To the Editor
Atopic dermatitis (AD) is a common chronic illness that often manifests in childhood as red, itchy, inflammatory, crusting skin lesions. However, the disease can occur at any age and waxes and wanes in severity. The most common genetic findings in children with AD are filaggrin (FLG; OMIM:135940) loss of function mutations (LOF) found on the gene’s third exon. . Europeans with the most severe and persistent forms of AD are more likely to have FLG LOF (Irvine et al., 2011, Margolis et al., 2012). Asians also have FLG LOF though many of their specific variants differ from those of Europeans (Wong et al., 2018). Past studies have claimed that FLG LOF variants are very rare, if not absent, in those of African ancestry (Thawer-Esmail et al., 2014, Winge et al., 2011, Margolis et al., 2014, Margolis et al., 2020a, Margolis et al., 2020b, Margolis et al., 2018). More recent studies of African-Americans indicate that while the prevalence of variants is much less than that of European ancestry, FLG LOF is not as rare as previously noted (Margolis et al., 2019). In fact, recent data from public access databases that used whole exome/genome approaches reveal more than 100 FLG LOF variants in those of African ancestry (https://gnomad.broadinstitute.org/). Oddly, gnomAD shows FLG to have many more distinct variants than other closely located genes in the epidermal differentiation complex found on chromosome 1 such as filaggrin-2, hornerin and trichohyalin (see supplement). The most common FLG mutations in those of African ancestry differ from those observed in European as well as Asian populations. Regardless of these newer findings, it has been suggested that AD outcomes in African Americans can be explained by population admixture between European and African ancestries. The goal of our study was to determine whether African Americans (AA) and European Americans (EA) are similar to each other and also to reference populations of Africans and Europeans, respectively, in terms of overall FLG variation.
Our primary data come from the Pediatric Elective Eczema Registry (PEER), which is a prospective 10-year observational cohort. Details regarding this study can be found in Margolis et al. (2012). We obtained FLG data on African and European ancestry participants from phase 3 of the 1000 Genomes Project of the International Genome Sample Resource (IGSR); we refer to this sample as 1000 Genomes (1000G). Data from PEER were initially analyzed to assess differences in the total number of FLG variants by ethnicity (European or African) using linear regression or negative binomial regression (IRR), as appropriate.
To explore genetic similarity between subjects within the FLG locus, we employed logistic principal component analysis (PCA) on FLG SNP observations using the approach by Landgraf & Lee (2015) in R (Landgraf and Lee, 2015). Their method extends the original PCA formulation of Pearson (1901) by constructing a projection of data that is closest to the original with respect to minimizing Bernoulli deviance (Pearson, 1901). This dimensionality reduction approach extracts information in the SNP data and summarizes that variation into a few principal components allowing for easier visualization of underlying patterns. Using the first and second principal components which account for a large portion of the variability in FLG SNPs, we assessed clustering of observations for the PEER population (both AA and EA) and combined with data from 1000G. We also conducted PCA on a separate cohort, Genetic of Atopic Dermatology (GAD), to verify that our results were not sample-specific.
The number of FLG synonymous and non-synonymous variants (excluding LOF variants) ranged from 2 to 109 per person in the PEER population with mean38.3 (sd: 33.4) and median of 49 (IQR: 5, 60). The number of FLG variants per person was greater in AA (mean: 54.7, sd: 31.8) than EA (mean: 24.7, sd: 24.7), IRR 2.0 (95% confidence interval: 1.7, 2.3; p=0.0009). For all FLG LOF variants, a composite was constructed (no variant, a single variant, two or more variants). In AA, the mean number of FLG SNPs was 55.1 for those with no LOF variants, 43.9 for those with one LOF variant, and 28.5 for those with two or more. The mean numbers for the EA group were 26.0, 23.0, and 18.2 for the three composite categories, respectively. Thus, there appears to be an inverse relationship between number of SNPs and presence of LOF variants.
Figure 1(a) presents the first two principal components based on 484 FLG SNPs for the 741 AD subjects. We distinguished between AA and EA to compare and contrast their respective genetic variations. For the most part, FLG variation is differs between AA and EA. To better understand the FLG variation of AA AD patients, we combined the PEER data with subjects who were African (population code: AFR) or European (population code: EUR) in 1000G (combined N=1905). 335 FLG SNPs noted in both PEER and 1000G were evaluated. Figure 1(b) shows that AA in PEER tend to cluster with Africans from 1000G, while EA in PEER tend to cluster with Europeans in 1000G. All humans originated from the same ancestral root, and as would be expected, some clusters include both ethnicities. The observed nonoverlap (clusters containing only AA or only EA in Figure 1(a)) suggests that FLG variation in AA is not explained by genetic admixture with EA in the United States. A comparison of patients with FLG LOF and those without is presented in the supplement.
Figure 1:
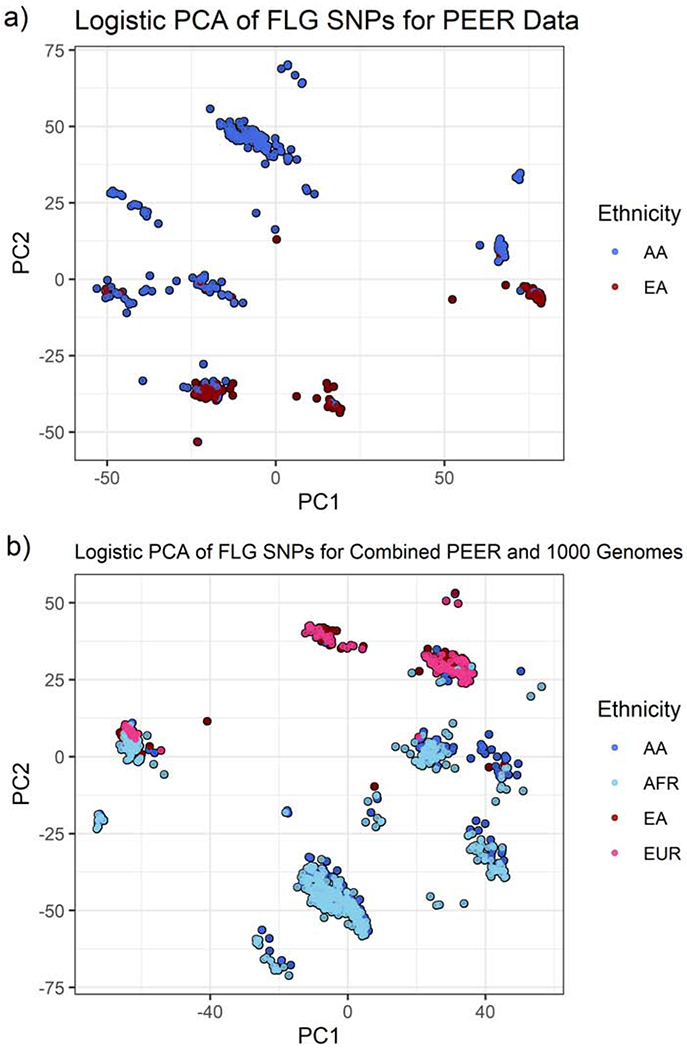
PCA plots showing clustering by ethnicity/ancestry for PEER cases based on FLG variation. (a) 484 FLG SNPs for subjects with AD in the PEER cohort and (b) 335 FLG SNPs in common to the PEER and 1000 Genomes data sets.
To replicate our results, a similar analysis was conducted for subjects with AD in the GAD cohort, which includes 412 subjects with 579 FLG SNPs. Figure 2(a) again indicates that there are differences in the genetic variation of FLG between AA and EA. Comparison of the GAD cohort to 1000G shows that AA from the GAD cohort and Africans from 1000G are more likely to cluster together (Figure 2(b)).
Figure 2:
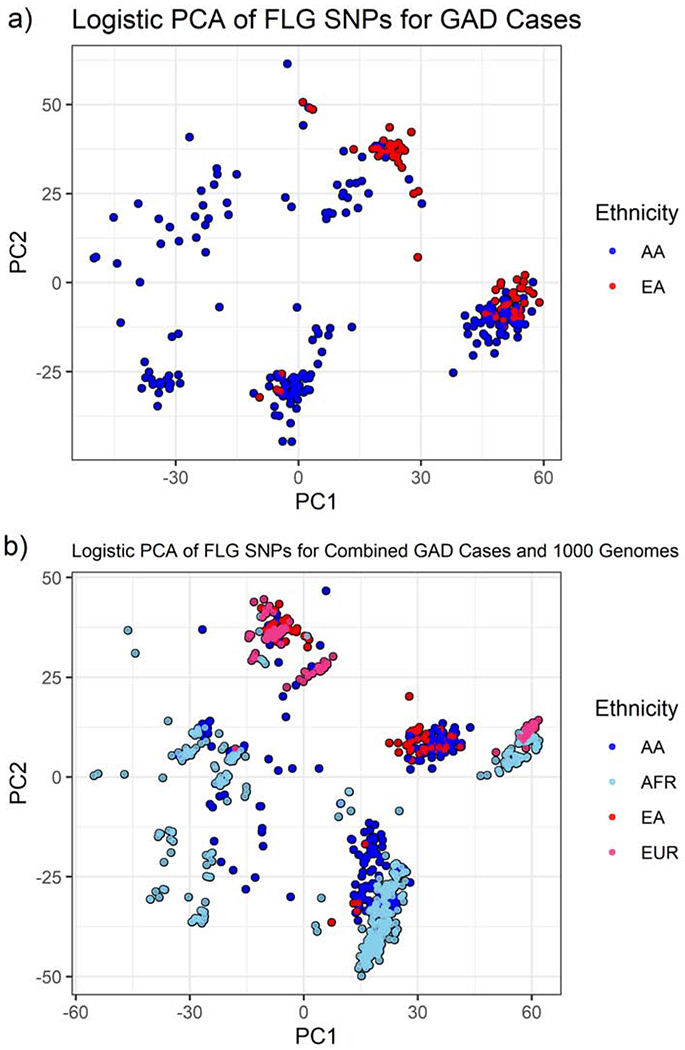
PCA plots showing clustering by ethnicity/ancestry for GAD cases based on FLG variation. (a) 579 FLG SNPs for subjects with AD in the GAD cohort and (b) 205 FLG SNPs in common to the GAD and 1000 Genomes data sets.
The number of FLG variants per individual appears to be different between EA and AA. While FLG LOF variation is associated with AD, there appears to be an inverse association between total FLG variation and the presence of FLG LOF. FLG variation seen in those of African ancestry (includes AA) is different from those of European ancestry (includes EA). Our team as well as other investigators have shown that FLG variation differs by ancestry and may be shaped by genetic drift as well as selection (Eaaswarkhanth et al., 2016). Our results indicate that it is unlikely that the risk for or persistence and severity of atopic dermatitis in AA as compared to EA is due to admixture of the FLG gene from Europeans to Africans. With respect to FLG variation, it appears that AA map to individuals of African ancestry rather than individuals of European ancestry. Because AD is more common in AA than in EA, other genic or environmental differences may account for AD disease mechanisms.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported in part by grants from the National Institutes for Health (NIAMS) R01-AR060962 (PI: Margolis). The PEER study is funded as the Atopic Dermatitis Registry by Valeant Pharmaceuticals International (PI: Margolis).
Abbreviations:
- FLG
Filaggrin
- LOF
Loss of function
- PEER
Pediatric Elective Eczema Registry
- AA
African Americans
- EA
European Americans
- PCA
Principal Component Analysis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Work was done in Philadelphia, PA 19104, USA.
CONFLICT OF INTEREST STATEMENT
With respect to this study, the authors report no conflicts of interest.
DATA AVAILABILITY STATEMENT
Datasets related to this article can be found at http://dx.doi.org/10.17632/wwz8h4yc6p.1, hosted at Mendeley Data (Zhu, Yaqian (2020), “FLG and FLG2 SNPs”, Mendeley Data, V1, doi: 10.17632/wwz8h4yc6p.1).We have no ownership over data from the 1000 Genomes Project or gnomAD, the data sets from which may be accessed online.
REFERENCES
- Eaaswarkhanth M, Xu D, Flanagan C, Rzhetskaya M, Hayes MG, Blekhman R, et al. Atopic Dermatitis Susceptibility Variants in Filaggrin Hitchhike Hornerin Selective Sweep. Genome Biol Evol 2016;8(10):3240–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine AD, McLean WH, Leung DY. Filaggrin mutations associated with skin and allergic diseases. [Review]. New England Journal of Medicine 2011;365(14):1315–27. [DOI] [PubMed] [Google Scholar]
- Landgraf AJ, Lee Y. Dimensionality Reduction for Binary Data through the Projection of Natural Parameters. https://arxivorg/abs/151006112 2015. [Google Scholar]
- Margolis DJ, Apter AJ, Gupta J, Hoffstad O, Papadopoulos M, Campbell LE, et al. The persistence of atopic dermatitis and filaggrin (FLG) mutations in a US longitudinal cohort. J Allergy Clin Immunol 2012;130(4):912–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis DJ, Gupta J, Apter AJ, Hoffstad O, Papadopoulos M, Rebbeck TR, et al. Exome sequencing of filaggrin and related genes in African-American children with atopic dermatitis. J Invest Dermatol 2014;134(8):2272–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis DJ, Mitra N, Berna R, Hoffstad O, Kim BS, Yan A, et al. Associating filaggrin copy number variation and atopic dermatitis in African-Americans: Challenges and opportunities. Journal of Dermatological Science 2020a;98(1):58–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis DJ, Mitra N, Gochnauer H, Wubbenhorst B, D’Andrea K, Kraya A, et al. Uncommon Filaggrin Variants Are Associated with Persistent Atopic Dermatitis in African Americans. J Invest Dermatol 2018;138(7):1501–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis DJ, Mitra N, Wubbenhorst B, D’Andrea K, Kraya AA, Hoffstad O, et al. Association of Filaggrin Loss-of-Function Variants With Race in Children With Atopic Dermatitis. JAMA Dermatol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis DJ, Mitra N, Wubbenhorst B, Nathanson KL. Filaggrin sequencing and bioinformatics tools. Arch Dermatol Res 2020b;312(2):155–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson K On lines and planes of closest fit to systems of points in space. Lond Edinb Dublin Philos Mag J Sci 1901;2(11):559–72. [Google Scholar]
- Thawer-Esmail F, Jakasa I, Todd G, Wen Y, Brown SJ, Kroboth K, et al. South african amaXhosa patients with atopic dermatitis have decreased levels of filaggrin breakdown products but no loss-of-function mutations in filaggrin. J Allergy Clin Immunol 2014;133(1):280–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winge M, Bilcha K, Liedén A, Shibeshi D, Sandilands A, Wahlgren CF, et al. Novel filaggrin mutation but no other loss-of-function variants found in Ethiopian patients with atopic dermatitis. British Journal of Dermatology (2011); 165: 1074–1080. [DOI] [PubMed] [Google Scholar]
- Wong X, Denil S, Foo JN, Chen H, Tay ASL, Haines RL, et al. Array-based sequencing of filaggrin gene for comprehensive detection of disease-associated variants. J Allergy Clin Immunol 2018;141(2):814–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


