Abstract
Purpose
This study aimed at identifying the main antimicrobial resistance of ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Escherichia coli) pathogens in a Romanian infectious diseases hospital. This antimicrobial resistance is a global threat, having high rates of multidrug resistance and limited treatment options.
Patients and Methods
This retrospective study (2016–2020) assessed the antimicrobial resistance of ESKAPE pathogens isolated from the patient’s biological samples. The microbiological diagnosis was performed by classical culture methods. The antimicrobial susceptibility analysis used the Kirby–Bauer disk-diffusion method and the method of minimum inhibiting concentration with the automated Vitek, according to the CLSI (Clinical and Laboratory Standards Institute) standards.
Results
Included in this study were 4293 bacterial isolates: 67% Gram-negative bacilli, 31% Gram-positive cocci and 2% other morphotinctorial bacteria. ESKAPE pathogens were found in 97% of the bacterial isolates strains; E. coli (38.26%) and Staphylococcus aureus (26%) were the most prevalent. Most bacterial strains were isolated from urine cultures (45.6%), skin and soft tissue secretions/collections (35.9%) and also blood cultures (4.2%). Increased antimicrobial resistance was observed for methicillin-resistant Staphylococcus aureus (MRSA)s, extended spectrum beta-lactamase producing (ESBL) Enterobacterales, carbapenem-resistant (CR) Pseudomonas aeruginosa, Acinetobacter baumannii and Klebsiella pneumoniae. No vancomycin resistance was found for Enterococcus faecium. The highest prevalence rates of multidrug resistance were found in methicillin-resistant Staphylococcus aureus (86.6%), Acinetobacter baumannii (36.8%), Pseudomonas aeruginosa (29.1%) and Klebsiella pneumoniae (24.4%).
Conclusion
ESKAPE pathogens are frequently isolated in the infectious diseases hospital, with main antimicrobial resistance: ESBL, MRSA and CR. The local antimicrobial resistance pattern is essential in updating the local protocols and for appropriately prescribing antibiotics. Streamlining microbiological diagnosis and aligning with the European standards for antimicrobial susceptibility testing are necessary steps in harmonizing the regional network for good antimicrobial resistance control practices.
Keywords: antimicrobial drug resistance, hospital, Romania, ESKAPE
Introduction
Antimicrobial resistance (AMR) is one of the most concerning public health problems globally, a fact that is due to its increasing impact on morbidity, mortality and care costs. Every year, over 700,000 deaths are associated with AMR.1,2 It is estimated that in the European Union (EU), in the year 2050, over 33,000 deaths will be caused by an antibiotic-resistant bacteria.3 AMR is a consequence of the natural evolution and adapting processes of bacteria, accelerated by the selection pressure which stems from the inadequate or excessive use of antibiotics.4 Multidrug resistance (MDR) is extending continuously across the globe and it is a challenge in treating infections, making it necessary to use the reserve antibiotics which can have higher cost-to-benefit ratios and a decreased security profile.5,6 Among MDR germs, “the ESKAPE pathogens” had the biggest impact on health care-associated infections, a group of six germs which have the capacity to elude the biocidal activity of antibiotics: Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Escherichia coli.5–13 The ESKAPE group is distinguished by pathogenic, transmission and resistance traits, which are represented by enzyme inactivation, target change, cell permeability alteration through porin loss or by increasing the expression of efflux pumps, and mechanical protection through biofilm synthesis.5,6 The antiseptic activity of antibiotics can sometimes also contribute to disturbances in the skin microbiome of the face, even that which is linked to endosymbiotes; on the other hand, dermatology is a field which frequently makes use of topical glucocorticoids (as in psoriasis) which register many side effects and have the downside of predisposing the skin to possibly severe infections, some with MDR agents. Likewise, this antibacterial role can also be exerted on germs that are isolated from various isolated skin and mucous membrane diseases, or from systemic ones, either in vivo or in vitro. In the continuous fight against such Gram-negative pathogens, interleukin-6 (IL-6) seems to have an inhibiting effect, expressing antibacterial properties, an intriguing subject which summons further research.14–18 The development of new antibiotics is far from keeping the pace with MDR progression rate. Thus, preserving the available antibiotics’ actions through the rational use of antibiotics is a measure adopted as an objective within the programs of antibiotic stewardship and it is mandatory concerning the quality standards of hospital activity, in accordance with Global Action Plan on Antimicrobial Resistance developed by the World Health Organization (WHO) and also with the regulations of the European Antimicrobial Resistance Surveillance Network (EARS-Net), which has Romania as a member.3,19,20 The current COVID-19 (Coronavirus disease of 2019) pandemic context has changed the priorities of the medical activity by influencing antibiotic use with unknown consequences in the long run.21
This study aimed to evaluate the evolution of the main antimicrobial resistance issues, as part of the stewardship antibiotic program in an infectious diseases hospital which is representative for the south-east region of Romania.
Patients and Methods
The current study was conducted in accordance with the Declaration of Helsinki. It is a retrospective type of study, extending from 2016 to 2020, that was done by analyzing the annual reports of the microbiology laboratory, within the antibiotic stewardship activities controlled by law in Romania,22,23 in a specialized infectious disease and dermatovenereology hospital with 160 beds in the south-east region of this country. Community infections, along with medical assistance associated infections (as transfers from other multidisciplinary hospitals), were admitted.
This study evaluated the antibiotic sensibility of bacteria isolated from biological products taken for diagnostic purposes from hospitalized patients. The bacterial isolates were identified by using classical bacteriology methods for microbial culture, on solid media agar-based, followed by biochemical (multi-testing and automated ones – Vitek 2 Compact) and specific antigen testing of the obtained colonies. Duplicate samples were excluded. Antibiotic sensibility was tested by using the Kirby–Bauer disk diffusion-method and the minimal inhibiting concentration method – Vitek 2 Compact, the resistance to a certain antibiotic being interpreted according to CLSI (Clinical and Laboratory Standards Institute) standards which are renewed annually.24–30 ESBL (extended spectrum beta-lactamase) producing Enterobacteriaceae strains were identified by using the test of double diffusion, with the help of cefotaxime in combination with inhibitory clavulanic acid and the Vitek 2 Compact Software.25,31 MRSA (methicillin-resistant Staphylococcus aureus) was taken into consideration when the cefoxitin disk diffusion test was over 22 mm.
MDR was defined by being non-susceptible to at least 1 agent from at least 3 antimicrobial categories.32 The following antibiotic classes were used: penicillins (PEN – penicillin, AMC – amoxicillin clavulanate), cephalosporins (FEP – cefepime, CTX – cefotaxime, CAZ – ceftazidime, CXM – cefuroxime, TZP – piperacillin tazobactam), carbapenems (ERT – ertapenem, IPN – imipenem, MEM – meropenem), fluoroquinolones (CIP – ciprofloxacin, LVX – levofloxacin), aminoglycosides (AMK – amikacin, GM – gentamicin, TOB – tobramycin) and tetracycline (TET – tetracycline, DOX – doxycycline), sulfonamide (SXT – sulfamethoxazole-trimethoprim).
The resistance analysis was focused on the ESKAPE germs which are monitored globally and regionally within the surveillance networks, and which are in the spotlight for stewardship antibiotic programs: Enterococcus spp., Staphylococcus aureus, Klebsiella spp., Acinetobacter spp., Pseudomonas spp., Enterobacter spp.
Results
The Evolution of Hospital Indicators
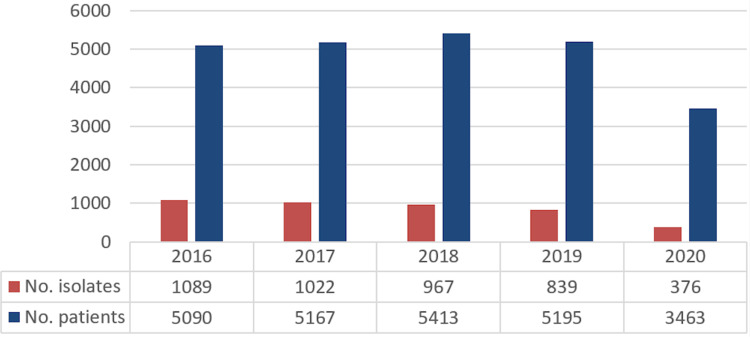
During 2016 and 2020, 24,328 patients were hospitalized from which 4293 bacterial strains were isolated. Presented in dynamic, the number of isolated strains diminished progressively from 2016 to 2019, having an even more abrupt decrease in the pandemic year of 2020, when 90% of the hospitalized patients had COVID-19 infection. In comparison to 2016, the number of admissions reduced by 33.4% in 2020, while the number of bacterial strains isolated decreased by 65%, meaning an isolation rate of almost 50% lower, in the context of a reduced diversity of analyzed biological products in viral-infected COVID-19 admitted patients, either as a suspected or confirmed infection (Figure 1).
Figure 1.
Annual evolution of the bacterial strain isolates and number of patients.
The Types of Isolated Bacterial Species (Isolates)
During 2016 and 2020, 4293 bacterial strains were isolated, 67% being Gram-negative bacilli, 31% Gram-positive cocci and 2% other bacteria.
The ESKAPE group made up 97% (4166/4293) of the bacterial strains isolated from 2016 to 2020 (Table 1). On the order of frequency, E. coli strains were most prevalent (38.26%), along with S. aureus (26%), followed by Klebsiella spp. (9.55%), Pseudomonas spp. (8.78%), Proteus spp. (5.71%), Salmonella spp. (3.5%), Enterococcus spp. (3.55%). Enterobacter spp., other Enterobacteriaceae (Serratia spp., Citrobacter spp., Morganella spp., Providencia spp.) and Acinetobacter spp., played a limited part in the ESKAPE group. The annual frequency of the isolated strains was a relatively constant one in those 5 years (Table 2). Thus being said, in the pandemic year of 2020 there was a slight decreasing rate in the numbers of staphylococci and enterococci, while Acinetobacter spp. and Pseudomonas spp. registered a slight increase.
Table 1.
Annual Frequency of ESKAPE Group Species
| Genera | No-2016 | No-2017 | No-2018 | No-2019 | No-2020 | Total | % |
|---|---|---|---|---|---|---|---|
| Enterococcus spp. | 62 | 27 | 39 | 12 | 8 | 148 | 3.55% |
| Staphylococcus spp. | 282 | 229 | 249 | 236 | 91 | 1087 | 26% |
| Klebsiella spp. | 96 | 94 | 92 | 75 | 41 | 398 | 9.55% |
| Acinetobacter spp. | 5 | 5 | 5 | 9 | 4 | 28 | 0.67% |
| Pseudomonas spp. | 88 | 83 | 67 | 82 | 46 | 366 | 8.78% |
| Enterobacteriaceae | |||||||
| Escherichia coli | 372 | 414 | 383 | 313 | 112 | 1594 | 38.26% |
| Proteus spp. | 65 | 68 | 54 | 29 | 22 | 238 | 5.71% |
| Enterobacter spp. | 25 | 22 | 22 | 18 | 13 | 100 | 2.40% |
| Salmonella spp. | 35 | 42 | 31 | 25 | 13 | 146 | 3.50% |
| Other Enterobacteriaceae | 16 | 21 | 9 | 12 | 3 | 61 | 1.46% |
| Total | 1089 | 1022 | 967 | 839 | 376 | 4293 | 100% |
Note: % annual frequency.
Table 2.
ESKAPE Distribution According to the Biological Product Analyzed (2016–2020)
| Urine Culture | Wound or Abscess | Blood Culture | Catheter Culture | Others | TOTAL | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | N | % | N | % | ||
| Enterococcus spp. | 125 | 84.4% | 16 | 10.8% | 4 | 2.7% | 3 | 2.0% | 0 | 0 | 148 |
| Enterococcus spp. untyped | 106 | 86.8% | 12 | 9.8% | 2 | 1.6% | 2 | 1.6% | – | – | 122 |
| Enterococcus faecium | 0 | – | 1 | – | 1 | – | 0 | – | – | – | 2 |
| Enterococcus faecalis | 19 | 79.1% | 3 | 12.5% | 1 | – | 1 | – | – | – | 24 |
| Staphylococcus spp. | 6 | 0.5% | 741 | 68.1% | 88 | 8.1% | 35 | 3.2% | 217 | 19.9% | 1087 |
| Staphylococcus aureus | 5 | 0.5% | 724 | 74.8 | 24 | 2.4% | 23 | 2.3% | 197 | 20.3% | 968 |
| Klebsiella spp. | 244 | 61.3% | 80 | 20.1% | 18 | 4.5% | 0 | 0 | 56 | 14.4% | 398 |
| Klebsiella pneumoniae | 159 | 77.9% | 52 | 25.4% | 13 | 6.3% | 0 | 0 | 34 | 16.6% | 258 |
| Acinetobacter spp. | 3 | 10.7% | 11 | 39.2% | 2 | 7.1% | 2 | 7.1% | 10 | 35.7% | 28 |
| Acinetobacter baumannii | 2 | 10.5% | 10 | 52.6% | 2 | 10.5% | 2 | 10.5% | 3 | 15.7% | 19 |
| Pseudomonas spp. | 44 | 12% | 299 | 81.7% | 0 | 0 | 0 | 0 | 23 | 6.2% | 366 |
| Pseudomonas aeruginosa | 39 | 19.1% | 151 | 74.0% | 0 | 0 | 0 | 0 | 14 | 6.8% | 204 |
| Enterobacteriaceae | |||||||||||
| Escherichia coli | 1346 | 84.4% | 98 | 6.14% | 53 | 3.3% | 0 | 0 | 97 | 6% | 1594 |
| Proteus spp. | 82 | 34.4% | 145 | 60.9% | 3 | 1.2% | 1 | 0.4% | 7 | 2.9% | 238 |
| Enterobacter spp. | 38 | 38.0% | 50 | 50.0% | 2 | 2% | 0 | 0 | 10 | 10% | 100 |
| Salmonella spp. | 0 | 0 | 0 | 0 | 3 | 2% | 0 | 0 | 143 | 97.9% | 146 |
| Other Enterobacteriaceae | 12 | 19.6% | 26 | 42.6% | 2 | 3.2% | 0 | 0 | 21 | 34.4% | 61 |
| Total | 1900 | 45.6% | 1466 | 35.89% | 175 | 4.2% | 41 | 0.9% | 561 | 13.4% | 4166 |
Most isolated bacterial strains were obtained from urine cultures (45.6%), from cutaneous secretions/collections (35.89%) and blood cultures (4.2%), and rarely from other biological products, such as sputum, otic, ocular, genital, pharyngeal or nasal secretions, vomiting, fecal matter, cerebrospinal fluid (Table 2). Strains isolated (isolates) from catheter cultures registered a frequency of 0.9%, and represented either contaminations or health care associated infections.
AMR of ESKAPE Germs
Enterococcus faecium
One hundred and forty-eight strains of Enterococcus spp. were isolated, but species identification was done from 2019, only for 26 cases, from which 2 were E. faecium and 24 E. faecalis. Ciprofloxacin (CIP) sensitivity was low in all species, ranging between 44.4% and 55.2%. E. faecalis’s sensitivity for ampicillin (AMP) was of 95.7%. VAN (vancomycin) and LNZ (linezolid) sensitivity was proven both for E. faecis and E. faecalis that were identified from 2019 to 2020, but during 2017 to 2018, 5 unspecified strains of Enterococcus spp. VAN-R (vancomycin-resistant) were isolated from urine cultures (Table 3).
Table 3.
Susceptibility and MDR of Gram-Positive Cocci from ESKAPE Group
| OXA | AMP | CC | CIP | ERY | GM* | LNZ | TEC | VAN | SXT | MDR | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Enterococcus spp. untyped | IR | 80 | 55.2 | – | – | 100 | – | 93.3 | – | 0 | |
| Enterococcus faecium | IR | IR | IR | 50 | IR | IR | 100 | – | 100 | IR | 0 |
| Enterococcus faecalis | IR | 95.7 | IR | 44.4 | IR | IR | 100 | – | 100 | IR | 0 |
| Staphylococcus aureus | 64.3 | IR | 47.9 | 72.6 | 32.9 | 77.6 | 99.7 | 87.8 | 85.7 | 86 | 41.8 |
| MSSA | – | IR | 62.6 | 86.8 | 47.8 | 89.2 | 99.7 | 88.9 | 77.8 | 90.4 | 16.5 |
| MRSA | – | IR | 21.1 | 46.9 | 5.7 | 56 | 99.6 | 86.4 | 100 | 78.3 | 86.6 |
Note: *Except for high level.
Abbreviations: OXA, oxacillin; AMP, ampicillin; CC, clindamycin; CIP, ciprofloxacin; ERY, erythromycin; GM, gentamicin; LNZ, linezolid; TEC, teicoplanin; VAN, vancomycin; SXT, sulfamethoxazole-trimethoprim; MDR, multidrug resistance; MSSA, methicillin sensible Staphylococcus aureus; MRSA, methicillin resistant Staphylococcus aureus.
Staphylococcus aureus
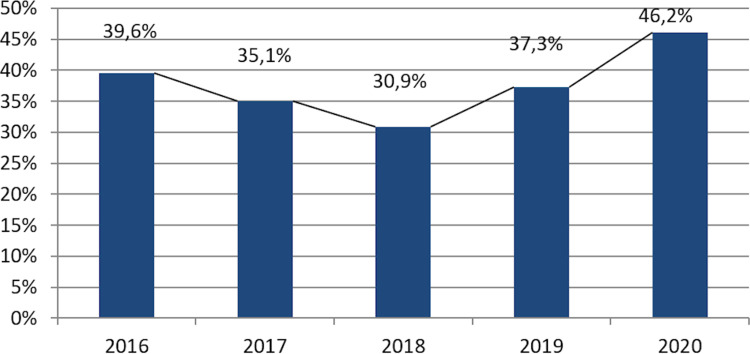
Staphylococcus aureus makes up 89.0% from staphylococcal isolates, most of them being obtained from infections located in the skin or soft tissues. The rate of MRSA had a decreasing tendency from 2016 to 2018, followed by an increase from 201 to 2020, when it reached its peak level of 46.2% (Figure 2). Methicillin resistance was correlated with a significantly increased rate of resistance as compared to other antibiotics, including VAN-R and MDR (Table 3).
Figure 2.
MRSA rate progression (2016–2020).
Klebsiella pneumoniae
Klebsiella pneumoniae sensitivity is reduced in more than one strains for SXT, CIP, GM, MEM, but especially for beta-lactamines, registering a 7.6% resistance to carbapenems and a 24.4% MDR. ESBL frequency varied between 33% (2018) and 20% (2019), with an increasing trend in the pandemic year of 2020 (Table 4).
Table 4.
Susceptibility and MDR of Gram-Negative Bacilli from the ESKAPE Group
| AMC | CXM | CTX | CAZ | FEP | TZP | ERT | MEM | CIP | GM | SXT | MDR | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acinetobacter baumannii | IR* | NT# | NT | 27.3 | 38.5 | 66.7 | IR* | 57.1 | 47.4 | 55.6 | NT | 36.8 |
| Pseudomonas aeruginosa | IR* | NT/IR* | NT/IR* | 66.5 | 60.3 | 73.7 | IR* | 55.2 | 64.8 | 61.6 | NT/IR* | 29.1 |
| Klebsiella pneumoniae | 50.4 | 63.7 | 66.1 | 22.2 | 38.7 | 72.5 | 43.3 | 73.2 | 63 | 74.1 | 60.4 | 24.4 |
| Enterobacteriaceae | ||||||||||||
| Escherichia coli | 60.2 | 85.2 | 87.3 | NT | 77.8 | 91.6 | 95.7 | 96 | 75.3 | 91.5 | 64.3 | 9.8 |
| Proteus spp. | 57 | 68.4 | 82.3 | NT | 66.3 | 93.8 | 98 | 100 | 73.8 | 68.5 | 44.3 | 15.1 |
| Enterobacter spp. | 15.7 | 34.9 | 70 | NT | 81.8 | 87 | 80 | 73.3 | 82.2 | 78.5 | 70 | 14.7 |
| Salmonella spp | 75 | NT | 100** | NT | NT | NT | 100 | 100 | 77.2 | 77.8 | 95 | 4 |
| Other Enterobacteriaceae | 14.9 | 43.5 | 63.3 | NT | 50 | 89.3 | 87.5 | 73.3 | 72.5 | 75.6 | 62 | 22.2 |
Notes: **For Salmonella spp. 1st and 2nd generation cephalosporins may appear to be as such in vitro, but are not effective clinically and should not be reported as susceptible.
Abbreviations: *IR, intrinsic resistance; #NT, not tested; AMC, amoxicillin clavulanate; CXM, cefuroxime; CTX, cefotaximue; CAZ, ceftazidime; FEP, cefepime; TZP, piperacillin- tazobactam; ERT, ertapenem; MEM, meropenem; CIP, ciprofloxacin; GM, gentamicin; SXT, sulfamethoxazole-trimethoprim; MDR, multidrug resistance.
Acinetobacter baumannii
Nineteen strains of Acinetobacter baumannii were identified, being isolated from cutaneous infections, urine cultures, blood cultures, other airway secretions, and also two from catheter cultures. Significant resistant strains were found for cephalosporins, quinolones, and aminoglycosides, with a rate of 36.8% MDR. From 2016 to 2019, almost half of Acinetobacter baumannii strains were found to be resistant to carbapenems (8/17), but in 2020, only two strains were isolated, both sensitive to meropenem.
Pseudomonas aeruginosa
Two hundred and four strains of Pseudomonas aeruginosa were identified, most of them from skin infections, with significant resistance to all tested antibiotics, with a carbapenems-resistance of 44.8% and an MDR of 29.1%. Annual variations in carbapenems resistance were registered, with an increasing rate from 9% in 2016, to 60.8% in 2019, and again a decrease to 31.1% in the pandemic year of 2020.
Enterobacteriaceae
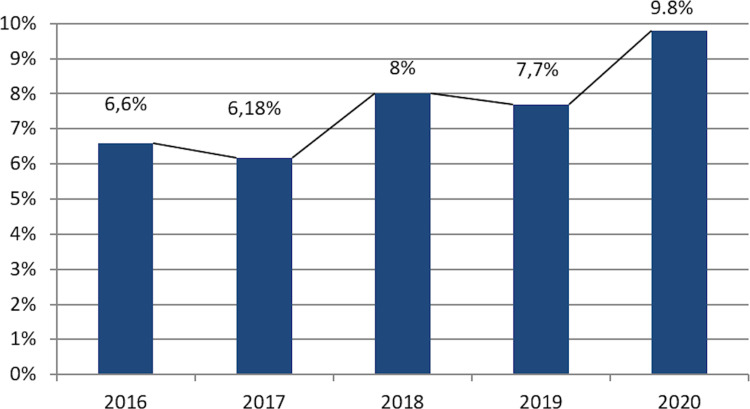
Each year, E. coli was the most frequent germ isolated in the microbiology laboratory, 84% of cases being found in urine cultures. E. coli frequency was of 9.8%, while ESBL was maintained in between 6% and 8% during the timeframe between 2016 and 2020, increasing in the pandemic year of 2020 (Figure 3).
Figure 3.
Escherichia coli ESBL+ progression rate (2016–2020).
From 2016 to 2020, MDR prevalence for Proteus spp. was of 15.1%, similar to that of ESBL, of 15.7%. Observations were made and it was found that ESBL prevalence is more elevated in comparison to MDR for Enterobacter spp., 14.7% versus 21.3% and for other Enterobacteriaceae (Citrobacter spp., Morganella spp., Serratia spp.), 22.2% versus 28.8%, with an increasing rate in 2020.
Salmonellas maintain their sensitivity to cephalosporins, carbapenems; 95% of cases are sensitive to SXT, while AMP resistance develops in one in four Enterococcus strains, and CIP and GM resistance develops in 22% of isolates.
Discussion
The pandemic year 2020 had a particular profile of bacterial reporting and antibiotic resistance in the infectious diseases hospital by reducing the number of isolates analyzed. Enterococcus faecium and Acinetobacter baumannii did not raise any resistance problems during this year, and Pseudomonas aeruginosa strains with CR decreased. In the same timeframe, the frequency of MRSA, ESBL and MDR resistance increased for Enterobacter spp. and Klebsiella pneumoniae.
Each year, between 2016 and 2020, bacteria genre from the ESKAPE pathogens group made up the majority of the hospital's isolated germ distribution. The laboratory’s difficulties concerning microbiological diagnosis by classical methods and changes in CLSI antibiotic sensitivity testing standards by annual update have a limiting effect in the multi-annual analysis of hospital data. The low number of blood cultured isolated strains in the course of invasive infections and the different standards used for reporting in the European space – the European Committee on Antimicrobial Susceptibility Testing (EUCAST) – also limit the possibility of comparing the obtained data with the level of ESKAPE resistance within the surveillance networks from the EU, part of which many multi-disciplinary hospitals from Romania are.3,33–35
The resistance profile for the ESKAPE germs isolated in the infectious diseases hospital from various biological products did not register significant statistical annual variations during the analyzed timeframe, although increasing tendencies in MRSA, ESBL and MDR rates were observed. VAN-R and CR prevalence is still low, but the inconsistent identification of species from these bacterial genes and the low number of strains analyzed annually do not support an adequate statistical analysis.
Romanian national data concerning AMR progression inside hospitals are limited to reports of blood culture strains in invasive infections, within the ECDC (European Centre for Disease Prevention and Control) network, which is done only by several multidisciplinary university hospitals which are not representative regionally. According to these data, Romania is one of the leading countries in Europe concerning AMR of invasive infections isolated bacteria: Klebsiella pneumoniae intermediary resistance to carbapenems 35.9%, Pseudomonas aeruginosa CR 52%, Acinetobacter spp. multi-resistant to carbapenems, aminoglycosides and quinolones 82.9%, Staphylococcus aureus MRSA 50.5%, Enterococcus faecium VAN-R 39%.3,34 AMR is an increasing concern not only in Europe, but in the Middle-East countries as well; studies carried out by Abbasi et al and Mansour et al have reported frequent MDR concerning E. coli and coagulase-negative staphylococci in Iran. E. coli MDR strains were found in high frequency (96.9%), mostly being resistant to AMP, cephalosporins and co-trimoxazole; coagulase-negative staphylococci also registered a high MDR frequency, being found resistant to 8 antibiotics, with the highest resistance rate to ERY (84.1%).36,37
A study done by the European Public Health Alliance identifies Romania to be “in the red zone” in what concerns the excessive use of antibiotics and the prevalence of antibiotic resistant bacteria, to which insufficient funding, insufficient medical equipment and technologies and the lack of qualified medical personnel contribute.38 Although these challenges are common to other European states, there are major differences between the resources and capacities of the national health systems, thus explaining “the red zones” within the ECDC network.39 Furthermore, these resources have an uneven distribution nationally, a fact reflected in the AMR data between different regions of Romania.34 Such challenges are present in the Romanian health system not only concerning MDR but also cancer diagnosis and treatment, associated high-risk diseases and treatment side effect risk, being major stressors not only for treating physicians but also for the patients, whom not rarely are of young age (such being the case of spiradenocarcinoma or AIDS (acquired immunodeficiency syndrome) patients) or can develop side effects even to commonly used drugs (such as beta-blockers); adequate treatment is thus an issue with consequent severe psychological impact.40–43
ESKAPE AMR evaluated in this study is lower than other resistances reported nationally in invasive infections, but it reflects their prevalence and persistence within the community. Pursuit for superbug surveillance in the infectious diseases hospital must develop by increasing the quality and accuracy of the identifying methods used, by adopting the EUCAST standards, increasing the clinical vigilance for infectious diagnosis and justified use of antibiotics.
Conclusion
ESKAPE pathogens are the majority of germs isolated in the infectious diseases hospital, the main AMR problems being ESBL and MRSA. Recognition of AMR is essential in understanding this phenomenon with the aim of updating the local protocols for antibiotic prescription. Streamlining the microbiological diagnosis and aligning with the European antimicrobial susceptibility testing are necessary steps in harmonizing the regional network for good AMR control practices.
Funding Statement
No funding was received for this article.
Abbreviations
AMR, antimicrobial resistance; EU, the European Union; MDR, multidrug resistance; ESKAPE pathogens – Enterococcus faecium – Staphylococcus aureus – Klebsiella pneumonia
Acinetobacter baumannii – Enterobacter spp; IL-6, interleukin-6; WHO, World Health Organization; EARS-Net, The European Antimicrobial Resistance Surveillance Network; COVID-19, Coronavirus Disease of 2019; CLSI, Clinical and Laboratory Standards Institute; ESBL, Extended-Spectrum Beta-lactamase Producers Enterobacteriaceae; MRSA, Methicillin resistant Staphylococcus aureus; PEN, penicillin; AMC, amoxicillin clavulanate; FEP, cefepime; CTX, cefotaxime; CAZ, ceftazidime; CXM, cefuroxime; TZP, piperacillin-tazobactam; ERT, ertapenem; IPN, imipenem; MEM, meropenem; CIP, ciprofloxacin; LVX, levofloxacin; AMK, amikacin; GM, gentamicin; TOB, tobramycin; TET, tetracycline; DOX, doxycycline; SXT, sulfamethoxazole-trimethoprim; AMP, ampicillin; VAN, vancomycin; LNZ, linezolid; VAN – R, vancomycin-resistant; OXA, oxacillin; CC, clindamycin; ERY, erythromycin; TEC, teicoplanin; MSSA, Methicillin Sensible Staphylococcus aureus; EUCAST, European Committee on Antimicrobial Susceptibility Testing; CR, carbapenem resistant; ECDC, the European Center for Diseases Control.
Data Sharing Statement
Not applicable.
Ethics Approval and Informed Consent
The Ethics Committee of the “Sf. Cuvioasa Parascheva” Hospital for Infectious diseases approved this study with the decision number 32 from 16.03.2021. Patient informed consent was granted and signed into the personal medical chart.
Consent for Publication
Consent for publication is granted by the Ethics Committee of the “Sf. Cuvioasa Parascheva” Hospital for Infectious diseases approved this study with the decision number 32 from 16.03.2021.
Author Contributions
MA, GG, EN, AVI, GL, SF, MCV and ALT have made substantial contributions to this article regarding its conception, study design, execution, and acquisition of data, analysis and interpretation. All authors have drafted, substantially revised and critically reviewed this article. All authors have agreed on the journal to which the article will be submitted and agreed to take responsibility and be accountable for the contents of the article. All authors reviewed and agreed on all versions of the article before submission, during revision, the final version accepted for publication, and any significant changes introduced at the proofing stage.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.The World Bank. By 2050, drug-resistant infections could cause global economic damage on par with 2008 financial crisis; 2016. Available from: https://www.worldbank.org/en/news/press-release/2016/09/18/by-2050-drug-resistant-infections-could-cause-global-economic-damage-on-par-with-2008-financial-crisis. Accessed January14, 2021.
- 2.Gajdács M, Albericio F. Antibiotic resistance: from the bench to patients. Antibiotics (Basel). 2019;8(3):129. doi: 10.3390/antibiotics8030129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.European Centre for Disease Prevention and Control. Antimicrobial resistance in the EU/EEA (EARS-Net) - annual epidemiological report for 2019; 2020. Available from: https://www.ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2019. Accessed March01, 2021.
- 4.Gatt YE, Margalit H. Common adaptive strategies underlie within-host evolution of bacterial pathogens. Mol Biol Evol. 2021;38(3):1101–1121. doi: 10.1093/molbev/msaa278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santajit S, Indrawattana N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed Res Int. 2016;2016:2475067. doi: 10.1155/2016/2475067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tatu AL, Merezeanu N, Pântea O, et al. Resistance features of Pseudomonas aeruginosa strains isolated from patients with infectious complications of cardiovascular surgery. Biointerface Res Appl Chem. 2017;7(2):2004–2008. [Google Scholar]
- 7.Sarshar M, Behzadi P, Scribano D, Palamara AT, Ambrosi C. Acinetobacter baumannii: an ancient commensal with weapons of a pathogen. Pathogens. 2021;10(4):387. doi: 10.3390/pathogens10040387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Senobar Tahaei SA, Stájer A, Barrak I, Ostorházi E, Szabó D, Gajdács M. Correlation between biofilm-formation and the antibiotic resistant phenotype in staphylococcus aureus isolates: a laboratory-based study in Hungary and a review of the literature. Infect Drug Resist. 2021;14:1155–1168. doi: 10.2147/IDR.S303992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juhász J, Ligeti B, Gajdács M, et al. Colonization dynamics of multidrug-resistant Klebsiella pneumoniae are dictated by microbiota-cluster group behavior over individual antibiotic susceptibility: a metataxonomic analysis. Antibiotics (Basel). 2021;10(3):268. doi: 10.3390/antibiotics10030268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller WR, Munita JM, Arias CA. Mechanisms of antibiotic resistance in enterococci. Expert Rev Anti Infect Ther. 2014;12(10):1221–1236. doi: 10.1586/14787210.2014.956092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behzadi P, Baráth Z, Gajdács M. It’s not easy being green: a narrative review on the microbiology, virulence and therapeutic prospects of multidrug-resistant pseudomonas aeruginosa. Antibiotics (Basel). 2021;10(1):42. doi: 10.3390/antibiotics10010042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gajdács M, Bátori Z, Ábrók M, Lázár A, Burián K. Characterization of resistance in gram-negative urinary isolates using existing and novel indicators of clinical relevance: a 10-year data analysis. Life (Basel). 2020;10(2):16. doi: 10.3390/life10020016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gheorghe I, Tatu AL, Lupu I, et al. Molecular characterization of virulence and resistance features in Staphylococcus aureus clinical strains isolated from cutaneous lesions in patients with drug adverse reactions. Rom Biotechnol Lett. 2017;22(1):12321–12327. [Google Scholar]
- 14.Tatu AL, Nwabudike LC. Rosacea-like demodicosis (but not primary demodicosis) and papulo pustular rosacea may be two phenotypes of the same disease-a microbioma, therapeutic and diagnostic tools perspective. J Eur Acad Dermatol Venereol. 2019;33(1):e46–e47. doi: 10.1111/jdv.15166 [DOI] [PubMed] [Google Scholar]
- 15.Pricop R, Cristea VC, Gheorghe I, Tatu AL, Mihaescu G, Chifiriuc MC. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) reveals the anaerobic Slakia exigua as unique etiology of a dental abscess. Biointerface Res Appl Chem. 2017;7(2):1995–1997. [Google Scholar]
- 16.Niculet E, Chioncel V, Elisei AM, et al. Multifactorial expression of IL‑6 with update on COVID‑19 and the therapeutic strategies of its blockade (Review). Exp Ther Med. 2021;21:263. doi: 10.3892/etm.2021.9693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niculet E, Bobeica C, Tatu AL. Glucocorticoid-induced skin atrophy: the old and the new. Clin Cosmet Investig Dermatol. 2020;13:1041–1050. doi: 10.2147/CCID.S224211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niculet E, Radaschin DS, Nastase F, et al. Influence of phytochemicals in induced psoriasis (Review). Exp Ther Med. 2021;20:3421–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.World Health Organization. Global action plan on antimicrobial resistance, 2015. World Health Organization; 2021. Available from: https://www.who.int/antimicrobial-resistance/global-action-plan/en/. Accessed February12, 2021. [DOI] [PubMed] [Google Scholar]
- 20.Doron S, Davidson LE. Antimicrobial stewardship. Mayo Clin Proc. 2011;86(11):1113–1123. doi: 10.4065/mcp.2011.0358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashiru-Oredope D, Kerr F, Hughes S, et al. Assessing the impact of COVID-19 on antimicrobial stewardship activities/programs in the United Kingdom. Antibiotics. 2021;10:110–122. doi: 10.3390/antibiotics10020110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spitalului Clinic Județean de Urgență „Pius Brînzeu”, Timișoara. Ordinul MS nr. 1101/2016 privind aprobarea Normelor de supraveghere, prevenire și limitare a infecțiilor asociate asistenței medicale în unitățile sanitare. Monitorul Oficial, Partea I nr. 791 din 07 octombrie 2016 [Romanian; national law – in-hospital medical assistance infection surveillance, preventing and limiting norms]; 2021. Available from: https://www.hosptm.ro/files/juridic/2016/ordinul_nr._1101_din_07.10.2016.pdf. Accessed February15, 2021.
- 23.Van Dijck C, Vlieghe E, Cox JA. Antibiotic stewardship interventions in hospitals in low-and middle-income countries: a systematic review. Bull World Health Organ. 2018;96(4):266–280. doi: 10.2471/BLT.17.203448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anikst V, Becket S, Tsan A. Ensuring reliable antimicrobial susceptibility test (AST) results when using an automated AST system. CLSI AST News Update. 2020;5(1):5–8. [Google Scholar]
- 25.Leber AL. Clinical Microbiology Procedures Handbook. 4th ed. Washington, DC: ASM Press; 2016. [Google Scholar]
- 26.Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 26th ed. CLSI Supplement M100. Wayne PA: Clinical and Laboratory Standards Institute; 2016. [Google Scholar]
- 27.Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 27th ed. CLSI Supplement M100. Wayne PA: Clinical and Laboratory Standards Institute; 2017. [Google Scholar]
- 28.Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. CLSI Supplement M100. Wayne PA: Clinical and Laboratory Standards Institute; 2018. [Google Scholar]
- 29.Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 29th ed. CLSI Supplement M100. Wayne PA: Clinical and Laboratory Standards Institute; 2019. [Google Scholar]
- 30.Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. CLSI Supplement M100. Wayne PA: Clinical and Laboratory Standards Institute; 2020. [Google Scholar]
- 31.Sahni RD, Mathai D, Sudarsanam TD, et al. Extended-spectrum beta-lactamase producers: detection for the diagnostic laboratory. J Glob Infect Dis. 2018;10(3):140–146. doi: 10.4103/jgid.jgid_49_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 33.Kassim A, Omuse G, Premji Z, Revathi G. Comparison of clinical laboratory standards institute and European committee on antimicrobial susceptibility testing guidelines for the interpretation of antibiotic susceptibility at a University teaching hospital in Nairobi, Kenya: a cross-sectional study. Ann Clin Microbiol Antimicrob. 2016;15:21. doi: 10.1186/s12941-016-0135-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Centrul National de Supraveghere si Control al Bolilor Transmisibile. Popescu GA, Șerban R, Niculcea A. CARMIN-ROM 2017 (Consumul de antibiotice, Rezistența microbiană și Infecții asociate asistenței medicale (Nosocomiale) în România - 2017), Institutul Național de Sănătate Publică; 2019. Available from: https://www.cnscbt.ro/index.php/analiza-date-supraveghere/infectii-nosocomiale-1/1309-consumul-de-antibiotice-rezistenta-microbiana-si-infectii-asociate-asistentei-medicale-nosocomiale-in-romania-2017/file. Acessed January5, 2021.
- 35.Erdem H, Puca E, Ruch Y, et al. Portraying infective endocarditis: results of multinational ID-IRI study. Eur J Clin Microbiol Infect Dis. 2019;38(9):1753–1763. doi: 10.1007/s10096-019-03607-x [DOI] [PubMed] [Google Scholar]
- 36.Amin M, Sirous M, Javaherizadeh H, et al. Antibiotic resistance pattern and molecular characterization of extended-spectrum β-lactamase producing enteroaggregative Escherichia coli isolates in children from southwest Iran. Infect Drug Resist. 2018;11:1097–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abbasi Montazeri E, Seyed-Mohammadi S, Asarehzadegan Dezfuli A, et al. Investigation of SCCmec types I-IV in clinical isolates of methicillin-resistant coagulase-negative staphylococci in Ahvaz, Southwest Iran. Biosci Rep. 2020;40(5):BSR20200847. doi: 10.1042/BSR20200847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.European Public Health Alliance. In the Red Zone – antimicrobial Resistance: lessons from Romania. Brussels; 2017. Available from: https://epha.org/wp-content/uploads/2017/06/In-the-red-zone-EPHA.pdf. Acessed February15, 2021. [Google Scholar]
- 39.World Health Organization. Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug resistant bacterial infections, including tuberculosis; 2017. Available from: https://www.who.int/medicines/areas/rational_use/PPLreport_2017_09_19.pdf?ua=1. Accessed February12, 2021.
- 40.Rebegea L, Firescu D, Baciu G, Ciubara A. Psycho-Oncology Support. BRAIN. 2019;10(3):77–88. [Google Scholar]
- 41.Rebegea LF, Firescu D, Dumitru M, Pătraşcu A. Skin spiradenocarcinoma - case presentation. Rom J Morphol Embryol. 2016;57(1):327–330. [PubMed] [Google Scholar]
- 42.Tatu AL, Elisei AM, Chioncel V, Miulescu M, Nwabudike LC. Immunologic adverse reactions of β-blockers and the skin. Exp Ther Med. 2019;18(2):955–959. doi: 10.3892/etm.2019.7504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Draganescu M, Baroiu L, Iancu A, et al. Perspectives on skin disorder diagnosis among people living with HIV in southeastern Romania. Exp Ther Med. 2021;21:97. doi: 10.3892/etm.2020.9529 [DOI] [PMC free article] [PubMed] [Google Scholar]