Abstract
Background
Soil salinity has been one of the biggest hurdles in achieving better crop yield and quality. Plant growth-promoting rhizobacteria (PGPR) are the symbiotic heterogeneous bacteria that play an important role in the recycling of plant nutrients through phytostimulation and phytoremediation. In this study, bacterial isolates were isolated from salt-polluted soil of Jhajjar and Panipat districts of Haryana, India. The potential salt-tolerant bacteria were screened for their PGPR activities such as phosphate solubilization, hydrogen cyanide (HCN), indole acetic acid (IAA) and ammonia production. The molecular characterization of potent isolates with salt tolerance and PGPR activity was done by 16S rDNA sequencing.
Results
Eighteen soil samples from saline soils of Haryana state were screened for salt-tolerant bacteria. The bacterial isolates were analyzed for salt tolerance ranging from 2 to 10%. Thirteen isolates were found salt tolerant at varied salt concentrations. Isolates HB6P2 and HB6J2 showed maximum tolerance to salts at 10% followed by HB4A1, HB4N3 and HB8P1. All the salt-tolerant bacterial isolates showed HCN production with maximum production by HB6J2. Phosphate solubilization was demonstrated by three isolates viz., HB4N3, HB6P2 and HB6J2. IAA production was maximum in HB4A1 (15.89) and HB6P2 (14.01) and least in HB4N3 (8.91). Ammonia production was maximum in HB6P2 (12.3) and least in HB8P1 (6.2). Three isolates HB6J2, HB8P1 and HB4N3 with significant salt tolerance, and PGPR ability were identified through sequencing of amplified 16SrRNA gene and were found to be Bacillus paramycoides, Bacillus amyloliquefaciens and Bacillus pumilus, respectively.
Conclusions
The salt-tolerant plant growth-promoting rhizobacteria (PGPR) isolated from saline soil can be used to overcome the detrimental effects of salt stress on plants, with beneficial effects of physiological functions of plants such as growth and yield, and overcome disease resistance. Therefore, application of microbial inoculants to alleviate stresses and enhance yield in plants could be a low cost and environmental friendly option for the management of saline soil for better crop productivity.
Supplementary Information
The online version contains supplementary material available at 10.1186/s43141-021-00186-3.
Keywords: Ammonia, HCN, Indole acetic acid, Phosphate solubilization, PGPR, Salt tolerant
Background
The world population is estimated approximately 7.8 billion. This is projected to rise up to 9.7 billion by 2050. The increase in the world’s population has increased the demand of food products [1, 2]. However, crop production per unit of land cultivated is unequal to meet the desired demand of food. The change in climate, loss of soil structure, nutrient degradation, draught and soil salinity are the major factors behind the decreased crop yield [2–4]. A worldwide loss of 50% land of the total land mass has been estimated by Food and Agricultural Organization (FAO) by the year 2050. Excess salt concentration in soil has negative impacts on plant growth and metabolism [5]. The salinity stress may also lead to generation of free radicals such as superoxide ions, hydrogen peroxide (H2O2), and singlet oxygen, decrease in plant defensive enzymes, imbalance in sodium hemostasis, decreased iron uptake, phenols and other trace elements [6–8].
In the recent time, various approaches have been applied to solve the problem of soil salinity and acidity [9–12]. There are some alternative methods available for retrieval of salt-affected soils such as phytoremediation and bioremediation [13–15]. The plant growth-promoting rhizobacteria (PGPR) are the heterogenous bacteria which are well known for their beneficial activities. There are a number of rhizobacteria such as genera of Alcaligenes, Pseudomonas, Azospirillum, Bacillus, Klebsiella, Azotobacter, Enterobacter, Burkholderia, Arthrobacter and Serratia that aid in plant growth through various mechanisms [16–18]. These bacteria act as biofertilizers and play an important role in the recycling of plant nutrients which help in phytostimulation and phytoremediation [19]. In the present study, bacteria were isolated from saline soils and screened for salt tolerance and PGP activities. PGP bacteria not only increase production of exopolysaccharides, siderophores, alter pH, modify toxic metals and solubilize phosphorus but also help in evacuating stress-alleviating metabolite 1-aminocyclopropane-1-carboxylic acid deaminase. They also play an important role in the secretion of indole-3-acetic acid (IAA), cytokinin and gibberellins and the development of antibiotic resistance [20–26].
Methods
Collection of soil samples
A total of 18 soil samples were collected from saline soils of Jhajjar and Panipat districts of Haryana. Ten out of eighteen soil samples were collected from the waterlogged saline area of the Jhajjar district of Haryana (28.37° North, 76.39° East). The remaining eight soil samples were collected from ICAR-CSSRI Nain farm, Panipat, Haryana (29.39° North, 76.97° East) (Fig. 1). All samples were collected in sterile polybags and divided in two parts, one for chemical analysis and second for isolation of PGP bacteria. Second part of the soil was stored at 4 °C till isolation.
Fig. 1.
Saline soil fields of Jhajjar, Haryana, India (a and b) and Panipat, Haryana (c and d)
Analysis of soil samples for pH and electrical conductivity
Soil samples were sieved by a 2-mm sieve and dried before analysis. Soil pH and electrical conductivity (EC) in soil:water ratios of 1:2 was determined as per the standard method described by Jackson [27].
Isolation and screening of bacteria for salt tolerance from saline soils
The isolation of salt-tolerant bacteria was done from the soil samples on different types of selective media such as Ashbys Mannitol Agar for Azotobacter and nonselective media such as Tryptone soya agar, Pikovasky’s Agar for phosphate-solubilizing bacteria and Jenson media for nitrogen-fixing bacteria [28]. The serial dilutions of the samples were plated on selected media and incubated for 48–72 h at 25 ± 2 °C. The pure culture of bacteria was obtained by repeated sub-culturing. The isolates were tested for their salt tolerance ability by spot inoculating the isolates on nutrient broth containing different concentrations of NaCl (2%, 4%, 6%, 8% and 10%), incubated at 25 ± 2 °C for 5 days and observed for growth.
Characterization of salt-tolerant bacterial isolates for plant growth-promoting activity
The isolates were further screened for PGP activities such as phosphate solubilization, production of indole acetic acid, hydrogen cyanide (HCN) and ammonia.
Phosphate solubilization
Phosphorus solubilizing activity of isolates was determined qualitatively according to the method described by Nautiyal [29]. Pikovskaya’s agar medium containing calcium tri phosphate (0.5%) as the inorganic form of phosphate was used in assay, 2.5 μl of the bacterial culture (O.D. 600) was streaked on the plates. All the plates were incubated at 28 °C for 4–5 days. Transparent halo zone around the bacterial colony indicates the phosphate solubilizing activity of the bacterial isolates.
Production of indole acetic acid (IAA)
Bacterial isolates were inoculated in nutrient broth enriched with tryptophan (1–2%) and incubated for 24 h at 28 °C on rotary shaker. Cultures were centrifuged at 10,000 rpm for 15 min. Salkowski’s reagent was mixed with supernatant, and incubated for 25 min at room temperature. Development of the pink color indicated IAA production and the quantitative estimation of IAA were performed by using the method described by Gordon and Weber [30].
Production of HCN
The salt-tolerant bacterial isolates were streaked on nutrient Agar medium containing 4.4 g/l glycine. Filter paper discs were dipped in 0.5% picric acid prepared in 2% sodium carbonate. The discs were placed in the lid of each petri plate and sealed. All the plates were incubated for 4 days at 28 °C. Color change of the filter paper from deep yellow to orange and orange to brown indicated the production of HCN by the bacterial isolate [31].
Production of ammonia
The salt-tolerant bacterial isolates were cultured in peptone water at 30 °C for 4 days. One milliliter of Nessler’s reagent was added to each tube. Development of faint yellow color indicated small amounts of ammonia production, and deep yellow to brownish color indicated maximum ammonia production [32].
Molecular characterization of bacterial isolates
Isolation of genomic DNA
Bacterial genomic DNA was isolated by taking 24-h-old broth culture (10 ml) of bacterial isolates. The broth culture was centrifuged at 2000 rpm for 10 min, and supernatant was discarded. One milliliter of freshly prepared extraction buffer was added to the pellet, and the resulting solution was transferred in a 2-ml Eppendorf tube, incubated at 65 °C in a water bath for 30 min, and equal volume of ice-cold solution of phenol: chloroform: isoamyl alcohol (25:24:1) was added to it. The solution was mixed well and centrifuged at 10,000 rpm for 10 min at 4 °C, and the upper aqueous layer was transferred to a new Eppendorf tube. Equal volume of ice-cold solution of phenol: chloroform: isoamyl alcohol (25:24:1) was added to this aqueous layer, and the step was repeated 3–4 times. In the final aqueous layer, the ice-cold absolute alcohol was added in excess to precipitate the bacterial gemomic DNA and the solution was centrifuged at 10,000 rpm at 4 °C for 10 min. The supernatant was discarded, and the pellet was washed with 100 μl of 70% (v/v) ethanol and centrifuged at 10,000 rpm for 15 min at 4 °C, the supernatant was discarded, and the pellet was dissolved in 50 μl of TE buffer (pH 8.0) and stored at – 20 °C for further studies.
Amplification and sequencing of 16S rRNA
Universal primers were used for the amplification of the 16S rRNA sequence of the selected salt-tolerant PGP isolates. The amplified product was purified and sequenced from Eurofins Genomics India Pvt. Ltd, Bengaluru. The sequence obtained were analysed and identified using BLAST search and were compared against bacterial 16S rRNA sequence available on NCBI database. The sequences were aligned by using Clustal W 1.74 followed by construction of neighbour joining phylogenetic tree, using MEGA4.
Results
Isolation of bacteria from saline soils of Haryana, India
In the present study, a total of eighteen soil samples from saline soils of Jhajjar and Panipat districts of Haryana state were screened for salt-tolerant bacteria (Fig. 1). Soil samples were evaluated for their pH and electrical conductivity. In Jhajjar district’s soil samples, electrical conductivity was maximum in HB5 (16.56) and minimum in HB9 (5.40). The pH of different soil samples varied from 8.61 (HB8) to 6.50 (HB1). In Panipat district’s soil samples, electrical conductivity was maximum in PS5 (10.01) and minimum in PS4 (2.33), whereas pH value ranged from 9.0 (PS5) to 8.21 (PS4) (Table 1). A total of 81 bacterial isolates were isolated from ten soil samples collected from salt-affected land of Jhajjar (Table 2). Sixteen isolates each were isolated from soil samples HB4 and HB6, whereas none of the bacteria were isolated from HB1. Maximum number of isolates was isolated on nutrient agar plates and least on Kings B agar media (Table 2). Twenty-six bacterial isolates from eight soil samples were isolated from soil samples of Panipat district (Table 3). Four isolates were isolated from each of PS2, PS4 and PS5, three isolated from PS5, PS6, PS7 and PS8 and two isolated from PS1. Maximum numbers of isolates were isolated on nutrient agar plates and least numbers on Ashby’s agar (Table 3).
Table 1.
Soil samples collected from Jhajjar (serial nos. 1 to 10) and Panipat districts (serial nos. 11–18) of Haryana, India along with their EC and pH analysis
| Sr. no. | Soil sample | Electrical conductivity (EC) dS/m | pH |
|---|---|---|---|
| 1 | HB1 | 13.84 | 6.50 |
| 2 | HB2 | 14.26 | 6.65 |
| 3 | HB3 | 13.96 | 6.72 |
| 4 | HB4 | 15.32 | 6.90 |
| 5 | HB5 | 16.56 | 6.84 |
| 6 | HB6 | 11.30 | 7.5 |
| 7 | HB7 | 10.92 | 8.12 |
| 8 | HB8 | 12.70 | 8.61 |
| 9 | HB9 | 5.40 | 7.9 |
| 10 | HB10 | 10.62 | 8.27 |
| 11 | PS1 | 5.04 | 8.90 |
| 12 | PS2 | 4.74 | 8.96 |
| 13 | PS3 | 3.04 | 8.24 |
| 14 | PS4 | 2.33 | 8.21 |
| 15 | PS5 | 10.01 | 9.00 |
| 16 | PS6 | 7.90 | 8.80 |
| 17 | PS7 | 12.10 | 8.86 |
| 18 | PS8 | 11.53 | 8.97 |
Table 2.
Details of bacteria isolated from soil samples of Jhajjar, Haryana, India
| S. no. | Soil sample | Growth of bacterial isolates on different media used | No. of bacterial isolates | |||||
|---|---|---|---|---|---|---|---|---|
| Nutrient agar (N) |
Jensen’s agar (J) |
Pikovaskaya agar (P) |
King’s B agar (K) |
Ashby’s agar (A) |
Triple sugar agar (T) |
|||
| 1 | HB1 | Undetected | Undetected | Undetected | Undetected | Undetected | Undetected | 0 |
| 2 | HB2 | Undetected | Undetected | HB2P2 | Undetected | Undetected | HB2T1 | 2 |
| 3 | HB3 |
HB3N1, HB3N2 HB3N3, HB3N4 |
HB3J1, HB3J2 |
HB3P1 HB3P2 |
Undetected | HB3A1 | HB3T1 | 10 |
| 4 | HB4 |
HB4N1, HB4N2 HB4N3, HB4N4 HB4N5 |
HB4J1, HB4J2 HB4J3, HB4J4 |
Undetected | Undetected | HB4A1 |
HB4T1, HB4T2 HB4T3, HB4T4 HB4T5, HB4T6 |
16 |
| 5 | HB5 | HB5N1, HB5N2 | Undetected | Undetected | Undetected | Undetected | HB5T1, HB5T2 | 4 |
| 6 | HB6 |
HB6N1, HB6N2 HB6N3 |
HB6J1, HB6J2 |
HB6P1, HB6P2 HB6P3 |
Undetected | HB6A1, HB6A2 |
HB6T1, HB6T2 HB6T3, HB6T4 HB6T5, HB6T6 |
16 |
| 7 | HB7 |
HB7N1, HB7N2 HB7N3 |
Undetected | HB7P1 | Undetected | HB7A1 |
HB7T1, HB7T2 HB7T3 |
8 |
| 8 | HB8 |
HB8N1, HB8N2 HB8N3 |
HB8J1, HB8J2 | HB8P1 | Undetected | HB8A1, HB8A2 | HB8T1, HB8T2 | 10 |
| 9 | HB9 |
HB9N1 HB9N2 |
HB9J1 HB9J2 |
HB9P1 HB9P2 |
Undetected |
HB9A1, HB9A2 HB9A3, HB9A4 |
HB9T1 | 11 |
| 10 | HB10 | HB10N1, HB10N2 | HBI0J1, HBI0J2 | Undetected | Undetected | Undetected | Undetected | 4 |
| Total no. of isolates | 24 | 14 | 10 | 0 | 11 | 22 | 81 | |
Table 3.
Details of bacteria isolated from soil samples of Panipat, Haryana, India
| Sr. no. | Soil samples | Growth of bacterial isolates on different media used | No. of bacterial isolates | |||||
|---|---|---|---|---|---|---|---|---|
| Nutrient agar (N) |
Jensen’s agar (J) |
Pikovaskaya agar (P) |
King’s B agar (K) |
Ashby’s agar (A) |
Triple sugar agar (T) |
|||
| 1 | PS1 | PS1N1 | Undetected | Undetected | PS1K1 | Undetected | Undetected | 2 |
| 2 | PS2 | PS2N2 | PS2J2 | Undetected |
PS2K1 PS2K2 |
Undetected | Undetected | 4 |
| 3 | PS3 | PS3N1 | PS3P1 | Undetected | PS3K1 | Undetected | Undetected | 3 |
| 4 | PS4 | PS4N1 | Undetected | PS4P1 | PS4K1 | Undetected | PS4T1 | 4 |
| 5 | PS5 | PS5N1 | Undetected | PS5P1 | PS5K1 | PS5A1 | Undetected | 4 |
| 6 | PS6 | PS6N1 | PS6J1 | Undetected | PS6K1 | Undetected | Undetected | 3 |
| 7 | PS7 | PS7N1 | Undetected | Undetected | PS7K1 | Undetected | PS7T1 | 3 |
| 8 | PS8 | PS8N1 | PS8J1 | PS8P1 | Undetected | Undetected | Undetected | 3 |
| Total no. of isolates | 8 | 4 | 3 | 8 | 1 | 2 | 26 | |
Screening of bacteria isolates for salt tolerance and PGPR potential
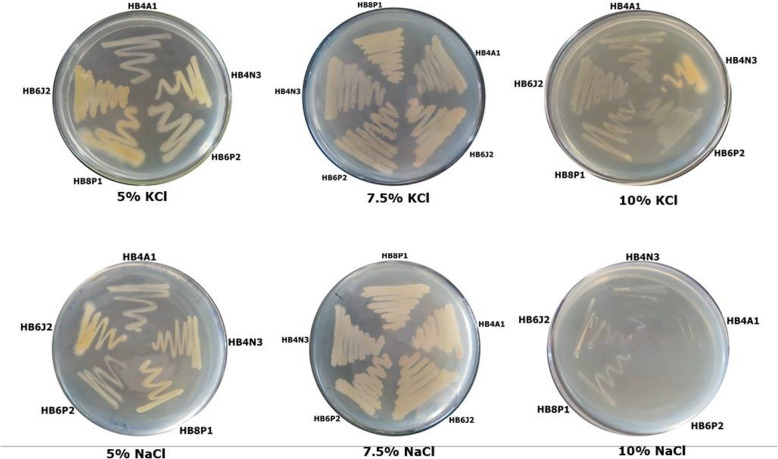
A total of thirteen isolates were found salt tolerant at varied salt concentrations. Isolates HB6P2 and HB6J2 have shown maximum tolerance to salts at 10% concentration followed by HB4A1, HB4N3 and HB8P1. All the isolates have shown salt tolerance up to 7.5% salt concentration; however, only five isolated bacteria were able to grow at 10% concentration (Fig. 2). The salt-tolerant bacteria were further screened for the ability to produce various plant growth-promoting traits such as HCN production, phosphate solubilization, IAA and ammonia production (Additional file 1: Fig. S1 and S2; Table 4). All the salt-tolerant bacteria demonstrated PGPR activities (Table 4). The phosphate solubilization was demonstrated by three salt-tolerant bacterial isolates viz; HB4N3, HB6P2 and HB6J2. The IAA production was maximum in HB4A1 (15.89) and HB6J2 (15.84) and least in HB4N3 (8.91). Ammonia production was maximum in HB6P2 (12.3) and least in HB8P1 (6.2) (Table 4).
Fig. 2.
Growth of bacterial isolates HB6J2, HB8P1, HB4A1, HB4N3 and Hb6P2 on media containing 5%, 7.5% and 10% KCl, and NaCl, respectively
Table 4.
Plant growth promoting (PGPR) activities of salt tolerant bacterial isolates
| Sr. no. | Isolate | Growth on media with 10% KCl & NaCl | HCN production activity | Phosphate solubilization activity | Production of IAA/ ammonia (in ppm) | |
|---|---|---|---|---|---|---|
| IAA | Ammonia | |||||
| 1 | HB4A1 | + | + | Undetected | 15.89 | 9.1 |
| 2 | HB4N3 | + + | + | + | 8.91 | 10.5 |
| 3 | HB6P2 | ++ | + | + | 14.01 | 12.3 |
| 4 | HB6J2 | ++ | +++ | + | 15.89 | 9.1 |
| 5 | HB8P1 | + | + | Undetected | 9.57 | 6.2 |
+: low; ++: medium; +++: high
Molecular identification of salt-tolerant bacterial isolates with PGPR activities
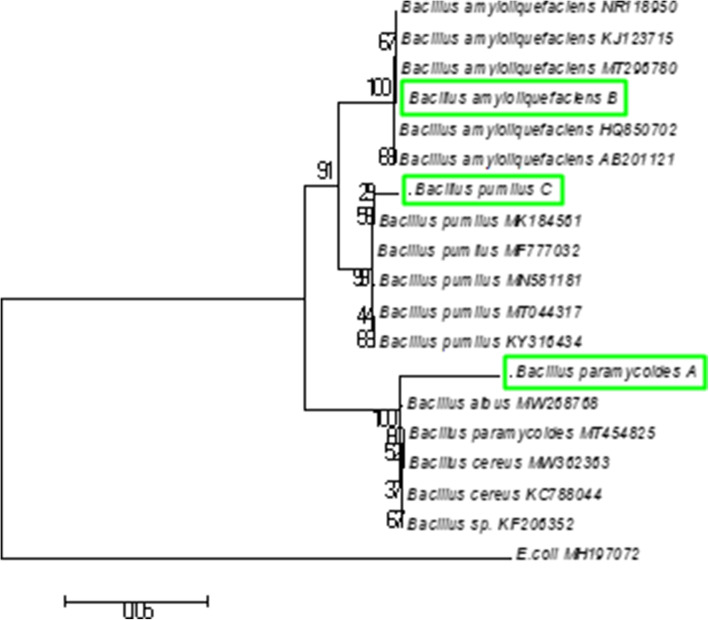
Identification of the salt-tolerant PGPR isolates was based on PCR amplification of 16S rRNA gene sequences The PGPR isolates HB6J2, HB8P1 and HB4N3 were further selected for molecular characterization based on their promising salt tolerance and plant growth-promoting properties. The 16S rRNA gene of the selected isolates was successfully amplified using PCR, and approximately 1500 bp of the amplified products were sequenced (Fig. 3). The BLAST-N comparison of the searched sequences in the NCBI nucleotide database revealed 99.87% similarity of the isolate HB6J2 with Bacillus paramycoides M9a1a (NCBI accession number: MT454825.1), 99.30% similarity of HB8P1 with Bacillus amyloliquefaciens strain K-8 (NCBI accession number: MT296780.1) and 98.88% similarity of HB4N3 with Bacillus pumilus strain EE107-PS (NCBI accession number: MN581181.1) (Additional file 1: Fig S3-S5). The phylogenetic tree of the selected bacterial isolates was constructed by using the neighbor-joining method (Fig. 4).
Fig. 3.
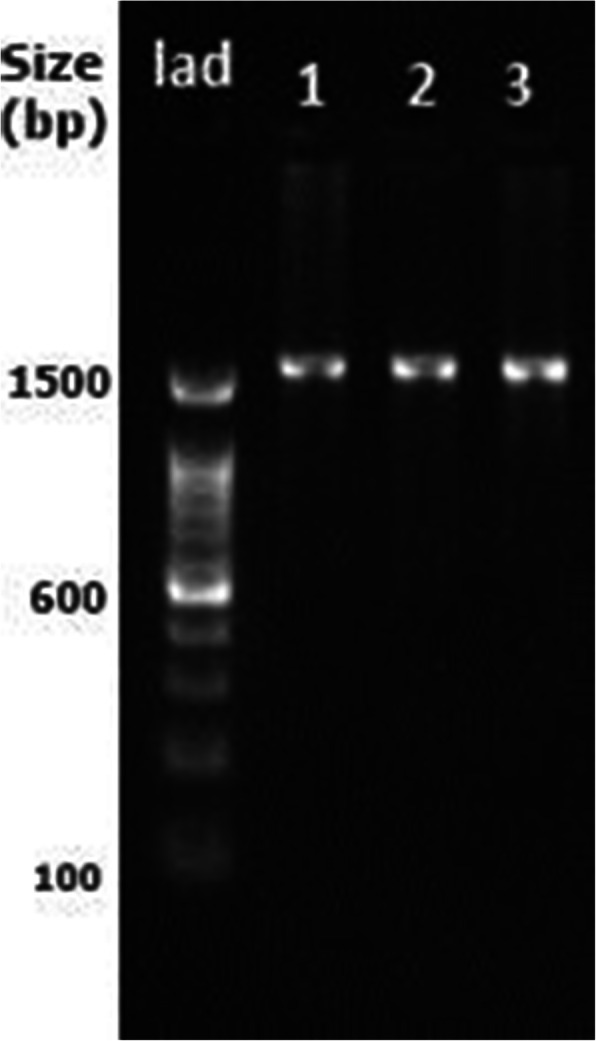
PCR amplified product of 16S rDNA of bacterial isolates. (Lad: molecular size marker 100 bp; lane 1: isolate HB6J2; lane 2: HB8P1; lane 3: HB4N3)
Fig. 4.
Phylogenetic tree constructed by the neighbor-joining method derived from analysis of the 16S rRNA gene sequence of salt-tolerant PGPR bacterial isolates and related sequences obtained from NCBI. Scale bar, 0.05 substitutions per nucleotide position. Bacillus paramycoides A (HB6J2), Bacillus amylofaciens B (HB8P1) and Bacillus pumilus C (HB4N3)
Discussion
The increase in soil salinization is one of the most common environmental threats for crop yield and quality [33, 34]. This problem has been regarded as one of the major hurdles in crop productivity in arid and semiarid regions [35, 36]. Sodic and saline soil is reducing the cultivable area for agriculture by 1–2% every year, thereby reducing food production [37, 38]. In the present study, all the soil samples collected from Panipat district are alkaline compared with soil samples of Jhajjar, while most of the Jhajjar soil samples have higher electrical conductivity than soil samples of Panipat (Table 1). The alkaline nature of the soil may affect the nutritional availability for plant growth. These variations in pH and EC might be responsible for the poor soil structure and infertility of the sample collection sites [2].
In the present study, a total of thirteen isolates were found salt tolerant at different salt concentrations. Isolates HB6P2 and HB6J2 have shown maximum tolerance to salts at 10% concentration followed by HB4A1, HB4N3 and HB8P1. In a similar study conducted by Mahmood and co-workers (2019), isolated, screened and characterized rhizosphere bacteria from the common ice plant Mesembryanthemum crystallinum L. In their study, 80 isolated strains out of 152 demonstrated tolerance to soil salinity. Bacterial strains of Streptomyces sp. PR-3 and Bacillus sp. PR-6 were effective against soil salinity (< 1250 mM NaCl) [39]. Zerrouk et al. [40], evaluated the capacity of Pseudomonas plecoglossicida strain Pp20 to mitigate the damages on maize roots caused by salt and aluminum. These workers found that Pp20 had the ability to grow at varied NaCl concentrations ranging from 50 to 600 mM. Their findings revealed a positive impact on stem weight, seminal roots, lateral roots and root length [41]. Similarly, study conducted by Ma and co-workers (2019), demonstrated the impact of Pseudomonas libanensis TR1 on Helianthus annuus and P. libanensis in exhibiting high resistance against saline stress (8%) [40]. However, in our study, all the isolated bacteria have shown salt tolerance up to 7.5% salt concentration, and only five isolates were able to grow at 10% concentration (Fig. 2). Several other workers tested Bacillus SB1 and Halobacillus SB2 strains for salt tolerance studies in combination with metals such zinc, aluminium and lead in the growth of Arachis hypogaea L under saline stress [42]. However, in our study, we did not perform salt tolerance studies in combination with metals.
The primary concern regarding saline soil is its impact on plant growth. Excessive salt concentration (more than 200 mM) may inhibit plant growth [43, 44]. In the present study, the bacterial isolates were further tested for their tolerance to salts at different salt concentrations ranging from 5 to 10% concentration (Fig. 2). The salt-tolerant bacteria were further screened for the ability to produce various plant growth-promoting traits such as HCN production, phosphate solubilization, IAA and ammonia production, and in the present study, all the salt-tolerant bacteria demonstrated PGPR activities (Table 4). In a similar study, conducted by Noori et al. [45], also isolated rhizobial and non-rhizobial drought and salinity-tolerant bacteria from the surface sterilized root nodules of alfalfa, grown in saline soils, and evaluated the effects of potential isolates on plant growth under salt stress. These workers co-inoculated alfalfa plant with bacterial strains such as Klebsiella sp. A36, K. cowanii A37, and rhizobial strain S. meliloti ARh29. In their results, it was demonstrated that Klebsiella sp. A36, Kcowanii A37 could deliver plant nitrogen and upsurge plant growth indices without rhizobial bacteria and nitrogen [46]. In the present study, phosphate solubilization was demonstrated by three salt-tolerant bacterial isolates viz; HB4N3, HB6P2 and HB6J2 (Table 4). In a similar study, Bacillus sp. (SB1) and Halobacillus sp. (SB2) isolated from groundnut rhizosphere had shown the ability to overcome the salt and metal stress [42]. In another study, the soil salinity mitigation by Streptomyces sp. strain PR-3 was done through phosphorus solubilization. These workers also demonstrated siderophore production by Bacillus sp. strain PR-6 and PR-3 and IAA production by PR-6 strain and suggested that these bacteria have the ability to promote growth in the common ice-plant [45].
Komaresofla et al. [47], demonstrated improved growth and salinity tolerance of the halophyte Salicornia sp. by co-inoculation with endophytic and rhizosphere bacteria. These workers evaluated drought tolerance of salt-tolerant isolates by performing 1-aminocyclopropane-1-carboxylate (ACC)-deaminase, IAA production and phosphate solubilization assays [47]. In the present study, all the isolated salt-tolerant bacteria have shown HCN production with maximum production by HB632 isolate. It was revealed that plant growth-promoting microbial isolates demonstrated escalation in ascorbate peroxidase (APX), superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX) and antioxidative enzymes under saline environment [48]. Sarkar et al. [48], studied halotolerant Enterobacter sp. strain P53 inoculated with rice seedlings. In their study, the bacteria demonstrated production of IAA, HCN, siderophore and antioxidant activity under salt stress [39]. We also obtained similar findings in our study; all the salt-tolerant bacterial isolates showed PGPR activities (Table 4).
The isolates HB6J2, HB8P1 and HB4N3 with significant salt-tolerant and PGPR ability were sequenced for amplified 16SrRNA gene and identified as Bacillus paramycoides, Bacillus amyloliquefaciens and Bacillus pumilus respectively. In a similar study conducted by Sultana et al. [49], three salt-tolerant bacterial isolates viz; Bacillus aryabhattai, Achromobacter denitrificans, and Ochrobactrum intermedium were identified through comparison of 16S rRNA gene sequences. These bacteria exhibited high atmospheric nitrogen fixation, phosphate solubilization, and indoleacetic acid production at concentration of 200 mmol/l salt [49]. In another study conducted by Tripathi et al. [50] studied the diversity of salt-tolerant bacteria present in the rhizosphere of Oryza sativa. These workers isolated fourteen bacterial isolates showing tolerance to 3% NaCl; however, these workers used restriction patterns produced by amplified 16S rDNA after digestion with restriction enzymes such as Sau3AI, AluI and RsaI for molecular characterization of bacterial isolates. Also, biodiversity among the strains was analyzed through the application of random amplified polymorphic DNA RAPD technique [50]. The salt-tolerant bacteria with PGPR activities may prove beneficial in the management of salt-affected agricultural fields for crop improvement. Alternatively, halophilic bacteria and their genes can be mined for salt-tolerant PGPR activities or salt tolerance traits can be transferred to crop plants [51–53].
Conclusion
The continuous increase in the population has also created more demand of food and hence agriculture productivity. Soil salinity has been one of the largest barriers in achieving better crop yield and quality. The much cost-effective methods such as application of microbes having PGPR activities can enhance plant growth, speed up seed germination, improve seedling emergence and protect plants. In the present study, a total of 18 soil samples from saline soils of Haryana state were screened for salt-tolerant bacteria. The bacterial isolates were further tested for their tolerance to different salt concentrations, and a total of thirteen isolates were found salt tolerant at varied salt concentrations. All the isolated salt-tolerant bacteria were further tested for the ability to produce PGPR activities such as phosphate solubilization, HCN, IAA and ammonia production. All the salt-tolerant bacteria demonstrated significant PGPR activities. The three isolates viz; HB6J2, HB8P1 and HB4N3 with good PGPR activities were identified, though sequencing of amplified 16SrRNA gene were found to be Bacillus paramycoides, Bacillus amyloliquefaciens and Bacillus pumilus respectively. The plant growth-promoting rhizobacteria (PGPR) isolated from saline soil can overcome the detrimental effects of salt stress on plants. Also, PGPR bacteria can positively impact physiological functions of plants such as growth and yield and overcome disease resistance. Therefore, application of microbial inoculants to alleviate stresses and enhance yield in plants could be a low-cost and environmentally friendly option for the management of saline soil for better crop productivity.
Supplementary Information
Additional file 1: Fig. S1 a. Formation of clear zone around the isolate HB6P2 represents the positive phosphate solubilization activity while isolates b. HB4A1 c. HB3A1 d. HB5N2 e. HB8P1 found negative for phosphate solubilization activity. Fig. S2 Change in the color of filter paper on the lid of the plate from deep yellow to orange -brown represents the positive HCN production. Positive HCN production by isolates a. HB6J2 b. HB6P2 c. negative HCN control (un-inoculated). Fig. S3 BLAST- N of PCR amplified 16Sr RNA gene sequence of HB6J2 with published sequences of NCBI database. Fig. S4 BLAST-N of PCR amplified 16Sr RNA gene sequence of HB8P1 with published sequences of NCBI database. Fig. S5 BLAST-N of PCR amplified 16Sr RNA gene sequence of HB4N3 with published sequences of NCBI database.
Acknowledgements
Authors would like to deeply acknowledge Vice Chancellor, Shoolini University, Bajhol, Solan, Himachal Pradesh, India for providing inspiration and infrastructural support for this study and all the members of ICAR-Central Soil Salinity Research Institute (CSSRI), Karnal for support.
Abbreviations
- PGPR
Plant growth-promoting rhizobacteria
- HCN
Hydrogen cyanide
- IAA
Indole acetic acid
- ICAR-CSSRI
Indian Council of Agricultural Research–Central Soil Salinity Research Institute
Authors’ contributions
ASh performed the experiments and drafted the manuscript; KD, ASo and MC conceived the study and participated in its design and coordination and helped to finalize the manuscript. The authors read and approved the final manuscript.
Funding
No funding was received for this study. CSSRI, Karnal, Haryana and Shoolini University, Solan, HP, India provided infrastructural and consumables support for the study.
Availability of data and materials
Additional data are provided as supplementary material.
Declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Arti Sharma, Email: sharmaarti2007@gmail.com.
Kamal Dev, Email: kamaldevbhardwaj1969@gmail.com.
Anuradha Sourirajan, Email: asourirajan@gmail.com.
Madhu Choudhary, Email: madhucssri@gmail.com.
References
- 1.Mesa-Marín J, Mateos-Naranjo E, Rodríguez-Llorente ID, Pajuelo E, Redondo-Gómez S. 15 synergic effects of rhizobacteria: increasing use of halophytes in a changing world. Halophytes and climate change: adaptive mechanisms and potential uses. 2019. p. 240. [Google Scholar]
- 2.Arti DK, Choudhary M, Sourirajan A. Salt tolerant bacteria for crop improvement in saline agriculture fields: development, challenges and opportunities. Plant Archives. 2020;20:7139–7155. [Google Scholar]
- 3.Khan N, Bano A, Rahman MA, Guo J, Kang Z, Babar MA. Comparative physiological and metabolic analysis reveals a complex mechanism involved in drought tolerance in chickpea (Cicer arietinum L.) induced by PGPR and PGRs. Sci Rep. 2019;9:1–19. doi: 10.1038/s41598-019-38702-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma Y, Rajkumar M, Oliveira RS, Zhang C, Freitas H. Potential of plant beneficial bacteria and arbuscular mycorrhizal fungi in phytoremediation of metal-contaminated saline soils. J Hazard Mater. 2019;379:120813. doi: 10.1016/j.jhazmat.2019.120813. [DOI] [PubMed] [Google Scholar]
- 5.El-Ramady H, Alshaal T, Abdelrahman H, El-Hady O (2019) Future soil issues. In: The soils of Egypt. Springer, Cham, Germany, pp 215–224
- 6.Mesa-Marín J, et al. PGPR reduce root respiration and oxidative stress enhancing spartina maritima root growth and heavy metal rhizoaccumulation. Front Plant Sci. 2018;9:1500. doi: 10.3389/fpls.2018.01500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korres NE, Varanasi VK, Slaton NA, Price AJ, Bararpour T (2019) Effects of salinity on rice and rice weeds: short-and long-term adaptation strategies and weed management. In: Advances in Rice Research for Abiotic Stress Tolerance. Woodhead Publishing: Sawston, UK, pp 159-176
- 8.Tully KL, Weissman D, Wyner WJ, Miller J, Jordan T. Soils in transition: saltwater intrusion alters soil chemistry in agricultural fields. Biogeochemistry. 2019;142:339–356. doi: 10.1007/s10533-019-00538-9. [DOI] [Google Scholar]
- 9.Costa SF, Martins D, Agacka-Mołdoch M, Czubacka A, de Sousa AS (2018) Strategies to alleviate salinity stress in plants. In: Salinity Responses and Tolerance in Plants. In: Salinity responses and tolerance in plants. Springer Nature: Berlin/Heidelberg, Germany, pp 307–337
- 10.Acuña Rodriguez IS, Hansen H, Gallardo J, Atala C, Molina-Montenegro MA. Antarctic extremophiles: biotechnological alternative to crop productivity in saline soils. Front Bioeng Biotechnol. 2019;7:22. doi: 10.3389/fbioe.2019.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jo S, Pak SH. The use of the freshwater snail Ampullaria tischbeini (Dohrn) as a biological control agent for remediation of salt-affected soil. Arch Agron Soil Sci. 2019;65:1677–1687. doi: 10.1080/03650340.2019.1575508. [DOI] [Google Scholar]
- 12.Gangwar P, Singh R, Trivedi M, Tiwari RK (2020) Sodic soil: management and reclamation strategies. In: Environmental concerns and sustainable development. Springer, Singapore, pp 175–190
- 13.Agrawal R, Verma A, Satlewal A (2018) Bioprospecting PGPR microflora by novel immunobased techniques. In: Crop improvement through microbial biotechnology. Oxford: Elsevier, pp 465–478
- 14.Kumar A, Verma JP (2019) The role of microbes to improve crop productivity and soil health. In: Ecological wisdom inspired restoration engineering. Springer Nature, Singapore, pp 249–265
- 15.Mishra P, Singh PP, Singh SK, Verma H (2019) Sustainable agriculture and benefits of organic farming to special emphasis on PGPR. In: Role of Plant Growth Promoting Microorganisms in Sustainable Agriculture and Nanotechnology. Elsevier, Woodhead publishing, Sawston, Cambridge, UK, pp 75-87
- 16.Kenneth OC, Nwadibe EC, Kalu AU, Unah UV (2018) Plant growth promoting rhizobacteria (PGPR): a novel agent for sustainable food production. Am J Agric Biol Sci 14 : 35-54
- 17.Meena VS (2018) Role of rhizospheric microbes in soil. Springer, Heidelberg Germany, Volume 1
- 18.Yasmeen T et al (2019) Ameliorative capability of plant growth promoting rhizobacteria (PGPR) and arbuscular mycorrhizal fungi (AMF) against salt stress in plant. In: Plant abiotic stress tolerance. Springer: Cham, Germany, pp 409–448
- 19.Zhang S, Fan C, Wang Y, Xia Y, Xiao W, Cui X (2018) Salt-tolerant and plant-growth-promoting bacteria isolated from high-yield paddy soil. Can J Microbiol 64:968–978 [DOI] [PubMed]
- 20.Paul B, Mohanta S, Deb B, Nath M (2019) In silico 16s rRNA phylogenetic analysis of extremely tolerant and resistant plant growth promoting rhizobacteria. J Glob Biosci 8:6021–6033
- 21.Mousavi SM, Motesharezadeh B, Hosseini HM, Alikhani H, Zolfaghari AA (2018) Root-induced changes of Zn and Pb dynamics in the rhizosphere of sunflower with different plant growth promoting treatments in a heavily contaminated soil. Ecotoxicol Environ Saf 147:206–216 [DOI] [PubMed]
- 22.Kadmiri IM, Chaouqui L, Azaroual SE, Sijilmassi B, Yaakoubi K, Wahby I (2018) Phosphate-solubilizing and auxin-producing rhizobacteria promote plant growth under saline conditions. Arab J Sci Eng 43:3403–3415
- 23.Bharti N, Barnawal D (2019) Amelioration of salinity stress by PGPR: ACC deaminase and ROS scavenging enzymes activity. In: PGPR amelioration in sustainable agriculture. Elsevier, Woodhead publishing, Sawston, Cambridge, UK,pp 85–106
- 24.Yousef NM. Capability of plant growth-promoting rhizobacteria (PGPR) for producing indole acetic acid (IAA) under extreme conditions. Eur J Biol Res. 2018;8:174–182. [Google Scholar]
- 25.Barnawal D, Singh R, Singh RP (2019) Role of plant growth promoting rhizobacteria in drought tolerance: regulating growth hormones and osmolytes. In: PGPR amelioration in sustainable agriculture. Elsevier BV: Amsterdam, The Netherlands, pp 107–128
- 26.Li J, Liu M. Biological features and regulatory mechanisms of salt tolerance in plants. J Cell Biochem. 2019;120:10914–10920. doi: 10.1002/jcb.28474. [DOI] [PubMed] [Google Scholar]
- 27.Jackson ML. Soil chemical analysis. New Delhi: Prentice Hall of India Pvt. Ltd; 1973. pp. 38–56. [Google Scholar]
- 28.Rajput LU, Imran A, Mubeen FA, Hafeez FY. Salt-tolerant PGPR strain Planococcus rifietoensis promotes the growth and yield of wheat (Triticum aestivum L.) cultivated in saline soil. Pak J Bot. 2013;45(6):1955–1962. [Google Scholar]
- 29.Nautiyal CS. An efficient microbiological growth medium for screening phosphorus solubilizing microorganisms. FEMS Microbiol Lett. 1999;170:2017–2021. doi: 10.1111/j.1574-6968.1999.tb13383.x. [DOI] [PubMed] [Google Scholar]
- 30.Gordon SA, Weber RP. Colorimetric estimation of indoleacetic acid. Plant Physiol. 1951;26(1):192–195. doi: 10.1104/pp.26.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bakker AW, Schipperes B. Microbial cyanide production in the rhizosphere in relation to potato yield reduction and Pseudomoas spp. - mediated plant growth stimulation. Soil Biol Biochem. 1987;19:451–457. doi: 10.1016/0038-0717(87)90037-X. [DOI] [Google Scholar]
- 32.Cappuccino JC, Sherman N. Microbiology: a laboratory manual. 3. New York: Benjamin/cummings Pub. Co.; 1992. pp. 125–179. [Google Scholar]
- 33.Rahman M, et al. Salinization in large river deltas: drivers, impacts and socio-hydrological feedbacks. Water Security. 2019;6:100024. doi: 10.1016/j.wasec.2019.100024. [DOI] [Google Scholar]
- 34.Tyerman SD, et al. Energy costs of salinity tolerance in crop plants. New Phytol. 2019;221:25–29. doi: 10.1111/nph.15555. [DOI] [PubMed] [Google Scholar]
- 35.Dong Y, Jiang C, Suri MR, Pee D, Meng L, Goldstein RER. Groundwater level changes with a focus on agricultural areas in the mid-Atlantic region of the United States, 2002–2016. Environ Res. 2019;171:193–203. doi: 10.1016/j.envres.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 36.Ivushkin K, et al. UAV based soil salinity assessment of cropland. Geoderma. 2019;338:502–512. doi: 10.1016/j.geoderma.2018.09.046. [DOI] [Google Scholar]
- 37.Etesami H, Beattie GA. Mining halophytes for plant growth-promoting halotolerant bacteria to enhance the salinity tolerance of non-halophytic crops. Front Microbiol. 2018;9:148. doi: 10.3389/fmicb.2018.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayes S, et al. Soil salinity limits plant shade avoidance. Curr Biol. 2019;29:1669–1676. doi: 10.1016/j.cub.2019.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarkar A, et al. A halotolerant Enterobacter sp. displaying ACC deaminase activity promotes rice seedling growth under salt stress. Res Microbiol. 2018;169:20e32. doi: 10.1016/j.resmic.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 40.Zerrouk IZ, Rahmoune B, Khelifi L, Mounir K, Baluska F, Ludwig-Müller J. Algerian Sahara PGPR confers maize root tolerance to salt and aluminum toxicity via ACC deaminase and IAA. Acta Physiol Plant. 2019;41:91. doi: 10.1007/s11738-019-2881-2. [DOI] [Google Scholar]
- 41.Mahmood A, Amaya R, Turgay OC, Yaprak AE, Taniguchi T, Kataoka R. High salt tolerant plant growth promoting rhizobacteria from the common ice-plant Mesembryanthemum crystallinum L. Rhizosphere. 2019;9:10–17. doi: 10.1016/j.rhisph.2018.10.004. [DOI] [Google Scholar]
- 42.Ma Y, Freitas H, Vosatka M. Beneficial microbes alleviate climatic stresses in plants. Front Plant Sci. 2019;10:595. doi: 10.3389/fpls.2019.00595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mukhtar S, Mirza BS, Mehnaz S, Mirza MS, Mclean J, Malik KA. Impact of soil salinity on the microbial structure of halophyte rhizosphere microbiome. World J Microbiol Biotechnol. 2018;34:136. doi: 10.1007/s11274-018-2509-5. [DOI] [PubMed] [Google Scholar]
- 44.Shahid SA, Zaman M, Heng L (2018) Introduction to soil salinity, sodicity and diagnostics techniques. In: Guideline for salinity assessment, mitigation and adaptation using nuclear and related techniques. Springer, Cham, Germany, pp 1–42
- 45.Noori F, Etesami H, Zarini HN, Khoshkholgh-Sima NA, Salekdeh GH, Alishahi F. Mining alfalfa (Medicago sativa L.) nodules for salinity tolerant non-rhizobial bacteria to improve growth of alfalfa under salinity stress. Ecotoxicol Environ Saf. 2018;162:129–138. doi: 10.1016/j.ecoenv.2018.06.092. [DOI] [PubMed] [Google Scholar]
- 46.Banik A, Pandya P, Patel B, Rathod C, Dangar M. Characterization of halotolerant, pigmented, plant growth promoting bacteria of groundnut rhizosphere and its in-vitro evaluation of plant-microbe protocooperation to withstand salinity and metal stress. Sci Total Environ. 2018;630:231–242. doi: 10.1016/j.scitotenv.2018.02.227. [DOI] [PubMed] [Google Scholar]
- 47.Komaresofla BR, Alikhani HA, Etesami H, Khoshkholgh-Sima NA. Improved growth and salinity tolerance of the halophyte Salicornia sp. by co-inoculation with endophytic and rhizosphere bacteria. Appl Soil Ecol. 2019;138:160–170. doi: 10.1016/j.apsoil.2019.02.022. [DOI] [Google Scholar]
- 48.Hmaeid N, Wali M, Mahmoud OM-B, Pueyo JJ, Ghnaya T, Abdelly C. Efficient rhizobacteria promote growth and alleviate NaCl-induced stress in the plant species Sulla carnosa. Appl Soil Ecol. 2019;133:104–113. doi: 10.1016/j.apsoil.2018.09.011. [DOI] [Google Scholar]
- 49.Sultana S, Paul SC, Parveen S, Alam S, Rahman N, Jannat B, Hoque S, Rahman MT, Karim MM (2019) Isolation and identification of salt-tolerant plant-growth-promoting rhizobacteria and their application for rice cultivation under salt stress. Can J Microbiol 66: 144-160 [DOI] [PubMed]
- 50.Tripathi AK, Verma SC, Ron EZ. Molecular characterization of a salt-tolerant bacterial community in the rice rhizosphere. Res Microbiol. 2002;153(9):579–584. doi: 10.1016/S0923-2508(02)01371-2. [DOI] [PubMed] [Google Scholar]
- 51.Gupta S, Sharma P, Dev K, Srivastava M, Sourirajan A (2015) A diverse group of halophilic bacteria exist in Lunsu, a natural salt water body of Himachal Pradesh, India. SpringerPlus 4:274 [DOI] [PMC free article] [PubMed]
- 52.Vaidya S, Dev K, Sourirajan A (2018) Distinct osmoadaptation strategies in the strict halophilic and halotolerant bacteria isolated from Lunsu salt water body of North West Himalayas. Curr Microbiol 75:888-895 [DOI] [PubMed]
- 53.Gupta S, Sharma P, Dev K, Sourirajan A (2020) Isolation of gene conferring salt tolerance from halophilic bacteria of Lunsu, Himachal Pradesh, India. J Genet Eng Biotechnol 18:57 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1 a. Formation of clear zone around the isolate HB6P2 represents the positive phosphate solubilization activity while isolates b. HB4A1 c. HB3A1 d. HB5N2 e. HB8P1 found negative for phosphate solubilization activity. Fig. S2 Change in the color of filter paper on the lid of the plate from deep yellow to orange -brown represents the positive HCN production. Positive HCN production by isolates a. HB6J2 b. HB6P2 c. negative HCN control (un-inoculated). Fig. S3 BLAST- N of PCR amplified 16Sr RNA gene sequence of HB6J2 with published sequences of NCBI database. Fig. S4 BLAST-N of PCR amplified 16Sr RNA gene sequence of HB8P1 with published sequences of NCBI database. Fig. S5 BLAST-N of PCR amplified 16Sr RNA gene sequence of HB4N3 with published sequences of NCBI database.
Data Availability Statement
Additional data are provided as supplementary material.