Abstract
Natural products and their derivatives offer a rich source of chemical and biological diversity; however, traditional engineering of their biosynthetic pathways to improve yields and access to unnatural derivatives requires a precise understanding of their enzymatic processes. High-throughput screening platforms based on allosteric transcription-factor based biosensors can be leveraged to overcome the screening bottleneck to enable searching through large libraries of pathway/strain variants. Herein, the development and application of engineered allosteric transcription factor-based biosensors is described that enable optimization of precursor availability, product titers, and downstream product tailoring for advancing the natural product bioeconomy. We discuss recent successes for tailoring biosensor design, including computationally-based approaches, and present our future outlook with the integration of cell-free technologies and de novo protein design for rapidly generating biosensor tools.
Graphical Abstract
Introduction
Natural products represent privileged scaffolds for the development of pharmaceuticals, as well as numerous high-value commodity and industrial chemicals in the growing bioeconomy. Industrial strategies to produce these secondary metabolites (e.g. alkaloids, phenylpropanoids, polyketides, and terpenoids) combine various approaches including chemical synthesis, biocatalysis, and extraction from natural sources [1]. As industries shift towards increasingly environmentally sustainable and cost-effective methods, it is no surprise that the enzyme industry reported a total added value of $106 billion to the US economy in 2016 [2].
Microbial metabolism has been harnessed to provide access to natural products and their analogues via chemo- and regioselective modification of complex organic scaffolds that is otherwise often challenging to achieve through traditional synthetic methods. However, engineering of enzymes, metabolic pathways, and microbial hosts are often required to optimize substrate scope, cofactor recycling, catalytic rate, substrate tolerance, and productivity within stringent industrial conditions [1]. The framework of synthetic biology’s iterative “design-build-test” cycle has enabled rapid engineering of microbial metabolism, leading to significant improvements in the design and construction of libraries. However, screening these libraries for variants with desired properties remains a critical bottleneck and highlights the need for high-throughput approaches that provide a rapid link between genotype and phenotype [3,4]. Genetically-encoded biosensors based on allosteric transcription factors (aTFs) that detect the structure and report the titer of a given natural product allow for the programmable high-throughput screening and selection of genetic constructs, host strains, and experimental growth conditions (Fig. 1).
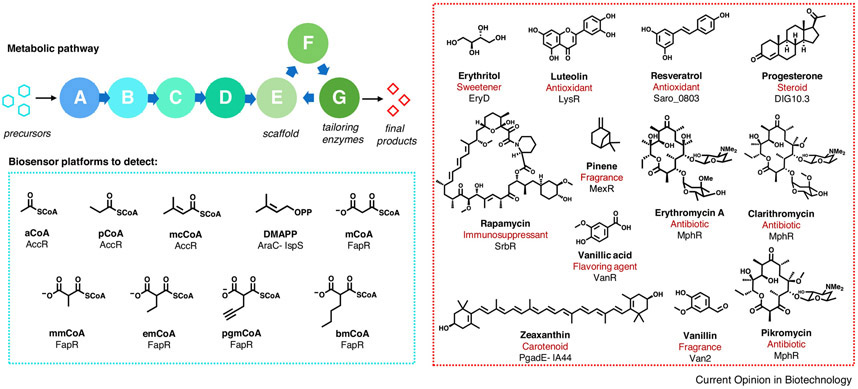
Figure 1.
Coupling genetically-encoded aTF biosensor platforms to natural product biosynthesis and engineering. Natural product biosynthetic logic is frequently divided into precursor generation, scaffold assembly, and tailoring, thereby providing several entry points for detection by aTFs. A variety of biosynthetic precursors (outlined in blue) and mature natural products (outlined in red), have been detected by aTF-based biosensors.
Ubiquitous in nature, aTFs activate or repress transcriptional machinery to regulate gene expression in response to environmental stimuli. The ligand-binding domain (LBD) of metabolite-responsive aTFs selectively bind effector molecules, which results in an allosterically induced conformational change that modulates transcription through the interactions of the aTF DNA-binding domain (DBD) and its cognate DNA operator sequence (Fig. 2a). Characterization of these proteins has resulted in the development of diverse aTF-based tools. The properties of these regulatory factors, including their inducer specificity as well as their various transfer functions as defined by the Hill equation, including sensitivity (K1/2), dynamic range (ON-OFF), and cooperativity (n), can be tailored to yield an optimized biosensor. aTFs respond to effector structures and are therefore completely agnostic to biological activity or mechanism of action of the natural product. Coupled with the ability to engineer aTFs with exquisite effector selectivity, aTFs present a unique opportunity to develop biosensors for a variety of natural products with molecular precision sufficient to guide the regulation and engineering of complex biosynthetic systems. Recent reports have leveraged these platforms for dynamic pathway regulation to balance the flux of intermediates and final products. In tandem with strain development, modularized gene expression, dynamic regulation, and other metabolic engineering strategies, engineered enzymes can be exploited to produce high-value natural products and their derivatives [5]. Herein, this review highlights the advancements in biosensor-guided approaches to natural product engineering over the last two years, with a special emphasis on aTF platforms for optimizing precursors, pathways, final titers, and post-assembly tailoring. Finally, we reflect on the future outlook of these tools.
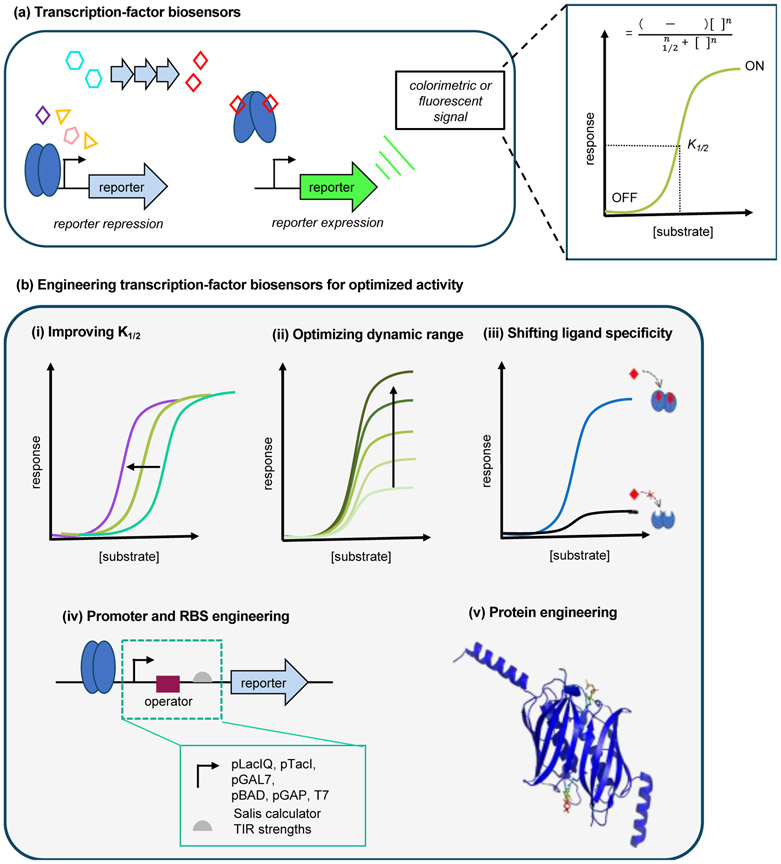
Figure 2.
Engineering and optimizing transcription factor-based biosensors. (a) Genetic circuit of a biosensor platform whereby the aTF regulates the expression of downstream genes by binding to its cognitive operator sequence. In the presence of an inducer, the system will derepress, allowing for the expression of a reporter signal. The corresponding dose response curve describing the response as a function of effector concentration is used to evaluate the biosensor transfer functions, as defined by the Hill equation. (b) aTF biosensor platforms can be tailored for a preferred function, including (i) sensitivity, (ii) dynamic range, (iii) effector specificity, (iv) control over transcription and translation via promoter and RBS engineering, (v) tailored substrate selectivity and/or promiscuity via protein engineering.
Development and Engineering of aTF-Based Biosensors
While approaches to mine and develop endogenous aTFs into biosensors have been successful, there are a limited number of characterized aTFs known in nature with substrate scopes that are relevant to natural products (Table 1). As such, recent studies have employed genomic sequencing to expand the biosensor platform repertoire by identifying previously uncharacterized aTFs for specific final products, including the phenylpropanoid resveratrol [6], and the steroid progesterone [7]. However, when a suitable native aTF cannot be identified, the ligand specificity of an aTF can be expanded via directed evolution to create designer biosensor platforms (Fig. 2b) by coupling rounds of mutagenesis [8-11] with high-throughput techniques such as fluorescence activated cell sorting (FACS) or antibiotic selection [6,12-14]. For example, Kasey et al engineered MphR, a promiscuous macrolide sensing aTF, to expand its promiscuity towards various natural and non-natural macrolides that were otherwise poor effectors of the wild-type MphR [12]. Similarly, a chimeric LysR biosensor was developed to selectively detect luteolin from three closely related flavonoids, naringenin, apigenin, and luteolin, by exploring a variety of chimeric detector-effector pairs [10]. The novel chimeric biosensor displayed stringent specificity for luteolin and is the first reported luteolin-specific transcriptional biosensor in Escherichia coli (E. coli) [10]. The protocatechuic acid (PCA) biosensor PcaV was evolved to alter its ligand specificity towards vanillin and other close aromatic aldehydes, generating the Van2 biosensor [11]. Mutational analysis revealed that the combination of mutations M113S/N114A was sufficient for the vanillin specificity, while I110V played an important role for the reduction of basal expression and stabilization [11]. The dynamic range of the Van2 biosensor with vanillin was 7.7-fold, while being unresponsive to the parental aromatic acid, PCA [11].
Table 1.
Examples of aTFs that respond to diverse natural products
| aTF | EFFECTOR | NATURAL PRODUCT CLASS |
|---|---|---|
| HcaR | Trans-cinnamic acid [15] | Phenyl-propanoid; Used in manufacture of flavors, dyes and pharmaceuticals |
| ItcR | Itaconic acid [16] | Dicarboxylic acid; Building block for resins, paints, plastics and synthetic fibers |
| AraC | Ectoine [17], triacetic acid lactone [18], mevalonate [19] | Carboxamidine, polyketide byproduct, terpene precursor |
| BenM | Cis,cis-muconic acid [20] | Dicarboxylic acid; Used for the manufacture of plastics |
| FdeR | Naringenin [21] | Flavonoid |
| QdoR | Quercetin and kaempferol [21] | Flavonoid |
| NitR | Caprolactam [22] | Lactam; Used in the manufacture of plastics |
| AvaR1/AvaR2 | Avenolide [23] | Avenolide is a signaling molecule for avermectin biosynthesis |
| JadR1/JadR2 | Chloroamphenicol and jadomycin B [24] | Polyketides |
| ChnR | ε-caprolactam, δ-valerolactam, and butyrolactam [25] | Lactam biosynthesis |
| AccR | Acetyl-, propionyl, methylcrotonyl-, malonyl-, and methylmalonyl-CoA [26] | Extender units for the biosynthesis of fatty acids and Polyketides |
| MphR | Erythromycin, azithromycin, clarithromycin and roxithromycin [12,27] | Polyketides |
| TtgR | Naringen, phloretin and genistein [28] | Flavonoids |
| TetR | Tetracycline, simocyclinone D8, actinorhodin [29] | Tetracyclines |
| QacR | Vanillin [30] | Terpene |
| OtrR | Oxytetracycline [31] | Tetracycline |
| CtcS | Chlortetracycline [32] | Tetracycline |
| SrbR | Rapamycin [33] | Polyketide |
| MexR | Pinene [34] | Terpene |
Tailoring the sensitivity and dynamic range of aTFs often relies on regulating the intracellular concentration of the aTF through the engineering of cis-regulatory components, such as the promoter or ribosome binding site (RBS), which control the rate of transcription and translation, respectively. Engineering of these components can be accomplished by semi-rational design with tools like ‘De Novo DNA’, an open-access online RBS and operon calculator that predicts translation initiation rates [35]. Alternatively, this can be achieved by random mutagenesis and screening. For instance, the dynamic range of MphR, a macrolide-sensing aTF, was improved 10-fold with randomized mutagenesis of the protein’s RBS sequence [12]. Dynamic range can also be fine-tuned by engineering the promoter sequence to reduce or eliminate undesired background noise, leaky expression, and varied limit of detection [36]. Beyond engineering of the repressor module, the sensitivity of an aTF biosensor can also be modulated by altering the location of the operator sequence within the promoter region of the reporter module [34]. However, mechanistic understanding of the interactions between aTFs and their corresponding regulatory components are typically not fully characterized or standardized. By leveraging a combination of RBS and promoter engineering, transcription and translation are controlled at the most fundamental level, enabling better control and success of aTF-biosensing platforms.
Computational Approaches to Biosensor Design
The success of aTF engineering via random or semi-random mutagenesis notwithstanding, computational platforms have leveraged statistical and mathematical modeling to create designer aTF-based biosensors and have tuned their transfer functions within complex gene regulatory networks. Recently, ‘design of experiments’, a statistical modeling system using structured, multivariate experimentation, was leveraged to map gene expression levels and tailor the Hill parameters of a protocatechuic acid biosensor [37]. Similarly, mathematical models have been applied to customize biosensor components to alter aTF biosensor activation thresholds, sensitivities, selectivity’s and dynamic ranges [38-40]. Quantitative modeling strategies, such as those developed by Swank et al, have been applied to characterize a library of synthetic transcription factors and their corresponding promoters, which were subsequently used to engineer and build de novo transcriptional regulatory networks [41].
Recent advances in biosensor engineering to create designer biosensors have also been accessed via structural modeling in silico [30]. Computational LBDs for de novo aTFs are expected to spur the next generation of biosensor development by providing exquisite affinity and selectivity for specific target molecules that model the same characteristics as naturally occurring aTFs [42]. As a result of improved computing power and the low cost of DNA synthesis, computationally designed de novo proteins are expected to revolutionize the next generation of biosensor development as they explore the full sequence space of possible amino acids [43]. For example, a small molecule biosensor was constructed from the computationally designed digoxigenin LBD [44]. Conditionally destabilized mutations in the digoxigenin LBD created a functional biosensor that is unstable unless bound to its new target effector, progesterone, thereby transforming a de novo LBD into a biosensor platform with unavailable or unknown aTFs [44]. It is anticipated that computational protein design and mathematical modeling will improve access to a broader range of molecular sensors, including those for natural products.
Biosensor-Guided Optimization of Precursor Biosynthesis
Small molecule primary metabolites are commonly leveraged as building blocks for the assembly of secondary metabolites; however, precursor deficiency can limit the productivity of natural product biosynthetic pathways [45] (Fig. 3a). Leveraging both native and engineered aTFs, several biosensor platforms have been developed for precursor detection as critical tools towards the long-term goals of optimizing metabolism and evolving de novo pathways for the production of natural products. For example, given the clinical significance of polyketides, there has been much interest in applying biosensor platforms to addressing long standing problems related to polyketide synthetic biology. Catalyzed by polyketide synthases (PKSs), polyketides are biosynthesized from the decarboxylative Claisen condensation of acyl-CoA building blocks. The potential modularity and versatility of such building blocks for accessing new-to-nature polyketides is driving development of biosensors that could be used to regulate their biosynthesis and to guide high-throughput engineering. Several aTFs utilize acyl-CoAs as effectors including AccR, which recognizes acetyl-, propionyl-, and methylcrotonyl-CoA [26], and FapR, which can detect its native malonyl-CoA (mCoA) as well as various C2-derivatives [46]. FapR-based biosensors have been used to produce oscillators that simultaneously regulate mCoA production via the upregulation of acetyl-CoA carboxylase and fatty acid biosynthesis [47]. Moreover, the newly identified promiscuity of this biosensor [46] could be leveraged for the directed evolution of de novo pathways to mCoA derivatives for precursor-directed biosynthesis of polyketide natural products.
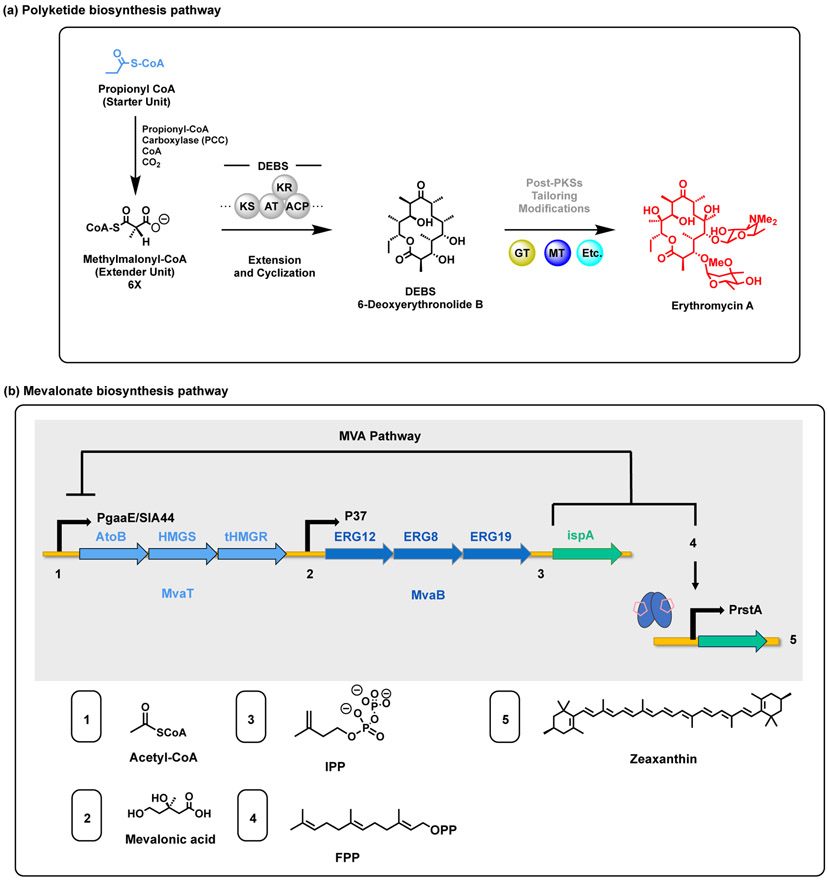
Figure 3.
Basic mechanism of (a) polyketide biosynthesis catalyzed by modular Type I PKSs and (b) the mevalonate pathway for hemiterpene production.
Isoprenoids are derived from the isomeric precursors, dimethylallyl pyrophosphate (DMAPP) and isopentenyl pyrophosphate (IPP), to yield >80,000 unique compounds. Although no native transcription factor biosensor platforms have been identified for DMAPP, a fusion between isoprene synthase, IspS, and the DBD of AraC was developed as a DMAPP responsive regulator of the pBAD promoter [48]. Extending IspS applications, an isoprene biosensor was developed that allows monitoring the intracellular concentration of isoprene in single bacterial cells by modifying the TbuT transcription factor with a detection limit of 0.1 mM. The fluorescence signal of isoprene producing E. coli correlated to the amount of produced bioisoprene [49]. Isoprene biosensors could be leveraged for high-throughput screening of isoprenoid biosynthesis and potentially be used for the design of artificial hemiterpene biosynthetic pathways [50].
Dynamic Metabolic Control for Pathway Optimization
Metabolic engineering can alter pathway flux to optimize cellular processes for improved titers of relevant natural products. However, the resulting imbalance of stringently regulated metabolites often imposes inherent challenges including the production of toxic intermediates, reduced catalytic efficiency, and inhibition of downstream pathways. Dynamic pathway regulation can be implemented to mitigate these effects through pathway-specific biosensor-based strategies [51,52], pathway-independent circuits [53], or a combination of these strategies [54]. While metabolic engineering for pathway optimization has been well explored, the following examples demonstrate the importance of implementing biosensors for improved dynamic control.
Introducing the mevalonate (MVA) pathway into a heterologous host often results in a flux imbalance of the isoprenoid precursors (IPP) and farnesyl pyrophosphate (FPP), which are toxic upon accumulation [55]. Previously, the expression levels of the ‘top’ three genes in the MVA (MvaT) (Fig. 3b) pathway have been balanced using tunable intergenic regions (TIGR), increasing mevalonate production by 7-fold [56]. Thus, in order to balance the ‘lower’ part of the MVA pathway (MvaB) (Fig. 3b) and to increase the available FPP, a TIGR library was constructed and screened using an FPP sensor plasmid, in which Green Fluorescence Protein (GFP) was under the control of the E. coli rstA promoter [55]. The rstA promoter responds to the accumulation FPP to regulate the expression of zeaxanthin thus creating a crosslink between the cell’s metabolic state and the pathway expression of the desired product [55]. To further reduce the metabolic burden, the TIGR optimized MvaB pathway plasmid and MvaT pathway were assembled on a single plasmid which resulted in a ~28% higher zeaxanthin production [55]. Furthermore, the PgadE/ IA44 sensor, which are downregulated by FPP and IPP respectively, were utilized to dynamically control the TIGR-mediated MVA pathway to prevent a toxic accumulation of precursors and further improve the zeaxanthin production by 1.6- and 1.7-fold [55].
Microbial production of fatty acid hydrocarbons, such as 1-alkene, which can be biosynthesized from activated fatty acids by polyketide synthases or from free fatty acids via cytochrome P450 enzymes, offer a sustainable opportunity for biofuels and olefins [57]. Recently, a Saccharomyces cerevisiae strain was engineered to improve the production of 1-alkenes by eliminating competing fatty acid consumption pathways and introducing the desaturase-like enzyme UndB from Pseudomonas fluorescens (PfUndB) [57]. These cumulative mutations resulted in up to ~29 fold-improvement of 1-alkene production [57]. To dynamically balance cell growth and product formation, PfUndB expression was regulated by the GAL7 promoter, which functions under low glucose levels, thereby enabling a distinct growth phase and a production phase that resulted in 100% higher 1-alkene titers [57]. Furthermore, FapR-based platforms have been used to dynamically control polyketide biosynthesis. For example, 6-methylsalicylic acid synthase (6-MSA) production capacity was increased 260% through the use of a hybrid yeast Prm1-FapR protein to regulate the malonyl-CoA dependent repression of fatty acid biosynthesis and the activation of the 6-MSA pathway genes, respectively [58].
Dynamic control has also been applied to improving titers via pathway independent regulatory elements, such as quorum-sensing (QS) circuits. A novel QS-based CRISPRi (EQCi) circuit, cell density-dependent gene regulation, was utilized in the rapamycin-producing strain Streptomyces rapamycinicus to autonomously and dynamically regulate multiple gene targets at once [33]. The EQCi circuit was designed to utilize the srbA promoter (srbAp) from S. rapamycinicus, which is under control of SrbR, to drive dCas9 expression [33]. At high cell density of S. rapamycinicus, the SrbR reaches a threshold in which it will no longer bind to srbAp, allowing for dCas9 expression, thereby switching the EQCi circuit on [33]. Furthermore, to improve rapamycin titers, three essential pathways were downregulated using this novel circuit resulting in the highest reported rapamycin titer of 1836 ±191 mg/L, an increase of ~660% compared to the wild-type producing strain [33]. Additionally, components from the lux and esaR QS systems were combined in E. coli to produce a single circuit to dynamically regulate a gene of interest [59]. This system was applied to control the flux through the naringenin biosynthetic pathway and found that down-regulation of endogenous mCoA genes resulted in a 140% higher titer [59]
Biosensor-Guided Enhancements of Final Product Titers
Biosensors can also be utilized to circumvent screening bottlenecks for natural product pathway and enzyme engineering by providing a precise high-throughput tool for final product detection and titer quantification, e.g. erythritol, a secondary metabolite, using the engineered aTF, EryD [60]. This platform was coupled with automatic strain mutagenesis and screening to characterize more than 1152 strains in one week. The top producing engineered strain produced more than 148 g/L erythritol and is the first reported work to develop EryD as a high throughput biosensor.
Similarly, a rational circuit design has been coupled with selection to increase cellular tolerance to toxic products. The MexR transcriptional repressor, derepressed in the presence of pinene, was employed to regulate expression of the AcrAB-TolC efflux pump, which provides tolerance to toxic compounds, such as the terpene pinene, but can inhibit cell growth when overexpressed, thus creating a synthetic feedback loop [34]. Subsequently, a synthetic promoter library containing MexR binding sites was created to drive the expression of GFP [34]. In addition, the mexA-mexR binding sequence was isolated and incorporated into different regions of the library [34]. To add feedback, the variants from the promoter library were used to control the expression of the acrAB pump in varying levels of pinene [34]. In order to determine which MexR promoter class led to pinene tolerance and responded by turning on the efflux pumps, next generation sequencing was conducted post selection [34]. Under pinene treatment, there was a strong selective pressure for MexR binding sites that were immediately upstream of the acrAB gene in addition to a convergence of promoter sequences which suggested that their goal of using feedback control to balance pinene and pump toxicity was successful [34].
Biosensors for Detecting Natural Product Tailoring Modifications
Following the biosynthesis of the natural product core structure, enzymatic tailoring including phosphorylation, methylation, hydroxylation, and glycosylation, are frequently carried out to optimize bioactivity of the mature natural product [61]. Often, the remarkable flexibility of downstream enzymes that tailor natural product scaffolds, including methyltransferases (MTs), hydroxylases, and glycosyltransferases, enables molecular diversity. Highly specific engineered aTFs for a modified target compound could be leveraged to detect natural product tailoring steps which can then be targeted via a biosensor-guided approach to identify new biocatalysts or to evolve existing ones. The application of a vanillate biosensor for reporting the regioselective methylation of catechol exemplifies this vision. The VanR-VanO vanillate sensor system was engineered to selectively detect vanillate but no other methylated regioisomers or biosynthetic precursors [62]. Subsequently, the biosensor was used to identify the conversion from protocatechuate to vanillate catalyzed by prospective O-MTs. First, deletion of the methionine biosynthesis regulator MetJ improved the conversion of protocatechuate to vanillate by increasing the pool of the methyl donor, S-adenosylmethionine (SAM). Remarkably, using the biosensor, three previously uncharacterized O-MTs were identified that supported the conversion of protocatechuate to vanillate [62]. As achieved in the vanillate example, a single aTF-based biosensor to report natural product tailoring needs to discriminate between the acceptor substrate and the tailored product. This could be done by leveraging the aforementioned strategies to engineer the required selectivity into a given aTF scaffold. Alternatively, a pair of ratiometric biosensors could be used for this purpose. For example, a FRET-based ratiometric sensor that incorporates the TetR repressor protein enabled correlation of fluorescence and tetracycline concentration allowing for accurate quantification enabling homogeneous assays without washing steps [63].
Although biosensors that directly detect the tailored product are likely preferred, aTFs with the required selectivity might not always be available. The development of biosensors that detect a cofactor, precursor, or byproduct of the tailoring reaction have also begun to be investigated for their potential utility. An example of this is the development of a genetically encoded ratiometric biosensor for NADH/NAD+ based on the redox-responsive transcription factor Rex [64]. The sensor was successfully deployed in a proof-of-principle high-throughput screen to enrich high NADH mutant strains that were diluted 10,000-fold in wild-type cells.
Conclusions and Future Outlook
Traditional engineering strategies for improving natural product titers and accessing non-natural designer compounds are often hampered by low-throughput screening technologies. Yet, the emergence and development of genetically-encoded biosensors have allowed for the rapid screening of potentially millions of variants to enhance pathway and enzymatic efficiencies. Subsequently, robust high-throughput screening platforms based on aTFs have enabled the detection of diverse natural product classes and their small molecule building blocks which has spurred the application of directed evolution and metabolic engineering of natural product biosynthetic pathways. We envision that these strategies will be particularly valuable to design aTFs with a defined linear range of detection in order to engineer prototype microbial strains for the biosynthesis of natural products, their analogues, and precursors.
Despite their utility, the discovery, characterization, and optimization of aTF-based biosensor platforms remains a critical bottleneck to their industrial application. aTF-based biosensors are limited in terms of their adaptability within host microbes to modulate proteins or complex gene networks. The transcription factors themselves must properly fold, maintain solubility in vivo and be amenable to engineering efforts. Indeed, effectors of transcription factors must be cell-wall permeable, readily available for binding, non-toxic, and stable inside the cell. Furthermore, it can be difficult to identify well-characterized aTFs that fit specific needs (e.g., specific effector or biosynthetic system). Even when a promising candidate aTF is identified, it can be difficult to identify a corresponding RBS and promoter system to make a functional gene circuit, given the diversity of microbial regulatory mechanisms [67]. However, as emerging technologies are developed, biosensor engineering is expected to progress towards more rapid, rational, and computational-based design for the facile development of highly targeted biosensors for a specific molecule of interest [66]. For example, the use of cell-free technology allows for quick prototyping of gene circuits for in vitro detection of molecules, or for future in vivo biosensor operation.
Recent advances utilizing cell-free transcription-translation to rapidly characterize novel biosensor components and aTF effector promiscuity now offer prototyping capabilities to quickly overcome these challenges, specifically issues with effector permeability and toxicity [27,65]. The application of directed evolution to tailor the effector selectivity of aTFs continues to provide biosensors with exquisite detection capabilities. We expect future trends in biosensor design to apply new, open-access computational tools that enable rational engineering of the intricate conformational dynamics of aTFs via homology modeling and molecular docking, which would otherwise be arduous using traditional engineering strategies. Foremost, de novo protein design is set to provide customizable aTFs biosensors from the start that can be applied across diverse classes of molecules. The use of these tools has rapidly accelerated engineering efforts to enable the generation of smart libraries to effectively find active-site pockets or to simply generate highly targeted biosensors for a specific molecule of interest. Together, these technologies underlie the foundations of screening throughput and genetic diversity, which are at the forefront of evolutionary biosensor engineering. Herein, we highlight our vision for biosensor-guided approaches towards natural products and their derivatives to include directed evolution of aTFs, advances in cell-free methods to characterize them, new applications of computational tools, and the deployment of engineered aTF biosensors to address longstanding problems in natural product biocatalysis.
Acknowledgements
Financial support is provided in part by the National Institutes of Health grants GM124112 (G.J.W), T32GM133366 (M.M), an NIH Pre-doctoral Biotechnology Traineeship NIH T32 GM008776 (J.M.G) and the Thomas Lord Distinguished Professorship Endowment (G.J.W). The authors also thank Alexandra A. Malico and Lindsay Nichols for assisting with this manuscript.
Footnotes
Declaration of interests
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Galanie S, Entwistle D, Lalonde J: Engineering biosynthetic enzymes for industrial natural product synthesis. Nat Prod Rep 2020, doi: 10.1039/c9np00071b. [DOI] [PubMed] [Google Scholar]
- 2.Daystar J, Handfield RB, Golden JS, and TEM: An Economic Impact Analysis of the U.S. Biobased Products Industry: 2018 Update. 2018. [Google Scholar]
- 3.Carpenter AC, Paulsen IT, Williams TC: Blueprints for biosensors: Design, limitations, and applications. Genes 2018, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng F, Tang XL, Kardashliev T: Transcription factor-based biosensors in high-throughput screening: Advances and applications. Biotechnol J 2018, 13. [DOI] [PubMed] [Google Scholar]
- 5.Li C, Zhang R, Wang J, Wilson LM, Yan Y: Protein engineering for improving and diversifying natural product biosynthesis. Trends Biotechnol 2020, 38:729–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun H, Zhao H, Ang EL: A new biosensor for stilbenes and a cannabinoid enabled by genome mining of a transcriptional regulator. ACS Synth Biol 2020, 9:698–705.* Genomic sequencing can expand the biosensor platform repertoire by identifying aTFs for a specific final product. This technique was utilized to identify and subsequently engineer a novel stilbene-responsive biosensor for the selective detection of resveratrol and resveratrol analog production
- 7.Grazon C, Baer RC, Kuzmanović U, Nguyen T, Chen M, Zamani M, Chern M, Aquino P, Zhang X, Lecommandoux S, et al. : A progesterone biosensor derived from microbial screening. Nat Commun 2020, 11:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Snoek T, Chaberski EK, Ambri F, Kol S, Bjørn SP, Pang B, Barajas JF, Weiner DH, Jensen MK, Keasling JD: Evolution-guided engineering of small-molecule biosensors. Nucleic Acids Res 2020, 48:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jha RK, Narayanan N, Pandey N, Bingen JM, Kern TL, Johnson CW, Strauss CEM, Beckham GT, Hennelly SP, Dale T: Sensor-enabled alleviation of product inhibition in chorismate pyruvate-lyase. ACS Synth Biol 2019, 8:775–786. [DOI] [PubMed] [Google Scholar]
- 10.de Paepe B, Maertens J, Vanholme B, de Mey M: Chimeric LysR-type transcriptional biosensors for customizing ligand specificity profiles toward flavonoids. ACS Synth Biol 2019, 8:318–331. [DOI] [PubMed] [Google Scholar]
- 11.F.M. L, Currin A, Dixon N: Directed evolution of the PcaV allosteric transcription factor to generate a biosensor for aromatic aldehydes. J Biol Eng 2019, 13:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kasey CM, Zerrad M, Li Y, Cropp TA, Williams GJ: Development of transcription factor-based designer macrolide biosensors for metabolic engineering and synthetic biology. ACS Synth Biol 2018, 7:227–239.**This paper demonstrates a powerful platform for sensing complex products of polyketide biosynthesis. MphR variants were identified which displayed improved sensitivity towards macrolides that were otherwise poor inducers of the wild-type MphR biosensor.
- 13.Thompson MG, Pearson AN, Barajas JF, Cruz-Morales P, Sedaghatian N, Costello Z, Garber ME, Incha MR, Valencia LE, Baidoo EEK, et al. : Identification, characterization, and application of a highly sensitive lactam biosensor from Pseudomonas putida. ACS Synth Biol 2020, 9:53–62. [DOI] [PubMed] [Google Scholar]
- 14.Kortmann M, Mack C, Baumgart M, Bott M: Pyruvate carboxylase variants enabling improved lysine production from glucose identified by biosensor-based high-throughput fluorescence-activated cell sorting screening. ACS Synth Biol 2019, 8:274–281. [DOI] [PubMed] [Google Scholar]
- 15.Flachbart LK, Sokolowsky S, Marienhagen J: Displaced by deceivers: Prevention of biosensor cross-talk is pivotal for successful biosensor-based high-throughput screening campaigns. ACS Synth Biol 2019, 8:1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanko EKR, Minton NP, Malys N: A transcription factor-based biosensor for detection of itaconic acid. ACS Synth Biol 2018, 7:1436–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen W, Zhang S, Jiang P, Yao J, He Y, Chen L, Gui X, Dong Z, Tang SY: Design of an ectoine-responsive AraC mutant and its application in metabolic engineering of ectoine biosynthesis. Metab Eng 2015, 30:149–155. [DOI] [PubMed] [Google Scholar]
- 18.Tang SY, Qian S, Akinterinwa O, Frei CS, Gredell JA, Cirino PC: Screening for enhanced triacetic acid lactone production by recombinant Escherichia coli expressing a designed triacetic acid lactone reporter. J Am Chem Soc 2013, 135:10099–10103. [DOI] [PubMed] [Google Scholar]
- 19.Tang SY, Cirino PC: Design and application of a mevalonate-responsive regulatory protein. Angew Chemie Int Ed 2011, 50:1084–1086. [DOI] [PubMed] [Google Scholar]
- 20.Wang G, Øzmerih S, Guerreiro R, Meireles AC, Carolas A, Milne N, Jensen MK, Ferreira BS, Borodina I: Improvement of cis.cis-muconic acid production in Saccharomyces cerevisiae through biosensor-aided genome engineering. ACS Synth Biol 2020, 9:634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siedler S, Stahlhut SG, Malla S, Maury JÔ, Neves AR: Novel biosensors based on flavonoid-responsive transcriptional regulators introduced into Escherichia coli. Metab Eng 2014, 21:2–8. [DOI] [PubMed] [Google Scholar]
- 22.Yeom SJ, Kim M, Kwon KK, Fu Y, Rha E, Park SH, Lee H, Kim H, Lee DH, Kim DM, et al. : A synthetic microbial biosensor for high-throughput screening of lactam biocatalysts. Nat Commun 2018, 9:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu J, Chen Z, Li J, Wen Y: AvaR1, a butenolide-type autoregulator receptor in Streptomyces avermitilis, directly represses avenolide and avermectin biosynthesis and multiple physiological responses. Front Microbiol 2017, 8:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sekurova ON, Zhang J, Kristiansen KA, Zotchev SB: Activation of chloramphenicol biosynthesis in Streptomyces venezuelae ATCC 10712 by ethanol shock: Insights from the promoter fusion studies. Microb Cell Fact 2016, 15:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Barajas JF, Burdu M, Ruegg TL, Dias B, Keasling JD: Development of a transcription factor-based lactam biosensor. ACS Synth Biol 2017, 6:439–445. [DOI] [PubMed] [Google Scholar]
- 26.Lyu M, Cheng Y, Han X, Wen Y, Song Y, Li J, Chen Z: AccR, a TetR family transcriptional repressor, coordinates short-chain acyl coenzyme A homeostasis in Streptomyces avermitilis. Appl Environ Microbiol 2020, 86:e00508–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung JK, Alam KK, Verosloff MS, Capdevila DA, Desmau M, Clauer PR, Lee JW, Nguyen PQ, Pastén PA, Matiasek SJ, et al. : Cell-free biosensors for rapid detection of water contaminants. Nat Biotechnol 2020, doi: 10.1038/s41587-020-0571-7.** This cell-free system was used to monitor water quality with whole cell biosensors. A group of programmable aTFs sensed a range of toxic contaminants, such as metals and antibiotics, within minutes, demonstrating the system’s simplified detection response and potential field use for communities at risk of unsafe drinking water.
- 28.Tera W, Felipe A, Segura A, Rojas A, Ramos J: Antibiotic-dependent induction of Pseudomonas putida DOT-T1E TtgABC efflux pump is mediated by the drug binding repressor TtgR. Antimicrob Agents Chemother 2003, 47:3067–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuthbertson L, Nodwell JR: The TetR family of regulators. Microbiol Mol Biol Rev 2013, 77:440–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Los Santos ELC, Meyerowitz JT, Mayo SL, Murray RM: Engineering transcriptional regulator effector specificity using computational design and in vitro rapid prototyping: Developing a vanillin sensor. ACS Synth Biol 2016, 5:287–295. [DOI] [PubMed] [Google Scholar]
- 31.Zhang W, Ames BD, Tsai SC, Tang Y: Engineered biosynthesis of a novel amidated polyketide, using the malonamyl-specific initiation module from the oxytetracycline polyketide synthase. Appl Environ Microbiol 2006, 72:2573–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu T, Cheng X, Liu Y, Deng Z, You D: Deciphering and engineering of the final step halogenase for improved chlortetracycline biosynthesis in industrial Streptomyces aureofaciens. Metab Eng 2013, 19:69–78. [DOI] [PubMed] [Google Scholar]
- 33.Tian J, Yang G, Gu Y, Sun X, Lu Y, Jiang W: Developing an endogenous quorum-sensing based CRISPRi circuit for autonomous and tunable dynamic regulation of multiple targets in Streptomyces. Nucleic Acids Res 2020, doi: 10.1093/nar/gkaa602.** A novel QS-based CRISPRi (EQCi) circuit was utilized in a rapamycin-producer strain that resulted in the highest rapamycin titers reported to date. This EQCi-based system is notable due to its ability to improve rapamycin titers without affecting cell-growth.
- 34.Siu Y, Fenno J, Lindle JM, Dunlop MJ: Design and selection of a synthetic feedback loop for optimizing biofuel tolerance. ACS Synth Biol 2018, 7:16–23. [DOI] [PubMed] [Google Scholar]
- 35.Salis HM: The ribosome binding site calculator. Methods Enzymol 2011, 498:19–42. [DOI] [PubMed] [Google Scholar]
- 36.Wan X, Marsafari M, Xu P: Engineering metabolite-responsive transcriptional factors to sense small molecules in eukaryotes: Current state and perspectives. Microb Cell Fact 2019, 18:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berepiki A, Kent R, MacHado LFM, Dixon N: Development of high-performance whole cell biosensors aided by statistical modeling. ACS Synth Biol 2020, 9:576–589.*A ‘design of experiments’ was used to map gene expression levels for optimization of poorly characterized transcription factors and metabolic pathways. This experimentation and statistical modeling design allowed researchers to minimally explore experimental space to determine the importance of specific factors for optimization of their aTF-based biosensor.
- 38.Gonzalez-Flo E, Alaball ME, Macla J: Two-component biosensors: Unveiling the mechanisms of predictable tunability. ACS Synth Biol 2020, 9:1328–1335. [DOI] [PubMed] [Google Scholar]
- 39.Ho JCH, Pawar S v., Hallam SJ, Yadav VG: An improved whole-cell biosensor for the discovery of lignin-transforming enzymes in functional metagenomic screens. ACS Synth Biol 2018, 7:392–398. [DOI] [PubMed] [Google Scholar]
- 40.Nadler DC, Morgan SA, Flamholz A, Kortright KE, Savage DF: Rapid construction of metabolite biosensors using domain-insertion profiling. Nat Commun 2016, 7:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swank Z, Laohakunakorn N, Maerkl SJ: Cell-free gene-regulatory network engineering with synthetic transcription factors. Proc Natl Acad Sci 2019, 116:5892–5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tinberg CE, Khare SD, Dou J, Doyle L, Nelson JW, Schena A, Jankowski W, Kalodimos CG, Johnsson K, Stoddard BL, et al. : Computational design of ligand-binding proteins with high affinity and selectivity. Nature 2013, 501:212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang PS, Boyken SE, Baker D: The coming of age of de novo protein design. Nature 2016, 537:320–327. [DOI] [PubMed] [Google Scholar]
- 44.Feng J, Jester BW, Tinberg CE, Mandell DJ, Antunes MS, Chari R, Morey KJ, Rios X, Medford JI, Church GM, et al. : A general strategy to construct small molecule biosensors in eukaryotes. elife 2015, 4:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pickens LB, Tang Y, Chooi YH: Metabolic engineering for the production of natural products. Annu Rev Chem Biomol Eng 2011, 2:211–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalkreuter E, Keeler AM, Malico AA, Bingham KS, Gayen AK, Williams GJ: Development of a genetically encoded biosensor for detection of polyketide synthase extender units in Escherichia coli. ACS Synth Biol 2019, 8:1391–1400.* This paper focused on the development of a FapR biosensor platform, in vivo and in vitro, for the detection of malonyl-CoA and its derivatives towards non-natural polyketides.
- 47.Xu P, Gu Q, Wang W, Wong L, Bower AGW, Collins CH, Koffas MAG: Modular optimization of multi-gene pathways for fatty acids production in E. coli. Nat Commun 2013, 4:1408–1409. [DOI] [PubMed] [Google Scholar]
- 48.Liu CL, Cai JY, Bi HR, Tan TW: A novel DMAPP-responding genetic circuit sensor for high-throughput screening and evolving isoprene synthase. Appl Microbiol Biotechnol 2018, 102:1381–1391. [DOI] [PubMed] [Google Scholar]
- 49.Kim SK, Kim SH, Subhadra B, Woo SG, Rha E, Kim SW, Kim H, Lee DH, Lee SG: A genetically encoded biosensor for monitoring isoprene production in engineered Escherichia coli. ACS Synth Biol 2018, 7:2379–2390. [DOI] [PubMed] [Google Scholar]
- 50.Lund S, Hall R, Williams GJ: An artificial pathway for isoprenoid biosynthesis decoupled from native hemiterpene metabolism. ACS Synth Biol 2019, 8:232–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao J, Li C, Zhang Y, Shen Y, Hou J, Bao X: Dynamic control of ERG20 expression combined with minimized endogenous downstream metabolism contributes to the improvement of geraniol production in Saccharomyces cerevisiae. Microb Cell Fact 2017, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang Y, Lin Y, Wang J, Wu Y, Zhang R, Cheng M, Shen X, Wang J, Chen Z, Li C, et al. : Sensor-regulator and RNAi based bifunctional dynamic control network for engineered microbial synthesis. Nat Commun 2018, 9:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gupta A, Reizman IMB, Reisch CR, Prather KLJ: Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat Biotechnol 2017, 35:273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doong SJ, Gupta A, Prather KLJ: Layered dynamic regulation for improving metabolic pathway productivity in Escherichia coli. Proc Natl Acad Sci 2018, 115:2964–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen HJ, Cheng BY, Zhang YM, Tang L, Li Z, Bu YF, Li XR, Tian GQ, Liu JZ: Dynamic control of the mevalonate pathway expression for improved zeaxanthin production in Escherichia coli and comparative proteome analysis. Metab Eng 2016, 38:180–190. [DOI] [PubMed] [Google Scholar]
- 56.Pfleger BF, Pitera DJ, Smolke CD, Keasling JD: Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat Biotechnol 2006, 24:1027–1032. [DOI] [PubMed] [Google Scholar]
- 57.Zhou YJ, Hu Y, Zhu Z, Siewers V, Nielsen J: Engineering 1-alkene biosynthesis and secretion by dynamic regulation in yeast. ACS Synth Biol 2018, 7:584–590. [DOI] [PubMed] [Google Scholar]
- 58.Wen J, Tian L, Liu Q, Zhang Y, Cai M: Engineered dynamic distribution of malonyl-CoA flux for improving polyketide biosynthesis in Komagataella phaffii. J Biotechnol 2020, 320:80–85. [DOI] [PubMed] [Google Scholar]
- 59.Dinh C v, Prather KLJ: Development of an autonomous and bifunctional quorum-sensing circuit for metabolic flux control in engineered Escherichia coli. Proc Natl Acad Sci 2019, 116:25562–25568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qiu X, Xu P, Zhao X, Du G, Zhang J, Li J: Combining genetically-encoded biosensors with high throughput strain screening to maximize erythritol production in Yarrowia lipolytica. Metab Eng 2020, 60:66–76. [DOI] [PubMed] [Google Scholar]
- 61.Rix U, Fischer C, Remsing LL, Rohr J: Modification of post-PKS tailoring steps through combinatorial biosynthesis. Nat Prod Rep 2002, 19:542–580. [DOI] [PubMed] [Google Scholar]
- 62.Kunjapur AM, Prather KLJ: Development of a vanillate biosensor for the vanillin biosynthesis pathway in E. coli. ACS Synth Biol 2019, 8:1958–1967.* An E. coli promoter library was used to improve the biosensor dynamic range 14-fold, producing a robust sensor for vanillin biosynthesis. The selectivity of the sensor was leveraged to identify O-methyltransferase candidates for the biosynthesis of the target natural product.
- 63.Nguyen TT, Chern M, Baer RC, Galagan J, Dennis AM: A förster resonance energy transfer-based ratiometric sensor with the allosteric transcription factor TetR. Small 2020, 1907522:16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Y, Landick R, Raman S: A regulatory NADH/NAD+ redox biosensor for bacteria. ACS Synth Biol 2019, 8:264–273. [DOI] [PubMed] [Google Scholar]
- 65.Voyvodic PL, Pandi A, Koch M, Conejero I, Valjent E, Courtet P, Renard E, Faulon JL, Bonnet J: Plug-and-play metabolic transducers expand the chemical detection space of cell-free biosensors. Nat Commun 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang W, Lai LH: Computational design of receptor and sensor proteins with novel functions. Nature 2003, 25:185–190. [DOI] [PubMed] [Google Scholar]
- 67.De Paepe B, Peters G, Coussement P, Maertens J, De Mey M: Tailor-made transcriptional biosensors for optimizing microbial cell factories. J Ind Microbiol Biot 2017, 44: 623–645. [DOI] [PubMed] [Google Scholar]