Abstract
Despite advances in melanoma treatment, more than 70% of patients with distant metastasis die within 5 years. Proactive treatment of early melanoma to prevent metastasis could save lives and reduce overall healthcare costs. Currently, there are no treatments specifically designed to prevent early melanoma from progressing to metastasis. We used the Connectivity Map (cMap) to conduct an in silico drug screen and identified HMGCR inhibitors (statins) as a drug class that might prevent melanoma metastasis. To confirm the in vitro effect of statins, RNA-sequencing was completed on A375 cells after treatment with fluvastatin to describe changes in the melanoma transcriptome. Statins induced differential expression in genes associated with metastasis and used in commercially available prognostic tests for melanoma metastasis. Finally, we completed a chart review of 475 melanoma patients. Patients taking statins were less likely to have metastasis at the time of melanoma diagnosis in both univariate and multivariate analysis (24.7% taking statins vs 37.6% not taking statins, ARR = 12.9%, p=0.038). These findings suggest that statins might be useful as a treatment to prevent melanoma metastasis. Prospective trials are required to verify our findings and to determine the mechanism of metastasis prevention.
Keywords: metastatic melanoma, fluvastatin, gene expression, computational drug screen
Introduction
Metastasis is the primary driver of cancer mortality.(Dillekås et al., 2019; Zbytek et al., 2008) Despite recent advances in treatment, patients with metastatic melanoma survive on average less than two years after diagnosis.(Kandel et al., 2018; Larkin et al., 2019; Robert et al., 2019) In addition, the cost of treating metastatic disease has increased significantly.(Kandel et al., 2018) Preventing early cutaneous melanomas from progressing to metastasis may decrease healthcare costs and save lives.
Melanomas at high risk of metastasis can be identified by their genetic expressio signature.(Gerami et al., 2015b, 2015a; Kashani-Sabet et al., 2017, 2009; Zager et al., 2018) Retrospective and prospective studies have identified a 28-gene expression signature as an independent predictor of metastasis.(Gerami et al., 2015b, 2015a; Greenhaw et al., 2020; Zager et al., 2018) The immediate clinical utility of these tests is controversial, and the functional role of these genes in metastasis remains elusive. However, the fact that these gene signatures identify melanomas with up to 22-fold higher odds of recurrence or metastasis means they might yield insights into the metastatic process and could even lead to potential therapies.(Chan and Tsao, 2020; Grossman et al., 2020)
The Connectivity Map (cMap) is a publicly available database maintained by the Broad Institute that contains microarray gene expression measurements from over 27,000 pharmaceutical compounds.(Lamb, 2007; Lamb et al., 2006) This database can be queried to identify drugs that induce expression signatures either similar to or opposed to a specified profile. By screening the database for compounds that induce genetic expression directly opposed to a disease signature, the database has been successfully used to identify drugs for computational drug repurposing (the process of discovering new indications for existing drugs).(Chen et al., 2017; Sirota et al., 2011)
We used the cMap to screen for drugs that reverse a high-risk genetic expression profile of melanoma that has been validated in clinical samples.(Gerami et al., 2015a; Greenhaw et al., 2018; Zager et al., 2018) We identified 3-Hydroxy-3-Methylglutaryl-CoA Reductase (HMGCR) inhibitors (statins) as candidate agents to oppose the high-risk melanoma gene expression profile. Previous studies on statins in melanoma have focused on initiation or primary prevention and have had mixed results. Two large cardiovascular trials demonstrated reduction in melanoma incidence with statin use, but this effect was not observed in the Women’s Health Initiative or two Dutch epidemiologic studies.(Jagtap et al., 2012; Rubins et al., 1999; Splichal et al., 2003) Two meta-analyses also demonstrated no reduction in melanoma incidence with statin use, and a randomized controlled trial of lovastatin for melanoma prevention did not identify any significant decreases in melanocytic atypia or other melanoma initiation markers.(Bonovas et al., 2010; Freeman et al., 2006; Linden et al., 2014) Recently, a Mendelian randomization analysis using the UK Biobank demonstrated that individuals with variants in the HMGCR region, which represent proxies for statin use, had decreased overall cancer risk but did not reach statistical significance for any site-specific cancers.(Carter et al., 2020)
While results have been equivocal in melanoma initiation, there is more consistent evidence that statins may prevent melanoma progression and metastasis. Both in vitro and animal models have demonstrated potential mechanisms by which statins could prevent melanoma metastasis through decreasing tumor cell migration, decreasing cell adhesion, and increasing immunogenicity.(Collisson et al., 2003; Kidera et al., 2010; Pich et al., 2013; Zanfardino et al., 2013) One study observed decreased Breslow depth and metastasis rate with statin use, but the decrease in metastasis was not statistically significant in multivariable analysis.(Koomen et al., 2007) Another population based study on all-cause mortality in melanoma patients found a trend towards decreased hazard of death particularly in men, but did not reach statistical significance.(Livingstone et al., 2014)
Here we present an in silico drug screen that suggests statins may modify genetic expression correlated with metastasis. We then conduct next generation RNA sequencing to characterize the direct effects of clinically relevant doses of fluvastatin on the melanoma transcriptome in vitro. Finally, we explore the association of statin use with metastasis in a retrospective cohort of cutaneous melanoma from our institution.
Results
Identification of statins as a potential treatment for metastasis prevention
We hypothesized that compounds that induced gene expression signatures opposite to that of metastatic melanoma would prevent metastasis. A search of the cMap database identified piroxicam, sotalol, acyclovir, zalcitabine, and simvastatin as potential therapeutic agents (Table 1). The ability of a drug to shift a gene expression signature is measured by its connectivity score (tau), which is a standardized measure ranging from −100 to 100. For each drug in the list of query results, the score corresponds to the fraction of reference gene sets that affect the 28 genes more strongly. The reference gene sets are generated from all reference signatures of drugs in the cMap database. A score of 90 indicates that only 10% of reference gene sets showed stronger effects. In general, a tau of 90 or higher is considered strong and should be considered as hypotheses for further study. Simvastatin had a score of 91. We also checked the score of all statins combined, to ensure that these drugs as a class had a consistent effect. The statin class score was 84.9, which is strong for a class of drugs averaged together. Statins were selected for further study because of their proven long-term tolerability, benign side effect profile suitable for the intended clinical use as a preventive drug, and possible melanoma chemopreventive effects published in the literature.
Table 1.
Candidate Drugs.
| Drug | Tau Score | Drug Class | Class Score |
|---|---|---|---|
| Piroxicam | 99.93 | Cyclooxygenase Inhibitor | −33.86 |
| Sotalol | 99.47 | Beta-Adrenergic Receptor Antagonist | −13.36 |
| Acyclovir | 99.44 | DNA Polymerase Inhibitor | 23.83 |
| Zalcitabine | 99.12 | Nucleoside Reverse Transcriptase Inhibitor | 97.48 |
| Simvastatin | 91.05 | HMGCR Inhibitors (Statins) | 84.94 |
Fluvastatin alters the genetic expression profile of melanoma
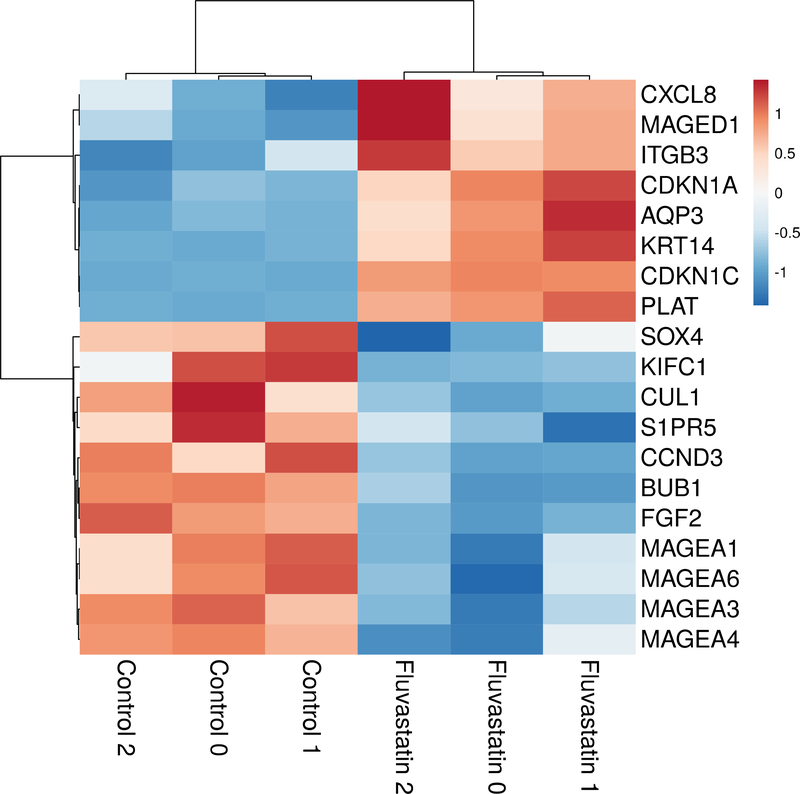
The cMap data is derived from treatment of cell lines with statins at 10 μM concentrations, which is above the maximal tolerated human dose.(López-Aguilar et al., 1999; Tse et al., 1992) Thus, we sought to characterize the effect of statins on the melanoma transcriptome at clinically tolerable doses. We used RNA-sequencing to measure gene expression of A375 melanoma cells before and after treatment with fluvastatin. The A375 cell line was specifically chosen since it has moderate metastatic potential and has been used in previous mechanistic studies of statins. Fluvastatin was chosen because of its lipophilicity (allowing extrahepatic distribution), benign side effect profile, and excellent bioavailability. We found 2615 differentially expressed genes (Figure 1).
Figure 1.
Heat map of differentially expressed genes related to metastasis.
Fluvastatin significantly affected the expression of genes previously shown to be involved in metastasis including MAGEA1, MAGEA3, MAGEA4, MAGEA6, MAGED1, SOX4, BUB1, and KIFC1.(Brasseur et al., 1995; Jafarnejad et al., 2013; Li et al., 2015; Riker et al., 2008; Weon and Potts, 2015) Genes that drive lymphangiogenesis were also found be significantly decreased by fluvastatin treatment including FGF2, S1PR5, and TGFBRAP1 (Table 2).(Cao et al., 2012; Huang et al., 2013; James et al., 2013)
Table 2.
Genes of interest differentially expressed after fluvastatin treatment.
| Gene symbol | log2fold change | q-value |
|---|---|---|
| MAGEA1 | −0.4357 | 0.0018 |
| MAGEA3 | −0.4574 | 0.0018 |
| MAGEA4 | −0.2919 | 0.0407 |
| MAGEA6 | −0.4529 | 0.0018 |
| MAGED1 | 0.7222 | 0.0018 |
| SOX4 | −0.4139 | 0.0018 |
| BUB1 | −0.7319 | 0.0018 |
| KIFC1 | −0.4213 | 0.0355 |
| FGF2 | −0.5791 | 0.0018 |
| S1PR5 | −1.3212 | 0.0292 |
| TGFBRAP1 | −0.3389 | 0.0381 |
| CDKN1A | 1.3053 | 0.0018 |
| CDKN1C | 3.2643 | 0.0018 |
| CUL1 | −0.3847 | 0.0062 |
| CCND3 | −0.7194 | 0.0089 |
| ITGB3# | 0.5304 | 0.027 |
| AQP3* | 3.2162 | 0.0018 |
| KRT14* | 2.8530 | 0.0018 |
| PLAT# | 1.7389 | 0.0018 |
| CXCL8# | 0.8218 | 0.0018 |
Castle Biosciences profile
SkylineDx profile
Consistent with the predicted effect on melanoma metastasis but not initiation, fluvastatin significantly altered the expression of genes included in melanoma prognostic tests (DecisionDx-Melanoma, Castle; Merlin Assay, SkylineDx), but did not alter expression of any genes used in a diagnostic test that distinguishes melanoma from nevi (myPath Melanoma, Myriad Genetics).(Clarke et al., 2015) Genes included in these assays that were significantly shifted are presented in Table 2 (ITGB3, PLAT, CXCL8, AQP3, and KRT14). This suggests that the effect of statins is specific to progression and metastasis rather than tumor initiation.
Gene ontology (GO) analysis of differentially expressed genes suggested significant enrichment of genes involved in the biological processes of cell proliferation, regulation of cell proliferation, tissue development, response to stimulus, and cell communication (all p<0.05). The molecular functions represented included signaling receptor activity, molecular transducer activity, receptor ligand activity, receptor regulator activity, and transmembrane receptor protein serine/threonine kinase binding (all p<0.05). Lastly, the cellular components represented included plasma membrane, cell periphery, extracellular matrix, plasma membrane part, and extracellular region (all p<0.05).
Patients taking statins have significantly lower incidence of metastasis at diagnosis
To evaluate the clinical impact of statin use on melanoma metastasis, a retrospective cohort of 475 patients with melanoma were reviewed, of which 311 patients met inclusion criteria. The mean age was 64.7 years. The mean Breslow depth in patients taking statins was 3.32 mm, as compared to 2.48 mm in those not taking statins (p=0.038) (Table 3). Regional or distant metastasis (defined as a positive completion lymph node dissection or distant metastasis detected on imaging) was identified at diagnosis in 24.7% of patients taking statins and 37.6% of patients not taking statins (p=0.038). This result was significant in multivariate analysis, after controlling for age, Breslow depth, ulceration, and mitotic rate (p=0.016) (Table 4).
Table 3.
Demographics of patients taking and not taking statin at time of biopsy.
| Taking statin (n=77) | Not taking statin (n=234) | p-value | |
|---|---|---|---|
| Mean age (years), n | 72.9 | 61.5 | <0.001 |
| Mean Breslow depth (mm), n | 3.32 | 2.48 | 0.038 |
| Ulceration, n (%) | |||
| Yes | 26 (33.8) | 79 (33.8) | |
| No | 51 (66.2) | 155 (66.2) | 0.999 |
| Mitotic Rate (mitoses/mm2), n (%) | |||
| 0 | 9 (11.7) | 28 (11.9) | |
| 1–5 | 56 (72.7) | 181 (77.4) | |
| 6–10 | 5 (6.5) | 21 (9.0) | |
| >10 | 7 (9.1) | 4 (1.7) | 0.015 |
| Metastasis detected during initial workup, n (%) | |||
| Yes | 19 (24.7) | 88 (37.6) | |
| No | 58 (75.3) | 146 (62.4) | 0.038 |
Table 4.
Multivariate analysis for factors that predispose to metastasis at initial workup.
| Factor | Odds Ratio (95% CI) | p-value |
|---|---|---|
| Age (years) | 0.98 (0.97 – 1.00) | 0.01 |
| Depth (mm) | 1.12 (1.00 – 1.24) | 0.04 |
| Ulceration Present | 1.44 (0.81 – 2.55) | 0.22 |
| Dermal Mitoses (per sq mm) | 1.02 (0.95 – 1.10) | 0.61 |
| Histologic Type | ||
| Nodular | 1 | - |
| Superficial Spreading | 0.93 (0.47 – 1.81) | 0.82 |
| Lentigo Maligna | 0.27 (0.08 – 0.88) | 0.03 |
| Other | 0.81 (0.39 – 1.67) | 0.57 |
| Taking a Statin | 0.48 (0.25 – 0.94) | 0.03 |
Discussion
Computational prediction has been successful in the past for identifying repurposing opportunities.(Chen et al., 2017; Menden et al., 2019; Sirota et al., 2011) Here we used an in silico screen to identify FDA-approved drugs that induce a genetic profile opposed to a validated gene expression profile that predicts melanoma metastasis. We selected statins for further investigation based on their long record of safety, their benign side effect profile consistent with use as a preventive drug, and the literature suggestive of their potential activity in melanoma chemoprevention. Since the in silico screen uses data from experiments at doses above the maximal tolerated human serum concentrations, we proceeded to verify the efficacy of statins in appropriately shifting the melanoma transcriptome at clinically achievable doses (3 μM).
We found that fluvastatin caused significant changes in the melanoma transcriptome and affected genes specific to melanoma metastasis at doses below the maximally tolerated dose.(López-Aguilar et al., 1999; Tse et al., 1992) At these doses, fluvastatin did not affect the expression of genes that are used to differentiate nevi from melanoma, consistent with prior clinical trial results demonstrating no effect of statins on the progression of dysplastic nevi to melanoma.(Linden et al., 2014) However, fluvastatin influenced the expression of genes used to measure risk of metastasis in commercially available tests, suggesting that the effect of statins is specific to melanoma progression and metastasis rather than melanoma initiation. This data also implies that history of statin use may be an important factor in interpreting the results of these prognostic tests. Since the drug concentrations we used were lower than those in cMap (therapeutic rather than supratherapeutic) and because we used RNA-seq rather than microarray, there were fewer changes in the 28-gene expression profile than initially predicted by cMap. We found that our RNA-seq validation experiments also revealed changes in genes outside of commercial tests that are known to influence metastatic potential and melanoma development.
Our results suggest potential mechanisms for the effect of statins on melanoma metastasis. Regulation of the G1/S transition appeared to be affected by statin treatment. CDKN1A (p21) and CDKN1C (Kip2), which inhibit cell cycle progression in G1 and S phase, are typically suppressed in melanoma, but had significantly increased expression after fluvastatin exposure.(Jalili et al., 2012; Yang et al., 2020) CUL1 (Cullin 1), which promotes G1 to S phase transition and drives melanoma proliferation, was significantly decreased after fluvastatin treatment.(Chen and Li, 2010) In addition, increased expression of CCND3 (Cyclin D3) has been shown to decrease survival and promote early relapse in melanoma.(Florenes et al., 2000) In our study, we found that CCND3 expression was significantly decreased after fluvastatin treatment. KIFC1, a gene important in centrosome clustering, is overexpressed in primary and uveal melanoma cell lines, as well as in breast and lung cancers.(Pannu et al., 2015) Our study demonstrated that fluvastatin decreased expression of KIFC1. Previous studies have demonstrated that metabolic differences in melanoma cells result in differences in metastatic potential.(Tasdogan et al., 2020) SLC16A1 (MCT1), which has metabolic functions in lactate transport, has been demonstrated to be an oncogene in malignant melanomas and neuroblastomas and drives melanoma metastasis.(Avitabile et al., 2020) We observed SLC16A1 downregulation after fluvastatin exposure, but did not achieve statistical significance after correction for multiple hypothesis testing (fold change = 0.797, q=0.062). Lymphangiogenesis is thought to be involved in both metastasis and immune regulation of the tumor microenvironment.(Lane et al., 2018; Lund et al., 2016b, 2016a, 2012) FGF2, S1PR5, and TGFBRAP1 are all involved in lymphangiogenesis and were downregulated by statin treatment. Finally, the MAGE gene family has been demonstrated to be expressed in a wide variety of malignancies, including melanoma and is associated with increased invasion and metastasis.(Barrow et al., 2006; Brasseur et al., 1995) We observed decreased expression of MAGEA1, MAGEA3, MAGEA4, and MAGEA6 with fluvastatin treatment.
A prior study found that atorvastatin decreases isoprenylation of RhoC, thereby decreasing migration and invasion in a Matrigel transwell assay of A375 cells, and metastasis in a mouse model.(Collisson et al., 2003) We chose the A375 cell line to build on this prior literature, and our data suggests that statins may also affect lymphangiogenesis, cell cycle regulation, and metabolism to reduce metastasis. The effect on cell cycle regulation identified in this study may be particularly relevant for treatment of familial melanomas induced by CDKN2A mutations.(Aspinwall et al., 2008; Goldstein et al., 2007, 2006; Leachman et al., 2009)
We considered the possibility that statins induce gene expression changes that are correlated with metastasis but are not causative of metastasis. If this were true, statin use should not be correlated with the risk of metastasis. Thus, we proceeded to investigate the association between statin use and metastasis in a retrospective cohort of melanoma patients. We found that patients taking statins at the time of biopsy were significantly less likely to have metastasis at the time of melanoma diagnosis as compared to those not taking statins, thereby suggesting that statins may be protective against melanoma metastasis. Statin use remained the strongest independent predictor of metastasis after correction for other prognostic factors including depth, ulceration, mitoses, and age. We considered the possibility that statins may simply be a marker of better access to healthcare resulting in earlier melanoma diagnosis. However, the statin group in our cohort actually had thicker primary melanomas with higher mitotic count indicating later diagnosis. The fact that these patients still had fewer metastases despite significantly worse primary tumors is remarkable.
This study has certain limitations. Our data is retrospective and correlative; despite controlling for all the prognostic factors available, it is possible that there are confounding variables beyond our knowledge such as differences in patient behavior or access to healthcare. We do not have enough follow up data to determine for certain whether patients on statins have better future outcomes, though the reduction in metastases at diagnosis is promising. In addition, our study cohort is from a single tertiary referral center, and thus may be biased towards larger effect size than might be seen in a population based study. Our results need to be confirmed with prospective data and potentially a clinical trial. In addition, further study of the mechanisms by which statins may affect metastasis is needed to determine causality and may reveal other drug targets.
By leveraging a validated prognostic genetic expression signature, we were able to identify statins as a potential preventive therapy for melanoma metastasis, describe the effects of fluvastatin on the melanoma transcriptome, and demonstrate clinical activity in a retrospective cohort. Since the discovery of statins as a potential prevention for metastasis was based on an existing commercial test, future clinical trials may be able to elegantly select the specific subset of patients who are most likely to benefit. Finally, as other genetic profiles are discovered, this tailored approach may identify additional drugs for prevention or treatment of metastasis.
Methods
In silico selection of candidates for metastasis prevention
The Connectivity Map (cMap) Query Tool (https://clue.io/query) was used to conduct an in silico drug screen. The input query consisted of the 28 gene expression profile from a commercially available prognostic test that predicts melanoma metastasis annotated by the desired change in expression (up or down).(Gerami et al., 2015b) Each compound and the corresponding drug class were scored for their ability to oppose the high-risk melanoma gene expression profile using the cMap connectivity score (tau), a standardized measure ranging from −100 to 100. The top 10% of drug candidates were then evaluated for FDA approval status and overall safety profile.
Characterization of transcriptome in human melanoma cells exposed to fluvastatin
We proceeded to characterize the effect of statins on the melanoma transcriptome using a well-established melanoma cell line. Human melanoma cell line A375 (a generous gift of Dr. John Letterio, CWRU) were maintained in standard growth media consisting of RPMI1640 + 10% FBS + 2 mM glutamine and grown in a 5%CO2 humidified atmosphere at 37°C. Cells were tested bi-annually and shown to be negative for mycoplasma contamination using the Mycoplasma Detection kit (MycoAlert™, Lonza, Basel, Switzerland). For gene expression studies A375 cells were seeded at 2.5E6 per 10cm2 dish and allowed to adhere overnight. Test samples (in triplicate) were treated with fluvastatin (purchased from MilliporeSigma, St Louis, Mo) at 3 μM concentration for 24 hours then harvested for RNA extraction using the RNeasy Plus Mini Kit (Cat. #74134, Qiagen, Germantown, MD) as per manufacturer’s instructions. Untreated control cells grown side by side were used as the reference control for differential expression analysis. RNA was quantified using the Qubit Broad Range RNA kit (Catalog #Q10210; Thermo Fisher Scientific, Waltham, MA) and diluted to 50ng/ul for RNA sequencing.
Sequencing reads generated from the Illumina HiSeq platform were assessed for quality and trimmed for adapter sequences using TrimGalore! v0.4.2 (Babraham Bioinformatics), a wrapper script for FastQC and cutadapt. Reads that passed quality control were subsequently aligned to the human reference genome (GRCh38) using STAR aligner v2.5.1. Sequence alignment was guided using the GENCODE annotation for hg38. The aligned reads were analyzed for differential expression using Cufflinks v2.2.1, a RNASeq analysis package which reports the fragments per kilobase of exon per million fragments mapped (FPKM) for each gene. Differential analysis report was generated using the cuffdiff command performed in a pairwise manner for each group. Differential genes were identified using a significance cutoff of q-value < 0.05. The differential expression profiles were then used as input in iPathwayGuide (AdvaitaBio) for pathway analysis.
Multivariate analysis of the effect of statin use on metastasis incidence
To further understand how statins affect melanoma metastasis rates in the clinical setting, we performed a retrospective chart review of patients diagnosed with melanoma in the dermatopathology archive at our tertiary medical center from January 1, 2007 through December 31, 2017. This study was IRB approved. Patients with a histopathological diagnosis of melanoma with Breslow depth greater than 0.8mm or with ulceration were included. Patients with greater than 3 primary melanomas were excluded to avoid confounding by patients with a germline predisposition to melanoma. Data collected included age at diagnosis of primary melanoma; sex; race; immunosuppression status (transplant, HIV, hematologic malignancy); statin use at the time of biopsy; histologic type; Breslow depth; ulceration; mitotic rate; tumor-infiltrating lymphocytes; regression; sentinel lymph node biopsy (SLNB) results; complete lymph node dissection (CLND) results; and presence of metastasis at diagnosis. Univariate and multivariate analyses using logistic regression were performed to determine the relationship of statin use to presence of metastasis at diagnosis, controlled for Breslow depth, ulceration status, and mitotic rate (glm function, R version 4.0.2). A p-value of at least 0.05 was considered significant.
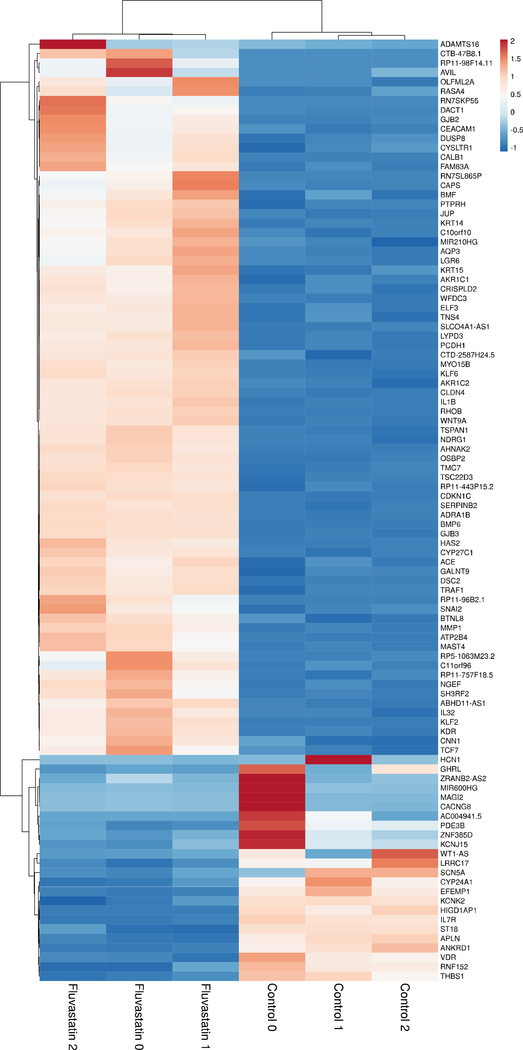
Figure 2.
Heatmap of top 100 differentially expressed genes after fluvastatin treatment.
Acknowledgements:
The authors would like to acknowledge Jennifer Arnold, Simone Edelheit, and Alexander Miron for technical assistance, Amanda Davies for regulatory guidance, and John Pounardjian for assistance with funding.
Funding Sources:
This publication was made possible by funding from the Case Comprehensive Cancer Center and from the Clinical and Translational Science Collaborative of Cleveland, UL1TR002548 from the National Center for Advancing Translational Sciences (NCATS) component of the National Institutes of Health and NIH Roadmap for Medical Research. This research was also supported by the Translational Research Shared Resource and the Genomics Shared Resource of the Case Comprehensive Cancer Center (P30 CA043703). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the funding institutions. RPK acknowledges support from the Melanoma Research Alliance, American Cancer Society, Cancer Research Institute, DOD PCRP award (W81XWH-17-1-0098) and DOD PRCRP award (W81XWH-17-1-0514).
Footnotes
Conflicts of Interest: WYY has received research funding from SkylineDx.
IRB approval status: Approved by University Hospitals IRB STUDY20200545
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Availability Statement
Datasets related to this article can be found at https://www.ncbi.nlm.nih.gov/geo/, hosted at the NCBI Gene Expression Omnibus.
References
- Aspinwall L, Leaf S, Dola E, Kohlmann W, Leachman SA. CDKN2A/p16 Genetic Test Reporting Improves Early Detection Intentions and Practices in High-Risk Melanoma Families. Cancer Epidemiol Biomarkers Prev 2008;17:1510–9. doi: 10.1158/1055-9965.EPI-08-0010. [DOI] [PubMed] [Google Scholar]
- Avitabile M, Succoio M, Testori A, Cardinale A, Vaksman Z, Lasorsa VA, et al. Neural crest-derived tumor neuroblastoma and melanoma share 1p13.2 as susceptibility locus that shows a long-range interaction with the SLC16A1 gene. Carcinogenesis 2020. doi: 10.1093/carcin/bgz153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrow C, Browning J, MacGregor D, Davis ID, Sturrock S, Jungbluth AA, et al. Tumor antigen expression in melanoma varies according to antigen and stage. Clin Cancer Res 2006. doi: 10.1158/1078-0432.CCR-05-1544. [DOI] [PubMed] [Google Scholar]
- Bonovas S, Nikolopoulos G, Filioussi K, Peponi E, Bagos P, Sitaras NM. Can statin therapy reduce the risk of melanoma? A meta-analysis of randomized controlled trials. Eur J Epidemiol 2010. doi: 10.1007/s10654-009-9396-x. [DOI] [PubMed] [Google Scholar]
- Brasseur F, Rimoldi D, Liénard D, Lethé B, Carrel S, Arienti F, et al. Expression of MAGE genes in primary and metastatic cutaneous melanoma. Int J Cancer 1995. doi: 10.1002/ijc.2910630313. [DOI] [PubMed] [Google Scholar]
- Cao R, Ji H, Feng N, Zhang Y, Yang X, Andersson P, et al. Collaborative interplay between FGF-2 and VEGF-C promotes lymphangiogenesis and metastasis. Proc Natl Acad Sci U S A 2012. doi: 10.1073/pnas.1208324109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter P, Vithayathil M, Kar S, Potluri R, Mason A, Larsson S, et al. Predicting the effect of statins on cancer risk using genetic variants: a Mendelian randomization study in UK Biobank. MedRxiv 2020. doi: 10.1101/2020.02.28.20028902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan WH, Tsao H. Consensus, Controversy, and Conversations about Gene Expression Profiling in Melanoma. JAMA Dermatology 2020. doi: 10.1001/jamadermatol.2020.1730. [DOI] [PubMed] [Google Scholar]
- Chen B, Ma L, Paik H, Sirota M, Wei W, Chua M-S, et al. Reversal of cancer gene expression correlates with drug efficacy and reveals therapeutic targets. Nat Commun 2017;8:16022. doi: 10.1038/ncomms16022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Li G. Increased Cul1 expression promotes melanoma cell proliferation through regulating p27 expression. Int J Oncol 2010. doi: 10.3892/ijo-00000786. [DOI] [PubMed] [Google Scholar]
- Clarke LE, Warf MB, Flake DD, Hartman A-R, Tahan S, Shea CR, et al. Clinical validation of a gene expression signature that differentiates benign nevi from malignant melanoma. J Cutan Pathol 2015;42:244–52. doi: 10.1111/cup.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collisson EA, Kleer C, Wu M, Abhijit De, Gambhir SS, Merajver SD, et al. Atorvastatin prevents RhoC isoprenylation, invasion, and metastasis in human melanoma cells. Mol Cancer Ther 2003. [PMC free article] [PubMed] [Google Scholar]
- Dillekås H, Rogers MS, Straume O. Are 90% of deaths from cancer caused by metastases? Cancer Med 2019;8:5574–6. doi: 10.1002/cam4.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florenes VA, Faye RS, Maelandsmo GM, Nesland JM, Holm R. Levels of cyclin d1 and d3 in malignant melanoma: Deregulated cyclin D3 expression is associated with poor clinical outcome in superficial melanoma. Clin Cancer Res 2000. [PubMed] [Google Scholar]
- Freeman SR, Drake AL, Heilig LF, Graber M, McNealy K, Schilling LM, et al. Statins, fibrates, and melanoma risk: A systematic review and meta-analysis. J Natl Cancer Inst 2006. doi: 10.1093/jnci/djj412. [DOI] [PubMed] [Google Scholar]
- Gerami P, Cook RW, Russell MC, Wilkinson J, Amaria RN, Gonzalez R, et al. Gene expression profiling for molecular staging of cutaneous melanoma in patients undergoing sentinel lymph node biopsy. J Am Acad Dermatol 2015a. doi: 10.1016/j.jaad.2015.01.009. [DOI] [PubMed] [Google Scholar]
- Gerami P, Cook RW, Wilkinson J, Russell MC, Dhillon N, Amaria RN, et al. Development of a prognostic genetic signature to predict the metastatic risk associated with cutaneous melanoma. Clin Cancer Res 2015b. doi: 10.1158/1078-0432.CCR-13-3316. [DOI] [PubMed] [Google Scholar]
- Goldstein AM, Chan M, Harland M, Gillanders EM, Hayward NK, Avril MF, et al. High-risk melanoma susceptibility genes and pancreatic cancer, neural system tumors, and uveal melanoma across GenoMEL. Cancer Res 2006. doi: 10.1158/0008-5472.CAN-06-0494. [DOI] [PubMed] [Google Scholar]
- Goldstein AM, Chan M, Harland M, Hayward NK, Demenais F, Bishop DT, et al. Features associated with germline CDKN2A mutations: A GenoMEL study of melanoma-prone families from three continents. J Med Genet 2007. doi: 10.1136/jmg.2006.043802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhaw BN, Covington KR, Kurley SJ, Yeniay Y, Cao NA, Plasseraud KM, et al. Molecular risk prediction in cutaneous melanoma: A meta-analysis of the 31-gene expression profile prognostic test in 1,479 patients. J Am Acad Dermatol 2020. doi: 10.1016/j.jaad.2020.03.053. [DOI] [PubMed] [Google Scholar]
- Greenhaw BN, Zitelli JA, Brodland DG. Estimation of prognosis in invasive cutaneous melanoma: An independent study of the accuracy of a gene expression profile test. Dermatologic Surg 2018. doi: 10.1097/DSS.0000000000001588. [DOI] [PubMed] [Google Scholar]
- Grossman D, Okwundu N, Bartlett E, Marchetti M, Othus M, Coit D, et al. Prognostic gene expression profiling in cutaneous melanoma: identifying the knowledge gaps and assessing the clinical benefit. JAMA Dermatology 2020. doi: 10.1001/jamadermatol.2020.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WC, Nagahashi M, Terracina KP, Takabe K. Emerging role of Sphingosine-1-phosphate in Inflammation, cancer, and lymphangiogenesis. Biomolecules 2013. doi: 10.3390/biom3030408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafarnejad SM, Ardekani GS, Ghaffari M, Martinka M, Li G. Sox4-mediated Dicer expression is critical for suppression of melanoma cell invasion. Oncogene 2013. doi: 10.1038/onc.2012.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagtap D, Rosenberg CA, Martin LW, Pettinger M, Khandekar J, Lane D, et al. Prospective analysis of association between use of statins and melanoma risk in the Women’s Health Initiative. Cancer 2012. doi: 10.1002/cncr.27497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalili A, Wagner C, Pashenkov M, Pathria G, Mertz KD, Widlund HR, et al. Dual suppression of the cyclin-dependent kinase inhibitors CDKN2C and CDKN1A in human melanoma. J Natl Cancer Inst 2012. doi: 10.1093/jnci/djs373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James JM, Nalbandian A, Mukouyama suke Y. TGFβ signaling is required for sprouting lymphangiogenesis during lymphatic network development in the skin. Dev 2013. doi: 10.1242/dev.095026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel M, Allayous C, Dalle S, Mortier L, Dalac S, Dutriaux C, et al. Update of survival and cost of metastatic melanoma with new drugs: Estimations from the MelBase cohort. Eur J Cancer 2018. doi: 10.1016/j.ejca.2018.09.026. [DOI] [PubMed] [Google Scholar]
- Kashani-Sabet M, Nosrati M, Miller JR, Sagebiel RW, Leong SPL, Lesniak A, et al. Prospective validation of molecular prognostic markers in cutaneous melanoma: A correlative analysis of E1690. Clin Cancer Res 2017. doi: 10.1158/1078-0432.CCR-17-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashani-Sabet M, Venna S, Nosrati M, Rangel J, Sucker A, Egberts F, et al. A multimarker prognostic assay for primary cutaneous melanoma. Clin Cancer Res 2009. doi: 10.1158/1078-0432.CCR-09-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidera Y, Tsubaki M, Yamazoe Y, Shoji K, Nakamura H, Ogaki M, et al. Reduction of lung metastasis, cell invasion, and adhesion in mouse melanoma by statin-induced blockade of the Rho/Rho-associated coiled-coil-containing protein kinase pathway. J Exp Clin Cancer Res 2010. doi: 10.1186/1756-9966-29-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koomen ER, Joosse A, Herings RMC, Casparie MK, Bergman W, Nijsten T, et al. Is statin use associated with a reduced incidence, a reduced Breslow thickness or delayed metastasis of melanoma of the skin? Eur J Cancer 2007. doi: 10.1016/j.ejca.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Lamb J. The Connectivity Map: a new tool for biomedical research. Nat Rev Cancer 2007;7:54–60. doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science (80-) 2006;313:1929–35. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Lane RS, Femel J, Breazeale AP, Loo CP, Thibault G, Kaempf A, et al. IFNγ-activated dermal lymphatic vessels inhibit cytotoxic T cells in melanoma and inflamed skin. J Exp Med 2018. doi: 10.1084/jem.20180654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin J, Chiarion-Sileni V, Gonzalez R, Grob J-J, Rutkowski P, Lao CD, et al. Five-Year Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N Engl J Med 2019;381:1535–46. doi: 10.1056/NEJMoa1910836. [DOI] [PubMed] [Google Scholar]
- Leachman SA, Carucci J, Kohlmann W, Banks KC, Asgari MM, Bergman W, et al. Selection criteria for genetic assessment of patients with familial melanoma. J Am Acad Dermatol 2009. doi: 10.1016/j.jaad.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Krahn JM, Flake GP, Umbach DM, Li L. Toward predicting metastatic progression of melanoma based on gene expression data. Pigment Cell Melanoma Res 2015. doi: 10.1111/pcmr.12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden KG, Leachman SA, Zager JS, Jakowatz JG, Viner JL, McLaren CE, et al. A randomized, double-blind, placebo-controlled phase II clinical trial of lovastatin for various endpoints of melanoma pathobiology. Cancer Prev Res (Phila) 2014;7:496–504. doi: 10.1158/1940-6207.CAPR-13-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingstone E, Hollestein LM, van Herk-Sukel MPP, van de Poll-Franse L, Joosse A, Schilling B, et al. Statin use and its effect on all-cause mortality of melanoma patients: A population-based Dutch cohort study. Cancer Med 2014. doi: 10.1002/cam4.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Aguilar E, Sepúlveda-Vildósola AC, Rivera-Márquez H, Cerecedo-Diaz F, Valdez-Sánchez M, Villasis-Keever MA. Security and Maximal Tolerated Doses of Fluvastatin In Pediatric Cancer Patients. Arch Med Res 1999;30:128–31. doi: 10.1016/S0188-0128(98)00018-9. [DOI] [PubMed] [Google Scholar]
- Lund AW, Duraes VF., Hirosue S, Raghavan VR, Nembrini C, Thomas SN, et al. VEGF-C Promotes Immune Tolerance in B16 Melanomas and Cross-Presentation of Tumor Antigen by Lymph Node Lymphatics. Cell Rep 2012. doi: 10.1016/j.celrep.2012.01.005. [DOI] [PubMed] [Google Scholar]
- Lund AW, Medler TR, Leachman SA, Coussens LM. Lymphatic vessels, inflammation, and immunity in skin cancer. Cancer Discov 2016a. doi: 10.1158/2159-8290.CD-15-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund AW, Wagner M, Fankhauser M, Steinskog ES, Broggi MA, Spranger S, et al. Lymphatic vessels regulate immune microenvironments in human and murine melanoma. J Clin Invest 2016b. doi: 10.1172/JCI79434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menden MP, Wang D, Mason MJ, Szalai B, Bulusu KC, Guan Y, et al. Community assessment to advance computational prediction of cancer drug combinations in a pharmacogenomic screen. Nat Commun 2019. doi: 10.1038/s41467-019-09799-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannu V, Rida PCG, Ogden A, Turaga RC, Donthamsetty S, Bowen NJ, et al. HSET overexpression fuels tumor progression via centrosome clustering-independent mechanisms in breast cancer patients. Oncotarget 2015. doi: 10.18632/oncotarget.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pich C, Teiti I, Rochaix P, Mariamé B, Couderc B, Favre G, et al. Statins reduce melanoma development and metastasis through MICA overexpression. Front Immunol 2013. doi: 10.3389/fimmu.2013.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riker AI, Enkemann SA, Fodstad O, Liu S, Ren S, Morris C, et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genomics 2008. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert C, Grob JJ, Stroyakovskiy D, Karaszewska B, Hauschild A, Levchenko E, et al. Five-Year Outcomes with Dabrafenib plus Trametinib in Metastatic Melanoma. N Engl J Med 2019;381:626–36. doi: 10.1056/NEJMoa1904059. [DOI] [PubMed] [Google Scholar]
- Rubins HB, Robins SJ, Collins D, Fye CL, Anderson JW, Elam MB, et al. Gemfibrozil for the secondary prevention of coronary heart disease in men with low levels of high-density lipoprotein cholesterol. N Engl J Med Pre-proof 1999. doi: 10.1056/NEJM199908053410604. [DOI] [PubMed] [Google Scholar]
- Sirota M, Dudley JT, Kim J, Chiang AP, Morgan AA, Sweet-Cordero A, et al. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci Transl Med 2011;3:96ra77. doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splichal JE, Stamm JA, Ornstein DL. The statins: Multifunctional antithrombotic and antineoplastic drugs. Semin Thromb Hemost 2003. doi: 10.1055/s-2003-40964. [DOI] [PubMed] [Google Scholar]
- Tasdogan A, Faubert B, Ramesh V, Ubellacker JM, Shen B, Solmonson A, et al. Metabolic heterogeneity confers differences in melanoma metastatic potential. Nature 2020;577:115–20. doi: 10.1038/s41586-019-1847-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse FLS, Jaffe JM, Troendle A. Pharmacokinetics of Fluvastatin After Single and Multiple Doses in Normal Volunteers. J Clin Pharmacol 1992;32:630–8. doi: 10.1002/j.1552-4604.1992.tb05773.x. [DOI] [PubMed] [Google Scholar]
- Weon JL, Potts PR. The MAGE protein family and cancer. Curr Opin Cell Biol 2015. doi: 10.1016/j.ceb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Yan Z, Hu F, Wei W, Sun Z, Xu W. Silencing of microRNA-517a induces oxidative stress injury in melanoma cells via inactivation of the JNK signaling pathway by upregulating CDKN1 C. Cancer Cell Int 2020. doi: 10.1186/s12935-019-1064-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zager JS, Gastman BR, Leachman S, Gonzalez RC, Fleming MD, Ferris LK, et al. Performance of a prognostic 31-gene expression profile in an independent cohort of 523 cutaneous melanoma patients. BMC Cancer 2018. doi: 10.1186/s12885-018-4016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanfardino M, Spampanato C, De Cicco R, Buommino E, De Filippis A, Baiano S, et al. Simvastatin reduces melanoma progression in a murine model. Int J Oncol 2013. doi: 10.3892/ijo.2013.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbytek B, Carlson JA, Granese J, Ross J, Mihm MC, Slominski A. Current concepts of metastasis in melanoma. Expert Rev Dermatol 2008;3:569–85. doi: 10.1586/17469872.3.5.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Datasets related to this article can be found at https://www.ncbi.nlm.nih.gov/geo/, hosted at the NCBI Gene Expression Omnibus.