Abstract
This study aims to quantify antibiotic consumption for suspected respiratory tract superinfections in COVID-19 patients, while investigating the associated drivers of antibiotic prescribing in light of the current signs of antibiotic overuse. Adult patients with a positive COVID-19 diagnosis admitted to a Belgian 721-bed university hospital were analyzed retrospectively (March 11th–May 4th, 2020), excluding short-term admissions (< 24 h). Antibiotic prescriptions were analyzed and quantified, using Defined Daily Doses (DDD) per admission and per 100 bed days. Possible drivers of antibiotic prescribing were identified by means of mixed effects logistic modelling analysis with backwards selection. Of all included admissions (n = 429), 39% (n = 171) were prescribed antibiotics for (presumed) respiratory tract superinfection (3.6 DDD/admission; 31.5 DDD/100 bed days). Consumption of beta-lactamase inhibitor–penicillin combinations was the highest (2.55 DDD/admission; 23.3 DDD/100 bed days). Four drivers were identified: fever on admission (OR 2.97; 95% CI 1.42–6.22), lower SpO2/FiO2 ratio on admission (OR 0.96; 95% CI 0.92–0.99), underlying pulmonary disease (OR 3.04; 95% CI 1.12–8.27) and longer hospital stay (OR 1.09; 95% CI 1.03–1.16). We present detailed quantitative antibiotic data for presumed respiratory tract superinfections in hospitalized COVID-19 patients. In addition to knowledge on antibiotic consumption, we hope antimicrobial stewardship programs will be able to use the drivers identified in this study to optimize their interventions in COVID-19 wards.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11739-021-02790-0.
Keywords: Antimicrobial stewardship, COVID-19, Respiratory tract infection, Pneumonia, Co-infection, Superinfection
Introduction
Antibiotic overconsumption and misuse are associated with detrimental effects, including the occurrence of multiple adverse drug events. Consequences of inappropriate antibiotic therapy involve direct patient harm resulting in organ dysfunction and/or infection with multi-drug resistant organisms. Long-term effects concern the increase in antimicrobial resistance (AMR) on a global level [1].
Since the emergence of the Coronavirus-19 infectious disease (COVID-19) pandemic, a rising number of reports have underlined the risk of rising AMR due to antibiotic overuse in COVID-19 patients. In addition, many physicians and pharmacists involved in antimicrobial stewardship had to shift their activities to the containment of the COVID-19 crisis [2–5].
In contrast to this observed overconsumption of antibiotics, very few bacterial superinfections have been documented in COVID-19 patients in the literature, especially in patients admitted to COVID wards and in the first days of admission. Bacterial superinfections ranged between 4.7% and 14.6% for hospital-acquired superinfections or superinfections in the intensive care unit (ICU) [6–9]. In contrast, higher numbers of bacterial superinfections have been reported with other viral infections, such as influenza, ranging between 11 and 35% of all influenza-related admissions [10]. The reason for this difference in susceptibility for bacterial superinfection in COVID-19 versus influenza is still unclear.
To date, no detailed quantitative data and few data concerning drivers of antibiotic prescriptions for COVID-19 respiratory superinfections have been published [11]. This retrospective study investigates the use of antibiotics for suspicion of COVID-19 respiratory tract superinfection in a quantitative manner and analyses the drivers associated with initiation of antibiotic treatment.
Materials and methods
All patients aged 18 years or older and admitted between March 11th and May 4th 2020 in the UZ Brussel, a Belgian university hospital comprising 721-beds, in the context of a COVID-19 infection, were suitable for this retrospective analysis. COVID-19 infection was diagnosed either by clinical suspicion with positive SARS-CoV-2 polymerase chain reaction (PCR) on nasopharyngeal swab or by clinical suspicion together with strongly suggestive chest CT scan. Patients admitted for less than 24 h were excluded from this retrospective cohort analysis. Readmissions were included in this retrospective analysis, without reporting stable demographic variables more than once in the result section. The data were extracted from the patient’s electronic medical file following a structured registration method. These data included demographics, comorbidities, information related to the patient’s hospital stay (e.g., length of stay and survival), symptoms, laboratory results, severity of COVID-19 (amount of oxygen needed, mechanical ventilation, ICU hospitalization), microbiological data and data related to antibiotic prescriptions. The need for supplemental oxygen was documented by calculating the pulsed finger saturation to fraction of inspired oxygen ratio (SpO2/FiO2), a validated surrogate marker for partial pressure of oxygen to fraction of inspired oxygen ratio among mechanically ventilated patients with acute respiratory distress (ARDS), but also a predictor of ARDS [12].
Fever was defined as tympanic temperature above 38 °C. A respiratory sample with significant result was defined as respiratory samples with culture results positive for a respiratory tract pathogen, in the presence of leukocytes on semi-quantitative analysis and without contamination by epithelial cells. Mycoplasma pneumoniae respiratory tract (super)infections were identified by the presence of compatible clinical signs and symptoms together with a fourfold increase in the specific antibody titre or the presence of a positive PCR on sputum or oropharyngeal swab.
The primary outcome was the quantity of antibiotic prescriptions for a suspected or confirmed bacterial respiratory tract co-infection in COVID-19 patients. Antibiotic prescriptions belonging to the ‘Anatomical Therapeutic Chemical’ classification system group J01 [13] were identified using an automated search engine, incorporated in the electronic hospital information system. Based on the careful examination of the patient’s record by an infectious disease specialist (see list in Supplementary Material 1), only antibiotic prescriptions targeting (presumed) respiratory tract superinfections were included. Azithromycin was excluded from analysis when it was used for its presumed immunomodulatory role [14]. Erythromycin was excluded because of its frequent use as a gastroprokinetic in the ICU. Non-respiratory antibiotics were registered descriptively, but not reported for the purpose of this study. For patients with a suspicion of respiratory tract superinfection, the Daily Defined Dose (DDD) was calculated as defined by the World Health Organization [13], both for single antibiotic formulations as well as for the total amount of administered antibiotics. Subsequently, we converted these numbers into two indicators: DDDs/admission and DDDs/100 hospital days.
Possible drivers of antibiotic prescription for a presumed or confirmed respiratory tract superinfection were investigated as secondary endpoints. The following variables of interest were included in the final analysis: demographics (age, sex, body mass index), laboratory variables (neutrophilic count, C-reactive protein (CRP), lymphopenia (< 1000 lymphocytes/mm3) and ferritin level at admission), the presence of fever at admission, symptoms present before or at admission (dyspnoea, cough, chest pain and fever), comorbidities (pre-existing lung disease, immunosuppressed state, congestive heart failure or ischaemic heart disease, diabetes mellitus, Charlson Comorbidity Index (CCI) [15], markers of severe disease (SpO2/FiO2 at admission, oxygen need at admission, quick Sequential Organ Failure assessment (qSOFA) score [16] and need for mechanical ventilation), the presence of positive respiratory microbiological samples, ICU stay during admission and duration of admission. Quantification of antibiotic prescriptions and analysis of the drivers was performed separately for the ICU and ward admissions, to avoid selection bias.
Data analysis
Descriptive statistics were applied to characterize the cohort, reporting percentages and medians with interquartile ranges (IQR) for patients on an internal medicine COVID-19 ward and patients with an ICU admission during their hospital stay. Baseline characteristics were compared between those two groups using a Chi-square or Fisher Exact test for dichotomous variables and Mann–Whitney U testing for continuous variables. p values were adjusted for multiple testing. The hospital pharmacist (SW) calculated the DDDs for each antibiotic. In a final model to identify hidden drivers, a mixed effects logistic regression was performed with a random intercept per admission to deal with clustering within patients. First, separate mixed effects logistic regression models were fitted for the prescription of antibiotics in function of the location where the patient was treated with antibiotics (ward/ICU) together with each previously defined variable of interest. Since patients, treated on the ward and in ICU, have a significantly different disease process, this was a first factor to adjust for. Variables of interest with a Wald test significance of less than 0.10 were added to the semi-final model. Before the semi-final model was built up, the authors tried to eliminate parts of the multicollinearity by including only the most interesting parameter in the semi-final model from a clinical perspective. Afterwards, all parameters of interest that were not highly correlated, together with the location of the patient (ward/ICU), were added in a semi-final model. Lastly, a backwards selection procedure was performed on this semi-final model with the final selected variables. Multicollinearity among variables was taken care of by looking at correlations between variables based on the likelihood of the models. The results with a p value of < 0.05 were considered statistically significant.
Ethical considerations
The study was approved by the local Ethical Committee prior to data collection (B.U.N. 1432020000092) and was carried out in accordance with the ethical principles for medical research involving human subjects established by Helsinki's Declaration, protecting the privacy of all participants, as well as the confidentiality of their personal information. Because of the retrospective character of this study, an informed consent waiver was obtained by the local ethics committee (Commissie Medische Ethiek, UZ Brussel).
Results
A total of 891 admissions were eligible for inclusion. After exclusion of patients with a rejected COVID-19 diagnosis or a hospital stay less than 24 h, 429 admissions were included in the retrospective cohort. Of these admissions, 336/429 (78%) were exclusively on a COVID-19 ward and 93/429 (22%) were at least partly hospitalized on COVID ICU.
Comorbidities predisposing for poor COVID-19 outcome [11] were distributed as follows: 12% of all admitted patients had an already existing pulmonary condition, 23% had diabetes mellitus, 15% had a history of ischaemic heart disease or heart failure and 14% were immune depressed. The median CCI was 1 (IQR 1). The median body mass index (BMI) was 26 (IQR 6). Patients had a median qSOFA score of 0 (IQR 3) and 1 (IQR 2) in, respectively, ward and ICU admissions. The median overall SpO2/FiO2 ratio at admission was 357 (IQR 124). For COVID-19 ward admissions, this ratio was comparable to the overall ratio, namely 350 (IQR 146), whereas it was lower, namely 296 (IQR 171) for ICU admissions. Mechanical ventilation was indicated in 8% of the admissions. The overall mortality rate was 15% (9% for patients exclusively hospitalized on a ward and 37% for ICU patients). Comparison of baseline characteristics showed in patients with an ICU stay significantly more fever at admission as well as higher CRP, white blood cell count and ferritin levels compared to patients solely admitted on the ward. SpO2/FiO2 at admission and the lowest SpO2/FiO2 rate during the total stay was also lower in ICU patients, while the rate of mechanical ventilation need was higher. All patient characteristics are presented in Table 1.
Table 1.
Patient characteristics
| Ward (n = 336) | ICU (n = 93) | p value | Total (n = 429) | |
|---|---|---|---|---|
| Demographics | ||||
| Age (years); (median, IQR) | 64 (25) | 64 (18) | 0.959 | 64 (27) |
| Sex (male); (n, %) | 187 (56) | 58 (62) | 0.247 | 245 (57) |
| BMIa (kg/m2); (median, IQR) | 26 (6) | 26 (6) | 0.156 | 26 (6) |
| Symptoms at admission (n, %) | ||||
| Fever or history of fever | 189 (58) | 52 (56) | 0.923 | 241 (61) |
| Fever at day 1 of hospitalization | 221 (68) | 73 (79) | 0.029* | 294 (70) |
| Dyspnoea | 183 (56) | 61 (66) | 0.060 | 244 (58) |
| Cough | 187 (57) | 48 (52) | 0.395 | 235 (56) |
| Thoracic pain | 55 (17) | 17 (18) | 0.734 | 72 (17) |
| Laboratory findings (median, IQR; except for lymphopenia) | ||||
| White blood cell count (/mm3) | 7150 (4525) | 7750 (4850) | 0.045 | 6800 (4400) |
| Neutrophil count (/mm3) | 5335 (4081) | 5986 (5078) | 0.093 | 5113 (3796) |
| Lymphocyte count (/mm3) | 995 (602) | 963 (528) | 0.357 | 1015 (606) |
| Ferritin (mcg/L) | 661 (1055) | 806 (1200) | 0.020* | 608 (1016) |
| CRPb (mg/dL) | 88 (140) | 139 (150) | < 0.001* | 71 (128) |
| Lymphopenia (< 1000/mm3); (n, %) | 202 (50) | 41 (44) | 0.221 | 239 (49) |
| Comorbidities | ||||
| CCIc (median, IQR) | 1 (3) | 1 (3) | 0.402 | 1 (3) |
| Chronic pulmonary disease (n, %) | 39 (12) | 11 (12) | 0.984 | 50 (12) |
| Ischaemic/congestive heart disease (n, %) | 49 (15) | 14 (15) | 0.976 | 63 (15) |
| Diabetes mellitus (n, %) | 69 (21) | 28 (30) | 0.069 | 97 (23) |
| Immune suppression (n, %) | 46 (14) | 16 (17) | 0.394 | 62 (14) |
| qSOFA score (median, IQR) | 0 (3) | 1 (2) | 0.124 | 1 (3) |
| Mechanical ventilation need (n, %) | NAd | 33 (35) | – | 33 (8) |
| (SpO2/FiO2 × 100) at admission (median, IQR) | 357 (124) | 296 (171) | < 0.001* | 350 (146) |
| (SpO2/FiO2 × 100) mine (median, IQR) | 321 (123) | 148 (29) | < 0.001* | 291 (192) |
| Mortality (n, %) | 30 (9) | 34 (37) | < 0.001* | 64 (15) |
IQR interquartile range
aBMI body mass index
bCRP C-reactive protein. Laboratory findings were collected at day 0 or 1 of hospitalization on a COVID 19 ward or ICU ward. If not available on day 0 or 1, the first available ferritin levels were collected
cCCI Charlson comorbidity index
dNA
eSpO2/FiO2 × 100 min = the lowest value of the SpO2/FiO2 rate during the total stay on a COVID-19 ward or in the ICU
*p values < 0.05
A total of 323 respiratory samples were collected, of which 81% originating from patients admitted on the ICU. From all included admissions, 14% had at least one respiratory sample showing a significant result (6.7% on the ward, 37% in the ICU). Detailed microbiological data are shown in Table 2. In respiratory samples, the most frequently isolated bacteria were methicillin-sensitive Staphylococcus aureus (20% of the significantly positive samples) and gram-negative bacteria, namely Klebsiella pneumoniae, Pseudomonas aeruginosa and Escherichia coli in, respectively, 10%, 9.6% and 8.7% of the significantly positive respiratory samples. Bacteraemia of respiratory origin was solely identified on the ICU (n = 7). One case of Clostridioides difficile colitis was documented. There were no co-infections with influenza A or B.
Table 2.
Microbiological data
| Ward | ICU | Total | |
|---|---|---|---|
| General data (n, % per admission) | |||
| Total amount of respiratory samples | 61 (15) | 262 (281) | 323 (75) |
| Total amount of respiratory samples with a significant result | 33 (8) | 82 (88) | 115 (27) |
| At least one respiratory sample | 71 (18) | 54 (58) | 125 (29) |
| At least one respiratory sample with a significant result | 27 (6.8) | 34 (37) | 61 (14) |
| At least one episode of bacteraemia | 8 (2.0) | 13 (14) | 21 (5.0) |
| Bacteraemia of respiratory origin | 0 (0) | 7 (7.5) | 7 (1.6) |
| Clostridioides Difficile colitis | 0 (0) | 1 (1.0) | 1 (0.2) |
| Positive influenza A or B PCR | 0 (0) | 0 (0) | 0 (0) |
| Mycoplasma spp. Infection | 3 (0.8) | 2 (2.2) | 5 (1.2) |
| ESBLa in significant respiratory sample | 3 (0.9) | 5 (5.4) | 8 (1.9) |
| CPEb in significant respiratory sample | 1 (0.3) | 2 (2.2) | 3 (0.7) |
|
Causal significant respiratory organisms isolated from the respiratory tract (% expressed in function of the total amount of respiratory samples with significant result) | |||
| Aerobic, gram positive cocci | 10 (30) | 21 (26) | 31 (27) |
| S. aureus MSSAc | 7 (21) | 16 (20) | 23 (20) |
| S. aureus MRSAd | 1 (3.0) | 0 (0) | 1 (0.9) |
| S. agalactiae | 1 (3.0) | 4 (4.8) | 5 (4.3) |
| S. pneumoniae | 1 (3.0) | 1 (1.2) | 2 (1.7) |
| Aerobic, gram negative bacilli, Enterobacteriaceae | 17 (52) | 35 (43) | 52 (45) |
| Citrobacter sp. | 4 (12) | 0 (0) | 4 (3.5) |
| E. cloacae | 2 (6.0) | 7 (8.5) | 9 (7.8) |
| E. coli | 4 (12) | 6 (7.3) | 10 (8.7) |
| K. oxytoca | 1 (3.0) | 1 (1.2) | 2 (1.7) |
| K. pneumoniae | 2 (6.0) | 10 (12) | 12 (10) |
| K. aerogenes | 1 (3.0) | 6 (7.3) | 7 (6.0) |
| Morganella spp. | 0 (0) | 1 (1.2) | 1 (0.9) |
| P. mirabilis | 1 (3.0) | 2 (2.4) | 3 (2.6) |
| Serratia spp. | 2 (6.0) | 2 (2.4) | 4 (3.5) |
| Aerobic, gram negative bacilli, non-enterobacteriaceae | 1 (3.0) | 4 (4.9) | 5 (4.3) |
| H. influenzae | 1 (3.0) | 4 (4.9) | 5 (4.3) |
| Aerobic, gram negative bacilli, non-fermenter | 5 (15) | 19 (23) | 24 (21) |
| A. baumanii | 2 (6.0) | 0 (0) | 2 (1.7) |
| B. cepacia | 0 (0) | 1 (1.2) | 1 (0.9) |
| Pseudomonas aeruginosa | 1 (3.0) | 10 (12) | 11 (9.6) |
| S. maltophilia | 1 (3.0) | 5 (6.0) | 6 (5.2) |
| Achromobacter spp. | 1 (3.0) | 0 (0) | 1 (0.9) |
| Other | 0 (0) | 3 (3.7) | 3 (2.6) |
aESBL extended spectrum beta-lactamase
bCPE carbapenemase producing Enterobacteriaceae
cMSSA methicillin-sensitive Staphylococcus aureus
dMRSA methicillin-resistant Staphylococcus aureus
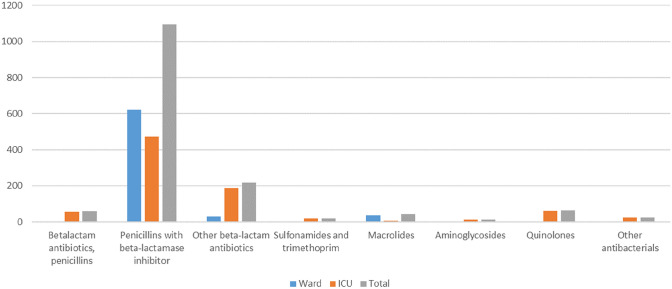
In contrast, 39% of the included admissions were prescribed an antibiotic in the context of a suspected or confirmed respiratory tract infection (59% ward patients, 41% ICU patients). As shown in Table 3, the main driver for antibiotic prescribing was a radiological consolidation (51%), followed by a positive microbiological sample (22%). Patients had a median antibiotic treatment duration of five days (IQR 5). The median lag time before antibiotic initiation was seven days, which lengthened up to 10 days for patients with an ICU stay (Fig. 1, Supplementary Material 4, 5). DDD/admission and DDD/100 hospital bed days were 3.6 and 31.5, respectively. Detailed DDD information on a molecule level is added in Supplementary Material 2. As azithromycin was only prescribed as an immunomodulatory drug (for a total of 0.2 DDD/admission, in 12 different patients), all associated prescriptions were excluded from the analysis. Both on the ward and in the ICU, a penicillin combined with a beta-lactamase inhibitor were most frequently initiated (89.5% of all antibiotic consumption), mainly amoxicillin–clavulanic acid and piperacillin–tazobactam (2.55 DDD/admission; 23.3 DDD/100 hospital bed days). In the ICU, the proportion of penicillin antibiotics decreased (85.4%) as other types of antibiotics were also initiated. The ICU consumption comprised 54.9% of overall antibiotic use in the studied population. A shift towards the category ‘other beta-lactam antibiotics’ is noted in the ICU population, where more third-generation cephalosporins were prescribed (especially ceftazidime: 5 versus 0 admissions; ceftriaxone: 10 versus 6 admissions), carbapenems (meropenem: 17 versus 0 admissions) or monobactams (aztreonam: 3 versus 0 admissions), compared with the COVID-19 ward group. Additionally, the proportion of antibiotics administered by intravenous injection or infusion was three times higher than oral antibiotics (Supplementary Material 6). In 72 admissions (17%), patients received at least one antibiotic prescription for another reason than for a respiratory tract superinfection.
Table 3.
Information on antibiotic treatment; antibiotic classes are presented according to the Anatomical Therapeutic Chemical (ATC)-classification system (J01 level 2) [13]
| Ward | ICU | Total | |
|---|---|---|---|
| Reasons for initiation of antibiotic treatment (n, %) | |||
| Empirically | 10 (10) | 0 (0) | 8 (5) |
| COPDa | 2 (2) | 6 (8) | 8 (5) |
| Positive microbiological sample | 25 (25) | 11 (16) | 36 (22) |
| Radiological consolidation | 47 (47) | 38 (55) | 85 (51) |
| Persistent/rising fever or inflammation | 3 (3) | 9 (13) | 12 (7) |
| Persistent/rising oxygen need | 2 (2) | 1 (1) | 3 (2) |
| Otherb | 11 (11) | 4 (6) | 15 (9) |
| Information on AB treatment, (median, IQR) | |||
| Number of AB treatments per admission | 1 (0) | 2 (2) | 1 (1) |
| Days of AB treatment per admission | 5 (3) | 8 (9) | 5 (5) |
| Lag time before AB initiation per admission | 6 (6) | 10 (8.8) | 7 (8) |
| Total DDDc (n, %) | 690.7 (45.1) | 841.8 (54.9) | 1532.5 (100) |
| Beta-lactam antibiotics, penicillins | 1.2 (0.2) | 56.8 (6.7) | 58 (3.8) |
| Beta-lactam antibiotics, penicillin, with inhibitor | 622 (90.2) | 473.6 (56.3) | 1095.6 (71.5) |
| Other beta-lactam antibiotics | 30 (4.4) | 188.2 (22.4) | 218.2 (14.2) |
| Sulphonamides and trimethoprim | NAd | 18.5 (2.2) | 18.5 (1.2) |
| Macrolides (azithromycin excluded) | 35.5 (5.1) | 6.5 (0.8) | 42 (2.7) |
| Aminoglycosides | NA | 12.8 (1.5) | 12.8 (0.8) |
| Quinolones | 2 (0.3) | 61.2 (7.3) | 63.2 (4.1) |
| Other antibacterials | NA | 24.2 (2.9) | 24.2 (1.6) |
| DDD/admission | |||
| Betalactam antibiotics, penicillins | 0.004 | 0.611 | 0.135 |
| Penicillins with beta-lactamase inhibitor | 1.851 | 5.092 | 2.554 |
| Other beta-lactam antibiotics | 0.089 | 2.024 | 0.509 |
| Sulphonamides and trimethoprim | 0.199 | 0.043 | |
| Macrolides (azithromycin excluded) | 0.106 | 0.070 | 0.098 |
| Aminoglycosides | 0.138 | 0.030 | |
| Quinolones | 0.006 | 0.658 | 0.147 |
| Other antibacterials | 0.260 | 0.056 | |
| DDD/100 bed days | |||
| Betalactam antibiotics, penicillins | 0.357 | 2.793 | 1.191 |
| Penicillins with beta-lactamase inhibitor | 185.119 | 23.284 | 22.506 |
| Other beta-lactam antibiotics | 8.929 | 9.253 | 4.482 |
| Sulphonamides and trimethoprim | 0.910 | 0.380 | |
| Macrolides (azithromycin excluded) | 10.565 | 0.320 | 0.863 |
| Aminoglycosides | 0.629 | 0.263 | |
| Quinolones | 0.595 | 3.009 | 1.298 |
| Other antibacterials | 1.190 | 0.497 | |
aCOPD chronic obstructive pulmonary disease
bOther reasons: e.g. filthy pulmonary secretions, hypotension/sepsis, rising procalcitonin
cDDD defined daily doses
dNA not applicable
Fig. 1.
Total consumption (expressed in DDD) of antibiotics for respiratory tract infection
In a first step of the model building, all variables were modelled one by one, complemented by whether the patient was hospitalized in the ICU, in function of the antibiotic prescription. In this phase, laboratory values (white blood cell count, neutrophil count, CRP level at admission), presence of at least one respiratory microbiological sample, comorbidity of earlier pulmonary disease, the need for oxygen at admission, SpO2/FiO2 parameters (measured at admission and maximum value), need for mechanical ventilation and length of stay (at the COVID-19 ward and at the ICU) were withheld. In the final mixed effects logistic regression, multicollinearity among the included variables was avoided by looking at the correlation between variables (see graphical representation of correlation matrix in Supplementary Material 3). In this model (Table 4), we found insufficient evidence to assume there is a difference in antibiotic prescription occurrence between patients who stayed on a regular ward exclusively versus those who were admitted to the ICU during their stay (p = 0.108). The presence of fever at admission increased the risk for antibiotic prescribing (OR 2.97; 95% CI 1.42–6.22), as well as pre-existing lung disease (OR 3.04; 95% CI 1.12–8.27). Every increase of 1000 neutrophils/mcL elevated the probability for an antibiotic prescription for a potential respiratory superinfection with 14% (OR 1.14, 95% CI 1.04–1.26). The SpO2/FiO2 ratio was negatively correlated with antibiotic prescribing (OR 0.96; 95% CI 0.92–0.99). Every additional hospital bed day increased the likelihood of antibiotic prescribing with about 9% (OR 1.09; 95% CI 1.03–1.16).
Table 4.
Mixed effects logistic regression analysis of potential drivers associated with antibiotic prescribing
| Variable | OR (95% confidence interval) | p value |
|---|---|---|
| Patient admitted to | 0.108 | |
| Regular ward exclusively | Ref | |
| ICU | 1.96 (0.86–4.48) | |
| Presence of fever at admission | 0.004* | |
| No | Ref | |
| Yes | 2.97 (1.42–6.21) | |
| History of pulmonary disease | 0.030* | |
| No | Ref | |
| Yes | 3.04 (1.12–8.27) | |
| Neutrophil count (per neutrophil increase in 1000 units/mcL) | 1.14 (1.04–1.26) | 0.007* |
| SpO2/FiO2 at admission (per rise of 10 units) | 0.96 (0.93–0.99) | 0.019* |
| Length of stay (per supplementary day) | 1.09 (1.03–1.16) | 0.002* |
*p < 0.05 is considered statistically significant
Discussion
Concerns have been raised about an expected ‘COVID-19 related’ increase in AMR due to both antibiotic overuse as well as a shift of antimicrobial stewardship team (AST) activities to the containment of the COVID-19 pandemic [2–5, 17, 18]. However, the rate of bacterial superinfections in patients affected with COVID-19 appears to be much lower to what is observed in other viral pneumonias, such as influenza [6, 7, 19–22]. To our knowledge, the current study is the first to investigate antibiotic prescription for suspected respiratory tract superinfections in admitted COVID-19 patients, in a quantitative and standardized way. This retrospective observational study adds to the current well of evidence on antibiotic prescribing for (presumed) respiratory tract superinfections in COVID-19 patients.
Overall, in our study population, the COVID-19 patients with an ICU stay showed more clinical signs of severe disease: higher inflammatory blood markers at admission (CRP, white blood cell count and ferritin), higher rate of fever at admission and higher oxygen need, which resulted in higher mortality as compared to patients solely admitted to the COVID-19 ward. This is in line with several observational COVID-19 studies [23, 24]. At least one respiratory sample was available in no more than 29% of the admissions (18% forward admissions, 58% for ICU admissions). Compared with a recent systematic review by Langsbury et al., we report higher rates of Staphylococcus aureus (23% versus 2%), lower rates of Haemophilus influenzae (5% versus 12%) and much lower rates of Mycoplasma pneumoniae in our study (5% versus 42%). However, Lansbury et al. defined a single positive IgM titre as diagnostic for Mycoplasma pneumoniae infection, whereas clinical probability, PCR and titres kinetics were taken into consideration in our study [7]. Staphylococcus aureus and Streptococcus pneumoniae infections are known to be the most frequently reported bacteria associated with influenza infections [10]. The higher Staphylococcus aureus rate was confirmed in our study. The fact that a higher Streptococcus pneumoniae rate was not found in this study could be explained by higher rates of pneumococcal vaccination in this older patient group and perhaps by an underdiagnosis since pneumococcal urinary antigen tests are not used in our centre.
Antibiotics were initiated in 39% of the admissions for the treatment of (presumptive) respiratory tract infections. This still seems rather low, in comparison to rates of antibiotic prescription ranging from 56 to 95% in other studies, including a recent systematic review of 30 studies with 3834 patients [6, 7, 9]. In our retrospective cohort, a total of 3.6 DDDs/admission and 31.5 DDDs/100 hospital days were counted, which means that approximately 1 in 3 admitted patients received one DDD of an antibiotic (e.g., 3 g of amoxicillin–clavulanic acid) during every single day of admission. Using DDDs per 100-bed-days instead of antibiotic prescriptions rates offers the opportunity to consider the length of antibiotic treatment with respect to the length of the patient’s hospital stay, ruling out differences in hospitalization durations as a confounding factor. In DDD registration, the administration route and drug class are considered as well. This method of quantifying antibiotic consumption allows the option of benchmarking. As we are the first to generate these data, we are not able to compare these data to those of other institutions.
It is remarkable that most prescribed antibiotics were a penicillin with beta-lactamase inhibitor, especially for the wards (90% versus 56% for ICU prescriptions). This could be explained by the fact that more empiric treatments were started and continued on the ward, while a more targeted antibiotic approach was performed in the ICU, due to the higher availability of (significant) respiratory samples. In a recent international survey investigating the antibiotic prescribing habits of physicians involved in the care of COVID-19 patients, most participants agreed that atypical germs and Staphylococcus aureus are the most important germs to cover in the context of COVID-19 [17]. However, our microbiological analysis did not show high rates of atypical germs. Nonetheless, the participants of that survey reported a high use of penicillin with beta-lactamase inhibitor combinations, which is in accordance with our findings. In contrast to Vaughn et al. [25], where 26% of the administered antibiotics included an anti-MRSA coverage, only 1% of our administered antibiotic doses covered MRSA. This is probably due to the low prevalence of MRSA in our setting (2.5% of all tested invasive isolates [26]) and to the fact that we did not include anti-MRSA antibiotics which were prescribed for other reasons than for (presumed) respiratory tract infections. The amount of intravenously (IV) administered antibiotic doses was almost three times higher as compared to the orally (PO) administered doses, which could partially be explained by the number of intubated patients on the ICU. However, even patients exclusively admitted to the ward received more IV than PO administrations (IV/PO ratio of 1.36). The latter observation highlights the need for a specific intervention in our institution. The median antibiotic duration of 5 days appears to be short, especially when compared with the suggested 7–14 days of antibiotic duration for nosocomial pneumonia in various international guidelines [27–30]. On the other hand, 5 days is sufficient for the treatment of community-acquired pneumonia [31]. Our findings are concurrent with Chen et al., who also described a median treatment duration of 5 days (IQR 3–7) [32]. Both shorter (median 4 days, IQR 2–5) and longer (median 7 days, IQR 4–9) courses [33] are reported as well. Uncertainties in bacterial superinfection diagnostics in COVID-19 patients could have resulted in impulsive antibiotic prescribing in cases of doubt. Fortunately, short treatment courses may also be indicative for de-escalation and interruption of antibiotic courses when judged unnecessary.
In the driver analysis, the presence of fever at admission, low SpO2/FiO2, pre-existing pulmonary disease and high neutrophil counts together with a longer length of stay were significantly associated with the administration of at least one antibiotic prescription during admission. Besides a prolonged hospital stay, which enhances the risk of nosocomial infection and a high leukocyte count at admission, none of the reported variables has been proven to be associated with a higher rate of bacterial superinfection in COVID-19 patients [25, 34]. As the presence of fever and low SpO2/FiO2 at admission are both, respectively, a diagnostic and a prognostic criterion of bacterial pneumonia [35], it is comprehensible to find these as possible drivers of antibiotic prescription. However, these two variables are bad distinguishers between viral and bacterial pneumonia [36]. The presence of pre-existing pulmonary disease at admission has been proven to be associated with a higher risk of bacterial pneumonia [37]. Nevertheless, it is questionable if COVID-19 mediated chronic obstructive pulmonary disease (COPD) exacerbations should be treated with antibiotics. In a similar multicentre study, Vaughn et al. described the following variables as a predictor of antibiotic therapy in COVID-19 patients: age, BMI, disease severity (e.g., respiratory support and sepsis), the presence of a lobar infiltrate and admission to a for-profit hospital [25]. The association between age or BMI and antibiotic treatment was not confirmed in our study. This could be explained by the fact that the study only found a very small difference in BMI [rate ratio (RR) 0.99; 95% CI 0.99–1.00] and age (RR 1.04; 95% CI 1.01–1.08) between both groups for a larger cohort (n = 1705). Moreover, in our study, we focussed on antibiotic prescribing during the entire hospital stay whereas Vaughn et al. evaluated the first two days of admission. Moreover, the authors did not exclusively focus on antibiotics started for respiratory tract infections, which is in contrast with our analysis. We decided not to analyze the presence of a lobar infiltrate as a potential predictor of antibiotic prescription, as other authors concluded that the presence of a lobar infiltrate is not predictive for bacterial co-infection [25]. Even though 51% of the admissions in our study received antibiotics because of a dense consolidation or worsening medical imagery, the presence of denser radiological consolidations is common in the later presentations of COVID-19, without the presence of bacterial co-infection [38–40].
This study has its limitations, primarily due to its retrospective character. Since we performed chart review for data collection, precise documentation of the reason for antibiotic prescribing is difficult. According to Belgian guidelines, it is compulsory to document the indication of every antibiotic prescription in the patient record [41]. However, the full work up for the diagnosis of bacterial superinfection is challenging in COVID-19 patients, mainly because of insufficient specificity of clinical parameters, like inflammatory markers, radiological consolidations, fever and procalcitonin in this setting [31, 42–44]. Further studies are needed to identify robust markers of bacterial superinfection in patients admitted with a viral respiratory tract infection such as COVID-19. For this reason, we did not evaluate the appropriateness of the antibiotic prescriptions. The effect of a learning curve, with a possible decrease of antibiotic prescriptions over time, was not considered in the prescription analysis. Therefore, comparative (prospective) studies in a similar setting are needed to evaluate this plausible effect. Additionally, it should be considered that in 17% of the admissions, patients received antibiotics for other reasons than for a respiratory tract superinfection. It is plausible that there might have been an underestimation of the rate of antibiotic prescriptions for suspected respiratory tract superinfections since these non-respiratory antibiotics may also cover suspected respiratory pathogens.
We have not been able to compare our data on antibiotic consumption to other data—yet due to the absence of quantitative analyses concerning antimicrobial prescriptions for this indication. Moreover, we were unable to compare our data with a historical influenza cohort, as quantitative data regarding antibiotic DDDs in the context of influenza (super)infection have not been published so far. This is precisely why we think the current study is of interest for healthcare professionals involved in antibiotic stewardship. We recommend that the ASTs should have increased awareness for COVID-19 related antibiotic strategies, using tailored interventions to support the judicious use of antibiotics. Our driver analysis provides different relevant patient and disease-related variables that ASTs can use to identify those patients most at risk for having an antibiotic therapy for a respiratory tract superinfection. By using standardized data about prescribed daily doses of antibiotics as an indicator of antibiotic consumption, stewardship related interventions can be evaluated and benchmarked [2].
Conclusion
Our study offers a quantitative and standardised benchmark regarding antibiotic prescriptions for suspected respiratory tract superinfections in patients admitted for COVID-19 and analyses the main drivers of antibiotic prescriptions. As bacterial superinfections are rare in COVID-19 patients: further prospective (interventional) studies are needed to substantiate AST interventions to help reduce antibiotic overuse and to counter the worldwide rise of AMR.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contributions
JVL and SW are both first authors and equally shared the workload: study concept, study design, data collection, data analysis and interpretation, writing and revision. JK, MC and MM: data collection. LS and PL: study design and revision. SVL: data analysis and revision. DP, ED and PL: revision. SDA contributed to the study concept, the study design, writing and revision.
Funding
No funding was used nor demanded.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
Not applicable.
Declarations
Conflict of interest
J. Van Laethem, S. Wuyts, J. Koulalis, M. Colman, S. Van Laere, M. Moretti, L. Seyler, P. Lacor, E. De Waele, D. Pierard, SD. Allard declare not having any kind of conflict of interest.
Ethics approval and consent to participate
The study was approved by the local Ethical Committee (Commissie Medische Ethiek (O.G. 016), Reflectiegroep Biomedische Ethiek, UZ Brussel) prior to data collection (B.U.N. 1432020000092) and was carried out in accordance with the ethical principles for medical research involving human subjects established by Helsinki's Declaration, protecting the privacy of all participants, as well as the confidentiality of their personal information. Because of the retrospective character of this study, an informed consent waiver was obtained by the local Ethics committee (Commissie Medische Ethiek, UZ Brussel).
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
J. Van Laethem and S. Wuyts contributed equally to this work.
Change history
5/13/2023
A Correction to this paper has been published: 10.1007/s11739-023-03302-y
References
- 1.Tamma PD, Avdic E, Li DX, Dzintars K, Cosgrove SE. Association of adverse events with antibiotic use in hospitalized patients. JAMA Intern Med. 2017;177(9):1308–1315. doi: 10.1001/jamainternmed.2017.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spernovasilis NA, Kofteridis DP. COVID-19 and antimicrobial stewardship: what is the interplay? Infect Control Hosp Epidemiol. 2020 doi: 10.1017/ice.2020.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miranda C, Silva V, Capita R, Alonso-Calleja C, Igrejas G, Poeta P. Implications of antibiotics use during the COVID-19 pandemic: present and future. J Antimicrob Chemother. 2020 doi: 10.1093/jac/dkaa350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rawson TM, Moore LSP, Castro-Sanchez E, Charani E, Davies F, Satta G, Ellington MJ, Holmes AH. COVID-19 and the potential long-term impact on antimicrobial resistance. J Antimicrob Chemother. 2020;75(7):1681–1684. doi: 10.1093/jac/dkaa194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antimicrobial resistance in the age of COVID-19 (2020). Nat Microbiol 5(6):779. 10.1038/s41564-020-0739-4(Editorial) [DOI] [PubMed]
- 6.Garcia-Vidal C, Sanjuan G, Moreno-García E, Puerta-Alcalde P, Garcia-Pouton N, Chumbita M, Fernandez-Pittol M, Pitart C, Inciarte A, Bodro M, Morata L, Ambrosioni J, Grafia I, Meira F, Macaya I, Cardozo C, Casals C, Tellez A, Castro P, Marco F, García F, Mensa J, Martínez JA, Soriano A. Incidence of co-infections and superinfections in hospitalized patients with COVID-19: a retrospective cohort study. Clin Microbiol Infect. 2020 doi: 10.1016/j.cmi.2020.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lansbury L, Lim B, Baskaran V, Lim WS. Co-infections in people with COVID-19: a systematic review and meta-analysis. J Infect. 2020;81(2):266–275. doi: 10.1016/j.jinf.2020.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hughes S, Troise O, Donaldson H, Mughal N, Moore LSP. Bacterial and fungal coinfection among hospitalized patients with COVID-19: a retrospective cohort study in a UK secondary-care setting. Clin Microbiol Infect. 2020;26(10):1395–1399. doi: 10.1016/j.cmi.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langford BJ, So M, Raybardhan S, Leung V, Westwood D, MacFadden DR, Soucy JR, Daneman N. Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin Microbiol Infect. 2020 doi: 10.1016/j.cmi.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein EY, Monteforte B, Gupta A, Jiang W, May L, Hsieh Y-H, Dugas A. The frequency of influenza and bacterial coinfection: a systematic review and meta-analysis. Influenza Other Respir Viruses. 2016;10(5):394–403. doi: 10.1111/irv.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet (London, England) 2020;395(10229):1054–1062. doi: 10.1016/s0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Festic E, Bansal V, Kor DJ, Gajic O. SpO2/FiO2 ratio on hospital admission is an indicator of early acute respiratory distress syndrome development among patients at risk. J Intensive Care Med. 2015;30(4):209–216. doi: 10.1177/0885066613516411. [DOI] [PubMed] [Google Scholar]
- 13.World Health Organization (WHO) collaborating centre for drugs statistics methodology (2020) DDD and ATC-classification. WHO collaborating centre for drugs statistics methodology. https://www.whocc.no/atc_ddd_index/. Accessed 11 Nov 2020
- 14.Gautret P, Lagier JC, Parola P, Hoang VT, Meddeb L, Mailhe M, Doudier B, Courjon J, Giordanengo V, Vieira VE, Tissot Dupont H, Honoré S, Colson P, Chabrière E, La Scola B, Rolain JM, Brouqui P, Raoult D. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020;56(1):105949. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40(5):373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 16.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche J-D, Coopersmith CM, Hotchkiss RS, Levy MM, Marshall JC, Martin GS, Opal SM, Rubenfeld GD, van der Poll T, Vincent J-L, Angus DC. The third international consensus definitions for sepsis and septic shock (sepsis-3) JAMA. 2016;315(8):801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beović B, Doušak M, Ferreira-Coimbra J, Nadrah K, Rubulotta F, Belliato M, Berger-Estilita J, Ayoade F, Rello J, Erdem H. Antibiotic use in patients with COVID-19: a 'snapshot' infectious diseases international research initiative (id-iri) survey. J Antimicrob Chemother. 2020;75(11):3386–3390. doi: 10.1093/jac/dkaa326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ginsburg AS, Klugman KP. COVID-19 pneumonia and the appropriate use of antibiotics. Lancet Glob Health. 2020;8(12):e1453–e1454. doi: 10.1016/S2214-109X(20)30444-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furukawa D, Graber CJ. Antimicrobial stewardship in a pandemic: picking up the pieces. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huttner BD, Catho G, Pano-Pardo JR, Pulcini C, Schouten J. COVID-19: don't neglect antimicrobial stewardship principles! Clin Microbiol Infect. 2020;26(7):808–810. doi: 10.1016/j.cmi.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karami Z, Knoop BT, Dofferhoff ASM, Blaauw MJT, Janssen NA, van Apeldoorn M, Kerckhoffs APM, van de Maat JS, Hoogerwerf JJ, Ten Oever J. Few bacterial co-infections but frequent empiric antibiotic use in the early phase of hospitalized patients with COVID-19: results from a multicentre retrospective cohort study in The Netherlands. Infect Dis (Lond) 2020 doi: 10.1080/23744235.2020.1839672. [DOI] [PubMed] [Google Scholar]
- 22.Bassetti M, Kollef MH, Timsit JF. Bacterial and fungal superinfections in critically ill patients with COVID-19. Intensive Care Med. 2020;46(11):2071–2074. doi: 10.1007/s00134-020-06219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pouw N, van de Maat J, Veerman K, Ten Oever J, Janssen N, Abbink E, Reijers M, de Mast Q, Hoefsloot W, van Crevel R, Slieker K, van Apeldoorn M, Blaauw M, Dofferhoff A, Hoogerwerf J. Clinical characteristics and outcomes of 952 hospitalized COVID-19 patients in The Netherlands: a retrospective cohort study. PLoS ONE. 2021;16(3):e0248713. doi: 10.1371/journal.pone.0248713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu C, Chen X, Cai Y, Xia J, Zhou X, Xu S, Huang H, Zhang L, Zhou X, Du C, Zhang Y, Song J, Wang S, Chao Y, Yang Z, Xu J, Zhou X, Chen D, Xiong W, Xu L, Zhou F, Jiang J, Bai C, Zheng J, Song Y. Risk Factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020;180(7):934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaughn VM, Gandhi T, Petty LA, Patel PK, Prescott HC, Malani AN, Ratz D, McLaughlin E, Chopra V, Flanders SA. Empiric Antibacterial therapy and community-onset bacterial co-infection in patients hospitalized with covid-19: a multi-hospital cohort study. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.European Centre for Disease Prevention and Control (2020) Antimicrobial resistance in the EU/EEA (EARS-Net). Annual Epidemiological Report 2019. Stockholm
- 27.Kalil AC, Metersky ML, Klompas M, Muscedere J, Sweeney DA, Palmer LB, Napolitano LM, O'Grady NP, Bartlett JG, Carratalà J, El Solh AA, Ewig S, Fey PD, File TM, Jr, Restrepo MI, Roberts JA, Waterer GW, Cruse P, Knight SL, Brozek JL. Management of adults with hospital-acquired and ventilator-associated pneumonia: 2016 clinical practice guidelines by the infectious diseases society of America and the American Thoracic Society. Clin Infect Dis. 2016;63(5):e61–e111. doi: 10.1093/cid/ciw353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torres A, Niederman MS, Chastre J, Ewig S, Fernandez-Vandellos P, Hanberger H, Kollef M, Li Bassi G, Luna CM, Martin-Loeches I, Paiva JA, Read RC, Rigau D, Timsit JF, Welte T, Wunderink R. International ERS/ESICM/ESCMID/ALAT guidelines for the management of hospital-acquired pneumonia and ventilator-associated pneumonia: guidelines for the management of hospital-acquired pneumonia (HAP)/ventilator-associated pneumonia (VAP) of the European Respiratory Society (ERS), European Society of Intensive Care Medicine (ESICM), European Society of Clinical Microbiology and Infectious Diseases (ESCMID) and Asociación Latinoamericana del Tórax (ALAT) Eur Respir J. 2017 doi: 10.1183/13993003.00582-2017. [DOI] [PubMed] [Google Scholar]
- 29.Dimopoulos G, Poulakou G, Pneumatikos IA, Armaganidis A, Kollef MH, Matthaiou DK. Short- vs long-duration antibiotic regimens for ventilator-associated pneumonia: a systematic review and meta-analysis. Chest. 2013;144(6):1759–1767. doi: 10.1378/chest.13-0076. [DOI] [PubMed] [Google Scholar]
- 30.Pugh R, Grant C, Cooke RP, Dempsey G. Short-course versus prolonged-course antibiotic therapy for hospital-acquired pneumonia in critically ill adults. Cochrane Database Syst Rev. 2015 doi: 10.1002/14651858.CD007577.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Metlay JP, Waterer GW, Long AC, Anzueto A, Brozek J, Crothers K, Cooley LA, Dean NC, Fine MJ, Flanders SA, Griffin MR, Metersky ML, Musher DM, Restrepo MI, Whitney CG. Diagnosis and treatment of adults with community-acquired pneumonia. An official clinical practice guideline of the American Thoracic Society and Infectious Diseases Society of America. Am J Respir Crit Care Med. 2019;200(7):e45–e67. doi: 10.1164/rccm.201908-1581ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet (London, England) 2020;395(10223):507–513. doi: 10.1016/s0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu J, Liu J, Zhao X, Liu C, Wang W, Wang D, Xu W, Zhang C, Yu J, Jiang B, Cao H, Li L. Clinical characteristics of imported cases of coronavirus disease 2019 (COVID-19) in Jiangsu province: a multicenter descriptive study. Clin Infect Dis. 2020;71(15):706–712. doi: 10.1093/cid/ciaa199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cook DJ, Walter SD, Cook RJ, Griffith LE, Guyatt GH, Leasa D, Jaeschke RZ, Brun-Buisson C. Incidence of and risk factors for ventilator-associated pneumonia in critically ill patients. Ann Intern Med. 1998;129(6):433–440. doi: 10.7326/0003-4819-129-6-199809150-00002. [DOI] [PubMed] [Google Scholar]
- 35.Woodhead M, Blasi F, Ewig S, Huchon G, Ieven M, Ortqvist A, Schaberg T, Torres A, van der Heijden G, Verheij TJ. Guidelines for the management of adult lower respiratory tract infections. Eur Respir J. 2005;26(6):1138–1180. doi: 10.1183/09031936.05.00055705. [DOI] [PubMed] [Google Scholar]
- 36.Jain S, Self WH, Wunderink RG, Fakhran S, Balk R, Bramley AM, Reed C, Grijalva CG, Anderson EJ, Courtney DM, Chappell JD, Qi C, Hart EM, Carroll F, Trabue C, Donnelly HK, Williams DJ, Zhu Y, Arnold SR, Ampofo K, Waterer GW, Levine M, Lindstrom S, Winchell JM, Katz JM, Erdman D, Schneider E, Hicks LA, McCullers JA, Pavia AT, Edwards KM, Finelli L. Community-acquired pneumonia requiring hospitalization among U.S. adults. N Engl J Med. 2015;373(5):415–427. doi: 10.1056/NEJMoa1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Almirall J, Serra-Prat M, Bolíbar I, Balasso V. Risk factors for community-acquired pneumonia in adults: a systematic review of observational studies. Respiration. 2017;94(3):299–311. doi: 10.1159/000479089. [DOI] [PubMed] [Google Scholar]
- 38.Wong HYF, Lam HYS, Fong AH, Leung ST, Chin TW, Lo CSY, Lui MM, Lee JCY, Chiu KW, Chung TW, Lee EYP, Wan EYF, Hung IFN, Lam TPW, Kuo MD, Ng MY. Frequency and distribution of chest radiographic findings in patients positive for COVID-19. Radiology. 2020;296(2):E72–e78. doi: 10.1148/radiol.2020201160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanne JP, Little BP, Chung JH, Elicker BM, Ketai LH. Essentials for radiologists on COVID-19: an update-radiology scientific expert panel. Radiology. 2020;296(2):E113–e114. doi: 10.1148/radiol.2020200527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan F, Ye T, Sun P, Gui S, Liang B, Li L, Zheng D, Wang J, Hesketh RL, Yang L, Zheng C. Time course of lung changes at chest ct during recovery from coronavirus disease 2019 (COVID-19) Radiology. 2020;295(3):715–721. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balligand E, Costers M, Van Gastel E (2014) Belgian antibiotic policy coordination committee beleidsnota legislatuur 2014–2019. Online beleidsdocument: https://overlegorganen.gezondheid.belgie.be/nl/documenten/beleidsnota-bapcoc-2014-2019
- 42.Lam SW, Bauer SR, Fowler R, Duggal A. Systematic review and meta-analysis of procalcitonin-guidance versus usual care for antimicrobial management in critically ill patients: focus on subgroups based on antibiotic initiation, cessation, or mixed strategies. Crit Care Med. 2018;46(5):684–690. doi: 10.1097/ccm.0000000000002953. [DOI] [PubMed] [Google Scholar]
- 43.Vanhomwegen C, Veliziotis I, Malinverni S, Konopnicki D, Dechamps P, Claus M, Roman A, Cotton F, Dauby N. Procalcitonin accurately predicts mortality but not bacterial infection in COVID-19 patients admitted to intensive care unit. Ir J Med Sci. 2021 doi: 10.1007/s11845-020-02485-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.May M, Chang M, Dietz D, Shoucri S, Laracy J, Sobieszczyk ME, Uhlemann AC, Zucker J, Kubin CJ. Limited utility of procalcitonin in identifying community-associated bacterial infections in patients presenting with coronavirus disease 2019. Antimicrob Agents Chemother. 2021 doi: 10.1128/aac.02167-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Not applicable.