Abstract
The development of an effective vaccine against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiologic agent of coronavirus disease 2019 (COVID-19), is a global priority. Here, we compare the protective capacity of intranasal and intramuscular delivery of a chimpanzee adenovirus-vectored vaccine encoding a prefusion stabilized spike protein (chimpanzee adenovirus [ChAd]-SARS-CoV-2-S) in Golden Syrian hamsters. Although immunization with ChAd-SARS-CoV-2-S induces robust spike-protein-specific antibodies capable of neutralizing the virus, antibody levels in serum are higher in hamsters vaccinated by an intranasal compared to intramuscular route. Accordingly, against challenge with SARS-CoV-2, ChAd-SARS-CoV-2-S-immunized hamsters are protected against less weight loss and have reduced viral infection in nasal swabs and lungs, and reduced pathology and inflammatory gene expression in the lungs, compared to ChAd-control immunized hamsters. Intranasal immunization with ChAd-SARS-CoV-2-S provides superior protection against SARS-CoV-2 infection and inflammation in the upper respiratory tract. These findings support intranasal administration of the ChAd-SARS-CoV-2-S candidate vaccine to prevent SARS-CoV-2 infection, disease, and possibly transmission.
Keywords: SARS-CoV-2, Syrian hamster, intranasal immunization, ChAd vectored vaccine
Graphical abstract
Bricker et al. show that a single intranasal dose of a SARS-CoV-2 vaccine, a chimpanzee adenoviral vector expressing the spike protein, induces neutralizing antibodies and protects Syrian hamsters against SARS-CoV-2 infection in both the upper and lower respiratory tracts.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) initiated a global pandemic in 2019, leading to millions of confirmed positive cases of coronavirus disease 2019 (COVID-19) and an estimated case-fatality rate of 2%–3% and infection fatality rate of 0.68% (Cao et al., 2020; Dong et al., 2020; Meyerowitz-Katz and Merone, 2020). The elderly, immunocompromised, and those with an underlying illness, including obesity, diabetes, hypertension, and chronic lung disease, are at greater risk of severe disease and death from SARS-CoV-2 (Mudatsir et al., 2020; Zhou et al., 2020). Antiviral therapies and vaccines are urgently needed to curb the spread of the virus and reduce infection and disease in the population.
The Golden Syrian hamster (Mesocricetus auratus) is one of several COVID-19 animal models (Chan et al., 2020; Cleary et al., 2020; Imai et al., 2020; Muñoz-Fontela et al., 2020; Osterrieder et al., 2020; Pandey et al., 2020; Rosenke et al., 2020b; Sia et al., 2020). Hamsters are naturally susceptible to SARS-CoV-2 infection, and intranasal inoculation results in mild-to-moderate disease, including labored breathing, signs of respiratory distress, ruffled fur, weight loss, and hunched posture (Imai et al., 2020; Sia et al., 2020). Aged and male hamsters develop more severe disease, mimicking COVID-19 in humans (Osterrieder et al., 2020). In hamsters, SARS-CoV-2 primarily infects the upper and lower respiratory tracts, although viral RNA and antigen has been detected in other tissues (e.g., intestines, heart, and olfactory bulb). The peak of virus replication occurs between days 2 and 3 post-infection (dpi) and is cleared by 14 dpi in surviving animals. Histopathological analysis of infected hamsters shows multifocal interstitial pneumonia characterized by pulmonary consolidation starting as early as 2 dpi. Inflammation is associated with leukocyte infiltration, comprised primarily of macrophages and neutrophils, and an increase in type I and III interferon (IFN) and other pro-inflammatory cytokine and chemokines (Boudewijns et al., 2020; Felipe et al., 2020). High-resolution computed tomography scans show airway dilation and consolidation in the lungs of infected hamsters (Imai et al., 2020). SARS-CoV-2-induced lung pathology in hamsters appears driven by immune pathology, as lung injury is reduced in STAT2 −/− hamsters despite an increase in viral burden and tissue dissemination (Boudewijns et al., 2020).
The SARS-CoV-2 hamster model has been used to study the efficacy of several drugs and candidate vaccines. Hydroxychloroquine had no impact on infectious virus titers and disease, whereas favipiravir reduced viral burden only when high doses were used (Driouich et al., 2020; Kaptein et al., 2020a, 2020b; Rosenke et al., 2020a). Several candidate vaccines also have been tested. Yellow fever 17D-vectored and adenovirus (Ad)26-vectored SARS-CoV-2 vaccine candidates conferred protection against SARS-CoV-2 challenge in hamsters (Felipe et al., 2020; Tostanoski et al., 2020). Hamsters immunized intramuscularly (IM) with Ad26-vectored prefusion-stabilized spike (S) protein sustained less weight loss and fewer SARS-CoV-2-infected cells in the lungs at 4 dpi (Tostanoski et al., 2020). Syrian hamsters immunized twice by intraperitoneal injection with YF17D vector expressing the S protein of SARS-CoV-2 showed reduced viral burden, inflammatory gene expression, and pathology in the lung (Felipe et al., 2020). Other vectored vaccines, including a Newcastle Disease virus-S (NDV-S) and vesicular stomatitis virus-S (VSV-S) vaccine delivered IM, also protected Syrian hamsters from SARS-CoV-2 infection (Sun et al., 2020; Yahalom-Ronen et al., 2020). Alternative routes of administration have not been tested in hamsters.
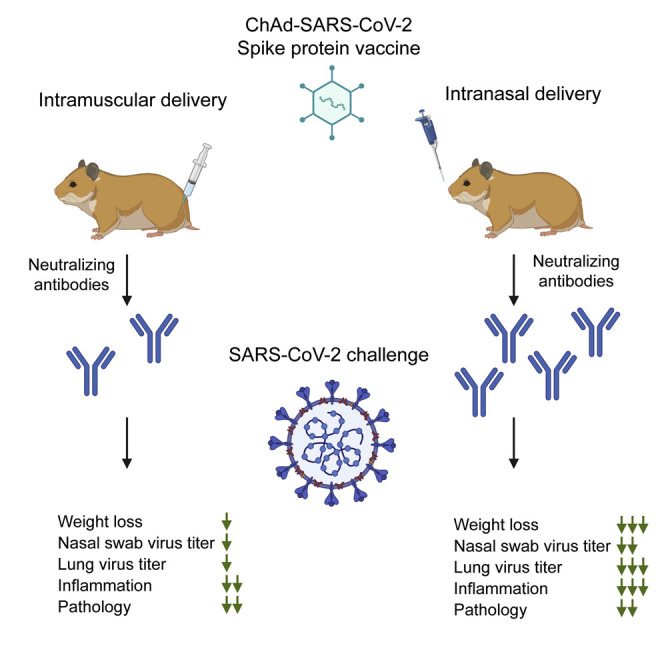
Here, we tested the efficacy of a chimpanzee adenovirus (ChAd)-vectored vaccine expressing a prefusion-stabilized version of the S protein of SARS-CoV-2 (ChAd-SARS-CoV-2-S) (Hassan et al., 2020) in Syrian hamsters following IM or intranasal (IN) delivery. A single dose of the vaccine induced a robust S-protein-specific antibody response capable of neutralizing SARS-CoV-2, with IN delivery inducing approximately 6-fold higher antibody titers than IM delivery. Upon challenge, the ChAd-SARS-CoV-2-S-immunized animals had less infectious virus and viral RNA in the lungs and nasal swabs, and this was associated with reduced pathology and numbers of viral-infected cells in the lungs at 3 dpi. The upper respiratory tract, i.e., the nasal cavity, of the hamsters demonstrated reduced pathology and SARS-CoV-2-infected cells only after IN immunization with ChAd-SARS-CoV-2-S. Collectively, these data show differences in protection mediated by the same vaccine when alternative routes of immunization are used and support intranasal vaccine delivery for optimal protection against SARS-CoV-2 challenge.
Results
Development of the SARS-CoV-2 hamster model
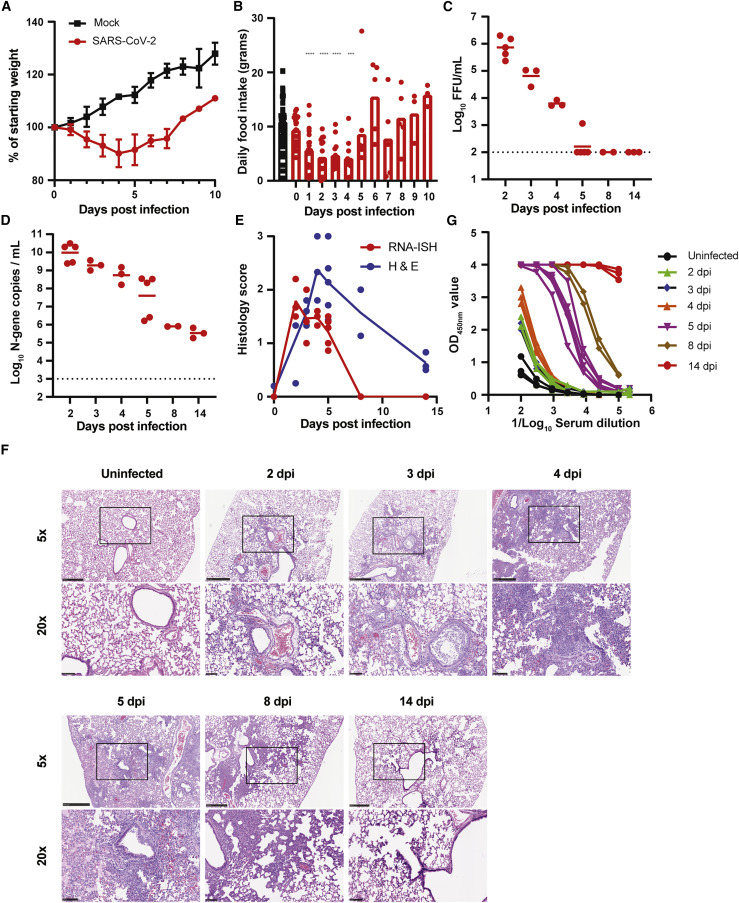
To establish the utility of the hamster model in our hands, we inoculated twenty-one 5- to 6-week-old male hamsters IN with 2 × 105 plaque forming units (PFUs) of a fully infectious SARS-CoV-2 isolate in 100 μL of PBS. A control group of three 5- to 6-week-old male hamsters was inoculated with PBS. Mock-infected animals continued to gain weight at a rate of ∼2.4 g or ∼3% per day (Figure 1 A). In contrast, SARS-CoV-2-inoculated hamsters began to lose weight at 2 dpi, and this continued through days 4 to 5, at which point the animals had lost approximately 10% of their body weight (Figure 1A). This decrease was associated with a reduction in food intake between 1 and 4 dpi (Figure 1B).
Figure 1.
Development of the SARS-CoV-2 hamster model
(A) Mean ± standard deviation (SD) weight loss or weight gain of uninfected (n = 3) or SARS-CoV-2 infected (n = 21).
(B) Daily food intake of uninfected and infected hamsters. Data points for the uninfected hamsters (n = 3 per day) were recorded for 14 days and plotted. For infected hamsters, food intake 1–10 dpi was recorded (∗∗∗∗p < 0.0001 and ∗∗∗p < 0.001 by ANOVA with Dunnett’s multiple comparison against the uninfected hamsters).
(C) Infectious virus titer was quantified by FFA from homogenates of the left lung lobe at indicated time points. Each dot is an individual hamster, and bars indicate median values (dotted line is the limit of detection).
(D) Lung viral RNA was quantified in the left lung lobe at indicated time points after infection. Each dot is an individual hamster, and bars indicate median values (dotted line is the limit of detection).
(E) Pathology score of the lungs from infected hamsters. <10% affected = 1, >10% but <50% = 2, and >50% = 3. Each lobe was scored, and the average score was plotted per animals. The solid line is the average score per day for RNA in situ hybridization (red line and dots) or inflammation (blue line and dots).
(F) Representative images at 5× and 20× magnification of H&E staining of SARS-CoV-2-infected hamsters sacrificed at different time points after inoculation (n = 5 for 2 dpi, n = 3 for 3 dpi, n = 3 for 4 dpi, n = 5 for 5 dpi, n = 2 for 8 dpi, n = 3 for 14 dpi, and n = 3 for uninfected). The scale bar represents 1 mm.
(G) Serum S-protein-specific IgG(H+L) responses in SARS-CoV-2-infected hamsters. Each color is a different day after infection.
(C and D) Bars indicate median values, and dotted lines are the LOD of the assays. See also Figure S1.
At indicated time points after infection, hamsters were sacrificed and tissues were collected for analysis of viral burden, histology, and serological response. The left lung lobe was collected, homogenized, and used to quantify SARS-CoV-2 N-gene copy number and infectious virus titer by qPCR and focus-forming assay (FFA), respectively. Infectious virus titers peaked at 2 dpi with 8 × 105 focus forming units/mL (FFUs/mL), and levels declined to low or undetectable by 5 dpi (Figure 1C). SARS-CoV-2 N-gene copy number also peaked at 2 dpi at 1010 copies per μL and gradually declined to 105 to 106 copies by 8–14 dpi (Figure 1D). The remainder of the lung tissue was fixed in formalin, embedded, and sectioned for viral RNA in situ hybridization (ISH) and hematoxylin and eosin (H&E) staining. SARS-CoV-2 viral RNA was detected by RNA-ISH at 2–5 dpi (Figures 1E and S1) and was no longer detectable by 8 dpi. Viral RNA was localized to both airway and alveolar epithelial cells (Figure S1). Infection was accompanied by immune cell infiltration in peribronchiolar and adjacent alveolar locations from 2 to 8 dpi (Figures 1E and 1F), a pattern that is consistent with bronchopneumonia. The immune cell infiltration was associated with alveolar edema, exudate, tissue damage, and intraparenchymal hemorrhage (Figures 1E and 1F). Each of these features of histopathology were markedly decreased by 14 dpi (Figure 1F).
Serum samples were assayed for the presence of antibodies specific for purified, recombinant S protein by ELISA. Low or undetectable antibody responses were detected through 4 dpi (Figure 1G). By day 5, S-specific immunoglobulin G (IgG)(H+L) responses were detected in all five animals, and the serum antibody titer further increased between 8 and 14 dpi.
ChAd-vectored vaccine elicits robust antibody responses against SARS-CoV-2 in hamsters
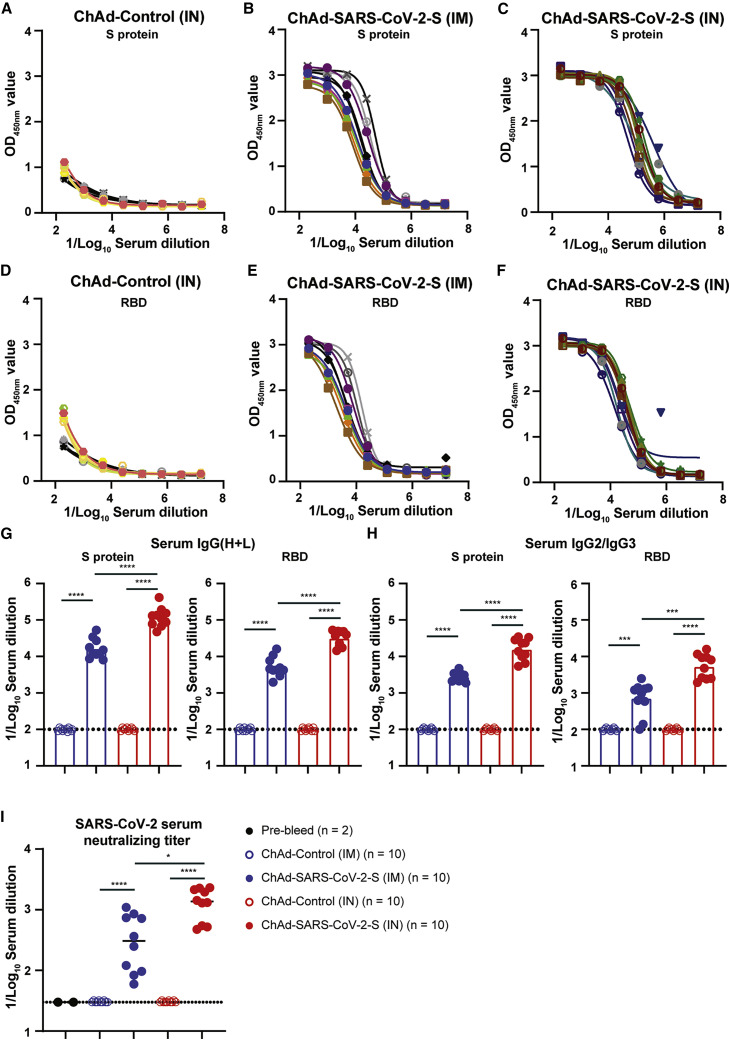
We assessed the immunogenicity of a replication-incompetent ChAd vector encoding a prefusion-stabilized, full-length sequence of ChAd-SARS-CoV-2-S (Hassan et al., 2020) in Golden Syrian hamsters. We used a ChAd vector without a transgene (ChAd-control) as a control. Groups of ten 5- to 6-week-old male hamsters were immunized once via IN or IM route with 1010 virus particles of ChAd-control or ChAd-SARS-CoV-2-S. Serum was collected prior to immunization or 21 days after, and antibody responses were evaluated by ELISA against purified recombinant S and receptor binding domain (RBD) proteins. Immunization with ChAd-SARS-CoV-2-S induced high levels of anti-S and anti-RBD IgG(H+L) and IgG2/IgG3 antibodies 21 days later, whereas low or undetectable levels of S- and RBD-specific antibodies were present in samples from ChAd-control-immunized animals (Figures 2 A–2F and S2). The antibody response was significantly higher after IN than IM immunization (5- to 7-fold; p < 0.0001 for anti-S and anti-RBD, respectively; Figures 2G and 2H). Serum samples also were tested for neutralization of infectious SARS-CoV-2 by focus-reduction neutralization test (FRNT). As expected, pre-immunization sera or sera from hamsters immunized with ChAd-control did not inhibit virus infection (Figure 2I). In contrast, sera from animals immunized with ChAd-SARS-CoV-2-S neutralized infectious virus with geometric mean titers (GMTs) of 1:1,217 and 1:276 for IN and IM immunization routes, respectively (Figure 2I).
Figure 2.
Humoral immune response following IN and IM immunization
(A–C) Anti-S-protein-specific serum IgG(H+L) titers in hamsters immunized IN with ChAd-control (A) or with ChAd-SARS-CoV-2-S IM (B) or IN (C). Each line is an individual animal.
(D–F) Receptor binding domain (RBD)-specific serum IgM titers in hamsters immunized IN with ChAd-control (D) or with ChAd-SARS-CoV-2-S IM (E) or IN (F). Each line is an individual animal.
(G and H) IC50 values for S-protein-specific or RBD-specific IgG(H+L) (G) or IgG2/IgG3 (H) serum antibodies in hamsters vaccinated IM (blue symbols) or IN (red symbols) with ChAd-control (open symbols) or ChAd-SARS-CoV-2-S (closed symbols). ∗∗∗∗p < 0.0001 and ∗∗∗p < 0.001 by Mann-Whitney test with a Bonferroni correction for multiple comparisons.
(I) SARS-CoV-2 serum neutralizing titer, measured by FRNT, in hamsters vaccinated IM or IN with ChAd-control or ChAd-SARS-CoV-2-S. ∗∗∗∗p < 0.0001 and ∗p < 0.05 by Mann-Whitney test with a Bonferroni correction for multiple comparisons.
(G–I) Bars indicate median values, and dotted lines are the LOD of the assays. See also Figures S2, S3, and S7.
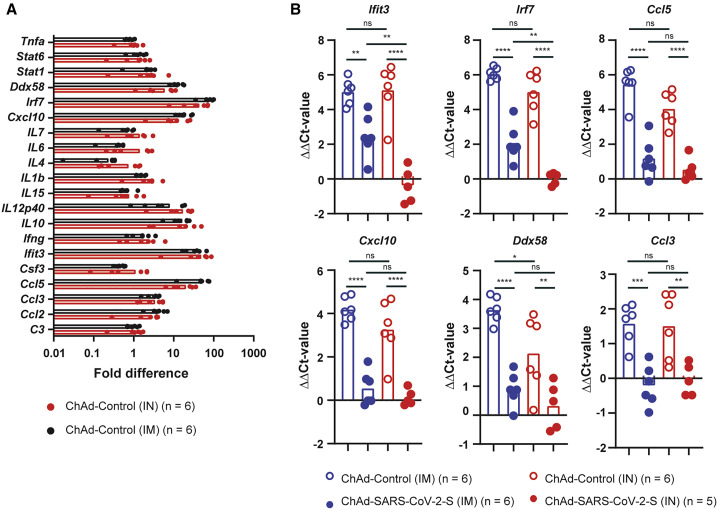
Because the ChAd-SARS-CoV-2-S vaccine was more immunogenic after IN administration, we quantified the expression of the spike gene and several inflammatory host genes in the lungs and muscle 3 days after immunization with the ChAd-SARS-CoV-2-S vaccine. Spike gene expression was detected in the lung and muscle of IN and IM immunized animals, respectively, but not in the control tissues. No significant difference in Spike gene expression between the lung and muscle was detected (p > 0.5; Figure S3). The expression of Irf7 and Ccl5 was increased in muscle of IM-immunized, but not in the lung of IN-immunized, animals (p < 0.05; Figure S3). No changes in the expression of Cxcl10, Ddx58, or Ccl3 were observed in either the lung or muscle in either group (Figure S3).
Immunization with ChAd-vectored vaccine protects hamsters from SARS-CoV-2 challenge
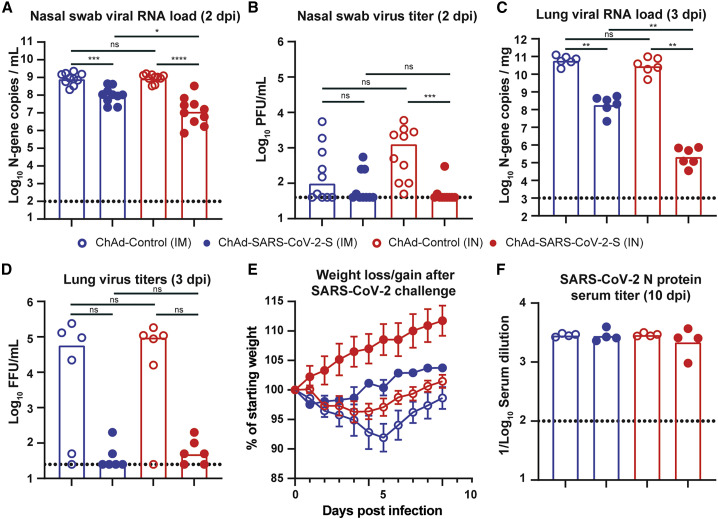
We next evaluated the protective effect of the ChAd vaccines in the hamster SARS-CoV-2 challenge model. Golden Syrian hamsters immunized with ChAd-SARS-CoV-2-S or ChAd-control were challenged IN with 2.5 × 105 PFUs of SARS-CoV-2, and 2 dpi, a nasal swab was collected for viral burden analysis by qPCR and plaque assay. The N-gene copy number in the ChAd-control-immunized animals was ∼109 copies per mL in both the IM and IN control groups (Figure 3 A). Immunization with ChAd-SARS-CoV-2-S reduced the N-gene copy number by 100-fold in the IN- (107/mL) and 10-fold in the IM (108/mL)-immunized animals (p < 0.0001 and p < 0.001 respectively; Figure 3A). N-gene copy number was significantly lower in the IN- than IM-immunized animals (p < 0.05; Figure 3A). At 2 dpi, infectious virus was detected by plaque assay in 4 of 20 ChAd-SARS-CoV-2-S-immunized animals and 15 of 20 ChAd-control-immunized animals (Figure 3B). At 3 dpi, six hamsters per group were sacrificed, and lungs were collected for viral burden analysis (left lobe) by qPCR or FFA or for histology (other lung lobes). In the control groups, we detected 109−1010 copies of N-gene per mg of lung homogenate, and the mean infectious titer was 6 × 104 FFU/mL. No difference was observed between the two control groups. Immunization with ChAd-SARS-CoV-2-S vaccine significantly reduced the N-gene copy number (p < 0.01; Figure 3C) and infectious titer in both the IN- and IM-immunized group (Figure 3D). A comparison between IN and IM immunization revealed significantly lower N-gene copies per mg (788-fold; p < 0.01; Figure 3C), but not in infectious virus titer (p = 0.5; Figure 3D), in the lungs of IN-immunized animals. The remaining four animals per group were monitored for weight loss for 10 days. The ChAd-control-immunized animals lost an average of 4% and 8% of their starting body weight (Figure 3E). Immunization with ChAd-SARS-CoV-2-S attenuated weight loss after SARS-CoV-2 challenge in both groups (p < 0.01; Figure 3E), with a possibly greater effect following IN immunization. To assess whether the ChAd-SARS-CoV-2-S vaccine induced sterilizing immunity by either of the immunization routes, we collected the serum 10 dpi to test for the presence of antibodies against recombinant N by ELISA. A robust anti-N IgG(H+L) (Figure S4) and IgG2/3 (Figure 3F) antibody response was detected in all ChAd-control and ChAd-SARS-CoV-2-S-immunized animals.
Figure 3.
IN immunization offers superior protection against challenge with SARS-CoV-2
(A–D) 28 days after a single IM (blue symbols) or IN (red symbols) vaccination with ChAd-control (open symbols) or ChAd-SARS-CoV-2-S (closed symbols), hamsters were challenged with 2.5 × 105 PFUs of SARS-CoV-2, and nasal swabs (A and B) and lungs (C and D) were collected for analysis of viral RNA levels by qPCR (A and C) and infectious virus by plaque assay (B and D). ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, and ns, not significant by Mann-Whitney test with a Bonferroni correction for multiple comparisons.
(E) Mean ± SD of weight loss or gain in SARS-CoV-2-challenged hamsters (n = 4 per group).
(F) SARS-CoV-2 N protein serum titer, measured by ELISA, in hamsters vaccinated IM or IN with ChAd-control or ChAd-SARS-CoV-2-S.
(A–D and F) Bars indicate median values, and dotted lines are the limit of detection of the assays. See also Figures S4 and S7.
Immunization with ChAd-vectored vaccine minimizes lung pathology in hamsters
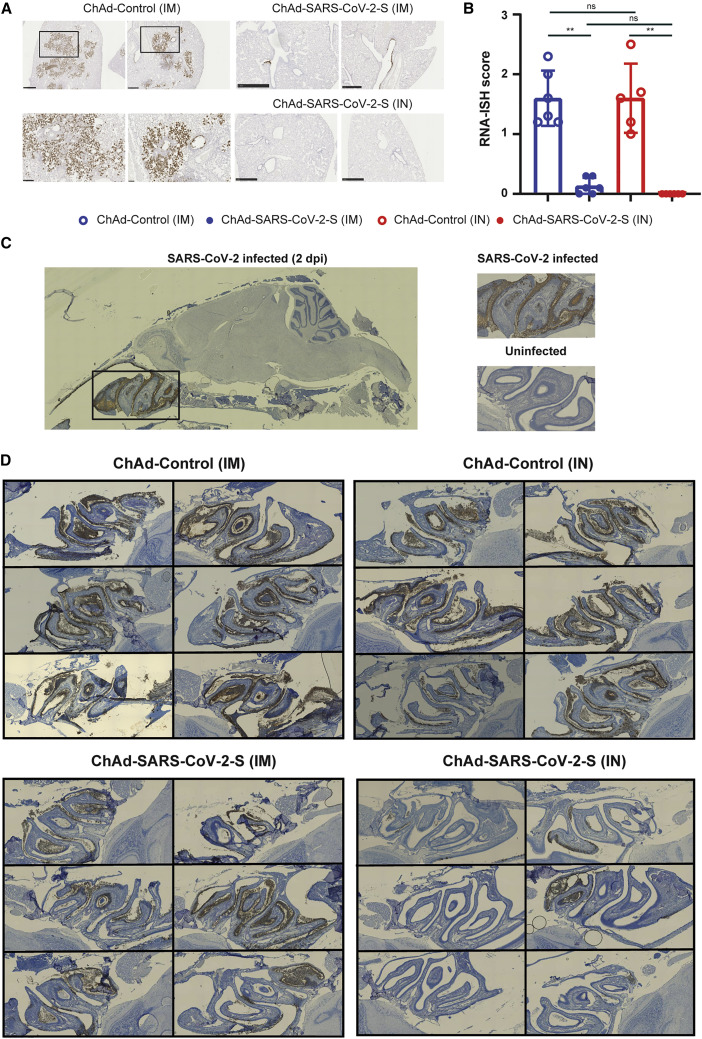
To support these findings, we performed RNA-ISH and H&E staining on sections from formalin-fixed lung tissues from immunized hamsters. RNA-ISH detected viral RNA in all animals immunized with ChAd-control vaccine (Figures 4 A, 4B, and S5). On average, ∼20% of the section was positive for SARS-CoV-2 RNA. The presence of viral RNA was associated with inflammation, tissue damage, and bronchopneumonia, as evidenced by immune cell infiltration around bronchioles, alveolar edema, fluid exudates, and intraparenchymal hemorrhage (Figure S5). In contrast, sections from animals immunized IM with ChAd-SARS-CoV-2-S contained no or few SARS-CoV-2-positive cells by RNA-ISH and inflammation was greatly reduced (p < 0.01; Figures 4A, 4B, and S6). No SARS-CoV-2-positive cells were detected following IN immunization with ChAd-SARS-CoV-2-S (p < 0.01; Figures 4A, 4B, and S6).
Figure 4.
IN immunization offers superior protection against SARS-CoV-2 infection of the nasal epithelium
(A) RNA in situ hybridization (ISH) for SARS-CoV-2 viral RNA in hamster lung sections. Representative images at 5× and 20× magnification of the ChAd-control (IM), ChAd-SARS-CoV-2-S (IM), and ChAd-SARS-CoV-2-S (IN) sections. The scale bar represents 1 mm.
(B) Comparison of RNA-ISH staining between groups of hamsters. Each lobe was scored according to the following system, <10% RNA-positive = 1, >10% but <50% RNA-positive = 2, and >50% RNA-positive = 3, and the average score was plotted per animals. ∗∗p < 0.01 and ns by Mann-Whitney U test with a Bonferroni correction for multiple comparisons.
(C) Representative images of sagittal sections of hamster heads infected with SARS-CoV-2 for 2 days or uninfected control. RNA-ISH was performed on the sections, and SARS-CoV-2 viral RNA was detected in the nasal turbinate.
(D) Detection of SARS-CoV-2 viral by RNA-ISH in sagittal sections of hamster heads from the immunized and SARS-CoV-2-challenged animals.
See also Figures S5 and S6.
An ideal SARS-CoV-2 vaccine would confer protection against disease and prevent virus infection and transmission. We hypothesized that IN delivery of the vaccine could provide superior protection in the upper respiratory tract compared to IM delivery. Hamster heads were collected 3 dpi and fixed in formalin. Following decalcification and embedding, sagittal sections were obtained and RNA-ISH was performed (Figure 4C). SARS-CoV-2 RNA was detected in the nasal cavity and ethmoturbinates of all 12 hamsters immunized IN or IM with ChAd-control (Figure 4D, left panel). No difference in viral RNA staining was observed between the two groups. Animals immunized IM with ChAd-SARS-CoV-2-S contained fewer SARS-CoV-2-positive cells and less cellular debris than ChAd-control-vaccinated animals (Figure 4D). However, animals immunized IN with ChAd-SARS-CoV-2-S had the fewest number of SARS-CoV-2-positive cells, and cellular debris was further reduced (Figure 4D). Collectively, these studies show that the ChAd-SARS-CoV-2-S vaccine is highly protective in the hamster model of COVID-19, and IN delivery of this vaccine provides superior protection against upper respiratory tract infection.
Inflammatory gene expression is reduced after SARS-CoV-2 challenge in the ChAd-SARS-CoV-2-immunized hamsters
Lung pathology after SARS-CoV-2 infection appears to be driven by inflammation (Boudewijns et al., 2020). Thus, a successful vaccine should reduce or eliminate the inflammatory response after infection or challenge with SARS-CoV-2. The inflammatory response was evaluated in the ChAd-control- and ChAd-SARS-CoV-2-S-immunized hamsters 3 days after challenge with SARS-CoV-2. RNA was extracted from the tissue homogenates and analyzed by qRT-PCR using 22 different validated primer-probe sets specific for two housekeeping genes (β2 m and Rpl18) and 20 different innate and inflammatory host genes (Table S1). Compared to five naive animals, expression of 8/20 inflammatory host genes (Ccl2, Ccl3, Cxcl10, Ddx58, Ifit3, IL10, IL12p40, and Irf7 increased >2-fold in the ChAd-control-immunized and SARS-CoV-2-challenged animals; Figure 5 A). A significant increase in gene expression was observed for Ccl3, Ifit3, Cxcl10, and Irf7 in both ChAd-control-IN and ChAd-control-IM animals (p < 0.05; Figure 5A). ChAd-SARS-CoV-2-S immunization significantly reduced inflammatory gene expression after SARS-CoV-2 challenge (Figure 5B) with the expression of a subset of host genes, such as Ccl5, Ccl3, and Cxcl10, near normal levels. A comparison in host gene expression between IM and IN immunization identified Irf7 and Ifit3 as two host genes whose expression was significantly (p < 0.01) lower in the IN- compared to IM-immunized animals (Figure 5B). These data suggest that IN delivery of the ChAd-vectored SARS-CoV-2-S vaccine provides greater protection against SARS-CoV-2 infection, inflammation, and disease in hamsters.
Figure 5.
ChAd-SARS-CoV-2 immunization ameliorates inflammatory gene expression following SARS-CoV-2 challenge
Inflammatory gene-expression (n = 22) was quantified by RT-PCR in RNA extracted from lung homogenates 3 dpi (primer and probe sequences are in Table S1).
(A) Fold increase in gene expression for ChAd-control-immunized (IN in red and IM in black) and SARS-CoV-2-challenged hamsters.
(B) ΔΔCt values for Ifit3, Irf7, Ccl5, Cxcl10, Ddx58, and Ccl3 in ChAd-control- (open symbols) and ChAd-SARS-CoV-2-S (closed symbols)-immunized and SARS-CoV-2-challenged animals 3 dpi. ns, ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, and ∗∗p < 0.01 by one-way ANOVA with a Šidák correction for multiple comparisons. Each dot is an individual animal from two experiments. Bars indicate average values.
See also Figure S7.
Discussion
Effective vaccines against SARS-CoV-2 are needed to combat the devastating pandemic. In this study, we evaluated IN and IM delivery of a ChAd-vectored vaccine expressing a prefusion stabilized S protein of SARS-CoV-2 in the Syrian hamster challenge model. A single dose of ChAd-SARS-CoV-2 induced S- and RBD-specific serum antibodies capable of neutralizing SARS-CoV-2. Antibody responses were higher after IN than IM immunization. Following challenge with a high dose of SARS-CoV-2, IN and IM immunization reduced infectious virus titers and viral RNA levels in the lungs and nasal swabs, albeit the effect was greater following IN immunization. Immunization with ChAd-SARS-CoV-2 also reduced weight loss, lung pathology, and inflammatory gene expression in the lungs of the animals with a greater effect again seen after IN immunization. Finally, IN immunization protected the upper respiratory tract of hamsters, whereas IM immunization did not. Combined, these studies demonstrate that a single dose of ChAd-SARS-CoV-2-S vaccine delivered IN provides better protection than IM immunization against SARS-CoV-2 challenge in Syrian hamsters.
At least four different virally vectored vaccines have been tested in Syrian hamsters (Sun et al., 2020; Tostanoski et al., 2020). A single dose of IN-delivered ChAd-SARS-CoV-2-S vaccine induced serum antiviral neutralizing titers of around 1:1,217, which is at least several fold higher than IM delivery of Ad26-S (1:360) and VSV-S or intraperitoneal delivery of Y17F-S (1:630). Furthermore, IN delivery of ChAd-SARS-CoV-2-S protected the upper respiratory tract against infection with SARS-CoV-2 and no weight loss was detected after virus challenge. This contrasts with the other vaccine candidates, where the vaccinated hamsters lose between 0% and 5% of their body weight after challenge.
The reason for the higher antibody responses after IN versus IM immunization currently is not known. Spike gene expression 3 days after IN or IM immunization was similar in the lung and muscle tissue, respectively, suggesting that the amount of spike protein produced is equal between the two sites. IN immunization also did not induce a more robust innate immune response, associated with stronger adaptive immune responses, compared to IM immunization. The most striking difference between IN and IM immunization is the enhanced protection of the upper respiratory tract infection, with minimal or no viral RNA detected in the nasal olfactory neuroepithelium, which expresses known receptors for SARS-CoV-2 (Chen et al., 2020). This effect may be due to the induction of local S-protein-specific immunity capable of neutralizing virus in the upper respiratory tract. In mice, IN immunization induced robust S-specific IgA antibodies (Hassan et al., 2020), whereas IM immunization did not. Anti-hamster-IgA secondary antibodies currently are not commercially available. Nonetheless, we would expect to find similar IgA responses that can neutralize incoming virus. As a result of the superior protection of the nasal cavity and upper respiratory tract, IN delivery of ChAd-SARS-CoV-2-S may offer protection against both infection and transmission of SARS-CoV-2. Aside from the specific immunity, IN delivery of the vaccine could provide some low-level non-specific protection against infection. However, no increase in the expression of Irf7, Ccl5, Cxcl10, Ddx58, or Ccl3 was detected 3 dpi in the lungs of IN-immunized animals (Figure S3). The expression of some (Ccl5 and Ddx58; Figure 5), but not all, pro-inflammatory genes trended lower in the lungs of animals inoculated IN with ChAd-control compared to the IM-vaccinated group. Similarly, IN-inoculated animals lost less weight compared to the IM-vaccinated animals (p > 0.05; one-way ANOVA with multiple comparisons).
Sera from the vaccinated hamsters were tested against several SARS-CoV-2 variants of concern, including B.1.1.7 and B.1.351 (Chen et al., 2021). No difference in serum neutralizing activity was detected between the parental WA1 isolate and the B.1.1.7 variant of SARS-CoV-2. In contrast, serum neutralizing activity against viruses containing a Spike protein with the E484K/N501Y/D614G or K417N/E484K/N501Y/D614G mutations or the entire spike protein of the B.1.351 variant of SARS-CoV-2 was significantly reduced. However, because viral vectored vaccines are better at inducing T-cell-mediated immunity, these vaccines may still confer substantial protection against variants in vivo.
IN immunization offers many benefits over more traditional approaches (Yusuf and Kett, 2017). Besides the ease of administration and lack of needles, IN delivery is associated with mucosal immune responses, including the production of IgA and stimulation of T and B cells in the nasopharynx-associated lymphoid tissue (Lycke, 2012). Influenza virus vaccines are the only licensed IN vaccines to date for individuals over the age of 2 and less than 50 years old. Live-attenuated influenza virus vaccine (LAIV) is considered safe and efficacious. The exception to this was the 2013–2014 and 2015–2016 season, when the vaccine was not effective against one of the four components (Parker et al., 2019). IN delivery of several viral-vectored vaccines has been evaluated in pre-clinical animal models. A single dose of ChAD-vectored vaccine against the Middle Eastern respiratory syndrome (MERS) virus protected hDPP4 knockin mice and rhesus macaques from MERS challenge (Jia et al., 2019; van Doremalen et al., 2020). Similarly, a parainfluenza-virus-5-vectored vaccine expressing the S protein of MERS protected mice from MERS (Li et al., 2020). A replication-incompetent recombinant serotype 5 adenovirus, Ad5-S-nb2, carrying a codon-optimized gene encoding S protein protected rhesus macaques from SARS-CoV-2 challenge (Feng et al., 2020). Besides coronaviruses, IN-delivered, adenoviral-vectored vaccines protected non-human primates from Ebola virus (Richardson et al., 2013). Importantly, in that study, protection occurred in the presence of existing adenovirus-specific immunity. Besides the many advantages of IN vaccines, IN delivery of a replication-defective adenovirus-5-vectored vaccine caused infection of olfactory nerves in mice (Lemiale et al., 2003). In humans, IN delivery of a non-replicating, adenovirus-vectored influenza vaccine was well tolerated and immunogenic (Van Kampen et al., 2005).
The pathogenesis following SARS-CoV-2 infection is mediated in part by a pathological inflammatory immune response (Meyerowitz-Katz and Merone, 2020). To evaluate the efficacy of this vaccine on reducing this part of the syndrome, we quantified changes in gene expression of 22 different hamster inflammatory and immune genes. Eight out of the 22 showed demonstrated a >2-fold increase in gene expression, with a clear enrichment for type I and III IFN-induced genes. The expression of several other hamster host genes, including IFN-γ, interleukins (ILs) (IL-10 is an exception), tumor necrosis factor alpha (TNF-α), and complement factors, did not increase after infection. This lack of expression may be due to the time of organ collection (3 dpi), when the inflammatory response is still developing. The lack of IFN-γ could be explained by the increase in IL-10 expression or the early time point evaluated that precedes influx of natural killer (NK) cells and activated T cells.
Correlates of immune protection and SARS-CoV-2-associated disease were investigated in our cohort of hamsters. Virus neutralization in serum correlated better with RBD-specific antibody levels (r = 0.83; p < 0.0001) than S-protein-specific responses (r = 0.31; p > 0.05; Figure S7). Of the three humoral response parameters, the virus neutralization serum titer (FRNT; IC50) correlated best with weight loss 3 dpi (r = 0.59; p < 0.0001; Figure S7). Weight loss at 3 dpi was strongly associated with viral RNA levels (r = −0.68; p < 0.001) and infectious virus load (r = −0.62; p < 0.01; Figure S7) in the lungs, but not in the nasal swabs. Finally, inflammatory host gene expression (Ifit3 and Cxcl10) correlated with RNA levels in the lungs (r = 0.68 and 0.84, respectively; p < 0.001) and serum virus neutralization titers (r = −0.70 and −0.64, respectively; p < 0.01). These analyses suggest that RBD-specific, but not S-specific, serum antibody and virus neutralization titers are important parameters of protection against SARS-CoV-2 and COVID-19 and that high antibody levels are associated with protection from infection and inflammation.
Overall, our studies in hamsters demonstrate that IN delivery of the ChAd-SARS-CoV-2-S vaccine confers protection against SARS-CoV-2 challenge. Protection is associated with lower virus levels in the lungs and upper and lower respiratory tracts, no weight loss, and reduced inflammation in the lungs. These findings support further pre-clinical and clinical studies investigating the vaccine efficacy of IN-delivered vaccines against SARS-CoV-2.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-SARS-CoV-2 mAb 1C02 | Gift from Dr. Ellebedy, Washington University in St Louis. | 1C02 |
| anti-hamster-IgG(H+L)-HRP | Southern Biotech | Cat#6061-05 |
| anti-hamster-IgG2/IgG3-HRP | Southern Biotech | Cat#1935-05 |
| anti-human-IgG-HRP | Sigma | Cat#S7900 |
| HRP-conjugated goat anti-human IgG | Sigma | Cat#A6029 |
| anti-SARS-CoV-2 mAb CR3022 | Yuan et al., 2020 | N/A |
| Bacterial and virus strains | ||
| SARS-CoV-2 (strain 2019 n-CoV/USA_WA1/2020) | CDC/BEI Resources | Cat#NR52281 |
| ChAd-SARS-CoV-2-S | Hassan et al., 2020 | N/A |
| ChAd-Control | Hassan et al., 2020 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant Spike protein of SARS-CoV-2 | Hassan et al., 2020 | N/A |
| Recombinant Receptor Binding Domain of Spike protein of SARS-CoV-2 | Hassan et al., 2020 | N/A |
| Recombinant Nucleoprotein of SARS-CoV-2 | Hassan et al., 2020 | N/A |
| TMB substrate | Vector laboratories | Cat#SK4400 |
| Critical commercial assays | ||
| RNA isolation kit | Omega Bio-Tek | Cat#R6834-01 |
| TaqMan Fast Universal PCR master mix | Thermo Scientific | Cat#4352042 |
| PrimeTime Gene Expression Master Mix | Integrated DNA technologies | Cat#1055772 |
| RNAscope® 2.5 HD Assay - BROWN | Advanced Cell Diagnostics | Cat#322310 |
| Experimental models: cell lines | ||
| Vero E6 | ATCC | Cat#CRL-1586 |
| MA-104 | ATCC | Cat#CRL-2378 |
| Experimental models: organisms/strains | ||
| LVG Golden Syrian Hamster | Charles Rivers Laboratories | Crl:LVG(SYR) |
| Oligonucleotides | ||
| SARS-CoV-2 N F: 5′-ATGCT GCAATCGTGCTACAA-3′ |
Integrated DNA technologies | N/A |
| SARS-CoV-2 N R: 5′-GACT GCCGCCTCTGCTC-3′ |
Integrated DNA technologies | N/A |
| SARS-CoV-2 N Probe: 5′-/56-FAM/TCAAGGAAC/ZEN/AACATTGCCAA/3IABkFQ/-3′ | Integrated DNA technologies | N/A |
| SARS-CoV-2 RNA ISH probe (S gene) | Advanced Cell Diagnostics | Cat#4848561 |
| SARS-CoV-2 S codon optimized F: 5′-GGCAACAGGAATGCGAAATG-3′ | IDT Technologies | N/A |
| SARS-CoV-2 S codon optimized R: 5′-TGAAACCGTACCAACCATCC-3′ | Integrated DNA technologies | N/A |
| SARS-CoV-2 S codon optimized probe: 5′-AACCCGCTATTGCAAATATGC-3′ | Integrated DNA technologies | N/A |
| SARS-CoV-2 5′ UTR F: 5′-ACTGT CGTTGACAGGACACG-3′ |
Integrated DNA technologies | N/A |
| SARS-CoV-2 5′ UTR R: 5′-AACAC GGACGAAACCGTAAG-3′ |
Integrated DNA technologies | N/A |
| SARS-CoV-2 5′ UTR PROBE: 5′-/56-FAM/CGTCTATCT/ZEN/TCTGCAGGCTG/3′-IABkFQ | Integrated DNA technologies | N/A |
| Software and algorithms | ||
| GraphPad Prism | GraphPad | v9.0 (https://www.graphpad.com:443/) |
| Nanozoomer Digital Pathology | Hamamatsu | v2 (https://www.hamamatsu.com/us/en/product/type/U12388-01/index.html) |
| BioSpot analyzer | Cellular Technology Limited | N/A |
| AxioImager Z2 system | Zeiss | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the Lead Contact, Adrianus C.M. Boon (jboon@wustl.edu).
Materials availability
All requests for resources and reagents should be directed to the Lead Contact author. This includes viruses, vaccines, and primer-probe sets. All reagents will be made available on request after completion of a Materials Transfer Agreement.
Data and code availability
All data supporting the findings of this study are available within the paper and are available from the corresponding author upon request. This paper does not include original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Cells and viruses
SARS-CoV-2 (strain 2019-nCoV/USA-WA1/2020) was obtained from the US Centers for Disease Control (CDC) and propagated on MA-104 monkey kidney cells, and the virus titer was determined by focus forming and plaque assays. The virus stock was sequenced by next-generation sequencing, and the spike protein sequence was identical to the original WA1 isolate. However, approximately 50% of the sequences, contained a 30 nucleotide deletion at the furin-cleavage of the spike protein. Vero E6 (CRL-1586, American Type Culture Collection (ATCC), were cultured at 37°C in Dulbecco’s Modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 10 mM HEPES, and 100 U/ml of penicillin–streptomycin. MA-104 cells (CRL-2378, ATCC) were cultured in Medium 199, supplemented with 5% FBS, 100 U/ml of penicillin–streptomycin, and 1 m L Amphotericin B. All work with infectious SARS-CoV-2 was performed in Institutional Biosafety Committee approved BSL3 and A-BSL3 facilities at Washington University School of Medicine using appropriate positive pressure air respirators and protective equipment.
Antibodies
The anti-S protein monoclonal antibody 1C02 was a gift from Dr. Ellebedy at Washington University.
Hamster experiments
Animal studies were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee at the Washington University School of Medicine (assurance number A3381–01). Five-week old male hamsters were obtained from Charles River Laboratories and housed at Washington University. Five days after arrival, a pre-immunization serum sample was obtained, and the animals (n = 10 per group) were vaccinated via intranasal (IN) or intramuscular (IM) route with 1010 viral particles of a chimpanzee adenovirus vector expressing a pre-fusion stabilized spike (S) protein of SARS-CoV-2 (ChAd-SARS-CoV-2-S (Hassan et al., 2020)) or a control chimpanzee adenovirus vector (ChAd-Control) in 100 μL of phosphate buffered saline (PBS). Twenty-one days later, a second serum sample was obtained, and the animals were transferred to the enhanced biosafety level 3 laboratory. One day later, the animals were challenged via IN route with 2.5 × 105 PFU of SARS-CoV-2. Animal weights were measured daily for the duration of the experiment. Two days after challenge, a nasal swab was obtained. The swab was moistened in 1.0 mL of serum-free media and used to rub the outside of the hamster nose. The swab was placed into the vial containing the remainder of the 1.0 mL of media, vortexed, and stored for subsequent virological analysis. Three days after challenge, a subset of animals was sacrificed, and their lungs were collected for virological and histological analysis. The left lobe was homogenized in 1.0 mL DMEM, clarified by centrifugation (21,000 x g for 5 minutes) and used for viral titer analysis by quantitative RT-PCR using primers and probes targeting the N gene or the 5′ UTR region, and by focus forming assay (FFA). From these same animals, we also collected serum for antibody analysis and heads for histological analysis. The remaining animals were sacrificed at 10 dpi, and serum was collected for analysis of antibody against the nucleoprotein (N protein) of SARS-CoV-2.
Method details
Spike protein and host response analysis after IM or IN immunization
Six-to-seven week old male Golden Syrian hamsters (n = 4-5 per group) were vaccinated via intranasal or intramuscular route with 1010 particles of ChAd-SARS-CoV-2-S in 100 μL of PBS. Three days later, animals were sacrificed, and the lung and muscle were homogenized in 1.0 or 2.0 mL of tissue culture fluid respectively, and clarified by centrifugation (21,000 x g for 5 minutes). RNA was extracted from 100 μL of homogenized tissue, eluted in 50 μL and used for qPCR analysis. Complementary DNA was synthesized using random hexamers and Superscript III (Thermo Scientific) with the addition of RNase inhibitor according to the manufacturer’s protocol. The expression of the SARS-CoV-2 spike gene and six inflammatory hamster host genes (Ccl3, Ccl5, Cxcl10, Ddx58, and Irf7) was quantified using TaqMan Fast Universal PCR master mix (Thermo Scientific) with validated primers/probe sets (Table S1). The results were normalized to Rpl18 expression levels. The primer probe sequences for the codon optimized Spike gene are: forward primer GGCAACAGGAATGCGAAATG, probe AACCCGCTATTGCAAATATGC, reverse primer TGAAACCGTACCAACCATCC. The difference in Ct-value between the gene or interest and Rpl18 (ΔCt) were compared between immunizations and route of administration.
Virus titration assays
FFA were performed on Vero-E6 cells in a 96-well plate. Lung tissue homogenates were serially diluted 10-fold, starting at 1:10, in cell infection medium (DMEM + 2% FBS + L-glutamine + penicillin + streptomycin), and 100 μL of the diluted virus was added to two wells per dilution per sample. After 1 h at 37°C, the inoculum was aspirated, the cells were washed with PBS, and a 1% methylcellulose overlay in infection medium was added. Positive and negative controls were included in every assay. Twenty-four hours after virus inoculation, the cells were fixed with formalin, and infected cells were detected by the addition of 100 μL of 1:1000 diluted anti-S protein monoclonal antibody (1C02, gift from Dr. Ellebedy at Washington University) in permeabilization buffer (1x PBS, 2% FBS, 0.2% saponin (Sigma, Cat #S7900)) for 1 h at 20°C or overnight at 4°C, followed by an anti-human-IgG-HRP antibody (Sigma, Cat. #A6029) in permeabilization buffer for 1 h at 20°C. The assay was developed using TMB substrate (Vector laboratories, SK4400) for 5-10 min at 20°C. The assay was stopped by washing the cells with water. The number of foci per well were counted on the BioSpot analyzer (Cellular Technology Limited) and used to calculate the focus forming units/mL (FFU/mL).
Plaque assays were performed on Vero E6 cells in 24-well plates. Nasal swabs or lung tissue homogenates were serially diluted 10-fold, starting at 1:10, in cell infection medium (DMEM + 2% FBS + L-glutamine + penicillin + streptomycin). Two hundred and fifty microliters of the diluted virus were added to a single well per dilution per sample. After 1 h at 37°C, the inoculum was aspirated, the cells were washed with PBS, and a 1% methylcellulose overlay in MEM supplemented with 2% FBS was added. Seventy-two hours after virus inoculation, the cells were fixed with 4% formalin, and the monolayer was stained with crystal violet (0.5% w/v in 25% methanol in water) for 1 h at 20°C. The number of plaques were counted and used to calculate the plaque forming units/mL (PFU/mL).
To quantify viral load in nasal swabs and lung tissue homogenates, RNA was extracted using RNA isolation kit (Omega). SARS-CoV-2 RNA levels were measured by one-step quantitative reverse transcriptase PCR (qRT-PCR) TaqMan assay as described previously (Hassan et al., 2020). A SARS-CoV-2 nucleocapsid (N) specific primers/probe set (L primer: ATGCTGCAATCGTGCTACAA; R primer: GACTGCCGCCTCTGCTC; probe: 5′-FAM/TCAAGGAAC/ZEN/AACATTGCCAA/3′-IABkFQ) or 5′ UTR specific primers/probe set (L primer: ACTGTCGTTGACAGGACACG; R primer: AACACGGACGAAACCGTAAG; probe: 5′-FAM/CGTCTATCT/ZEN/TCTGCAGGCTG/3′-IABkFQ). Viral RNA was expressed as (N) gene or 5′ UTR copy numbers per mg for lung tissue homogenates or mL for nasal swabs, based on a standard included in the assay, which was created via in vitro transcription of a synthetic DNA molecule containing the target region of the N gene and 5′-UTR region.
ELISA
Purified viral antigens (S, RBD, or N) (Hassan et al., 2020) were coated onto 96-well Maxisorp clear plates at 2 μg/mL in 50 mM Na2CO3 pH 9.6 (70 μL) or PBS (50 μL) overnight at 4°C. Coating buffers were aspirated, and wells were blocked with 200 μL of 1X PBS + 0.05% Tween-20 + 5% BSA + 0.02% NaN3 (Blocking buffer, PBSTBA) or 1X PBS + 0.05% Tween-20 + 10% FCS (PBSTF) either for 2 h at 20°C, 1 h at 37°C or overnight at 4°C. Heat-inactivated serum samples were diluted in PBSTBA or PBSTF in a separate 96-well polypropylene plate. The plates then were washed thrice with 1X PBS + 0.05% Tween-20 (PBST), followed by addition of 50 μL of respective serum dilutions. Sera were incubated in the blocked ELISA plates for at least 1 h at room temperature. The ELISA plates were again washed thrice in PBST, followed by addition of 50 μL of 1:1000 anti-hamster-IgG(H+L)-HRP (Southern Biotech Cat. #6061-05) in PBST or PBSTF or 1:1000 anti-hamster-IgG2/IgG3-HRP in PBST or PBSTF (Southern Biotech Cat. #1935-05). Plates were incubated at room temperature for 1-2 h, washed thrice in PBST and 50 μL of 1-Step Ultra TMB-ELISA was added (Thermo Fisher Scientific, Cat. #34028). Following a 12 to 15-min incubation, reactions were stopped with 50 μL of 2 M H2S04. The absorbance of each well at 450 nm was read (Synergy H1 or Epoch) within 2 min of addition of H2S04.
SARS-CoV-2 neutralization assay
Heat-inactivated serum samples were diluted 1:10 fold serially and incubated with 102 FFU of SARS-CoV-2 for 1 h at 37°C. The virus-serum mixtures were added to Vero-E6 cell monolayers in 96-well plates and incubated for 1 h at 37°C. Subsequently, cells were overlaid with 1% (w/v) methylcellulose in MEM supplemented with 2% FBS. Plates were incubated for 30 h before fixation using 4% PFA in PBS for 1 h at 20°C. Cells were washed and then sequentially incubated with anti-SARS-CoV-2 CR3022 mAb (Yuan et al., 2020) (1 μg/mL) and a HRP-conjugated goat anti-human IgG (Sigma, Cat#A6029) in PBS supplemented with 0.1% (w/v) saponin and 0.1% BSA. TrueBlue peroxidase substrate (KPL) was used to develop the plates before counting the foci on a BioSpot analyzer (Cellular Technology Limited).
Histology and RNA in situ hybridization
The lungs and heads from SARS-CoV-2 infected and control hamsters were fixed in 10% formalin for seven days. Lungs were embedded in paraffin and sectioned before hematoxylin and eosin staining (H & E) and RNA in situ hybridization (RNA-ISH) to detect SARS-CoV-2 RNA. Following formalin fixation, heads were decalcified in 0.5 M EDTA for seven days, cryoprotected in three exchanges of 30% sucrose for three days, and then embedded in O.C.T. compound before RNA-ISH. RNA-ISH was performed using a probe against the S gene of SARS-CoV-2 (V-nCoV2019-S, Cat #848561) with the RNAscope® 2.5 HD Assay—BROWN (ACDBio, Cat#322310) according to the manufacturers’ recommendations. Lung slides were scanned using the Hamamatsu NanoZoomer slide scanning system and head sections were imaged using the Zeiss AxioImager Z2 system. Lung sections were scored according to a previous publication (Imai et al., 2020) (< 10% affected lung tissue = 1, > 10% but < 50% affected area = 2, > 50% affected area = 3).
Host response gene analysis
RNA extracted from hamster lung tissue homogenates was used to synthesize cDNA using random hexamers and Superscript III (Thermo Scientific) with the addition of RNase inhibitor according to the manufacturer’s protocol. The expression of 22 inflammatory host genes was determined using PrimeTime Gene Expression Master Mix (Integrated DNA Technologies) with primers/probe sets specific for C3, C5, Ccl2, Ccl3, Ccl5, Csf3, Cxcl10, Ddx58, Ifit3, Ifng, Irf7, IL1b, IL4, IL5, IL6, IL7, IL10, IL12p40, IL15, Stat1, Stat6, and Tnfa and results were normalized to Rpl18 and B2m levels. The primers and probes were derived from previous publications (Zivcec et al., 2011) or developed in-house (see Table S1). Fold change was determined using the 2-ΔΔCt method comparing immunized and SARS-CoV-2 challenged hamsters to naive controls.
Quantification and statistical analysis
Statistical significance was assigned when P values were < 0.05 using GraphPad Prism version 9.0. Tests, number of animals (n), median values, and statistical comparison groups are indicated in the Figure legends. Analysis of weight change was determined by two-way ANOVA. Changes in functional parameters or immune parameters were compared to isotype-treated animals and were analyzed by one-way ANOVA with Sidak, Dunnett’s or Bonferonni test for multiple comparisons. We used a Pearson correlation analysis to immune correlates of vaccine-mediated protection SARS-CoV-2.
Acknowledgments
We acknowledge Krzysztof Hyrc for help with the Hamamatsu NanoZoomer slide scanning system. We also thank Amy Dillard for help with the serum collections from the animals and Ali Ellebedy and Jackson Turner for help with antibody ELISA and recombinant protein. We thank Drs. Susan Cook and Ken Boschert for support of the COVID-19 research. This study was funded by NIH contracts and grants (R01 AI157155, 75N93019C00062, U01 AI151810, HHSN272201400018C, and HHSN272201700060C). J.B.C. is supported by a Helen Hay Whitney Foundation postdoctoral fellowship.
Author contributions
T.L.B. and T.L.D. performed the animal experiments. A.O.H. upscaled and purified the ChAd vectors. A.O.H., A.C.M.B., and J.B.C. performed the serological analysis. H.H.H., T.L.B., and T.L.D. performed viral load analysis by qRT-PCR, focus forming, or plaque assay. T.L.D. developed and validated primer-probes sets for Syrian hamster genes and performed the host gene expression assays. M.J.H. performed the histology analysis. A.S., X.J., and R.K. performed the histology and RNA-ISH on the hamster heads. A.C.M.B. performed RNA-ISH on the hamster lungs. A.L.B. and J.B.C. provided qPCR reagents and protocols. Y.-N.D., H.Z., L.J.A., and D.H.F. provided recombinant proteins for serological analysis. M.S.D. and A.C.M.B. provided supervision and acquired funding. A.C.M.B. wrote the initial draft, with the other authors providing editorial comments.
Declaration of interests
M.S.D. is a consultant for Inbios, Vir Biotechnology, NGM Biopharmaceuticals, and Carnival Corporation and on the Scientific Advisory Board of Moderna and Immunome. The Diamond laboratory has received unrelated funding support in sponsored research agreements from Moderna, Vir Biotechnology, and Emergent BioSolutions. The Boon laboratory has received unrelated funding support in sponsored research agreements from AI Therapeutics, GreenLight Biosciences Inc., AbbVie Inc., and Nano targeting & Therapy Biopharma Inc. M.S.D. and A.O.H. have filed a disclosure with Washington University for possible commercial development of ChAd-SARS-CoV-2. A.C.M.B. is a recipient of a licensing agreement with Abbvie Inc. for commercial development of SARS-CoV-2 monoclonal antibody (mAb).
Published: July 20, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109400.
Supplemental information
References
- Boudewijns R., Thibaut H.J., Kaptein S.J.F., Li R., Vergote V., Seldeslachts L., Van Weyenbergh J., De Keyzer C., Bervoets L., Sharma S. STAT2 signaling restricts viral dissemination but drives severe pneumonia in SARS-CoV-2 infected hamsters. Nat. Commun. 2020;11:5838. doi: 10.1038/s41467-020-19684-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Hiyoshi A., Montgomery S. COVID-19 case-fatality rate and demographic and socioeconomic influencers: worldwide spatial regression analysis based on country-level data. BMJ Open. 2020;10:e043560. doi: 10.1136/bmjopen-2020-043560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.-W., Zhang A.J., Yuan S., Poon V.K.-M., Chan C.C.-S., Lee A.C.-Y., Chan W.-M., Fan Z., Tsoi H.-W., Wen L. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020;71:2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Shen W., Rowan N.R., Kulaga H., Hillel A., Ramanathan M., Jr., Lane A.P. Elevated ACE-2 expression in the olfactory neuroepithelium: implications for anosmia and upper respiratory SARS-CoV-2 entry and replication. Eur. Respir. J. 2020;56:2001948. doi: 10.1183/13993003.01948-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary S.J., Pitchford S.C., Amison R.T., Carrington R., Robaina Cabrera C.L., Magnen M., Looney M.R., Gray E., Page C.P. Animal models of mechanisms of SARS-CoV-2 infection and COVID-19 pathology. Br. J. Pharmacol. 2020;177:4851–4865. doi: 10.1111/bph.15143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driouich J.-S., Cochin M., Lingas G., Moureau G., Touret F., Petit P.R., Piorkowski G., Barthélémy K., Coutard B., Guedj J. Favipiravir antiviral efficacy against SARS-CoV-2 in a hamster model. bioRxiv. 2020 doi: 10.1038/s41467-021-21992-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felipe L.S., Vercruysse T., Sharma S., Ma J., Lemmens V., van Looveren D., Arkalagud Javarappa M.P., Boudewijns R., Malengier-Devlies B., Kaptein S.F. A single-dose live-attenuated YF17D-vectored SARS-CoV2 vaccine candidate. bioRxiv. 2020 doi: 10.1038/s41586-020-3035-9. [DOI] [PubMed] [Google Scholar]
- Feng L., Wang Q., Shan C., Yang C., Feng Y., Wu J., Liu X., Zhou Y., Jiang R., Hu P. An adenovirus-vectored COVID-19 vaccine confers protection from SARS-COV-2 challenge in rhesus macaques. Nat. Commun. 2020;11:4207. doi: 10.1038/s41467-020-18077-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A.O., Kafai N.M., Dmitriev I.P., Fox J.M., Smith B.K., Harvey I.B., Chen R.E., Winkler E.S., Wessel A.W., Case J.B. A single-dose intranasal ChAd vaccine protects upper and lower respiratory tracts against SARS-CoV-2. Cell. 2020;183:169–184.e13. doi: 10.1016/j.cell.2020.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. USA. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W., Channappanavar R., Zhang C., Li M., Zhou H., Zhang S., Zhou P., Xu J., Shan S., Shi X. Single intranasal immunization with chimpanzee adenovirus-based vaccine induces sustained and protective immunity against MERS-CoV infection. Emerg. Microbes Infect. 2019;8:760–772. doi: 10.1080/22221751.2019.1620083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaptein S.J., Jacobs S., Langendries L., Seldeslachts L., ter Horst S., Liesenborghs L., Hens B., Vergote V., Heylen E., Maas E. Antiviral treatment of SARS-CoV-2-infected hamsters reveals a weak effect of favipiravir and a complete lack of effect for hydroxychloroquine. bioRxiv. 2020 [Google Scholar]
- Kaptein S.J.F., Jacobs S., Langendries L., Seldeslachts L., Ter Horst S., Liesenborghs L., Hens B., Vergote V., Heylen E., Barthelemy K. Favipiravir at high doses has potent antiviral activity in SARS-CoV-2-infected hamsters, whereas hydroxychloroquine lacks activity. Proc. Natl. Acad. Sci. USA. 2020;117:26955–26965. doi: 10.1073/pnas.2014441117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemiale F., Kong W.P., Akyürek L.M., Ling X., Huang Y., Chakrabarti B.K., Eckhaus M., Nabel G.J. Enhanced mucosal immunoglobulin A response of intranasal adenoviral vector human immunodeficiency virus vaccine and localization in the central nervous system. J. Virol. 2003;77:10078–10087. doi: 10.1128/JVI.77.18.10078-10087.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Li Z., Wohlford-Lenane C., Meyerholz D.K., Channappanavar R., An D., Perlman S., McCray P.B., Jr., He B. Single-dose, intranasal immunization with recombinant parainfluenza virus 5 expressing Middle East respiratory syndrome coronavirus (MERS-CoV) Spike protein protects mice from fatal MERS-CoV infection. MBio. 2020;11:e00554-20. doi: 10.1128/mBio.00554-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lycke N. Recent progress in mucosal vaccine development: potential and limitations. Nat. Rev. Immunol. 2012;12:592–605. doi: 10.1038/nri3251. [DOI] [PubMed] [Google Scholar]
- Meyerowitz-Katz G., Merone L. A systematic review and meta-analysis of published research data on COVID-19 infection fatality rates. Int. J. Infect. Dis. 2020;101:138–148. doi: 10.1016/j.ijid.2020.09.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudatsir M., Fajar J.K., Wulandari L., Soegiarto G., Ilmawan M., Purnamasari Y., Mahdi B.A., Jayanto G.D., Suhendra S., Setianingsih Y.A. Predictors of COVID-19 severity: a systematic review and meta-analysis. F1000Res. 2020;9:1107. doi: 10.12688/f1000research.26186.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Fontela C., Dowling W.E., Funnell S.G.P., Gsell P.S., Riveros-Balta A.X., Albrecht R.A., Andersen H., Baric R.S., Carroll M.W., Cavaleri M. Animal models for COVID-19. Nature. 2020;586:509–515. doi: 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterrieder N., Bertzbach L.D., Dietert K., Abdelgawad A., Vladimirova D., Kunec D., Hoffmann D., Beer M., Gruber A.D., Trimpert J. Age-dependent progression of SARS-CoV-2 infection in Syrian hamsters. Viruses. 2020;12:779. doi: 10.3390/v12070779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey K., Acharya A., Mohan M., Ng C.L., Reid S.P., Byrareddy S.N. Animal models for SARS-CoV-2 research: a comprehensive literature review. Transbound. Emerg. Dis. 2020 doi: 10.1111/tbed.13907. Published online October 31, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker L., Ritter L., Wu W., Maeso R., Bright H., Dibben O. Haemagglutinin stability was not the primary cause of the reduced effectiveness of live attenuated influenza vaccine against A/H1N1pdm09 viruses in the 2013-2014 and 2015-2016 seasons. Vaccine. 2019;37:4543–4550. doi: 10.1016/j.vaccine.2019.06.016. [DOI] [PubMed] [Google Scholar]
- Richardson J.S., Pillet S., Bello A.J., Kobinger G.P. Airway delivery of an adenovirus-based Ebola virus vaccine bypasses existing immunity to homologous adenovirus in nonhuman primates. J. Virol. 2013;87:3668–3677. doi: 10.1128/JVI.02864-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenke K., Jarvis M.A., Feldmann F., Schwarz B., Okumura A., Lovaglio J., Saturday G., Hanley P.W., Meade-White K., Williamson B.N. Hydroxychloroquine prophylaxis and treatment is ineffective in macaque and hamster SARS-CoV-2 disease models. JCI Insight. 2020;5:143174. doi: 10.1172/jci.insight.143174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenke K., Meade-White K., Letko M.C., Clancy C., Hansens F., Liu Y., Okumura A., Tang-Huau T.L., Li R., Saturday G. Defining the Syrian hamster as a highly susceptible preclinical model for SARS-CoV-2 infection. bioRxiv. 2020 doi: 10.1080/22221751.2020.1858177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia S.F., Yan L.M., Chin A.W.H., Fung K., Choy K.T., Wong A.Y.L., Kaewpreedee P., Perera R.A.P.M., Poon L.L.M., Nicholls J.M. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., McCroskery S., Liu W.C., Leist S.R., Liu Y., Albrecht R.A., Slamanig S., Oliva J., Amanat F., Schaefer A. A Newcastle disease virus (NDV) expressing membrane-anchored spike as a cost-effective inactivated SARS-CoV-2 vaccine. bioRxiv. 2020 doi: 10.3390/vaccines8040771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tostanoski L.H., Wegmann F., Martinot A.J., Loos C., McMahan K., Mercado N.B., Yu J., Chan C.N., Bondoc S., Starke C.E. Ad26 vaccine protects against SARS-CoV-2 severe clinical disease in hamsters. Nat. Med. 2020;26:1694–1700. doi: 10.1038/s41591-020-1070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Haddock E., Feldmann F., Meade-White K., Bushmaker T., Fischer R.J., Okumura A., Hanley P.W., Saturday G., Edwards N.J. A single dose of ChAdOx1 MERS provides protective immunity in rhesus macaques. Sci. Adv. 2020;6:eaba8399. doi: 10.1126/sciadv.aba8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kampen K.R., Shi Z., Gao P., Zhang J., Foster K.W., Chen D.T., Marks D., Elmets C.A., Tang D.C. Safety and immunogenicity of adenovirus-vectored nasal and epicutaneous influenza vaccines in humans. Vaccine. 2005;23:1029–1036. doi: 10.1016/j.vaccine.2004.07.043. [DOI] [PubMed] [Google Scholar]
- Yahalom-Ronen Y., Tamir H., Melamed S., Politi B., Shifman O., Achdout H., Vitner E.B., Israeli O., Milrot E., Stein D. A single dose of recombinant VSV-ΔG-spike vaccine provides protection against SARS-CoV-2 challenge. bioRxiv. 2020 doi: 10.1038/s41467-020-20228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuf H., Kett V. Current prospects and future challenges for nasal vaccine delivery. Hum. Vaccin. Immunother. 2017;13:34–45. doi: 10.1080/21645515.2016.1239668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivcec M., Safronetz D., Haddock E., Feldmann H., Ebihara H. Validation of assays to monitor immune responses in the Syrian golden hamster (Mesocricetus auratus) J. Immunol. Methods. 2011;368:24–35. doi: 10.1016/j.jim.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and are available from the corresponding author upon request. This paper does not include original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.