Abstract
The prevalence of autoimmune diseases is on the rise globally. Currently, autoimmunity presents in over 100 different forms and affects around 9% of the world’s population. Current treatments available for autoimmune diseases are inadequate, expensive, and tend to focus on symptom management rather than cure. Clinical trials have shown that live helminthic therapy can decrease chronic inflammation associated with inflammatory bowel disease and other gastrointestinal autoimmune inflammatory conditions. As an alternative and better controlled approach to live infection, we have identified and characterized two peptides, Acan1 and Nak1, from the excretory/secretory component of parasitic hookworms for their therapeutic activity on experimental colitis. We synthesized Acan1 and Nak1 peptides from the Ancylostoma caninum and Necator americanus hookworms and assessed their structures and protective properties in human cell–based assays and in a mouse model of acute colitis. Acan1 and Nak1 displayed anticolitic properties via significantly reducing weight loss and colon atrophy, edema, ulceration, and necrosis in 2,4,6-trinitrobenzene sulfonic acid–exposed mice. These hookworm peptides prevented mucosal loss of goblet cells and preserved intestinal architecture. Acan1 upregulated genes responsible for the repair and restitution of ulcerated epithelium, whereas Nak1 downregulated genes responsible for epithelial cell migration and apoptotic cell signaling within the colon. These peptides were nontoxic and displayed key immunomodulatory functions in human peripheral blood mononuclear cells by suppressing CD4+ T cell proliferation and inhibiting IL-2 and TNF production. We conclude that Acan1 and Nak1 warrant further development as therapeutics for the treatment of autoimmunity, particularly gastrointestinal inflammatory conditions.
Keywords: peptides, parasite, disulfide, peptide chemical synthesis, autoimmune disease, colitis, protein structure
Abbreviations: CBA, cytometric bead array; CTV, CellTrace Violet; DC, dendritic cell; DE, differential expression; FC, fold change; IBD, inflammatory bowel disease; MW, molecular weight; PBMC, peripheral blood mononuclear cell; PI, phorbol 12-myristate 12-acetate and ionomycin; QIMRB, QIMR Berghofer Medical Research Institute; RP-HPLC, reversed phase HPLC; SPPS, solid phase peptide synthesis; TNBS, 2,4,6-trinitrobenzene sulfonic acid; UC, ulcerative colitis
There are more than 100 autoimmune conditions described with an additional 40 diseases suspected (1). For unknown reasons, autoimmune incidence is rising annually in both developing and industrialized countries, currently affecting 3% to 12% of individuals, depending on the country (2). Treatment is costing more than $100 billion dollars annually just in the United States (NIAID, 2020). The vast majority of autoimmune conditions are incurable, and new treatments are urgently needed. Helminth therapy has been shown to be a promising treatment option for many autoimmune diseases like inflammatory bowel disease (IBD). Clinical trials for diseases such as ulcerative colitis (UC) and Crohn’s disease have found that treatment with a low-dose inoculation of helminths has overall successfully improved patient Disease Activity Index (3, 4). However, helminthic therapy is controversial and the idea of live infection will be difficult to standardize and accept for a significant subset of patients. Therefore, the utilization of the immunomodulatory proteins and peptides secreted by helminths have become a more attractive target for drug development (5, 6, 7).
Helminths produce and secrete immunomodulatory proteins, glycoproteins, and small-molecular-weight (MW) compounds from their oral openings or outer surfaces to avoid detection and expulsion from the host (8). A small number of these molecules have been characterized and reported to have benefits in treating allergic or autoimmune diseases (5, 6, 9). Although many studies focused on higher-MW proteins (>5 kDa), there are limited examples in the literature about lower-MW products (1–5 kDa). Anti-inflammatory protein-2 is an ∼19-kDa protein shown to generate a proregulatory imprint on mesenteric lymphoid tissues that results in long-term protection against allergic asthma (5). The protein has promising translational properties as anti-inflammatory protein-2 can modulate costimulatory molecules on human dendritic cells (DCs) obtained from human peripheral blood mononuclear cells (PBMCs) in vitro and suppress allergen-specific T cell proliferation. AcK1 and BmK1 are two low-MW peptides (∼4 kDa each) found in the excretory/secretory component of the hookworm Ancylostoma caninum and the human filarial nematode Brugia malayi, respectively (10). Both peptides were found to suppress mouse T cell proliferation in vitro and suppress mitogen stimulation in T cells (10). Ack1 and BmK1 contain a ShKT domain, which characteristically adopts a fold comprising two almost perpendicular stretches of helices that are interconnected by three disulfide bonds, stabilizing the structure (11). The ShKT domain was originally identified in a toxin isolated from the Stichodactyla helianthus sea anemone (12). The ShK toxin blocks voltage-gated potassium channels that are crucial for the activation of terminally differentiated effector memory T cells (13). Many potassium channel–blocking peptides contain a critical conserved “functional dyad” consisting of a lysine and a neighboring aromatic or aliphatic residue (14). ShK possesses the functional dyad, made up of a lysine and a tyrosine residue, which are proposed to contribute to activity (15). A derivative of the ShK toxin, dalazatide, has recently undergone a randomized phase 1b trial for the treatment of the autoimmune disease plaque psoriasis (16). The study reported that dalazatide was well tolerated and improved skin lesions by modulating T cell surface and activation marker expression, as well as inhibiting mediators of inflammation within the blood (NCT02435342).
Transcriptomic characterization of the human hookworm, Necator americanus, has shown that the ShKT domain family of proteins are one of the most abundantly expressed protein families in adult hookworms (17). Therefore, we proposed that one or more hookworm-secreted peptides may have immunomodulatory activity similar to other known immunomodulatory proteins of the ShKT domain family. Here we describe two low-MW peptides identified from the N. americanus and A. caninum hookworms that possess ShKT domains. Acan1 was identified from EST databases through custom blast algorithms using specific cysteine configurations. Phylogenetic analysis was used to map Nak1, previously identified in another study (10). Both peptides form the characteristic three intramolecular disulfide bonds and contain multiple helical segments within their three-dimensional structure similar to those in the ShKT family. In this study, the therapeutic efficacy of Acan1 and Nak1 was determined in vivo using a colitis model and in in vitro models using primary human immune cells. Peptide stability was additionally determined using human serum and simulated human intestinal and gastrointestinal fluid. Collectively, these data show promise for the two new immune-modulating proteins.
Results
Identification of ShKT domain–containing worm sequences
Using a custom algorithm, seven peptides comprising sequences with a plurality of suitably spaced cysteine residues were identified in the transcriptomes of A. caninum and N. americanus. These peptides contain the cysteine configuration of the ShK toxin (Fig. 1). To determine sequence identity, optimal alignment of amino acid and/or nucleotide sequences was conducted by computerized implementations of algorithms, i.e., BLAST. The Nak1 and Nak2 peptides were previously identified by Chhabra, et al. (10). Each peptide is shown to possess the conserved cysteines (highlighted in red), and the putative disulfide bond connections are shown by the black lines. Only Acan1 was found to possess the functional dyad (highlighted in green and purple) that is responsible for the ShK toxin activity.
Figure 1.
Aligned sequences of helminth peptides from Ancylostoma caninum and Necator americanus and the ShK toxin. Conserved cysteine residues are shown in red, with the black line highlighting the disulfide bond connection (I–VI, II–IV, III–V). The critical lysine (K) residue is in green, and neighboring aromatic/aliphatic residues are highlighted in purple.
Synthesis and folding of helminth peptides
The linear hookworm peptides were synthesized on rink amide resin using solid phase peptides synthesis (SPPS). The sequences of Name1 and Name2 could not be synthesized in full using SPPS, and therefore, a native chemical ligation approach was used to successfully produce the Name2 peptide. This was achieved by ligating the N- and C-terminal segments together at the third cysteine residue of the sequence. Name1 was unable to be synthesized. The disulfide bonds of each peptide were formed by random oxidative folding in aqueous buffer, which was optimized through multiple different buffer conditions. All peptides were most amenable to folding in 0.1 M ammonium bicarbonate at room temperature with the aid of both reduced (2 mM) and oxidized (0.5 mM) glutathione (pH 8.2). During random oxidation of Name3 and Name4, the N-terminal glutamine was found to cyclize to form pyroglutamic acid. Both Name3 and Name4, with and without pyroglutamic acid, were used for further experimentation, as they were unable to be separated via reversed phase (RP)-HPLC. Peptide fractions with >95% purity were used for experimentation (Fig. S1). Acan1 and Nak1 were chosen of the seven peptide homologs for further immunomodulation experimentation, as they were shown to be the most suppressive in an initial cytometric bead array (CBA) screen of proinflammatory cytokines (Fig. S2).
Structural analysis using NMR spectroscopy
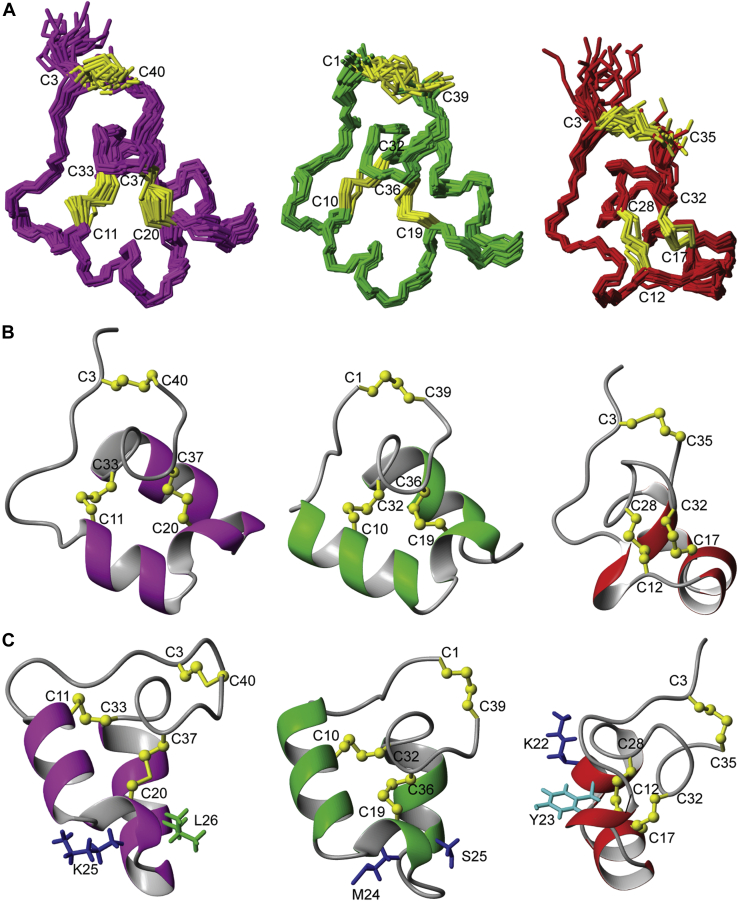
Folding of the synthetic peptides was initially examined using one-dimensional 1H NMR spectroscopy. These spectra confirmed that each peptide has a well-defined structure based on the quality of the data in terms of signal dispersion and peak linewidth. Further analyses were performed to determine the three-dimensional (3D) structures of the hookworm peptides Acan1 and Nak1, in order to compare them with ShK (18) (Fig. 2). Two-dimensional TOCSY and NOESY spectra allowed for sequential manual assignment (Fig. S3). The 3D structures for Acan1 and Nak1 were calculated using simulated annealing and structural restraints (Table S1) derived from the NMR data, and both peptides were shown to resemble that of ShK (Fig. 2A). The structure calculations with no disulfide bonds were performed, revealing well-defined structural ensembles (Fig. S4). Structures were then calculated for all 15 possible disulfide bond isomers for both peptides, with the native disulfide connectivity generating the lowest target function average (Table S2). Although the helminth peptides adopt a predominantly helical structure with a short 310 helix separating the two α-helices, the locations and lengths of the helices differ slightly between each peptide (Fig. 2B). In ShK, two short α-helices encompass residues 14 to 19 and 21 to 25 and a 310 helix is located between residues 9 and 11. The α-helices in Acan1 are located between residues 11 and 17, and 23 and 32 with the 310 helix between 19 and 21, whereas in Nak1, α-helices are located between residues 7 and 15, and 25 and 31 and a 310 helix is located between 18 and 20. Despite the similarities in 3D structures, only Acan1 was found to possess the functional dyad that is crucial for the activity of ShK on the Kv1.3 channels (Fig. 2C). The side chains of Lys22 (blue) and Tyr23 (cyan) in ShK are located on a short helical region orientating outward. Compared with ShK, the side chains of the residues in the dyad in Acan1 are shown to possess a different orientation, appearing to angle away from each other.
Figure 2.
Nuclear magnetic resonance structures of Acan1 (purple), Nak1 (green), and ShK (red).A, the 20 lowest energy structures are represented in stick format. B, the lowest energy structures are in ribbon representation. C, comparison of the functional dyad found in Acan1 (purple) and ShK (red). The lysine and neighboring aromatic tyrosine/amphipathic leucine that make up the functional dyad are shown. Nak1 (green) does not possess the functional dyad. Cysteine residues are labeled, and conserved disulfide bond connections are in yellow. Structure images were produced in MOLMOL.
Acan1 inhibits human Kv1.3 channels
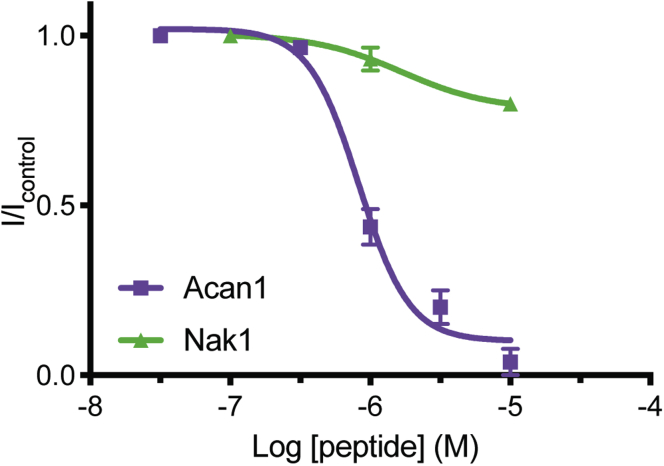
To determine whether Acan1 and Nak1 are able to inhibit human Kv1.3 channel, a two-electrode voltage clamp electrophysiology was used to examine their effects on channels expressed in Xenopus laevis oocytes (Fig. 3). ShK was used as a control and found to inhibit Kv1.3 with an IC50 of 57 pM (data not shown), replicating the low picomolar values described in the previous literature (19). Acan1 inhibited the channel with an IC50 value of 829 nM, a concentration 15,000 times higher than the positive control, ShK. Nak1 was found to have little efficacy on blocking the channel, likely due to the lack of functional dyad within the sequence.
Figure 3.
Voltage-dependent activity of Kv1.3 channels expressed in Xenopus laevis oocytes. Two-electrode voltage clamp electrophysiology showed Acan1 (purple) inhibited human Kv1.3 expressed in X. laevis oocytes with a 15,000-fold less IC50 value of 829.85 nM than the positive control ShK (IC50 of 57.4 pM, data not shown). Nak1 (green) had little efficacy and potency on Kv1.3, tested up to 10 μM. Data show mean ± SEM of a representative experiment, n = 5.
Acan1 and Nak1 display high biological stability
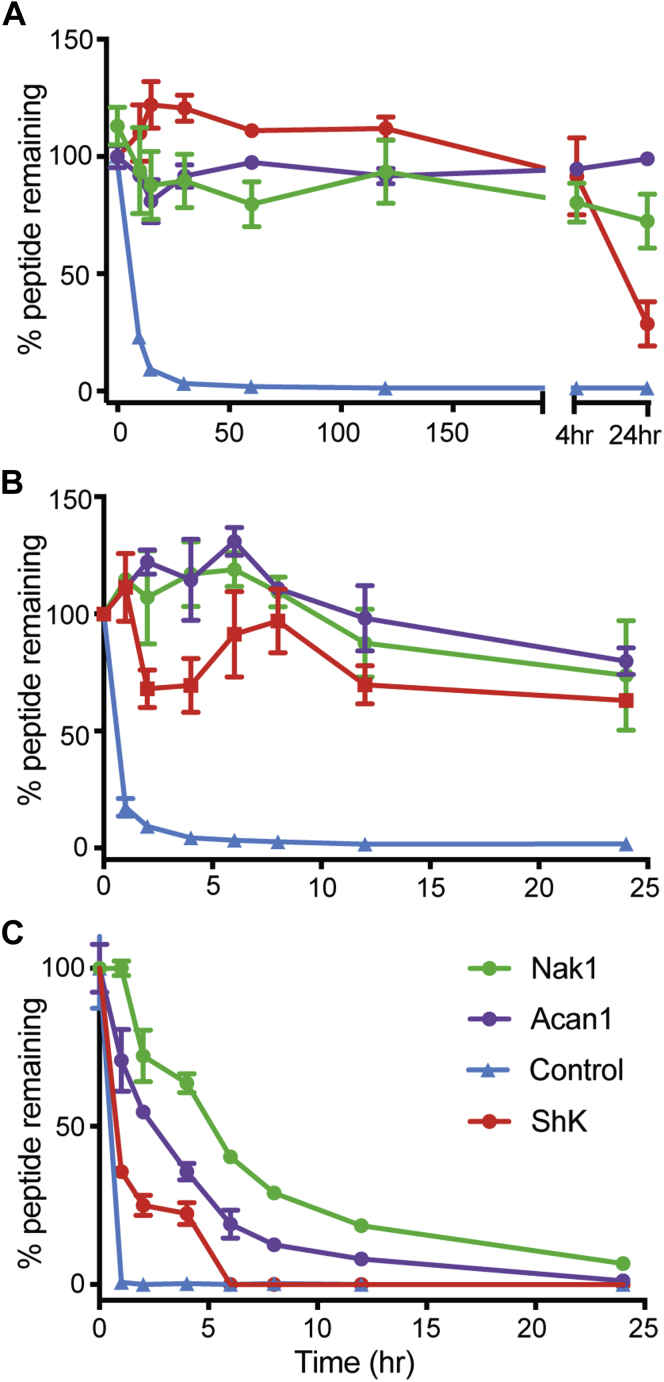
The stability of each peptide was determined via LC/MS quantification during 24-h incubations in 100% human serum, and porcine simulated gastrointestinal and intestinal fluid (Fig. 4). Both Acan1 (purple) and Nak1 (green) were found to be stable in 100% human serum over the time course, whereas ShK (red) began to degrade after the 4 h time point (Fig. 4A). In simulated gastrointestinal fluid, all peptides were shown to be stable (Fig. 4B). In simulated intestinal fluid, ShK (red) was completely degraded after 6 h with a half-life of 0.8 h. By contrast, Acan1 (purple) and Nak1 (green) did not fully degrade until 24 h and had a half-life of 2.4 and 4.4 h, respectively (Fig. 4C). The linear control peptide R3 B1-22R I15A (20) (blue) was found to be fully degraded within the first hour of each experiment. The lower stability of ShK demonstrates that the disulfide bonds alone are not sufficient to confer high stability. It is likely that the stability of the hookworm peptides in hostile protease-rich environments may have resulted from evolutionary adaptations by helminths that can live in the small intestine for decades.
Figure 4.
Stability of peptides Acan1 and Nak1 to proteolysis. Stability assays using (A) serum, (B) simulated porcine gastrointestinal fluid, and (C) simulated porcine intestinal fluid of synthetic peptides Acan1 (purple), Nak1 (green), and ShK (red). The percentage of peptide remaining in all three environments was assessed by liquid chromatography mass spectrometry. The hookworm peptides show better stability than ShK. The positive control linear peptide (blue), degraded within the first 30 min. Data show mean ± SEM of a representative experiment, n = 3.
Acan1 and Nak1 potently suppress proinflammatory cytokines and T cell proliferation
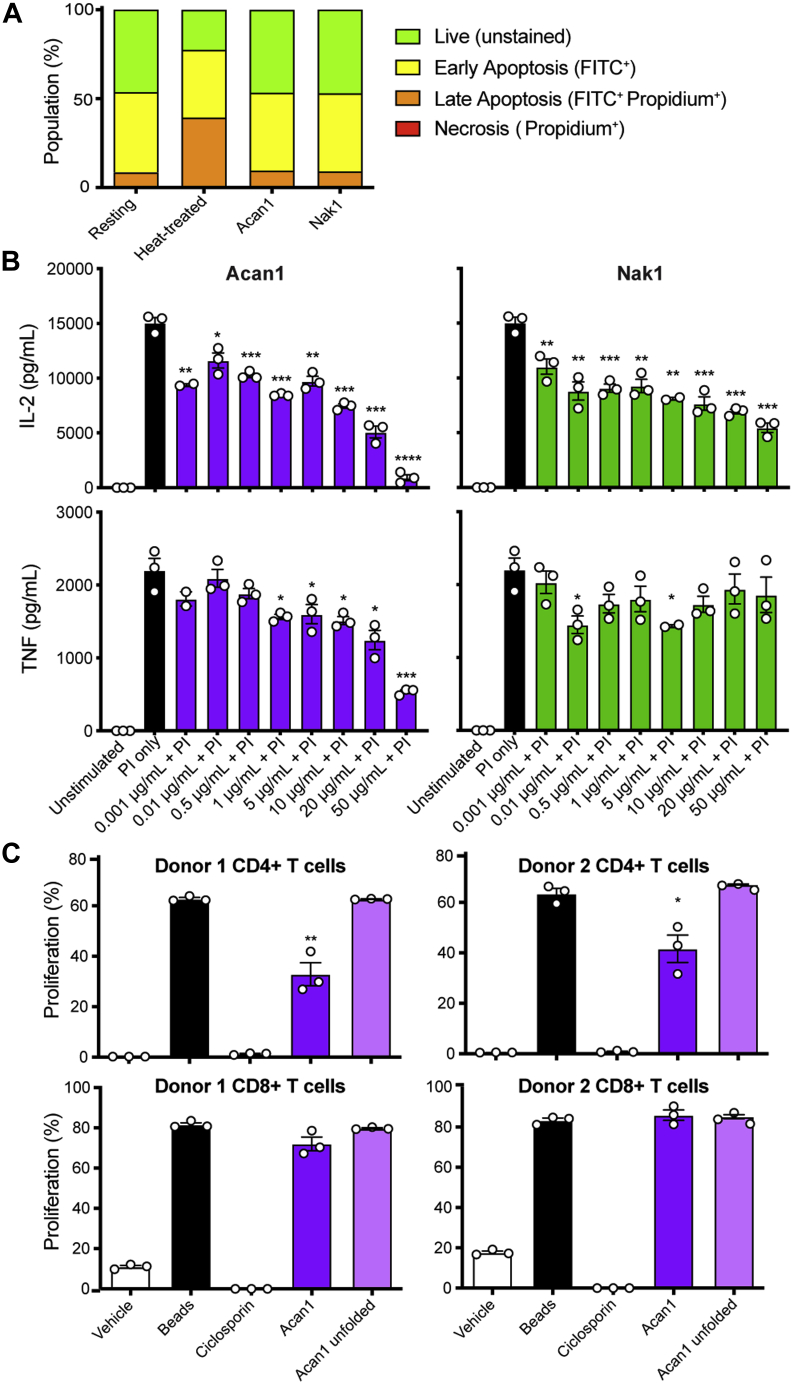
Owing to their origin and similar structural scaffold of helminth peptides Acan1 and Nak1 to the ShK toxin, it was hypothesized that both peptides would have modulatory effects on human T cells. To investigate this hypothesis, the potential toxicity of Acan1 and Nak1 to human PBMC was first assessed by quantifying cell surface phosphatidylserine following peptide treatment. To do this, apoptotic cell populations were identified using flow cytometry following Annexin V-FITC and propidium iodide staining (Fig. S5). Cells treated with either synthetic peptide (10 μg/ml) retained a similar percentage of live cell (unstained) populations as the untreated control, whereas the heat-treated cells (positive control) showed an increase in earlyFITC+ to lateFITC+Propidium+ apoptotic populations (Fig. 5A). Consequently, the anti-inflammatory potential of Acan1 and Nak1 was determined using a cytokine suppression assay. In preliminary intracellular staining assays using human PBMC, it was revealed that Nak1 did not inhibit LPS-induced productions of IL-6, IL-8, or MIP-1β in classical (CD14+) or intermediate (CD14+/CD16+) monocytes after 24-h treatment (Fig. S6). Additional preliminary data had shown that Acan1 did not significantly alter the viability/proliferation of metabolically active cells in a mixed lymphocyte reaction following 4 days of LPS stimulation and peptide treatment (Fig. S7). It has been reported that ShK principally inhibits T cell function through ion channel modulation and calcium-dependent signaling (21). Accordingly, human PBMCs were activated with phorbol 12-myristate 12-acetate and ionomycin (PI) to increase intracellular Ca2+ levels and induce T cell cytokine production. Activated cells were then treated with peptide for 24 h and IL-2 and TNF measured by CBA (Fig. 5B). Both Acan1 and Nak1 suppressed PI-induced IL-2 release in a dose-dependent manner, whereas Acan1 additionally decreased TNF secretion. Next, given the observed suppression of proinflammatory TNF by Acan1, the antiproliferative capacity of the peptide was assessed on MACS-purified CD3+ T cells. CellTrace Violet (CTV)-stained T cells were activated via TCR stimulation using anti-CD3/CD28 beads and treated with 10 μg/ml ciclosporin (positive control), 100 μg/ml Acan1, or 100 μg/ml unfolded Acan1 (negative control) for 4 days. Following incubation, T cell division was evaluated by flow cytometry and CTV fluorescence (Fig. S8). Folded but not unfolded Acan1 suppressed CD4+ T cell proliferation in two different donors, albeit to a lesser extent than the calcineurin inhibitor, ciclosporin (Fig. 5C). Of interest, Acan1 showed no significant effect on CD8+ T cell division. Collectively, these data suggest a CD4+ helper T cell modulatory mechanism for Acan1 but requires further investigation.
Figure 5.
Acan1 and Nak1 modulate cytokine production in human immune cells.A, apoptosis population percentage in human peripheral blood mononuclear cells when incubated with synthetic hookworm peptides (10 μg/ml). Peptides were incubated for 4 h at 37 °C/10% CO2. Heat-treated positive control was placed in a heating block for 20 min at 56 °C. Apoptosis was measured using Annexin V-FITC and propidium iodide staining and analyzed by flow cytometry. B, culture supernatant concentrations of TNF and IL-2 assessed by cytometric bead array from human peripheral blood mononuclear cells stimulated with phorbol 12-myristate 12-acetate (50 ng/ml) + ionomycin (1 μg/ml) (PI) for 24 h. Data show mean ± SEM of a representative experiment out of three, with n = 3. Unpaired Student t test performed comparing PI versus Acan1 or Nak1 + PI. C, CD4+ and CD8+ T cell proliferation percentage assessed by CTV assay from MACS-purified CD3+ human T cells stimulated with anti-CD3/CD28 beads and incubated with hookworm peptide (100 μg/ml) for 4 days at 37 °C/10% CO2 before being analyzed by flow cytometry. Ciclosporin (10 μg/ml) was used as a positive control. Data show mean ± SEM of a representative experiment, n = 3. Unpaired Student t test was performed comparing anti-CD3/CD28 beads with Acan1 or Acan1 (linear); ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗p < 0.0001.
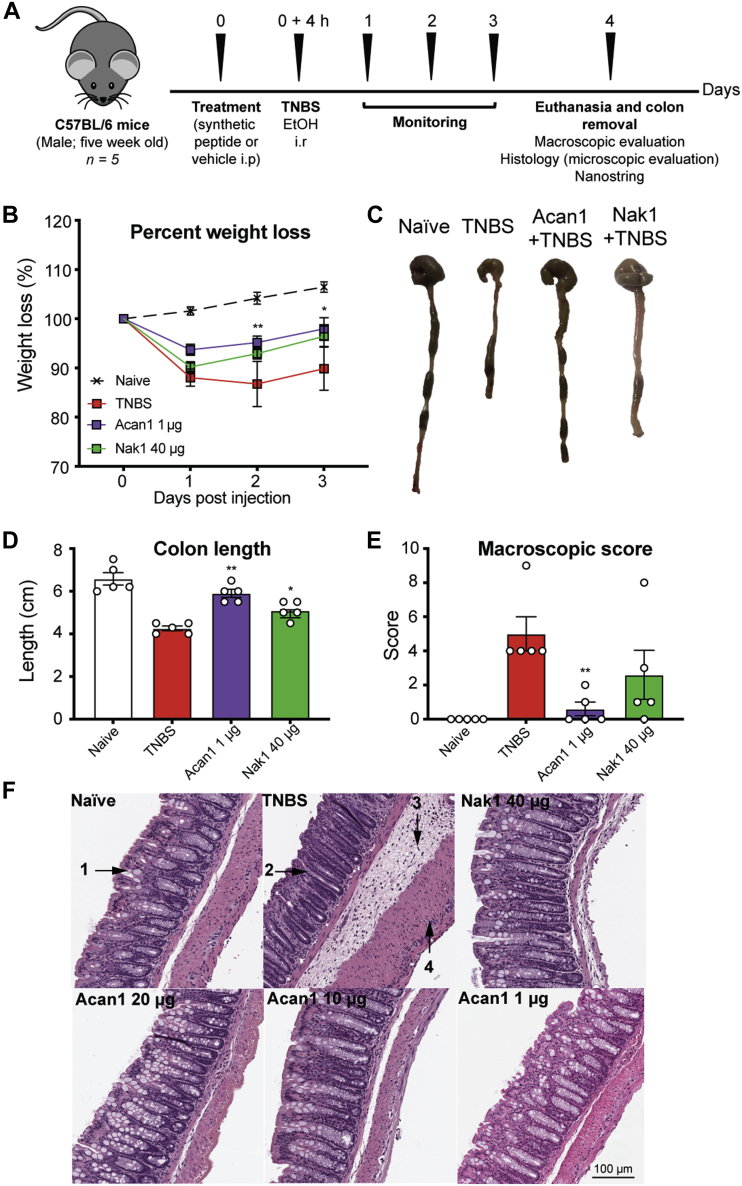
Synthetic helminth peptides protect against TNBS-induced intestinal inflammation
To determine the therapeutic potential of synthetic hookworm peptides in a model of acute inflammation, mice were treated once prophylactically with varying doses of Acan1 (20, 10, 5, 1, and 0.1 μg) or Nak1 (40, 20, 10, 5, 1, and 0.1 μg) and assessed over 4 days (Fig. 6A). Over the course of the study, the vehicle (2,4,6-trinitrobenzene sulfonic acid [TNBS]) group experienced a 15% weight loss (Fig. 6B). Mice treated with 1 μg of Acan1 (0.05 mg/kg−1) via the intraperitoneal (i.p.) route displayed significantly reduced weight loss at day 3 (p < 0.05), compared with the vehicle control (Fig. 6B), but no significant improvement in weight loss was observed in the Nak1-treated group. Colon pathology was assessed on day 4 (Fig. 6C) and was consistent with enhanced protection against TNBS-induced colitis. Acan1-treated mice had visibly longer and normal-looking colons when compared with the vehicle control group (p = 0.0079), which manifested shortened, thickened, and erythematous tissue. Nak1-treated mice were also found to have significantly longer colons compared with disease control (p = 0.0238) (Fig. 6D). The colons were scored macroscopically based on the following parameters: adhesion, edema, wall thickening, and ulceration (Fig. 6E). It was found that mice treated with Acan1 displayed a significant reduction (p = 0.008) in the overall macroscopic pathology score. Macroscopic histological observations (Fig. 6F) of naive mouse colon displayed normal colon tissue architecture. This included the anatomy of the lamina propria, gastric mucosa, goblet cells, and colonic musculature. Colons of vehicle-treated mice exhibited extensive histological damage and severe lesions, including thickening of the lamina propria and colon wall, evidence of edema and mucosal erosion with destruction of goblet cells. Mice treated with either Acan1 or Nak1 had healthy colons, possessing architecture comparable with the naive (healthy control) group, with normal crypts and goblet cells, good mucosal integrity, and no ulceration. Together, these results show that hookworm-derived peptides, particularly Acan1 and to a lesser extent Nak1, can suppress TNBS-induced intestinal inflammation in vivo.
Figure 6.
Effects of hookworm-derived peptides in the 2,4,6-trinitrobenzenesulfonic acid (TNBS) mouse model of colitis.A, schematic overview of the experimental strategy followed. Five-week-old C57BL/6 male mice were analyzed to study the effect of different synthetic hookworm peptide doses in the intestine of mice with TNBS-induced colitis. Hookworm peptide was dosed 4 h before the administration of TNBS. Mice were monitored daily for weight loss and general well-being over 4 days before they were euthanized and their colons removed. Colons were measured in length; adhesion, edema, mucosal wall thickening, and ulceration were scored on a scale of 0 to 3, with 3 indicating the highest degree of damage. Colons were then opened up longitudinally and washed in PBS, and a 1-cm section from the distal colon was fixed in 4% paraformaldehyde. Histological micrographs of hematoxylin and eosin–stained tissue (×200) obtained from a representative mouse from each group were observed for epithelial changes (presence of goblet cells, hyperplasia, erosion, cell infiltrate and mucosal architecture). A section of colon from a representative mouse from each group was also used to assess the gene expression profile via Nanostring. B, body weight was recorded daily for the indicated groups. Data show mean ± SEM of a representative experiment out of three with n = 5. Two-way ANOVA with Dunnett’s multiple comparisons test was used to compare each group with vehicle over time. C and D, colons were removed and measured on day 4. E, macroscopic scoring of colon pathology; adhesion, edema, mucosal wall thickening, and ulceration were scored on a scale of 0 to 3, with 3 indicating the highest degree of damage. F, data show histological micrographs of hematoxylin and eosin–stained tissue (×200) obtained from a representative mouse from each group. Colons were assessed for epithelial changes (presence of goblet cells (1), hyperplasia (2), erosion, cell infiltrate (3, 4) and mucosal architecture. Data show mean ± SEM of a representative experiment with n = 5. Mann–Whitney U test was performed comparing each group with vehicle; ∗p < 0.05; ∗∗p < 0.01.
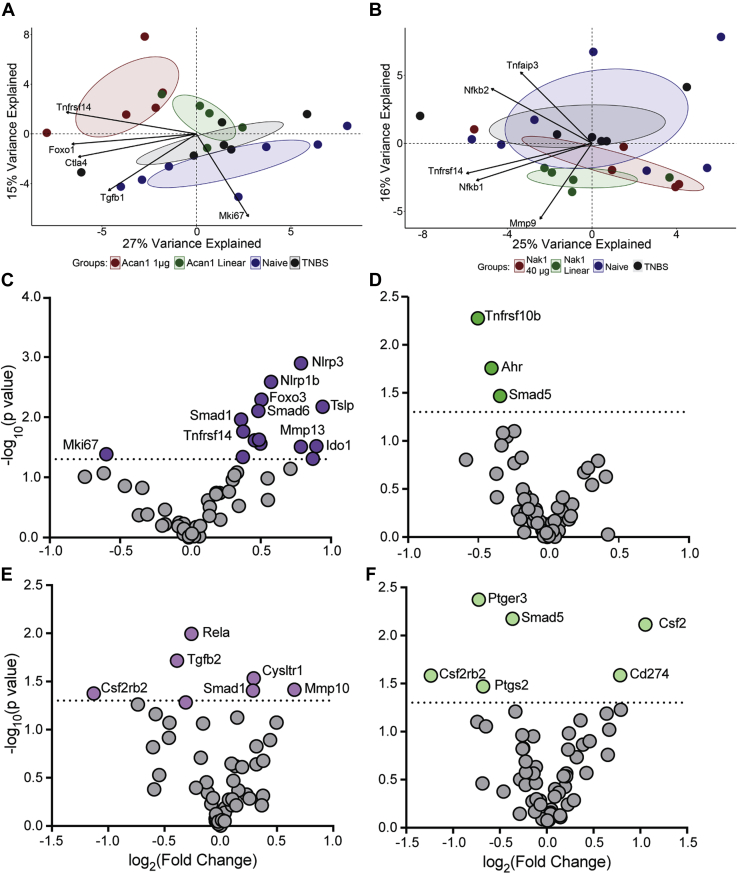
Acan1 induces the upregulation of immune regulatory and tissue repair genes
We next performed transcriptomics on the colons of each mouse cohort using a custom 57 gene NanoString panel that targeted genes critical in inflammation, immune regulation, gut barrier homeostasis, epithelial homeostasis, and tissue destruction. A multivariate analysis principal component analysis was performed to identify genes that contribute the most to the largest sources of variation within the dataset (Fig. 7). No significant differences between the contributions of the groups were identified along PC1 (x-axis) for both Acan1 (Fig. 7A) and Nak1 (Fig. 7B). However, a significant difference between the contribution of groups alongside PC2 was observed between Acan1 and naive (p = 0.00182), Acan1 and vehicle (p = 0.04369), and naive and Acan1 (linear) (p = 0.0363), as well as along PC2 between Nak1 (linear) and vehicle (p = 0.0382). Together the two principal components captured 42% of the variation in the datasets for Acan1 and 41% for Nak1. The genes with the largest contributions to PC1 and PC2 variation are shown by the arrows. These included Tnfrsf14, Foxo1, Ctla4, Tgfb1, and Mki67 (Fig. 7A) and Tnfaip3, Nfkb2, Tnfrsf14, Nfkb1, and Mmp9 (Fig. 7B). We next visualized differential gene expression (DE) between treatment and vehicle control using volcano plots. This was achieved by pairing the log2 fold change (FC) and relative p value for each gene within each comparison. As shown, when comparing the Acan1 cohort with the vehicle cohort a significant DE of 16 genes was identified, characterized largely by positive FC (Fig. 7C). Unlike Acan1, when comparing the Nak1 group with the vehicle control group, significant p values were identified in only three genes and all were characterized by negative FC (Fig. 7D). Our controls, the linear Acan1 (Fig. 7E) and Nak1 (Fig. 7F) versus the vehicle control, showed insignificant DE of genes and very low significant values from a small cluster of genes (Fig. 7E). These data are further validation that the folded version of both peptides is required for activity.
Figure 7.
A principal component analysis plot of gene expression profiles between treatment groups. Each dot represents an observation for each treatment projected onto PC1 (x-axis) and PC2 (y-axis). A, significant differences were determined between the contribution of groups along PC2 in the following comparisons: Acan1 and naive, Acan1 and vehicle, and naive and Acan1 (linear) along the PC2 axis. B, significant differences between the contribution of groups along PC2 were identified in the following comparisons: Nak1 (linear) and vehicle along PC2. The arrows show the strongest loadings, i.e., contribution of the original variables to the principal components. No significant difference was found along PC1. Volcano plot of gene expression analyzed in the colon obtained from a representative mouse from each group (C) Acan1, (D) Nak1, (E) Acan1 linear, and (F) Nak1 linear. Significantly expressed genes are shown in color. X-axis values are log2(fold change) and y-axis are the −log10 of p value. The horizontal dashed line represents a p value of 0.05. Nonsignificant genes are shown by gray dots.
Discussion
The well-characterized TNBS-induced colitis mouse model is a widely used in vivo model that shares many clinical, histological, and biochemical features with human UC (22). An initial TNBS-induced colitis mouse model was used to screen the candidate hookworm-derived peptides Acan1 and Nak1 for their dosage and efficacy (Fig. S9). Following the pilot study, dose responses of both hookworm peptides were determined. Our study showed that, in this model of experimental colitis, synthetic hookworm peptide, Acan1, significantly protected mice against TNBS-induced weight loss, as well as the principal features of colitis. Both Acan1 and Nak1 promoted retention of healthy gastrointestinal architecture after TNBS administration with normal crypts and goblet cell anatomy, mucosal integrity and absence of ulceration, necrosis, edema, and colon shortening. Although colons from the 40-μg Nak1 (2 mg/kg−1) group appeared to resemble that of the naive group, the required dosage to obtain efficacy was much higher than that of Acan1 (0.05 mg/kg−1). Taking whole colon RNA, gene expression of each treatment group was analyzed. Several genes were found to have positive FC in expression when comparing Acan1 with the vehicle cohort. Many of these genes are found to be mediators involved in mucosal barrier repair (Nlrp3) (23, 24), repair and restitution of ulcerated epithelium (mmp, smad) (25), cell proliferation (wnt5a, ido1) (26), and regulation and differentiation of immune cells (tslp (27), csf2 (28, 29), cd274 (29), tnfrsf14 (30)). The mechanism by which Nak1 produces its effect is more difficult to interpret, with a negative FC found in three significant genes. These findings suggest that the efficacy of the hookworm in treating IBD can be related to the peptides secreted into the gut by the parasite. However, mice and humans are significantly different in genetics, physiology, and anatomy and do not always corelate in pharmaceutical testing. Our results prove that the living parasite itself is not required to achieve IBD suppression and highlights that immune-modulating peptides derived from the hookworms can protect against a model of colitis.
Attenuation of inflammation in the mouse model with treatment of the hookworm peptides may also be linked to the decrease in proinflammatory cytokines and CD4+ T cells. In vitro experiments revealed that Acan1 and Nak1 were nontoxic and did not induce apoptosis as determined using human PBMC. Acan1 and Nak1 were found to decrease IL-2 production in a dose-dependent manner, suggesting an immunosuppressive function. This was further confirmed following mitogen stimulation, where Acan1 impaired the proliferation of CD4+ T cells but not that of CD8+ T cells. A dysregulated T cell response is often implicated as a key player of chronic inflammation in IBD (30, 31). Mounting evidence shows that CD4+ T cells are the main driver of IBD pathology (32, 33). The inhibitory activity of Acan1 on human T cell proliferation in vitro suggests that this therapeutic peptide could effectively antagonize the inflammatory cascade and potentially prevent relapse. In addition, Acan1 was found to decrease TNF production, an integral player in the pathogenesis of IBD (34).
The three-dimensional structure of both Acan1 and Nak1, determined by NMR spectroscopy, are well defined. Based on the activity observed in the TNBS mouse model, the folded structure of the hookworm peptide is required for its attenuation of the disease. Both Acan1 and Nak1 share the same disulfide framework as the known immunomodulator ShK; however, their amino acid homology is less than 30%. Like ShK, Acan1 also possesses the functional dyad located within the second helix, which is responsible for ShK’s ability to block the pore of the Kv1.3 channel. However, in Acan1 the aromatic tyrosine is replaced with an aliphatic leucine and its side chain can be seen in a different orientation in the dyad, compared with ShK. This may be responsible for the lower potency of Acan1 blocking the potassium (Kv1.3) channel. Many studies have demonstrated that the expression of T cell Kv1.3 channel increases in active UC and a higher expression is correlated with both endoscopic and histological degree of inflammation (35, 36, 37). Reduced activity of this channel implies that Acan1 may be of therapeutic utility in inflammatory and autoimmune diseases. As predicted, up to 10 μM of Nak1 was found to have little activity on the channel, theorized to be as a result of the lack of functional dyad.
The excretory/secretory component secreted by parasitic worms contains a vast array of proteins that can influence the immune response of its mammalian host (9). A significant number of clinical trials have shown the beneficial effects of hookworm therapy for the treatment of several autoimmune diseases, including IBD, type 1 diabetes mellitus, and psoriasis (9). Here, we show that the use of small-MW synthetic hookworm-derived peptides, rather than live infection, is able to induce protection in a mouse model of colitis and promote tissue repair and restitution. Acan1 and Nak1 were shown to influence the human immune response through suppression of cytokine production and T cell proliferation. Acan1 was found to have a more pronounced immunomodulatory effect that was directly translatable in a human in vitro system. Taken together, this study demonstrates that Acan1 has anti-inflammatory properties that warrant further mode of action investigation and preclinical development.
Experimental procedures
Animals
All animal experiments were conducted in accordance with the James Cook University Animal Ethics Committee (Ethics approval Number A2481). Five-week-old male C57BL/6 were purchased from the Animal Research Centre and were housed in specific pathogen-free conditions. Mice received food and water ad libitum.
X. laevis oocyte surgeries were performed in accordance with the guidelines from the Australian code of practice for the care and use of animals for scientific purposes, 8th edition, 2013. The protocol was approved by the Anatomical Biosciences group of the Animal Ethics Committee at The University of Queensland (Approval Number: QBI/AIBN/087/16/NHMRC/ARC).
Discovery of ShKT domain peptides from hookworms
Transcriptomic studies identified several transcripts encoding the ShKT domain family in N. americanus (17). Using an iterative approach, these ShK-like sequences from N. americanus were used in 1) BLASTx (NCBI, www.ncbi.nIm.nih.gov) and BLASTn (EMBL-EBI Parasite Genome Blast Server, www.ebi.ac.uk) searches of N. americanus and A. caninum transcriptome sequences; and 2) a simple text search for the pattern “C[ˆC]{4,8}C[ˆC]{5,10}C[ˆC]{10,12}C[ˆC]{3,6}C[ˆC]{2,6}C[ˆC]{0,5}\\∗” in N. americanus and A. caninum mRNA transcripts translated in six reading frames. Sequences identified were manually aligned with known ShK-like sequences, and if judged to be a ShK-like peptide, they were in turn used in new searches of the same database.
Peptide synthesis and purification
Peptides were synthesized using SPPS on an automated peptide synthesizer (CS Bio Pty Ltd) using fluorenylmethoxycarbonyl chemistry on rink amide resin (0.25 mmol scale). Amino acids (4 eq.) were activated in 0.5 M 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate and N,N-diisopropylethylamine (8 eq.) in N,N’-dimethylformamide. Peptides were cleaved from the resin using TFA/water/triisopropylsilane and 2,2’-(ethylenedioxy)diethanethiol (92.5:2.5:2.5:2.5) for 2 h, precipitated with cold diethyl ether, filtered, then dissolved in 50% acetonitrile, 0.05% TFA and subsequently lyophilized. The resulting crude peptides were purified with RP-HPLC on a C-18 preparative column (Phenomenex Jupiter 250 × 21.2 mm) using a 1% gradient of solvent B (solvent A: 0.05% TFA; solvent B: 90% acetonitrile, 0.05% TFA). Peptides were oxidized at a concentration of 0.25 mg/ml in 0.1 M ammonium bicarbonate with reduced (2 mM) and oxidized (0.5 mM) glutathione at room temperature (pH 8) for 48 h. Oxidized peptides were purified using RP-HPLC and the mass analyzed by MS in positive ion mode using an ABSciex API 2000TM. To prevent intramolecular disulfide bond formation, linear hookworm peptides were synthesized as described above and pure peptide was dissolved in 10 M guanidine hydrochloride and 0.2 M Tris hydrochloride solution at a concentration of 100 μM, with an addition of 20× molar excess of 1,4-dithiothreitol (DTT). The peptides were incubated for 1 h at 37 °C. Following incubation, a solution of 200 mM iodoacetamide, 10 M guanidine hydrochloride, and 1 M Tris hydrochloride solution (pH 8) was added to the reaction and left to incubate in the dark for 1 h at 37 °C. After the reaction, 20 ml of 1% aqueous TFA was added and the sample was directly subjected to RP-HPLC using 1% buffer B (90% acetonitrile, 0.05% TFA) 8 ml gradient method on a C-18 preparative column (Phenomenex Jupiter 250 × 21.2 mm). Pure linear peptides were purified using RP-HPLC and the mass analyzed by MS in positive ion mode using an ABSciex API 2000TM.
NMR spectroscopy
Lyophilized peptides were resuspended to a final concentration of 0.5 mM in 90% water and 10% deuterium oxide (2H2O) at pH ∼3.5. Two-dimensional data sets including 1H TOCSY, 1H NOESY, 15N-1H heteronuclear single quantum coherence, and 1H-13C heteronuclear single quantum coherence spectra were acquired at 298 K using a Bruker Avance 600-MHz spectrometer with a cryoprobe. Topspin 4.0.3 (Bruker) was used for data processing, with data referenced to the solvent signal at 4.77 ppm. The program CARA (38) was used to analyze and assign the data using sequential assignment strategies (39). Structural coordinates were deposited in the Protein Data Bank.
Structure calculations
Structures were determined using previously reported protocols (40). The peak volumes of the cross peaks within the NOESY spectrum were used to derive interproton distance restraints. The φ (C1-N-Cα-C) and ψ (N-Cα-C-N1+) backbone dihedral angles were predicted from the generation of torsion angle likelihood obtained from shift and sequence similarity (TALOS-N) (41). Hydrogen bond donors were identified by 2H2O exchange experiments and acceptors identified by preliminary structure calculations. Within the program CYANA (42), 50 structures were calculated by torsion angle simulated annealing using automatic assignments of NOESY data and translation into distance restraints. To confirm disulfide bond configuration, we first calculated the structure of each peptide with disulfide bond restraints and then calculated all possible disulfide bond connections in cyana. The ShK disulfide bond connectivity gave the lowest target function average of all the isomers and allowed the greatest number of NOEs to be assigned. Therefore, the disulfide bond configuration (Acan1: 3–40, 11–33, and 20–37, Nak1: 1–39, 10–32, and 19–36) was enforced by distance restraints during calculation. Final structures were calculated using water refinement within the CNS program using simulated annealing and energy-minimization protocols in explicit water (43). The final ensemble of 20 structures were chosen based on final energies and stereochemistry as assessed by MolProbity (44). Structures were visualized using MolMol (45). The structures, NMR restraints, and chemical shift information have been submitted to the Protein Data Bank, and the accession codes are 6DRI for Acan1 and 7L2G for Nak1.
Stability assays
The stability of the peptides was tested in 100% human male AB plasma for a period of 24 h. Briefly, peptides were incubated in 100% serum or pure water at 37 °C with a final concentration of 40 μM. Aliquots (40 μl) were taken at 0 h, 5 min, 10 min, 15 min, 30 min, 1 h, 2 h, 4 h, 6 h, 8 h, and 24 h. The aliquots of serum were quenched with 40 μl of 6 M urea and incubated for 10 min at 4 °C. Following incubation, aliquots were then quenched with 40 μl of 20% trichloroacetic acid and incubated for 10 min at 4 °C to precipitate serum proteins. The water samples also received the same treatment. Peptide samples were then centrifuged at 14,000g for 10 min, and 80 μl of the supernatant was analyzed by LC-MS (15 μl injection and multiple reaction monitoring protocol).
Two-electrode voltage clamp
X. laevis oocytes were obtained from surgery and human Kv1.3–containing plasmid was linearized with NotI enzyme and transcribed for cRNA injections as described (46). Membrane currents were recorded in ND96 solution (in mM: 96 NaCl, 2 KCl, 1.8 CaCl2, 2 MgCl2 and 5 Hepes; pH 7.45) at room temperature (18–22 °C) under voltage clamp (Axoclamp 900A). Currents were elicited by a 200-ms depolarization pulse to +10 mV from a holding voltage of −90 mV with leak and background conductance subtracted by addition of agitoxin-2 after recordings. Data were digitized at 5000 Hz and filtered at 0.01 Hz with pClamp software. Peak currents were analyzed to obtain normalized concentration–response data that were fitted with the Hill equation to calculate the half-maximal response. All data represent at least five independent oocytes for a given experiment.
Human immune cell functional testing
Healthy donor blood was studied using protocols carried out in accordance with guidelines and regulations under QIMR Berghofer Medical Research Institute (QIMRB) (HREC P2058). All methods and human participants involved in the study were approved by the QIMRB HREC. Informed consent was obtained from all participants in the study. The study was performed according to the rules of the Declaration of Helsinki of 1975. PBMCs were separated, as described (47). Briefly, PBMCs were isolated by Ficoll-Paque PLUS density gradient centrifugation and cryopreserved in R10 medium (RPMI-1640 containing 10% fetal calf serum) supplemented with 10% dimethyl sulfoxide.
Apoptosis assay
For the apoptosis assay, 2 × 105 cells were seeded and incubated with Acan1 and Nak1 (10 μg/ml) for 4 h at 37 °C/10% CO2. A positive control was generated by placing 2 × 105 cells in a heating block for 20 min at 56 °C. Cells were then stained using Annexin V-FITC and propidium iodide according to manufacturer’s instructions (BD Biosciences) and analyzed by flow cytometry (BD LSRFortessa) within 1 h.
Intracellular cytokine staining assay
For the intracellular cytokine staining assay, PBMCs were seeded at 1 × 106 cells per well and incubated for 4 h at 37 °C/10% CO2 with unstimulated, LPS alone (10 ng/ml), or Nak1 (100 μg/ml) or ShK peptide (10 μg/ml) with LPS supplemented with Brefeldin A (GolgiPlug) (1 μl/ml) and monenisin (GolgiStop) (0.1 μl/ml) (BD Biosciences). The cells were then washed and incubated at 4 °C for 30 min with fluorescently labeled mAbs specific for cell surface markers CD14-APC and CD16-PB (BioLegend). Cells were washed and then fixed and permeabilized with Cytofix/Cytoperm fixation/permeabilization solution (BD Pharmingen) at 4 °C for 20 min. Next, cells were washed in Perm/Wash (BD Pharmingen), incubated with fluorochrome-conjugated anti-cytokine antibody (IL-6, IL-8, and MIP-1β) at 4 °C for 30 min, washed with Perm/Wash, resuspended in PBS, and analyzed on a BD LSRFortessa.
Mixed lymphocyte reaction
CD14+ cells were isolated from whole PBMCs using CD14 MicroBeads (Miltenyi Biotec) as per manufacturer’s instructions. Monocyte purity was evaluated by staining against T cell marker (CD3-PE and CD14-FITC) and over 96% purity was obtained as determined using flow cytometry. Isolated monocytes were then stimulated with granulocyte-macrophage colony stimulating factor and IL-4 (1 μl/ml) for 5 days at 37 °C/10% CO2. On day 5 the DC phenotype was confirmed by staining with CD11c-Blue violet, CD14-FITC, and live dead and analyzed by flow cytometry (BD LSRFortessa). If CD14 negative, cells were seeded at 5 × 104 cells per well and incubated with LPS alone (100 ng/ml) or Acan1 (100 μg/ml) with LPS overnight at 37 °C/10% CO2. On day 6, the plate was incubated at 4 °C for 30 min to prevent DCs from sticking to the bottom of the well and 1 × 104 cells per well were transferred to a new plate. T cells were isolated from human whole PBMCs using the Pan T Cell negative isolation kit (Miltenyi Biotec) as per manufacturer’s instructions. Over 96% purity was obtained as determined using flow cytometry. Isolated T cells were then added to each well (1 × 105 per well) and incubated for 5 days at 37 °C/10% CO2. On day 11, cells were treated with the CellTiter-Glo Luminescent Cell Viability Assay as per the manufacturer’s instructions and luminescence (RLU) was analyzed using luminometer BioTek H4 Hybrid Microplate Reader.
Cytometric bead array
For the CBA, 1 × 105 whole PMBCs were seeded, stimulated with phorbol 12-myristrate 12-acetate (50 ng/ml) and ionomycin (1 μg/ml), and incubated with various concentrations of Acan1 or Nak1 for 24 h at 37 °C/10% CO2. Levels of IL-2 and TNF-α cytokines from culture supernatants were quantified using a CBA (BD Biosciences) with high-throughput sampler according to the manufacturer’s instructions. Data were acquired on a BD LSRFortessa and the data analyzed using BD FCAP Array software version 3.
CellTrace Violet T cell proliferation assay
For the T cell proliferation assay, T cells were isolated from human whole PBMCs using the Pan T Cell negative isolation kit (Miltenyi Biotec) as per manufacturer’s instructions. Over 96% purity was obtained as determined using flow cytometry. Cells were stained with CTV according to the user guide (Thermo Fisher) and seeded at 1 × 105 cells density with Dynabeads Human T-Activator CD3/CD28 beads (Thermo Fisher Scientific). Cells were incubated with Acan1, Acan1 linear (as peptide control) 100 μg/ml, ciclosporin 10 μg/ml or Dynabeads beads alone as positive controls, for 4 days at 37 °C/10% CO2. Proliferation was analyzed by flow cytometry using a BD LSRFortessa (BD Bioscience), and the data were analyzed using FlowJo software (FlowJo LLC).
TNBS model of colitis
Mice were divided randomly into each group. Chemically synthesized hookworm peptides, Acan1 and Nak1, were administered once via the i.p. route in sterile phosphate buffered saline at indicated doses (Fig. S9). Four hours later, mice were anesthetized with xylazine (5 mg kg−1, Rompun 2% Bayer) and ketamine (50 mg kg−1, Ketavest; Pfizer Inc). TNBS was prepared by dissolving 2.5 mg in 50% ethanol. Once unresponsive, mice received an enema with a 125-mg kg−1 dose of TNBS using a lubricated 20-G soft catheter as described (48, 49). Animals were monitored daily for weight loss and general well-being over 3 days. Mice were weighed daily and their overall appearance (piloerection), mobility, and fecal consistency/bleeding were quantified. On day 4 following colitis induction, colons (from cecum to rectum) were collected, quantified, and assessed macroscopically. Clinical changes were scored as described (22). Briefly, clinical pathology including adhesion, edema, wall thickening, and ulceration was scored from 0 to 3, with 3 corresponding to most severe disease. This was for a maximum total score of 12 as described (7). Mouse colons were also assessed histologically for inflammation and gene expression as described (6).
Gene expression analysis
Total RNA was extracted and purified using RNAzol RT (Astral Scientific) according to manufacturer’s instructions. A custom CodeSet panel that included 57 genes related to immune inflammation and regulation, gut barrier, epithelium and tissue destruction, and three housekeeping genes (Table S3) was run according to manufacturer’s instructions (NanoString Technologies). mRNA levels were normalized by the geometric mean of three housekeeping genes (Actb, Gusb, and HprtI). Transcriptomic analysis was performed using the nCounter analysis system. The samples were processed with the fully automated nCounter Prep Station the following day. The normalized data were then visualized and analyzed with R studio software (version 3.4.1).
Statistical analyses
All data were analyzed with GraphPad Prism 7.0 (GraphPad Software). Body weight values were analyzed using two-way analysis of variance (ANOVA) followed by the Dunnett’s multiple comparison test. Comparison for colon length and macroscopic score were analyzed using a Mann–Whitney U test (unpaired, nonparametric). Cytokine concentrations and T cell proliferation were analyzed using an unpaired student t test. A principal component analysis was used as a multivariate analysis of gene expression in each treatment group. An ANOVA analysis was used to identify significant differences in the gene expression profiles of each treatment along principal components 1 and 2. The log2(FC) was calculated by performing log2 ratio of treatment versus vehicle control and compared within a volcano plot. All results represent a mean ± SEM. p Values of <0.05 were considered significant.
Data availability
Structural co-ordinates, NMR restraints, and chemical shift information have been submitted to the RCSB Protein Data Bank, accession codes 6DRI (Acan1) and 7L2G (Nak1). All other data are either presented in the article, are in the supporting information, or are available from the corresponding author upon request.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We would like to thank Dr Yide Wong for his assistance and guidance with the human cell assays.
Author contributions
T. B. S., S. N., B. C.-A., J. P. M., K. J. R., A. L., J. J. M., and R. J. C. conceptualization; T. B. S., A. L., J. J. M., and R. J. C. resources; T. B. S., T. S. W., K. T., R. Y. M. R., K. J. R., A. L., J. J. M., and R. J. C. formal analysis; T. B. S., K. J. R., A. L., J. J. M., and R. J. C. supervision; T. B. S., J. P. M., K. J. R., J. J. M., and R. J. C. funding acquisition; T. B. S., S. N., B. C.-A., T. S. W., K. T., R. Y. M. R., O. L. H., V. P. L., J. P. M., K. J. R., A. L., and R. J. C. investigation; T. B. S., S. N., and B. C.-A. methodology; T. B. S., S. N., B. C.-A., and R. J. C. writing–original draft; T. B. S., J. J. M., and R. J. C. project administration; T. B. S., S. N., B. C.-A., T. S. W., K. T., R. Y. M. R., J. P. M., K. J. R., A. L., J. J. M., and R. J. C. writing–review and editing.
Funding and additional information
This work was supported by grants from the Australian Postgraduate Award (T. B. S., B. C. -A., T. S. W., R. Y. M. R.), Australian Research Council Future Fellowship (R. J. C.; grant number FT100100476 and K. J. R.; grant number FT130100890), Australian Infectious Disease Research Centre (R. J. C., J. J. M., and J. P. M.), NCI, National Institutes of Health (J. P. M.; grant number R01CA155297), National Health and Medical Research (NHMRC) Career Development Fellowship (J. J. M. and J. P. M.; grant numbers 1031652 and 1051627), and senior principal research fellowship (A. L.; grant 1117504), and Children’s Hospital Foundation (S. N.; RCP10317). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Peter Cresswell
Contributor Information
John J. Miles, Email: john.miles@jcu.edu.au.
Richard J. Clark, Email: richard.clark@uq.edu.au.
Supporting information
References
- 1.Luo X., Miller S.D., Shea L.D. Immune tolerance for autoimmune disease and cell transplantation. Annu. Rev. Biomed. Eng. 2016;18:181–205. doi: 10.1146/annurev-bioeng-110315-020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bach J.-F. The hygiene hypothesis in autoimmunity: The role of pathogens and commensals. Nat. Rev. Immunol. 2018;18:105–120. doi: 10.1038/nri.2017.111. [DOI] [PubMed] [Google Scholar]
- 3.Summers R.W., Elliott D.E., Urban J.F., Thompson R.A., Weinstock J.V. Trichuris suis therapy for active ulcerative colitis: A randomized controlled trial. Gastroenterology. 2005;128:825–832. doi: 10.1053/j.gastro.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Croese J., O'Neil J., Masson J., Cooke S., Melrose W., Pritchard D., Speare R. A proof of concept study establishing Necator americanus in Crohn's patients and reservoir donors. Gut. 2006;55:136–137. doi: 10.1136/gut.2005.079129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Navarro S., Pickering D.A., Ferreira I.B., Jones L., Ryan S., Troy S., Leech A., Hotez P.J., Zhan B., Laha T., Prentice R., Sparwasser T., Croese J., Engwerda C.R., Upham J.W. Hookworm recombinant protein promotes regulatory T cell responses that suppress experimental asthma. Sci. Transl. Med. 2016;8 doi: 10.1126/scitranslmed.aaf8807. [DOI] [PubMed] [Google Scholar]
- 6.Ferreira I.B., Pickering D.A., Troy S., Croese J., Loukas A., Navarro S. Suppression of inflammation and tissue damage by a hookworm recombinant protein in experimental colitis. Clin. Transl. Immunol. 2017;6 doi: 10.1038/cti.2017.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreira I., Smyth D., Gaze S., Aziz A., Giacomin P., Ruyssers N., Artis D., Laha T., Navarro S., Loukas A., McSorley H.J. Hookworm excretory/secretory products induce interleukin-4 (IL-4)+ IL-10+ CD4+ T cell responses and suppress pathology in a mouse model of colitis. Infect. Immun. 2013;81:2104–2111. doi: 10.1128/IAI.00563-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mulvenna J., Hamilton B., Nagaraj S.H., Smyth D., Loukas A., Gorman J.J. Proteomics analysis of the excretory/secretory component of the blood-feeding stage of the hookworm, ancylostoma caninum. Mol. Cell. Proteomics. 2009;8:109. doi: 10.1074/mcp.M800206-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Smallwood T.B., Giacomin P.R., Loukas A., Mulvenna J.P., Clark R.J., Miles J.J. Helminth immunomodulation in autoimmune disease. Front. Immunol. 2017;8:453. doi: 10.3389/fimmu.2017.00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chhabra S., Chang S.C., Nguyen H.M., Huq R., Tanner M.R., Londono L.M., Estrada R., Dhawan V., Chauhan S., Upadhyay S.K., Gindin M., Hotez P.J., Valenzuela J.G., Mohanty B., Swarbrick J.D. Kv1.3 channel-blocking immunomodulatory peptides from parasitic worms: Implications for autoimmune diseases. FASEB J. 2014;28:3952–3964. doi: 10.1096/fj.14-251967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dauplais M., Lecoq A., Song J., Cotton J., Jamin N., Gilquin B., Roumestand C., Vita C., de Medeiros C.L., Rowan E.G., Harvey A.L., Menez A. On the convergent evolution of animal toxins. Conservation of a diad of functional residues in potassium channel-blocking toxins with unrelated structures. J. Biol. Chem. 1997;272:4302–4309. doi: 10.1074/jbc.272.7.4302. [DOI] [PubMed] [Google Scholar]
- 12.Castañeda O., Sotolongo V., Amor A.M., Stöcklin R., Anderson A.J., Harvey A.L., Engström Å., Wernstedt C., Karlsson E. Characterization of a potassium channel toxin from the Caribbean sea anemone Stichodactyla helianthus. Toxicon. 1995;33:603–613. doi: 10.1016/0041-0101(95)00013-c. [DOI] [PubMed] [Google Scholar]
- 13.Norton R.S., Pennington M.W., Wulff H. Potassium channel blockade by the sea anemone toxin ShK for the treatment of multiple sclerosis and other autoimmune diseases. Curr. Med. Chem. 2004;11:3041–3052. doi: 10.2174/0929867043363947. [DOI] [PubMed] [Google Scholar]
- 14.Mouhat S., De Waard M., Sabatier J.M. Contribution of the functional dyad of animal toxins acting on voltage-gated Kv1-type channels. J. Pept. Sci. 2005;11:65–68. doi: 10.1002/psc.630. [DOI] [PubMed] [Google Scholar]
- 15.Pennington M.W., Mahnir V.M., Krafte D.S., Zaydenberg I., Byrnes M.E., Khaytin I., Crowley K., Kem W.R. Identification of three separate binding sites on SHK toxin, a potent inhibitor of voltage-dependent potassium channels in human T-lymphocytes and rat brain. Biochem. Biophys. Res. Commun. 1996;219:696–701. doi: 10.1006/bbrc.1996.0297. [DOI] [PubMed] [Google Scholar]
- 16.Tarcha E.J., Olsen C.M., Probst P., Peckham D., Muñoz-Elías E.J., Kruger J.G., Iadonato S.P. Safety and pharmacodynamics of dalazatide, a Kv1.3 channel inhibitor, in the treatment of plaque psoriasis: A randomized phase 1b trial. PLoS One. 2017;12 doi: 10.1371/journal.pone.0180762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cantacessi C., Mitreva M., Jex A.R., Young N.D., Campbell B.E., Hall R.S., Doyle M.A., Ralph S.A., Rabelo E.M., Ranganathan S., Sternberg P.W., Loukas A., Gasser R.B. Massively parallel sequencing and analysis of the Necator americanus transcriptome. PLoS Negl. Trop. Dis. 2010;4 doi: 10.1371/journal.pntd.0000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tudor J.E., Pallaghy P.K., Pennington M.W., Norton R.S. Solution structure of ShK toxin, a novel potassium channel inhibitor from a sea anemone. Nat. Struct. Biol. 1996;3:317–320. doi: 10.1038/nsb0496-317. [DOI] [PubMed] [Google Scholar]
- 19.Pennington M.W., Chang S.C., Chauhan S., Huq R., Tajhya R.B., Chhabra S., Norton R.S., Beeton C. Development of highly selective Kv1.3-blocking peptides based on the sea anemone peptide ShK. Mar. Drugs. 2015;13:529–542. doi: 10.3390/md13010529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haugaard-Kedstrom L.M., Lee H.S., Jones M.V., Song A., Rathod V., Hossain M.A., Bathgate R.A.D., Rosengren K.J. Binding conformation and determinants of a single-chain peptide antagonist at the relaxin-3 receptor RXFP3. J. Biol. Chem. 2018;293:15765–15776. doi: 10.1074/jbc.RA118.002611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fung-Leung W.-P., Edwards W., Liu Y., Ngo K., Angsana J., Castro G., Wu N., Liu X., Swanson R.V., Wickenden A.D. T cell subset and stimulation strength-dependent modulation of T cell activation by Kv1.3 blockers. PLoS One. 2017;12 doi: 10.1371/journal.pone.0170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shepherd C., Giacomin P., Navarro S., Miller C., Loukas A., Wangchuk P. A medicinal plant compound, capnoidine, prevents the onset of inflammation in a mouse model of colitis. J. Ethnopharmacol. 2018;211:17–28. doi: 10.1016/j.jep.2017.09.024. [DOI] [PubMed] [Google Scholar]
- 23.Zaki M.H., Boyd K.L., Vogel P., Kastan M.B., Lamkanfi M., Kanneganti T.-D. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 2010;32:379–391. doi: 10.1016/j.immuni.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirota S.A., Ng J., Lueng A., Khajah M., Parhar K., Li Y., Lam V., Potentier M.S., Ng K., Bawa M., McCafferty D.M., Rioux K.P., Ghosh S., Xavier R.J., Colgan S.P. NLRP3 inflammasome plays a key role in the regulation of intestinal homeostasis. Inflamm. Bowel Dis. 2011;17:1359–1372. doi: 10.1002/ibd.21478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sedda S., Marafini I., Dinallo V., Di Fusco D., Monteleone G. The TGF-β/smad system in IBD pathogenesis. Inflamm. Bowel Dis. 2015;21:2921–2925. doi: 10.1097/MIB.0000000000000542. [DOI] [PubMed] [Google Scholar]
- 26.Spadoni I., Iliev I.D., Rossi G., Rescigno M. Dendritic cells produce TSLP that limits the differentiation of Th17 cells, fosters Treg development, and protects against colitis. Mucosal Immunol. 2012;5:184–193. doi: 10.1038/mi.2011.64. [DOI] [PubMed] [Google Scholar]
- 27.Morelli A.E., Thomson A.W. Tolerogenic dendritic cells and the quest for transplant tolerance. Nat. Rev. Immunol. 2007;7:610–621. doi: 10.1038/nri2132. [DOI] [PubMed] [Google Scholar]
- 28.Bogunovic M., Ginhoux F., Helft J., Shang L., Hashimoto D., Greter M., Liu K., Jakubzick C., Ingersoll M.A., Leboeuf M., Stanley E.R., Nussenzweig M., Lira S.A., Randolph G.J., Merad M. Origin of the lamina propria dendritic cell network. Immunity. 2009;31:513–525. doi: 10.1016/j.immuni.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varol C., Vallon-Eberhard A., Elinav E., Aychek T., Shapira Y., Luche H., Fehling H.J., Hardt W.-D., Shakhar G., Jung S. Intestinal lamina propria dendritic cell subsets have different origin and functions. Immunity. 2009;31:502–512. doi: 10.1016/j.immuni.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 30.Maynard C.L., Weaver C.T. Intestinal effector T cells in health and disease. Immunity. 2009;31:389–400. doi: 10.1016/j.immuni.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marsal J., Agace W.W. Targeting T-cell migration in inflammatory bowel disease. J. Intern. Med. 2012;272:411–429. doi: 10.1111/j.1365-2796.2012.02588.x. [DOI] [PubMed] [Google Scholar]
- 32.Imam T., Park S., Kaplan M.H., Olson M.R. Effector T helper cell subsets in inflammatory bowel diseases. Front. Immunol. 2018;9:1212. doi: 10.3389/fimmu.2018.01212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smids C., Horjus Talabur Horje C.S., Drylewicz J., Roosenboom B., Groenen M.J.M., van Koolwijk E., van Lochem E.G., Wahab P.J. Intestinal T cell profiling in inflammatory bowel disease: Linking T cell subsets to disease activity and disease course. J. Crohns Colitis. 2017;12:465–475. doi: 10.1093/ecco-jcc/jjx160. [DOI] [PubMed] [Google Scholar]
- 34.Levin A.D., Wildenberg M.E., van den Brink G.R. Mechanism of action of anti-TNF therapy in inflammatory bowel disease. J. Crohns Colitis. 2016;10:989–997. doi: 10.1093/ecco-jcc/jjw053. [DOI] [PubMed] [Google Scholar]
- 35.Hansen L.K. The role of T cell potassium channels, KV1.3 and KCa3.1, in the inflammatory cascade in ulcerative colitis. Dan. Med. J. 2014;61 [PubMed] [Google Scholar]
- 36.Kazama I. Roles of lymphocyte Kv1.3-channels in gut mucosal immune system: Novel therapeutic implications for inflammatory bowel disease. Med. Hypotheses. 2015;85:61–63. doi: 10.1016/j.mehy.2015.03.023. [DOI] [PubMed] [Google Scholar]
- 37.Mei Y., Ye S., Han W., Yang Q., Hu C., Liu X., Mei Q., Xu J. Clinical significance of potassium channel and NLRP3 expression in platelets of active ulcerative colitis. Inflamm. Bowel Dis. 2019;25:e115–e116. doi: 10.1093/ibd/izz109. [DOI] [PubMed] [Google Scholar]
- 38.Keller R. Cantina Verlag; Goldau, Switzerland: 2004. The Computer Aided Resonance Assignment Tutorial. [Google Scholar]
- 39.Wüthrich K. Wiley; New York, NY: 1986. NMR of Proteins and Nucleic Acids. [Google Scholar]
- 40.Schroeder C.I., Rosengren K.J. Three-dimensional structure determination of peptides using solution nuclear magnetic resonance spectroscopy. In: Priel A., editor. Snake and Spider Toxins: Methods and Protocols. Springer US; New York, NY: 2020. pp. 129–162. [DOI] [PubMed] [Google Scholar]
- 41.Shen Y., Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR. 2013;56:227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Güntert P. Protein NMR Techniques. Springer; Totowa, NJ: 2004. Automated NMR structure calculation with CYANA; pp. 353–378. [DOI] [PubMed] [Google Scholar]
- 43.Brünger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 44.Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X., Murray L.W., Arendall W.B., III, Snoeyink J., Richardson J.S. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koradi R., Billeter M., Wuthrich K. Molmol: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 46.Cristofori-Armstrong B., Soh M.S., Talwar S., Brown D.L., Griffin J.D.O., Dekan Z., Stow J.L., King G.F., Lynch J.W., Rash L.D. Xenopus borealis as an alternative source of oocytes for biophysical and pharmacological studies of neuronal ion channels. Sci. Rep. 2015;5:14763. doi: 10.1038/srep14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lepletier A., Lutzky V.P., Mittal D., Stannard K., Watkins T.S., Ratnatunga C.N., Smith C., McGuire H.M., Kemp R.A., Mukhopadhyay P., Waddell N., Smyth M.J., Dougall W.C., Miles J.J. The immune checkpoint CD96 defines a distinct lymphocyte phenotype and is highly expressed on tumor-infiltrating T cells. Immunol. Cell Biol. 2019;97:152–164. doi: 10.1111/imcb.12205. [DOI] [PubMed] [Google Scholar]
- 48.Scheiffele F., Fuss I.J. Induction of TNBS colitis in mice. Curr. Protoc. Immunol. 2002 doi: 10.1002/0471142735.im1519s49. Chapter 15, Unit 15.19. [DOI] [PubMed] [Google Scholar]
- 49.Wirtz S., Neufert C., Weigmann B., Neurath M.F. Chemically induced mouse models of intestinal inflammation. Nat. Protoc. 2007;2:541–546. doi: 10.1038/nprot.2007.41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structural co-ordinates, NMR restraints, and chemical shift information have been submitted to the RCSB Protein Data Bank, accession codes 6DRI (Acan1) and 7L2G (Nak1). All other data are either presented in the article, are in the supporting information, or are available from the corresponding author upon request.