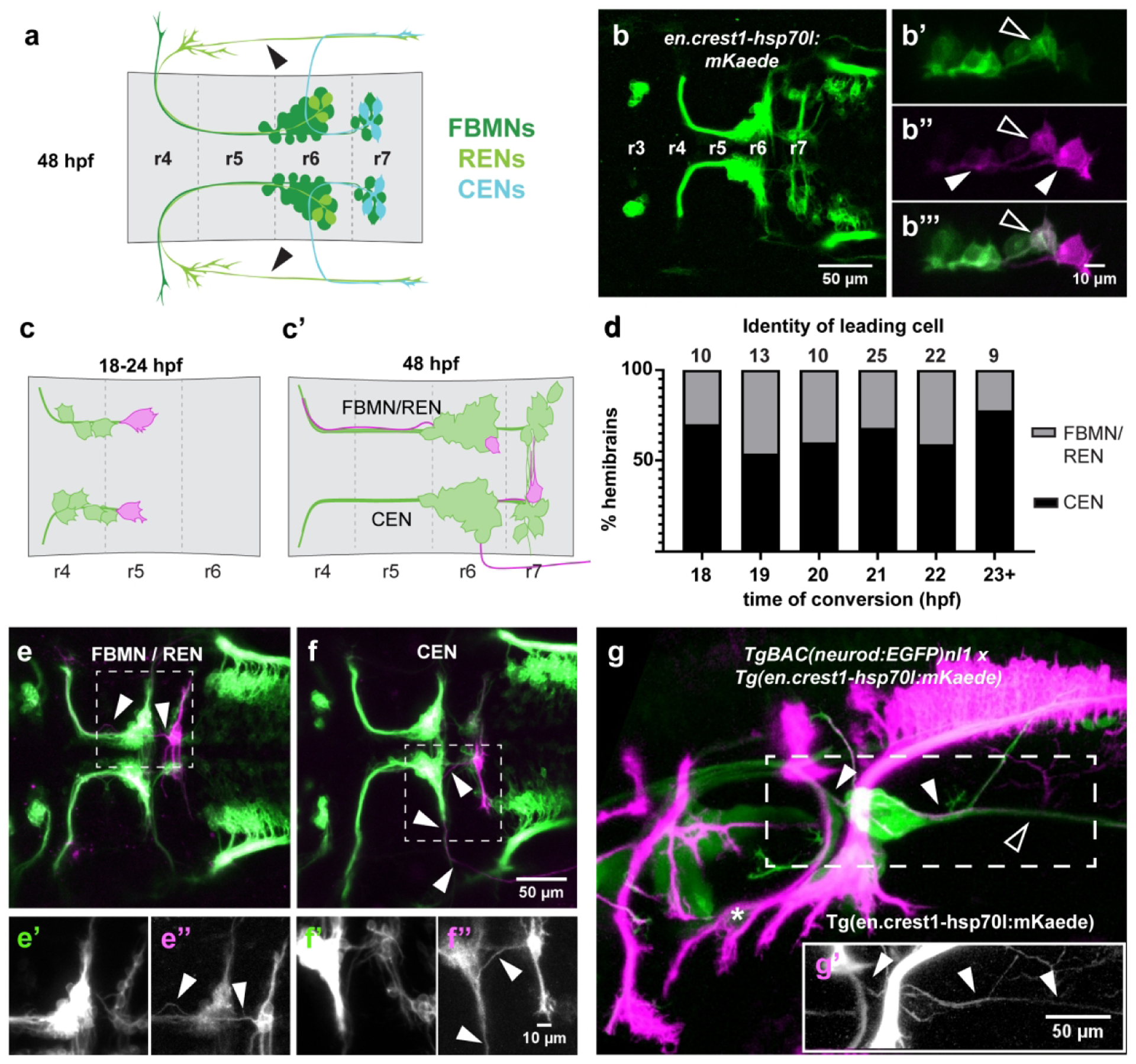
(a) Schematic of FBMN, REN, and CEN somas and axon morphologies at 48 hpf. The REN projection crossing the otic vesicle (arrowheads) is not visible in all specimens. (b) The newly-generated Tg(en.crest1-hsp70l:mKaede) line uses the islet1 zCREST1 enhancer (Uemura et al., 2005) to drive expression of the photoconvertible protein Kaede in a subset of cranial efferent neurons. (b’-b”’) Single-cell labeling via photoconversion of Kaede from green to red. A fully converted leading cell and its trailing axon (closed arrowheads) are identified by the presence of red and absence of green protein. Contrast with partial conversion of a follower, in which green protein remains (open arrowheads). (c) Schematic of photoconversion experiments targeting leading mKaede-expressing neurons between 18–24 hpf and (c’) screening criteria for axon morphology of different cell types at 48 hpf. (d) CENs are present in the leading position in over 50% of embryos at every time point between 18–24 hpf. (e) Axon morphology typical of FBMN/RENs (arrowheads); (e’-e”) insets of boxed area show separated green and red channels, respectively. (f) Axon morphology typical of CENs (arrowheads); (f’-f”) insets of boxed area show separated green and red channels, respectively. (g-g’) Double transgenic line with converted Tg(en.crest1-hsp70l:mKaede) and Tg-BAC(neurod:EGFP)nl1 demonstrates that efferent projections leaving r6 (red; closed arrowheads) fasciculate with the sensory afferent projections to the lateral line (green; open arrowheads). Glossopharyngeal motor neurons (nIX) are indicated by an asterisk.
The rostral efferent neurons (RENs) are located in r6 and project their axons anteriorly to r4 where they exit the hindbrain together with the main fascicle of the FBMNs. The caudal efferent neurons (CENs) cluster loosely in r7 and project their axons anteriorly to r6, where they turn laterally and exit the hindbrain together with the glossopharyngeal nerve (nIX) (Chandrasekhar, 2004; Higashijima et al., 2000). Although both groups of rhombencephalic efferents cluster into larger nuclei with the FBMNs, zebrafish OENs are a fairly rare cell type: there are 2–3 RENs per fish, and the CENs are only slightly more numerous at 3–5 per specimen (Metcalfe et al., 1985). Single hindbrain OENs project either to the anterior or posterior LLS, with RENs displaying a preference for innervating the anterior LLS and CENs more commonly projecting to the posterior LLS (Bricaud et al., 2001). RENs typically also innervate the ear (Metcalfe et al., 1985; Sapede et al., 2005).
Little is known about the migration of the OENs. RENs migrate from r4 to r6 with the FBMNs, and they are at least partially responsive to Sdf1a gradients (Sapède et al., 2005). There is indirect evidence that CENs migrate from r6 to r7 after 24 hpf, but whether they perform additional movements before 24 hpf is unknown (Sapède et al., 2005). Even less is known about OEN birthplace and specification. Experiments in mouse have shown that the vestibulocochlear efferent neurons are specified alongside the FBMNs by Hoxb1 function in r4 (Goddard et al., 1996; Pata et al., 1999; Studer et al., 1996), but the exact timing of their birth and their molecular divergence from FBMNs is unknown. Despite previous analyses of zebrafish hoxb1a gene function (McClintock et al., 2002; Rohrschneider et al., 2007), a role for Hox genes in OEN development has not been investigated, largely due to the absence of molecular markers to distinguish OENs from FBMNs.
In this study, we provide an in depth analysis of the development of zebrafish rhombencephalic OENs, allowing comparison of their properties with those of the better described FBMN population. Our study was facilitated by establishment of a new Tg(en.crest1-hsp70l:mKaede) transgenic line, which allows for labeling of single efferent neurons and their projections via photoconversion. We find that OENs migrate concurrently with FBMNs beginning at ~18 hpf, and they reach their final destinations by 48 hpf. Using time-lapse microscopy and cell tracking methods, we demonstrate that the CENs are born in r5, in a region spatially distinct from the more anterior origin of RENs and FBMNs. We also demonstrate that similar to FBMNs, migration of RENs depends upon function of the r4-expressed Hox gene, hoxb1a. However, while FBMN migration is additionally dependent on the function of the Hoxb1a downstream effector gene pk1b, REN migration is pk1b-independent. This finding reveals that RENs and FBMNs are subject to distinct molecular control. Finally, we investigate physical interactions between FBMNs and OENs during their migration. We show that r4-derived FBMN/RENs and r5-derived CENs can make contact across the r4/r5 border. However, neuron ablation experiments suggest that these interactions are not necessary for successful migration of FBMNs or OENs.
2. Materials and Methods
2.1. Transgenic lines and fish husbandry
Zebrafish (Danio rerio) were maintained according to IACUC-approved protocols. Embryos were raised in E3 solution (in mM: 5.0 NaCl, 0.17 KCl, 0.33 CaCl2, 0.33 MgSO4) at 21.5–28.5 °C and staged following standard morphological criteria (Kimmel et al., 1995). Specimens analyzed at stages later than 24 hours post-fertilization (hpf) were treated with 0.2 mM 1-phenyl 2-thiourea (PTU; Sigma) to inhibit melanin synthesis. Transgenic and mutant lines used in this study include Tg(en.crest1-hsp70l:mRFP)ch102 (Mapp et al., 2010), Tg(h2az2a:h2az2a-GFP) (Pauls et al., 2001), TgBAC(neurod:EGFP)nl1 (Obholzer et al., 2008), Tg(isl1:GFP) (Higashijima et al., 2000), and pk1bf122 (Mapp et al., 2011).
A new transgenic line, Tg(en.crest1-hsp70l:mKaede)ch104, was established as follows: Primers containing SalI (FOR 5’ TTAGTCGACATGAGTCTGATTAAACCAGAAAT 3’) and BamHI (REV 5’ TTCACACACGAGAGGACTGGATCCATT 3’) restriction sites were used to PCR amplify a CaaX-modified variant of the photoconvertible protein Kaede (Ando et al., 2002), kindly provided by Drs. Clare Buckley and Jon Clarke. The part of the primer sequence complementary to the CaaX motif is indicated in bold. The resulting sequence was cloned downstream of the zCREST1 enhancer (a subset of the isl1 regulatory sequence) and hsp70l minimal promoter in a plasmid containing Tol2 transposon sequences (Kawakami & Shima, 1999; Mapp et al., 2011; Uemura et al., 2005). We injected this construct as circular DNA into single-cell *AB embryos at a concentration of 80 ng/μl, together with 80 ng/μl of capped Tol2 transposase mRNA transcribed using the MEGAscript SP6 Kit (Ambion) according to manufacturer’s instructions. Injected embryos were raised to adulthood. The progeny of these fish were screened for fluorescence and a single adult founder, selected for bright transgene expression, was outcrossed to generate a stable transgenic line.
2.2. Generation of 3D-printed embryo molds
Three separate molds were designed to hold 8–10 embryos at the 18, 24, or 48 hpf stages, using TinkerCad and rendered in MeshLab. They were then 3D printed using acrylonitrile butadiene styrene (ABS) at the Polsky Exchange Fabrication Lab. Imprints of these molds were made with 3% agarose dissolved in E3, providing a semi-rigid structure that enabled rapid and reproducible orientation of embryos with the hindbrain in dorsal view for photoconversion experiments. Cast designs are open-source and hosted for download at the NIH 3D Print Exchange, accessible via https://3dprint.nih.gov/users/beiriger. [Code will be made available on publication of the manuscript.]
2.3. Photoconversions and data analysis
Photoconversions were performed on a Zeiss LSM 710 upright confocal microscope (Carl Zeiss Microscopy, Thornwood, NY) using a 40x/1.0 W Plan-Apochromat objective. A region of interest (ROI) was selected and repeatedly scanned with 405 nm light until the majority of green fluorescent Kaede protein in a given structure had been converted by photo-cleavage to the red fluorescent form (Ando et al., 2002). It should be noted that despite careful ROI selection, the close proximity of individual neurons can on occasion lead to partial photoconversion of Kaede in adjacent neurons, due to scattering of UV light within the tissue. For these reasons, converted neurons were identified not only based on the strength of signal in the red channel, but also the absence of signal in the green channel. At 48 hpf, confocal stacks were collected from converted specimens, and the location and morphology of red-labelled neurons was recorded.
2.4. Single-plane illumination microscopy (SPIM) and cell tracking
Zebrafish embryos were staged to 10 hpf and mounted in Fluorostore Fractional FEP Tubing (F018153-5) using a modified multilayer technique (Kaufmann et al., 2012). Embryos were immobilized using 0.3% agarose (Invitrogen UltraPure Agarose #16500) dissolved in E3 medium and 0.2 mg/ml tricaine, and the FEP tubing was capped with a 1.2% agarose plug. Embryos were incubated at 28.5°C during data collection. Images were captured with a Zeiss Lightsheet Z.1 single-plane illumination microscope (Carl Zeiss Microscopy, Thornwood, NY) with tandem PCO.edge sCMOS cameras (PCO.Imaging, Kelheim, Germany) and Zeiss Zen imaging software. A 20x/1.0 long working distance detection objective was used alongside a pair of 10x/0.2 dry illumination objectives, and the excitation sheet was narrowed to 2.0 μm. Volumes were acquired every two minutes between 11–23 hpf, with 10 ms exposure per slice for both green (488 nm, 7.5%) and red (561 nm, 7.0%) channels.
Cell tracks were manually reconstructed using the mTrackJ plugin in FIJI (Meijering et al., 2012). Data were exported to a custom Python script for plotting of cell movements and birthplaces. The code is open-source and available for download from https://github.com/aebeiriger/plot-tracks. [Code will be made available on publication of the manuscript.] This script includes functions to plot tracking data and cell birthplaces, smooth random cell movements, and register cell locations to the tip of the notochord. Since the majority of cell births occurred between 13–14 hpf, we used morphological landmarks from the 13–14 hpf embryo to plot cell tracks. Embryos were incubated in a 1X working solution of CellMask Deep Red plasma membrane stain (ThermoFisher, C10046) for one hour, and washed in E3 for 15 minutes prior to imaging. We took detailed measurements from the resulting images. The anterior limit of the notochord lies at the r3/4 boundary between 13–14 hpf, providing an anatomical landmark that allows us to compare birthplaces between embryos. Rhombomere widths were estimated based on published expression patterns of Krox20, a marker of r3 and r5, in 14 hpf embryos (Ma et al., 2014; Prince et al., 1998) and confirmed by measurement of CellMask labeled live specimens.
2.5. Cell ablation experiments
Cell ablation experiments were performed as previously described (Wanner & Prince, 2013), using an inverted Leica SP5 Tandem Scanner Spectral 2-photon confocal microscope (Leica Microsystems, Inc., Buffalo Grove, IL). Phenotypes were scored on a Zeiss LSM 710 upright confocal microscope (Carl Zeiss Microscopy, Thornwood, NY) using a 40x/1.0 W Plan-Apochromat objective. We note that the configurations of these particular microscopes produce mirror images; thus care was taken to ensure phenotypes were correctly assigned to right- and left-hand sides of the specimen before quantification. Ablation phenotypes were quantified with one-way ANOVA tests performed using GraphPad Prism version 7.00 for Windows, GraphPad Software, La Jolla California USA, www.graphpad.com.
Quantifications were corrected for the number of CENs in r6 as follows: in each embryo with no ablation or an r4-only ablation, 2 or 3 cells were subtracted from the r6 totals; in each embryo with an r5-only or two-cell ablation, 1 or 2 cells were subtracted from r6 totals. The higher number was used only in rare cases where 10 or more neurons were visible in r6. These corrections are based on the number of CENs we found per hemibrain in the time-lapse experiments described above. We assumed that a single CEN was ablated in the r5-only and two-cell ablation conditions.
2.6. Morpholino injections
Morpholino oligonucleotides (MOs) used in this study have been previously described. These include start-codon targeting morpholinos to hoxb1a (McClintock et al., 2002) and splice-blocking morpholinos targeting pk1b (Rohrschneider et al., 2007). All morpholinos were synthesized by Genetools, LLC and solubilized in water to a stock concentration of 10 ng/nl. Single-cell embryos were injected at a concentration of 1 ng/embryo for hoxb1a knockdowns and 2.5 ng/embryo for pk1b knockdowns. We did not observe any off-target phenotypes in morpholino-injected embryos.
3. Results
3.1. A new Kaede transgenic line allows visualization of single efferent neurons and their projections
We began our analysis of the zebrafish rhombencephalic octavolateral efferent neurons (OENs) by developing a new transgenic tool that facilitates single cell labeling of Islet1-positive (Isl1(+)) neurons. The new line, Tg(en.crest1-hsp70l:mKaede), uses the zCREST1 enhancer element from the islet1 locus (Uemura et al., 2005) to drive a membrane-targeted variant of the Kaede fluorophore in zebrafish cranial efferent neurons (Figure 1b). Exposure to UV light cleaves Kaede protein, converting it from green to red fluorescence. A CaaX motif was added at the C-terminal end of the molecule, ensuring the red fluorescent variant of the protein remains membrane bound and permitting visualization of fine cellular projections from individual neurons (Figure 1b’–b’”). After photoconversion, red fluorescent Kaede remains visible for around 3 days, allowing us to identify the same cell through multiple developmental stages.
Importantly, the Tg(en.crest1-hsp70l:mKaede) line is extremely bright and accrues fluorescence earlier than the Tg(en.crest1-hsp70:mRFP) and Tg(isl1:GFP) lines used in our previous studies (Higashijima et al., 2000; Mapp et al., 2010). As a consequence, cranial efferent neurons become visible at slightly earlier stages in the Tg(en.crest1-hsp70l:mKaede) line. This includes the facial branchiomotor neurons (FBMNs), which we can reliably detect by 17 hpf, an hour earlier than with the previous transgenic lines. We will return to this point as we compare findings in this study with those reported previously.
3.2. OENs migrate concurrently with FBMNs through the hindbrain
We first sought to establish the relative timing of FBMN and OEN migration. Using our new Tg(en.crest1-hsp70l:mKaede) line, we investigated the Isl1(+) neurons that migrate through r5 and r6 in a chain-like manner. In a series of experiments, we photoconverted individual leading (most posteriorly located) neurons, in embryos staged between 18 and 24 hpf (Figure 1c). We then screened for red fluorescence at 48 hpf, scoring the identity of converted cells in each hemibrain based on their axon morphology (Figure 1c’).
Morphological features typical of the FBMNs and the REN population of OENs are shown in Figure 1e–e”, where a converted cell projects its axon anteriorly to exit the hindbrain from r4. Conversely, axons of the CEN population of OENs project anteriorly for a shorter distance, before exiting the hindbrain from r6 (Figure 1f–f”). For the purposes of quantitative analysis FBMNs and RENs were grouped into a single category, since individual RENs displayed considerable variation in their axon branch points and therefore could not be reliably distinguished from FBMNs. We found that the leading neurons ultimately displayed CEN morphology in more than 50% of the hemibrains assayed (Figure 1d), independent of the stage at which photoconversion was performed. Additionally, leading neurons and most of their immediate followers terminated migration in r7, regardless of cell identity (Supplemental Figure 1a–c). In many cases, leading cells in each hemibrain of a single embryo also displayed differing identities (Supplemental Figure 1d–f).
Other cell populations are known to use the r6 exit point, namely the glossopharyngeal (nIX) motor neurons. Projections of the nIX are labeled by Tg(en.crest1-hsp70l:mKaede), but these turn rostrally and route under the otic vesicle after leaving r6 (asterisk; Figure 1g) rather than turning caudally and projecting down the tail of the fish as is typical for the CENs. To affirm that Isl1(+) neurons that project caudally from the r6 exit point are indeed CENs, we crossed fish carrying Tg(en.crest1-hsp70l:mKaede) with fish of the TgBAC(neurod:EGFP) line. TgBAC(neurod:EGFP) labels sensory afferent projections, including those of the posterior lateral line (PLL, Obholzer et al., 2008). We carried out whole-embryo photoconversions so that all Isl1(+) efferents were labeled in red, in contrast with the Nrd(+) sensory afferent projections expressing EGFP. In double-transgenic embryos, a subset of Isl1(+) axons (converted, red) fasciculate with Nrd(+) sensory afferent projections to the PLL (closed arrowhead, green) and project together down the tail of the fish (Figure 1g–g’). This common axon routing pattern confirms that the Isl1(+) neurons that project posteriorly from the r6 exit point are indeed CENs.
Together, these results demonstrate that OENs migrate concurrently with FBMNs, beginning as early as 18 hpf. Additionally, the leading, or most posteriorly-located, Isl1(+) cell is often a CEN rather than an FBMN or REN. This finding raises the intriguing question of whether the previously described ‘pioneer’ neuron (Wanner and Prince, 2013) could be a CEN rather than an FBMN, a topic we return to below. Finally, CEN axons have established their characteristic morphology by 48 hpf, using the r6 exit point and sending their growth cones posteriorly along the same axon tract as sensory projections of the PLL.
3.3. CENs do not share a common spatial origin with FBMNs
To examine the initial birthplaces of OENs, we performed a series of backtracking experiments designed to visualize OEN and FBMN progenitor divisions. We created a double transgenic line by crossing fish carrying Tg(en.crest1-hsp70l:mRFP), which labels cranial efferents with RFP, with fish carrying Tg(h2az2a:h2az2a-GFP), which labels all cell nuclei via expression of a GFP-tagged histone protein (Pauls et al., 2001). Using this double transgenic, we performed single-plane illumination microscopy (SPIM) to generate detailed time-lapse movies for cell lineage reconstruction (Movie 1). Data acquisition was started at 11.5 hpf, during early stages of neurulation, and stopped close to 24 hpf, after Isl1(+) neurons had begun to migrate into r6. Because expression of Tg(en.crest1-hsp70l:mRFP) can only be reliably detected after 18 hpf, we identified FBMNs and OENs at the end of each time-lapse movie and used nuclear Tg(h2az2a:h2az2a-GFP) to track these cells back through time to their birthplaces.
In Figure 2, we provide an overview of all cell tracks and birthplaces. As expected, the vast majority of tracks begin in r4, where hoxb1a function has been shown to confer FBMN identity (McClintock 2002). Furthermore, most Isl1(+) neurons move posteriorly after their births, reflecting the well-described tangential migration of the FBMNs (Figure 2a). However, in each specimen, a subset of Isl1(+) neurons was born in r5 (Figure 2b), posterior to the domain of elevated hoxb1a expression. Detailed measurements of rhombomere width during early neurulation, and rhombomere placement relative to morphological landmarks such as the notochord, support this r5 origin (Supplemental Figure 2a–b). Across three embryos, r5-derived neurons made up roughly 20% of all tracked cells (Figure 2c). After 22 hpf, the r5-derived neurons consistently sent cellular projections out of the r6 exit point (Figure 2d, arrowheads), as would be expected of CENs.
Figure 2: Caudal efferent neurons (CENs) do not share a common developmental origin with FBMNs.
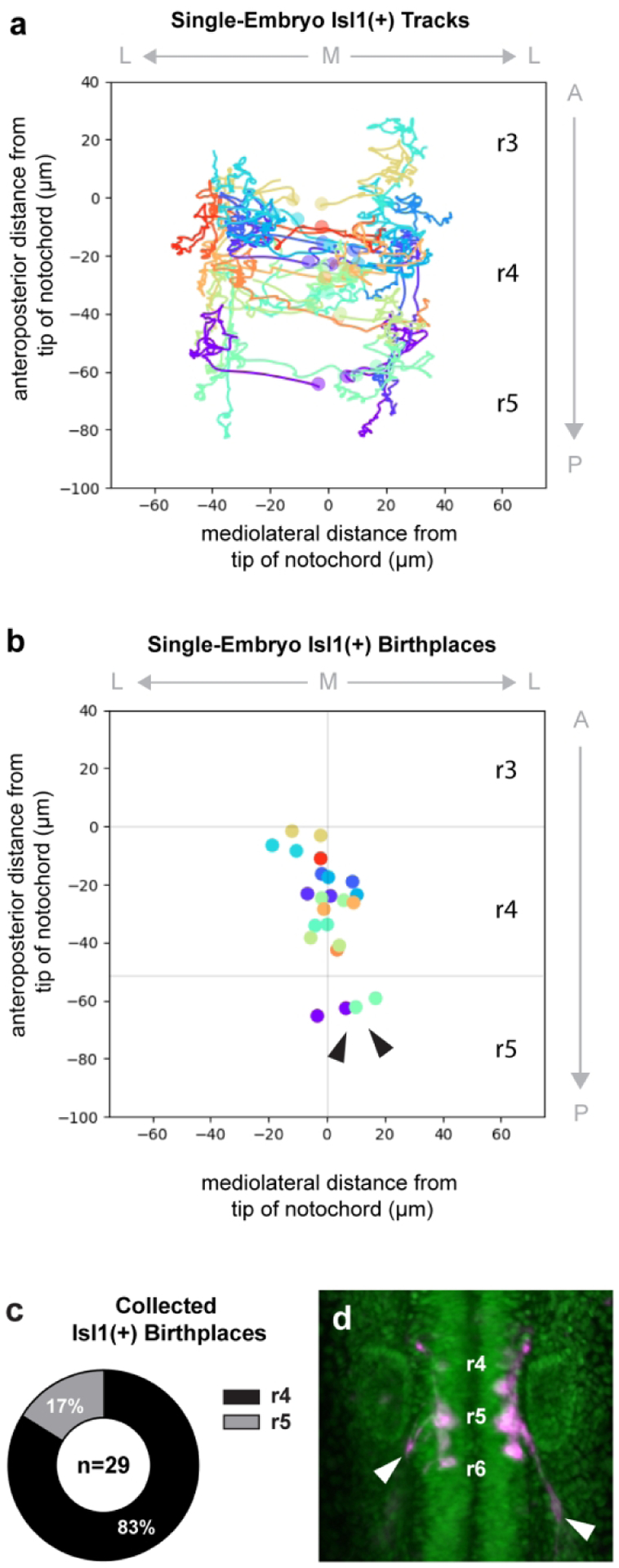
Cell lineages were reconstructed by tracking the nuclei of neurons carrying both Tg(h2az2a:h2az2a-GFP) and Tg(en.crestl1-hsp70l:mRFP). (a) Cell tracks from one specimen are graphed on an anatomical plot, where the origin is placed at the tip of the notochord. Each lineage is represented by a different color, and circles indicate cell position at the time of division. (b) Birthplaces of all tracked neurons from the same specimen are plotted; sister cells are color-coded by lineage. Two progenitor divisions occur close to the midline in r5 rather than r4, giving rise to Isl1(+) CENs (arrowheads). (c) Aggregated data from three embryos show that 17% of all Isl1(+) cells tracked between 13–24 hpf are born in r5. Here, n refers only to Isl1(+) neurons and excludes Isl1(−) sister cells. (d) Movie still from a SPIM time-lapse of the Tg(h2az2a:h2az2a-GFP) and Tg(en.crestl1-hsp70l:mRFP) double transgenic line. By 22 hpf, neurons born in r5 have sent out projections characteristic of CENs (arrowheads).
However, other cell populations such as the aforementioned glossopharyngeal (nIX) motor neurons are also known to use the r6 exit point. Therefore, we sought to confirm the identity of neurons using the r6 exit point at these early stages by performing a series of axon conversions. We photoconverted the lateral projections in 20 hpf Tg(en.crest1-hsp70l:mKaede) embryos, allowing the converted Kaede protein to diffuse back into the cell soma (Figure 3a–a’). Every cell converted in this manner displayed CEN-like morphology at 48 hpf. These neurons localized to r7, sent contralateral projections across the midline, and projected their axon through the r6 exit point and posteriorly towards the lateral line (Figure 3a”, n=8/8). This is a markedly different projection pattern from nIX neurons, which turn anteriorly after exiting the brain at the r6 exit point. We do note that the timing of CEN axon outgrowth varies between hemibrains (Supplemental Figure 3). Taken together, the axon conversions and our backtracking experiments indicate that CENs have a more posterior segmental origin than the FBMNs and RENs. Additionally, they demonstrate that CENs can be reliably identified by their laterally projecting axons during early stages of migration.
Figure 3: CENs send axons towards the r6 exit point during early stages of migration.
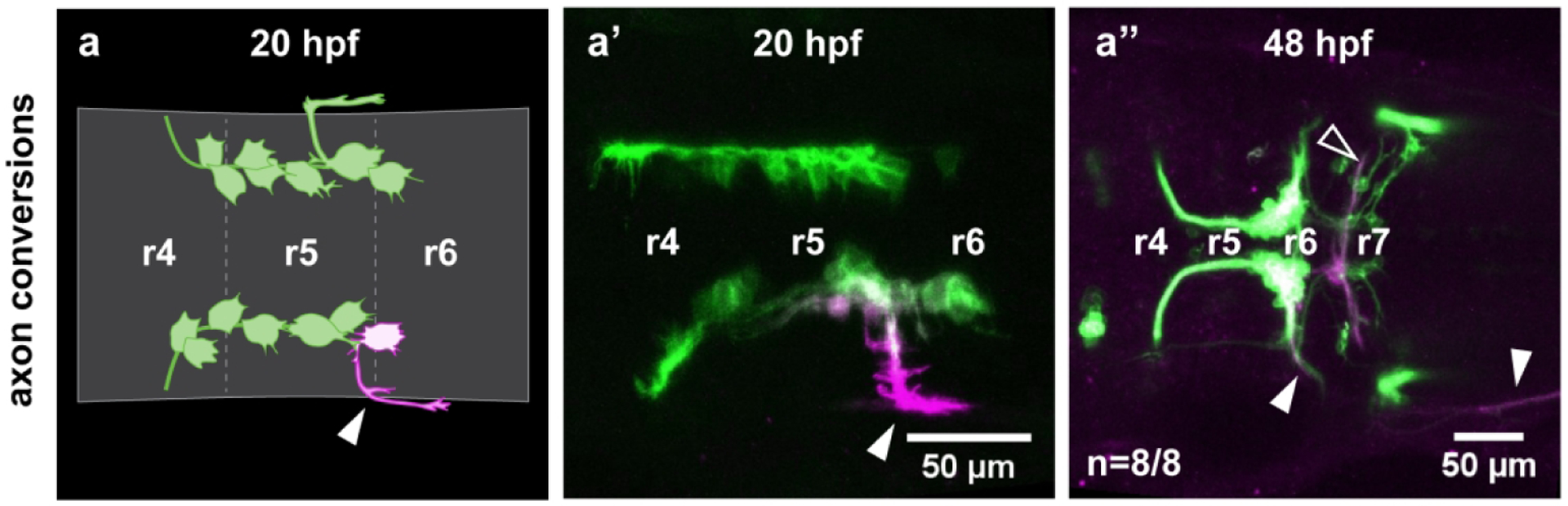
Axon conversions were performed in Tg(en.crest1-hsp70l:mKaede) embryos to determine the identity of laterally-projecting neurons. (a-a’) Photoconversion of lateral projections (closed arrowhead) at 20 hpf allows converted protein to diffuse back into the cell soma. (a”) Neurons that send out lateral projections at 20 hpf display morphology typical of CENs at 48 hpf, including contralateral dendritic projections (open arrowhead) and posteriorly-routed axons (closed arrowhead; n=8/8).
Despite their different segmental origins, FBMNs and both populations of OENs are born at similar developmental stages. Neurons arising from r4, comprising both FBMNs and RENs, were born continuously during neural plate and keel stages, between 11 and 14 hpf (n=24/29). The r5-derived CENs were born closer to 12 hpf (n=5/29). These data reflect only a subset of Isl1(+) neurons in each fish, since some later-born neurons do not accrue enough fluorescent protein to be detectible at 24 hpf. However, we can conclude that the divisions from which FBMNs and OENs arise occur during early neurulation, and while their birthdates are very similar, the r5-derived CENs are born slightly later than the earliest born r4-derived efferents.
3.4. Hoxb1a function is required for REN but not CEN migration
As a second approach to investigating the separate segmental origins of RENs and CENs, we examined the role of hoxb1a in each cell type. The hoxb1a transcript is expressed at high levels in a characteristic r4 ‘stripe’, as well as at much lower levels more posteriorly (Prince et al., 1998). When Hoxb1a function is disrupted, FBMNs fail to migrate out of r4 and instead display characteristics reminiscent of the r2-derived trigeminal (nV) motor neurons, a phenotype consistent with a classic anteriorizing homeotic transformation (McClintock et al., 2002). Given that FBMNs and RENs are born together in r4, we expected that knockdown of hoxb1a would also affect REN migration. By contrast, as hoxb1a is not expressed at significant levels in r5, we hypothesized the r5-born CENs might migrate normally to their typical r7 location in the absence of Hoxb1a function.
To test this model, we performed Hoxb1a morpholino knockdown experiments as previously described (McClintock et al., 2002). In wild-type embryos, REN axons branch extensively upon reaching the otic vesicle, while CENs project posteriorly towards the lateral line as previously described (Figure 4a–a’). In Hoxb1a-deficient embryos, the CENs appear undisturbed, migrating normally to r7 and successfully routing their axons towards the lateral line (Figure 4b–b’; n=32/32). Meanwhile, as we previously reported (McClintock et al., 2002), r4-derived Isl1(+) neurons fail to migrate posteriorly (Figure 4b–b’; n=32/32). Interestingly, in Hoxb1a morphants no RENs were found in r6, providing additional support for a model in which RENs share characteristics with FBMNs. Not only do FBMNs and RENs arise together in r4, but both cell types require Hoxb1a function in order to migrate.
Figure 4: Rostral efferent neurons (RENs) are specified by hoxb1a in r4, but do not need pk1b to migrate.
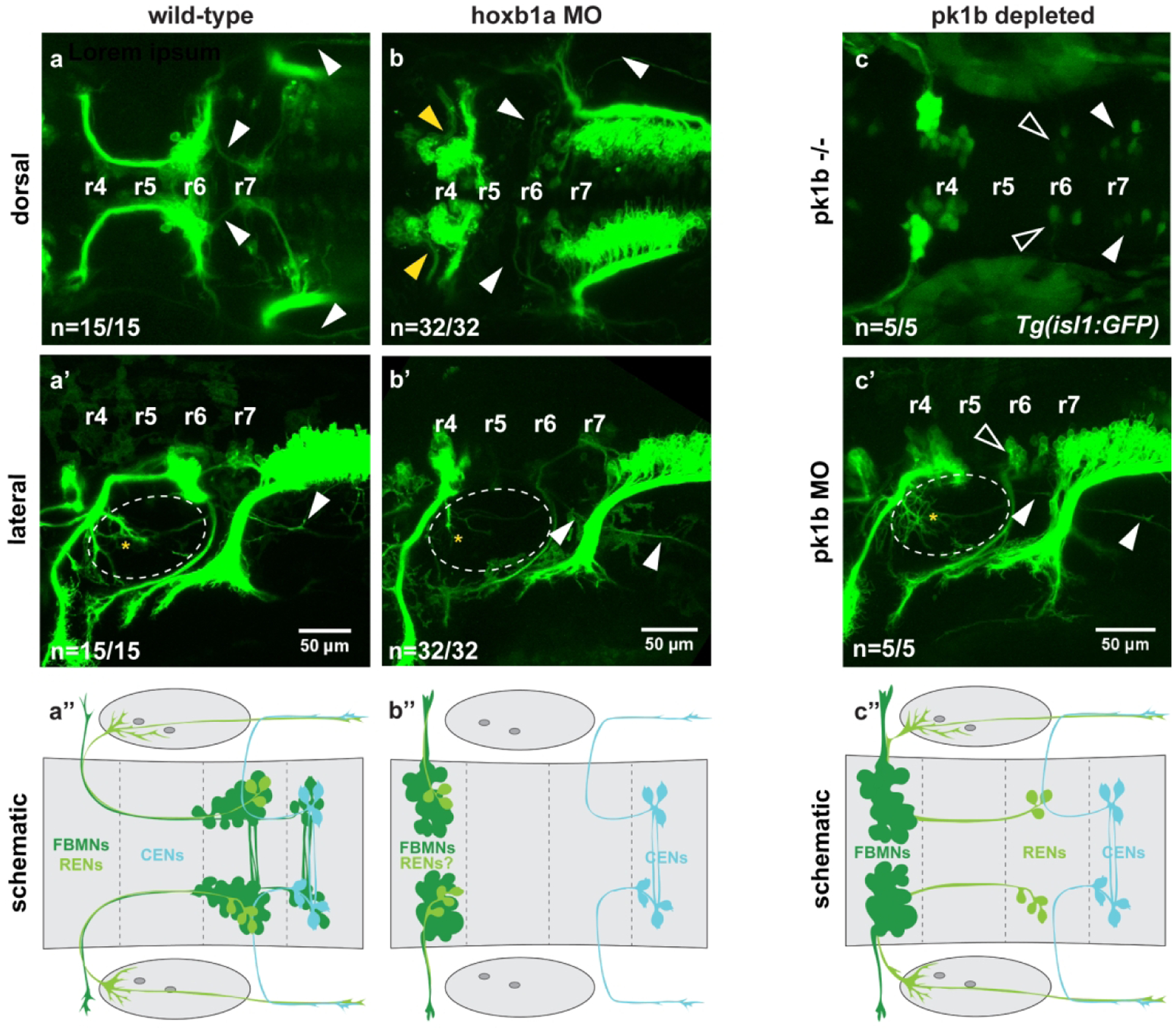
(a-a’) Wild-type Tg(en.crest1-hsp70l:mKaede) embryos show characteristic projections of RENs (yellow asterisk) and CENs (arrowheads) in dorsal and lateral views (n=15/15 embryos). (a”) Schematic indicates wild-type cell soma locations. (b-b”) Morpholino knockdown of hoxb1a in the Tg(en.crest1-hsp70l:mKaede) transgenic background results in a complete block to FBMN migration (yellow arrowheads); notably, RENs are absent from r6 but CENs still migrate successfully (arrowheads) (n=32/32 embryos). Otic vesicles are indicated with a dotted line. (c) In pk1b homozygous mutants, RENs (open arrowheads) and CENs (closed arrowheads) are still able to migrate, indicating that the absence of r6-localized RENs in hoxb1a depleted fish is not due to downstream effects on pk1b expression (n=5/5 embryos). Transgenic background is Tg(isl1:GFP). (c’) Morpholino knockdown of pk1b in Tg(en.crest1-hsp70l:mKaede) fish demonstrates that projections of RENs (asterisk) and CENs (closed arrowhead) are undisturbed in the absence of Pk1b (n=5/5 embryos). Otic vesicles are indicated with a dotted line. (c”) Schematic indicates soma and projection locations of all three cell types in Pk1b-depleted embryos.
3.5. OEN migration is independent of Pk1b function
We next sought to determine whether OEN migration relies on the function of pk1b, a downstream target of Hoxb1a (Rohrschneider et al., 2007). Not only is pk1b expressed specifically in the FBMNs, but it also functions cell-autonomously within these neurons to facilitate their migration (Mapp et al., 2011; Mapp et al., 2010; Rohrschneider et al., 2007). Since our knockdown results demonstrated that CEN migration is independent of Hoxb1a, we predicted that CEN migration was similarly unlikely to rely on Pk1b function. To test this prediction, we examined pk1bfh122/fh122 homozygous mutants in the Tg(isl1:GFP) background, which provides a cytoplasmic label of Isl1(+) neurons. We found that in pk1b mutant embryos, r6 and r7 each contained 6–10 Isl1(+) cells. The neurons in r7 likely correspond to CENs, while the r6 cells are likely to be RENs (Figure 4c, n=5/5), suggesting that both classes of hindbrain OENs migrate independently of Pk1b. Consistent with this interpretation, pk1b morpholino knockdown in Tg(en.crest1-hsp70l:mKaede) embryos confirms that neurons localized in r6 and r7 display axon morphology characteristic of the REN and CEN classes of OENs (Figure 4c’, n=5/5). The successful migration of RENs in the absence of Pk1b function indicates that despite sharing a spatial origin with FBMNs, they nevertheless rely on different molecular mechanisms to migrate.
3.6. The leading CEN does not act as a pioneer neuron
In a previous study, we described the unique ability of the leading FBMN to pioneer a route through the neuroepithelium for its followers (Wanner & Prince, 2013). Here, we revisit the pioneer hypothesis in light of the experimental findings presented above on OEN birth and migration. Our original pioneer ablation experiments demonstrated that the pioneer performs its function at 18 hpf, around the time that migrating FBMNs first cross the r4/5 border (Wanner & Prince, 2013). Given our new evidence that CENs begin migrating as early as 18 hpf and are often found leading the migrating chain of Isl1(+) neurons, we sought to investigate whether the pioneer neuron might be a CEN. Similarly, we wanted to address whether contact between an r4-derived FBMN/REN and an r5-derived CEN might provide a mechanism for pioneer activity.
To determine when contact first occurs between FBMN/RENs and CENs, we analyzed cellular protrusions in a Tg(en.crest1-hsp70l:mKaede) embryo. Our time-lapse data reveal that r4-derived FBMN/RENs make transient contacts with r5-localized CENs at 18 hpf. Between 18–19.5 hpf, contact between the two cells persisted for an average of three frames (24 minutes). Eventually, these contacts appear to be stabilized, with FBMN/REN protrusions contacting branches of the CEN axonal arbor across the r4/5 border (Movie 2, Figure 5a–a’”). After 19.5 hours, the duration of contacts between an FBMN/REN and CEN approximately tripled, reaching an an average of 9 frames (72 minutes). We note that several branches of the CEN axon used as early points of contact by FBMN/RENs are later retracted as the CEN soma migrates towards the posterior (Supplemental Figure 4).
Figure 5: Interactions between neuron classes and their impact on neuronal migration.
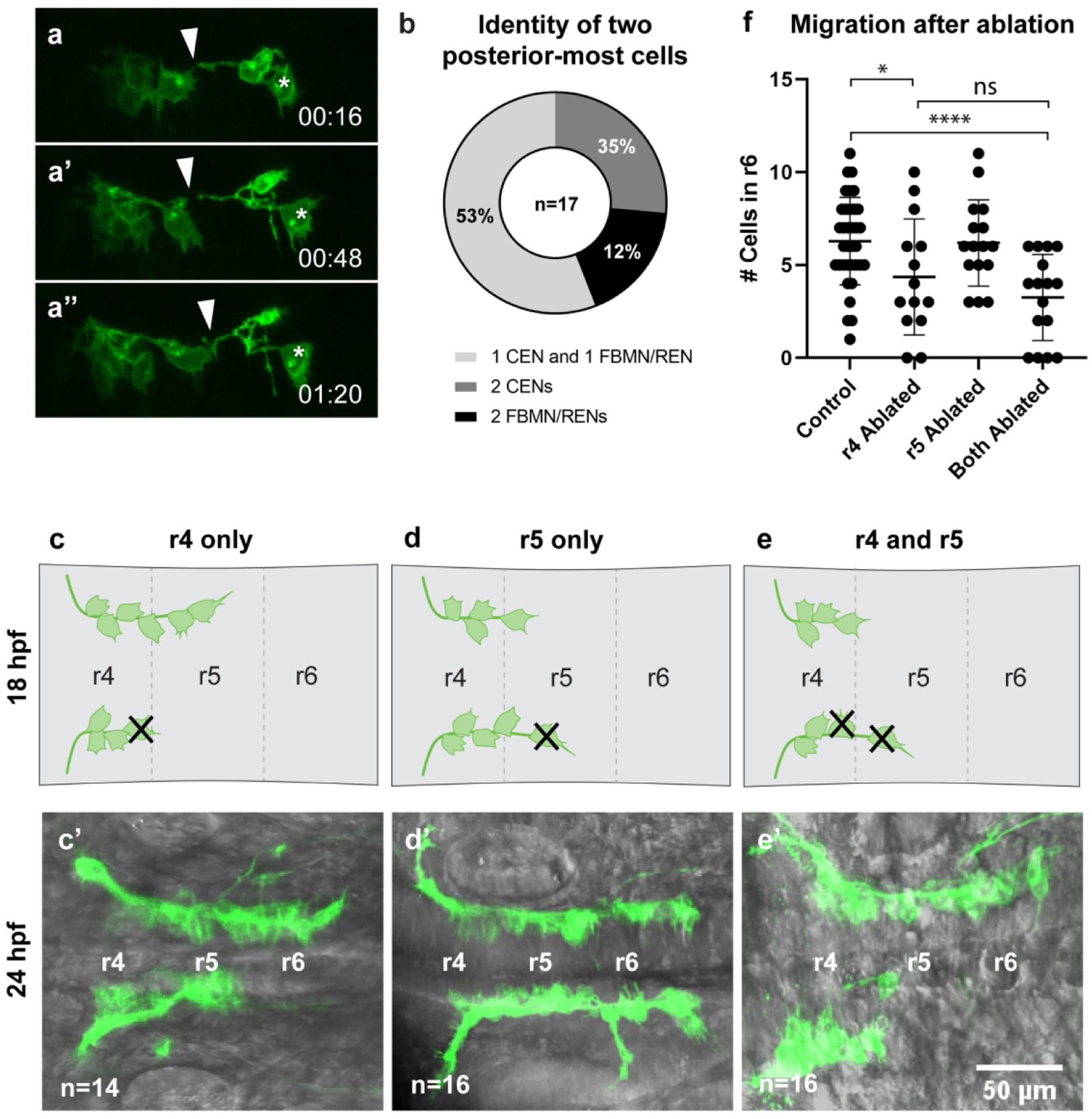
(a-a”’) Time-lapse stills of embryos carrying Tg(en.crest1-hsp70l:mKaede), with time points as indicated in minutes. Time-lapse analysis begins at 18hpf. FBMN/RENs make transient contacts (arrowheads) with CENs (asterisks) during migration which are later stabilized. (b) Quantification of two-cell conversions indicates that one CEN and one FBMN/REN typically occupy the leading position together at 18 hpf. (c-c’) Ablation of a leading neuron localized in r4 was performed at 18 hpf, and neuronal migration assayed at 24 hpf. (d-d’) Ablation of a leading neuron localized in r5 was performed at 18 hpf, and neuronal migration assayed at 24 hpf. (e-e’) Ablation of two leading neurons - one localized in r4 and one in r5 - was performed at 18 hpf, and neuronal migration assayed at 24 hpf. (f) Number of neurons located in r6 after each class of ablation. Ablation of single leading neurons in r4 led to a significant decrease in the number of neurons reaching r6 in comparison to unablated controls (*P<0.05, n=14), while ablation of a pair of leading neurons led to a highly significant block in migration in comparison to unablated controls (****P<0.0001, n=16). There is not a statistically significant difference between the number of neurons reaching r6 in single r4-localized ablations and double r4/r5-localized ablations.
If contact between these two cell types is necessary for FBMN/RENs to cross the r4/5 border, we would expect to find FBMN/RENs and CENs consistently localizing together at the leading position at 18 hpf. We selected 18 hpf specimens in which neurons carrying Tg(en.crest1-hsp70l:mKaede) had reached or just crossed the r4/5 border and photoconverted two leading migratory neurons per hemibrain. Two rounds of photoconversion were performed: the first at 18 hpf, when Kaede protein had just begun to accrue, and the second at 20 hpf, when each red-labeled neuron was again exposed to UV light to boost the red fluorescent signal. At 48 hpf, we observed that a majority of hemibrains displayed one converted FBMN/REN and one converted CEN (53%, n=34, Figure 5b). These results leave open the possibility of a mechanism in which observed pioneer activity is influenced by the leading FBMN/REN reaching across the r4/5 boundary to contact a CEN.
To test whether this contact is necessary for migration, we next performed a series of ablation experiments, similar in design to those we previously reported (Wanner & Prince, 2013). In 18 hpf specimens, we ablated cells based on their precise location in the hindbrain, which varies both between embryos and between hemibrains at the equivalent stage (schematized in Figure 5c–e). In specimens where all observable Isl1(+) neurons were still localized in r4, and thus had not yet migrated across the r4/5 boundary, we ablated a single leading neuron (Figure 5c). Based on our photoconversion analyses, in this experimental paradigm the ablated neuron was either an FBMN or a REN. As shown in Figure 5c’, and summarized in Figure 5f, the ablation of a single r4-localized FBMN/REN significantly abrogated migration (P<0.05), consistent with our previously reported findings (Wanner & Prince, 2013). In those specimens where one Isl1(+) neuron was located in r5, we ablated either one or two leading neurons (Figure 5d,e). In these experiments, the leading neuron could represent either an r4-derived FMBN/REN that had just crossed the r4/5 boundary, or an r5-derived CEN. As shown in Fig. 5d’ and summarized in Fig. 5f the ablation of a single neuron in r5 had no discernible effect on migration, with the resulting range of phenotypes resembling those of unablated control specimens (P=0.89). Interestingly, in those cases where we ablated two leading neurons in a single hemibrain we found significant abrogation of migration (Figure 5e’, f) in comparison to controls (P<0.0005). However, this effect was not significantly different from the outcome of ablation of a single r4-localized neuron.
We note that the ablations performed in this study resulted in a less severe phenotype (i.e. more Isl1(+) neurons visible in r6) than our previously published 18 hpf ablation experiments (Wanner & Prince, 2013). We suspect that this discrepancy is related to use of the new Tg(en.crest1-hsp70l:mKaede) transgene, which is brighter than the Tg(islet1:GFP) and Tg(en.crest1-hsp70l:mRFP) transgenes we used previously. In our raw data, r6-localized neurons are often visible even after ablation of a single pioneer in r4, leading us to suspect that the èxtra’ r6-localized cells are CENs that migrate independently of the pioneer neuron. If we correct for the presence of CENs in all ablation conditions reported here, the resulting phenotypes more closely resemble those described in our previously published work (Supplemental Figure 5; Wanner & Prince, 2013).
These experiments clarify and extend our previously published work on pioneer activity (Wanner & Prince, 2013). Our results confirm that the pioneer is 1) active at 18 hpf and 2) r4-derived, from which we conclude that the pioneer is a FBMN or REN. As ablation of a single r5-localized neuron had no measurable consequence for migration, and at least in some cases that neuron will have been a FBMN/REN, we conclude further that the pioneer neuron becomes dispensable for posterior movement once it has crossed the r4/5 border. In other cases the single r5 neuron will have been a CEN, allowing us to conclude that the first CEN to migrate in r5 does not have pioneer function. However, the results of our two-neuron ablation experiments leave open the possibility that the presence of a CEN has some influence on FBMN/REN migration. While two-cell ablations result in a severe block to migration, the bulk of this effect likely stems from removal of an r4-derived pioneer rather than an r5-localized CEN. Instead of requiring contact with a CEN to migrate, we suggest that the FBMN/REN pioneer may use such contacts as just one of several mechanisms that enable it to cross the r4/5 border.
4. Discussion
This study provides the first comprehensive description of the birthplaces and early movements of rhombencephalic OENs. In our single-cell backtracking experiments, we demonstrate that OENs arise in two distinct locations: RENs are born in r4 with the FBMNs, while CENs are born in r5. Accordingly, REN soma location and axon morphology depend on expression of hoxb1a in r4, while CEN identity is independent of hoxb1a. We observe that the migration of both OEN populations occurs between 18 and 48 hpf, concurrent with migration of the FBMNs. Unlike the FBMNs, however, neither RENs nor CENs need Pk1b function to migrate. CENs also appear to migrate independently of the pioneer neuron. Together, these results suggest that OENs migrate through the hindbrain using a different set of mechanisms than those employed by FBMNs.
While rhombencephalic OENs have been examined in a variety of aquatic organisms (reviewed by Blaxter, 1987), previous studies were limited by a lack of specific markers for this group of sensory efferents. Previously, researchers identified OENs via backfilling from the periphery, restricting observation of this cell type to developmental stages occurring after its projections reached terminal end organs (Bricaud et al., 2001; Metcalfe et al., 1985; Sapède et al., 2005). Here, we circumvent this limitation by photoconverting single Isl1(+) neurons starting at 18 hpf and using the persistent red form of Kaede to identify OENs based on axon morphology. We demonstrate that RENs and CENs both begin their posterior migration by 18 hpf, making their tangential migration into r6 and r7 concurrent with that of the FBMNs. As previously observed, RENs form a tight cluster with the major motor nucleus of the FBMNs upon reaching r6, while CENs cluster with the minor motor nucleus of the FBMNs in anterior r7 (Bricaud et al., 2001; Metcalfe et al., 1985).
Our reconstructions of Isl1(+) cell birthplaces from time-lapse data also support an r5 origin for CENs. Isl1(+) cells have been backtracked in only one other study to date, which focused on the mediolateral rather than anteroposterior spread of FBMN birthplaces in Cdh2-deficient embryos (Stockinger et al., 2011). Insights into the segmental origins of Isl1(+) neurons have otherwise been gained largely through genetic manipulations (Gavalas et al., 2003; Jungbluth et al., 1999; McClintock et al., 2002) or extrapolated from backfilling experiments (Sapède et al., 2005). Interestingly, Sapède and colleagues do posit that RENs and CENs have separate segmental origins, using cell soma locations at 24 and 72 hpf to conclude that CENs are born in r6 and later migrate to r7 (Sapède et al., 2005). Here, we find via cell tracking that CENs are born in r5 between 13–14 hpf, although we could directly visualize the birthplaces of only a limited number of CENs (n=6) due to the technical difficulty of our backtracking experiments. Crucially, we were also able to photoconvert a significant number of CENs in r5 between 18–20 hpf (n=18/33), a stage at which we can confidently identify rhombomere location based on morphological criteria (e.g. Figure S1d), adding additional support to an r5 origin for this cell type. Together, these experiments place the origin of the CENs further anterior than previously described.
Through our time-lapse experiments, we also revealed new details about the early movements and projection patterns of CENs. We find that migratory CENs begin sending projections laterally while still in r5, despite the fact that mature CENs use the r6 exit point. Immature CENs project their axons laterally at a fairly acute angle, in contrast to the gently curving genu displayed by the mature neuron (Metcalfe et al., 1985). It is possible that the CEN axon undergoes some level of axon remodeling between 18 and 48 hpf, and we have found preliminary evidence that several early branches of the CEN axon are later withdrawn (Figure S4, Movie 2). We suggest that the CEN axon may be remodeled in response to signals from the PLL afferent projections, which enter the hindbrain at the r6 exit point around 22.6 hpf (Zecca et al., 2015). However, this phenomenon warrants further study.
These separate segmental origins are also corroborated by our knockdown experiments, in which we demonstrate that disruption of hoxb1a in r4 impacts REN, but not CEN, development. Experiments in multiple species including chick, mouse, and zebrafish have previously shown that Hoxb1 function is required both for the migration of FBMNs and the proper fasciculation of their projections (Gavalas et al., 2003; Goddard et al., 1996; Jungbluth et al., 1999; McClintock et al., 2002). Of particular relevance to our study is the observation that Hoxb1 also impacts development of the vestibulocochlear efferent neurons in both chick and mouse (Bell et al., 1999; Goddard et al., 1996). Our hoxb1a knockdowns in zebrafish support a model in which RENs, like vestibulocochlear efferents in amniotes, rely on hoxb1a to migrate.
To date, many authors have speculated that OENs are evolutionarily derived from FBMNs (Bruce et al., 1997; Fritzsch & Elliott, 2017; Meredith & Roberts, 1986; Tiveron et al., 2003). Indeed, ablation of the inner ear afferents in mouse causes prospective efferents to re-route their axons in an FBMN-like manner (Fritzsch et al., 1999; Ma et al., 2000). While our hoxb1a knockdowns and backtracking results support this interpretation of REN origins, CENs are likely derived from a different population of neurons within the branchiomotor column. Recent studies have lent credence to the idea that FBMNs are not the only motor neurons capable of routing to sensory hair cells, as motor neuron populations in the spinal cord and trunk can also re-route to innervate ectopic ears in Xenopus laevis embryos (Elliott & Fritzsch, 2010; Elliott et al., 2013). Sapède and colleagues (Sapède et al., 2005) argue that CENs may have arisen as a subset of the glossopharyngeal motor neurons (nIX), which migrate from r6 to r7. Based on our OEN birthplace reconstructions, we can instead suggest that the CENs may have arisen as a subset of the r5-derived abducens motor neurons (nVI), which despite being somatic motor neurons are cholinergic like the FBMNs (Rodella et al., 1996). While nVI are not labeled by available Islet1 transgenes, they do express Islet1 protein as assayed by immunolabeling (Chandrasekhar et al., 1997; Beiriger, unpublished). Additional work will be needed to establish the relationship between these r5 and r6 motor neuron populations and the CENs.
Despite their common origins, our analysis of Pk1b-deficient embryos suggests that RENs and FBMNs begin to take on distinct identities soon after they are born. Pk1b is a core component of the planar cell polarity (PCP) pathway that also acts as a nuclear translocator of RE1-silencing transcription factor, or Rest (Mapp et al., 2011; Mapp et al., 2010). In FBMNs, Rest is required to repress terminal maturation genes during migration, keeping the neurons in an immature state until they reach r6 and r7 (Love & Prince, 2015). Here, we show that unlike FBMNs, RENs can migrate successfully to r6 in the absence of Pk1b. Moreover, Pk1b-deficient FBMNs are unable to follow RENs into r6, indicating that these two cell populations do not migrate collectively. These results support previous suggestions that FBMNs and RENs become molecularly distinct before the onset of migration, and that their separate identities are reinforced through integration with the correct end organs (Pata et al., 1999; Simmons, 2002). However, the exact timing and mechanism of their divergence remains unknown.
Additional differences between FBMN and OEN migration are evident in their reliance on a pioneer. In a previous study, we demonstrated that the first Isl1(+) neuron to cross the r4/5 border acts as a pioneer neuron and is necessary to lead initial posterior migration of the FBMNs. Further, we established that the role of the pioneer is time-dependent, since leading cell ablations performed after 19 hpf did not impact FBMN migration (Wanner & Prince, 2013). Here, we add a spatial dimension to our understanding of the pioneer: ablation of a single neuron in r4 results in a severe migration defect, while ablation of a single neuron in r5 has no significant effect. We consider these conditions comparable to our previously published 18 hpf and 19 hpf ablations, respectively. Our results demonstrate that only an r4-derived neuron can act as a pioneer, and raise the additional question of whether the pioneer is an FBMN or a REN. We consider it more likely that the pioneer is an FBMN based on the outcome of our pk1b-knockdown experiments. If the pioneer were a REN, we would expect the FBMNs to follow it into r6 even in the absence of Pk1b function. This does not occur; instead, the FBMNs remain clustered in r4 in pk1b-deficient embryos despite the successful posterior migration of the RENs.
While our two-cell ablations suggest that contacts between FBMN/RENs and CENs across the r4/5 border may not be essential to migration, our approach is limited by the lack of CEN-specific markers. Our membrane targeted transgene allows us to identify CENs based on their placement in r5 and lateral axon projections, but this distinction becomes much harder to make once FBMN/RENs reach r5 and lie in close proximity to the CENs. As a result, we are unable to ablate all CENs and therefore cannot rule out the possibility that FBMN/RENs establish contact with CENs that begin their migration after our ablations have taken place. Moreover, our birthplace data demonstrates that CENs are present in r5 starting at 14–15 hpf, even though they do not express Tg(en.crest1-hsp70l:mKaede) transgene until 18 hpf. Therefore, we also cannot rule out the possibility that FBMN/RENs are able to derive a migratory cue from CENs at these early stages, before our ablations take place.
Interestingly, the contacts we have documented between FBMN/RENs and CENs may help explain “escaper” phenotypes in Cdh2-deficient embryos. In wild-type embryos, Cdh2 both helps to maintain neuroepithelial cohesion and also appears to stabilize contacts between Isl1(+) neurons during their migration. Loss of these cell-cell interactions after global knockdown of Cdh2 causes FBMNs to cluster aberrantly in r4 and r5 (Stockinger et al., 2011; Wanner & Prince, 2013). However, small groups of Isl(+) neurons can occasionally escape to r6 in Cdh2-deficient embryos (Stockinger et al., 2011; Wanner & Prince, 2013). Our present study suggests that the transient contacts that r4-derived FBMN/RENs appear to make with CENs across the r4/5 border may, if stabilized, allow some of these r4-derived neurons to migrate into r6 with the CENs even in Cdh2-deficient specimens. Alternately, all of these “escaper” neurons could be r5-derived CENs that migrate into r6 independently of Cdh2-mediated contacts with the FBMN/RENs. In a recent study, Rebman and colleagues (Rebman et al., 2016) also observed escaper cells after expression of a dominant-negative Cdh2 construct specifically in Isl(+) neurons. Replication of these results in embryos expressing a membrane-targeted transgene would provide insight into the projection patterns of escapers, and clarify whether CENs do indeed migrate independently of Cdh2.
In this study, we have identified significant differences between the disposition and migration of FBMNs and OENs. However, many of the molecular differences between OENs and related motor neuron populations remain uncharacterized. Several studies have provided preliminary insights: dendritic projections of zebrafish OENs cross the midline in a manner dependent on activity of the guidance molecule Netrin and its receptor DCC (Suli et al., 2006), while proper axon routing of the related contralateral vestibuloacoustic efferents (CVAs) in amniotes relies on the GATA family of transcription factors (Karis et al., 2001; Pata et al., 1999). Additional work will be needed to identify the mechanisms that uniquely drive OEN migration and guide their projections towards the proper end organs. Nevertheless, our findings have uncovered several unique qualities of the OENs, providing a better understanding of this rare cell type.
Supplementary Material
Supplemental Figure 1: Leading neurons terminate their migration in r7. (a) The transgenic line Tg(en.crest1-hsp70l:Kaede) expresses a cytoplasmic variant of Kaede in all cranial efferent neurons, shown here at 48 hpf. (a-a’’’) Photoconversion of Kaede protein in the leading neuron (closed arrowhead) from green to red fluorescence at 18 hpf. A small amount of off-target photoconversion can be seen in the immediate follower (open arrowhead). (b) Schematic of leading and following cell conversions at 18 hpf. Leading neurons have already migrated into r5 at the time of conversion. All followers converted in this experiment were within 1–3 cell lengths of the leader. (c) Location of converted leaders and followers at 48 hpf. Leaders and their immediate followers are both more likely to terminate migration in r7. (d) Location of leading neurons in Tg(en.crest1-hsp70l:mKaede) background at the time of photoconversion (17–18 hpf). (e) The converted neuron in the right hemibrain shows trailing axon morphology typical of an FBMN or REN at 19 hpf. (f) In the left hemibrain, the converted neuron displays a laterally projecting axon morphology atypical of FBMNs or RENs. Neurons projecting laterally in this manner become CENs, as described in Figure 3.
Supplemental Figure 2: Morphological landmarks during early neurulation allow for comparison of cell birthplaces between embryos. (a) Single z-slices showing brightfield and (d) CellMask membrane label in a typical 13 hpf embryo. Yellow line indicates the r3/4 border, which lies over the anterior tip of the notochord at 13 hpf. Rhombomeres are roughly 50 μm in width.
Supplemental Figure 3: Photoconversion of CEN axons shows variation in timing of outgrowth between hemibrains. Embryos staged to 20 hpf can exhibit CEN axon outgrowth (arrowheads) on either one or both sides of the brain. Here, CEN axons were converted once a clearly visible growth cone could be seen projecting laterally away from the midline. Green neurons posterior to the CENs are likely to be FBMNs that have passed the CENs during their migration. (a-b) The majority of specimens showed left-right asymmetry in the timing of CEN axon outgrowth between hemibrains (n=8/12), with a bias towards earlier outgrowth on the left (n=7/8). (c) In a subset of specimens, CEN axons were visible on both sides of the brain at 20 hpf (n=4/12). Otic vesicle is designated as ov.
Supplemental Figure 4: CEN axons may undergo remodeling during early development. Time-lapse stills reveal retraction of several small, anterior branches of the CEN axon between 18–19.5 hpf. (a-d) Yellow arrowhead indicates membrane protrusions of the CEN axon that are subsequently retracted in the following frames.
Supplemental Figure 5: Transgene brightness influences perceived severity of ablation results. The number of neurons in r6 was corrected to account for earlier visibility of CENs in Tg(en.crest1-hsp70l:mKaede) fish in comparison to Tg(en.crest1-hsp70l:mRFP) fish used in our previous study (Wanner and Prince, 2013). (a) Between 1–3 cells were subtracted from the quantification results for each embryo (see Methods). (b) The number of cells in r6 was binned for each embryo and the distribution of phenotypes was plotted using the methodology in Wanner and Prince, 2013. These results better approximate the distribution of phenotypes in our previously published ablation experiments. However, there is wider variation in the extent of Isl1(+) neuron migration in the Tg(en.crest1-hsp70l:mKaede) controls quantified here versus the Tg(Isl1:GFP) and Tg(en.crest1-hsp70l:mRFP) controls quantified in our previously published work.
Movie 1: SPIM time-lapse captures early neurulation and Isl1(+) cell migration between 12–22.5 hpf.
Movie 2: Interactions between FBMN/RENs and CENs.
In this study, we provide an overview of rhombencephalic octavolateral efferent neuron (OEN) development in the zebrafish. OENs are born in two groups: the rostral (REN) group is born in r4 with the facial branchiomotor neurons (FBMNs), while the caudal (CEN) group is born in r5. From there, the OENs migrate alongside the FBMNs to r6 and r7, where they cluster into bilateral nuclei. However, we identify several key differences between the mechanisms OENs and FBMNs use to migrate.
Acknowledgments
We thank Vytas Bindokas, Christine Labno, and Hari Shroff for their expert advice on imaging methods. We gratefully acknowledge Anita Ng for providing expert fish care, and Clare Buckley and Jon Clarke for providing us with a Kaede construct. We thank Emre Sevgen for assistance with data analysis in Python, and Teodora Szasz of the University of Chicago Research Computing Center for advice on image processing. We are grateful to Sarah Wanner, Gordon Kindlmann, Cliff Ragsdale, Edwin (Chip) Ferguson, Sally Horne-Badovinac, Robert Ho, and members of the Prince and Ho labs for helpful discussions. This work was supported by a National Science Foundation (NSF) award 1528911 to VEP and by the NSF Graduate Research Fellowship Program Grant #DGE-1144082 to AB. This project was also supported by the National Center for Advancing Translational Sciences (NCATS) of the National Institutes of Health (NIH) through Grant Number 5UL1TR002389 that funds the Institute for Translational Medicine (ITM). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or NSF. Finally, this work benefitted from the resources of the ZFIN database (zfin.org).
Supported by NSF Grant Nos. 1528911 and DGE-1144082 and NIH Grant No. 5UL1TR002389.
Footnotes
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflict of Interest
The authors have no known or potential conflicts of interest
5. References
- Ando R, Hama H, Yamamoto-Hino M, Mizuno H, & Miyawaki A (2002). An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proceedings of the National Academy of Sciences of the United States of America, 99(20), 12651–12656. 10.1073/pnas.202320599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CC (1981). Central distribution of octavolateral afferents and efferents in a teleost (mormyridae). Journal of Comparative Neurology, 195(3), 391–414. 10.1002/cne.901950303 [DOI] [PubMed] [Google Scholar]
- Bell E, Wingate RJT, & Lumsden A (1999). Homeotic transformation of rhombomere identity after localized Hoxb1 misexpression. Science, 284(5423), 2168–2171. 10.1126/science.284.5423.2168 [DOI] [PubMed] [Google Scholar]
- Blaxter JHS (1987). Structure and Development of the Lateral Line. Biological Reviews, 62(4), 471–514. 10.1111/j.1469-185x.1987.tb01638.x [DOI] [Google Scholar]
- Bricaud O, Chaar V, Dambly-Chaudire C, & Ghysen A (2001). Early efferent innervation of the zebrafish lateral line. Journal of Comparative Neurology, 434(3), 253–261. 10.1002/cne.1175 [DOI] [PubMed] [Google Scholar]
- Bruce LL, Kingsley J, Nichols DH, & Fritzsch B (1997). The Development of Vestibulocochlear Efferents and Cochlear Afferents in Mice. ~ Pergamon Int. J. Devl Neuroscience (Vol. 15). [DOI] [PubMed] [Google Scholar]
- Chandrasekhar A (2004). Turning Heads: Development of Vertebrate Branchiomotor Neurons. Developmental Dynamics, 229(1), 143–161. 10.1002/dvdy.10444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekhar A, Kimmel C, & Kuwada JY (1997). Development of branchiomotor neurons in zebrafish. Retrieved from https://www.researchgate.net/publication/14002514 [DOI] [PubMed]
- Elliott KL, & Fritzsch B (2010). Transplantation of Xenopus laevis ears reveals the ability to form afferent and efferent connections with the spinal cord. The International Journal of Developmental Biology, 54(10), 1443–1451. 10.1387/ijdb.103061ke [DOI] [PubMed] [Google Scholar]
- Elliott KL, Houston DW, & Fritzsch B (2013). Transplantation of Xenopus laevis Tissues to Determine the Ability of Motor Neurons to Acquire a Novel Target. PLoS ONE, 8(2), e55541. 10.1371/journal.pone.0055541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flock A, & Wersgll J (1962). A study of the orientation of the sensory hairs of the receptor cells in the lateral line organ of fish, with special reference to the function of the receptors. Journal of Cell Biology. 10.1083/jcb.15.1.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Pirvola U, Ylikoski J, Fritzsch B, Pirvola U, & Ylikoski J (1999). Making and breaking the innervation of the ear: neurotrophic support during ear development and its clinical implications. Tissue Res (Vol. 295). [DOI] [PubMed] [Google Scholar]
- Fritzsch Bernd, & Elliott KL (2017). Evolution and development of the inner ear efferent system: Transforming a motor neuron population to connect to the most unusual motor protein via ancient nicotinic receptors. Frontiers in Cellular Neuroscience, 11. 10.3389/fncel.2017.00114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavalas A, Ruhrberg C, Livet J, Henderson CE, & Krumlauf R (2003). Neuronal defects in the hindbrain of Hoxa1, Hoxb1 and Hoxb2 mutants reflect regulatory interactions among these Hox genes. 10.1242/dev.00802 [DOI] [PubMed]
- Goddard JM, Rossel M, Manley NR, & Capecchi MR (1996a). Mice with targeted disruption of Hoxb-1 fail to form the motor nucleus of the VIIth nerve. Development, 122(10), 3217–3228. [DOI] [PubMed] [Google Scholar]
- Goddard JM, Rossel M, Manley NR, & Capecchi MR (1996b). Mice with targeted disruption of Hoxb-1 fail to form the motor nucleus of the VIIth nerve. [DOI] [PubMed]
- Grant PK, & Moens CB (2010). The neuroepithelial basement membrane serves as a boundary and a substrate for neuron migration in the zebrafish hindbrain. Neural Development, 5(1), 9. 10.1186/1749-8104-5-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleckmann H, Niemann U, and B. F. (1991). Peripheral and Central Aspects of the Acoustic and Lateral Line System of a Bottom Dwelling Catfish, Ancistrus sp. Journal of Comparative Neurology. [DOI] [PubMed] [Google Scholar]
- Hellniann B, & Fritzsch B (1996). Neuroanatomical and histochemical evidence for the presence of common lateral line and inner ear efferents and of efferents to the basilar papilla in a frog, xenopus laevis. Brain, Behavior and Evolution, 47(4), 185–194. 10.1159/000113238 [DOI] [PubMed] [Google Scholar]
- Higashijima SI, Hotta Y, & Okamoto H (2000). Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the Islet-1 promoter/enhancer. Journal of Neuroscience, 20(1), 206–218. 10.1523/jneurosci.20-01-00206.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungbluth S, Bell E, & Lumsden A (1999). Motor neuron identity and Hox genes. [DOI] [PubMed]
- Karis A, Pata I, Van Doorninck JH, Grosveld F, De Zeeuw CI, De Caprona D, & Fritzsch B (2001). Transcription factor GATA-3 alters pathway selection of olivocochlear neurons and affects morphogenesis of the ear. Journal of Comparative Neurology, 429(4), 615–630. [DOI] [PubMed] [Google Scholar]
- Kaufmann A, Mickoleit M, Weber M, & Huisken J (2012). Multilayer mounting enables long-term imaging of zebrafish development in a light sheet microscope. Development, 139(17), 3242–3247. 10.1242/dev.082586 [DOI] [PubMed] [Google Scholar]
- Kawakami K, & Shima A (1999). Identification of the Tol2 transposase of the medaka fish Oryzias latipes that catalyzes excision of a nonautonomous Tol2 element in zebrafish Danio rerio. Gene, 240(1), 239–244. 10.1016/S0378-1119(99)00444-8 [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, & Schilling TF (1995). Stages of embryonic development of the zebrafish. Developmental Dynamics, 203(3), 253–310. 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Kok FO, Taibi A, Wanner SJ, Xie X, Moravec CE, Love CE, … Sirotkin HI (2012). Zebrafish rest regulates developmental gene expression but not neurogenesis. Development, 139(20), 3838–3848. 10.1242/dev.080994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love CE, & Prince VE (2015). Rest represses maturation within migrating facial branchiomotor neurons. Developmental Biology, 401(2), 220–235. 10.1016/j.ydbio.2015.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma LH, Grove CL, & Baker R (2014). Development of oculomotor circuitry independent of hox3 genes. Nature Communications, 5, 4221. 10.1038/ncomms5221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Anderson DJ, & Fritzsch B (2000). Neurogenin 1 null mutant ears develop fewer, morphologically normal hair cells in smaller sensory epithelia devoid of innervation. JARO - Journal of the Association for Research in Otolaryngology, 1(2), 129–143. 10.1007/s101620010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapp OM, Walsh GS, Moens CB, Tada M, & Prince VE (2011). Zebrafish Prickle1b mediates facial branchiomotor neuron migration via a farnesylation-dependent nuclear activity. Development, 138(10), 2121–2132. 10.1242/dev.060442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapp Oni M., Wanner SJ, Rohrschneider MR, & Prince VE (2010). Prickle1b mediates interpretation of migratory cues during zebrafish facial branchiomotor neuron migration. Developmental Dynamics, 239(6), 1596–1608. 10.1002/dvdy.22283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock JM, Kheirbek MA, & Prince VE (2002). Knockdown of duplicated zebrafish hoxb1 genes reveals distinct roles in hindbrain patterning and a novel mechanism of duplicate gene retention. Development, 129, 2339–2354. [DOI] [PubMed] [Google Scholar]
- Meijering E, Dzyubachyk O, & Smal I (2012). Methods for cell and particle tracking. In Methods in Enzymology (Vol. 504, pp. 183–200). Academic Press Inc. 10.1016/B978-0-12-391857-4.00009-4 [DOI] [PubMed] [Google Scholar]
- Meredith GE, & Roberts BL (1986). Central organization of the efferent supply to the labyrinthine and lateral line receptors of the dogfish. Neuroscience, 17(1), 225–233. 10.1016/0306-4522(86)90238-1 [DOI] [PubMed] [Google Scholar]
- Meredith GE, & Roberts BL (1987). Distribution and morphological characteristics of efferent neurons innervating end organs in the ear and lateral line of the european eel. Journal of Comparative Neurology, 265(4), 494–506. 10.1002/cne.902650404 [DOI] [PubMed] [Google Scholar]
- Metcalfe WK, Kimmel CB, & Schabtach E (1985). Anatomy of the Posterior Lateral Line System in Young Larvae of the Zebrafish, 389. [DOI] [PubMed] [Google Scholar]
- Obholzer N, Wolfson S, Trapani JG, Mo W, Nechiporuk A, Busch-Nentwich E, … Nicolson T (2008). Vesicular glutamate transporter 3 is required for synaptic transmission in zebrafish hair cells. Journal of Neuroscience, 28(9), 2110–2118. 10.1523/JNEUROSCI.5230-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pata I, Studer M, van Doorninck JH, Briscoe J, Kuuse S, Engel JD, … Karis A (1999). The transcription factor GATA3 is a downstream effector of Hoxb1 specification in rhombomere 4. [DOI] [PubMed] [Google Scholar]
- Pauls S, Geldmacher-Voss B, & Campos-Ortega JA (2001). A zebrafish histone variant H2A.F/Z and a transgenic H2A.F/Z:GFP fusion protein for in vivo studies of embryonic development. Development Genes and Evolution, 211(12), 603–610. 10.1007/s00427-001-0196-x [DOI] [PubMed] [Google Scholar]
- Prince VE, Moens CB, Kimmel CB, & Ho RK (1998). Zebrafish hox genes: expression in the hindbrain region of wildtype and mutants of the segmentation gene, valentino. Retrieved from http://dev.biologists.org/content/develop/125/3/393.full.pdf [DOI] [PubMed]
- Rebman JK, Kirchoff KE, & Walsh GS (2016). Cadherin-2 Is Required Cell Autonomously for Collective Migration of Facial Branchiomotor Neurons. PLOS ONE, 11(10), e0164433. 10.1371/journal.pone.0164433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts BL, & Meredith GE (1989). The Efferent System. In The Mechanosensory Lateral Line (pp. 445–459). Springer; New York. 10.1007/978-1-4612-3560-6_22 [DOI] [Google Scholar]
- Rodella L, Rezzani R, Gioia M, & Bianchi R (1995). Immunohistochemical Study of the Neurons Projecting to the Flocculus in the Abducens Nucleus of the Rat: A Double-Labelling Study. Journal of Anatomy, 152(4), 262–262. [Google Scholar]
- Rohrschneider MR, Elsen GE, & Prince VE (2007). Zebrafish Hoxb1a regulates multiple downstream genes including prickle1b. Developmental Biology, 309(2), 358–372. 10.1016/j.ydbio.2007.06.012 [DOI] [PubMed] [Google Scholar]
- Rossel M, Dambly-chaudie C, & Ghysen A (2005). Role of SDF1 chemokine in the development of lateral line efferent and facial motor neurons, (Track II), 2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryugo DK, & Fay RR (Eds.). (2010). Auditory and Vestibular Eff erents. Retrieved from https://link-springer-com.proxy.uchicago.edu/book/10.1007/978-1-4419-7070-1
- Sapède D, Rossel M, Dambly-Chaudière C, & Ghysen A (2005). Role of SDF1 chemokine in the development of lateral line efferent and facial motor neurons. Proceedings of the National Academy of Sciences of the United States of America, 102(5), 1714–1718. 10.1073/pnas.0406382102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons DD (2002). Development of the Inner Ear Efferent System across Vertebrate Species. J Neurobiol, 53, 228–250. 10.1002/neu.10130 [DOI] [PubMed] [Google Scholar]
- Stockinger P, Maitre J-L, & Heisenberg C-P (2011). Defective neuroepithelial cell cohesion affects tangential branchiomotor neuron migration in the zebrafish neural tube. Development. 10.1242/dev.071233 [DOI] [PubMed] [Google Scholar]
- Studer M, Lumsden A, Ariza-McNaughton L, Bradley A, & Krumlauf R (1996). Altered segmental identity and abnormal migration of motor neurons in mice lacking Hoxb-1. Nature, 384(6610), 630–634. 10.1038/384630a0 [DOI] [PubMed] [Google Scholar]
- Suli A, Mortimer N, Shepherd I, & Chien C. Bin. (2006). Netrin/DCC signaling controls contralateral dendrites of octavolateralis efferent neurons. Journal of Neuroscience, 26(51), 13328–13337. 10.1523/JNEUROSCI.2858-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiveron MC, Pattyn A, Hirsch MR, & Brunet JF (2003). Role of Phox2b and Mash1 in the generation of the vestibular efferent nucleus. Developmental Biology, 260(1), 46–57. 10.1016/S0012-1606(03)00213-6 [DOI] [PubMed] [Google Scholar]
- Uemura O, Okada Y, Ando H, Guedj M, Higashijima SI, Shimazaki T, … Okamoto H (2005). Comparative functional genomics revealed conservation and diversification of three enhancers of the isl1 gene for motor and sensory neuron-specific expression. Developmental Biology, 278(2), 587–606. 10.1016/j.ydbio.2004.11.031 [DOI] [PubMed] [Google Scholar]
- Walsh GS, Grant PK, Morgan JA, & Moens CB (2011). Planar polarity pathway and Nance-Horan syndrome-like 1b have essential cell-autonomous functions in neuronal migration. Development, 138(14), 3033–3042. 10.1242/dev.063842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner SJ, & Prince VE (2013). Axon tracts guide zebrafish facial branchiomotor neuron migration through the hindbrain. Development, 140(4), 906–915. 10.1242/dev.087148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner Sarah J., Saeger I, Guthrie S, & Prince VE (2013). Facial motor neuron migration advances. Current Opinion in Neurobiology, 23(6), 943–950. 10.1016/j.conb.2013.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakaria S, Mao Y, Kuta A, Ferreira De Sousa C, Gaufo GO, McNeill H, … Francis-West PH (2014). Regulation of neuronal migration by Dchs1-Fat4 planar cell polarity. Current Biology, 24(14), 1620–1627. 10.1016/j.cub.2014.05.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecca A, Dyballa S, Voltes A, Bradley R, & Pujades C (2015). The order and place of neuronal differentiation establish the topography of sensory projections and the entry points within the hindbrain. Journal of Neuroscience, 35(19), 7475–7486. 10.1523/JNEUROSCI.3743-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Leading neurons terminate their migration in r7. (a) The transgenic line Tg(en.crest1-hsp70l:Kaede) expresses a cytoplasmic variant of Kaede in all cranial efferent neurons, shown here at 48 hpf. (a-a’’’) Photoconversion of Kaede protein in the leading neuron (closed arrowhead) from green to red fluorescence at 18 hpf. A small amount of off-target photoconversion can be seen in the immediate follower (open arrowhead). (b) Schematic of leading and following cell conversions at 18 hpf. Leading neurons have already migrated into r5 at the time of conversion. All followers converted in this experiment were within 1–3 cell lengths of the leader. (c) Location of converted leaders and followers at 48 hpf. Leaders and their immediate followers are both more likely to terminate migration in r7. (d) Location of leading neurons in Tg(en.crest1-hsp70l:mKaede) background at the time of photoconversion (17–18 hpf). (e) The converted neuron in the right hemibrain shows trailing axon morphology typical of an FBMN or REN at 19 hpf. (f) In the left hemibrain, the converted neuron displays a laterally projecting axon morphology atypical of FBMNs or RENs. Neurons projecting laterally in this manner become CENs, as described in Figure 3.
Supplemental Figure 2: Morphological landmarks during early neurulation allow for comparison of cell birthplaces between embryos. (a) Single z-slices showing brightfield and (d) CellMask membrane label in a typical 13 hpf embryo. Yellow line indicates the r3/4 border, which lies over the anterior tip of the notochord at 13 hpf. Rhombomeres are roughly 50 μm in width.
Supplemental Figure 3: Photoconversion of CEN axons shows variation in timing of outgrowth between hemibrains. Embryos staged to 20 hpf can exhibit CEN axon outgrowth (arrowheads) on either one or both sides of the brain. Here, CEN axons were converted once a clearly visible growth cone could be seen projecting laterally away from the midline. Green neurons posterior to the CENs are likely to be FBMNs that have passed the CENs during their migration. (a-b) The majority of specimens showed left-right asymmetry in the timing of CEN axon outgrowth between hemibrains (n=8/12), with a bias towards earlier outgrowth on the left (n=7/8). (c) In a subset of specimens, CEN axons were visible on both sides of the brain at 20 hpf (n=4/12). Otic vesicle is designated as ov.
Supplemental Figure 4: CEN axons may undergo remodeling during early development. Time-lapse stills reveal retraction of several small, anterior branches of the CEN axon between 18–19.5 hpf. (a-d) Yellow arrowhead indicates membrane protrusions of the CEN axon that are subsequently retracted in the following frames.
Supplemental Figure 5: Transgene brightness influences perceived severity of ablation results. The number of neurons in r6 was corrected to account for earlier visibility of CENs in Tg(en.crest1-hsp70l:mKaede) fish in comparison to Tg(en.crest1-hsp70l:mRFP) fish used in our previous study (Wanner and Prince, 2013). (a) Between 1–3 cells were subtracted from the quantification results for each embryo (see Methods). (b) The number of cells in r6 was binned for each embryo and the distribution of phenotypes was plotted using the methodology in Wanner and Prince, 2013. These results better approximate the distribution of phenotypes in our previously published ablation experiments. However, there is wider variation in the extent of Isl1(+) neuron migration in the Tg(en.crest1-hsp70l:mKaede) controls quantified here versus the Tg(Isl1:GFP) and Tg(en.crest1-hsp70l:mRFP) controls quantified in our previously published work.
Movie 1: SPIM time-lapse captures early neurulation and Isl1(+) cell migration between 12–22.5 hpf.
Movie 2: Interactions between FBMN/RENs and CENs.


