Abstract
Background:
Liquid biopsies are increasingly tested in patients with colorectal cancer to assess tumor burden, response to therapy, and prognosis. The significance of liquid biopsy results after resection of colorectal liver metastases (CLM) is not well-defined.
Study Design:
Sixty-three patients undergoing CLM resection between 2016–2018 had plasma drawn postoperatively for liquid biopsy evaluation. Next-generation sequencing analysis was performed to detect somatic mutations in 70 genes.
Results:
Liquid biopsy after CLM resection was positive in 42 of 63 patients (67%). Eleven patients (18%) had 1 gene mutation, 14 (22%) had 2–3 mutations, and 17 (27%) had ≥ 4 mutations. The most common mutation was APC, detected in 32 (76%) patients, followed by TP53 (74%) and KRAS (38%). Two-year OS from date of liver resection was significantly worse among patients with a positive liquid biopsy (70% versus 100%, p = 0.005), particularly for those with ≥ 4 gene mutations detected, whose 2-year OS was 41%. Sixteen of the 63 patients underwent serial liquid biopsies, resulting in 100 liquid biopsy with matched serum CEA and computed tomography (CT) scan results. Metastases were identified in 74 CT scans, which correlated with positive liquid biopsy in 77% of samples (p < 0.001) and CEA > 3 ng/ml in 45% of samples (p < 0.22).
Conclusion:
Liquid biopsy results provide complementary information to serum CEA and CT imaging about disease burden and prognosis. A positive liquid biopsy after CLM resection is associated with worse OS, particularly when multiple gene mutations are detected.
Keywords: Liquid biopsy, liver metastases, colorectal cancer, cell-free DNA, hepatectomy, circulating tumor DNA
PRECIS
After resection of colorectal liver metastases, detection of a somatic mutation in a 70-gene liquid biopsy panel correlates with worse overall survival, particularly when multiple gene mutations are identified. A positive liquid biopsy correlates with radiologic evidence of disease recurrence.
INTRODUCTION
Colorectal cancer is the second leading cause of cancer-related mortality in the United States, and most patients who die of colorectal cancer will have liver metastases.(1) Surgical resection of colorectal liver metastases (CLM) remains the gold standard treatment and is associated with a 5-year overall survival (OS) rate of 58%.(2) However, up to 70% of patients will suffer disease recurrence after hepatic resection.(3) Strategies to reduce recurrence rates include perioperative chemotherapy, targeted agents, and biomarkers to stratify prognosis and predict response to therapy.(4-6) Specifically, microsatellite instability and RAS mutation status predict response to immunotherapy and anti-epidermal growth factor receptor (EGFR) agents, respectively.(1)
Currently, biomarker detection in colorectal cancer depends upon genotyping of tissue biopsy or resection specimens. However, tissue biopsy requires a percutaneous or endoscopic procedure and only provides a single, static snapshot of a small fragment of tumor. Liquid biopsy is emerging as a promising, non-invasive alternative to tissue biopsy.(7) DNA present in the non-cellular portion of blood, or cell-free DNA (cfDNA), originates from normal tissue or tumor cells. Circulating tumor DNA (ctDNA) is cfDNA secreted into the bloodstream by tumor cells. Analysis of ctDNA, also termed liquid biopsy, has the potential to detect minimal residual disease, genotype tumors to direct targeted therapy, and track tumor evolution in real-time. In addition, liquid biopsy may provide a more comprehensive molecular profile of cancers than a single tissue biopsy.
In colorectal cancer, ctDNA detection rates have been shown to correlate with stage of disease, identified in 40% and > 90% of patients with stage I and IV disease, respectively.(8) Among patients undergoing CLM resection, preoperative detection of specific somatic gene mutations in peripheral blood is associated with worse disease-specific survival.(9) The purpose of this study was to evaluate a 70-gene liquid biopsy panel and its impact on OS after CLM resection, and correlations with postoperative computed tomography (CT) imaging and serum carcinoembryonic antigen (CEA) levels.
METHODS
Study population and follow-up
A prospectively maintained database was queried to identify 63 consecutive patients who underwent CLM resection at The University of Texas MD Anderson Cancer Center between January 2016 and November 2018, and had plasma drawn postoperatively for liquid biopsy analysis (Fig. 1).
Figure 1.
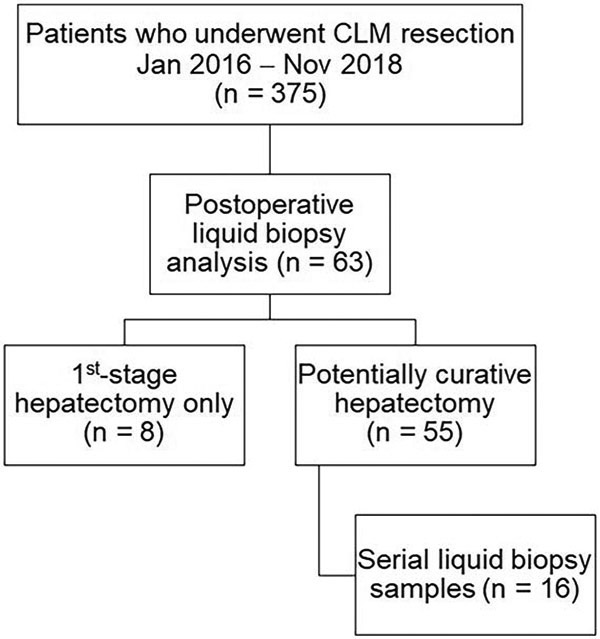
Selection of patients undergoing resection of colorectal liver metastases (CLM) and postoperative liquid biopsy analysis.
Major hepatectomy was defined as resection of ≥ 3 contiguous Couinaud segments.(10) Synchronous liver metastases were defined as metastases diagnosed within 6 months of primary tumor diagnosis. R1 resection margin was defined as tumor cells at the line of transection. Patients undergoing 1st-stage hepatectomy without completing the 2nd-stage were classified as having R2 resections. Number and size of liver metastases were recorded from surgical pathology reports or CT imaging.
Postoperatively, patients were followed with history and physical examination, serum CEA evaluation, and CT scans of the chest, abdomen, and pelvis every 3 months for the first 2 years after CLM resection. Sixteen patients underwent serial liquid biopsies, resulting in 100 samples with matched CEA and CT results. The study was approved by the Institutional Review Board.
Genetic analyses
A next-generation sequencing analysis to detect somatic mutations in 70 cancer-related genes was performed on ctDNA isolated from plasma in a Clinical Laboratory Improvement Amendments (CLIA)-certified molecular diagnostics laboratory, based on Guardant360®CDx technology (Guardant Health, Inc., Redwood City, CA).(11) The genes and exons tested are listed in eTable 1. The lower limit of detection was 0.3% mutation allelic frequency for 30 ng cfDNA and 1% for 5 ng cfDNA. A positive liquid biopsy was defined as one or more gene mutations in plasma.
eTable 1.
Genes and Exons Tested
| Gene | Exons tested |
|---|---|
| AKT1 | 3, 6 |
| ALK | 18-29 |
| APC | 2-16 |
| AR | ALL |
| ARAF | 7, 14 |
| ARID1A | 1-3, 6-8, 11-20 |
| ATM | 8, 55, 63 |
| BRAF | 1-15, 17-21 |
| BRCA1 | 2-23 |
| BRCA2 | 2-11, 13-27 |
| CCND1 | 1-2, 4-5 |
| CCND2 | 1-2, 5 |
| CCNE1 | 4-6, 8-12 |
| CDK4 | 2-8 |
| CDK6 | 2-8 |
| CDKN2A | ALL |
| CTNNB1 | 3 |
| DDR2 | 14-17 |
| EGFR | ALL |
| ERBB2 | ALL |
| ESR1 | 5, 6-8 |
| EZH2 | 16 |
| FBXW7 | 9-10, 12 |
| FGFR1 | 2-4, 6-8, 10-13, 15, 17-18 |
| FGFR2 | 2-13, 15-18 |
| FGFR3 | 2-18 |
| GNA11 | 5 |
| GNAQ | 5 |
| GNAS | 8, 9 |
| HNF1A | 3, 4 |
| HRAS | 2-5 |
| IDH1 | 4 |
| IDH2 | 4 |
| JAK2 | 14 |
| JAK3 | 13 |
| KIT | ALL |
| KRAS | 2-5 |
| MAP2K1 | 2, 3 |
| MAP2K2 | 2, 3 |
| MAPK1 | ALL |
| MAPK3 | ALL |
| MET | 2-9, 10-21 |
| MLH1 | 12 |
| MPL | 10 |
| MTOR | 2-30 |
| MYC | ALL |
| NF1 | 2-6, 8-14, 16-18, 20-21, 24-28, 30, 32-50, 52-55, 57-58 |
| NFE2L2 | 2 |
| NOTCH1 | 4, 8, 26, 34 |
| NOTCH2 | 1, 34 |
| NPM1 | 11 |
| NRAS | 2-5 |
| NTRK1 | 8-10, 12, 14-15 |
| NTRK3 | 16-17 |
| PDGFRA | 3-7, 9-12, 14-15, 18-20, 22-23 |
| PIK3CA | 2-21 |
| PTEN | 1-2, 4-9 |
| PTPN11 | 3, 13 |
| RAD51 | 2-3, 5-10 |
| RAF1 | 2-3, 5, 7-8, 10, 15-17 |
| RB1 | 1-4, 7-8, 10-11, 13, 16-23, 25-27 |
| RET | 9-12, 14-16 |
| ROS1 | 31-32, 34-36, 38 |
| SMAD4 | 3, 5-6, 8-12 |
| SMO | 5, 9 |
| STK11 | ALL |
| TERT | 1, 5 |
| TP53 | 2-11 |
| TSC1 | 15, 23 |
| VHL | ALL |
Next-generation sequencing was performed on tissue from primary tumors and/or liver metastases to detect somatic mutations in coding sequences of 128 cancer-related genes from DNA extracted from formalin-fixed paraffin-embedded tumor samples, as previously described. (12) Fourteen patients underwent analysis of 50 genes only, and 5 patients did not undergo tissue multigene panel testing.
Statistical analyses
Group comparisons were performed using chi-square tests for categorical variables and Mann–Whitney U test for continuous variables. OS was estimated using the Kaplan-Meier method and compared with the log-rank test. Factors significant on univariable analysis were entered into a multivariable Cox analysis of OS. All p values were 2-sided, and p < 0.05 was considered statistically significant. All analyses were performed with SPSS Statistics 23.0 (IBM Corp., Chicago, IL).
RESULTS
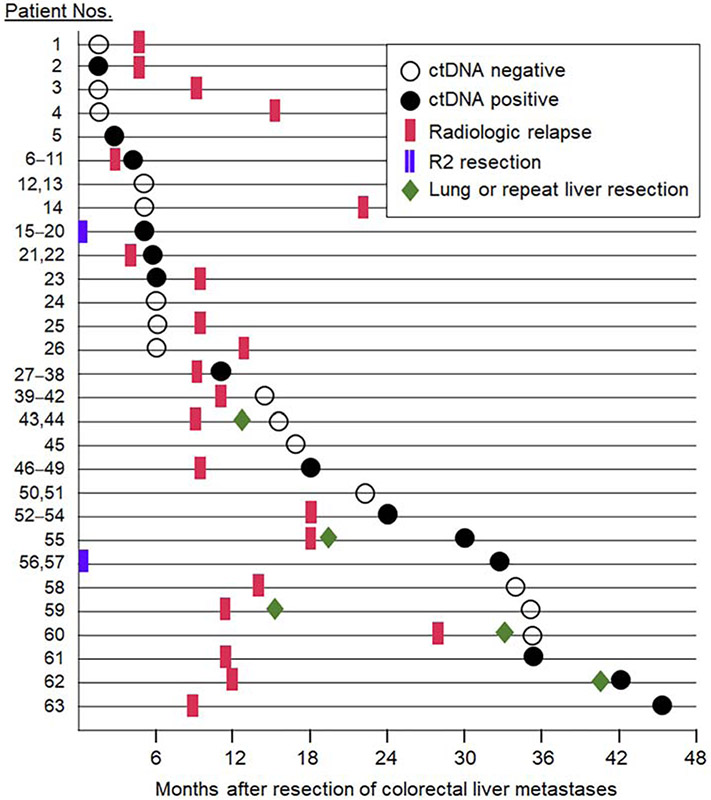
Clinicopathologic characteristics of the 63 patients are listed in Table 1. Median follow-up was 30 months (range, 9–53 months). Median time interval between hepatectomy and postoperative blood draw for liquid biopsy analysis was 13 months (range, 1–45 months) (Fig. 2). Most liquid biopsies were ordered by medical oncologists to detect minimal residual disease (21 patients), to guide systemic therapy (n = 19), including detection of resistance mutations to anti-EGFR therapy; for clinical trial screening (n = 13), and as baseline evaluation before surgery or ablation for lung or recurrent liver metastases (n = 10).
Table 1.
Patient and Tumor Characteristics (n = 63)
| Characteristic | N | % |
|---|---|---|
| Age, y, median (range) | 55 (30-82) | |
| Sex, n (%) | ||
| Male | 32 (51) | |
| Female | 31(49) | |
| Node-positive primary tumor, n (%) | 54 (86) | |
| Synchronous diagnosis of liver metastases, n (%) | 46 (73) | |
| Chemotherapy before liver resection, n (%) | 55 (87) | |
| Chemotherapy after liver resection, n (%) | 37 (59) | |
| Preoperative CEA, ng/mL, median (range) | 4.1 (0-1639) | |
| Extrahepatic metastases, n (%) | 21 (33) | |
| No. of liver metastases, median (range) | 3 (1-13) | |
| Size of largest liver metastasis, cm, median (range) | 2.5 (0.7-8.2) | |
| Surgical resection margin, n (%) | ||
| R0 | 39 (62) | |
| R1 | 16 (25) | |
| R2 (1st stage hepatectomy only) | 8 (13) | |
| Major hepatectomy, n (%) | 23 (37) | |
| Completed two-stage hepatectomy, n (%) | 6 (10) | |
| Disease recurrence, n (%) | 56 (89) | |
CEA, carcinoembryonic antigen
Figure 2.
Timeline of first postoperative liquid biopsy and radiologic relapse after resection of colorectal liver metastases.
Among the 21 patients with extrahepatic metastases, 12 patients underwent synchronous hepatectomy and resection of disease in the portal or retroperioneal lymph nodes (n = 6), lung (n = 2), peritoneum (n = 2), ovary (n = 1), or adrenal (n = 1). Unresected extrahepatic metastases included low-volume disease responding to chemotherapy in the lungs (n = 3) or retroperitoneal lymph nodes (n = 2); bone metastasis treated with radiation (n = 1), and 3 patients who underwent 1st-stage hepatectomy only.
First postoperative liquid biopsy and correlation with tumor tissue
The number of gene mutations detected in each plasma sample was no mutation in 21 patients (33%), one mutation in 11 patients (18%), 2–3 mutations in 14 patients (22%), and ≥ 4 mutations in 17 patients (27%). Among the 42 patients with positive liquid biopsy, the most common gene mutation was APC, detected in 32 patients (76%), followed by TP53 (74%) and KRAS (38%).
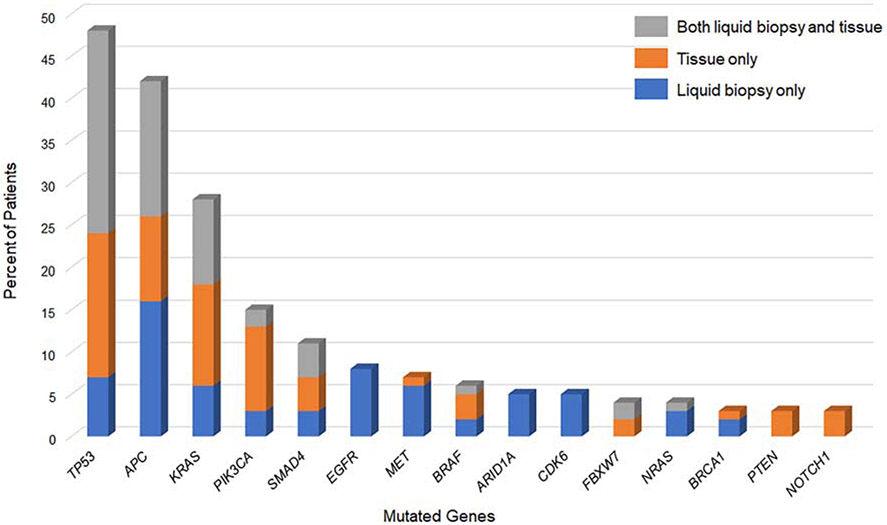
Multigene panel testing on the primary tumor and/or liver metastases was performed in 58 of the 63 patients. Figure 3 shows the distribution of somatic mutations identified in liquid biopsy only, tissue only, and both liquid biopsy and tissue. The most commonly mutated genes, TP53, APC, and KRAS, were identified in both liquid biopsy and tissue among 38%, 25%, and 16% of patients, respectively. A KRAS mutation was detected in tissue but not liquid biopsy in 2 patients. Conversely, KRAS or NRAS mutation was identified in liquid biopsy but not tissue in 8 patients. In these 8 patients, ctDNA analysis was performed > 1 year after tissue genotyping, and anti-EGFR therapy was administered in the interim. Four patients had a single gene mutation in liquid biopsy samples not identified in tissue; the 4 single gene mutations were RET, MTOR, EGFR, and GNAS.
Figure 3.
Prevalence of gene mutations in liquid biopsy and tumor tissue among 63 patients undergoing resection of colorectal liver metastases. Rare gene mutations, identified in < 5% of patients, not shown.
Association between liquid biopsy and survival
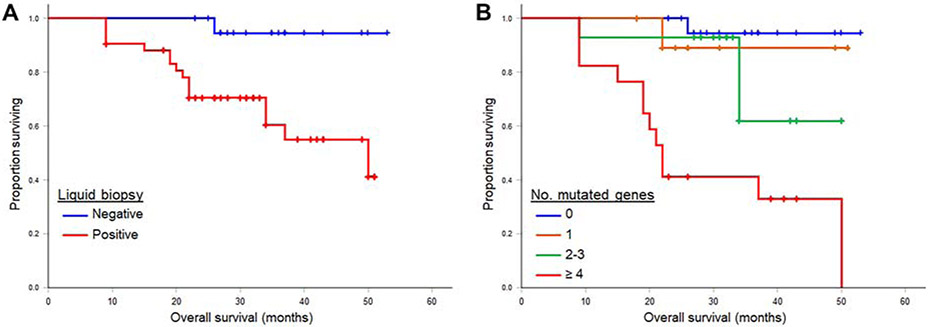
Two-year OS from date of liver resection was significantly lower among patients with a positive liquid biopsy (70% versus 100%, p = 0.005) (Fig. 4A). When further stratified by number of gene mutations, the presence of ≥ 4 mutations was associated with significantly lower 2-year OS of 41% (p < 0.001) (Fig. 4B). Results of univariable and multivariable analyses of the predictors of survival are shown in Table 2. Factors significantly associated with worse survival were resection margin and ≥ 4 gene mutations on liquid biopsy. Size of liver metastasis was marginally significant (p = 0.06). On multivariable analysis, resection margin and ≥ 4 gene mutations on liquid biopsy remained independent predictors of OS.
Figure 4.
Overall survival of 63 patient after resection of colorectal liver metastases, by (A) postoperative liquid biopsy status and (B) number of mutated genes in liquid biopsy samples.
Table 2.
Multivariable Overall Survival Analysis Of 63 Patients Undergoing Resection of Colorectal Liver Metastases
| Variable | 2-year overall survival, % |
Univariable p value |
Multivariab le p value |
Hazard ratio (95% CI) |
|---|---|---|---|---|
| Age | – | 0.48 | ||
| Sex | 0.88 | |||
| Male | 77 | |||
| Female | 84 | |||
| Primary tumor lymph node status | 0.82 | |||
| Positive | 81 | |||
| Negative | 75 | |||
| Synchronous CLM | 0.74 | |||
| Yes | 80 | |||
| No | 82 | |||
| Extrahepatic metastases | 0.57 | |||
| Yes | 83 | |||
| No | 77 | |||
| Preoperative CEA | – | 0.25 | ||
| Surgical margin | < 0.001 | |||
| R0 | 90 | Ref | ||
| R1 | 87 | 0.80 | ||
| R2 | 25 | 0.006 | 4.95 (1.58-15.52) | |
| Size of largest liver metastasis | – | 0.06 | 0.96 | |
| Number of liver metastases | 0.87 | |||
| Solitary | 83 | |||
| Multiple | 80 | |||
| Major hepatectomy | 0.61 | |||
| Yes | 88 | |||
| No | 78 | |||
| ≥ 4 gene mutations on liquid biopsy | < 0.001 | 0.001 | 6.60 (2.07-21.06) | |
| Yes | 41 | |||
| No | 96 |
CEA, carcinoembryonic antigen; CLM, colorectal liver metastasis; Ref, reference
Associations between first postoperative liquid biopsy and clinicopathologic factors
A positive postoperative liquid biopsy was not significantly associated with the following clinicopathologic factors: node-positive primary, number and size of liver metastases, CEA > 200 ng/ml, disease-free interval < 12 months between diagnosis of primary tumor and CLM, and extrahepatic disease. All 8 patients who underwent R2 resection had a positive liquid biopsy, including 6 with ≥ 4 gene mutations. Resection margin was significantly associated with the presence of ≥ 4 gene mutations on liquid biopsy, detected in 21% (8/39), 19% (3/16), and 75% (6/8) of patients with R0, R1, and R2 margins, respectively (p = .005).
First postoperative liquid biopsy and recurrence after CLM resection
Liquid biopsy results were not significantly associated with disease recurrence in the liver, lungs, or peritoneum. Among 11 patients with recurrence in abdominal and/or retroperitoneal lymph nodes, 8 patients had ≥ 4 gene mutations on liquid biopsy, compared with 3 patients with 1–3 gene mutations (p = .041).
Correlations between liquid biopsy, CEA, and CT imaging
Sixteen of the 63 patients underwent serial liquid biopsies, resulting in an additional 37 liquid biopsy samples. For all 63 patients, there were 100 liquid biopsy with matched serum CEA and CT results. Metastatic disease was identified in 74 of the 100 CT scans, either at the initial postoperative restaging scan or subsequent follow-up scans. CT evidence of disease correlated with positive liquid biopsy in 77% of samples (p < 0.001) and CEA > 3 ng/ml in 45% of samples (p = 0.22).
DISCUSSION
To our knowledge, the current study is the first to analyze the impact of a 70-gene liquid biopsy panel on patient survival after CLM resection and associations with CT imaging and serum CEA. We report that patients with a positive liquid biopsy after CLM resection had significantly worse 2-year OS than those with a negative liquid biopsy. The number of gene mutations detected in liquid biopsy samples correlated with survival, with lower OS observed with ≥ 4 mutations, compared with a single gene mutation. A positive liquid biopsy, but not elevated serum CEA, was significantly associated with CT evidence of metastatic disease.
Our study results showing worse survival with positive liquid biopsy after CLM resection are consistent with data from non-metastatic patients undergoing resection of stages I-III colorectal cancer, in whom ctDNA-positivity on postoperative day 30 conferred a 7-fold higher rate of disease relapse.(8) After CLM resection, Scholer and colleagues observed a 100% relapse rate for patients with positive liquid biopsy 3 months postoperatively, compared with 50% relapse with negative liquid biopsy.(13) In the current study, resection margin after CLM resection was significantly associated with ≥ 4 gene mutations on liquid biopsy, observed in 75% of patients with R2 margin, compared with 21% of patients with R0 margin. Taken together, these data suggest that after resection of localized and metastatic colorectal cancer, a positive liquid biopsy correlates with the presence of residual disease.
We observed that the presence of ≥ 4 gene mutations, occurring in 40% of patients with a positive liquid biopsy, was associated with significantly worse OS compared with a solitary gene mutation. The lower survival with multiple mutations in liquid biopsy samples is congruent with emerging data on the importance of co-mutations in tissue samples after CLM resection, particularly the co-occurrence of RAS, TP53, and SMAD4 mutations.(5, 14) Similarly, in a study of 298 patients with metastatic cancer, including liver, pancreas, and colorectal cancer, the presence of more than one nonsynonymous mutation in cfDNA correlated with significantly worse OS.(11) The inferior prognosis with multiple mutations is attributed to increased tumor heterogeneity and the emergence of subclones resistant to systemic therapy.
Our data demonstrate a significant correlation between CT evidence of metastatic disease and a positive liquid biopsy but not serum CEA levels. In a study of patients undergoing surgery for stages I–III colorectal cancer, serial liquid biopsies revealed disease recurrence up to 16.5 months earlier than standard-of-care radiologic imaging.(8) Given the small number of patients who underwent serial liquid biopsy in our study, the ability of liquid biopsy to detect early recurrence was not analyzed.
Until recently, the use of liquid biopsy was limited by the need to sequence known individual gene mutations present in tissue. In colorectal cancer, several genes have been identified that are recurrently somatically mutated, including TP53, RAS, and APC. In this analysis, we used a ctDNA detection technology that analyzes mutations in 70 genes. We observed 83% concordance in RAS mutation status between liquid biopsy and tissue. Prior reports have shown concordance rates in RAS mutation status between liquid biopsy and tissue ranging between 71% to 100%.(15, 16) In this study, 8 patients had RAS mutations detected in ctDNA but not tumor tissue. All 8 patients underwent liquid biopsy testing > 1 year after tissue genotyping and received anti-EGFR therapy in the interim. The emergence of RAS mutant subclones or new RAS mutations arising from ongoing mutagenesis are increasingly recognized as important causes of acquired resistance to anti-EGFR therapy.(17) Other causes of discordance between mutations detected in liquid biopsy versus tissue include tumor location, sample volume, amount of ctDNA, and assay sensitivity.(7) In our study, discordant results between liquid biopsy and tissue are partly attributable to varying levels of tumor burden among patients who underwent ctDNA analysis at different time intervals after hepatectomy.
In the current report, 4 patients had liquid biopsy samples with a single gene mutation that was not identified in tumor tissue. False-positive liquid biopsy results can result from a phenomenon known as clonal hematopoiesis of indeterminate potential (CHIP), which results in somatic mutations released by hematopoietic stem cells related to aging, and not colorectal cancer.(7) Thus, liquid biopsy results with a single gene mutation not identified in tissue should be interpreted with caution.
According to our study results, liquid biopsies are currently used to detect minimal residual disease, screen for clinical trials, and guide systemic therapy, including detection of RAS mutant clones driving acquired resistance to anti-EGFR therapy. Further research is needed to standardize ctDNA assays across institutions and to validate interpretation of results, particularly false-positive mutations. Future uses of ctDNA include counseling patients on adjuvant therapy after CLM resection and determining the extent and frequency of postoperative surveillance. In patients with negative radiologic findings, a positive liquid biopsy may signal the need for additional imaging or closer follow-up.
Limitations include the retrospective analysis of 63 patients who underwent postoperative ctDNA analysis, representing a small subset (17%) of the 375 patients undergoing CLM resection during the study period. Therefore, the study population was enriched for patients with suspected or radiologically-confirmed disease recurrence, and the time interval between CLM resection and liquid biopsy analysis varied widely, between 1–45 months. Prospective studies with specified time-points for blood collection are needed to assess ctDNA kinetics after CLM resection. Another limitation is analysis restricted to postoperative ctDNA detection. A prior study from our institution evaluated a 23-gene liquid biopsy panel among 54 patients treated with chemotherapy before CLM resection.(18) After neoadjuvant chemotherapy, the ctDNA mutation detection rate was 80%. In addition, a larger sample size is needed to determine associations between postoperative ctDNA and patterns of disease recurrence. Finally, we did not stratify liquid biopsies by variant allele frequency, which has been shown to correlate with tumor burden and survival.(11)
CONCLUSIONS
In this cohort of patients undergoing CLM resection, positive postoperative liquid biopsy was associated with significantly worse overall survival, particularly when ≥ 4 gene mutations were detected. A positive liquid biopsy correlated with CT evidence of disease recurrence. These data support continued investigation of liquid biopsy in CLM for non-invasive molecular profiling, detection of minimal residual disease, and prognostication.
Acknowledgments
Support: This work was supported by the National Institutes of Health / National Cancer Institute (P30-CA016672).
Footnotes
Presented virtually at the Western Surgical Association 128th Scientific Session, November 2020.
Disclosure Information: Nothing to disclose.
This work was presented at the Western Surgical Association 2020 Annual Meeting.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Chun YS, Passot G, Yamashita S, et al. Deleterious Effect of RAS and Evolutionary High-risk TP53 Double Mutation in Colorectal Liver Metastases. Ann Surg. 2019. May;269(5):917–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdalla EK, Vauthey JN, Ellis LM, et al. Recurrence and outcomes following hepatic resection, radiofrequency ablation, and combined resection/ablation for colorectal liver metastases. Ann Surg. 2004. June;239(6):818–25; discussion 25-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Jong MC, Pulitano C, Ribero D, et al. Rates and patterns of recurrence following curative intent surgery for colorectal liver metastasis: an international multi-institutional analysis of 1669 patients. Ann Surg. 2009. September;250(3):440–8. [DOI] [PubMed] [Google Scholar]
- 4.Nordlinger B, Sorbye H, Glimelius B, et al. Perioperative FOLFOX4 chemotherapy and surgery versus surgery alone for resectable liver metastases from colorectal cancer (EORTC 40983): long-term results of a randomised, controlled, phase 3 trial. Lancet Oncol. 2013. November;14(12):1208–15. [DOI] [PubMed] [Google Scholar]
- 5.Kawaguchi Y, Lillemoe HA, Panettieri E, et al. Conditional Recurrence-Free Survival after Resection of Colorectal Liver Metastases: Persistent Deleterious Association with RAS and TP53 Co-Mutation. J Am Coll Surg. 2019. September;229(3):286–94 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gruenberger T, Bridgewater J, Chau I, et al. Bevacizumab plus mFOLFOX-6 or FOLFOXIRI in patients with initially unresectable liver metastases from colorectal cancer: the OLIVIA multinational randomised phase II trial. Ann Oncol. 2015. April;26(4):702–8. [DOI] [PubMed] [Google Scholar]
- 7.Heitzer E, Haque IS, Roberts CES, Speicher MR. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. 2019. February;20(2):71–88. [DOI] [PubMed] [Google Scholar]
- 8.Reinert T, Henriksen TV, Christensen E, et al. Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients With Stages I to III Colorectal Cancer. JAMA Oncol. 2019. May 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bidard FC, Kiavue N, Ychou M, et al. Circulating Tumor Cells and Circulating Tumor DNA Detection in Potentially Resectable Metastatic Colorectal Cancer: A Prospective Ancillary Study to the Unicancer Prodige-14 Trial. Cells. 2019. May 28;8(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Couinaud C [Liver lobes and segments: notes on the anatomical architecture and surgery of the liver ]. Presse Med. 1954. May 5;62(33):709–12. [PubMed] [Google Scholar]
- 11.Pairawan S, Hess KR, Janku F, et al. Cell-free Circulating Tumor DNA Variant Allele Frequency Associates with Survival in Metastatic Cancer. Clin Cancer Res. 2020. April 15;26(8):1924–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luthra R, Patel KP, Routbort MJ, et al. A Targeted High-Throughput Next-Generation Sequencing Panel for Clinical Screening of Mutations, Gene Amplifications, and Fusions in Solid Tumors. J Mol Diagn. 2017. March;19(2):255–64. [DOI] [PubMed] [Google Scholar]
- 13.Scholer LV, Reinert T, Orntoft MW, et al. Clinical Implications of Monitoring Circulating Tumor DNA in Patients with Colorectal Cancer. Clin Cancer Res. 2017. September 15;23(18):5437–45. [DOI] [PubMed] [Google Scholar]
- 14.Kawaguchi Y, Kopetz S, Newhook TE, et al. Mutation Status of RAS, TP53, and SMAD4 is Superior to Mutation Status of RAS Alone for Predicting Prognosis after Resection of Colorectal Liver Metastases. Clin Cancer Res. 2019. October 1;25(19): 5843–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thierry AR, Pastor B, Jiang ZQ, et al. Circulating DNA Demonstrates Convergent Evolution and Common Resistance Mechanisms during Treatment of Colorectal Cancer. Clin Cancer Res. 2017. August 15;23(16):4578–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toledo RA, Cubillo A, Vega E, et al. Clinical validation of prospective liquid biopsy monitoring in patients with wild-type RAS metastatic colorectal cancer treated with FOLFIRI-cetuximab. Oncotarget. 2017. May 23;8(21):35289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012. June 28;486(7404):532–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Overman MJ, Vauthey JN, Aloia TA, et al. Circulating tumor DNA (ctDNA) utilizing a high-sensitivity panel to detect minimal residual disease post liver hepatectomy and predict disease recurrence. J Clin Oncol. 2017; 35(15_suppl):3522. [Google Scholar]