Abstract
The mcr-1 gene mediating mobile colistin resistance in Escherichia coli was first reported in China in 2016 followed by reports among different species worldwide, especially in E. coli and Klebsiella. However, data on its transmission in Salmonella are still lacking. This study analyzed the antimicrobial resistance (AMR) profiles and the mcr-1 gene presence in 755 foodborne Salmonella from 26 provinces of mainland, China in 2016. Genomic features of two mcr-1-carrying isolates, genome sequencing, serotypes and further resistance profiles were studied. Among the 755 Salmonella tested, 72.6% were found to be resistant to at least one antimicrobial agent and 10% were defined as multi-drug resistant (MDR). Salmonella Derby CFSA231 and Salmonella Typhimurium CFSA629 were mcr-1-harboring isolates. Both expressed an MDR phenotype and included a single circular chromosome and one plasmid. Among the 22 AMR genes identified in S. Derby CFSA231, only the mcr-1 gene was localized on the IncX4 type plasmid pCFSA231 while 20 chromosomal AMR genes, including four plasmid-mediated quinolone resistance (PMQR) genes, were mapped within a 64 kb Salmonella genomic island (SGI) like region. S. Typhimurium CFSA629 possessed 11 resistance genes including an mcr-1.19 variant and two ESBL genes. Two IS26-flanked composite-like transposons were identified. Additionally, 153 and 152 virulence factors were separately identified in these two isolates with secretion system and fimbrial adherence determinants as the dominant virulence classes. Our study extends our concern on mcr-1-carrying Salmonella in regards to antimicrobial resistance and virulence factors, and highlight the importance of surveillance to mitigate dissemination of mcr-encoding genes among foodborne Salmonella.
Keywords: Salmonella, colistin, antimicrobial resistance (AMR), mcr-1, plasmids, China
Introduction
Salmonella, one of the top-ranking foodborne pathogens worldwide, is known to cause mild to severe foodborne infections, and has posed a significant public health challenge globally (Jazeela et al., 2020). Antimicrobial compounds are used to treat both human infections and food animal production and evidence suggests that antimicrobial use in food-producing animals contributes to resistance among foodborne Salmonella (Crump et al., 2011). This usage also increases the risk of failure when clinical treatment measures are platformed (Bai et al., 2016). On-going surveillance is a necessary step toward monitoring the emergence of multi-drug resistance (MDR) isolates of Salmonella.
Colistin (polymyxin E) is considered to be an antimicrobial agent of last-resort for treatment of MDR Gram-negative bacterial infections (Elbediwi et al., 2019). A plasmid-mediated polymyxin resistance mechanism MCR (mobile colistin resistance) and the first mcr-1 gene was reported in China in 2016 (Liu et al., 2016). The key point of this mechanism is that it encodes a phosphoethanolamine transferase and confers a transferable colistin resistance (Zhao et al., 2017), thus accelerating the therapeutic failure of colistin as a last-resort treatment option for many MDR Gram-negative bacteria (Janssen et al., 2020). Global reports of the identical mcr-1 gene among several different bacterial species were published shortly thereafter (Sun et al., 2018) along with various publications on related mcr-1 variants and more divergent mcr genes (mcr-2 ∼ mcr-10) (Kluytmans, 2017; Partridge et al., 2018; Osei Sekyere, 2019). According to the proposal for assignment of allele numbers for mcr genes and relevant variants (Partridge et al., 2018), mcr-1.27 and mcr-10 were designated as the latest mcr-1 variant and mcr gene, respectively. Fifteen bacterial genera have been reported to carry mcr genes to date, and with the majority within the Enterobacteriaceae (11/15) family. Enterobacteriaceae members Escherichia coli, Salmonella, Klebsiella, and Aeromonas of the Aeromonadaceae family are the most common bacteria from which mcr genes have been detected with the highest mcr gene prevalence reported in non-pathogenic E. coli (Elbediwi et al., 2019). China reported the highest number of mcr-positive strains in a recent meta-analysis (Elbediwi et al., 2019), and identified mcr-1 gene in Guangdong, Shanghai, Zhejiang, Hubei, Jiangsu, Sichuan, Shandong, Anhui, Chongqing, Hong Kong, and Taiwan. Based on literature review in public scientific databases, mcr-like genes are reported at lower rates in Salmonella when compared to E. coli, however there have been increased reported numbers of mcr-mediated colistin resistance in Salmonella spp. from humans, animals, and foods after 2016 (Li et al., 2016; Sun et al., 2018; Lima et al., 2019; Borowiak et al., 2020; Sia et al., 2020).
In this report, a surveillance of the overall antimicrobial resistance (AMR) features of 755 foodborne Salmonella isolates in mainland China in 2016 and an investigation of the genomic characteristics of AMR determinants and virulence factors (VFs) of the two mcr-1-habouring isolates is conducted to address this data gap.
Materials and Methods
Bacterial Isolates
A total of 755 foodborne Salmonella isolates were collected from 26 provinces across mainland China in 2016. Salmonella isolates were collected from broad food categories including: special nutritional products (powder infant formula, PIF), raw meat and meat products, aquatic and aquatic products, egg and egg products, soy products, frozen drinks, rice and flour products, nuts and seed products, beverages, cocoa products, and local foods among others.
Antimicrobial Susceptibility Testing (AST) and Screening of the mcr-1 Gene
All collected Salmonella were subjected to AST testing against a panel of compounds (Table 1) by broth micro-dilution using the Biofosun® Gram-negative panel which contained 10 classes (16 kinds) of drugs (Fosun Diagnostics, Shanghai, China). Data obtained was interpreted following recommendations described by Clinical and Laboratory Standards Institute guidelines (CLSI, M100-S28). Additionally, CLSI (M31-A3) and European Committee on Antimicrobial Susceptibility Testing (EUCAST, version 2018) documents were consulted when CLSI M100 standards were not available for some antimicrobial compounds. E. coli ATCCTM25922 was included as a reference strain.
TABLE 1.
Antimicrobial susceptibility of 755 foodborne Salmonella isolates against to a panel of antimicrobial agents.
| Antimicrobial class | Antimicrobial agent (abbreviation)a | Number of resistant isolates | Resistant rate (%) | Number of intermediate isolates | Intermediate rate (%) | Number of susceptible isolates | Susceptible rate (%) |
| Penicillins | Ampicillin (AMP) | 291 | 38.5 | 3 | 0.4 | 461 | 61.1 |
| β-Lactam combination agents | Ampicillin/sulbactam (SAM) | 263 | 34.8 | 32 | 4.2 | 460 | 60.9 |
| Cephalosporins | Cefotaxime (CTX) | 89 | 11.8 | 4 | 0.5 | 662 | 87.7 |
| Ceftazidime (CAZ) | 45 | 6.0 | 11 | 1.5 | 699 | 92.6 | |
| Cephalothin (KF) | 103 | 13.6 | 47 | 6.2 | 605 | 80.1 | |
| Cefepime (FEP) | 40 | 5.3 | 7 | 0.9 | 708 | 93.8 | |
| Carbapenems | Imipenem (IMP) | 0 | 0.0 | 0 | 0.0 | 755 | 100.0 |
| Meropenem (MEM) | 0 | 0.0 | 0 | 0.0 | 755 | 100.0 | |
| Aminoglycosides | Gentamicin (GEN) | 83 | 11.0 | 0 | 0.0 | 672 | 89.0 |
| Tetracyclines | Tetracycline (TET) | 358 | 47.4 | 11 | 1.5 | 386 | 51.1 |
| (Fluoro)Quinolones | Nalidixic (NAL) | 396 | 52.5 | – | – | 359 | 47.5 |
| Ciprofloxacin (CIP) | 161 | 21.3 | 289 | 38.3 | 305 | 40.4 | |
| Folate pathway inhibitors | Trimethoprim/sulfamethoxazole (SXT) | 179 | 23.7 | –e | –e | 576 | 76.3 |
| Phenicols | Chloramphenicol (CHL) | 187 | 24.8 | 87 | 11.5 | 481 | 63.7 |
| Florfenicol (FFC)b,c | 170 | 22.5 | 65 | 8.6 | 520 | 68.9 | |
| Polymyxin | Polymyxin E (Colistin, CT)c,d | 61 | 8.1 | –e | –e | 694 | 91.9 |
aInterpretation according to the CLSI guidelines M100-S28, 2018, for all drugs except FFC and CT; binterpretation according to the CLSI guidelines M31-A3, 2008; cused as a feed additive in animal production; dinterpretation according to EUCAST clinical breakpoints, 2018; eno break-point data.
All 755 foodborne Salmonella isolates were screened for the presence of mcr-1 gene by qPCR as described previously (Hu et al., 2019). Isolates that carry mcr-1 gene were selected for further analysis as described below.
Serotyping and Further AST
Serotypes of mcr-1 positive isolates were identified by both classical slide agglutination with commercialized antisera (SSI, Denmark) following the Kauffman White Le Minor scheme (WKLM, version 2007, 9th edition), and also by molecular serotyping with xMAP® Salmonella Serotyping Assay Kit (SSA, Cat No. AGSSA4502, Luminex, United States) following the manufacturer’s protocol. Extended AST tests using additional selected antimicrobial agents relevant to Enterobacteriaceae, including 13 classes (composing 27 compounds, Table 2), were carried out on the mcr-1 positive isolates. Extended Spectrum Beta-Lactamase (ESBL) phenotype testing was also performed according to the protocol and breakpoints described in CLSI (M100-S28). Klebsiella pneumoniae ATCCTM700603 was included as a suitable positive control.
TABLE 2.
Antimicrobial susceptibility of Salmonella isolate CFSA231 and CFSA629 to a further panel of antimicrobial agents and acquired antimicrobial resistance-encoding genes identified in the bacterial genome with online retrieval in Resfinder database.
| CFSA231 | CFSA629 | CFSA231 | CFSA629 | |||||||
| Antimicrobial class | Antimicrobial agent (abbreviation) | MIC (mg/L) | R/I/Sa | MIC (mg/L) | R/I/Sa | Resistance genes | Resistance genes | |||
|
or point mutation |
or point mutation |
|||||||||
| Chromosome | Plasmid | Chromosome | Plasmid | |||||||
| β-lactam combination agents | Ampicillin/sulbactam (SAM) | ≥32/16 | R | ≥32/16 | R | blaOXA–1 | blaTEM–1B | blaCTX–M–14 | ||
| Penicillins | Ampicillin (AMP) | ≥32 | R | ≥32 | R | |||||
| Cephalosporins | Cefotaxime (CTX) | 0.12 | S | 64 | R | |||||
| Cefotaxime + clavulanate (CTX + CLA) | 0.06/4 | – | 0.12/4 | – | ||||||
| Ceftazidime (CAZ) | 0.5 | S | 2 | S | ||||||
| Ceftazidime + clavulanate (CAZ + CLA) | 0.25/4 | – | 0.25/4 | – | ||||||
| Cephalothin (KF) | 2 | S | ≥32 | R | ||||||
| Cefoxitin (FOX) | 2 | S | 8 | S | ||||||
| Ceftriaxone (CRO) | 0.12 | S | ≥4 | R | ||||||
| Cefepime (FEP) | 2 | S | ≥16 | R | ||||||
| Carbapenems | Imipenem (IMP) | 0.25 | S | 0.25 | S | |||||
| Meropenem (MEM) | 0.03 | S | 0.03 | S | ||||||
| Ertapenem (ETP) | 0.015 | S | 0.5 | S | ||||||
| Monobactams | Aztreonam (ATM) | 0.06 | S | 4 | S | |||||
| Aminoglycosides | Gentamicin (GEN) | ≥16 | R | 8 | I | aac(3)-IV, aac(6′)-Iaa, aac(6′)-Ib-cr, aadA1, aadA2,aaaA2b, aadA8b, aph(3′)-Ia, aph(4)-Ia | aac(6′)-Iaa, aph(3′′)-Ib, aph(6)-Id | aac(3)-IV, aph(4)-Ia | ||
| Amikacin(AK) | 8 | S | 1 | S | ||||||
| Tetracyclines | Tetracycline (TET) | ≥16 | R | ≥16 | R | tetA | tetB | |||
| Tigecycline (TGC) | 0.12 | S | 0.25 | S | ||||||
| (Fluoro)Quinolones | Nalidixic acid (NAL) | ≥32 | R | ≥32 | R | ParC T57S; aac(6′)-Ib-cr, oqxA, oqxB, qnrS2 | GyrA D87Y | |||
| Ciprofloxacin (CIP) | 4 | R | 0.5 | I | ||||||
| Folate pathway inhibitors | Trimethoprim/sulfamethoxazole (SXT) | ≥8/152 | R | 0.25/4.75 | S | sul1, sul2, sul3, dfrA12 | sul2 | |||
| Trimethoprim(TMP) | ≥16 | R | 0.25 | S | ||||||
| Phenicols | Chloramphenicol (CHL) | ≥32 | R | 8 | S | catB3, cmlA1, floR | ||||
| Florfenicol (FFC)b,c | ≥16 | R | 8 | I | ||||||
| Nitrofurans | Nitrofurantoin (NIT) | 32 | S | 16 | S | |||||
| Polymyxins | Polymyxin E (Colistin, CT)c,d | 2 | S | 4 | R | mcr-1.1 | mcr-1.19 | |||
| Polymyxin B (PB)d | 4 | I | 4 | I | ||||||
| Fosfomycins | Fosfomycin (FOS) | –e | –e | –e | –e | fosA7 | fosA3 | |||
R, resistant; I, intermediate; S, susceptible; aR/I/S according to the CLSI guidelines M100-S28, 2018; bR/I/S according to the CLSI guidelines M31-A3, 2008; cused as a feed additive in animal production; dR/I/S according to EUCAST clinical breakpoints, 2018; eno data.
Plasmid Conjugal Transfer
The transfer ability and frequency of mcr-carrying plasmids was investigated by broth mating conjugation experiments with plasmid-free and sodium azide-resistant E. coli J53 as the recipient strain. The transconjugants were selected on MacConkey agar plates (Beijing Landbridge, China) supplemented with 100 mg/L sodium azide (Sigma–Aldrich) and 2 mg/L colistin (Sigma–Aldrich). Two different conjugation temperatures (30 and 37°C) were used for the transfer in this study. Transfer frequencies were calculated as the number of transconjugants obtained per recipient. Transfer of mcr-1 to transconjugants was confirmed by PCR (Liu et al., 2016). The colistin MIC value of the J53 and transconjugants were tested according to the description above.
DNA Extraction, Whole Genome Sequencing (WGS), Assembly and Annotation
DNA extraction and WGS were carried out for mcr-1-carrying isolates to obtain complete genomes. Briefly, a single colony for each isolate was cultured overnight in brain heart infusion (BHI) broth at 37°C. A TIANamp Bacterial DNA extraction kit (DP302, TIANGEN BIOTECH, Beijing, China) was used to extract the genomic DNA from each bacterial culture according to the manufacturer’s instructions, followed by a 10-kbp template library preparation step with PacBio® Template Prep Kit. Sequencing was performed commercially using SMRT® Pacific Biosciences RS II platform (Tianjin Biochip Corporation, Tianjin, China) with C4 sequencing chemistry and P6 polymerase within one SMRT®cell.
SMRT® Analysis v2.3.0 was used for demultiplexing, base calling, raw reads quality filtering, and de novo assembly according to RS Hierarchical Genome Assembly Process (HGAP) workflow v3.0. Subsequently, Consed software version 28.0 (Gordon and Green, 2013) was used to manually inspect and trim duplicate ends to generate single, complete and closed sequences for each chromosome and plasmid. The genomes assembled from PacBio data were then error corrected by Pilon software (version 1.23) (Walker et al., 2014) with Illumina MiSeq sequencing reads data, of which a library was prepared with a NEBNext® Ultra DNA Library Prep Kit for Illumina (NEB#E7370) followed by sonication fragmentation (350-bp insert), before being loaded on an Illumina HiSeq platform with PE 150 sequencing strategy (Novogene, Beijing, China) with a HiSeq X Ten Reagent Kit v2.5 (Illumina, San Diego, CA, United States). The corrected and assembled contigs were deposited in National Center for Biotechnology Information (NCBI) and automatically annotated using the NCBI Prokaryotic Genomes Automatic Annotation Pipeline (PGAP).
Genomic Information Mining
Plasmid replicon types (Inc groups) were identified through the center for genomic epidemiology (CGE) website with PlasmidFinder 2.0 (Carattoli et al., 2014). The predicted serotypes were confirmed and the multi-locus sequence typing (MLST) type were identified using Salmonella In Silico Typing Resource (SISTR) (Yoshida et al., 2016). CRISPR loci in the genomes were predicted using CRISPRfinder (Grissa et al., 2007). Similarly, prophage sequences were identified using the PHAge Search Tool Enhanced Release (PHASTER) (Arndt et al., 2016).
Assessment of Virulence
Seven extensively used Salmonella reference complete genomes LT2 (NC_003197), 14028s (NC_016856), DT104 (NC_022570), CT18 (NC_003198), Ty2 (NC_004631) and two hypervirulent isolates [D23580 (LS997973) and 4/74 (NC_016857)] (Canals et al., 2019), available from GenBank, were used to confirm and compare the presence of Salmonella pathogenicity islands (SPIs) and the potential virulence factors (VFs) with two mcr-1-carrying isolates in this study by SPIFinder (Roer et al., 2016) and Virulence factor database (VFDB) (Liu et al., 2019). To explore the prevalence of different VFs among various Salmonella, a heatmap was made with R and pheatmap, providing for a comparison of the presence or absence of different VFs among the above nine listed genomes and thirteen representative genomes already in the VFDB database with known VFs.
AMR Analysis
Antimicrobial resistance genes were identified through the Center for Genomic Epidemiology (CGE) website with ResFinder 3.0 (Zankari et al., 2012). DNA sequences of each identified AMR gene regions were selected for detailed BLAST analysis. A MUSCLE alignment was performed in Geneious prime software (version 2019.2.3) between mcr-1.1 (NG_050417.1) and the two mcr-1 genes sequences in this study. All genes, plasmids and chromosome sequences used in this study were managed and analyzed by Geneious.
A further comparative sequence alignments were performed in Geneious to identify the nucleotide polymorphism and related amino acid substitution sites compared with the original mcr-1.1 and all variants which could be found on NCBI to date (mcr-1.2 ∼ mcr-1.27). An unrooted rectangular cladogram tree was also generated for all currently known representatives of the MCR protein subgroups (MCR-1 through MCR-10) and related alleles or variants using the Geneious Tree Builder, with UPGMA tree build method and Jukes-Cator genetic distance model in Geneious software, followed by visualization on EvolView (Subramanian et al., 2019). To better understand the genetic environment of the mcr-1 locus on plasmids of mcr-carrying isolates, these sequences were extracted with Geneious, and compared and displayed using Easyfig v2.2.2 (Sullivan et al., 2011).
Genome Data Availability
The genome data of chromosome and plasmid sequences of the two mcr-1 gene positive Salmonella isolates was submitted to the NCBI nucleotide database under BioProject no. PRJNA498334 with Biosample no. of SAMN10291561 and SAMN10291586, and related accession numbers for chromosome and plasmid sequences were: CP033349, CP033350, CP033351, and CP033352.
Results
AST for 755 Foodborne Salmonella Isolates
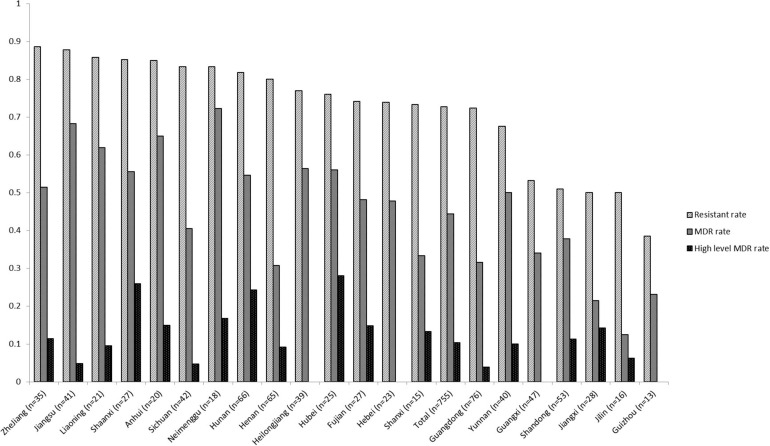
The percentages related to AMR for all 755 Salmonella isolates recovered from various foods are shown in Table 1. Among these, 206 isolates (27.3%) were susceptible to all antimicrobial agents and 549 (72.7%) exhibited resistance to at least one compound. The resistance rates were classified into three categories: (1) higher than 34%: NAL, TET, AMP, and SAM; (2) between 11.0 and 24.8%: CHL, SXT, FFC, CIP, KF, CTX and GEN; (3) lower than 10%: CT, CAZ, FEP, IMP, and MEM. Resistance to four cephalosporin-type compounds demonstrated a decreasing trend across the generations of this drug class (KF > CTX/CAZ > FEP). No isolate was resistant to carbapenem-type compounds (IMP and MEM). Sixty five (65/755, 8.6%) isolates were co-resistant to both cefotaxime and ciprofloxacin, two first-line antimicrobial agents used to treat human salmonellosis clinically. One hundred and twenty eight (17.0%), 86 (11.4%), 90 (11.9%), 79 (10.5%), 39 (5.2%), 49 (6.5%), 52 (6.9%), and 26 (3.4%) isolates were resistant to 1, 2, 3, 4, 5, 6, 7, and 8 classes of antimicrobial agents tested, respectively. In total, 134 different AMR profiles were recorded among 549 AMR Salmonella isolates. There were 335 isolates (44.4%) were classified as MDR (resistant to three or more classes of antimicrobial agents) and 78 isolates (10.3%) were identified as high level MDR (resistant to seven or eight classes of antimicrobials). Three isolates were co-resistant to 13 different antimicrobial (GEN-AMP-SAM-FEP-SXT-NAL-CHL-TET-CTX-FFC-CAZ-KF-CIP). Nineteen out of 26 provinces returned a resistant rate of higher than 50%, and 9 were recorded to exceed 80% with the highest of 88.6% (31/35, Zhejiang province). Ten of 26 had an MDR rate of no less than 50%, while 4 of them were found to exceed 60% (Inner Mongolia, Jiangsu, Anhui, and Liaoning). The level of MDR rates of Hubei, Shaanxi, and Hunan provinces were higher than 24% (Figure 1). The AMR data of 755 isolates were available in Supplementary data sheet 1.
FIGURE 1.
Antimicrobial resistance of foodborne Salmonella isolates recovered from different provinces in mainland China, 2016. [Multi-drug resistant (MDR), resistant to three or more classes of antimicrobial agents].
Serotyping, Further AST and Plasmid Conjugation for mcr-1 Gene Positive Isolates
The complete collection was tested for the presence of the mcr-1 gene by qPCR and two Salmonella isolates, CFSA231 and CFSA629, were positive with an mcr-1 gene detection rate of 0.26% (2/755) among Salmonella from mainland China, 2016. The two isolates were recovered from a pork dumpling sample (Huangshi, Hubei) and from an egg sample (Zhongshan, Guangdong), respectively. Based on both serotyping methods mentioned above, CFSA231 and CFSA629 were separately identified as Derby and Typhimurium serotype with the antigen formulas of 1,4,[5],12:f,g:- and 1,4,[5],12:i:1,2. Both isolates demonstrated an MDR phenotype to six or seven classes of antimicrobials but with some notable differences (Table 2). For instance, S. Derby CFSA231 expressed resistance to gentamicin, ciprofloxacin, folate pathway inhibitors and phenicols, while resistance against cephalosporins and colistin were observed for S. Typhimurium CFSA629. Based on the MIC value change for cefotaxime and ceftazidime in combination with clavulanate compared with when tested alone (Table 2), S. Typhimurium CFSA629 was confirmed as an ESBL positive strain whilst S. Derby CFSA231 was negative. mcr-harboring plasmids of both isolates could transfer into E. coli J53 at frequencies as below: pCFSA231, 1.5 × 10–7 (30°C) and 2.6 × 10–6 (37°C); pCFSA629, 2.1 × 10–4–2.0 × 10–2 (30°C), and 3.9 × 10–6 (37°C).
Genome Sequence Features, SPIs, Virulence Factors (VFs) and AMR Genes
Both genomes of S. Derby CFSA231 and S. Typhimurium CFSA629 consisted of a single circular chromosome and a circular plasmid. Details of the genomic features of the bacterial chromosomes and plasmids are shown in Table 3. CRISPR features and prophage information is available in Supplementary Tables 1, 2, respectively. Different serotypes showed different SPI genotypes, and SPI details are summarized in Table 4, all six S. Typhimurium strains contain the same SPI genotype independent of STs; Meanwhile, when compared with two S. Typhi (CT18 and Ty2), a small SPI type difference was noted and related to the presence of SPI-6, whilst these could be distinguished from other serotypes in having SPI-7 through SPI-14 and C63PI.
TABLE 3.
Genome sequence features for CFSA231 and CFSA629.
| Strain or plasmid name | Serotype, MLST type or plasmid replicon type | Number of reads | Mean read length (bp) | Coverage | Size (bp) | G + C content | Number of coding genes, pseudo genes and RNA genes |
| CFSA231 | Derby ST40 | 65,092 | 8,680 | 68.13x | 4,834,516 | 52.1% | 4,519; 134; 119 |
| pCFSA231 | IncX4 | 33,309 | 41.9% | ||||
| CFSA629 | Typhimurium ST34 | 54,855 | 9,083 | 57.39x | 4,999,270 | 52.1% | 4,937; 117; 125 |
| pCFSA629 | IncHI2A/IncHI2 | 210,674 | 45.2% |
TABLE 4.
Distribution of Salmonella pathogenicity island (SPIs) in seven representative genomes of Salmonella isolates (Identity threshold: 95%, minimum length: 60%).
| Salmonella strain | CFSA231 | CFSA629 | LT2 | 14028S | DT104 | D23580 | 4/74 | CT18 | Ty2 |
| Serotype | Derby | Typhimurium | Typhimurium | Typhimurium | Typhimurium | Typhimurium | Typhimurium | Typhi | Typhi |
| MLST type | ST40 | ST34 | ST19 | ST19 | ST19 | ST313 | ST19 | ST2 | ST1 |
| Accession number | CP033350.2 | CP033352.2 | NC_003197.2 | NC_016856.1 | NC_022570.1 | LS997973.1 | NC_016857.1 | NC_003198.1 | NC_004631.1 |
| SPI-1 | − | + | + | + | + | + | + | + | + |
| SPI-2 | + | + | + | + | + | + | + | + | + |
| SPI-3 | + | + | + | + | + | + | + | + | + |
| SPI-4 | + | + | + | + | + | + | + | + | + |
| SPI-5 | − | + | + | + | + | + | + | + | + |
| SPI-6 | − | − | − | − | − | − | − | + | − |
| SPI-7 | − | − | − | − | − | − | − | + | + |
| SPI-8 | − | − | − | − | − | − | − | + | + |
| SPI-9 | − | − | − | − | − | − | − | + | + |
| SPI-10 | − | − | − | − | − | − | − | + | + |
| SPI-11 | − | − | − | − | − | − | − | − | − |
| SPI-12 | − | − | − | − | − | − | − | + | + |
| SPI-13 | − | + | + | + | + | + | + | − | − |
| SPI-14 | − | + | + | + | + | + | + | − | − |
| C63PI | + | + | + | + | + | + | + | − | − |
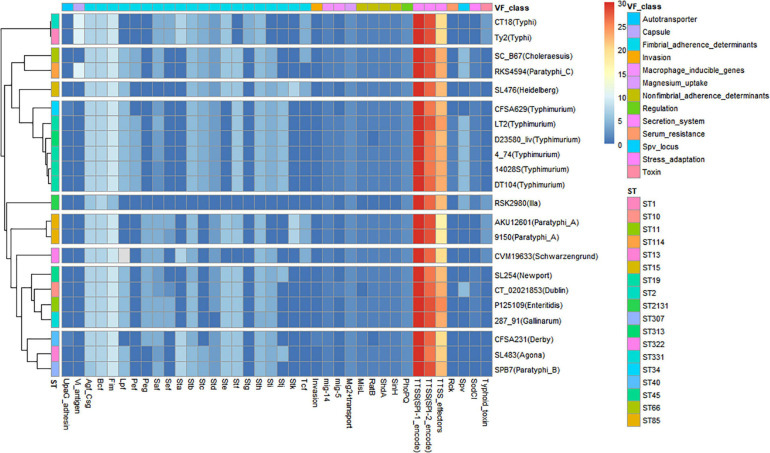
A VF heatmap was generated using R for CFSA231, CFSA629 and 20 additional Salmonella reference genomes with known VFs (Figure 2). It showed that the bacterial secretion system was the dominant VF class, and the top three VFs included the SPI-1 encoded Type Three Secretion System (T3SS), the SPI-2 encoded T3SS and T3SS effectors denoted by the red and yellow colors in the heatmap (Figure 2). The VF pattern of one Salmonella isolate appeared to be more closely related to isolates of the same serotype, rather than to the isolates of any other serotypes, such as S. Typhi, S. Typhimurium, and S. Paratyphi A. CFSA629 ST34 was distinguished from those of S. Typhimurium ST19 and ST313, with the main difference being the absence of the following VFs: Pef, Mig-5 and Spv. Six typhoidal Salmonella, including two S. Typhi and four S. Paratyphi were clustered into four distinct clades. As listed in Supplementary Table 3, the VF number of the 22 Salmonella genomes varied from 110 (S. enterica subsp. arizonae ser.62:z4,z23:– str. RSK2980) to 176 (S. Paratyphi C str. RKS4594). The annotated genes belonging to secretion system and fimbrial adherence virulence classes were the top two VFs for CFSA231 and CFSA629.
FIGURE 2.
Heatmap showing the relationship of 22 genomes/isolates including Salmonella Derby CFSA231 and Salmonella Typhimurium CFSA629 and 20 reference genomes based on their differential virulence factor classes. Numbers of virulence factors were represented by different colors, high number were marked with red squares (pixels) and low numbers were represented with blue squares.
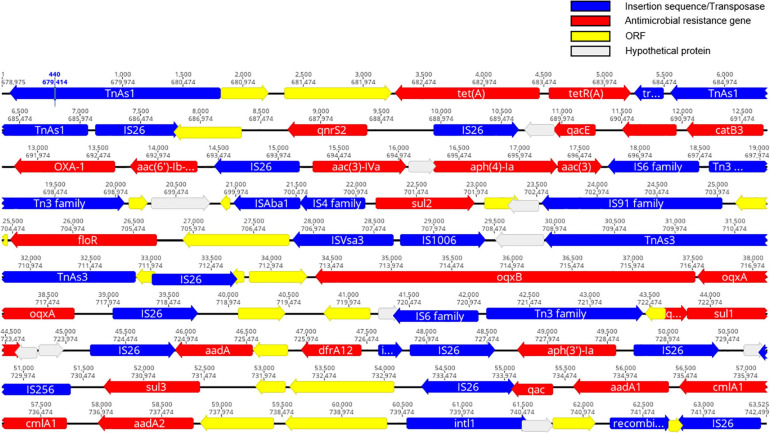
The resistance genotypes of CFSA231 and CFSA629 identified by Resfinder were shown in Table 2. In S. Derby CFSA231 the mcr-1 gene was located on an IncX4-type plasmid pCFSA231 along with a pap2-encoding gene distal to this site. We identified a number of similar well conserved plasmids differing by less than 4 nucleotides by BLAST on NCBI, and all of which were distributed in Enterobacteriaceae, with a high number of these (>80%) being associated with E. coli. Except mcr-1, all other 21 AMR-encoding genes identified in the genome of CFSA231, including one ESBL gene (blaOXA–1), were localized to the bacterial chromosome; one point mutation in the parC gene, resulting in an amino acid substitution (T57S) was identified. Four ciprofloxacin resistance-encoding genes [aac(6′)-Ib-cr, oqxA, oqxB, qnrS2], that are more commonly associated with plasmid-mediated quinolone resistance (PMQR), were mapped on the chromosome. These 21 genes were located within a 64-kbp locus that contained 83 CDS identified which encoded 23 transposases, 20 chromosomal antimicrobial resistant genes mediating resistance to nine drugs in this study, one class 1 integron gene, one recombinase gene and other genes encoding functional and hypothetical proteins (Figure 3). A query coverage value of 100% and identity of 100% were recorded with a single nucleotide difference identified between this locus and a similar arrangement reported earlier in a chicken-derived Salmonella isolate CD-SL01 recovered from chicken in China, 2015-2016 (NCBI accession number: CP028900.1). Both chromosomal genomes were compared using Geneious and the MAFFT Alignment programme with default settings, and were found to have an identity of 99.947% and 2,565 base/residue differences in total and CD-SL01 was devoid of any plasmids.
FIGURE 3.
A schematic illustration showing the structural of ∼64kb MDR gene cluster region on chromosome of CFSA231 (created using Geneious software). Antimicrobial resistance (AMR)-encoding genes are indicated in red boxes/arrows. Blue boxes/arrows denote transposon- and integron-associated genes. The individual open reading frame (ORF) are indicated with yellow boxes/arrows. The light gray boxes indicate hypothetical proteins.
In S. Typhimurium CFSA629, a mutation in gyrA giving rise to a D87Y substitution was found; six and five antimicrobial resistant genes were located on the chromosome and plasmid, respectively and two different ESBL genes (blaTEM–1B and blaCTX–M–14) mapped to them each. The mcr-1 gene was also located on plasmid, an IncHI2 type plasmid pCFSA629, which possessed a single copy of the ISApl1 element along with a gene encoding the PAP2 family protein flanking the left and the right side of the mcr-1 gene. There was two IS26 composite-like transposonal modules in this isolate which was rich in insertion sequences and transposons: one consisting of IS26-fosA3-IS1182-blaCTX–M–4-IS26 and another one was IS26-[aac(3)-IV]-[aph(4)-Ia]-IS6 family-Tn3-IS26.
Comparison and Relationship Between mcr Genotypes and MCR Variants
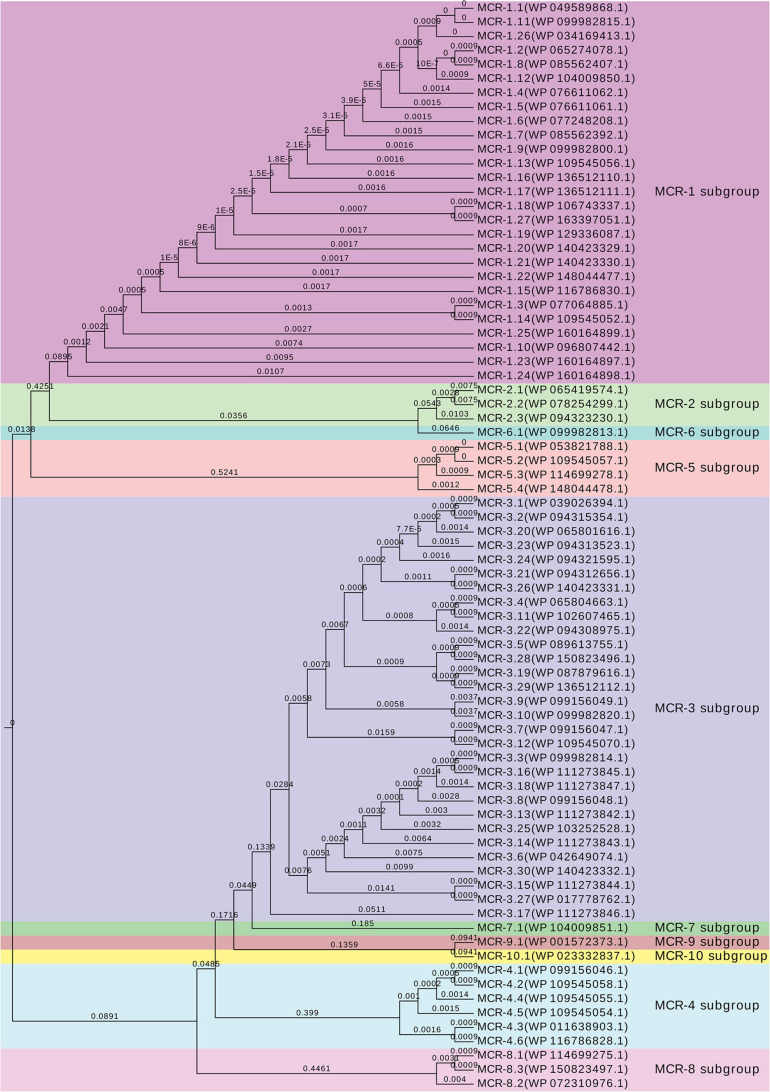
An mcr-1 variant was identified and confirmed on pCFSA629 by Geneious (Hu et al., 2021), and was assigned with allele number mcr-1.19 (MK490674.1, G1,534A) and corresponding protein MCR-1 allele number of MCR-1.19 (QBC35984.1, Val512-to-Ile) in GenBank. Compared with the mcr-1.1, no base mutations were observed in the mcr-1 gene on pCFSA231. The comparative alignment between 26 mcr-1 variants and mcr-1.1, as well as the amino acid differences among their related mcr-1 coding protein variants was shown in Table 5. One single nucleotide difference was observed among most mcr-1 genotypes (n = 21), and a dinucleotide difference existed in three of these variants. In terms of MCR-1.1 with 541 amino acids, a trinucleotide duplication was found in mcr-1.11, resulting in an additional amino acid for an MCR-1 variant of 542 amino acids, while mcr-1.15 and mcr-1.26 with 540 amino acids, arising from a single nucleotide polymorphism (SNP) located at initiation codon. It should be noted that three mcr-1 variants (mcr-1.10, mcr-1.23, and mcr-1.24) had more than 12 nucleotide differences relative to mcr-1.1, leading to at least 7 amino acid differences from MCR-1.1. The non-rooted rectangular cladogram showed two distinguished clusters among ten MCR protein sub-groups (Figure 4). In the case of the genetic distances within the MCR-1 subgroup, MCR-1.10, MCR-1.23, and MCR-1.24, which exhibited the most amino acid differences relative to MCR-1.1 were also the furthest in genetic distance from MCR-1.1, followed by MCR-1.25, MCR-1.14, MCR-1.3, and MCR-1.15.
TABLE 5.
Nucleotide/amino acid changes of mcr-1/MCR-1 alleles/variants compared with mcr-1.1/MCR-1.1 (the accession numbers emanated from Reference gene browser with gene family “mcr-1,” database version: 2020–05-04.1. Nucleotide and amino acid differences were identified by Geneious software).
| Allele/variant | RefSeq protein | Refseq nucleotide | GenBank protein | GenBank nucleotide | Nucleotide differences compared with mcr-1.1 | Amino acid differences compared with MCR-1.1 | First discovered host bacteria |
| mcr-1.1 | WP_049589868.1 | NG_050417.1 | AKF16168.1 | KP347127.1 | N/A | N/A | Escherichia coli |
| mcr-1.2 | WP_065274078.1 | NG_051170.1 | OBY14952.1 | LXQO01000025.1 | A8T | Gln3Leu | Klebsiella pneumoniae |
| mcr-1.3 | WP_077064885.1 | NG_052861.1 | ANJ15621.1 | KU934208.1 | AA111-112GG | Ile38Val | Escherichia coli |
| mcr-1.4 | WP_076611062.1 | NG_052664.1 | APM87143.1 | KY041856.1 | G1318A | Asp440Asn | Escherichia coli |
| mcr-1.5 | WP_076611061.1 | NG_052663.1 | APM84488.1 | KY283125.1 | C1354T | His452Tyr | Escherichia coli |
| mcr-1.6 | WP_077248208.1 | NG_052893.1 | AQK48217.1 | KY352406.1 | G1263A, G1607A | Arg536His | Salmonella enterica |
| mcr-1.7 | WP_085562392.1 | NG_054678.1 | AQQ11622.1 | KY488488.1 | G643A | Ala215Thr | Escherichia coli |
| mcr-1.8 | WP_085562407.1 | NG_054697.1 | AQY61516.1 | KY683842.1 | A8G | Gln3Arg | Escherichia coli |
| mcr-1.9a | WP_099982800.1 | NG_055582.1 | ASK38392.2 | KY964067.1 | T1238C | Val413Ala | Escherichia coli |
| mcr-1.10b | WP_096807442.1 | NG_055583.1 | ASK49940.1 | MF176238.1 | – | – | Moraxella sp. |
| mcr-1.11c | WP_099982815.1 | NG_055784.2 | ATM29809.1 | KY853650.2 | GTG19-21dup | Val7dup | Escherichia coli |
| mcr-1.12 | WP_104009850.1 | NG_056412.1 | BBB21811.1 | LC337668.1 | G9C | Gln3His | Escherichia coli |
| mcr-1.13 | WP_109545056.1 | NG_057466.1 | AVM85874.1 | MG384739.1 | G465A | Met155Ile | Escherichia coli |
| mcr-1.14 | WP_109545052.1 | NG_057460.1 | ARA74236.1 | KX443408.2 | AA111-112GG, G591A | Ile38Val, Met197Ile | Klebsiella pneumoniae |
| mcr-1.15d | WP_116786830.1 | NG_061610.1 | AXL06756.1 | MG763897.1 | AT1-2TA, C836A | Met1del, Thr279Lys | Klebsiella pneumoniae |
| mcr-1.16 | WP_136512110.1 | NG_064787.1 | QBG64271.1 | MK568462.1 | C952A | Arg318Ser | Escherichia coli |
| mcr-1.17 | WP_136512111.1 | NG_064788.1 | QBG64272.1 | MK568463.1 | G410C | Ser137Thr | Escherichia coli |
| mcr-1.18 | WP_106743337.1 | NG_064789.1 | – | PGLM01000025.1 | T25G | Tyr9Asp | Escherichia coli |
| mcr-1.19 | WP_129336087.1 | NG_065449.1 | QBC35984.1 | MK490674.1 | G1534A | Val512Ile | Salmonella enterica |
| mcr-1.20 | WP_140423329.1 | NG_065450.1 | SPQ84451.1 | LS398440.1 | A184C | Met62Leu | Escherichia coli |
| mcr-1.21 | WP_140423330.1 | NG_065451.1 | QCU55424.1 | MK965883.1 | C1234T | Pro412Ser | Escherichia coli |
| mcr-1.22 | WP_148044477.1 | NG_065944.1 | QDO71694.1 | MN017134.1 | C1277T | Ser426Phe | Escherichia coli |
| mcr-1.23e | WP_160164897.1 | NG_067235.1 | QHD57408.1 | MN873697.1 | – | – | Salmonella enterica |
| mcr-1.24f | WP_160164898.1 | NG_067236.1 | QHD64700.1 | MN879257.1 | – | – | Escherichia coli |
| mcr-1.25 | WP_160164899.1 | NG_067237.1 | QHD64702.1 | MN879259.1 | G41C, C565G | Ser14Thr, Leu189Val | Escherichia coli |
| mcr-1.26d | WP_034169413.1 | NG_068217.1 | NEU93872.1 | JAAGSA010000042.1 | T2C | Met1del | Escherichia coli |
| mcr-1.27 | WP_163397051.1 | NG_068218.1 | NEU89143.1 | JAAGSB010000042.1 | A26G | Tyr9Cys | Escherichia coli |
aUsing start codon that matches other mcr-1 genes rather than the one in the original INSDC entry. bDetails not listed for 39 nucleotide differences between mcr-1.10 and mcr-1.1 as well as 7 amino acid differences between MCR-1.10 and MCR-1.1. cDup, duplication of nucleotides/amino acids at positions indicated. dDel, deletion of amino acids at positions indicated. In comparison of mcr-1.1, the nucleotide at position 1 and 2 (AT) of mcr-1.15 mutate into TA, and also the nucleotide at position 2 (T) of mcr-1.26 mutate into C, resulting in a unavailable translation initiation codon of ATG at 1-3 and a retroposed codon from 4-6(ATG). eDetails not listed for 12 nucleotide differences between mcr-1.23 and mcr-1.1 as well as 9 amino acid differences between MCR-1.23 and MCR-1.1. fDetails not listed for 13 nucleotide differences between mcr-1.24 and mcr-1.1 as well as 10 amino acid differences between MCR-1.24 and MCR-1.1.
FIGURE 4.
Genetic relationships between MCR protein subgroups and related alleles. UPGMA rectangular cladogram tree of the deduced amino acid sequences of putative phosphoethanolamine transferase belonged to different subgroups was constructed using Geneious prime v2019.0.4 software with Jukes–Cantor genetic distance model, and visualized by EvolView online tool (v2.0, https://www.evolgenius.info/evolview/). NCBI accession numbers are listed following by the MCR alleles and available in GenBank.
Genetic Environment Context of mcr-1 Gene in pCFSA231 and pCFSA629
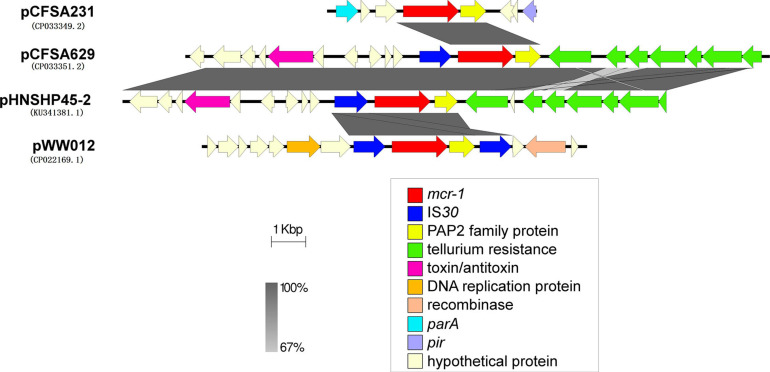
The mcr-1 locus of plasmid pCFSA231 and pCFSA629 were extracted and compared with the relevant mcr-1 locus on plasmids pHNSHP45-2 (KU341381.1) from E. coli and pWW012 (CP022169.1) from Salmonella (Figure 5). It showed that the gene structures near mcr-1 on plasmids varied but shared the same regions (genes encoding MCR-1 and PAP2 family proteins), with different presence of insertion sequences (ISs). pCFSA231 had no IS and pCFSA629 obtained only one ISApl1; a tellurium resistance gene cluster was located downstream PAP2 coding gene of pCFSA629 and pHNSHP45-2; pWW012, the mcr-1-carrying plasmid from our previous study, consisted of an IS-mcr-1-PAP2-IS module which is an ISApl1-flanked composite transposon (Tn6330).
FIGURE 5.
Genetic environments related to mcr-1 gene in bacterial plasmids. The figure was generated by Easyfig (v2.2.2). Plasmids marked with “pCFSA” were carried by two mcr-1 positive Salmonella isolates in this research, and plasmid pWW012 belonged to a previous research in our lab (accession number: CP022169), while pHNSHP45-2 (accession number: KU341381) belonged to Escherichia coli strain SHP45, which is the first isolate reported harboring mcr-1 gene. Confirmed and putative open reading frames (ORFs) are indicated by block arrows and their orientations with different colors, and arrow size is proportional to the predicted ORF length. mcr-1 gene is indicated by a red arrow, while genes encoding mobile elements (insertion sequence, IS) are indicated by blue arrows. Regions of homology between the plasmids ranging from 67 to 100% are indicated by the graded shaded regions between sequences.
Discussion
Salmonella are important foodborne zoonotic pathogens often linked to cases of gastroenteritis and bacteremia, and are one of the leading causes of global bacterial food poisoning worldwide (Threlfall, 2002). The antimicrobial resistance expressed by Salmonella is spreading in both developed and developing countries (Parisi et al., 2018). In this study, similar antimicrobial resistance for foodborne Salmonella isolates was observed with previous study in China (Hu et al., 2017). The AMR rate recorded against ciprofloxacin (21.3%) indicated a comparatively statistically significant increase when compared with the previous year (Hu et al., 2017). A potential explanation for the high ciprofloxacin intermediate rate in this study (38.3%) may be due to the updated and extended range of criteria in the CLSI guidelines, but this finding partly signal a continuously reduced susceptibility trend among foodborne Salmonella to ciprofloxacin in recent years. Gong et al. (2019) found that almost 90% of S. Indiana were resistant to both ciprofloxacin and cefotaxime and 8.6% of the tested isolates in this study demonstrated this feature of resistance, posing a serious threat to public health and ongoing surveillance is clearly necessary in regard to monitoring the emergence of resistance and identify transmission routes.
In this study different serotypes of Salmonella possessed different SPI and VF genotypes, and the impact of these differences for a comparison of serotype on virulence potential remains unclear. For example, the hypervirulent invasive non-typhoidal (iNTS) S. Typhimurium ST313 str. D23580 could not easily be distinguished from other S. Typhimurium of different ST types based on the VFs, since there are several mechanisms that could contribute to its pathogenicity in iNTS (Carden et al., 2015, 2017; Hammarlöf et al., 2018). Even within a serovar, differences in SPI content may also affect virulence potential such as the presence or absence of the avrA secreted effector gene in certain S. Montevideo lineages (Nguyen et al., 2018). In comparison to many studies of resistance prediction from AMR genes, increased emphasis or the joint on functional transcriptomics, proteomics and genomics techniques are necessary for virulence potential investigation/prediction and pathogenicity modeling.
In this study, two mcr-1-carrying foodborne Salmonella isolates were recovered among 755 strains (0.26%). According to our latest data related to more than 3,800 foodborne Salmonella recovered in mainland China between 2011 and 2019 (data not published), 14 mcr-1-harboring isolates were detected corresponding to a positivity rate of 0.4%, which could be regarded as a low prevalence rate. Similar results were reported previously for isolates cultured from clinical, food or food-producing animals (Carnevali et al., 2016; Cui et al., 2017; Lu et al., 2019; Luo et al., 2020). Susceptibility testing of CFSA231 and CFSA629 against colistin, recorded MIC values of 2 mg/L, are consistent with mcr-1 mediated low-level colistin resistance (2–8 mg/L) (Zhang et al., 2019). Acquisition of this gene could also facilitate further selection of chromosomal mutants in some cases, leading to high-level colistin resistance (HLCR) (Zhang et al., 2019). Thus screening and surveillance for the mcr-1 in bacteria of importance to human health is critical. Besides their distinct geographic and sample origins, the two mcr-1-positive Salmonella isolates in this study exhibited different MDR profiles. All 27 compounds tested in the extended AST tests in this study are listed in CLSI and EUCAST and are used in human and veterinary settings. An integrated One Health based surveillance system is crucial in tracking these developments, and it should be a more initiative monitoring model which could focus on the antimicrobial resistance dissemination among human beings, animals and environments at the same time (Luo et al., 2020).
The AMR determinants detected in this report were in coherence with the AMR patterns obtained by AST. In the case of S. Derby CFSA231 all resistance genes, except for mcr-1, were mapped to the chromosome, including four PMQR genes that are more commonly associated with plasmid. The 64 kbp putative SGI-like MDR region in this isolate was largely similar to the Salmonella Genomic Island 1 (SGI1) with an ACSSuT phenotype reported earlier (Mulvey et al., 2006), differing in the numbers of mobile genetic elements (MGEs) it contained. BLAST analysis of this locus identified genetically homologous regions in both chromosomal- and plasmid-based sequences in different species including Salmonella, E. coli, and K. pneumoniae, suggesting that this putative SGI1-like region might have already disseminated among the Enterobacteriaceae family, and this will cause a potential of a processed copy-out-paste-in transpositional event resulting in dissemination and stabilization of these related resistant genes. The corresponding resistance genes that zoonotic Salmonella have acquired are more commonly located on plasmids, in transposons, gene cassettes, or variants of the SGI1 and SGI2 loci (Michael and Schwarz, 2016), thus studies exploring the dissemination of this putative SGI1-like locus may provide further insights into its evolution (Michael and Schwarz, 2016).
The mcr-1 gene has been found in plasmids with different Inc types such as IncI2, IncHI1, IncHI2, IncP, IncX4, IncFI, and IncFIB (Zhi et al., 2016; Poirel et al., 2017). Based on the epidemiological study (Lu et al., 2019), the mcr-1-carrying IncHI2 type plasmid was originally identified in Salmonella isolated from diarrhoeal outpatients in Shanghai in 2014 and increasingly detected after the summer of 2015, representing the primary replicon type in 2016. It was reported that foods had played important roles in the expansion of mcr-1-carrying IncHI2 plasmids among different members of the Enterobacteriaceae family before 2016, and what is of increasing concern, with usage of antimicrobials other than polymyxins, co-selection of mcr-1 may happen due to the MDR phenotype and conjugative ability of the IncHI2 plasmids (Zhi et al., 2016). For instance, a Salmonella IncHI2 plasmid that predate the era of mass antibiotic usage has been sequenced with no detectable AMR genes (Nguyen et al., 2017), but modern IncHI2 plasmids encoding multiple AMR genes are predominant in MDR Salmonella (Chen et al., 2016). Thus, there is a potential for capture of mcr-1 in MDR Salmonella due to co-selection by other antimicrobials other than polymyxin. The prevalence of the mcr gene is higher in resistant bacterial isolates cultured from food-producing animals when compared to those cultured from humans. This may be due to the application of antimicrobials in agricultural for production purposes. Although use of colistin in agriculture production has been recently banned in China and Brazil, surveillance must be maintained as increasing clinical usage of polymyxins may also contribute to the risk of dissemination of mcr markers in nosocomial settings (Osei Sekyere, 2019).
The co-existence of plasmid-mediated mcr-1 and carbapenemase-encoding genes has been identified in Enterobacteriaceae and there are increasing reports of this co-occurrence worldwide. Isolates of both clinical and animal origin combining bla-encoding genes (such as blaNDM–1, blaNDM–5, blaNDM–9, blaOXA–48, blaKPC–2, and blaVIM–1) with mcr-1 gene may signal the risk of the emerging of strains expressing pan-drug resistance (PDR) (Yang et al., 2016; Lai et al., 2017; Wang et al., 2018; Le-Vo et al., 2019). It is important to note that the presence of mcr-1 gene on the chromosome in recent studies (Falgenhauer et al., 2016; Zurfluh et al., 2016; Yamaguchi et al., 2020) suggest that the mcr-1 gene could become more stable through vertical inheritance in mcr-1-carrying isolates.
The role of MGEs such as transposons has made an important contribution to the AMR rapid dissemination by horizontal gene transfer under selective pressure imposed by the antimicrobial usage (Snesrud et al., 2018). mcr-1 gene could be found in various combinations with one or two copies of ISApl1 or devoid of the IS element with different replicon types (Li et al., 2017). Snesrud et al. (2018) presented representative sequences of the four general mcr-1 structures identified to date: (a) composite transposon Tn6330 that is thought to mediate the initial mobilization of event; (b) a single-ended structure with a distal copy of ISApl1; (c) a structure lacking both copies of ISApl1; (d) and a single-ended structure with a proximal copy of ISApl1 only. We can imagine a genetic element with the loss of ISApl1 may provide a more stable mcr-1 state, especially when integrated in the chromosome. In this study, pCFSA231 and pCFSA629 are classified in the structures described in (c) and (b) above, respectively. They did not contain two ISs and the likelihood of losing or moving mcr-1 gene relatively decreased, however, our results still suggest that plasmids of various replicon types may contribute to the mcr-1 gene movement and global spread, accelerating the frequency of colistin resistance worldwide.
Conclusion
Colistin is an important antibacterial agent used for treating MDR Gram-negative bacteria infections. Intrinsic colistin resistance located on the chromosome was generally thought to be non-transferable until the detection of MCR, a transferable polymyxin resistance reported globally among various bacterial species with comparatively few descriptions in Salmonella. In the context of the rapid spreading trend of the clinical mcr-1-harboring Salmonella and continuous discoveries of novel mcr genes and related variants, we report on the AMR profiles of a set of foodborne Salmonella cultured from mainland China and describe the complete genomes of two mcr-1-positive Salmonella isolates, including a S. Typhimurium isolate with an mcr-1-variant. Improved surveillance is important for understanding the dissemination of mcr genes among foodborne Salmonella around the world.
Data Availability Statement
The genome data of chromosome and plasmid sequences of the two mcr-1 gene positive Salmonella isolates were submitted to the NCBI nucleotide database under BioProject numbers PRJNA498334 with Biosample No. of SAMN10291561 and SAMN10291586, and related accession numbers for chromosome and plasmid sequences were: CP033349, CP033350, CP033351, and CP033352.
Author Contributions
YH performed the literature search. YH, FL, and SF designed the research. YH, SN, WW, XG, YD, CL, and XC performed the experiments and collected the data. YH, SN, WW, and JX analyzed and interpreted the data and finished the figures and tables. YH and SN wrote the manuscript. FL and SF reviewed and edited the manuscript. All authors read and approved the manuscript. YH, SN, FL, and SF have accessed and verified the underlying data.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We sincerely thank all the participants who took part in this study.
Footnotes
Funding. This work was supported financially by National Key Research and Development Program of China (2018YFC1604303 and 2018YFC1603904), CAMS Innovation Fund for Medical Science (CIFMS 2019-12M-5-024), and China Food Safety Talent Competency Development Initiative: CFSA 523 Program. The sponsors of the study had not any role in study design, data collection, data analysis, interpretation and writing of the report. We have not been paid to write this article by a pharmaceutical company or other agency. We state that all authors had full access to the full data in the study and accept responsibility to submit for publication.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.636284/full#supplementary-material
References
- Arndt D., Grant J. R., Marcu A., Sajed T., Pon A., Liang Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44 W16–W21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L., Hurley D., Li J., Meng Q., Wang J., Fanning S., et al. (2016). Characterisation of multidrug-resistant Shiga toxin-producing Escherichia coli cultured from pigs in China: co-occurrence of extended-spectrum β-lactamase- and mcr-1-encoding genes on plasmids. Int. J. Antimicrob. Agents 48 445–448. 10.1016/j.ijantimicag.2016.06.021 [DOI] [PubMed] [Google Scholar]
- Borowiak M., Baumann B., Fischer J., Thomas K., Deneke C., Hammerl J. A., et al. (2020). Development of a Novel mcr-6 to mcr-9 Multiplex PCR and assessment of mcr-1 to mcr-9 Occurrence in colistin-resistant Salmonella enterica Isolates from environment, feed, animals and food (2011-2018) in Germany. Front. Microbiol. 11:80. 10.3389/fmicb.2020.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canals R., Hammarlöf D. L., Kröger C., Owen S. V., Fong W. Y., Lacharme-Lora L., et al. (2019). Adding function to the genome of African Salmonella Typhimurium ST313 strain D23580. PLoS Biol. 17:e3000059. 10.1371/journal.pbio.3000059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Zankari E., García-Fernández A., Voldby Larsen M., Lund O., Villa L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58 3895–3903. 10.1128/aac.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carden S., Okoro C., Dougan G., Monack D. (2015). Non-typhoidal Salmonella Typhimurium ST313 isolates that cause bacteremia in humans stimulate less inflammasome activation than ST19 isolates associated with gastroenteritis. Pathog. Dis. 73:ftu023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carden S. E., Walker G. T., Honeycutt J., Lugo K., Pham T., Jacobson A., et al. (2017). Pseudogenization of the secreted effector gene ssei confers rapid systemic Dissemination of S. Typhimurium ST313 within migratory dendritic cells. Cell Host Microbe 21 182–194. 10.1016/j.chom.2017.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnevali C., Morganti M., Scaltriti E., Bolzoni L., Pongolini S., Casadei G. (2016). Occurrence of mcr-1 in colistin-resistant Salmonella enterica Isolates recovered from humans and animals in Italy, 2012 to 2015. Antimicrob. Agents Chemother. 60 7532–7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Fang T., Zhou X., Zhang D., Shi X., Shi C. (2016). IncHI2 plasmids are predominant in antibiotic-resistant Salmonella Isolates. Front. Microbiol. 7:1566. 10.3389/fmicb.2016.01566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump J. A., Medalla F. M., Joyce K. W., Krueger A. L., Hoekstra R. M., Whichard J. M., et al. (2011). Antimicrobial resistance among invasive nontyphoidal Salmonella enterica isolates in the United States: national Antimicrobial resistance monitoring system, 1996 to 2007. Antimicrob. Agents Chemother. 55 1148–1154. 10.1128/aac.01333-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M., Zhang J., Gu Z., Li R., Chan E. W., Yan M., et al. (2017). Prevalence and molecular characterization of mcr-1-Positive Salmonella STRAINS RECOVERED FROM CLINICAL SPECIMEns in China. Antimicrob. Agents Chemother. 61:e02471-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbediwi M., Li Y., Paudyal N., Pan H., Li X., Xie S., et al. (2019). Global burden of colistin-resistant bacteria: mobilized colistin resistance genes study (1980-2018). Microorganisms 7:461. 10.3390/microorganisms7100461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falgenhauer L., Waezsada S. E., Gwozdzinski K., Ghosh H., Doijad S., Bunk B., et al. (2016). Chromosomal Locations of mcr-1 and blaCTX–M–15 in Fluoroquinolone-Resistant Escherichia coli ST410. Emerg. Infect. Dis. 22 1689–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J., Zeng X., Zhang P., Zhang D., Wang C., Lin J. (2019). Characterization of the emerging multidrug-resistant Salmonella enterica serovar Indiana strains in China. Emerg. Microb. Infect. 8 29–39. 10.1080/22221751.2018.1558961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D., Green P. (2013). Consed: a graphical editor for next-generation sequencing. Bioinformatics 29 2936–2937. 10.1093/bioinformatics/btt515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissa I., Vergnaud G., Pourcel C. (2007). CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35 W52–W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlöf D. L., Kröger C., Owen S. V., Canals R., Lacharme-Lora L., Wenner N., et al. (2018). Role of a single noncoding nucleotide in the evolution of an epidemic African clade of Salmonella. Proc. Natl. Acad. Sci. U.S.A. 115 E2614–E2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Fanning S., Gan X., Liu C., Nguyen S., Wang M., et al. (2019). Salmonella harbouring the mcr-1 gene isolated from food in China between 2012 and 2016. J. Antimicrob. Chemother. 74 826–828. 10.1093/jac/dky496 [DOI] [PubMed] [Google Scholar]
- Hu Y., He Y., Wang Y., Fanning S., Cui S., Chen Q., et al. (2017). Serovar diversity and antimicrobial resistance of non-typhoidal Salmonella enterica recovered from retail chicken carcasses for sale in different regions of China. Food Control 81 46–54. 10.1016/j.foodcont.2017.05.031 [DOI] [Google Scholar]
- Hu Y., Fanning S., Nguyen S. V., Wang W., Liu C., Cui X., et al. (2021). Emergence of a Salmonella enterica serovar Typhimurium ST34 isolate, CFSA629, carrying a novel mcr-1.19 variant cultured from egg in China. J. Antimicrobial Chemother. dkab090. 10.1093/jac/dkab090 [DOI] [PubMed] [Google Scholar]
- Janssen A. B., Hout D. V., Bonten M. J. M., Willems R. J. L., Schaik W. V. (2020). Microevolution of acquired colistin resistance in Enterobacteriaceae from ICU patients receiving selective decontamination of the digestive tract. J. Antimicrob. Chemother. 75 3135–3143. 10.1093/jac/dkaa305 [DOI] [PubMed] [Google Scholar]
- Jazeela K., Chakraborty A., Karunasagar I., Deekshit V. K. (2020). Nontyphoidal Salmonella: a potential anticancer agent. J. Appl. Microbiol. 128 2–14. 10.1111/jam.14297 [DOI] [PubMed] [Google Scholar]
- Kluytmans J. (2017). Plasmid-encoded colistin resistance: mcr-one, two, three and counting. Euro Surveillance 22:30588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C. C., Chuang Y. C., Chen C. C., Tang H. J. (2017). Coexistence of MCR-1 and NDM-9 in a clinical carbapenem-resistant Escherichia coli isolate. Int. J. Antimicrob. Agents 49 517–518. 10.1016/j.ijantimicag.2017.02.001 [DOI] [PubMed] [Google Scholar]
- Le-Vo H. N., Tran P. T., Le L., Matsumoto Y., Motooka D., Nakamura S., et al. (2019). Complex class 1 integron in a clinical Escherichia coli strain from vietnam carrying Both mcr-1 and blaNDM–1. Front. Microbiol. 10:2472. 10.3389/fmicb.2019.02472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Xie M., Zhang J., Yang Z., Liu L., Liu X., et al. (2017). Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J. Antimicrob.Chemother. 72 393–401. 10.1093/jac/dkw411 [DOI] [PubMed] [Google Scholar]
- Li X. P., Fang L. X., Song J. Q., Xia J., Huo W., Fang J. T., et al. (2016). Clonal spread of mcr-1 in PMQR-carrying ST34 Salmonella isolates from animals in China. Sci. Rep. 6:38511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima T., Domingues S., Da Silva G. J. (2019). Plasmid-mediated colistin resistance in Salmonella enterica: a review. Microorganisms 7:55. 10.3390/microorganisms7020055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Zheng D., Jin Q., Chen L., Yang J. (2019). VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 47 D687–D692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Y., Wang Y., Walsh T. R., Yi L. X., Zhang R., Spencer J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet. Infect. Dis. 16 161–168. 10.1016/s1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- Lu X., Zeng M., Xu J., Zhou H., Gu B., Li Z., et al. (2019). Epidemiologic and genomic insights on mcr-1-harbouring Salmonella from diarrhoeal outpatients in Shanghai, China, 2006-2016. EBioMedicine 42 133–144. 10.1016/j.ebiom.2019.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q., Wan F., Yu X., Zheng B., Chen Y., Gong C., et al. (2020). MDR Salmonella enterica serovar Typhimurium ST34 carrying mcr-1 isolated from cases of bloodstream and intestinal infection in children in China. J. Antimicrob. Chemother. 75 92–95. 10.1093/jac/dkz415 [DOI] [PubMed] [Google Scholar]
- Michael G. B., Schwarz S. (2016). Antimicrobial resistance in zoonotic nontyphoidal Salmonella: an alarming trend? Clin. Microbiol. Infect. 22 968–974. 10.1016/j.cmi.2016.07.033 [DOI] [PubMed] [Google Scholar]
- Mulvey M. R., Boyd D. A., Olson A. B., Doublet B., Cloeckaert A. (2006). The genetics of Salmonella genomic island 1. Microbes Infect. 8 1915–1922. 10.1016/j.micinf.2005.12.028 [DOI] [PubMed] [Google Scholar]
- Nguyen S. V., Harhay D. M., Bono J. L., Smith T., Fields P. I., Dinsmore B. A., et al. (2018). Comparative genomics of Salmonella enterica serovar montevideo reveals lineage-specific gene differences that may influence ecological niche association. Microb. Genomics 4:e000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen S. V., Harhay G. P., Bono J. L., Smith T. P., Harhay D. M. (2017). Genome sequence of the thermotolerant foodborne pathogen Salmonella enterica serovar senftenberg ATCC 43845 and phylogenetic analysis of loci encoding increased protein quality control mechanisms. mSystems 2:e00190-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osei Sekyere J. (2019). Mcr colistin resistance gene: a systematic review of current diagnostics and detection methods. MicrobiologyOpen 8:e00682. 10.1002/mbo3.682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi A., Crump J. A., Glass K., Howden B. P., Furuya-Kanamori L., Vilkins S., et al. (2018). Health outcomes from multidrug-resistant Salmonella Infections in high-income countries: a systematic review and meta-analysis. Foodb. Pathog. Dis. 15 428–436. 10.1089/fpd.2017.2403 [DOI] [PubMed] [Google Scholar]
- Partridge S. R., Di Pilato V., Doi Y., Feldgarden M., Haft D. H., Klimke W., et al. (2018). Proposal for assignment of allele numbers for mobile colistin resistance (mcr) genes. J. Antimicrob. Chemother. 73 2625–2630. 10.1093/jac/dky262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Jayol A., Nordmann P. (2017). Polymyxins: antibacterial activity. susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30 557–596. 10.1128/cmr.00064-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roer L., Hendriksen R. S., Leekitcharoenphon P., Lukjancenko O., Kaas R. S., Hasman H., et al. (2016). Is the evolution of Salmonella enterica subsp. enterica linked to restriction-modification systems? mSystems 1 e00009-e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia C. M., Greig D. R., Day M., Hartman H., Painset A., Doumith M., et al. (2020). The characterization of mobile colistin resistance (mcr) genes among 33?000 Salmonella enterica genomes from routine public health surveillance in England. Microb. Genomics 6:e000331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snesrud E., McGann P., Chandler M. (2018). The birth and demise of the ISApl1-mcr-1-ISApl1 composite transposon: the vehicle for transferable colistin resistance. mBio 9:e02381-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian B., Gao S., Lercher M. J., Hu S., Chen W. H. (2019). Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 47 W270–W275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. J., Petty N. K., Beatson S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27 1009–1010. 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Zhang H., Liu Y. H., Feng Y. (2018). Towards understanding MCR-like colistin resistance. Trends Microbiol. 26 794–808. 10.1016/j.tim.2018.02.006 [DOI] [PubMed] [Google Scholar]
- Threlfall E. J. (2002). Antimicrobial drug resistance in Salmonella: problems and perspectives in food- and water-borne infections. FEMS Microbiol. Rev. 26 141–148. 10.1111/j.1574-6976.2002.tb00606.x [DOI] [PubMed] [Google Scholar]
- Walker B. J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Liu Y., Zhang Q., Jin L., Wang Q., Zhang Y., et al. (2018). The prevalence of colistin resistance in Escherichia coli and Klebsiella pneumoniae isolated from food animals in China: coexistence of mcr-1 and blaNDM with low fitness cost. Int. J. Antimicrob. Agents 51 739–744. 10.1016/j.ijantimicag.2018.01.023 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T., Kawahara R., Hamamoto K., Hirai I., Khong D. T., Nguyen T. N., et al. (2020). High prevalence of colistin-resistant Escherichia coli with chromosomally carried mcr-1 in healthy residents in vietnam. mSphere 5:e00117-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R. S., Feng Y., Lv X. Y., Duan J. H., Chen J., Fang L. X., et al. (2016). Emergence of NDM-5- and MCR-1-Producing Escherichia coli Clones ST648 and ST156 from a Single Muscovy Duck (Cairina moschata). Antimicrob. Agents Chemother. 60 6899–6902. 10.1128/aac.01365-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida C. E., Kruczkiewicz P., Laing C. R., Lingohr E. J., Gannon V. P., Nash J. H., et al. (2016). The Salmonella in silico typing resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS One 11:e0147101. 10.1371/journal.pone.0147101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67 2640–2644. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhao D., Quan J., Hua X., Yu Y. (2019). mcr-1 facilitated selection of high-level colistin-resistant mutants in Escherichia coli. Clin. Microbiol. Infect. 25 517.e1–517.e4. [DOI] [PubMed] [Google Scholar]
- Zhao F., Feng Y., Lü X., McNally A., Zong Z. (2017). Remarkable diversity of Escherichia coli carrying mcr-1 from hospital sewage with the identification of two new mcr-1 variants. Front. Microbiol. 8:2094. 10.3389/fmicb.2017.02094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi C., Lv L., Yu L. F., Doi Y., Liu J. H. (2016). Dissemination of the mcr-1 colistin resistance gene. Lancet. Infect. Dis. 16 292–293. 10.1016/s1473-3099(16)00063-3 [DOI] [PubMed] [Google Scholar]
- Zurfluh K., Tasara T., Poirel L., Nordmann P., Stephan R. (2016). Draft genome sequence of Escherichia coli S51, a chicken isolate harboring a chromosomally encoded mcr-1 gene. Genome Announcements 4:e00796-16. 10.1128/genomeA.00796-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome data of chromosome and plasmid sequences of the two mcr-1 gene positive Salmonella isolates were submitted to the NCBI nucleotide database under BioProject numbers PRJNA498334 with Biosample No. of SAMN10291561 and SAMN10291586, and related accession numbers for chromosome and plasmid sequences were: CP033349, CP033350, CP033351, and CP033352.