SUMMARY
Even though SYK and ZAP70 kinases share high sequence homology and serve analogous functions, their expression in B- and T-cells is strictly segregated throughout evolution. Here, we identified aberrant ZAP70-expression as a common feature in a broad range of B-cell malignancies. We validated SYK as the kinase that sets the thresholds for negative selection of autoreactive and premalignant clones. When aberrantly expressed in B-cells, ZAP70 competes with SYK at the BCR-signalosome and redirects SYK from negative selection to tonic PI3K signaling, thereby promoting B-cell survival. In genetic mouse models for B-ALL and B-CLL, conditional expression of Zap70 accelerated disease onset, while genetic deletion impaired malignant transformation. Inducible activation of Zap70 during B-cell development compromised negative selection of autoreactive B-cells, resulting in pervasive autoantibody production. Strict segregation of the two kinases is critical for normal B-cell selection and represents a central safeguard against the development of autoimmune disease and B-cell malignancies.
eTOC Blurb
While SYK and ZAP70 are segregated in normal B- and T-lymphocytes, Sadras et al show aberrant ZAP70 coexpression is common across multiple B-cell malignancies. By competing with SYK, ZAP70 diverts BCR-signaling from NFAT to PI3K-activation, which disables mechanisms of negative selection of autoreactive or premalignant B-cell clones.
Graphical Abstract

INTRODUCTION
B-cells are subject to central tolerance checkpoints and stringently selected for intermediate levels of BCR-signaling strength (Muschen, 2018; Pelanda and Torres, 2012). According to a Goldilocks principle of B-cell selection, both attenuation below minimum thresholds (e.g. non-functional BCR) and excess above maximum thresholds (e.g. from an autoreactive BCR or oncogenic signaling molecule) trigger negative selection for elimination of defective or pathogenic B-cells.
SYK is the most proximal tyrosine kinase that initiates BCR-signaling to activate two diverging downstream signaling pathways, namely Ca2+ signaling via BLNK, BTK and PLCγ2 (Baba et al., 2001), or PI3K-signaling via phosphorylation of CD19 and PIK3AP1 (Okada et al., 2000). T-cells express a structurally similar tyrosine kinase, CD3ζ-chain-associated protein of 70 kDa (ZAP70) which plays an analogous role to SYK in initiating proximal signaling from the T-cell receptor (TCR). Compared to SYK, ZAP70 has intrinsically weaker tyrosine kinase activity (Konigsberger et al., 2012; Latour et al., 1996; Zoller et al., 1997). Supporting distinct roles for SYK and ZAP70 in B and T-cell ontogeny, Zap70−/− mice display arrested T-cell development (Negishi et al., 1995), while deletion of Syk causes a profound block in B-cell development (Cheng et al., 1995; Turner et al., 1995). These findings suggest that SYK and ZAP70 may be functionally adapted to lineage-specific contexts of B- and T-cell development, respectively.
A number of observations indicate that the segregation of SYK to B-cells and ZAP70 to T-cells is less stringent in lymphoid malignancies. For instance, high expression of SYK is detected in sub-groups of aggressive peripheral T-cell lymphomas (Feldman et al., 2008). Conversely, ZAP70 is expressed in about half of chronic lymphocytic leukemia (B-CLL) cases and has also been documented in other B-cell malignancies (Carreras et al., 2005; Crespo et al., 2006; Sup et al., 2004). In CLL, ZAP70+ cases have inferior clinical outcomes and express unmutated Ig VH region genes (Crespo et al., 2003; Durig et al., 2003; Wiestner et al., 2003) and typically express an autoreactive BCR (Duhren-von Minden et al., 2012; Herve et al., 2005). ZAP70 is phosphorylated following BCR activation in B-CLL cells (Chen et al., 2002; Gobessi et al., 2007), but little is known about how this kinase contributes to oncogenic BCR-signaling in B-CLL and why B-cells would acquire expression of an additional kinase. In this study, we demonstrate that expression of ZAP70 occurs broadly across multiple B-cell malignancies. In these diseases, ZAP70 diverts oncogenic BCR-signaling from Ca2+-NFAT signaling to PI3K-activation, hence escaping B-cell anergy and negative selection as the result of chronic BCR-stimulation. We propose that SYK-ZAP70 coexpression subverts mechanisms of negative selection (e.g. B-cell anergy) that would normally eliminate autoreactive and premalignant B-cells and thus enables persistent signaling from autoreactive BCRs or oncogenic activation of the BCR-signaling pathway.
RESULTS
ZAP70 is expressed in a broad range of B-cell malignancies
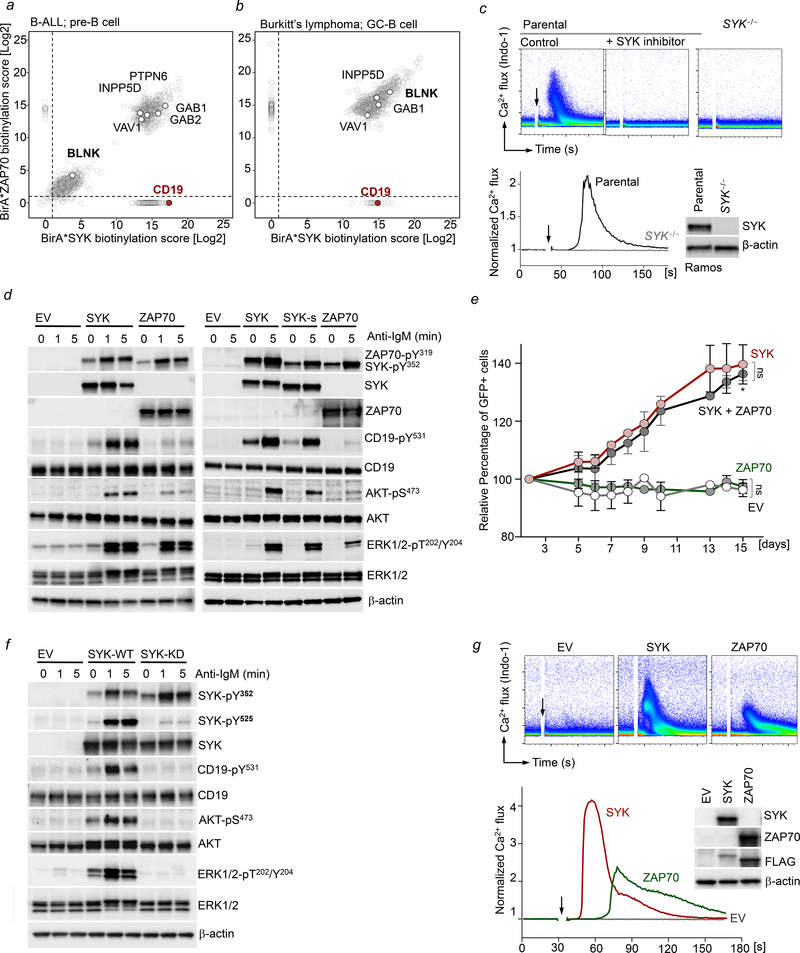
With the exception of thymocytes and some TCRγδ+ T-cells (Muro et al., 2018), SYK and ZAP70 are strictly segregated to B- and T-cells, respectively (Figure 1a, Table S1). Studying ZAP70 and SYK mRNA expression in B-cell malignancies, we found expression of ZAP70 in most B-ALL and mantle cell lymphoma (MCL) cases in addition to CLL (Figure 1b). ZAP70 mRNA levels were particularly high in TCF3-PBX1 and BCR-ABL1-rearranged B-ALL that are driven by active pre-BCR signaling (Geng et al., 2015) and its oncogenic mimic (Feldhahn et al., 2005a) (Figure S1a). Conversely, expression of ZAP70 was low in most cases of germinal-center (GC)- and post-GC B-cell malignancies (Figure 1b). These results were validated at the protein level by Western blot (Figure 1c, S1b). Similar to B-CLL, MCL is characterized by the accumulation of clonal, CD5+ autoreactive B-cells that express stereotyped BCRs that are skewed in favor of a small group of VH genes, including VH1–69 (Lai et al., 2006). ZAP70 mRNA expression was significantly higher in cases with unmutated VH region genes, mirroring the association between ZAP70 expression and unmutated VH regions in B-CLL (Figure S1c). For immunohistochemistry, adjacent tissue sections from biopsies of MCL, follicular lymphoma, DLBCL and classical Hodgkin’s disease were stained for expression of SYK and ZAP70 (Figure S2a). Multiplex staining for SYK and ZAP70 was performed to confirm coexpression of both kinases (Figure S2b).
Figure 1: ZAP70 is aberrantly expressed in a broad range of B-cell malignancies.
(a) Normalized mRNA expression of SYK and ZAP70 in normal hematopoietic populations. Complemented by Table S1. (b) Normalized mRNA expression of SYK and ZAP70 in B-cell malignancies from patients with: B-ALL (n=637), B-CLL (n=646), MCL (n=174), BL (n=120), DLBCL (n=392) and MM (n=113). Normal B- and T-cells are included as a reference. Dashed line indicates mean ZAP70 expression across B-cell populations. (c) SYK and ZAP70 protein levels were analyzed by Western blotting. Normal CD19+ B-cells and CD3+ T-cells were included as controls. (d-f) Single-cell (sc) protein analyses for SYK and ZAP70. Complemented by Table S2. Sc-WB analysis was performed on (d) normal peripheral blood pan-B (CD19+) and T-cells (CD3+), (e) purified leukemic populations from two different PDX samples of patients with BCR-ABL1 B-ALL, TCF3-PBX1 B-ALL, MCL, DLBCL and 2 primary B-CLL patient samples with unmutated or mutated BCRs, (f) Pro/Pre-B, Immature, and plasma cells purified from BM; CD5+ CD19+, new emigrant mature naïve, and memory B-cells purified from peripheral blood and germinal center B-cells purified from resected tonsillar tissue by flow sorting.
Single-cell protein analysis demonstrated concurrent SYK and ZAP70 expression in individual cells
To confirm concurrent expression of SYK and ZAP70 in single cells and to determine whether SYK and ZAP70 coexpressing cells represent clonal subpopulations, we developed a single-cell protein assay to study primary B-cell leukemia and lymphoma samples (Figure S3). Accordingly, strict segregation of SYK to B-cells and ZAP70 to T-cells was observed in normal lymphocytes (Figure 1d). We analyzed four primary B-CLL samples, carrying unmutated and mutated VH region genes and patient-derived xenografts (PDX) from primary B-ALL (BCR-ABL1 or TCF3-PBX1), MCL and DLBCL samples (Figure 1e). Clear SYK and ZAP70 coexpressing populations could be identified in the U-CLL, MCL and B-ALL samples encompassing >85% of tumor cells. Consistent with previous reports (Crespo et al., 2003), B-CLL patient samples carrying mutated VH region genes had less than 30% ZAP70+ cells, and DLBCL PDX samples expressed exclusively SYK (Figure 1e).
Single-cell characterization of ZAP70 protein expression during normal B-cell differentiation
Since ZAP70 expression was common in pre-GC but not post-GC B-cell tumors, we extended our analysis of ZAP70 to normal human B-cell development. Although ZAP70 expression was not detectable in peripheral blood CD19+ B-cells (Figure 1d), we observed distinct changes of ZAP70-expressing cells during B-cell development (Figure 1f, S4). Strikingly, ZAP70 was coexpressed at high levels in ~40% of pro- and pre-B-cells, while more than half of B-cell precursors exclusively expressed SYK. At subsequent stages of B-cell development, the percentage of dual kinase expressing cells sharply dropped to ~10% in immature B-cells and below detection limit at the time of bone marrow egress. ZAP70-expressing cells were rare among circulating new emigrant, mature naïve, CD5+ B-cells, and tonsillar germinal center B-cells (Figure 1f). Interestingly, both kinases were present in ~1% of memory B-cells and >20% of plasma cells that no longer express a BCR. Plasma cells even included a fraction of cells that only express ZAP70. Importantly, these findings together with the identification of distinct B-cell stage-specific subsets of SYK-ZAP70 coexpressing cells, highlight the unique resolution of single-cell protein analysis that would not be achievable with traditional Western blot techniques.
Interactome analysis of SYK and ZAP70 reveals kinase specific integration into B-cell networks
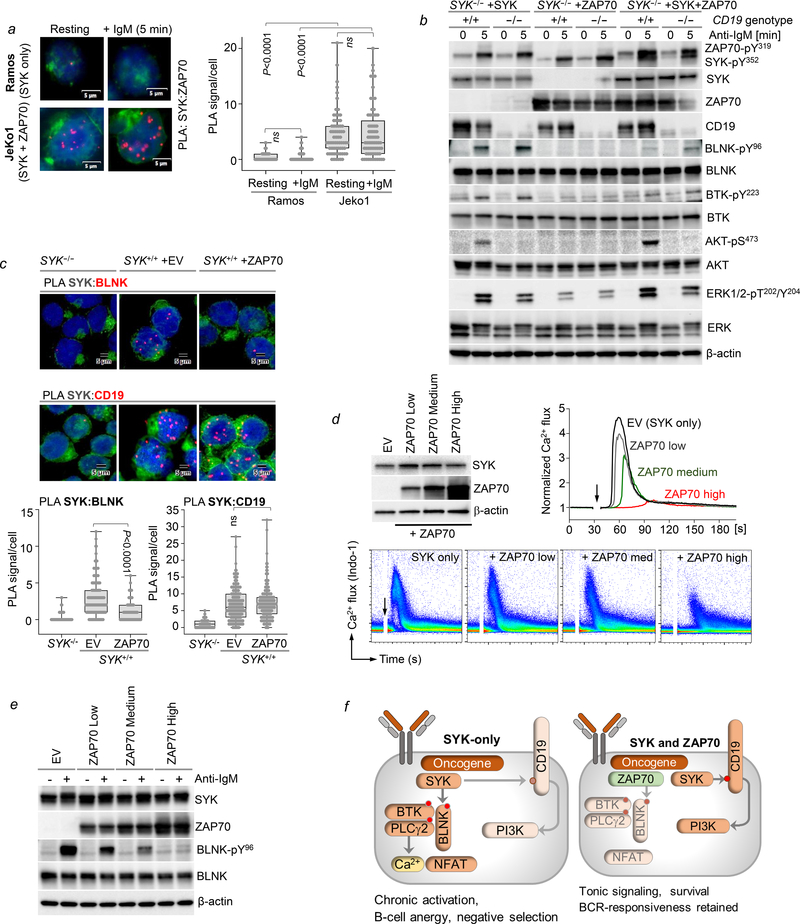
To identify SYK- and ZAP70-dependent signaling complexes, we performed a global interactome analysis in B-ALL and Burkitt’s lymphoma cells using BioID, a proximity-dependent biotinylation strategy that harnesses an engineered promiscuous bacterial biotin ligase, known as E. coli BirA R118G (BirA*) to identify proteins within an approximate radius of 10 nm from the active site of BirA* (Figure S5a–f). This analysis recovered multiple common interaction partners of SYK and ZAP70 (Figure 2a–b, Table S3–4), including BCR-associated adaptors (GAB1 and VAV1), phosphatases (INPP5D and PTPN6), and BLNK, a central scaffold that assembles BCR-downstream signaling molecules BTK-PLCγ2 to mediate intracellular calcium (Ca2+) signaling. Among the interactome proteins specific for ZAP70 were the RAC1-activated kinase PAK1, while the SYK-specific complexes included the BCR co-receptor molecule CD19 in both B-ALL and Burkitt’s lymphoma cells. We confirmed that total CD19 levels were comparable in the BioID lines expressing SYK or ZAP70 to rule out the possibility of altered CD19-protein expression as the reason for preferential interaction with SYK (Figure S5c, S5f). The identification of CD19 as an exclusive interaction partner for SYK but not ZAP70 is particularly important because CD19 carries the YXXM motif on its cytoplasmic tail, which is essential for initiation of PI3K signaling (Aiba et al., 2008; Beitz et al., 1999).
Figure 2: Interactome analyses of SYK and ZAP70 defines kinase-specific integration into B-cell networks.
BioID-interactome analysis of SYK and ZAP70 in Kasumi-2 B-ALL cells (a) and in Ramos lymphoma cells (b). Biotinylated peptide abundance ratios of SYK and ZAP70 BirA*-expressing cells are illustrated. (c) Single-cell clone SYK−/− derived Ramos cells were generated using RNP CRISPR mediated targeting of SYK. BCR-induced Ca2+ flux following IgM stimulation (10 mg/mL) was measured in parental Ramos cells +/− pre-treatment with the SYK inhibitor PRT062607 (5 μmol/L) and in Ramos SYK−/− cells using Indo-1. (d) SYK−/− Ramos cells were reconstituted with MSCV-IRES-GFP empty vector (EV), SYK (SYK or SYK-s) or ZAP70 constructs and analyzed for phosphorylation of BCR-activated substrates by WB. (e) SYK−/− Ramos cells reconstituted with EV, SYK, ZAP70 or SYK+ZAP70 were mixed 1:1 with untransduced SYK−/− cells, and changes in GFP were measured by flow cytometry over 15 days. Data are shown as mean ± standard error of mean (SEM) of two independent experiments. Statistical significance for day 15 was calculated using an unpaired Student’s t-test (*p<0.05). (f) Ramos SYK−/− cells were reconstituted with EV, SYK-wildtype (WT) or SYK-kinase dead (KD) constructs and analyzed for phosphorylation of BCR-activated substrates. (g) BCR-induced Ca2+ flux following IgM stimulation (10 μg/mL) was measured in SYK−/− Ramos cells reconstituted with EV, SYK or ZAP70 using Indo-1. Levels of SYK and ZAP70 were measured by Western blotting using antigen-specific antibodies, and a FLAG-antibody against each tagged protein (inset).
Unlike SYK, ZAP70 lacks the ability to initiate PI3K-activation via CD19
To stringently dissect the signaling capacity of SYK and ZAP70, we deleted SYK using Cas9-ribonucleoproteins (RNPs) in Ramos B-cells (Figure 2c). As expected, SYK−/− Ramos B-cells displayed impaired Ca2+ flux after BCR stimulation, and complete absence of CD19-YXXM phosphorylation and ERK activation (Figure 2c, Figure S5g). Importantly, reconstitution of SYK−/− cells with SYK but not an empty vector (EV) control, restored BCR-dependent Ca2+ flux (Figure S5h). In line with our interactome analyses, reconstitution of SYK−/− cells with an EV control, SYK or ZAP70 revealed that BCR-induced CD19-PI3K activation was only fully restored in cells reconstituted with SYK, despite similar phosphorylation of the activation interdomain residues in SYK (Y352) and ZAP70 (Y319) following BCR-stimulation (Figure 2d). This observation, together with the BioID results, suggests that SYK, but not ZAP70 can readily engage CD19 signaling upon short (Figure 2d) and prolonged (Figure S6a) BCR stimulation. Consistent results were also observed in mantle cell lymphoma cells (Jeko1) that endogenously express both SYK and ZAP70. In this setting, CRISPR-mediated deletion of SYK but not ZAP70 abrogated BCR-induced CD19-phosphorylation (Figure S6b).
One central feature that differentiates SYK from ZAP70, is the presence of a 23 amino-acid insert within the flexible linker region (interdomain B) which bridges the tandem-SH2 and the kinase domains (Turner et al., 2000). This same region is lacking in an alternatively spliced variant of SYK (SYK-s; (Rowley et al., 1995; Yagi et al., 1994). Given the possibility that this linker insert may provide essential docking sites, we compared the capacity of SYK-s, full length SYK and ZAP70 in promoting CD19-PI3K signaling (Figure 2d). Although marginally weaker than full-length SYK, cells expressing SYK-s showed, unlike ZAP70, robust phosphorylation of the CD19-YXXM motif and AKT phosphorylation. Hence, the 23 amino-acid insert in the SYK interdomain B likely does not account for the ability of SYK to engage CD19-dependent PI3K-activation.
Along with its role in amplifying BCR signals, the CD19-PI3K axis plays a critical function in mediating tonic survival signals in B-cells (He et al., 2018; Otero et al., 2001). In the absence of BCR-stimulation, Ramos cells expressing SYK, but not ZAP70, displayed basal levels of CD19 phosphorylation (Figure 2d, S6a). Therefore, we predicted that SYK but not ZAP70 may be able to sustain tonic signaling and provide a growth advantage over time. To test this hypothesis, Ramos SYK−/− cells (GFP-) were mixed in a 1:1 ratio with SYK−/− cells reconstituted with EV, SYK or ZAP70 (GFP+) (Figure 2e). While co-cultures containing EV and ZAP70 transduced cells showed no competitive advantage over SYK−/− cells, the SYK-expressing Ramos cells were enriched over a 15-day period.
SYK, together with Src-family kinases (SFKs), are critical for propagation of BCR-proximal signaling. Of note, several studies suggest that CD19 phosphorylation may be initiated directly by SFKs (Buhl et al., 1997; Fujimoto et al., 2000; Tedder et al., 1994). Hence while our results demonstrate a requirement for SYK in CD19-PI3K signaling downstream of the BCR, this does not exclude that SYK may be acting as a signaling adaptor (e.g. for SFKs), independent of its kinase function. To directly address this question, we tested the phosphorylation of CD19 and AKT in Ramos SYK−/− cells reconstituted with wild-type (WT) SYK or a kinase-dead (KD) form of SYK (SYKK402A). These experiments showed that phosphorylation of CD19-AKT was only observed in the presence of SYK-WT but not SYKK402A (Figure 2f), indicating that CD19-AKT phosphorylation is dependent on SYK kinase activity. In contrast to CD19, BLNK was identified as common interaction partner of both SYK and ZAP70 (Figure 2a–b). Given the role of BLNK as a central scaffold in the initiation of Ca2+ signaling, we tested BCR-induced Ca2+ release in the presence of SYK or ZAP70. Both SYK and ZAP70-expressing cells displayed Ca2+ release following BCR-stimulation (Figure 2g), which was dependent on BLNK (Figure S6c). However, in the sole presence of ZAP70, Ca2+ release was delayed and attenuated compared to cells expressing only SYK.
Coexpression of ZAP70 diverts BCR signaling from Ca2+-NFAT to PI3K-activation
Given that ZAP70 fails to activate PI3K signaling and elicits delayed Ca2+ signals at lower amplitude, we asked whether coexpression of ZAP70 may alter normal BCR signaling. Hence, we compared the BCR-induced signaling output of SYK−/− Ramos cells engineered to express SYK or ZAP70 alone or in combination. Compared to SYK alone, coexpression of ZAP70 with SYK markedly reduced BCR-dependent phosphorylation of BLNK, BTK, PLCγ1/2, the cascade that leads to Ca2+ release (Figure 3a). On the other hand, activation of the PI3K pathway was increased above activation by SYK alone (Figure 3b, S6d). We also studied the phosphorylation of the phosphatases SHP1 and SHIP1, which play key roles as negative feedback regulators of BCR signaling (Pauls and Marshall, 2017; Rhee and Veillette, 2012). However, phosphorylation of SHP1 and SHIP1 and kinase-domain phosphorylation of SYK (Y525/Y526) were largely unaltered in SYK and ZAP70 coexpressing cells (Figure S7a).
Figure 3: Coexpression of ZAP70 suppresses chronic autonomous BCR-signaling and NFAT-activation.
(a-c) Analysis of BCR-mediated signaling in SYK−/− Ramos cells reconstituted with EV, SYK, ZAP70 or SYK + ZAP70. (a-b) Phosphorylation of BCR-activated substrates was measured by Western blot following stimulation with IgM (10 μg/mL). (c) Ca2+ flux following IgM stimulation was measured using Indo-1.(d) Basal calcium oscillations were measured in Ramos lines expressing a fluorescent calcium reporter (GCaMP6s). Images were taken of each well at 40 second intervals for one hour in a 37°C chamber. Represented are 40 individual cells analyzed per condition. (e-f) NFAT localization was analyzed in parental Ramos cells with ZAP70 overexpression following IgM stimulation: (e) Immunofluorescence staining for NFAT1 was performed by incubating slides with NFAT1-primary antibody overnight, followed by 1h incubation with AF-647 secondary antibodies at room temperature. Bar graph shows mean ± standard error of mean from three independent experiments analyzed using QuPath. Statistical significance was calculated using an unpaired Student’s t-test. (f) Western blotting of sub-cellular fractions (cytoplasmic/C vs. nuclear/N) in parental Ramos cells with ZAP70 overexpression.
Consistent with reduced activation of the BLNK-BTK-PLCγ axis in Ramos cells expressing both SYK and ZAP70, Ca2+ flux was weaker and delayed after BCR-stimulation compared to the SYK-only cells (Figure 3c). These findings were confirmed in parental Ramos cells with enforced expression of ZAP70 (Figure S7b–c), and in two additional SYK−/− Ramos clones (Figure S8).
Besides Ca2+ release in response to BCR-stimulation, engineering of Ramos B-cells with a fluorescent-based calcium sensor (GCaMP6s) revealed autonomous Ca2+-oscillations under baseline conditions (Figure 3d). Strikingly, however, concurrent expression of ZAP70 with SYK almost entirely extinguished basal Ca2+ oscillations (Figure 3d). Importantly, the ability of ZAP70 coexpression to suppress BCR-induced Ca2+-release had profound effects on the Ca2+-decoding transcription factors NFAT1/2. As expected, prolonged BCR-stimulation resulted in massive nuclear accumulation of NFAT in SYK+/+ Ramos B-cells, which was sensitive to deletion of BLNK, or BTK-inhibition (Figure S9a). Coexpression of ZAP70 with SYK dramatically reduced BCR-induced NFAT nuclear localization (Figure 3e–f), whereas total NFAT levels were not altered (Figure S9c). Given the role of NFAT in B-cell anergy (Martinez et al., 2015; Yarkoni et al., 2010) and negative selection (Barrington et al., 2006; Goodnow et al., 2005; Healy et al., 1997), it is conceivable that ZAP70, by preventing nuclear NFAT accumulation, could enable autoreactive or premalignant cells to evade negative selection despite prolonged activation of the BCR-signaling pathway.
ZAP70 redirects SYK from BLNK to CD19-PI3K
Because ZAP70 interfered with SYK-mediated NFAT activation, we hypothesized that SYK and ZAP70 may be present within close proximity at the apex of BCR-initiated interactions and compete for common downstream substrates. We utilized proximity ligation assays (PLA) to monitor the proximity of SYK and ZAP70 in resting or BCR-stimulated MCL B-cells (Jeko1) with endogenous SYK and ZAP70 coexpression. Interestingly, SYK-ZAP70 PLA produced robust fluorescent puncta in SYK and ZAP70 coexpressing MCL cells compared to controls (Figure 4a). BCR-engagement did not significantly increase or decrease PLA signals, suggesting that SYK and ZAP70 are constitutively located within close proximity. SYK-ZAP70 complexes may qualitatively change B-cell signaling output, in line with markedly reduced phosphorylation of BLNK, BTK and PLCγ1/2 substrates and reduced Ca2+ flux, in favor of increased activity of the PI3K pathway (Figure 3a–b). For this reason, we examined in a genetic experiment how the two kinases, individually and in combination, engage the BLNK-BTK and PI3K-AKT signaling pathways in the presence and absence of CD19 (Figure 4b). Consistent with a critical role of CD19 in the initiation of PI3K signaling, genetic deletion of CD19 impaired SYK-dependent phosphorylation of AKT, while phosphorylation of BLNK and BTK remained intact (Figure 4b). Conversely, engineered coexpression of ZAP70 with SYK, significantly increased PI3K activity and AKT-phosphorylation, while phosphorylation of BLNK and BTK was reduced. Genetic deletion of CD19 in SYK-ZAP70 coexpressing B-cells also abrogated PI3K-AKT but not BLNK-BTK signaling (Figure 4b). In PLA experiments, we corroborated that engineered coexpression of ZAP70 significantly reduced interactions between SYK and BLNK but not interactions between SYK and CD19 (Figure 4c).
Figure 4: ZAP70 diverts SYK from BLNK-BTK to CD19-PI3K activation.
(a) Proximity-ligation assays (PLA) were performed using antibodies recognizing SYK and ZAP70 in parental Ramos (endogenous SYK only) and Jeko1 (endogenous SYK and ZAP70) using Duolink Red Detection reagents. Statistical significance was calculated using an unpaired Student’s t-test. (b) CD19-deletion was achieved in SYK−/− Ramos cells reconstituted with SYK, ZAP70 or SYK+ZAP70 using RNP CRISPR crRNAs targeting CD19. Phosphorylation of BCR-activated substrates was compared in cells in the presence or absence of CD19 following stimulation with IgM (10 μg/mL). (c) PLA was performed for SYK:BLNK and SYK:CD19 in parental Ramos cells with ZAP70 overexpression. PLA signal/cell was analyzed using BlobFinder software. Statistical significance was calculated using an unpaired Student’s t-test. (d-e) Parental Ramos cells were generated to express a gradient of ZAP70 expression: Zero (EV control), Low, Medium and High, and validated by Western blotting. (d) Ca2+ flux following IgM stimulation was measured in these cells using Indo-1. (e) Phosphorylation of BLNK was measured by Western blotting in these ZAP70-gradient cells following IgM stimulation. (f) Proposed model for impact of ZAP70-expression on BCR-signaling.
Our data suggests, that while ZAP70 can initiate signaling via the BLNK-BTK pathway, it is much less adept than SYK at phosphorylating BLNK and BTK, resulting in decreased activation of Ca2+-NFAT signaling (Figure 3a, c). To further asses the concept that expression of ZAP70 may act as a competitive inhibitor of SYK-mediated activation of the BLNK-Ca2+ cascade, we generated SYK+/+ Ramos lines with a gradient of ZAP70 expression levels (Figure 4d). Consistent with the scenario of competition between SYK and ZAP70 based on stoichiometry, ZAP70 reduced BCR-induced Ca2+-release and phosphorylation of BLNK in a dose-dependent manner (Figure 4d–e). In summary, we propose that ZAP70 redirects SYK activity towards CD19-PI3K activity while exerting a net dominant-negative effect on BLNK-BTK by competing with SYK for binding to these substrates (Figure 4f).
Expression of Zap70 in B-cells subverts mechanisms of negative selection
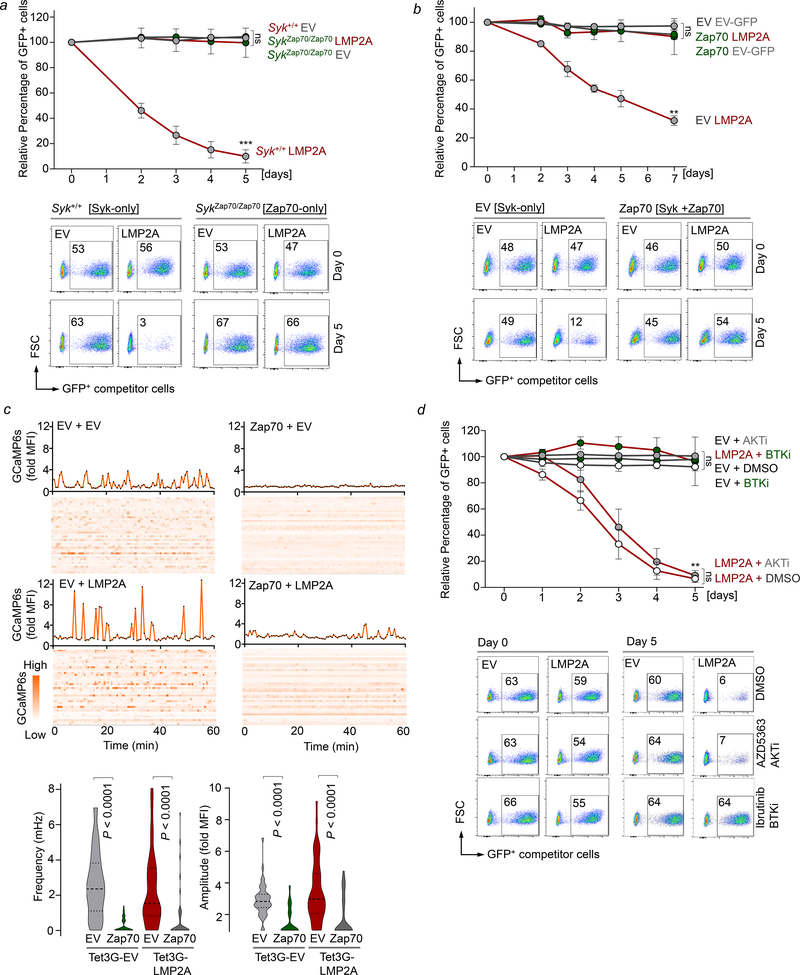
The oncogenic BCR-ABL1 kinase phosphorylates SYK and mimics constitutive activity of the BCR-pathway (Feldhahn et al., 2005a). We engineered expression of GFP-tagged phosphomimetic SYK or ZAP70 mutants (Brdicka et al., 2005) in BCR-ABL1-driven B-ALL cells. GFP+ clones carrying Syk-phosphomimetics were rapidly eliminated from cell culture, while Zap70-phosphomimetics did not impact B-ALL cells (Figure S9d). In an alternative approach, we studied the Epstein-Barr virus latent membrane protein 2A (LMP2A), which mimics oncogenic BCR-signaling by activating SYK (Lanier, 2006; Merchant et al., 2000; Thorley-Lawson, 2001). We confirmed that inducible activation of LMP2A results in hyperphosphorylation of Syk (Figure S9e). To compare the ability of SYK and ZAP70 to trigger clonal deletion, we studied LMP2A-activation in Syk-Zap70 knock-in kinase ‘switch’ mice (SykZap70/Zap70; (Konigsberger et al., 2012) in which Zap70 cDNA replaces exon 2 of Syk at its endogenous locus (Figure 5a). In Syk+/+ B-ALL cells, expression of GFP-tagged LMP2A induced acute cell death resulting in rapid elimination of GFP+ cells, however negative selection mechanisms were completely abolished in SykZap70Zap70 B-ALL cells (Figure 5a). Based on our biochemical experiments in human cells (Figure 3, 4c–e), we hypothesized that enforced expression of Zap70 may reduce BLNK-BTK and Ca2+-signaling output to protect from negative selection. To test this hypothesis, we expressed LMP2A in B-ALL cells either alone or in the presence of Zap70-coexpression. In the presence of SYK alone, expression of LMP2A induced rapid clearance, while coexpression of Zap70 compromised negative selection and enabled survival of GFP+ LMP2A-expressing cells (Figure 5b). Using a fluorescent-based calcium sensor (GCaMP6s), induction of LMP2A increased Ca2+-oscillations that were almost entirely abolished in the presence of Zap70 coexpression (Figure 5c). Together, these results suggest that rather than overall extent of SYK kinase activation, its selective engagement of either BLNK-BTK or PI3K-AKT pathways may determine the outcome of hyperactive BCR-signaling in autoreactive and premalignant B-cells. To directly test this hypothesis, we induced LMP2A expression in B-ALL cells that only expressed Syk in the presence and absence of Btk (ibrutinib) and Akt (AZD5363) inhibitors. Consistent with selective engagement of Blnk-Btk as the underlying mechanism, Syk-dependent negative selection was completely prevented by inhibition of Btk whereas inhibition of Akt had no effect (Figure 5d).
Figure 5: The ratio of SYK and ZAP70 expression sets thresholds for negative B-cell selection.
(a) Syk+/+ or SykZap70/Zap70 BCR-ABL1-driven B-ALL cells were transduced with GFP-tagged EV or LMP2A and relative changes of transduced (GFP+) cells were monitored by flow cytometry. Data are shown as mean ± standard error of mean (SEM) of three independent experiments. Statistical significance for day 5 was calculated using an unpaired Student’s t-test (*** p<0.001). (b) B-ALL cells with Zap70 overexpression were transduced with GFP-tagged EV or LMP2A and relative changes of transduced (GFP+) cells were monitored by flow cytometry. Data are shown as mean ± SEM of three independent experiments. Statistical significance for day 7 was calculated using an unpaired Student’s t-test (**p<0.01). (c) B-ALL cells carrying a fluorescent calcium reporter (GCaMP6s) were engineered to express a doxycycline-inducible LMP2A, or EV control, in the presence or absence of Zap70. Calcium oscillations were measured in these following addition of doxycyline (1μg/mL) to induce expression of LMP2A. Images were taken of each well at 40 second intervals for one hour in a 37°C chamber. 40 individual cells analyzed per condition are represented. Statistical significance of changes in frequency or amplitude of calcium oscillations was calculated using a Mann-Whitney test. (d) B-ALL cells pre-treated with control (DMSO), AKT inhibitor, AZD5363 (3 μM) or BTK inhibitor, ibrutinib (2.5 μM) for 16 hours were transduced with GFP-tagged EV or LMP2A and relative changes in percentages of transduced (GFP) cells were monitored by flow cytometry. Data are shown as mean ± SEM of two independent experiments. Statistical significance for day 5 was calculated using an unpaired Student’s t-test (**p<0.01).
A genetic model for B cell-specific expression of Zap70 in vivo
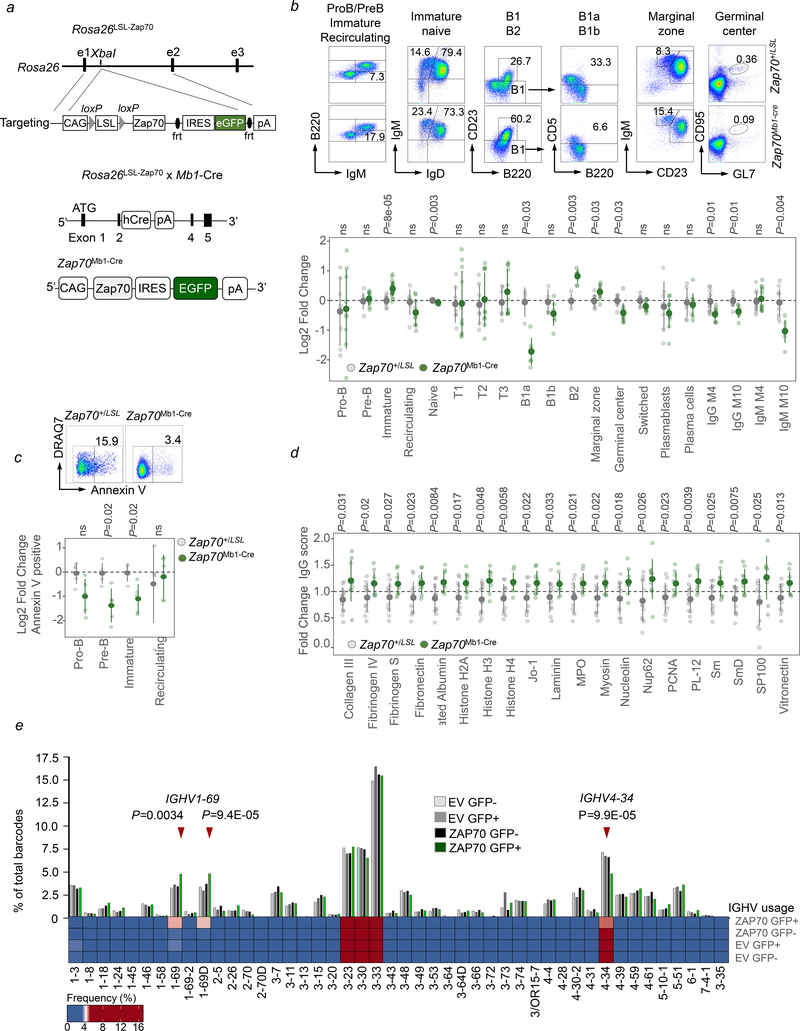
Our results suggest a previously unrecognized consequence of increased expression of ZAP70 in B-cells in altering signaling thresholds, which may impact mechanisms of normal B-cell selection. To validate this concept in vivo, we generated a genetically tractable model of B-cell-specific Zap70 expression. The murine Zap70 cDNA together with enhanced green fluorescent protein (eGFP) was introduced into the mouse Rosa26 locus preceded by a loxP-flanked STOP cassette (LSL; Rosa26LSL-Zap70 mice). Crossing Rosa26LSL-Zap70 mice to Mb1-Cre transgenic mice for B-cell-specific expression resulted in inducible expression of Zap70 from earliest stages of B-cell development in the offspring (Zap70Mb1-Cre mice; Figures 6a, S10a–c). As expected, GFP expression was detected in B-cells from Zap70Mb1-Cre mice from the pro-B stage, but not in bone marrow multilineage progenitors and mature T-cells (Figure S10a). Heterozygous Rosa26LSL-Zap70 mice were used as controls in all experiments. We first assessed B-cell development in primary and secondary lymphoid organs by flow cytometry. Compared to controls, Zap70Mb1-Cre mice had significantly increased proportions of bone marrow and splenic immature B-cells (Figure 6b). Interestingly, the fraction of marginal zone B-cells was increased at the expense of the B1a cell pool. Since strength of BCR-signaling determines the relative size of the B1 (high) and marginal zone (low) B-cell pools (Casola et al., 2004), these results are in line with reduced basal BCR-signaling activity in B-cells from Zap70Mb1-Cre mice.
Figure 6: B-cell-specific expression of Zap70 subverts negative selection of autoreactive B-cells.
(a) Construct of Rosa26LSL-Zap70 model and approach used to generate Zap70Mb1-Cre mice. (b) Summary of flow cytometry analyses showing representative FACS plots, and fold change in frequencies of B-cell subsets and sera IgG and IgM levels in Zap70Mb1-Cre (green) and control (gray) mice compared to average for control subsets. Medullar subsets, including plasmablasts and plasma cells: n=7 (control) and n=9 (Zap70Mb1-Cre), from 3 independent experiments; peritoneal subsets: n=4 (control) and n=7 (Zap70Mb1-Cre), from 3 independent experiments; splenic subsets: n=7 (control) and n=11 (Zap70Mb1-Cre), from 4 independent experiments. Serum immunoglobulins: M4, n=12 (control) and n=11 (Zap70Mb1-Cre); M14, n=10 (control) and n=7 (Zap70Mb1-Cre). Representative dot plots for each differentially regulated population is provided (top). Statistical significance was calculated using an ANOVA with Tukey’s post-test using R-statistical software. (c) Flow cytometry showing spontaneous early apoptosis of medullar B-cell subsets in Zap70Mb1-Cre mice (log2 fold change over control average). Statistical significance was calculated using an ANOVA with Tukey’s post-test. A representative dot plot is provided (top). n=5 (control) and n=5 (Zap70Mb1-Cre) from 2 independent experiments. (d) Sera of Zap70Mb1-Cre (green) and control mice (grey) was taken at 10-months of age and analyzed on the Autoantigen Microarray Panel I (94 antigens) from the University of Texas Southwestern Medical Center to obtain autoreactivity scores. Only antigens both clinically relevant in medicine and significantly different between control and Zap70Mb1-Cre mice are shown. n=12 (control) and n=9 (Zap70Mb1-Cre) for each antigen. Statistical significance was calculated using an ANOVA with Tukey’s post-test using R-statistical software. (e) Humanized mice were generated by injection of NSG-KitW41J/Tyr mice with cord-blood derived CD34+ progenitors transduced with GFP-tagged ZAP70 or empty vector (EV) lentivirus. GFP positive and GFP negative naïve B-cells were sorted from spleens of 6 month-old mice and sequenced for VH-repertoires. Bar-graph and heat map depict VH-usage in B-cells from the four groups. Statistical significance was calculated by χ-squared comparison of VH-usage in ZAP70-expressing (ZAP70 GFP+) B-cells to ZAP70 negative groups (EV GFP−, EV GFP+, ZAP70 GFP−) with multiple test correction using R-statistical software.
B-cell-specific expression of Zap70 impairs central tolerance and B-cell negative selection
Since negative selection of autoreactive B-cells mainly targets the immature B-cell compartment (Limnander et al., 2011), we hypothesized that impaired negative selection may be the reason for the observed expansion of immature B-cells in Zap70Mb1-Cre mice (Figure 6b). While control mice showed the expected increase in spontaneous apoptotic Annexin V+ cells among pre-B and immature cells compared to pro-B cells (Goodnow et al., 2005; Lu and Osmond, 2000), propensity to apoptosis was substantially reduced in pre-B-and immature B-cells from Zap70Mb1-Cre mice (Figure 6c). Based on these observations, we predicted that the reduced propensity to apoptosis and accumulation of immature B-cells may reflect defective central B-cell tolerance in Zap70Mb1-Cre mice. We therefore assessed ageing mice for the development of autoantibodies. Serum of 10 months-old Zap70Mb1-Cre mice and littermate controls was harvested for comparison of IgM and IgG autoreactivity scores against a panel of 94 autoantigens (Table S5). Zap70Mb1-Cre mice had a substantially increased burden of circulating IgM and IgG autoantibodies against 20 (21.3%) of the 94 tested autoantigens (Figure 6d). Among these 20 autoantigens, 12 are known to be associated with autoimmune diseases (i.e. Jo-1, MPO, PL-12, histone H2A, H3, H4, Sm, SmD, nucleolin, laminin, collagen III and fibronectin). Hence, Zap70 expression in B-cells in vivo undermines in B-cell tolerance and enables production of multiple autoantibodies.
ZAP70 expression in developing human B-cells selects for autoreactive VH1–69+ clones as in B-CLL and MCL
In line with defective B-cell tolerance in Zap70Mb1-Cre mice, we next tested whether ectopic expression of ZAP70 would be sufficient to alter mechanisms of B-cell selection during human B-lymphopoiesis. To answer this question, we generated humanized mice by injecting human CD34+ hematopoietic progenitor cells (HPCs) transduced with ZAP70-GFP lentivirus (or GFP-only empty vector control) into NSG-KitW41J/Tyr mice. Importantly, normal central B-cell tolerance barriers function normally in this system (Ippolito et al., 2012; Schickel et al., 2016), providing an ideal setting to understand the effect of ZAP70 expression on normal human B-cell development. For BCR analysis, naive human B-cells that developed from transduced (GFP+) or untransduced (GFP−) HPCs, were isolated from the spleens of 6-month old humanized mice. Single cell sequence analysis for naïve B-cells from all groups showed development of diverse BCR repertoires (Figure S10d–e, Table S6). Detailed analysis of IGHV sequences confirmed that B-cells from the three ZAP70-negative groups had very similar distributions (Figure 6e). The IGHV repertoire of ZAP70+ B-cells (ZAP70-GFP+) also largely resembled the distribution observed in ZAP70-negative groups. Strikingly, however, ZAP70+ B-cells showed a highly significant overrepresentation of IGHV1–69 gene segments and a reduced incidence of IGHV4–34-encoded BCRs (Figure 6e). These observations are particularly relevant in the context of B-CLL and MCL: IGHV1–69 is the most commonly used VH segment in B-CLL (Duke et al., 2003; Fais et al., 1998; Johnson et al., 1997) and is highly enriched in unmutated cases. Similarly, IGHV1–69 accounts for 52% of unmutated MCL cases (Lai et al, 2006). In contrast to IGHV1–69+, ZAP70+ human B-cells developing in NSG-KitW41J/Tyr mice showed a significant underrepresentation of IGHV4–34. Interestingly, usage of IGHV4–34 is common in B-CLL but occurs almost exclusively in indolent B-CLL, which lack ZAP70 expression and carry somatically mutated VH regions (Stamatopoulos et al., 2007; Xochelli et al., 2017). These results suggest that ZAP70 expression may be an early event in some cases of B-CLL and MCL pathogenesis, and selectively enable survival and clonal expansion of IGHV1–69 autoreactive, premalignant naïve B-cells.
Aberrant expression of Zap70 contributes to malignant B-cell transformation in vivo
In addition to B-CLL and MCL, we found aberrant coexpression of ZAP70 as a common feature of human B-ALL (Figure 1b–c). B-cell development in BCR-ABL1-transgenic mice (Heisterkamp et al., 1990) transitions a phenotypically normal pre-leukemic phase before giving rise to full-blown B-ALL. Comparing premalignant to overt leukemic phases, mRNA and protein levels of Syk were significantly decreased during transformation, whereas Zap70 mRNA and protein levels were increased (Figure 7a–b). To directly test whether aberrant coexpression of Zap70 facilitates malignant B-cell transformation, we utilized Cas9-RNP delivery of crRNAs targeting Zap70 in bone marrow pre-B cells derived from pre-leukemic BCR-ABL1-transgenic mice. Consistent with the concept that coexpression of Zap70 enables premalignant clones to evade Syk-mediated negative selection, these experiments showed that deletion of Zap70 significantly prolonged survival of recipient mice and latency to overt leukemic transformation in vivo (Figure 7c).
Figure 7: Zap70 prevents B-cell anergy to enable progression of B-ALL and B-CLL in vivo.
(a) mRNA expression of Syk or Zap70 determined by microarray analysis of 3 independent C57B6 or BCR-ABL1-tg mice which developed B-ALL (GSE110104). Statistical significance was calculated using an unpaired Student’s t-test (**p<0.01). (b) Western blot analysis of Syk and Zap70 was performed in purified CD19+IgM− B-cells from at least 3 healthy C57B6 or BCR-ABL1-tg mice which developed B-ALL. (c) pre-B cells from BCR-ABL1-tg pre-leukemic mice were electroporated with Cas9-crRNA-tracRNA complexes containing crRNAs targeting Zap70 or a non-targeting (NT) control and injected into sublethally irradiated recipient NSG mice (n=5 for NT, n=6 for Zap70-crRNA group). Statistical significance was calculated using Log-rank test. (d) Inset, Western blot analysis of Syk and Zap70 was performed on purified B-CLL cells (CD19+ CD5+) cells from spleens of Eμ-TCL1 and Eμ-TCL1 Syk+Zap70 mice. Bar graphs and representative FACS plots show circulating blasts (CD19+ CD5+) measured in the peripheral blood of 9-month-old mice. Statistical significance was calculated using unpaired Student’s t-test. (e) Overall survival was analyzed in cohort of Eμ-TCL1 (n=15) and Eμ-TCL1 Syk+/Zap70 (n=21) mice. Statistical significance was calculated by Log-rank test. (f) Nfat1 and Nfat2 expression was measured in cytoplasmic and nuclear fractions of B-CLL cells derived from Eμ-TCL1 and Eμ-TCL1 Syk+/Zap70 mice. (g) Immunofluorescence staining for Nfat1 was performed in purified B-CLL cells (CD19+CD5+) cells from spleens of sick Eμ-TCL1 and Eμ-TCL1 Syk+/Zap70 mice. Bar graph shows mean ± standard error of mean from three mice for each genotype, analyzed using QuPath. Statistical significance was calculated using an unpaired Student’s t-test. Western blot analysis of phospho-Syk/Zap70 (h) and pan tyrosine phosphorylation (4G10) (i) in lysates from Eμ-TCL1 and Eμ-TCL1 Syk+Zap70 B-cells treated with anti-IgM for 5 minutes. (j) Ca2+ flux was analyzed in B-CLL cells from Eμ-TCL1 and Eμ-TCL1 Syk+/Zap70 mice following anti-IgM stimulation using Indo-1. (k) RNA sequencing was performed on CLL cells purified from spleens of 4 Eμ-TCL1 and 4 Eμ-TCL1 Syk+/Zap70 mice GSEA analysis was performed using the DOSE package in R and compared to gene expression from Nfat2−/− Eμ-TCL1 CLL cells (Martinez et al., 2015).
To assess the contribution of aberrant Zap70 coexpression in the development of B-CLL, we crossed heterozygous Syk+/Zap70 knockin mice with Eμ- TCL1 B-CLL mice (Bichi et al., 2002), which develop fatal B-CLL disease starting from 12 months of age. Analysis of purified splenic B-CLL blasts (CD19+ CD5+) confirmed coexpression of Syk and Zap70 in Eμ-TCL1 × Syk+/Zap70 crosses, whereas Eμ-TCL1 mice only expressed Syk (Figure 7d). Wildtype, Eμ-TCL1, Syk+/Zap70 and Eμ-TCL1 × Syk+/Zap70 mice were closely monitored over a 450-day period, including monitoring of peripheral blood CD5+ B-cells by flow cytometry. A combined analysis of 9-month old mice revealed a significant increase in peripheral blood CD5+ B-cells in the Eμ-TCL1 × Syk+/Zap70 mice (Figure 7d). Accelerated expansion of CD5+ B-cells in Eμ-TCL1 × Syk+/Zap70 mice was paralleled by shortened disease-latency to fatal B-CLL and decreased overall survival (Figure 7e). Previous studies have shown that similar to human disease, CLL cells from the Eμ-TCL1 model have characteristic features of anergic B-cells (Marklin et al., 2017; Martinez et al., 2015), a state that reflects functional inactivation of autoreactive B-cell clones after chronic persistent BCR-stimulation. Mechanistically, anergy is induced by increased Ca2+-NFAT signaling (Dolmetsch et al., 1997). Given our findings that Zap70 suppressed autonomous and BCR-induced Ca2+-NFAT signaling in B-cells (Figure 3c–f, 4d, 5c), we predicted that Zap70 could enable premalignant B-CLL clones to evade anergy. Hence, we analyzed nuclear Nfat levels in purified B-CLL cells from terminally ill Eμ-TCL1 and Eμ-TCL1 × Syk+/Zap70 mice. While total levels of Nfat were similar in CD5+ B-CLL blasts from Eμ-TCL1 and Eμ-TCL1 × Syk+/Zap70 mice (Figure S10f), nuclear localization of Nfat was markedly reduced in CLL cells from Eμ-TCL1 × Syk+/Zap70 mice (Figure 7f–g). Furthermore, CLL cells from Eμ-TCL1 × Syk+/Zap70 retained responsiveness to BCR-stimulation in vitro (Figure 7h–j) and recapitulated the gene expression program of Nfat2-deficient CLL cells (Figure 7k, S10h).
DISCUSSION
We have previously shown that activation of SYK sets the threshold for negative selection and clonal deletion of B-cells (Chen et al., 2015; Shojaee et al., 2015; Shojaee et al., 2016). Thereby, hyperactivation of SYK, either downstream of an autoreactive BCR or a transforming oncogene triggers cell death for clearance of autoreactive or premalignant clones. Furthermore, genetic lesions and aberrant splicing of the SYK-downstream signaling molecules BLNK (Flemming et al., 2003; Jumaa et al., 2003) and BTK (Feldhahn et al., 2005b; Kersseboom et al., 2003) have been reported in B-ALL patients, suggesting that limiting SYK-downstream signaling via BLNK-BTK may contribute to leukemogenesis.
In tune with altered BCR-signaling strength and quality in SYK and ZAP70 coexpressing cells, not only BTK-inhibition (e.g. by ibrutinib) but also ZAP70-mediated diversion of BCR-signaling rescued autoreactive or premalignant B-cell clones from negative selection.
This was further supported by the phenotype of our new Zap70Mb1-Cre knockin mouse model designed to drive B-cell-specific coexpression of Zap70 in addition to Syk. Pre-B and immature B-cells in Zap70Mb1-Cre knockin mice had a substantially decreased apoptosis rate and increased autoantibody production against multiple self-antigens, which supports defective central tolerance. Interestingly, a recent study of a family with a hereditary autoimmune syndrome and compound heterozygous ZAP70 mutations resulting in a net gain of function suggested that increased activity of ZAP70 may subvert negative selection of autoreactive B-cells (Chan et al., 2016).
ZAP70 expression correlates with poor prognosis but is also associated with increased BCR-signaling in B-CLL (Crespo et al., 2003; Durig et al., 2003; Wiestner et al., 2003). On the surface, our findings that ZAP70 largely suppresses SYK-mediated phosphorylation of BLNK, BTK, PLCγ2 and Ca2+-signaling seem to contradict the consensus of increased BCR-activity in ZAP70+ B-CLL cells. However, reconciling our observations with previously published work, we showed that by suppressing chronic autonomous BCR-activity, ZAP70 enables B-cell clones to evade NFAT-mediated induction of an anergic state and to retain responsiveness upon BCR-engagement (Figure 3d, 7f–j). These findings match observations in patients: Compared to ZAP70+ cases, B-CLL cells lacking coexpression of ZAP70 are mostly anergic, have lost BCR-responsiveness and generally give rise to more indolent disease (Mockridge et al., 2007; Packham et al., 2014). In direct support for this concept, we demonstrate that Syk and Zap70 coexpression significantly accelerated development of B-CLL in vivo, consistent with significantly reduced levels of nuclear Nfat.
Limitations of this study
Our single-cell protein expression studies show that, in addition to CLL, coexpression of SYK and ZAP70 kinases occurs in other B-cell malignancies including MCL and B-ALL. However the molecular mechanisms that promote aberrant ZAP70 expression in these diseases remain unclear. We also identified expression of ZAP70 in some normal B-cell fractions, including early B-cell progenitors and a small number of memory B-cells, raising the intriguing possibility that these cells may represent the cell of origin of some ZAP70+ malignancies. In addition, our preliminary observations (Figure 7a) suggest that expression of ZAP70 may be a compensatory mechanism resulting from downregulation of SYK. However, additional studies will be required to determine whether ZAP70 expression in B-cell malignancies results directly from oncogenic signaling, altered levels of SYK, or whether it represents the malignant transformation of a ‘poised’ ZAP70-expressing normal B-cell.
Since constitutive, B-cell-specific (Mb1-Cre) expression of Zap70 affects normal development and selection of B-cells, it is possible that the observed differences in B-cell signaling are not only caused by Zap70 but also reflect the altered composition of the B-cell pool in these mice. While future studies of genetic mouse models based on inducible, rather than constitutive expression of Zap70 could elegantly address this limitation, we have addressed this issue in experiments that use enforced Zap70 expression in normal B-cells (Figure 5c) and biochemical studies of genetically engineered human B-cell lines (Figure 4).
Here, we propose that ZAP70 diverts BCR-signaling through competition with SYK for substrates such as BLNK, while leaving PI3K survival signals intact, which are downstream of SYK-CD19 specific interactions. We cannot exclude, however, that other BCR-proximal signaling molecules which were not explored in this study contribute to this phenotype. Nonetheless, our work reveals an unexpected mechanism, by which ZAP70, a kinase otherwise adapted to the cellular and signaling context of T-cells, alters the signaling and consequent functional outcome of B-cell selection.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Markus Muschen (markus.muschen@yale.edu).
Materials Availability
All cell lines, plasmids, and other stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement
Data and Code Availability
The autoantigen arrays and sequencing datasets generated in this paper have been deposited in the Gene Expression Omnibus (GEO) with the following accession numbers: autoantigen arrays from Zap70Mb1-Cre mice (GSE148797), single-cell V(D)J sequencing from humanized mouse (GSE149199) and RNA-sequencing from Eμ-TCL1 mice (GSE159908). For analysis of ZAP70 expression in B-cell malignancies published mRNA expression datasets were combined and normalized using MAS5 (see Table S7 for full information). Analysis of Syk and Zap70 expression in the BCR-ABL1-tg mouse model was obtained from GSE110104. Original data for figures in this paper is available at Mendeley Data (DOI:10.17632/sdkg5xfvkx.2).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human leukemia and lymphoma patient-derived xenografts and cell lines
CLL patient samples were obtained in compliance with the internal review boards of the Beckman Research Institute of City of Hope. Xenografts from B-ALL, MCL and DLBCL patient samples used in single-cell western experiments were propagated in NSG mice. The patient derived xenografts (PDXs) are part of a joint initiative with Dr. David Weinstock, Dana Farber Cancer Institute; (Townsend et al., 2016). Detailed information including cytogenetics and immunophenotype is available at http://www.proxe.org/. Likewise, detailed information on our leukemia xenograft repository for PDXs, including those which can be continuously cultured in vitro is available at http://lymphoblasts.org/. Full information for the patient samples used in this study can be found in Table S8. The human cell lines and PDXs were cultured in RPMI-1640 or Alpha-Mem (GIBCO) respectively, supplemented with GlutaMAX, 20% FBS, 100 IU/mL penicillin, 100 μg/mL streptomycin at 37 °C in a humidified incubator with 5% CO2. All human primary samples and cell lines were tested negative for mycoplasma by detection kit (MycoAlert PLUS, LONZA).
Primary hematopoietic cells
Healthy BM, peripheral blood and cord blood CD34+ samples were purchased from ALLCELLS. For single-cell westerns, Pro/Pre-B (CD19+CD38+CD10+CD27−IgM−), Immature (CD19+CD38+CD10+CD27−IgM+), and plasma cells (CD19+CD27+CD38+) were purified from BM, and pan-B (CD19+), pan-T (CD3+), CD5+, new emigrant (CD19+CD38+CD10+IgM+CD21+) mature naïve (CD19+CD38+CD10−IgM+CD21+), and memory B-cells (CD19+CD10−CD27+) were purified from peripheral blood by flow sorting. Centroblasts (CD19+CD38+CD77+) and centrocytes (CD19+CD38+CD77−) were purified from inflamed tonsils donated with informed consent from the Children’s Hospital Los Angeles (CHLA). All experiments were performed on cell populations isolated from at least two independent donors.
TMA analysis
Patient biopsies were banked in compliance with the internal review board of the Beckman Research Institute of City of Hope. Tissue microarrays (TMAs) were constructed with Formalin-fixed paraffin-embedded biopsies from patients diagnosed with follicular lymphoma, diffuse-large B-cell lymphoma, classical Hodgkin’s lymphoma and mantle cell lymphoma. Tissue sections were cut at 4 microns and processed on a Ventana Discovery Ultra IHC automated stainer (Ventana Medical Systems, Roche Diagnostics, Indianapolis, USA). The TMA slides were then incubated with anti-SYK (mouse mAb CST, 4D10) and anti-ZAP70 (rabbit mAb, 99F2) antibodies, followed by HQ secondary antibodies (DISCOVERY) and anti-HQ-HRP detection system (DISCOVERY). The stains were visualized with ChromoMap DAB Kit (DISCOVERY) and counterstained with hematoxylin (Ventana). Triple IHC stains were also performed on Ventana Discovery Ultra IHC Auto Stainer. The three antigens, ZAP70 (rabbit mAb, CST 99F2), SYK (mouse mAb CST, 4D10) and CD20 (mouse mAb, Ventana clone L26) were sequentially detected and heat inactivation was used to prevent antibody cross-reactivity between the same species. Following each primary antibody incubation, DISCOVERY anti-Rabbit HQ or NP or DISCOVERY anti-Mouse HQ or NP and DISCOVERY anti-HQ-HRP or anti-NP-AP were incubated. The stains were then visualized with DISCOVERY Yellow Kit, DISCOVERY Teal Kit and DISCOVERY Purple Kit, respectively, and counterstained with hematoxylin (Ventana).
Murine pre-B cell culture and generation of pre-B ALL cells
Bone marrow cells were harvested from 6- to 12-week-old mice by flushing cavities of femur and tibia with cold media and cells were filtered through a 70 μm mesh to generate single cell suspensions. Bone marrow cells were then cultured in Iscove’s modified Dulbecco’s medium (IMDM; GIBCO) with GlutaMAX containing 20% FBS, 50 μmol/mL 2-mercaptoethanol, 100 IU/mL penicillin, 100 μg/ml streptomycin. Cells were maintained in presence of 10 ng/mL of recombinant mouse IL-7 (mIL-7) (Peprotech) for at least 7 days to generate IL-7 dependent pre-B cells. To produce BCR–ABL1 leukemia cells, pre-B cells were retrovirally transduced with BCR-ABL1 and then mIL-7 was removed to promote the outgrowth of transformed cells.
Generation of Rosa26LSL-Zap70 mice
Total mRNA was extracted from BALB /c splenic T-cells using RNeasy Kit (Qiagen). cDNA synthesis was done using High Capacity Reverse Transcription Kit (Applied Biosystems). The coding sequence of murine Zap70 (1850 kb, NM14888.1) was amplified from cDNA using the following primers: Forward 5′- ATGCCCGATCCCGCGGCGCACCTGCCATTC-3′ and Reverse 5’-GCCACATGCAGCCTCGGCCACCT GTTC-3′. The PCR conditions were as follows: 94°C for 2 min; 18 cycles at 94°C for 30 s, 62°C for 30 s, 62°C for 2 min, 72°C for 3 min; final elongation at 72°C for 5 min. The PCR product was firstly cloned in the TA cloning vector (Invitrogen) then two Asc1 restriction sites were added by PCR using the following primers: Forward 5′- ATTGGCGCGCCTCATACGCCACCATGCCCGATCCCGCGG-3′ and Reverse 5′- ATTGGCGCGCCTCAGCCACATGCAGCCTCG-3′. The PCR conditions were as follows: 94°C for 2 min; 5 cycles at 94°C for 30 s, 52°C for 2 min, 72°C for 3 min; 13 cycles at 94°C for 30 s, 65°C for 2 min, 72°C for 3 min; final elongation at 72°C for 5 min. The PCR product was then cloned into the CTV Vector (Addgene) as previously described (Xiao et al., 2007; Xiao et al., 2008). The Rosa26LSL-Zap70 mouse line was established at the MCI/ ICS (Mouse Clinical Institute—Institut Clinique de la Souris, Illkirch, France). The linearized construct was electroporated in C57BL/6N mouse embryonic stem (ES) cells (ICS proprietary line). After neomycin selection, targeted clones were identified by long-range PCR and further confirmed by Southern blot with an internal (Neo) probe and a 5′ external probe. Two positive ES clones were validated by karyotype spreading and microinjected into C57BL/6N blastocysts. Resulting male chimeras were bred with wild-type C57BL/6N females. Germline transmission was achieved in the first litter. The presence of the transgene in the mice was assessed by PCR. Zap70Mb1-Cre mice were obtained by crossing heterozygous Rosa26LSL-Zap70 mice with Mb1-Cre mice allowing Zap70 overexpression specifically in B-cells. For all mice experiments, heterozygous Rosa26LSL-Zap70 mice (bearing the Zap70 transgene but without expression) were used as controls. All animals experiments were performed with the approval of the “Direction departementale des services veterinaires” (Strasbourg, France) and protocols were approved by the ethics committee (“Comité d’éthique en matière d’Experimentation Animale de Strasbourg,”CREMEAS; APAFIS#10771–2018011808141516).
Humanized mice and naïve B-cell isolation
Cord blood derived human CD34+ cells (ALLCELLS) were thawed and were lentivirally transduced with pCL6-ZAP70-GFP or an empty vector control. To obtain optimal transduction, CD34+ cells were maintained for 2 days in StemSPan SFEM II with hTPO (50 ng/mL), hSCF (50 ng/mL) and hAngpl5 (100 ng/mL). To generate humanized mice, 100, 000 transduced CD34+ cells were injected intravenously into 8 to 10 weeks old NOD/Scid/Il2rg/KitW41J/Tyr mice. Engraftment of human cells was confirmed in peripheral blood after 8 weeks. GFP positive and GFP negative naïve B-cells (CD19+CD10−CD27−) were sorted from spleens of 6 months-old mice and 10,000 single-cells per group (pooled from 3 independent mice) were sequenced for BCR-VDJ-repertoire using 10× Genomics.
Eμ-TCL1 in vivo CLL model
Eμ-TCL1 mice were crossed with heterozygous Syk+/Zap70 mice to generate an in vivo CLL model with Zap70 expression. Littermate Wt and Syk+Zap70 mice were maintained as controls. Eμ-TCL1 cohorts were monitored for onset of disease, and peripheral blood leukemic blasts were measured regularly by FACS. Mice were euthanized when terminally ill, ascertained by a hunched posture, difficulty breathing and inability to move. For experiments with CLL splenocytes, spleens from sick mice were negatively enriched for B-cells by MojoSort Mouse Pan B Cell Isolation Kit II (Miltenyi Biotec) and purity confirmed by flow cytometry. Kaplan-Meier survival analyses were performed using GraphPad Prism 7 (GraphPad Software Inc.) to compare overall survival (OS). Mantel–Cox log-rank test was used as statistical analysis using GraphPad Prism 7.
METHOD DETAILS
Quantitative RT-PCR for expression of Zap70
To assay expression of Zap70 mRNA in B-cells, GFP+ splenic B-cells from Zap70Mb1-Cre and control mice were purified by FACS sorting. Total RNA was isolated using the RNeasy Mini Kit (Qiagen) including the step with DNAse1, according to the manufacturer’s instructions. cDNA was synthesized with the High-Capacity Reverse Transcription Kit (Applied Biosystems). Real-time quantitative PCR was performed on 10ng of cDNA using Taqman Universal Mastermix (Applied Biosystems) and probes specific for Zap70, Hprt1 and Gapdh (Applied Biosystems). Each sample was amplified in triplicate in a StepOnePlus real-time PCR machine (Applied Biosystems). Both mouse Hprt and Gapdh were used as endogenous controls. The relative change in expression (fold values) was calculated using the 2−ΔΔC cycle method.
Retroviral constructs and transduction
Transfection of retroviral constructs was performed using Lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions. Retroviral supernatant was produced by co-transfecting HEK293FT cells with the plasmids pHIT60 (gag-pol) and pHIT123 (ecotropic envelope). Lentiviral supernatant was produced by co-transfecting HEK293FT cells with the plasmids pCDNL-BH and VSV-G or pAmpho. HEK293FT cells were cultured in high glucose Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) with GlutaMAX containing 10% fetal bovine serum, 100 IU/mL penicillin, 100 μg/mL streptomycin, 25 mmol/L HEPES, 1 mmol/L sodium pyruvate and 0.1 mmol/L non-essential amino acids. Regular medium was replaced after 16 hours by growth medium containing 10 mmol/L sodium butyrate for 8 hours. Twenty-four hours later, viral supernatant was collected and clarified through a 0.45 μm filter. For retroviral transductions, 2 mL of virus was loaded by centrifugation (2,000 g, 90 min at 32 °C) onto 50 μg/mL RetroNectin- (Takara) coated non-tissue six-well plates, followed by centrifugation of target cells (2 million/well) at 600 g for 30 min. For lentiviral transductions, cells were resuspended in viral supernatant and centrifuged onto RetroNectin coated non-tissue six-well plates at 600g for 30 min.
Western blotting
Cells were washed with ice-cold PBS and lysed in CelLytic buffer (Sigma-Aldrich) supplemented with 1× cOmplete Mini EDTA-free Protease Inhibitor cocktail (Calbiochem Millipore) and 1 mM PMSF (CST). 5–20 μg of protein mixture per sample was separated on midi precast gels (BioRad), transferred onto nitrocellulose membranes and blocked with 3% BSA before probing overnight with primary antibodies. Membranes were then incubated with alkaline-phosphatase conjugated secondary antibodies (Invitrogen) and analyzed with Chemi DocTM MP Imaging System (BioRad). For nuclear and cytoplasmic separation, lysates were prepared from 5–106 cells using the NE-PER Nuclear and Cytoplasmic Extraction Reagents kit from ThermoFisher according to manufacturer’s instructions.
BioID with BioSITe
Kasumi-2 and Ramos cells were transduced with GFP labelled SYK or ZAP70 constructs with N-terminal BirA* tags or a BirA* expressing vector control. Pure GFP+ populations were obtained by flow sorting. Efficient biotinylation in presence of SYK or ZAP70 was validated in whole cell lysates of cells cultured with 50 μM Biotin (Sigma) for 16 hours. Streptavidin-HRP (Thermo Scientific) was used to detect biotinylated proteins by western blotting. For BioSITe proteomics, samples were processed as previously described (Kim et al., 2018). In brief, following overnight incubation with 50 μM Biotin, 200–300 million cells were pelleted, washed with PBS and lysed using a lysis buffer containing 50 mM TEABC and 8 M urea. The protein concentration of samples was measured by BCA assay. Reduction and alkylation were performed by serial incubation with 10 mM DTT for 30 min and by 30 mM IAA for 30 min in the dark. Lysates were diluted to 2 M urea by adding three cell lysate volumes of 50 mM TEABC. The proteins were digested with trypsin (Worthington Biochemical Corporation Supplier Diversity Partner TRYPSIN TPCK TREATED) at a 1:20 ratio of trypsin to total protein at 37 °C overnight. For isolation of biotinylated peptides, 100μg of anti-biotin antibody (Bethyl) was coupled to 120 μL of protein G bead slurry overnight at 4 °C. Antibody-coupled beads were further washed with PBS once and BioSITe capture buffer (50 mM Tris, 150 mM NaCl, 0.5% Triton X-100) twice. Peptides were dissolved in 1 mL of BioSITe capture buffer and subsequently incubated with anti-biotin antibody-bound protein G beads for 2 hours at 4 °C. The bead slurry was sequentially washed two times with BioSITe capture buffer, two times with 50 mL of Tris, and two times with ultrapure water. Biotinylated peptides were eluted four times using elution buffer (80% acetonitrile and 0.2% trifluoroacetic acid in water). The eluent was further cleaned up using C18 reversed-phase column as previously described. The resulting peptides were fractionated and analyzed on an Orbitrap Fusion Lumos Tribrid Mass spectrometer coupled to the Easy-nLC 1200 nanoflow liquid-chromatography system (Thermo Fisher Scientific). Proteome Discoverer (v 2.1; Thermo Scientific) suite was used for quantitation and peptide identification as previously described (Kim et al., 2018).
Single-cell western blot analyses of SYK and ZAP70
For single-cell protein analyses, 100,000–500,000 cells were resuspended in 2 mL of 1× Suspension Buffer (Small scWest kit for small or standard cells, ProteinSimple). On each scWest chip, there is a 2×8 array of 400-well blocks (total 6400 wells) patterned into a pre-cast polyacrylamide gel. Prior to loading of cells, scWest chips were hydrated in 1× Suspension Buffer for at least 10 minutes at RT. Cells were then loaded onto each chip and allowed to settle for ~7–10 minutes while visualizing under a light microscope. Optimal cell loading of ~10–15% of wells assures that 2% or fewer wells will contain more than 1 cell. Following removal of unsettled cells by washing with 1× Suspension Buffer, cells were lysed (10 seconds), electrophorized (60 seconds, 240V) and immobilized with UV capture (240 seconds) using the Milo (ProteinSimple). Chips were then probed with SYK and ZAP70 primary antibodies (1:20) overnight at 4°C, followed by probing with fluorescent secondary antibodies the following day for 1 hour at room temperature. To verify each detected peak was associated with cell occupancy in each well, chips were subsequently probed for Histone H3 (1:30), followed by DNA staining using TOTO™-1 Iodide (514/533; ThermoFisher Scientific). Fluorescence intensities on chips were measured using the InnoScan 710 microarray scanner (Innopsys). Scout Software (ProteinSimple) was used for peak identification and data analysis, and each individual cell was inspected to verify that the signal was associated with a peak located at the correct migration size (distance from the well center).
Ribonucleoprotein CRISPR-mediated gene deletion
Chemically synthesized predesigned crRNAs and tracrRNAs (IDT technologies) reconstituted at 100 μM in Duplex Buffer (IDT technologies) were mixed 1:1 and annealed by incubation at 95 °C for 5 min. Recombinantly produced Cas9 (10 μg) was then mixed with crRNA-tracrRNA duplexes to produce RNA ribonucleoprotein (RNP) complexes. RNPs were freshly complexed prior to electroporation. Electroporation was performed using the Neon Transfection System (ThermoFisher) with 2–3 million cells combined with each RNP reaction. To generate single-cell clones, individual live cells were sorted into 96-well plates 72h after electroporation.
Flow cytometry
For cell surface staining, PBS washed cells were blocked with Fc blocker (BD Biosciences) for 10 min on ice and then stained with the indicated antibodies or isotype control for 30 min on ice. Cells were then washed and resuspended in chilled PBS containing 0.75 μg/mL of DAPI or DRAQ7 to exclude dead cells. For medullar Annexin V staining, cells were washed and incubated using Annexin V buffer (140 mM NaCl, 5 mM CaCl2, Hepes/NaOH 10 mM, pH 7.4), and DRAQ7 (Biolegend) to identify or exclude dead cells. For intracellular Zap70 staining, erythrocyte-depleted splenic cells were first stained with Fixable Viability Dye eFluor 780 (eBioscience) according to the manufacturer’s instructions. Cells were then fixed and permeabilized with Lysing solution (BD Biosciences) according to the manufacturer’s instructions, followed by staining with fluorescence-labeled antibody against Zap70 at room temperature for 40 min. Zap70 positivity was defined using isotypic control for each sample on splenic T-lymphocytes. For competitive growth assays, percentage of GFP+ cells was monitored by flow cytometry daily. All flow cytometry experiments were performed with the LSRFortessa X-20 or FACScalibur flow cytometers (BD Biosciences). Fluorescence-based cell sorting was performed on a FACSAria II (BD Biosciences). FACS data was analyzed with FlowJo software (FlowJo, LLC) or Kaluza Software (Beckman Coulter).
Intracellular calcium mobilization measurements
1–5 × 106 viable cells were incubated with 1.5 μM of the Ca2+-binding dye Indo-1 (Thermo Fisher Scientific) for 30 min at 37 °C in the dark. Cells were then washed twice with PBS and resuspended in PBS/0.1% BSA and maintained at 37 °C for 10–15 minutes before analyzing by flow cytometry. Basal Ca2+ was recorded for 30 seconds, and continued to be measured for up to 300 seconds following addition of stimulant. For BCR stimulation 10 μg/mL of anti-IgM (F(ab’)2 anti-human μ-chain, (Jackson Immunoresearch) was used. Indo-1 ratios were calculated using derived and kinetics parameters on FlowJo software (FlowJo, LLC), and normalized to basal calcium readings.
GCaMP6s Calcium oscillation experiments
Ramos cell lines expressing GCaMP6s-MSCV-IRES-puromycin were generated by retroviral transduction and selected in puromycin. Mouse B-ALL cells expressing GCaMP6s-MSCV-IRES-blasticidin were generated by retroviral transduction and selected in blasticidin. Equal number of cells were seeded in Cell-Tak (FisherScientific) coated 12-well microscopy plates in pre-warmed Live Cell Imaging Solution (Thermo Fisher Scientific) with 20% FBS by centrifugation at 400g for 5 minutes. Wells were imaged using the Widefield Zeiss Observer 7 microscope in a pre-warmed 37 °C chamber. Images were taken of each well with 60ms exposure time at 40 second intervals for one hour. At least 40 cells were analyzed per condition. Time-lapse image analysis was performed using Time Series Analyzer V3 plugin on Fiji software.
Proximity ligation assays
Cells were washed and settled on Shandon Single Cytoslide slides by cytospin at 450 rpm for 5 min. Cells were then fixed with 4% paraformaldehyde (CytoFIx, BD Biosciences) for 20 min at room temperature and then washed in PBS. Cellular membranes were labeled with 5 μg/ml wheat germ agglutinin (WGA) conjugated to Alexa Fluor 488 (Thermo Fisher Scientific) for 10 min at room temperature. Cells were then permeabilized with methanol (Perm Buffer III, BD Biosciences), washed in PBS and blocked with Duolink Blocking buffer (Sigma) for 30 min at 37°C. Primary antibodies were diluted 1:150 in Duolink Antibody Diluent (Sigma) and incubated overnight at 4°C. The following day, cells were washed with PLA Wash Buffer A (Sigma), followed by addition of the appropriate Duolink probes (Sigma), according to the manufacturer’s instructions. Cells were incubated for 1 hour at 37 °C, after which cells were washed in PLA Wash Buffer A. Ligation and amplification steps of PLA were performed using the Duolink in situ Detection Reagents Red kit (Sigma) according to the manufacturer’s instructions. Following the PLA, slides were dried and cells were mounted in Prolong Gold anti-fade mounting media with DAPI (ThermoFisher). Images were acquired with an Olympus IX3–55 and analyzed by CellSens imaging software (Olympus). PLA spots were counted in cell lines using Blobfinder software. One PLA dot was defined as a pixel size of 5 × 5. Statistical significance was calculated using unpaired Student’s t-test.
NFAT immunofluorescence staining and quantification
Cells were washed and settled on Shandon Single Cytoslide slides by cytospin at 450 rpm for 5 min. Cells were then fixed with 4% paraformaldehyde (CytoFIx, BD Biosciences) for 20 min at room temperature and then washed in PBS. Cells were then permeabilized with methanol (Perm Buffer III, BD Biosciences), washed in PBS and blocked with Blocking buffer (Sigma) for 30 min at room temperature. Slides were incubated overnight at 4°C with NFAT1 antibody (1:150) (CST). The following day, cells were washed three times with PBS and incubated for 1 hour at room temperature with Anti-rabbit IgG AF-647 secondary antibody (Invitrogen). Slides were washed with PBS, dried and mounted in Prolong Gold anti-fade mounting media with DAPI (ThermoFisher). Images were acquired with an Olympus IX3–55 and analyzed by CellSens imaging software (Olympus). Quantitative image analysis was conducted using QuPath 0.2.2 (Bankhead et al., 2017). Nuclei were first detected using QuPath’s implementation of StarDist, selecting the DAPI channel for nuclear detection. Objects too small to be cells were automatically removed. Next, manual curation of cells was conducted to remove incorrectly segmented objects. At least 50 cells were scored for each condition. Finally, the median nuclear Cy5 signal was used to determine positivity for NFAT1 within all cells.
Antibody Detection by ELISA
Total IgG or IgM sera levels were measured in serum from aging mice at 4 and 10 months of age. Absorbance was read at 492 nm with the Multiskan FC photometer and analyzed with SkanIt microplate reader software (Thermo Fisher, Waltham, Mass). The concentration of total serum immunoglobulin was evaluated by means of comparison with a standard curve using purified mouse IgG or IgM standards (Jackson ImmunoResearch, and Sigma respectively).
Autoantigen Array
Sera were harvested from aging control and Zap70Mb1-Cre mice at 10 months of age. IgG and IgM autoantigen arrays were performed using the Autoantigen Microarray Panel I at the Microarray Core Facility of the University of Texas Southwestern Medical Center. The autoantigens and controls on the autoantigen array are listed on the UTSW Microarray Core Facility core website (http://microarray.swmed.edu/products/product/autoantigen-microarray-panel-i/). The background-subtracted median signal intensity of each antigen was normalized using standard practices, then adjusted on corresponding total IgG and IgM sera level for each antigen and each mouse (adjusted score). Individual autoantigen arrays with array assay normalized and adjusted values for IgG and IgM are shown in Table S5 in the supplemental data. Statistical analysis was performed using R and difference in signal compared by two-tailed Welsh’s t-test. Values are plotted as fold change vs mean of each antigen. The autoantigen array accession number is GSE148797.
Single cell BCR sequencing
The single cell BCR-seq libraries were generated with the Chromium Single Cell 5’ Reagent Kits (10× Genomics) according to the manufacturer’s protocol. Cellular suspensions were loaded on a Chromium Controller instrument (10× Genomics) to generate single-cell gel bead-in-emulsions (GEMs). GEM-reverse transcriptions (GEM-RTs) were performed in a Veriti 96-well thermal cycler (Thermo Fisher Scientific). After RT, GEMs were harvested and the cDNAs were amplified and cleaned up with the SPRIselect Reagent Kit (Beckman Coulter). Indexed single cell BCR-seq libraries were constructed using the Chromium Single Cell V(D)J Enrichment Kit (10× Genomics) for enzymatic fragmentation, end-repair, A-tailing, adaptor ligation, ligation cleanup, sample index PCR, and PCR cleanup. The purity and library size were validated by capillary electrophoresis using a 2,100 Bioanalyzer (Agilent Technologies). The quantity was measured fluorometrically using a Qubit dsDNA HS Assay Kit from Invitrogen. The VDJ libraries were sequenced to a depth of ~5k reads per cell. Raw sequencing data were processed using the 10× Genomics’ Cell Ranger pipeline version 3.1.0 to generate FASTQ files and aligned to GRCh38-alts-ensembl-2.0.0 to generate VDJ count and identify CDR3 sequences. The functional VDJ clones (defined as “productive” by CellRanger) were retained for subsequent analysis. IGH, IGK and IGL were analyzed separately. VDJTools v1.1.7 (Shugay et al., 2015) was used to calculate diversity, V and J region usage, as well as creating various plots, including spectrum plot, and heatmaps. Χ2 test was calculated for significance of IGHV gene usage between ZAP70 positive vs negative samples. The single-cell BCR sequencing data accession number is GSE149199.
RNA-sequencing and gene expression analyses
Total mRNA was extracted from 1–10×106 CLL cells purified from spleens of 4 Eμ-TCL1 and 4 Eμ-TCL1 Syk+/Zap70 mice using Machery Magel RNA plus kit according to the manufacturer’s instructions. Sequencing was performed on an Illumina Hiseq 2500 instrument (Illumina, San Diego, CA, USA) using the TruSeq SE Cluster Kit V4-cBot-HS (Illumina) to generate 51 bp single-end read sequencing with v4 chemistry. Quality control of RNA-Seq reads was performed using FastQC. Raw sequence reads were mapped to the mouse genome (Gencode GRCm38 vM24) using STAR v2.5.3 (Dobin et al., 2013), and transcripts quantified with Salmon v.1.1.0 (Patro et al., 2017). Normalization and differential expression analysis were performed in R using DESeq2 v1.28.1. GSEA analysis was performed using the DOSE package in R, genes were ranked by log2 fold-change. The RNA-sequencing data accession number is GSE159908.
In vivo transplantation experiments
The indicated numbers of leukemia cells were injected into sublethally irradiated (190 cGy) NSG mice via the tail vain. 8- to 10-week-old female NSG mice were randomly allocated before injection. Mice were euthanized when they showed signs of leukemia burden including a hunched posture, weight loss and/or paralysis. Bone marrow and/or spleen cells were harvested to test for leukemic infiltration by flow cytometry. Kaplan-Meier survival analyses were performed using GraphPad Prism 7 (GraphPad Software Inc.) to compare overall survival (OS). Mantel–Cox log-rank test was used as statistical analysis using R-Statistical software.
Quantification and Statistical Analysis
The statistical details of experiments is described in corresponding figure legends and includes the type of statistical test, the value of n and dispersion and precision measures. Unless otherwise indicated, statistical analyses were performed using GraphPad Prism 7.
Supplementary Material
Table S1. SYK and ZAP70 expression in normal hematopoiesis. Related to Figure 1a.
Table S3. BioID interactome analysis in Kasumi-2 cells. Related to Figure 2a.
Table S4. BioID interactome analysis in Ramos cells. Related to Figure 2b.
Table S5. Autoantigen array in Zap70 knock-in mice. Related to Figure 6d.
Table S6. Single-cell BCR sequence analysis in humanized mice. Related to Figure 6e.
Table S7. Accession numbers for SYK and ZAP70 microarray datasets. Related to STAR methods.
Table S8. Primary and PDX derived patient information. Related to STAR Methods.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| 4G10 (phospho-tyrosine) | Millipore | 05-1050 |
| AKT-pS473 antibody | Cell Signaling | 9271 |
| AKT-pT308 antibody | Cell Signaling | 13038 |
| AKT antibody | Cell Signaling | 9272 |
| Anti-FLAG (M2) antibody | Sigma | 1804 |
| Anti-mouse IgG, AF-555 (Cy3) | Invitrogen | A31570 |
| Anti-rabbit IgG, AF-647 (Cy5) | Invitrogen | A31573 |
| β-actin antibody | Cell Signaling | 4970 |
| β-catenin antibody | BD Biosciences | 610154 |
| β-tubulin antibody | Cell Signaling | 2146 |
| BLNK-pY96 antibody | Cell Signaling | 3601 |
| BLNK-pT152 antibody | Cell Signaling | 49183 |
| BLNK antibody | Cell Signaling | 12168 |
| BTK-pT223 antibody | Cell Signaling | 5082 |
| BTK antibody | Cell Signaling | 8547 |
| CD19-pY531 antibody | Cell Signaling | 3571 |
| CD19 antibody | Cell Signaling | 90176 |
| ERK1/2-pT202/Y204 antibody | Cell Signaling | 4370 |
| ERK1/2 antibody | Cell Signaling | 9102 |
| FOXO1-pS256 antibody | Cell Signaling | 9641 |
| FOXO1 antibody | Cell Signaling | 2880 |
| H3 antibody | Cell Signaling | 4499 |
| NFAT1 antibody | Cell Signaling | 5861 |
| NFAT2 antibody | Cell Signaling | 8032 |
| PLCγ2-pY1217 antibody | Cell Signaling | 3871 |
| PLCγ2 antibody | Cell Signaling | 3872 |
| PLCγ1-pY783 antibody | Cell Signaling | 2821 |
| PLCγ1 antibody | Cell Signaling | 5690 |
| Streptavidin-HRP antibody | ThermoFisher | 21130 |
| SYK-pY352 ZAP70-pY319 antibody | Cell Signaling | 2717 |
| SYK antibody | Cell Signaling | 13198 |
| SYK antibody(scWB) | Cell Signaling | 80460 |
| TBP antibody | Cell Signaling | 8515 |
| ZAP70 antibody | Cell Signaling | 3165 |
| Biological samples | ||
| Normal bone marrow mononuclear cells | ALLCELLS | https://www.allcells.com/tissue-products/bone-marrow/ |
| Normal peripheral blood mononuclear cells | ALLCELLS | https://www.allcells.com/tissue-products/leukopak/ |
| Cord blood CD34+ progenitor Cells | ALLCELLS | https://www.allcells.com/tissue-products/cord-blood/ |
| PDX derived B-ALL, MCL and DLBCL samples | Weinstock Laboratory | https://www.proxe.org/; Table S8 |
| Primary CLL and B-cell lymphoma patient samples | City of Hope National Medical Center | Table S8 |
| Chemicals, peptides, and recombinant proteins | ||
| PRT062607 (P505-15) HCl (Syk inhibitor) | Selleckchem | S8032 |
| Ibrutinib (Btk inhibitor) | Selleckchem | S2680 |
| AZD5363 (Akt inhibitor) | Selleckchem | S8019 |
| Doxycycline | Clontech Laboratories | 631311 |
| Indo-1, AM | ThermoFisher | I1223 |
| AffiniPure F(ab’)2 Fragment Goat Anti-Human IgM | Jackson ImmunoResearch | 109-006-129 |
| Recombinant murine IL-7 | Peprotech | 217-17 |
| Recombinant human TPO | Peprotech | 300-18 |
| Recombinant human SCF | Peprotech | 300-07 |
| Recombinant human ANGPTL5 | Miltenyi Biotec | 130-096-126 |
| Critical commercial assays | ||
| Duolink™ In Situ Detection Reagents Red | Sigma | DUO92008 |
| Small scWest Kit | Protein Simple | K500 |
| NE-PER Nuclear and Cytoplasmic Extraction Kit | ThermoFisher | 78835 |
| Deposited data | ||
| Autoantigen arrays on Zap70Mb1-Cre mice | This paper | GSE148797 |
| Single-cell V(D)J sequencing from humanized mouse | This paper | GSE149199 |
| RNA-sequencing from Eμ-TCL1 mice | This paper | GSE159908 |
| Uncut immunoblots and microscopy images | This paper; Mendeley Data | DOI:10.17632/sdkg5xfvkx.2 |
| Experimental models: cell lines | ||
| KOPN8 | DMSZ | ACC 552 |
| SEM | DMSZ | ACC 546 |
| RS4;11 | DMSZ | ACC 508 |
| JFK51R | Müschen laboratory | N/A |
| REH | DMSZ | ACC 22 |
| ICN6 | Müschen laboratory | N/A |
| SUP-B15 | DMSZ | ACC 389 |
| Tom1 | DMSZ | ACC 578 |
| PDX2 | Müschen laboratory | N/A |
| BLQ5 | Müschen laboratory | N/A |
| 697 | DMSZ | ACC 42 |
| RCH-ACV | DMSZ | ACC 548 |
| Kasumi-2 | ATCC | ACC 526 |
| ICN-12 | Müschen laboratory | N/A |
| JEKO1 | DSMZ | ACC 533 |
| MINO | ATCC | ACC 687 |
| Z-138 | DSMZ | CRL-3001 |
| Raji | DSMZ | ACC 319 |
| Ramos | DSMZ | ACC 603 |
| Daudi | DSMZ | ACC 78 |
| Toledo | ATCC | CRL-2631 |
| OCI-LY3 | DSMZ | ACC 761 |
| OCI-LY7 | DSMZ | ACC 688 |
| SU-DHL-2 | ATCC | CRL-2956 |
| SU-DHL-4 | DSMZ | ACC 495 |
| L1236 | DSMZ | ACC 530 |
| L428 | DSMZ | ACC 197 |
| JJN3 | DSMZ | ACC 541 |
| LP1 | DSMZ | ACC 41 |
| U266 | DSMZ | ACC 9 |
| SFO11 | Müschen laboratory | N/A |
| Jurkat | DSMZ | ACC 281 |
| Experimental models: organisms/strains | ||
| Mouse: Mb1Cre | The Jackson Laboratory | JAX: 020505 |
| Mouse: Rosa26LSL-Zap70 | This paper | N/A |
| Mouse: Syk+/Zap70 | Konigsberger et al., 2012 | N/A |
| Mouse: Eμ-TCL1 | Bichi et al., 2002 | N/A |
| Mouse: NSG-Kit-W41J/Tyr | The Jackson Laboratory | JAX: 026622 |
| Mouse: BCR-ABL1-tg | >Heisterkamp et al., 1990 | N/A |
| Oligonucleotides | ||
| SYK crRNA AB (for human SYK deletion) | IDT | CTTTTTCGCAACATCACCC |
| Zap70 crRNA AA (for mouse Zap70 deletion) | IDT | ACGACAGCACGTAGCCGCCC |
| CD19 crRNA AB (for human CD19 deletion) | IDT | AGAGCTGAAGGACGATCGCC |
| BLNK crRNA AA (for human BLNK deletion) | IDT | TAGTCCCTTCGAGGAACACT |
| Recombinant DNA | ||
| MSCV ERT2-Puro | Müschen laboratory | N/A |
| MSCV Cre-ERT2-Puro | Müschen laboratory | N/A |
| MSCV-IRES-GFP (EV) | Müschen laboratory | N/A |
| MSCV-IRES-puro (EV) | Müschen laboratory | N/A |
| MSCV-IRES-mCherry (EV) | Müschen laboratory | N/A |
| MSCV-BirA*-GFP | Müschen laboratory | N/A |
| MSCV-BCR-ABL1 | Müschen laboratory | N/A |
| MSCV-TCF3-PBX1-GFP | Müschen laboratory | N/A |
| MSCV-SYK-GFP | Müschen laboratory | N/A |
| MSCV-SYK-s-GFP | Müschen laboratory | N/A |
| MSCV-SYKY348E/Y352E-GFP | Müschen laboratory | N/A |
| MSCV-SYKY348F/Y352F-GFP | Müschen laboratory | N/A |
| MSCV-SYK-KDK402A-GFP | Müschen laboratory | N/A |
| MSCV-BirA*SYK-GFP | This paper | N/A |
| MSCV-SYK-mCherry | This paper | N/A |
| MSCV-ZAP70-GFP | Müschen laboratory | N/A |
| MSCV-ZAP70Y315E/Y319E-GFP | Müschen laboratory | N/A |
| MSCV-ZAP70Y315F/Y319F-GFP | Müschen laboratory | N/A |
| MSCV-BirA*ZAP70-GFP | This paper | N/A |
| MSCV-ZAP70-mCherry | Müschen laboratory | N/A |
| MSCV-ZAP70-puro | Müschen laboratory | N/A |
| MSCV-GCaMPS6-puro | Müschen laboratory | N/A |
| MSCV-GCaMPS6-blast | Müschen laboratory | N/A |
| MSCV LMP2A-GFP | Müschen laboratory | N/A |
| pRetroX-TET3G-Neo | Müschen laboratory | N/A |
| pRetroX-TRE3G-LMP2A-puro | Müschen laboratory | N/A |
| pTOL-hCMV-TET3G-Hygro | Müschen laboratory | N/A |
| pCLIP-Cas9-TRE3G-ZsGreen | Müschen laboratory | N/A |
| pCLIP-hCMV-tRFP-gRNA | Müschen laboratory | N/A |
| pCL6-GFP (EV) | Müschen laboratory | N/A |
| pCL6-ZAP70-GFP | This paper | N/A |
| Software and algorithms | ||
| ImageJ | NIH | https://imagej.net/Fiji |
| Scout | Protein Simple | https://www.proteinsimple.com/scout |
| QuPATH | N/A | https://qupath.github.io/ |
| VDJTools v1.1.7 | Shugay et al., 2015 | N/A |
| GraphPad Prism 7 | GraphPad Software Inc. | https://www.graphpad.com/scientific-software/prism/ |
| R v4.0.3 | R Project for Statistical computing | https://www.r-project.org |
Highlights.
ZAP70 is aberrantly coexpressed with SYK in a broad range of B-cell malignancies
Forced Zap70 expression in vivo disrupts negative selection of autoreactive B-cells
ZAP70 diverts SYK from NFAT-dependent anergy to PI3K-dependent survival signaling
Zap70 prevents NFAT-dependent anergy and accelerates B-cell transformation
Acknowledgments.
We would like to thank Theresa Kadlecek and Arthur Weiss (UCSF), David Weinstock (Harvard University and DFCI) and Vaidehi Nagarajan (City of Hope) for their support and critical discussion, Lauren Kim-Sing, and current and former members of the Muschen laboratory, as well as A.S Korganow, F. Gros and L. Vallat and the former members of the UPR CNRS 3572 for their support and help with experimental work. We would also like to acknowledge Aimin Li and Michael Nelson for their technical support with TMAs and microscopy. Research in the Müschen laboratory is funded by the NIH through an NCI Outstanding Investigator Award R35CA197628, R01CA157644, R01CA213138 and P01CA233412 (to M.M.), the Howard Hughes Medical Institute HHMI-55108547 (to M.M.), the Arthur H. and Isabel Bunker Chair in Hematology (to M.M.), a Blood Cancer Discoveries Grant program through The Leukemia & Lymphoma Society, The Mark Foundation for Cancer Research, and The Paul G. Allen Frontiers Group and the V Foundation for Cancer Research T2018–003B (to M.M.). This work was supported by grant funding from the NIH-NIAID AI061093 and AI118855 and by a TIL grant from the Lupus Research Alliance to E. M. The generation of the Zap70Mb1-Cre knockin model was supported by the foundations “Alsace contre le Cancer”, “Groupe Pasteur Mutualité” and “CSL Behring”. T.S. is a fellow of the Lymphoma Research Foundation. M.M. is a Howard Hughes Medical Institute (HHMI) Faculty Scholar.
Footnotes
Conflict of interest statement.
E.M. is an advisor for and receives funding from AbbVie, Inc. Other authors declare that they have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aiba Y, Kameyama M, Yamazaki T, Tedder TF, and Kurosaki T (2008). Regulation of B-cell development by BCAP and CD19 through their binding to phosphoinositide 3-kinase. Blood 111, 1497–1503. [DOI] [PubMed] [Google Scholar]
- Baba Y, Hashimoto S, Matsushita M, Watanabe D, Kishimoto T, Kurosaki T, and Tsukada S (2001). BLNK mediates Syk-dependent Btk activation. Proc Natl Acad Sci U S A 98, 2582–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ, Coleman HG, et al. (2017). QuPath: Open source software for digital pathology image analysis. Sci Rep 7, 16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrington RA, Borde M, Rao A, and Carroll MC (2006). Involvement of NFAT1 in B cell selftolerance. J Immunol 177, 1510–1515. [DOI] [PubMed] [Google Scholar]
- Beitz LO, Fruman DA, Kurosaki T, Cantley LC, and Scharenberg AM (1999). SYK is upstream of phosphoinositide 3-kinase in B cell receptor signaling. J Biol Chem 274, 32662–32666. [DOI] [PubMed] [Google Scholar]
- Bichi R, Shinton SA, Martin ES, Koval A, Calin GA, Cesari R, Russo G, Hardy RR, and Croce CM (2002). Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc Natl Acad Sci U S A 99, 6955–6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brdicka T, Kadlecek TA, Roose JP, Pastuszak AW, and Weiss A (2005). Intramolecular regulatory switch in ZAP-70: analogy with receptor tyrosine kinases. Mol Cell Biol 25, 4924–4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhl AM, Pleiman CM, Rickert RC, and Cambier JC (1997). Qualitative regulation of B cell antigen receptor signaling by CD19: selective requirement for PI3-kinase activation, inositol-1,4,5-trisphosphate production and Ca2+ mobilization. J Exp Med 186, 1897–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreras J, Villamor N, Colomo L, Moreno C, Ramon y Cajal S, Crespo M, Tort F, Bosch F, Lopez-Guillermo A, Colomer D, et al. (2005). Immunohistochemical analysis of ZAP-70 expression in B-cell lymphoid neoplasms. J Pathol 205, 507–513. [DOI] [PubMed] [Google Scholar]
- Chan AY, Punwani D, Kadlecek TA, Cowan MJ, Olson JL, Mathes EF, Sunderam U, Fu SM, Srinivasan R, Kuriyan J, et al. (2016). A novel human autoimmune syndrome caused by combined hypomorphic and activating mutations in ZAP-70. J Exp Med 213, 155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Widhopf G, Huynh L, Rassenti L, Rai KR, Weiss A, and Kipps TJ (2002). Expression of ZAP-70 is associated with increased B-cell receptor signaling in chronic lymphocytic leukemia. Blood 100, 4609–4614. [DOI] [PubMed] [Google Scholar]
- Chen Z, Shojaee S, Buchner M, Geng H, Lee JW, Klemm L, Titz B, Graeber TG, Park E, Tan YX, et al. (2015). Signalling thresholds and negative B-cell selection in acute lymphoblastic leukaemia. Nature 521, 357–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AM, Rowley B, Pao W, Hayday A, Bolen JB, and Pawson T (1995). Syk tyrosine kinase required for mouse viability and B-cell development. Nature 378, 303–306. [DOI] [PubMed] [Google Scholar]
- Crespo M, Bosch F, Villamor N, Bellosillo B, Colomer D, Rozman M, Marce S, Lopez-Guillermo A, Campo E, and Montserrat E (2003). ZAP-70 expression as a surrogate for immunoglobulin-variable-region mutations in chronic lymphocytic leukemia. N Engl J Med 348, 1764–1775. [DOI] [PubMed] [Google Scholar]
- Crespo M, Villamor N, Gine E, Muntanola A, Colomer D, Marafioti T, Jones M, Camos M, Campo E, Montserrat E, et al. (2006). ZAP-70 expression in normal pro/pre B cells, mature B cells, and in B-cell acute lymphoblastic leukemia. Clin Cancer Res 12, 726–734. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolmetsch RE, Lewis RS, Goodnow CC, and Healy JI (1997). Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature 386, 855–858. [DOI] [PubMed] [Google Scholar]
- Duhren-von Minden M, Ubelhart R, Schneider D, Wossning T, Bach MP, Buchner M, Hofmann D, Surova E, Follo M, Kohler F, et al. (2012). Chronic lymphocytic leukaemia is driven by antigen-independent cell-autonomous signalling. Nature 489, 309–312. [DOI] [PubMed] [Google Scholar]
- Duke VM, Gandini D, Sherrington PD, Lin K, Heelan B, Amlot P, Mehta AB, Hoffbrand AV, and Foroni L (2003). V(H) gene usage differs in germline and mutated B-cell chronic lymphocytic leukemia. Haematologica 88, 1259–1271. [PubMed] [Google Scholar]
- Durig J, Nuckel H, Cremer M, Fuhrer A, Halfmeyer K, Fandrey J, Moroy T, Klein-Hitpass L, and Duhrsen U (2003). ZAP-70 expression is a prognostic factor in chronic lymphocytic leukemia. Leukemia 17, 2426–2434. [DOI] [PubMed] [Google Scholar]
- Fais F, Ghiotto F, Hashimoto S, Sellars B, Valetto A, Allen SL, Schulman P, Vinciguerra VP, Rai K, Rassenti LZ, et al. (1998). Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J Clin Invest 102, 1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldhahn N, Klein F, Mooster JL, Hadweh P, Sprangers M, Wartenberg M, Bekhite MM, Hofmann WK, Herzog S, Jumaa H, et al. (2005a). Mimicry of a constitutively active pre-B cell receptor in acute lymphoblastic leukemia cells. J Exp Med 201, 1837–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldhahn N, Rio P, Soh BN, Liedtke S, Sprangers M, Klein F, Wernet P, Jumaa H, Hofmann WK, Hanenberg H, et al. (2005b). Deficiency of Bruton’s tyrosine kinase in B cell precursor leukemia cells. Proc Natl Acad Sci U S A 102, 13266–13271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming A, Brummer T, Reth M, and Jumaa H (2003). The adaptor protein SLP-65 acts as a tumor suppressor that limits pre-B cell expansion. Nat Immunol 4, 38–43. [DOI] [PubMed] [Google Scholar]
- Fujimoto M, Fujimoto Y, Poe JC, Jansen PJ, Lowell CA, DeFranco AL, and Tedder TF (2000). CD19 regulates Src family protein tyrosine kinase activation in B lymphocytes through processive amplification. Immunity 13, 47–57. [DOI] [PubMed] [Google Scholar]
- Geng H, Hurtz C, Lenz KB, Chen Z, Baumjohann D, Thompson S, Goloviznina NA, Chen WY, Huan J, LaTocha D, et al. (2015). Self-enforcing feedback activation between BCL6 and pre-B cell receptor signaling defines a distinct subtype of acute lymphoblastic leukemia. Cancer Cell 27, 409–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobessi S, Laurenti L, Longo PG, Sica S, Leone G, and Efremov DG (2007). ZAP-70 enhances B-cell-receptor signaling despite absent or inefficient tyrosine kinase activation in chronic lymphocytic leukemia and lymphoma B cells. Blood 109, 2032–2039. [DOI] [PubMed] [Google Scholar]
- Goodnow CC, Sprent J, Fazekas de St Groth B, and Vinuesa CG (2005). Cellular and genetic mechanisms of self tolerance and autoimmunity. Nature 435, 590–597. [DOI] [PubMed] [Google Scholar]
- He X, Klasener K, Iype JM, Becker M, Maity PC, Cavallari M, Nielsen PJ, Yang J, and Reth M (2018). Continuous signaling of CD79b and CD19 is required for the fitness of Burkitt lymphoma B cells. EMBO J 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy JI, Dolmetsch RE, Timmerman LA, Cyster JG, Thomas ML, Crabtree GR, Lewis RS, and Goodnow CC (1997). Different nuclear signals are activated by the B cell receptor during positive versus negative signaling. Immunity 6, 419–428. [DOI] [PubMed] [Google Scholar]
- Heisterkamp N, Jenster G, ten Hoeve J, Zovich D, Pattengale PK, and Groffen J (1990). Acute leukaemia in bcr/abl transgenic mice. Nature 344, 251–253. [DOI] [PubMed] [Google Scholar]
- Herve M, Xu K, Ng YS, Wardemann H, Albesiano E, Messmer BT, Chiorazzi N, and Meffre E (2005). Unmutated and mutated chronic lymphocytic leukemias derive from self-reactive B cell precursors despite expressing different antibody reactivity. J Clin Invest 115, 1636–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippolito GC, Hoi KH, Reddy ST, Carroll SM, Ge X, Rogosch T, Zemlin M, Shultz LD, Ellington AD, Vandenberg CL, et al. (2012). Antibody repertoires in humanized NOD-scid-IL2Rgamma(null) mice and human B cells reveals human-like diversification and tolerance checkpoints in the mouse. PLoS One 7, e35497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TA, Rassenti LZ, and Kipps TJ (1997). Ig VH1 genes expressed in B cell chronic lymphocytic leukemia exhibit distinctive molecular features. J Immunol 158, 235–246. [PubMed] [Google Scholar]
- Jumaa H, Bossaller L, Portugal K, Storch B, Lotz M, Flemming A, Schrappe M, Postila V, Riikonen P, Pelkonen J, et al. (2003). Deficiency of the adaptor SLP-65 in pre-B-cell acute lymphoblastic leukaemia. Nature 423, 452–456. [DOI] [PubMed] [Google Scholar]
- Kersseboom R, Middendorp S, Dingjan GM, Dahlenborg K, Reth M, Jumaa H, and Hendriks RW (2003). Bruton’s tyrosine kinase cooperates with the B cell linker protein SLP-65 as a tumor suppressor in Pre-B cells. J Exp Med 198, 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, Cutler JA, Na CH, Reckel S, Renuse S, Madugundu AK, Tahir R, Goldschmidt HL, Reddy KL, Huganir RL, et al. (2018). BioSITe: A Method for Direct Detection and Quantitation of Site-Specific Biotinylation. J Proteome Res 17, 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konigsberger S, Prodohl J, Stegner D, Weis V, Andreas M, Stehling M, Schumacher T, Bohmer R, Thielmann I, van Eeuwijk JM, et al. (2012). Altered BCR signalling quality predisposes to autoimmune disease and a pre-diabetic state. EMBO J 31, 3363–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai R, Lefresne SV, Franko B, Hui D, Mirza I, Mansoor A, Amin HM, and Ma Y (2006). Immunoglobulin VH somatic hypermutation in mantle cell lymphoma: mutated genotype correlates with better clinical outcome. Mod Pathol 19, 1498–1505. [DOI] [PubMed] [Google Scholar]
- Lanier LL (2006). Viral immunoreceptor tyrosine-based activation motif (ITAM)-mediated signaling in cell transformation and cancer. Trends Cell Biol 16, 388–390. [DOI] [PubMed] [Google Scholar]
- Latour S, Chow LM, and Veillette A (1996). Differential intrinsic enzymatic activity of Syk and Zap-70 protein-tyrosine kinases. J Biol Chem 271, 22782–22790. [DOI] [PubMed] [Google Scholar]
- Lu L, and Osmond DG (2000). Apoptosis and its modulation during B lymphopoiesis in mouse bone marrow. Immunol Rev 175, 158–174. [PubMed] [Google Scholar]
- Marklin M, Heitmann JS, Fuchs AR, Truckenmuller FM, Gutknecht M, Bugl S, Saur SJ, Lazarus J, Kohlhofer U, Quintanilla-Martinez L, et al. (2017). NFAT2 is a critical regulator of the anergic phenotype in chronic lymphocytic leukaemia. Nat Commun 8, 755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez GJ, Pereira RM, Aijo T, Kim EY, Marangoni F, Pipkin ME, Togher S, Heissmeyer V, Zhang YC, Crotty S, et al. (2015). The transcription factor NFAT promotes exhaustion of activated CD8(+) T cells. Immunity 42, 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant M, Caldwell RG, and Longnecker R (2000). The LMP2A ITAM is essential for providing B cells with development and survival signals in vivo. J Virol 74, 9115–9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockridge CI, Potter KN, Wheatley I, Neville LA, Packham G, and Stevenson FK (2007). Reversible anergy of sIgM-mediated signaling in the two subsets of CLL defined by VH-gene mutational status. Blood 109, 4424–4431. [DOI] [PubMed] [Google Scholar]
- Muro R, Nitta T, Nakano K, Okamura T, Takayanagi H, and Suzuki H (2018). gammadeltaTCR recruits the Syk/PI3K axis to drive proinflammatory differentiation program. J Clin Invest 128, 415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschen M (2018). Autoimmunity checkpoints as therapeutic targets in B cell malignancies. Nat Rev Cancer 18, 103–116. [DOI] [PubMed] [Google Scholar]
- Negishi I, Motoyama N, Nakayama K, Nakayama K, Senju S, Hatakeyama S, Zhang Q, Chan AC, and Loh DY (1995). Essential role for ZAP-70 in both positive and negative selection of thymocytes. Nature 376, 435–438. [DOI] [PubMed] [Google Scholar]
- Okada T, Maeda A, Iwamatsu A, Gotoh K, and Kurosaki T (2000). BCAP: the tyrosine kinase substrate that connects B cell receptor to phosphoinositide 3-kinase activation. Immunity 13, 817–827. [DOI] [PubMed] [Google Scholar]
- Otero DC, Omori SA, and Rickert RC (2001). Cd19-dependent activation of Akt kinase in B-lymphocytes. J Biol Chem 276, 1474–1478. [DOI] [PubMed] [Google Scholar]
- Packham G, Krysov S, Allen A, Savelyeva N, Steele AJ, Forconi F, and Stevenson FK (2014). The outcome of B-cell receptor signaling in chronic lymphocytic leukemia: proliferation or anergy. Haematologica 99, 1138–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, and Kingsford C (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauls SD, and Marshall AJ (2017). Regulation of immune cell signaling by SHIP1: A phosphatase, scaffold protein, and potential therapeutic target. Eur J Immunol 47, 932–945. [DOI] [PubMed] [Google Scholar]
- Pelanda R, and Torres RM (2012). Central B-cell tolerance: where selection begins. Cold Spring Harb Perspect Biol 4, a007146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee I, and Veillette A (2012). Protein tyrosine phosphatases in lymphocyte activation and autoimmunity. Nat Immunol 13, 439–447. [DOI] [PubMed] [Google Scholar]
- Rowley RB, Bolen JB, and Fargnoli J (1995). Molecular cloning of rodent p72Syk. Evidence of alternative mRNA splicing. J Biol Chem 270, 12659–12664. [DOI] [PubMed] [Google Scholar]
- Schickel JN, Kuhny M, Baldo A, Bannock JM, Massad C, Wang H, Katz N, Oe T, Menard L, Soulas-Sprauel P, et al. (2016). PTPN22 inhibition resets defective human central B cell tolerance. Sci Immunol 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shojaee S, Caeser R, Buchner M, Park E, Swaminathan S, Hurtz C, Geng H, Chan LN, Klemm L, Hofmann WK, et al. (2015). Erk Negative Feedback Control Enables Pre-B Cell Transformation and Represents a Therapeutic Target in Acute Lymphoblastic Leukemia. Cancer Cell 28, 114–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shojaee S, Chan LN, Buchner M, Cazzaniga V, Cosgun KN, Geng H, Qiu YH, von Minden MD, Ernst T, Hochhaus A, et al. (2016). PTEN opposes negative selection and enables oncogenic transformation of pre-B cells. Nat Med 22, 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shugay M, Bagaev DV, Turchaninova MA, Bolotin DA, Britanova OV, Putintseva EV, Pogorelyy MV, Nazarov VI, Zvyagin IV, Kirgizova VI, et al. (2015). VDJtools: Unifying Post-analysis of T Cell Receptor Repertoires. PLoS Comput Biol 11, e1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatopoulos K, Belessi C, Moreno C, Boudjograh M, Guida G, Smilevska T, Belhoul L, Stella S, Stavroyianni N, Crespo M, et al. (2007). Over 20% of patients with chronic lymphocytic leukemia carry stereotyped receptors: Pathogenetic implications and clinical correlations. Blood 109, 259–270. [DOI] [PubMed] [Google Scholar]
- Sup SJ, Domiati-Saad R, Kelley TW, Steinle R, Zhao X, and Hsi ED (2004). ZAP-70 expression in B-cell hematologic malignancy is not limited to CLL/SLL. Am J Clin Pathol 122, 582–587. [DOI] [PubMed] [Google Scholar]
- Tedder TF, Zhou LJ, and Engel P (1994). The CD19/CD21 signal transduction complex of B lymphocytes. Immunol Today 15, 437–442. [DOI] [PubMed] [Google Scholar]
- Thorley-Lawson DA (2001). Epstein-Barr virus: exploiting the immune system. Nat Rev Immunol 1, 75–82. [DOI] [PubMed] [Google Scholar]
- Townsend EC, Murakami MA, Christodoulou A, Christie AL, Koster J, DeSouza TA, Morgan EA, Kallgren SP, Liu H, Wu SC, et al. (2016). The Public Repository of Xenografts Enables Discovery and Randomized Phase II-like Trials in Mice. Cancer Cell 30, 183. [DOI] [PubMed] [Google Scholar]
- Turner M, Mee PJ, Costello PS, Williams O, Price AA, Duddy LP, Furlong MT, Geahlen RL, and Tybulewicz VL (1995). Perinatal lethality and blocked B-cell development in mice lacking the tyrosine kinase Syk. Nature 378, 298–302. [DOI] [PubMed] [Google Scholar]
- Turner M, Schweighoffer E, Colucci F, Di Santo JP, and Tybulewicz VL (2000). Tyrosine kinase SYK: essential functions for immunoreceptor signalling. Immunol Today 21, 148–154. [DOI] [PubMed] [Google Scholar]
- Wiestner A, Rosenwald A, Barry TS, Wright G, Davis RE, Henrickson SE, Zhao H, Ibbotson RE, Orchard JA, Davis Z, et al. (2003). ZAP-70 expression identifies a chronic lymphocytic leukemia subtype with unmutated immunoglobulin genes, inferior clinical outcome, and distinct gene expression profile. Blood 101, 4944–4951. [DOI] [PubMed] [Google Scholar]
- Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, and Rajewsky K (2007). MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 131, 146–159. [DOI] [PubMed] [Google Scholar]
- Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, and Rajewsky K (2008). Lymphoproliferative disease and autoimmunity in mice with increased miR-17–92 expression in lymphocytes. Nat Immunol 9, 405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xochelli A, Baliakas P, Kavakiotis I, Agathangelidis A, Sutton LA, Minga E, Ntoufa S, Tausch E, Yan XJ, Shanafelt T, et al. (2017). Chronic Lymphocytic Leukemia with Mutated IGHV4–34 Receptors: Shared and Distinct Immunogenetic Features and Clinical Outcomes. Clin Cancer Res 23, 5292–5301. [DOI] [PubMed] [Google Scholar]
- Yagi S, Suzuki K, Hasegawa A, Okumura K, and Ra C (1994). Cloning of the cDNA for the deleted syk kinase homologous to ZAP-70 from human basophilic leukemia cell line (KU812). Biochem Biophys Res Commun 200, 28–34. [DOI] [PubMed] [Google Scholar]
- Yarkoni Y, Getahun A, and Cambier JC (2010). Molecular underpinning of B-cell anergy. Immunol Rev 237, 249–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller KE, MacNeil IA, and Brugge JS (1997). Protein tyrosine kinases Syk and ZAP-70 display distinct requirements for Src family kinases in immune response receptor signal transduction. J Immunol 158, 1650–1659. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. SYK and ZAP70 expression in normal hematopoiesis. Related to Figure 1a.
Table S3. BioID interactome analysis in Kasumi-2 cells. Related to Figure 2a.
Table S4. BioID interactome analysis in Ramos cells. Related to Figure 2b.
Table S5. Autoantigen array in Zap70 knock-in mice. Related to Figure 6d.
Table S6. Single-cell BCR sequence analysis in humanized mice. Related to Figure 6e.
Table S7. Accession numbers for SYK and ZAP70 microarray datasets. Related to STAR methods.
Table S8. Primary and PDX derived patient information. Related to STAR Methods.
Data Availability Statement
The autoantigen arrays and sequencing datasets generated in this paper have been deposited in the Gene Expression Omnibus (GEO) with the following accession numbers: autoantigen arrays from Zap70Mb1-Cre mice (GSE148797), single-cell V(D)J sequencing from humanized mouse (GSE149199) and RNA-sequencing from Eμ-TCL1 mice (GSE159908). For analysis of ZAP70 expression in B-cell malignancies published mRNA expression datasets were combined and normalized using MAS5 (see Table S7 for full information). Analysis of Syk and Zap70 expression in the BCR-ABL1-tg mouse model was obtained from GSE110104. Original data for figures in this paper is available at Mendeley Data (DOI:10.17632/sdkg5xfvkx.2).