Abstract
Objective
Autophagy and long noncoding RNAs (lncRNAs) have been the focus of research on the pathogenesis of melanoma. However, the autophagy network of lncRNAs in melanoma has not been reported. The purpose of this study was to investigate the lncRNA prognostic markers related to melanoma autophagy and predict the prognosis of patients with melanoma.
Methods
We downloaded RNA sequencing data and clinical information of melanoma from the Cancer Genome Atlas. The coexpression of autophagy-related genes (ARGs) and lncRNAs was analyzed. The risk model of autophagy-related lncRNAs was established by univariate and multivariate Cox regression analyses, and the best prognostic index was evaluated combined with clinical data. Finally, gene set enrichment analysis was performed on patients in the high- and low-risk groups.
Results
According to the results of the univariate Cox analysis, only the overexpression of LINC00520 was associated with poor overall survival, unlike HLA-DQB1-AS1, USP30-AS1, AL645929, AL365361, LINC00324, and AC055822. The results of the multivariate Cox analysis showed that the overall survival of patients in the high-risk group was shorter than that recorded in the low-risk group (p < 0.001). Moreover, in the receiver operating characteristic curve of the risk model we constructed, the area under the curve (AUC) was 0.734, while the AUC of T and N was 0.707 and 0.658, respectively. The Gene Ontology was mainly enriched with the positive regulation of autophagy and the activation of the immune system. The results of the Kyoto Encyclopedia of Genes and Genomes enrichment were mostly related to autophagy, immunity, and melanin metabolism.
Conclusion
The positive regulation of autophagy may slow the transition from low-risk patients to high-risk patients in melanoma. Furthermore, compared with clinical information, the autophagy-related lncRNA risk model may better predict the prognosis of patients with melanoma and provide new treatment ideas.
1. Introduction
Melanoma is a highly malignant tumor characterized by strong invasiveness and metastasis [1]. Although it accounts for a low proportion of skin cancer cases, it is associated with a high mortality rate compared with other skin cancers [2]. Its occurrence is mainly caused by long-term exposure to ultraviolet radiation, sunburn, physical stimulation, and other factors [3]. These risk factors can promote the malignant transformation of melanocytes. At present, the treatment of melanoma mainly involves surgery, radiotherapy, chemotherapy, and other means to kill tumor cells and promote tumor cell apoptosis [4–6]. However, the mechanism of melanoma is regulated by a complex molecular regulatory network; hence, the effectiveness of currently available treatments is limited. In addition, according to the literature, autophagy-related genes (ARGs) and TNM staging can predict the prognosis of patients with melanoma [7, 8]. However, due to the heterogeneity of tumors, TNM staging cannot accurately predict the prognosis of patients. Therefore, early diagnosis and treatment of patients with melanoma are particularly important.
Autophagy is a catabolic process capturing proteins or organelles that need to be degraded and maintaining cell stability through lysosomes [9]. At present, studies have shown that autophagy plays an important role in the formation of tumors [10]. In the early stages of cancer, autophagy can inhibit tumor growth [11]. However, with the development of cancer, cancer cells show a high degree of autophagy dependence, and autophagy plays a role in promoting the growth of such cells [12]. In addition, autophagy can be used as a prognostic marker for a variety of cancers, while inhibition or activation of autophagy can destroy tumor cells [13–15]. Therefore, autophagy also plays an important role in the diagnosis and treatment of cancer.
Long noncoding RNAs (lncRNAs) are noncoding protein RNAs, with a length > 200 bp. In recent years, lncRNAs have been identified as important factors in the regulation of cancer, autoimmune disease, cardiovascular disease, etc. [16]. They can affect gene expression by regulating biological processes, including the binding of transcription factors, regulation of chromatin structure, and induction of miRNA, and subsequently regulate the biological function of cancer cells [17]. Current studies have shown that lncRNAs account for a large proportion of melanoma genomes compared with genes that encode proteins [18]; however, the mechanism of melanoma regulation remains unclear. Moreover, lncRNAs can also be used as diagnostic and prognostic markers to predict the prognosis of patients with melanoma [19]. Therefore, the role of lncRNA in melanoma is self-evident.
At present, numerous studies have confirmed the role of autophagy in melanoma [20, 21]. The increase in autophagy promotes the progression of melanoma [21], but it can also inhibit its growth [22]. Moreover, these studies are based on ARGs to diagnose or predict the prognosis of patients with this disease. In addition, recent studies have found that lncRNAs can directly or indirectly regulate autophagy [23]. They can directly activate the protein that initiates autophagy or indirectly regulate the expression of ARGs through the competing endogenous RNA mechanism [24]. Therefore, the network between lncRNAs and ARGs may be the key to elucidating the autophagy mechanism of melanoma. However, there are few studies on autophagy-related lncRNAs in this setting, and fewer autophagy-related lncRNAs have been used to evaluate the prognosis of patients with melanoma. Therefore, it is important to clarify the autophagy mechanism of melanoma related to lncRNAs and identify new molecular therapeutic targets.
In this study, we downloaded melanoma expression data from the Cancer Genome Atlas (TCGA) and obtained seven lncRNAs related to autophagy. Further multivariate Cox regression analysis, construction of an autophagy-related coexpression network, and gene set enrichment analysis (GSEA) were performed. Most importantly, we constructed a risk model that can more accurately diagnose and predict the prognosis of patients than clinical information, suggesting that the autophagy-related lncRNA signature is a reliable predictor for patients with melanoma. The workflow of the specific analysis is shown in Supplementary Figure S1.
2. Materials and Methods
2.1. Data Preparation
We downloaded the FPKM data and related clinical information of 471 melanoma samples from TCGA (https://www.cancergenome.nih.gov/). Furthermore, the mRNA matrix and lncRNA matrix of melanoma were obtained. TCGA database is a public database containing 33 tumor samples and matching normal samples. At present, the research on ARGs is relatively perfect, and the Human Autophagy-dedicated Database (HADb) (http://www.autophagy.lu/) is the first public database dedicated to human autophagy [25]. Hence, we downloaded ARGs from the HADb.
2.2. Coexpression Analysis of Autophagy Genes
After obtaining the autophagy genes from the HADb website, we compared the mRNA matrix of melanoma with this gene set and further extracted the autophagy-related mRNA matrix of melanoma. Finally, the coexpression network of the mRNA matrix and lncRNA matrix related to melanoma autophagy was analyzed using the limma package of the R software (version 4.0.2). ∣Correlation coefficient | ≥5 and p value < 0.001 were the best screening criteria [26, 27].
2.3. Survival Analysis of lncRNAs Associated with Autophagy
Prior to the multivariate Cox analysis of lncRNAs, we evaluated the significance of lncRNAs in survival through single-factor Cox analysis and conducted a univariate Cox analysis of the subsequent lncRNAs. We combined the survival time and survival status of the clinical information with lncRNAs and subsequently performed the Kaplan–Meier survival analysis of the autophagy-related lncRNAs through the survival package of the R software. p values < 0.05 denoted statistically significant differences.
2.4. Cox Regression Model Analysis
We further screened lncRNAs that can be used for the prediction of prognosis of patients. We used the survival package of the R software to perform a stepwise Cox regression analysis of lncRNAs with survival significance and obtain a risk score. The patients were divided into high- and low-risk groups according to their risk score. At present, the combination of multiple genes has demonstrated potential for predicting the prognosis of patients. Thus, the lncRNAs were screened through survival analysis and receiver operating characteristic (ROC) curve analysis using the survivalROC package. Finally, based on the median value of the risk score, we drew the risk score, survival diagram, and heat map using the pheatmap package. To better predict the prognosis of patients, we used a survival package combined with clinical information of patients and the risk model constructed in this experiment for univariate and multivariate Cox analyses and obtained the related forest plots. Furthermore, the ROC curve related to clinical information was drawn to evaluate the prognostic value of the model. p value < 0.05.
2.5. Construction of a Coexpression Network
The Cytoscape software (version 3.7.2) was used to visualize the lncRNA-related autophagy coexpression networks [28]. The Sankey diagram can directly show the autophagy-related coexpression network of the high- and low-risk groups; hence, this diagram was constructed using the ggalluvial package of the R software. Hazard ratios (HR) > 1 and <1 indicated high and low risk, respectively.
2.6. GSEA
GSEA enables researchers to better understand the potential pathogenic mechanism in the high- and low-risk groups. Therefore, we analyzed the Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) of the high- and low-risk groups through GESA (version 4.0.3) [29]. Normalized enrichment score ≥ 1, nominal p value ≤ 0.05, and false discovery rate q value ≤ 0.25 denoted statistical significance.
3. Results
3.1. Coexpression Analysis of Autophagy Genes
We downloaded a total of 232 ARGs from the HADb database. In addition, by screening the mRNA matrix of TCGA, we obtained the mRNA expression matrix related to melanoma autophagy. Furthermore, we set ∣correlation coefficient | ≥5 and p value < 0.001 as the cut-off standards and used the limma package of the R software to analyze the coexpression of the mRNA matrix and lncRNA matrix of melanoma. Finally, we screened 89 mRNAs and 100 lncRNAs in patients with melanoma (Supplementary Figure S1).
3.2. Survival Analysis of lncRNAs Associated with Autophagy
Prior to the multivariate Cox regression analysis, we performed a univariate Cox survival analysis on 100 lncRNAs. Finally, the Kaplan–Meier survival analysis showed that 39 lncRNAs were closely related to the prognosis of patients with melanoma. The high expression of only two lncRNAs was significantly correlated with shorter overall survival (OS) in patients with melanoma, whereas the remaining 37 lncRNAs did not show such a relationship.
3.3. Multivariate Cox Regression Model Analysis
The combination of multiple genes can better predict the prognosis of patients. Thus, we conducted a stepwise Cox regression analysis of 37 lncRNAs with survival significance and finally obtained a gene combination (Figure 1, Table 1) composed of seven lncRNAs (HLA-DQB1 antisense RNA 1 (HLA-DQB1-AS1), USP30 antisense RNA 1 (USP30-AS1), AL645929, AL365361, long intergenic nonprotein coding RNA 520 (LINC00520), LINC00324, and AC055822). In the multivariate Cox regression analysis, patients were divided into high- and low-risk groups according to the median value of their risk score. According to the risk score, we used the survivalROC package to draw the survival curve (Figure 2(a)) and ROC curve (Figure 2(b)) of this gene combination. Survival results showed that the high expression of this gene combination was significantly associated with poor OS in patients at high risk. The AUC of 0.742 suggests that this gene combination may be used to predict the prognosis of patients with melanoma. The risk score results showed that the high-risk group had a higher risk coefficient than the low-risk group, rising from left to right as illustrated in Figure 2(c). Compared with the low-risk group, the survival diagram results showed that the high-risk group was characterized by more deaths and shorter survival time (Figure 2(d)). Finally, the expression of seven genes in the high- and low-risk groups was visualized using a heat map (Figure 2(e)).
Figure 1.
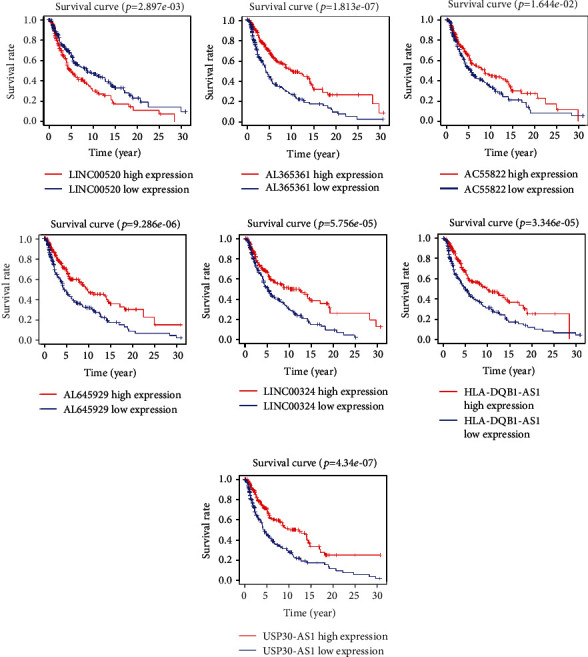
Survival analysis for the seven lncRNAs. lncRNA: long noncoding RNA.
Table 1.
Survival analysis for seven lncRNAs associated with autophagy. lncRNA: long noncoding RNA.
| Gene | KM | B | SE | HR | HR.95L | HR.95H | p value |
|---|---|---|---|---|---|---|---|
| HLA-DQB1-AS1 | 3.35E − 05 | -0.46 | 0.09 | 0.63 | 0.53 | 0.75 | 2.73E − 07 |
| USP30-AS1 | 4.34E − 07 | -0.50 | 0.09 | 0.61 | 0.51 | 0.72 | 2.22E − 08 |
| AL365361 | 1.81E − 07 | -0.49 | 0.11 | 0.62 | 0.50 | 0.76 | 1.11E − 05 |
| LINC00520 | 2.90E − 03 | 0.13 | 0.04 | 1.13 | 1.06 | 1.22 | 4.50E − 04 |
| LINC00324 | 5.76E − 05 | -0.74 | 0.15 | 0.48 | 0.35 | 0.64 | 1.42E − 06 |
| AC055822 | 1.64E − 02 | -0.47 | 0.16 | 0.62 | 0.46 | 0.84 | 2.29E − 03 |
Figure 2.
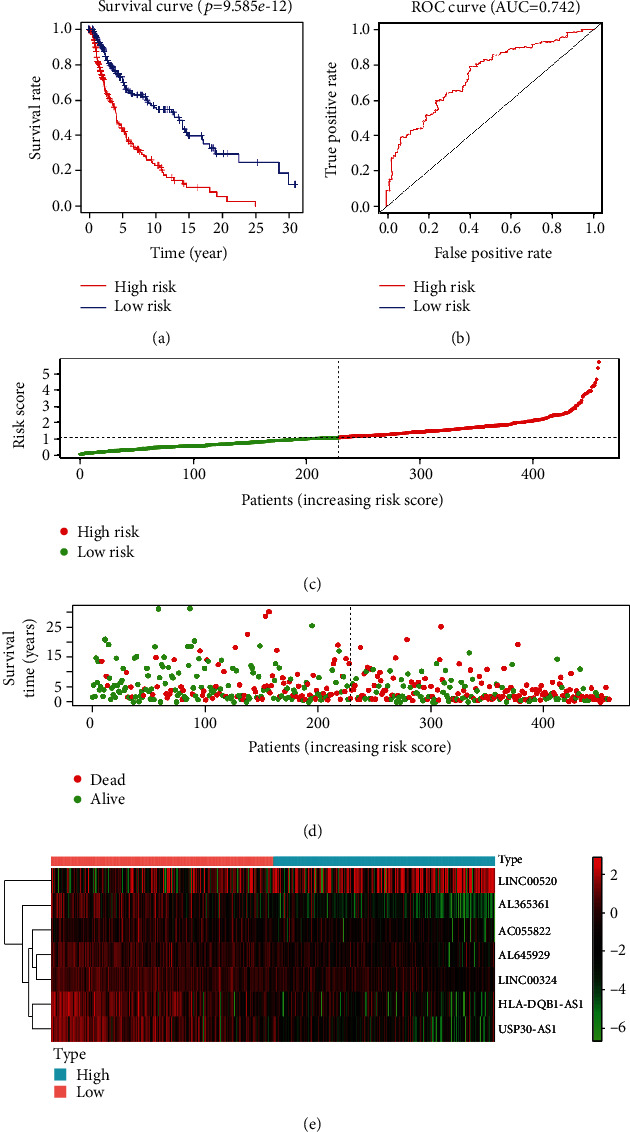
Multivariate Cox regression model analysis. (a) OS curves for the seven gene combinations in melanoma. (b) ROC curve and AUC. (c) Risk scores. Red indicates high risk. Green indicates low risk. (d) Survival diagram. Red nodes indicate death. Green indicates survival. (e) A heat map of seven genes. AUC: area under the curve; OS: overall survival; ROC: receiver operating characteristics.
In addition, to verify the higher accuracy of the risk model for predicting the prognosis of patients versus clinical information, we first performed a univariate Cox analysis based on clinical information, including sex, stage, T, M, and N. The forest plot results of the univariate analysis showed that stage, T, M, N, and risk score could be used to predict the prognosis of patients (Figure 3(a)). However, we used a multivariate Cox regression analysis and ROC curve to draw the clinical information and determine the index that may be used as the best independent prognostic factor. The forest plot results of the multivariate Cox analysis showed that T, N, and risk score could be used as independent prognostic factors (Figure 3(b)). However, the ROC results showed that the AUC of risk score, T, and N was 0.734, 0.707, and 0.658, respectively (Figure 3(c)). Therefore, the risk model we constructed is more accurate than T and N in predicting patient survival and is better than other traits (Table 2).
Figure 3.
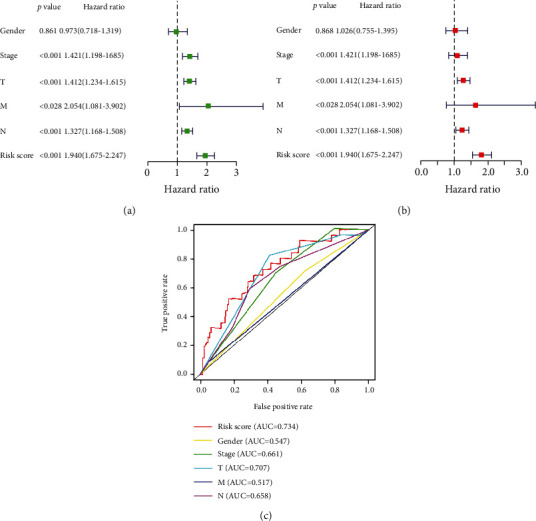
Independent prognostic analysis in melanoma: (a) forest plot of univariate Cox analysis; (b) forest plot of multivariate Cox analysis; (c) ROC curve and AUC. AUC: area under the curve; ROC: receiver operating characteristics.
Table 2.
Results of Cox analysis of the risk model and clinical data.
| Cox | ID | B | SE | z | HR | HR.95L | HR.95H | p value |
|---|---|---|---|---|---|---|---|---|
| UniCox | Gender | -0.03 | 0.16 | -0.18 | 0.97 | 0.72 | 1.32 | 8.61E − 01 |
| Stage | 0.35 | 0.09 | 4.03 | 1.42 | 1.20 | 1.68 | 5.52E − 05 | |
| T | 0.34 | 0.07 | 5.02 | 1.41 | 1.23 | 1.62 | 5.20E − 07 | |
| M | 0.72 | 0.33 | 2.20 | 2.05 | 1.08 | 3.90 | 2.80E − 02 | |
| N | 0.28 | 0.07 | 4.34 | 1.33 | 1.17 | 1.51 | 1.39E − 05 | |
| Risk score | 0.66 | 0.07 | 8.84 | 1.94 | 1.67 | 2.25 | 9.74E − 19 | |
|
| ||||||||
| MultiCox | Gender | 0.03 | 0.16 | 0.17 | 1.03 | 0.76 | 1.39 | 8.68E − 01 |
| Stage | 0.08 | 0.12 | 0.67 | 1.09 | 0.85 | 1.38 | 5.03E − 01 | |
| T | 0.24 | 0.07 | 3.37 | 1.27 | 1.11 | 1.46 | 7.46E − 04 | |
| M | 0.49 | 0.38 | 1.29 | 1.63 | 0.78 | 3.43 | 1.96E − 01 | |
| N | 0.21 | 0.08 | 2.66 | 1.23 | 1.06 | 1.43 | 7.78E − 03 | |
| Risk score | 0.59 | 0.08 | 7.55 | 1.81 | 1.55 | 2.11 | 4.21E − 14 | |
3.4. Construction of the Coexpression Network
We constructed an autophagy-related coexpression network of seven lncRNAs and 16 mRNAs using the Cytoscape software (Figure 4(a), Table 3). HR > 1 indicated a risk factor, whereas HR < 1 indicated a protective factor. We found that, in the coexpression network, only LINC00520 was a risk factor, whereas the other six lincRNAs were protective factors. Of note, the Sankey diagram yielded consistent results (Figure 4(b)).
Figure 4.
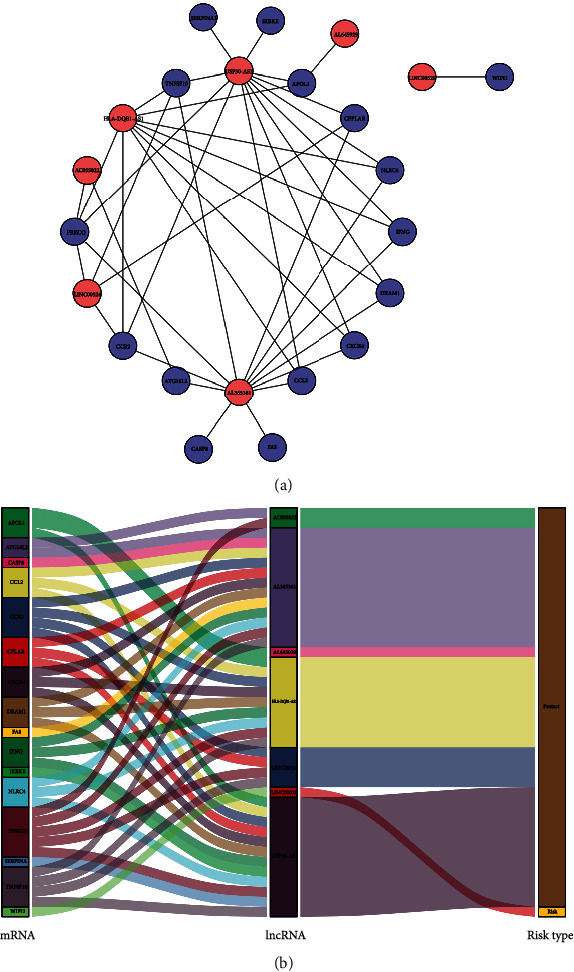
Coexpression network of autophagy. (a) Coexpression network constructed in Cytoscape. Red indicates lncRNAs. Blue indicates mRNAs. (b) Sankey diagram. lncRNA: long noncoding RNA; mRNA: messenger RNA.
Table 3.
Coexpression network of autophagy.
| ARGgene | lncRNA | Cor | p value |
|---|---|---|---|
| APOL1 | HLA-DQB1-AS1 | 0.55 | 4.38E − 39 |
| CCL2 | HLA-DQB1-AS1 | 0.53 | 1.83E − 35 |
| CCR2 | HLA-DQB1-AS1 | 0.60 | 4.04E − 48 |
| CXCR4 | HLA-DQB1-AS1 | 0.53 | 2.62E − 35 |
| DRAM1 | HLA-DQB1-AS1 | 0.54 | 2.73E − 36 |
| IFNG | HLA-DQB1-AS1 | 0.60 | 2.44E − 48 |
| NLRC4 | HLA-DQB1-AS1 | 0.55 | 5.89E − 39 |
| PRKCQ | HLA-DQB1-AS1 | 0.58 | 2.77E − 44 |
| TNFSF10 | HLA-DQB1-AS1 | 0.60 | 7.52E − 48 |
| APOL1 | USP30-AS1 | 0.78 | 3.73E − 98 |
| CCL2 | USP30-AS1 | 0.58 | 8.08E − 43 |
| CCR2 | USP30-AS1 | 0.78 | 3.82E − 96 |
| CFLAR | USP30-AS1 | 0.61 | 7.63E − 49 |
| CXCR4 | USP30-AS1 | 0.64 | 3.70E − 55 |
| DRAM1 | USP30-AS1 | 0.63 | 2.19E − 53 |
| IFNG | USP30-AS1 | 0.86 | 6.41E − 142 |
| IKBKE | USP30-AS1 | 0.54 | 2.51E − 36 |
| NLRC4 | USP30-AS1 | 0.66 | 5.48E − 61 |
| PRKCQ | USP30-AS1 | 0.79 | 4.51E − 100 |
| SERPINA1 | USP30-AS1 | 0.68 | 6.25E − 66 |
| TNFSF10 | USP30-AS1 | 0.70 | 8.92E − 72 |
| ATG16L2 | AL365361 | 0.59 | 6.67E − 45 |
| CASP8 | AL365361 | 0.52 | 7.13E − 34 |
| CCL2 | AL365361 | 0.53 | 3.50E − 36 |
| CCR2 | AL365361 | 0.83 | 1.05E − 121 |
| CFLAR | AL365361 | 0.68 | 2.52E − 65 |
| CXCR4 | AL365361 | 0.65 | 2.08E − 57 |
| DRAM1 | AL365361 | 0.53 | 4.36E − 36 |
| FAS | AL365361 | 0.51 | 1.55E − 32 |
| IFNG | AL365361 | 0.64 | 1.19E − 56 |
| NLRC4 | AL365361 | 0.67 | 1.03E − 62 |
| PRKCQ | AL365361 | 0.77 | 2.36E − 95 |
| TNFSF10 | AL365361 | 0.66 | 1.33E − 59 |
| CCR2 | LINC00324 | 0.54 | 1.32E − 37 |
| CFLAR | LINC00324 | 0.53 | 8.85E − 36 |
| PRKCQ | LINC00324 | 0.52 | 2.84E − 34 |
| TNFSF10 | LINC00324 | 0.52 | 1.68E − 34 |
| ATG16L2 | AC055822 | 0.60 | 2.04E − 47 |
| PRKCQ | AC055822 | 0.51 | 1.27E − 32 |
| APOL1 | AL645929 | 0.55 | 3.80E − 38 |
| WIPI1 | LINC00520 | 0.51 | 6.32E − 33 |
3.5. GSEA in Melanoma
We used GSEA to analyze the GO and KEGG and study the potential pathogenic mechanism of patients at high and low risks of melanoma. The results of the GO enrichment analysis showed that the BP of the high-risk group was mainly related to the metabolism of melanin. The GO of patients at low risk was related to the positive regulation of autophagy and the activation of the immune response (Figure 5(a)). The results of KEGG were related to the regulation of autophagy, antigen processing and presentation, and T cell receptor signaling pathway (Figure 5(b)). Therefore, we hypothesized that the potential pathogenesis of melanoma in patients at high and low risks is related to the regulation of autophagy and the immune system. It was further demonstrated that the immune response plays an important role in the autophagy of melanoma.
Figure 5.
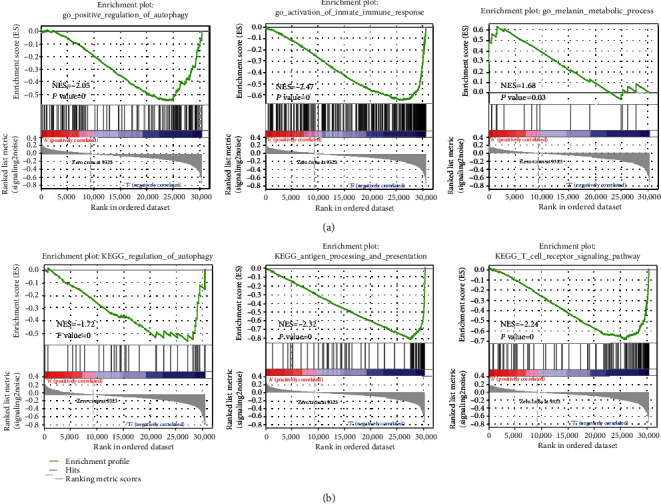
GSEA: (a) biological processes in melanoma; (b) KEGG analysis comparing the high- and low-risk groups. GSEA: gene set enrichment analysis; KEGG: Kyoto Encyclopedia of Genes and Genomes.
4. Discussion
Melanoma is a disease associated with high mortality and difficult to diagnose in the early stage [30]. Therefore, there is an urgent need for the identification of molecular markers for early diagnosis and treatment. At present, studies have found that autophagy is closely related to the occurrence of melanoma [31]. However, most studies tend to evaluate the prognostic and therapeutic role of ARGs in melanoma. Thus far, there are no investigations on the diagnostic and prognostic value of autophagy-related lncRNA in melanoma. Hence, it is urgent to clarify the molecular mechanism of autophagy-related lncRNAs in melanoma and identify new molecular therapeutic targets.
In this study, we constructed an autophagy-related coexpression network composed of seven lncRNA and 16 mRNA genes. Following GSEA, we found that the GO in the low-risk group was more enriched in the activation of the immune system and the positive regulation of autophagy, while the enrichment results of KEGG were similar. Therefore, we hypothesized that autophagy and immune system regulation may affect the transition from low risk to high risk in melanoma. Univariate Cox regression showed that, among the seven examined lncRNAs, only LINC00520 was a risk factor, and its high expression was significantly correlated with poor OS in patients at high risk of melanoma. The remaining six lncRNAs were protective factors. Further multivariate Cox analysis showed that the gene set composed of the seven lncRNAs had high accuracy in predicting the prognosis of patients in the high- and low-risk groups.
According to the literature, the expression of mRNA and protein of autophagy genes decreased with the development of melanoma compared with the normal melanocyte [32]. Beclin 1 (BECN1) is a gene that activates autophagy and programmatically inhibits tumor proliferation. Light chain 3 (LC3) is closely related to the formation of the autophagy membrane. Miracco et al. found that the expression of these two autophagy genes decreased with the progression of melanoma compared with a benign nevus [33]. The expression of these two genes decreased with the increase in tumor grade and lymph node metastasis. A study conducted by Ding et al. on invasive liver cancer yielded similar findings [34]. Beclin 1 is an essential molecule for the formation of autophagosomes. It can mediate the localization of other autophagy proteins to autophagy vesicles to regulate the formation and maturation of autophagosomes. And it can comediate autophagy with the interaction of ARGS in some diseases reported in this paper. Jounai et al. found that NLRP4 can act as a negative regulator of autophagy and inhibit the occurrence of autophagy by binding to beclin 1 [35]. Besides, most studies judge the occurrence of autophagy by the expression level of beclin 1 and LC3. Cui et al. used beclin 1 and LC3 as indicators of autophagy and found that silencing DRAM led to the downregulation of beclin 1 and LC3 [36]. Similarly, He et al. found that TNFSF10 can promote the expression of beclin 1 as an inducing factor of autophagy [37]. Therefore, our follow-up mechanism study will take beclin 1/LC3 as the detection standard to further explore the relationship between autophagy-related lncRNA and ARGS. Also, bioinformatics analysis performed by Chen et al. revealed that the biological processes of the clear cell renal cell carcinoma (ccRCC) low-risk group were related to the regulation of autophagy [38]. Li et al. also found that the positive regulation of autophagy and enrichment of immune activation pathways were more associated with patients in the low-risk group than those in the high-risk group [14]. Moreover, in our study, activation of the immune system also occurred in patients in the low-risk group. In addition, autophagy can promote the development of T and B cells in cancer and be used to present antigens to major histocompatibility complex class II molecules, which can be recognized by the immune system to play an antitumor role. A good prognosis in the low-risk group may be related to autophagy and activation of the immune system. Therefore, the reduction of autophagy may increase the risk of melanoma.
The WD repeat domain phosphoinositide interacting 1 (WIPI1) is a member of the WIPI family [39]. It is involved in the formation of autophagosomes and the fusion of autophagy lysosomes. Studies have reported that WIPI1 was highly expressed in both prostate cancer and melanoma [40, 41]. Henry et al. pointed out that WIPI1 was highly expressed in melanoma cells compared with normal melanocytes, and tumor proliferation can be inhibited by inhibiting its expression [42]. In our study, WIPI1 and LINC00520 were coexpressed. As an oncogene, LINC00520 was highly expressed in numerous types of cancers. In breast cancer, LINC00520 promotes the proliferation of tumor cells by regulating STAT3 [42]. LINC00520 was also highly expressed in melanoma. Overexpression of LINC00520 can promote the proliferation, invasion, and migration of melanoma. At present, there is no report on LINC00520 and autophagy in any type of tumor [43]. Therefore, further experimental verification is warranted. In our autophagy coexpression network, LINC00324 was coexpressed with C-C motif chemokine receptor 2 (CCR2), CASP8, and FADD-like apoptosis regulator (CFLAR) and protein kinase C theta (PRKCQ). CCR2 is a specific chemokine for monocyte chemotaxis, playing a role in resisting many types of tumors in the human immune system [44]. According to the literature, CCR2 can enhance the monitoring of the immune system and play a role in tumor resistance by triggering the TH1 response and recruiting CD8+ cells [45]. CFLAR is an antiapoptotic protein, and its function is mostly related to the autophagy of T cells. In the body, a decrease in this autophagy gene can affect the development of peripheral T cells and decrease the proliferation ability of peripheral T cells [46]. Dunkle and He found that the rate of apoptosis of peripheral T cells with CFLAR deficiency increased [47]. Following bioinformatics analysis of ARGs in ccRCC, Miracco et al. found that PRKCQ was associated with poor OS in patients with ccRCC and may be used to evaluate their prognosis [33]. In summary, these genes are often associated with immune response and autophagy in tumors. According to our study, coexpression of CCR2, CFLAR, PRKCQ, and LINC00324 was associated with autophagy in melanoma. In our risk model, LINC00324 was a protective factor in patients at low risk. This observation may be related to activation of the immune response and positive regulation of autophagy in patients at low risk. Besides, in our study, HLA-DQB1-AS1, USP30-AS1, AL645929, AL365361, and AC055822 were also protective factors in patients at low risk. Moreover, in the coexpression network, they were closely related to CCR2, CFLAR, and PRKCQ. Interestingly, Sun et al. found that USP30-AS1 was significantly correlated with CFLAR, CXCR4, DRAM1, IFNG, IKBKE, and TNFSF10 in bladder cancer. And USP30-AS1 is also a protective factor in the bladder [48]. This is similar to what we found in melanoma. Therefore, USP30-AS1 may participate in the process of autophagy by affecting ARGs. Surprisingly, except for the high expression of LINC00520 in melanoma, thus far, we have not observed the expression and mechanism of the other six lncRNAs in melanoma. Moreover, we did not find any data reporting the role of these seven lncRNAs in the autophagy mechanism of melanoma. Therefore, our study provides a new idea for revealing the autophagy mechanism of this disease.
In recent years, a variety of functions of lncRNA have been continuously discovered, among which the study of the ceRNA mechanism is more in-depth. According to literature reports, some lncRNA can affect cell autophagy through the ceRNA mechanism and then lead to the occurrence of a variety of cancers and diseases. Liu et al. found that lncRNAGAS5 promotes autophagy of the colorectal cancer cell line (CRC) and inhibits the migration and invasion of CRC cells by regulating miR-222 and affecting the autophagy-related gene PTEN [49]. In colorectal cancer cells, lncRNA SNHG6 targets miR-26a-5p to promote chemotherapy through autophagy induced by ULK1 [50]. At the same time, the ceRNA network analysis related to autophagy in hepatocellular carcinoma was also verified by Yang et al. [51]. In this study, we found that autophagy-related lncRNA may affect the development of melanoma through ARGs. With the continuous increase in high-throughput sequencing and various databases, the mechanism of ceRNA has been further explored and verified: for example, LnCeVar and LncACTdb2.0 [52, 53]. These databases are more focused on predicting the ceRNA dataset of lncRNA and more accurately predicting the relationship between lncRNA and the disease. Therefore, these studies will improve our understanding of coding and noncoding RNA in complex diseases and promote our understanding of the relationship between autophagy and lncRNA from the perspective of ceRNA. Therefore, the autophagy-related lncRNA network we constructed may also be related to the ceRNA mechanism, but this still needs further research.
Autophagy was closely related to the prognosis of patients with melanoma. Hence, it is crucial to identify accurate prognostic markers for patients with melanoma. By constructing the coexpression network of ARGs and lncRNAs in melanoma, we performed univariate and multivariate Cox regression analyses of lncRNAs and eventually included seven lncRNAs in the risk model. To verify the specificity and accuracy of our risk model, we also performed univariate and multivariate Cox analyses using the clinical information of patients with melanoma. The final results showed that, compared with other clinical information, our risk model can more accurately predict the prognosis of patients with melanoma and provide new ideas for the mechanism and treatment of this disease.
5. Conclusion
We constructed an autophagy-related coexpression network using autophagy-related mRNAs and lncRNAs. This will provide a basis for our subsequent research on the autophagy mechanism of melanoma. In addition, we constructed a risk model, which may provide new insight into the diagnosis and prognosis of patients with melanoma in the future versus clinical information. Further investigations are warranted to verify the present findings.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grant nos. 81660324 and 81660521); the Educational Research and Reform Project of Xinjiang Medical University, China (grant no. YG2019053); and the Key Discipline Construction of the 13th Five-Year Plan in Xinjiang, China—Plateau Discipline Project. Also, we have a registered preprint online (https://orcid.org/0000-0003-1356-6401).
Data Availability
All data analyzed during this study are included in this article.
Conflicts of Interest
The authors declare no conflict of interest.
Authors' Contributions
YD designed the study and wrote the manuscript. TL, ML, and TT contributed to data gathering and analysis. MZ and LC contributed to statistical analysis. SF contributed to overall supervision.
Supplementary Materials
Supplementary Figure S1: specific workflow.
References
- 1.Wei C. Y., Zhu M. X., Lu N. H., et al. Bioinformatics-based analysis reveals elevated MFSD12 as a key promoter of cell proliferation and a potential therapeutic target in melanoma. Oncogene. 2019;38(11):1876–1891. doi: 10.1038/s41388-018-0531-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng P. F. Medical bioinformatics in melanoma. Current Opinion in Oncology. 2018;30(2):113–117. doi: 10.1097/CCO.0000000000000428. [DOI] [PubMed] [Google Scholar]
- 3.Sun C., Wang L., Huang S., et al. Reversible and adaptive resistance to BRAF (V600E) inhibition in melanoma. Nature. 2014;508(7494):118–122. doi: 10.1038/nature13121. [DOI] [PubMed] [Google Scholar]
- 4.Roszik J., Markovits E., Dobosz P., et al. TNFSF4 (OX40L) expression and survival in locally advanced and metastatic melanoma. Cancer Immunology, Immunotherapy. 2019;68(9):1493–1500. doi: 10.1007/s00262-019-02382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin W. M., Fisher D. E. Signaling and immune regulation in melanoma development and responses to therapy. Annual Review of Pathology. 2017;12:75–102. doi: 10.1146/annurev-pathol-052016-100208. [DOI] [PubMed] [Google Scholar]
- 6.Kudchadkar R. R., Michielin O., van Akkooi A. C. J. Practice-changing developments in stage III melanoma: surgery, adjuvant targeted therapy, and immunotherapy. American Society of Clinical Oncology Educational Book. 2018;38:759–762. doi: 10.1200/EDBK_200241. [DOI] [PubMed] [Google Scholar]
- 7.Fujisawa Y., Yoshikawa S., Minagawa A., et al. Clinical and histopathological characteristics and survival analysis of 4594 Japanese patients with melanoma. Cancer Medicine. 2019;8(5):2146–2156. doi: 10.1002/cam4.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moya-Plana A., Mangin D., Dercle L., et al. Risk-based stratification in head and neck mucosal melanoma. Oral Oncology. 2019;97:44–49. doi: 10.1016/j.oraloncology.2019.07.026. [DOI] [PubMed] [Google Scholar]
- 9.Levy J. M. M., Towers C. G., Thorburn A. Targeting autophagy in cancer. Nature Reviews. Cancer. 2017;17(9):528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poillet-Perez L., White E. Role of tumor and host autophagy in cancer metabolism. Genes & Development. 2019;33(11-12):610–619. doi: 10.1101/gad.325514.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li P., He J., Yang Z., et al. ZNNT1 long noncoding RNA induces autophagy to inhibit tumorigenesis of uveal melanoma by regulating key autophagy gene expression. Autophagy. 2020;16(7):1186–1199. doi: 10.1080/15548627.2019.1659614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Peterson L. M., Guzman-Perez G., Krier C. R., Ahmad N. The sirtuin 6: an overture in skin cancer. Experimental Dermatology. 2020;29(2):124–135. doi: 10.1111/exd.14057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang T., Song X., Yang Y., et al. Autophagy and hallmarks of cancer. Critical Reviews in Oncogenesis. 2018;23(5-6):247–267. doi: 10.1615/CritRevOncog.2018027913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z., Dong K., Guo P., et al. Identification of autophagy-related genes and small-molecule drugs in esophageal carcinoma. Medical Science Monitor. 2020;26 doi: 10.12659/MSM.921855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu G., Pei F., Yang F., et al. Role of autophagy and apoptosis in non-small-cell lung cancer. International Journal of Molecular Sciences. 2017;18(2) doi: 10.3390/ijms18020367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar M. M., Goyal R. lncRNA as a therapeutic target for angiogenesis. Current Topics in Medicinal Chemistry. 2017;17(15):1750–1757. doi: 10.2174/156802661766616111644744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hombach S., Kretz M. Non-coding RNAs: classification, biology and functioning. Advances in Experimental Medicine and Biology. 2016;937:3–17. doi: 10.1007/978-3-319-42059-2_1. [DOI] [PubMed] [Google Scholar]
- 18.Sarkar D., Leung E. Y., Baguley B. C., Finlay G. J., Askarian-Amiri M. E. Epigenetic regulation in human melanoma: past and future. Epigenetics. 2015;10(2):103–121. doi: 10.1080/15592294.2014.1003746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu X., Zheng H., Tse G., Chan M. T., Wu W. K. Long non-coding RNAs in melanoma. Cell Proliferation. 2018;51(4) doi: 10.1111/cpr.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashrafizadeh M., Mohammadinejad R., Tavakol S., Ahmadi Z., Roomiani S., Katebi M. Autophagy, anoikis, ferroptosis, necroptosis, and endoplasmic reticulum stress: potential applications in melanoma therapy. Journal of Cellular Physiology. 2019;234(11):19471–19479. doi: 10.1002/jcp.28740. [DOI] [PubMed] [Google Scholar]
- 21.Liu H., He Z., Simon H. U. Targeting autophagy as a potential therapeutic approach for melanoma therapy. Seminars in Cancer Biology. 2013;23(5):352–360. doi: 10.1016/j.semcancer.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 22.Li X., Li Z., Li X., Liu B., Liu Z. Mechanisms of tanshinone II a inhibits malignant melanoma development through blocking autophagy signal transduction in A375 cell. BMC Cancer. 2017;17(1):p. 357. doi: 10.1186/s12885-017-3329-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frankel L. B., Lubas M., Lund A. H. Emerging connections between RNA and autophagy. Autophagy. 2017;13(1):3–23. doi: 10.1080/15548627.2016.1222992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang L., Wang H., Shen Q., Feng L., Jin H. Long non-coding RNAs involved in autophagy regulation. Cell Death & Disease. 2017;8(10, article e3073) doi: 10.1038/cddis.2017.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rahim S. A., Dirkse A., Oudin A., et al. Regulation of hypoxia-induced autophagy in glioblastoma involves ATG9A. British Journal of Cancer. 2017;117(6):813–825. doi: 10.1038/bjc.2017.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yi Y., Liu Y., Wu W., Wu K., Zhang W. Reconstruction and analysis of circ RNAmiRNAmRNA network in the pathology of cervical cancer. Oncology Reports. 2019;41(4):2209–2225. doi: 10.3892/or.2019.7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludwig N., Fehlmann T., Kern F., et al. Machine learning to detect Alzheimer's disease from circulating non-coding RNAs. Genomics, Proteomics & Bioinformatics. 2019;17(4):430–440. doi: 10.1016/j.gpb.2019.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J. P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Systems. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim S. Y., Lee J. H., Diefenbach R. J., Kefford R. F., Rizos H. Liquid biomarkers in melanoma: detection and discovery. Molecular Cancer. 2018;17(1):p. 8. doi: 10.1186/s12943-018-0757-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corazzari M., Fimia G. M., Lovat P., Piacentini M. Why is autophagy important for melanoma? Molecular mechanisms and therapeutic implications. Seminars in Cancer Biology. 2013;23(5):337–343. doi: 10.1016/j.semcancer.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Tang D. Y., Ellis R. A., Lovat P. E. Prognostic impact of autophagy biomarkers for cutaneous melanoma. Frontiers in Oncology. 2016;6:p. 236. doi: 10.3389/fonc.2016.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miracco C., Cevenini G., Franchi A., et al. Beclin 1 and LC3 autophagic gene expression in cutaneous melanocytic lesions. Human Pathology. 2010;41(4):503–512. doi: 10.1016/j.humpath.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 34.Ding Z. B., Shi Y. H., Zhou J., et al. Association of autophagy defect with a malignant phenotype and poor prognosis of hepatocellular carcinoma. Cancer Research. 2008;68(22):9167–9175. doi: 10.1158/0008-5472.CAN-08-1573. [DOI] [PubMed] [Google Scholar]
- 35.Jounai N., Kobiyama K., Shiina M., Ogata K., Ishii K. J., Takeshita F. NLRP4 negatively regulates autophagic processes through an association with beclin 1. Journal of Immunology. 2011;186(3):1646–1655. doi: 10.4049/jimmunol.1001654. [DOI] [PubMed] [Google Scholar]
- 36.Cui L., Song Z., Liang B., Jia L., Ma S., Liu X. Radiation induces autophagic cell death via the p 53/DRAM signaling pathway in breast cancer cells. Oncology Reports. 2016;35(6):3639–3647. doi: 10.3892/or.2016.4752. [DOI] [PubMed] [Google Scholar]
- 37.He W., Wang Q., Xu J., et al. Attenuation of TNFSF10/TRAIL-induced apoptosis by an autophagic survival pathway involving TRAF2- and RIPK1/RIP1-mediated MAPK8/JNK activation. Autophagy. 2012;8(12):1811–1821. doi: 10.4161/auto.22145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen M., Zhang S., Nie Z., Wen X., Gao Y. Identification of an autophagy-related prognostic signature for clear cell renal cell carcinoma. Frontiers in Oncology. 2020;10:p. 873. doi: 10.3389/fonc.2020.00873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ho H., Kapadia R., Al-Tahan S., Ahmad S., Ganesan A. K. WIPI1 coordinates melanogenic gene transcription and melanosome formation via TORC1 inhibition. The Journal of Biological Chemistry. 2011;286(14):12509–12523. doi: 10.1074/jbc.M110.200543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahmed M., Lai T. H., Zada S., et al. Functional linkage of RKIP to the epithelial to mesenchymal transition and autophagy during the development of prostate cancer. Cancers. 2018;10(8) doi: 10.3390/cancers10080273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D'Arcangelo D., Giampietri C., Muscio M., Scatozza F., Facchiano F., Facchiano A. WIPI1, BAG1, and PEX3 autophagy-related genes are relevant melanoma markers. Oxidative Medicine and Cellular Longevity. 2018;2018:12. doi: 10.1155/2018/1471682.1471682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henry W. S., Hendrickson D. G., Beca F., et al. LINC00520 is induced by Src, STAT3, and PI3K and plays a functional role in breast cancer. Oncotarget. 2016;7(50):81981–81994. doi: 10.18632/oncotarget.11962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luan W., Ding Y., Yuan H., et al. Long non-coding RNA LINC00520 promotes the proliferation and metastasis of malignant melanoma by inducing the miR-125b-5p/EIF5A2 axis. Journal of Experimental & Clinical Cancer Research. 2020;39(1):p. 96. doi: 10.1186/s13046-020-01599-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Braga T. T., Correa-Costa M., Silva R. C., et al. CCR2 contributes to the recruitment of monocytes and leads to kidney inflammation and fibrosis development. Inflammopharmacology. 2018;26(2):403–411. doi: 10.1007/s10787-017-0317-4. [DOI] [PubMed] [Google Scholar]
- 45.Kolattukudy P. E., Niu J. Inflammation, endoplasmic reticulum stress, autophagy, and the monocyte chemoattractant protein-1/CCR2 pathway. Circulation Research. 2012;110(1):174–189. doi: 10.1161/CIRCRESAHA.111.243212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He M. X., He Y. W. CFLAR/c-FLIPL: a star in the autophagy, apoptosis and necroptosis alliance. Autophagy. 2013;9(5):791–793. doi: 10.4161/auto.23785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dunkle A., He Y. W. Apoptosis and autophagy in the regulation of T lymphocyte function. Immunologic Research. 2011;49(1-3):70–86. doi: 10.1007/s12026-010-8195-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun Z., Jing C., Xiao C., Li T. An autophagy-related long non-coding RNA prognostic signature accurately predicts survival outcomes in bladder urothelial carcinoma patients. Aging. 2020;12(15):15624–15637. doi: 10.18632/aging.103718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu L., Wang H. J., Meng T., et al. lncRNA GAS5 inhibits cell migration and invasion and promotes autophagy by targeting mi R-222-3p via the GAS5/PTEN-signaling pathway in CRC. Molecular Therapy--Nucleic Acids. 2019;17:644–656. doi: 10.1016/j.omtn.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Wang X., Lan Z., He J., et al. lncRNA SNHG6 promotes chemoresistance through ULK1-induced autophagy by sponging miR-26a-5p in colorectal cancer cells. Cancer Cell International. 2019;19:p. 234. doi: 10.1186/s12935-019-0951-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang C., Wang Y., Xue W., Xie Y., Dong Q., Zhu C. Competing endogenous RNA (ceRNA) network analysis of autophagy-related genes in hepatocellular carcinoma. Pharmacogenomics and Personalized Medicine. 2020;13:445–462. doi: 10.2147/PGPM.S267563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang P., Li X., Gao Y., et al. LnCeVar: a comprehensive database of genomic variations that disturb ceRNA network regulation. Nucleic Acids Research. 2020;48(D1):D111–D117. doi: 10.1093/nar/gkz887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang P., Li X., Gao Y., et al. LncACTdb 2.0: an updated database of experimentally supported RNA interactions curated from low- and high-throughput experiments. Nucleic Acids Research. 2019;47(D1):D121–D127. doi: 10.1093/nar/gky1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1: specific workflow.
Data Availability Statement
All data analyzed during this study are included in this article.


