Abstract
Background: Acute myeloid leukemia (AML) is a group of heterogeneous hematologic malignancies correlates with poor prognosis. It is important to identify biomarkers for effective treatment of AML. Kinases participate in many regulatory pathways and biological activities in AML. Previous studies demonstrated that MAP4K1, a serine/threonine kinase, was associated with immune regulation and cancer progression. However, its role and mechanism in acute myeloid leukemia (AML) have not been explored.
Methods: RNA-seq profiling was performed for Homoharringtonine (HHT)-resistant and Homoharringtonine (HHT)-sensitive cell lines. Bioinformatic tools were used for differential analysis. Cell culture and transfection, Cell proliferation, apoptosis and Cell cycle assay, Quantitative RT-PCR, and Western blotting analysis were used to explore biological phenotypes in vitro.
Findings: We found that MAP4K1 was highly expressed in HHT-induced resistant AML cell lines. In addition, overexpression of MAP4K1 in AML cells induced resistance of AML cells against HHT. Not only that, the findings of this study showed that overexpression of MAP4K1 was an independent risk factor that predicts poor prognosis of AML. Further, In vitro studies showed that MAP4K1 modulated cell cycle through MAPK and DNA damage/repair pathways. Therefore, MAP4K1 is a potential target for developing therapies for AML.
Interpretation: This study demonstrates that MAP4K1 not only regulates HHT resistance but also independently predicts AML prognosis. In addition, understanding the regulatory mechanism of MAP4K1 reveals novel treatment strategies for resistant and refractory AML.
Fundings: This work was supported by the National Natural Science Foundation of China (NSFC) (Grant No.81800199, 81670124, 82070118) and the Natural Science Foundation of Zhejiang Province (LY20H080008).
Keywords: Acute myeloid leukemia, MAP4K1, Homoharringtonine resistance, Prognosis, DNA damage, MAPK
Research in context.
Evidence before this study
Acute myeloid leukemia (AML) is a group of heterogeneous hematologic malignancies with high incidence rate, high recurrence rate and high mortality. The emergence of drug resistance against conventional therapies limits the efficacy of these therapies resulting in poor patient outcomes. Therefore, it is important to identify novel therapeutic targets to improve the treatment efficacy. Previous studies demonstrated that MAP4K1, a serine/threonine kinase, is strongly implicated in immune regulation and cancer progression. However, the role of MAP4K1 in regulating AML progression and chemoresistance remains unknown.
Added value of this study
In this study, we developed three HHT-resistant strains for the first time and labeled them as MV4-11 R10, MV4-11 R30, MV4-11 R50. We found that the upstream MAPK gene, MAP4K1 was upregulated in all resistant strains via RNA-seq analysis. This study reveals, for the first time, the MAP4K1 regulates resistance against HHT and the prognosis of AML. In addition, MAP4K1 functions as a tumor promotor and drug mediator by modulating the DNA damage/repair system and MAPK pathway. More than that, two inhibitors, sunitinib and SP600125, were found to have good synergy with HHT, which may provide new insights into AML treatment.
Implications of all the available evidence
The findings of the present study suggest that MAP4K1 is an independent risk factor that influences the prognosis of AML and a regulator affects the development of drug resistance in AML. Therefore, MAP4K1 might represent a novel therapeutic target. The combination therapy proposed in this article may improve the sensitivity of AML to HHT.
Alt-text: Unlabelled box
1. Introduction
Acute myeloid leukemia (AML) is a group of heterogeneous hematologic malignancies characterized by numerous cytogenetic and molecular changes. AML is associated with high incidence rate, high recurrence rate and high mortality [1]. Nowadays, advances and development of targeted agents and modern immunotherapy have improved the responsiveness of AML patients. However, drug resistance against conventional therapies limits the efficacy of these therapies resulting in poor patient outcomes [2]. Therefore, it is important to identify novel therapeutic targets of AML to improve the prognosis of AML especially relapsed/refractory AML. Studies should be performed to explore the mechanisms of drug resistance to develop effective drugs against drug resistance AML.
Homoharringtonine (HHT) is a cytotoxic alkaloid originally extracted from Cephalotaxus hainanensis. HHT has significant activity against various leukemia types such as acute myeloid leukemia, chronic myeloid leukemia and myelodysplastic syndrome. Our research team has been investigating the effect, mechanism and clinical application of HHT in AML for several years. A prospective, multicenter phase 3 study showed that both the complete remission (73% vs 61%, p=0.00108) and the 3-year event-free survival (35.4% vs 23.1%) in HAA treatment group were better than that in patients who received DA (doxorubicin+arac) plan [3,4]. Therefore, HHT could be a potential drug for the treatment of AML because of its good single effect on AML cells as well as its potential to combine with other traditional drugs. However, along with the wide use of HAA and other HHT-based plans, the resistance of HHT has become a new challenge and still lacks research. Exploring the resistant mechanism and related targets must have great practical significance for AML patients who relied on HHT-based treatment.
In this study, we developed three resistant strains to HHT and labeled them as MV4-11 R10, MV4-11 R30, MV4-11 R50. Resistant strains were developed by long-term induction of increasing concentrations of HHT. Further, we used second-generation high-throughput sequencing technology to determine the expression level of genes before and after induction of resistance. The findings of this study showed that the MAPK pathway was upregulated in the three resistant strains with significantly high expression level of an upstream MAPK gene named MAP4K1. MAP4K1 knockdown increased sensitivity of AML cells to HHT whereas overexpression of MAP4K1 increased resistance of AML cells to HHT. These findings imply that MAP4K1 may play a key role in regulating HHT resistance of AML.
Kinases are involved in many regulatory pathways and biological activities. Kinases play essential roles in tumor development and chemotherapy resistance thus they are desirable targets for improving survival of cancer patients and overcoming drug resistance [5]. Mitogen-activated protein kinases (MAPKs) belong to serine/threonine kinase family and they modulate embryogenesis, cell differentiation, proliferation, and cell death pathways [6]. In addition, enzymes involved in these pathways are potential targets for the development of antitumor therapy [7,8]. Not only that, the MAP kinase signaling pathway is closely related to hematological malignancies [9]. MAP4K1 was originally cloned from human hematopoietic myeloid cells and always has a typically high expression in myeloid and lymphoid lineage cells. Studies have reported that the expression of MAP4K1 is associated with the pathogenesis of several cancer types. Yang, H.S., et, al. reported that inhibition of MAP4K1 suppressed colon cancer progression [10]. Furthermore, Wang, J., et, al. reported that HPK1 positive expression associated with longer overall survival in patients with estrogen receptor-positive invasive ductal carcinoma [11]. MAP4K1 affected cell-cycle progression and mediated Vitamin D resistance in AML cell lines [12]. However, previous studies have not explored the mechanism and role of MAP4K1 as an independent prognostic marker for AML and its role in regulating HHT resistance. Therefore, the functional role and mechanism of MAP4K1 in AML should be explored to provide information for developing novel effective therapies.
In the present study, we carried out in vitro and in vivo experiments to explore the role of MAP4K1 in regulating AML prognosis and its role in modulating drug resistance against HHT. The findings of this study show that MAP4K1 plays a key role in the pathogenesis of AML, patient survival and drug resistance. The possible regulatory mechanisms are discussed according to present data, bioinformatics and related literatures. We confirm that MAP4K1 mediates HHT resistance and is an independent risk factor for AML prognosis. These findings provide novel information for the development of AML therapies to overcome drug resistance and improve patient survival.
2. Methods
2.1. Cell culture and transfection
MV4-11 cell was treated with increasing concentrations of HHT (from 1nmol/L to 50nmol/L). Cells that grew normally in complete medium containing 10 nmol / L, 30 nmol / L, and 50 nmol / L HHT were preserved. The cells were defined as MV4-11 R10, MV4-11 R30, MV4-11 R50. The MV4-11 (CVCL_0064), MV4-11 R10, MV4-11 R30, MV4-11 R50, MOLM13 (CVCL_2119) and THP-1 (CVCL_0006), KG-1 (CVCL_1824), HL-60 (CVCL_A794), Kasumi (CVCL_0589) were cultured in IMDM medium (Gibco, REF: C12440500BT) and RPMI-1640 medium (Gibco, REF: C11875500BT) containing L-glutamine (Corning) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Gibco, USA, REF: 10099-141C) at 37°C in a humidified atmosphere with 5% CO2. For MAP4K1 knockdown, three shRNA sequences were designed and the first and third sequences were selected for subsequent experiments based on qPCR and western blot results. THP-1 and MV4-11 R50 were transfected with green fluorescent protein (GFP) containing shRNA lentiviral particles directed against human MAP4K1 or with the shScramble vector. For overexpression, the coding DNA sequence of MAP4K1 was cloned into the PCDH vector and then transfected into MV4-11 and THP-1. Cells were selected in culture media containing puromycin (1μg/ml, InvivoGen, Cat: ant-pr-1) for about 1-2 weeks and used for subsequent experiments. Sequences for MAP4K1 shRNA and non-silencing control shRNA were:
Sh1: 5’ - CCTGTATTCTCATAGCATCCT - 3’
Sh3: 5’ - GCCCTTCTCGTAAAGTTGTTC - 3’ shScramble vector: 5’-GCTTCGCGCCGTAGTCTTA - 3’
2.2. Reagents and antibodies
Homoharringtonine (HHT, Cat: HY-14944/CS-2872) was purchased from MCE, JNK inhibitor (SP600125, Cat: S1460) and HPK1 inhibitor (Sunitinib, Cat: S1042) were purchased from Selleck Chemicals (Houston, TX, USA). For western blotting, the following antibodies were obtained from Cell Signaling Technologies: HPK1 (4472, AB_2140826), P-JNK (4668, AB_823588), JNK (9252, AB_330894), P-C-JUN (ser63,2361, AB_490908), C-JUN (9165, AB_2130165), P-ATM (5883, AB_10835213), P-ATR (2853, AB_2290281), ATR (2790, AB_2227860), P-CHK1 (2348, AB_331212), P-CHK2 (2197, AB_2080501), CHK2 (2662, AB_2080793), P-H2AX (9718, AB_2118009), H2AX (7631, AB_10860771), P21 (2947, AB_823586), P27 (3686, AB_2077850), GAPDH (5174, AB_10622025). ATM (27156-1-AP, AB_2880780), CHK1 (25887-1-AP, AB_2880283), tubulin (66031-1-Ig, AB_11042766) were purchased from proteintech. A human CD45 antibody was purchased from MULTI SCIENCES (Cat: AH04504-100).
2.3. Cell line and antibody validation
Cell lines (THP-1, MV4-11, HL-60, Kasumi, KG-1, MOLM13) used for follow-up in vitro and in vivo experiments have been validated by the institute, STR profiling of these cell lines has been listed in Supplementary Data (Supplementary materials 3) as Part1. Mycoplasma testing has been performed. Cell lines used in this article are free of mycoplasma contamination and results have been listed in Supplementary Data (Supplementary materials 3) as Part1. All antibodies are commonly used antibodies, RRID tags of all antibodies can be obtained. Information of the relevant references for antibody application has been listed in Supplementary Data (Supplementary materials 3) as Part2.
2.4. Patient sample
BM samples were obtained from 178 AML patients with their written informed consent. Clinical data with detailed diagnosis and treatment information of 178 AML patients were obtained from medical records between March 2010 and June 2014. Mononuclear cells (MNCs) were isolated for use in subsequent experiments. WHO classification, conventional cytogenetic banding assay and molecular analyses were performed centrally in Zhejiang Institute of Hematology (ZIH), China as previously described in AML diagnosis [13]. Chromosomal abnormalities and the genetic mutations were explored in the First Affiliated Hospital of Zhejiang. The researchers performed the above cytogenetics analyses without knowledge of MAP4K1 expression profile and clinical outcome. Cord blood CD34-positive hematopoietic stem cells were provided by healthy donors. In this study, no patient had undergone allogeneic transplantation. Patients with secondary AML or acute promyelocytic leukemia were excluded. Cytogenetic groups of patients were classified as favorable, intermediate, and unfavorable risk according to the NCCN guideline [14]. Favorable subgroup comprised t(8;21)/AML1-ETO and inv16/CBFβ-MYH11. Adverse group consisted of t (9;22), inv(3)/t(3;3), -5, -7, del(5q), del(7p), 11q23 and complex translocations. Intermediate subtype contained cytogenetically normal and AML with other cytogenetic abnormalities. Patients received HAA (homoharringtonin 2 mg/m2/day for 7 days, cytarabine 100 mg/m2/day for 7 days and aclarubicin 20 mg/m2/day for 5 days), DA (daunorubicin 45 mg/m2/ day for 3 days and cytarabine 100 mg/m2/day for 7 days) and IA regimen (idarubicin 8-10 mg/m2/day for 3 days and cytarabine 150 mg/m2/day for 7 days) [4,15]. Ethical approval for the study was provided by the Ethics Committee of the First Affiliated Hospital, College of Medicine, Zhejiang University.
2.5. Primary patient cells
Five BM aspirates (not included in the 178 AML patients) were obtained from AML patients. Mononuclear cells (MNCs) were isolated through mononuclear cell separation medium and immediately used for subsequent experiments. Genetic mutations were determined by the First Affiliated Hospital of Zhejiang. Ethical approval for the study was provided by the Ethics Committee of the First Affiliated Hospital, College of Medicine, Zhejiang University.
2.6. In vivo studies
Severe immune-deficient strain NCG (NOD/ShiLtJGpt-Prkdcem26Cd52Il2rgem26Cd22/Gpt) mice were purchased from GemPharmatech Co, Ltd (Nanjing, China) and were raised in the Experimental Animal Center of Zhejiang Chinese Medicine university laboratory animal research center. Fifteen 5-week-old female NCG mice were randomly divided into 3 groups after being bred for one week. 1 × 106 THP-1 Luci cells transfected with shRNA-1, shRNA-3 and non-silencing control (NC) vector were injected into every group of mice respectively through the tail vein. Mice's body weight was determined once a week. Growth of the leukemia cells was monitored using an IVIS every week and the tumor burden was analyzed using GraphPad Prism 5 [two-tailed Student's t-tests]. Levels of human CD45+ cells in mice peripheral blood during the fourth week were determined by flow cytometry and analyzed using GraphPad Prism 5 [two-tailed Student's t-tests]. Mice were euthanized when they developed a bowed back and their lower limbs were paralyzed. The survival curve of mice was generated using GraphPad Prism 5, and the difference between groups was analyzed using two-tailed Student's t-tests. Data of 5 animals in each group were included in the final statistical analysis. Mice dying for nonrelated cancer causes were excluded from the studies (such as fights or infections). Each experimental group comprised mice derived from the different brood. All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee. All animal experiments were in accordance with animal use guidelines and ethical approval.
2.7. RNA-seq
RNA sequencing was performed using total RNA samples isolated from MV4-11, MV4-11 R10, MV4-11 R30, MV4-11 R50 cell lines using Illumina HiSeq X Ten platforms. Expression levels of mRNA in sequence data were calculated as RPKM (Reads Per Kilo-base per Million reads). RNA sequencing data was uploaded to NCBI's SRA, accession to cite for the SRA data: PRJNA664675.
2.8. MTS cell viability assay
General procedure: Cells were seeded into 24-well plates at a concentration of 100,000 cells per well with or without drugs. Cells from each well were homogeneously transferred into three wells, with a total volume of 100 μL for each well in 96-well plate and supplemented with 10 μL MTS (Promega CellTitre 96, Promega Corporation, Madison, WI, USA, REF: G3581) (5 mg/mL−1). Cells were incubated for 4h and absorbance was determined at 490 nm. For sunitinib and HHT combination effect assay, cells were first seeded in individual wells of a 24-well plate at a concentration of 100,000 cells per well with 1 mL of culture medium. Sunitinib (0, 5, 10, 20, 40 nM for MV4-11 R50 and 0, 1, 2, 4, 8 nM for MV4-11 and 0, 0.5, 1, 2, 4 uM for THP-1, HL-60, primary patient cells) as well as HHT (0, 10, 20, 40, 80 nM for MV4-11 R50, THP-1, HL-60 and primary patient cells and 0, 2.5, 5, 10, 20 nM for MV4-11 cells) were added and incubated for 48 h. For JNK inhibitor (SP600125) and HHT combination assay, cells were first seeded in individual wells of a 24-well plate at a concentration of 100,000 cells per well with 1 mL of culture medium. SP600125 (0, 5, 10, 20, 40 uM for MV4-11 R50, THP-1, MV4-11 and HL-60) and HHT (0, 10, 20, 40, 80 nM for MV4-11 R50, THP-1, HL-60 and 0, 2.5, 5, 10, 20 nM for MV4-11) were then added and incubated for 48 h. Cells from each well were homogeneously transferred into three wells with a total volume of 100 μL for each well in a 96-well plate and supplemented with 10 μL MTS incubated for 4 h before analysis. “Calcusyn” was used to simulate and calculate the combination index.
2.9. Cell apoptosis and cell cycle assay
Cell apoptosis assay (Cat: AP101) was carried out using a kit obtained from MULTI SCIENCES following the manufacturer's instructions. Collected cells were resuspended twice in PBS and AV and PI were added. Cells were incubated for 30 min at 37°C and analyzed by flow cytometer. Propidium iodide (PI) DNA staining (Cat: CCS012) obtained from MULTI SCIENCES was used for cell cycle assay. Collected cells were incubated for 30 min at 37°C and analyzed using FACScan™ flow cytometer (Becton Dickinson, San Diego, CA, USA).
2.10. Quantitative RT-PCR
Total RNA was extracted from cells using Trizol reagent (Invitrogen, USA, Cat: 9109). RNA was reverse transcribed to cDNA using a Reverse Transcription Kit (TaKaRa, Cat: RR036A-1). Quantitative real-time PCR was carried out using SYBR Green qPCR Master Mix (TaKaRa, Cat: RR420) and CFX96/384 Real-Time PCR detection systems (Bio-Rad, Hercules, CA). A total volume of 25 µl containing 1 µl of 100 ng/µl sample cDNA, 12.5 µl of 2 × PCR Mix, 1 µl of 0.5 µM of each primer and 10.5 µl of ddH2O was used for PCR reactions. Quantification was carried out using ∆∆CT method and the expression level of GAPDH was used as a control to normalize values across different target genes. The following primers were used for quantitative PCR:
MAP4K1 (forward 5′ - GTCGTGGACCCTGACATTTTC - 3′; reverse 5′ - CCTTAAAGACTTCCCCATACGTG - 3′) and
GAPDH (forward 5’ - GGAGCGAGATCCCTCCAAAAT - 3’; Reverse 5’ - GGCTGTTGTCATACTTCTCATGG - 3’).
2.11. Western blotting analysis
Cells were lysed using RIPA buffer (Thermo Fisher Scientific, USA, REF: 89900) supplemented with protease inhibitor and phosphatase inhibitor cocktail (Thermo Fisher Scientific, USA, REF: 1861280) on ice for 30 min. The cell lysate was centrifuged at 12000 g for 15 min at 4°C and the protein concentration of the cellular supernatant was determined using a BCA reagent (Thermo Fisher Scientific, USA, REF: reagent A-23228, reagent B-23224). Approximately 40-60 μg protein was loaded per well on 4-10% SDS-PAGE gel and transferred onto PVDF membrane (Millipore, Billerica, MA, USA) preactivated with methanol. Membranes were blocked using 5% nonfat milk for 1 h and incubated with primary antibodies overnight at 4°C. After incubation, membranes were washed thrice with TBST and incubated with secondary antibodies (Cell Signaling Technology) for 1 h at room temperature. Target proteins were then visualized using a ECL detection kit (Thermo Fisher Scientific, USA, REF: 34096) and analyzed using Image Lab™ software (Bio-Rad Laboratories, Hercules, CA, USA).
2.12. Statistics
2.12.1. Patient data analysis
Overall survival (OS) was defined as period between the date of diagnosis to death due to any cause. In this study, event free survival (EFS) was defined as the time from date of diagnosis to withdrawal from the study due to absence of complete remission (CR), relapse or death. Relapse-free survival (RFS) was defined as the time from the start of treatment to relapse or death from any cause. Association between MAP4K1 expression and OS, EFS, RFS was estimated using Kaplan-Meier method and log-rank test. Multivariate analysis was carried out using Cox regression model.
2.13. Experiments data analysis
All experiments were performed in triplicates. Data were analyzed with GraphPad Prism 5 and expressed as mean ± SEM. The two-tailed Student's t-tests was used to compare means between the two groups. P < 0.05 was considered statistically significant. Kaplan-Meier survival curve was generated using IBM SPSS Statistics 20 and log-rank test was used to calculate the P values. * P < 0.05, ** P < 0.01, *** P < 0.001.
2.14. Bioinformatic analysis
Differences in gene expression between normal individuals and patients were analysed using the “GEPIA” online tool [16,17]. DE Seq (1.18.0) R package was used for differential analysis of mRNAs in RNA-seq data. KEGG pathway enrichment analysis for these mRNAs was performed via the “Enrichr” online tool [18]. TCGA data were divided into high and low expression groups and differential Hallmarkers between the high and low expression groups were determined using GSEA [19].
2.15. Ethics statement
Humans: This study was approved by the Ethics Committee of the First Affiliated Hospital, College of Medicine, Zhejiang University (acceptance number: IIT20200523A). Written informed consents was obtained from all participants before enrollment. All experimental protocols and procedures were carried out following the ethical standards of the First Affiliated Hospital at which the studies were conducted. Five BM aspirates were collected in the Bone marrow department of the First Affiliated Hospital of Zhejiang University, with the approval of the hospital Ethical Committee and according to local legal and ethical regulations (acceptance number: IIT20200523A). All of the subjects were provided with written informed consent before enrolment. Animals: All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee (Approval No: IACUC-20200608-06). And all animal experiments were performed following animal use guidelines and ethical approval.
2.16. Role of funders
The funders were not involved in study design, data collecting, analysis, interpretation, decision to publish or writing of the manuscript. The corresponding author Jie Jin had full access to all the data in the study and had final responsibility for the decision to submit the manuscript for publication.
3. Results
1 MAP4K1 mRNA profile in HHT-sensitive MV4-11cells was different from the profile in HHT-resistant MV4-11 R10/R30/R50 cells
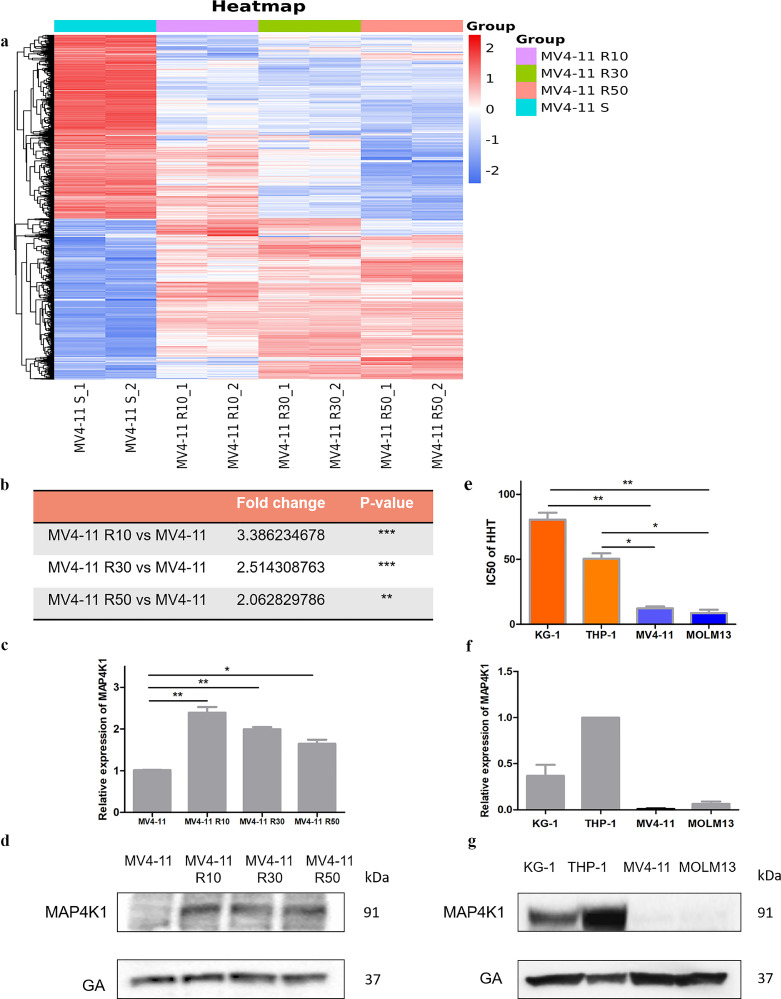
We established three HHT-resistant cell lines from MV4-11 to determine molecules associated with HHT-resistance. Resistance strains were developed by long-term culturing of MV4-11 cells in the presence of increasing concentrations of HHT. Three resistant strains named MV4-11 R10, MV4-11 R30, MV4-11 R50 which were induced by 10, 30, 50nM concentration of HHT were selected for RNA sequencing. Further, we analyzed the differential gene expression profiles between HHT-resistant cells and HHT-sensitive MV4-11 cells, to explore the mechanisms of HHT resistance (Fig. 1a, Table S1). HHT-resistant cells showed significantly higher expression levels of several genes compared with wild-type MV4-11. Notably, the MAP4K1 expression level was significantly higher in the three resistant cell lines compared with the levels in the wild-type MV4-11 (Fig. 1b). The result of qPCR and western blot confirmed the higher expression of MAP4K1 in a subset of HHT‐resistant cell lines (Fig. 1c-d). The same phenomena existed in Wild-type HHT-sensitive MV4-11, MOLM13 cell lines, and resistant cell lines THP-1, KG-1. We determined IC50 values of HHT in four AML cell lines (Fig. 1e) and analysed the expression levels of MAP4K1 in these cell lines using quantitative PCR and western blot (Fig. 1f-g). The data showed cells with high resistance to HHT had relatively high expression levels of MAP4K1 in both wild-type and constructed resistant cell lines.
Fig. 1.
a Gene expression levels between HHT-resistant cells and HHT-sensitive MV4-11 cells. b Fold change and P value of MAP4K1 expression in all three comparison groups (data from RNA-seq) show that MAP4K1 has a significantly high expression in all three resistant strains [two-tailed Student's t-tests; **P < 0.01, ***P < 0.001]. c MAP4K1 expression in all three HHT resistant strains and sensitive cell via q-PCR [two-tailed Student's t-tests; *P < 0.05, **P < 0.01]. d MAP4K1 expression in all three HHT resistant strains and sensitive cells was determined using western blot. e IC50 of HHT in the four cell lines (KG-1 and THP-1 are native resistant strains whereas MV4-11 and MOLM13 are more sensitive cells) [two-tailed Student's t-tests; *P < 0.05, **P < 0.01]. f mRNA expression level of MAP4K1 in KG-1, THP-1, MV4-11, MOLM13. g Expression of MAP4K1 in KG-1, THP-1, MV4-11, MOLM13 at protein level. Western Blot bands were derived from three separate experiments but only one representative loading control is shown. Each value indicates the mean ± SEM of three or more independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
2 MAP4K1 is an independent risk factor of AML
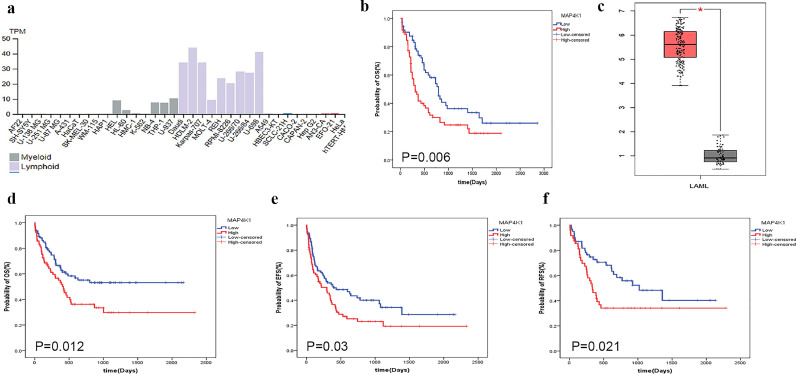
Drug resistance is a major challenge in clinical oncology as it limits the efficacy of drugs and affects tumor prognosis [20]. In this study, we explored the association of the expression level of MAP4K1 with AML progression based on online data and data from our own conhort. Data from Human Protein Atlas showed typically high expression levels of MAP4K1 in myeloid and lymphoid lineages (Fig. 2a). TCGA data showed that high expression level of MAP4K1 was associated with a poor prognosis of AML (Fig. 2b). Notably, the expression level of MAP4K1 in AML patients was more than 5.5-fold higher compared with expression levels of normal donors based on the online database “GEPIA” [log-rank tests; P-value < 0.01] (Fig. 2c). Further, the expression levels of MAP4K1 in BM samples collected from 178 AML patients in our cohort was were determined using quantitative-PCR. Interquartile range of MAP4K1 transcript level was from 0.23 to 1.03. The value at 50% was used as the cut-off value to group patients into high and low MAP4K1 expression. Numerical values 0.71 (0.54, 1.03) and 0.31 (0.23, 0.37) were the median and interquartile range of high and low MAP4K1 expression groups, respectively. Clinical characteristics of patients with high and low expression levels of MAP4K1 are shown in Table 1. Expression levels of MAP4K1 showed no significant correlation with variables such as sex, age, white blood cell counts (WBC), hemoglobin levels, percentage of bone marrow blasts, mutations in FLT3-ITD, CEBPA, NPM1, DNMT3A, IDH1, and IDH2 genes. MAP4K1 high expression group was significantly associated with shorter OS, EFS and RFS [log-rank tests; P-value = 0.012, 0.03, 0.021, respectively] (Fig. 2d–f). Therefore, the expression of MAP4K1 is a predictor of poor prognosis in AML patients. To further explore the prognostic role of MAP4K1 and exclude potential confounders factors, we constructed multivariable models of OS, EFS, and RFS after adjusting for known risk factors such as age, WBC, blast, karyotype-risk group, treatment protocols, and several mutations like NPM1, FLT3-ITD, DNMT3α, etc. The findings showed that the MAP4K1 expression was still an independent prognostic factor after adjusting for these factors (Table 2, 3, and 4). Analysis of the whole AML population showed that people with high MAP4K1 expression levels had a 1.686-fold higher OS risk (LR; P = 0.033), 1.786-fold higher EFS risk (LR; P = 0.009), and 2.369-fold higher RFS risk (LR; P = 0.005). Other negative prognostic factors included elder age (LR; OS: P = 0.063, RFS: P = 0.007), WBC count (LR; EFS: P = 0.059), BM blast (LR; OS: P = 0.039, RFS: P = 0.023), FLT3ITD mutation (LR; OS: P = 0.040), and presence of DNMT3A (LR; OS: P = 0.011, EFS: P = 0.001, RFS: P = 0.089).
Fig. 2.
a MAP4K1 expression in different lineages shows it is mainly expressed in the myeloid and lymphoid lineages (Figure was obtained from “The Human Protein Atlas”). b High expression of MAP4K1 was associated with poor prognosis of AML in TCGA data (red represents high expression) [log-rank tests; P-value = 0.006]. c MAP4K1 expression level in the AML patients was more than 5.5-fold higher compared with the level in the normal donors (red represents AML patient) (Figure was obtained from “GEPIA”). d MAP4K1 high expression group of the current study cohort was significantly associated with shorter OS (red represents high expression) [log-rank tests; P-value = 0.012]. e MAP4K1 high expression group in the current study cohort was significantly associated with shorter EFS (red represents high expression) [log-rank tests; P-value = 0.03]. f MAP4K1 high expression group in the current study cohort was significantly associated with shorter RFS (red represents high expression) [log-rank tests; P-value = 0.021] (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article..
Table 1.
Characteristics of AML patients by high and low MAP4K1 expression.
| Variables | MAP4K1 expression |
P-value | |
| Low expression (N=89) | High expression(N=89) | ||
| Number(%) | 89 (50%) | 89 (50%) | |
| Male,n(%) | 53 (58.9) | 45 (50.0) | 0.13 |
| Age,median(IQR),years | 54.00 (34.50, 61.00) | 50.00 (42.00, 59.00) | 0.678 |
| BM blast, median(IQR),%1 | 66.00 (43.75, 82.00) | 69.00 (49.00, 84.00) | 0.433 |
| WBC, median(IQR),ⅹ109/L2 | 18.40 (5.15, 70.15) | 13.70 (3.85, 100.75) | 0.271 |
| HB, median(IQR),g/L3 | 80.00 (66.00, 100.00) | 78.00 (64.10, 100.00) | 0.106 |
| PLT,median(IQR),ⅹ109/L4 | 45.00 (23.50, 78.00) | 42.00 (21.00, 94.50) | 0.694 |
| FAB classification,n(%)5 | 0.996 | ||
| M0 | 5 (5.6) | 2 (2.2) | |
| M1 | 3 (3.4) | 3 (3.4) | |
| M2 | 73 (80.9) | 75 (84.3) | |
| M3 | 4 (4.5) | 4 (4.5) | |
| M4 | 0 (0.0) | 0 (0.0) | |
| M5 | 0 (0.0) | 0 (0.0) | |
| M6 | 0 (0.0) | 0 (0.0) | |
| Karyotype risk, n (%) | 0.834 | ||
| Favorable | 3 (3.4) | 3 (3.4) | |
| Intermediate | 77 (86.5) | 78 (87.6) | |
| Unfavorable | 9 (10.1) | 8 (9) | |
| Genes mutations,n(%) | |||
| FLT3ITD | 17 (19.1) | 19 (21.3) | 0.704 |
| CEBPA | 24 (73.0) | 21 (23.6) | 0.790 |
| NPM1 | 27 (30.3) | 23 (25.8) | 0.395 |
| DNMT3A | 6 (6.7) | 13 (14.6) | 0.068 |
| IDH1 | 5 (5.6) | 12 (13.5) | 0.074 |
| IDH2 | 8 (9.0) | 10 (11.2) | 0.602 |
| MAP4K1,median(IQR) | 0.31 (0.23, 0.37) | 0.71 (0.54, 1.03) | 0.037 |
| Treatment protocols6 | 0.335 | ||
| DA | 18 (20.3) | 15 (16.9) | |
| IA | 46 (51.7) | 43 (48.3) | |
| HAA | 25 (28) | 31 (34.8) | |
Abbreviations:1BM, bone marrow; 2WBC, white blood cell; 3HB, hemoglobin; 4PLT, platelet counts; 5FAB, French–American–British classification systems. 6 The protocols used for induction therapy in different groups including donorubicin/Ara-C (DA)-based treatment group, idarubicin/Ara-C (IA)-based, and homoharringtonine/Ara-C/aclarubicin (HAA)-based treatment group.
Table 2.
Multivariable analysis for clinical outcome for patienets with AML(OS).
| Overll survival |
||
| Variables | P-value | HR (95%CI) |
| MAP4K1 expression (High vs Low) | 0.033 | 1.686(1.043,2.726) |
| Age | 0.063 | 1.016(0.999,1.032) |
| WBC | 0.525 | 1.001(0.997,1.006) |
| BM blast | 0.039 | 1.013(1.001,1.026) |
| Karyotype-risk group | ||
| Intermediate vs. favorable | 0.646 | 0.757(0.230,2.491) |
| Poor vs. favorable | 0.584 | 1.546(0.324,7.375) |
| Gene mutation | ||
| NPM1 | 0.243 | 0.700(0.385,1.273) |
| FLT3ITD | 0.040 | 1.747(1.026,2.975) |
| DNMT3a | 0.011 | 2.211(1.201,4.071) |
| IDH1 | 0.839 | 1.110(0.405,3.042) |
| IDH2 | 0.677 | 1.183(0.536,2.612) |
| Treatment protocols | ||
| IA vs.DA | 0.929 | 1.032(0.513,2.078) |
| HAA vs.DA | 0.823 | 0.910(0.397,2.084) |
HR hazard ratio;CI confidence interval;
*Multivariate analysis were performed by Cox proportional hazards models with the backward likehood stepwise procedures.
Table 3.
Multivariable analysis for clinical outcome for patienets with AML(EFS).
| Event-free survival |
||
| Variables | P-value | HR (95%CI) |
| MAP4K1 expression (High vs Low) | 0.009 |
1.786(1.159,2.752) |
| Age | 0.946 | 1.001(0.983,1.019) |
| WBC | 0.059 | 1.003(1.000,1.007) |
| BM blast | 0.423 | 1.005(0.993,1.016) |
| Karyotype-risk group | ||
| Intermediate vs. favorable | 0.325 | 0.590(0.207,1.688) |
| Poor vs. favorable | 0.966 | 1.030(0.264,4.022) |
| Gene mutation | ||
| NPM1 | 0.839 | 1.058(0.616,1.815) |
| FLT3ITD | 0.574 | 1.169(0.678,2.016) |
| DNMT3a | 0.001 | 2.480(1.432,4.295) |
| IDH1 | 0.459 | 0.715(0.294,1.740) |
| IDH2 | 0.247 | 1.511(0.751,3.038) |
| Treatment protocols | ||
| IA vs.DA | 0.384 | 0.773(0.433,1.379) |
| HAA vs.DA | 0.120 | 0.579(0.291,1.153) |
HR hazard ratio;CI confidence interval;
*Multivariate analysis were performed by Cox proportional hazards models with the backward likehood stepwise procedures.
Table 4.
Multivariable analysis for clinical outcome for patienets with AML(RFS).
| Relapse-free survival |
||
| Variables | P-value | HR (95%CI) |
| MAP4K1 expression (High vs Low) | 0.005 | 2.369(1.302,4.311) |
| Age | 0.007 | 1.034(1.009,1.058) |
| WBC | 0.370 | 1.003(0.997,1.008) |
| BM blast | 0.023 | 1.017(1.002,1.031) |
| Karyotype-risk group | ||
| Intermediate vs. favorable | 0.846 | 1.122(0.350,3.596) |
| Poor vs. favorable | 0.672 | 1.228(0.474,3.184) |
| Gene mutation | ||
| NPM1 | 0.898 | 0.951(0.441,2.050) |
| FLT3ITD | 0.414 | 1.360(0.650,2.848) |
| DNMT3a | 0.089 | 2.035(0.896,4.620) |
| IDH1 | 0.809 | 1.165(0.337,4.022) |
| IDH2 | 0.327 | 0.364(0.048,2.745) |
| Treatment protocols | ||
| IA vs.DA | 0.111 | 0.372(0.111,1.254) |
| HAA vs.DA | 0.390 | 0.366(0.037,3.613) |
HR hazard ratio;CI confidence interval;
*Multivariate analysis were performed by Cox proportional hazards models with the backward likehood stepwise procedures.
3 MAP4K1 is an independent risk factor of AML in vivo
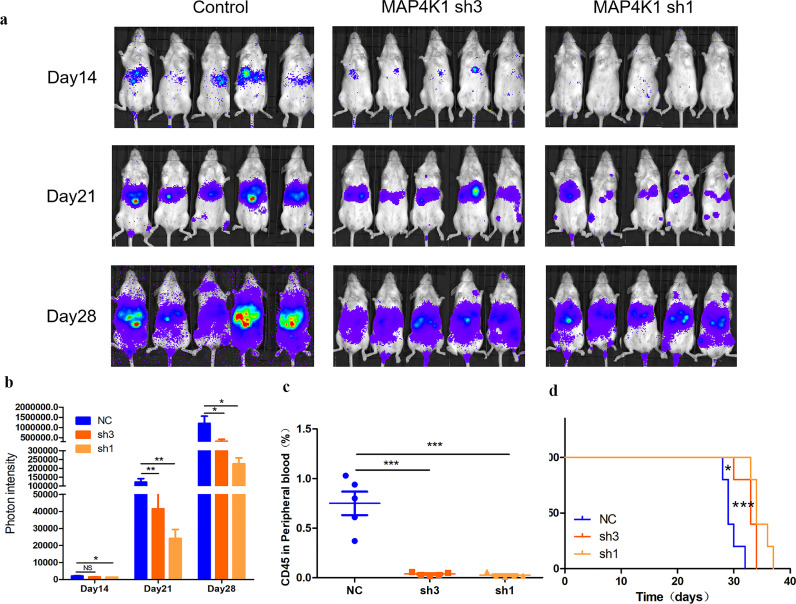
Findings from the previous section showed that MAP4K1 played a pro-leukemia role at the patient level. Therefore, we explored the effects of MAP4K1 in vivo by constructing an immunodeficient mice model. A total of 15 mice were divided into three groups. Mice in the three groups were injected with THP-1 Luci cells treated with MAP4K1 control, sh3, and sh1 vector, respectively. Progression of tumor load in mice was monitored for 4 weeks using bioluminescence imaging (Fig. 3a-b). The rate of human CD45+ cells in mice peripheral blood was detected by flow cytometry in the fourth week to evaluate the tumor load in the three groups (Fig. 3c). In MAP4K1 shRNA groups, tumor load was significantly lower compared with the tumor load of control groups. In addition, estimation of survival showed that MAP4K1 knockdown improved the survival time of mice (Fig. 3d). In summary, in vivo experiments showed that the expression of MAP4K1 was a prognostic factor in AML patients.
Fig. 3.
a Progression of the tumor load was followed up to 4 weeks by bioluminescence imaging of the three groups (MAP4K1 control, sh3, sh1, n = 5, each). b Photon intensity of the mice tumor load of the three group (n = 5, each) [two-tailed Student's t-tests; *P < 0.05, **P < 0.01]. c Levels of human CD45+ cells in mice peripheral blood during the fourth week were detected by flow cytometry of the three group (n = 5, each) [two-tailed Student's t-tests; ***P < 0.001]. d Survival time of the three group (n = 5, each) [two-tailed Student's t-tests; *P < 0.05, ***P < 0.001].
4 Changes in MAP4K1 expression levels modulate the growth of AML cells through inhibition of G0 / G1 phase
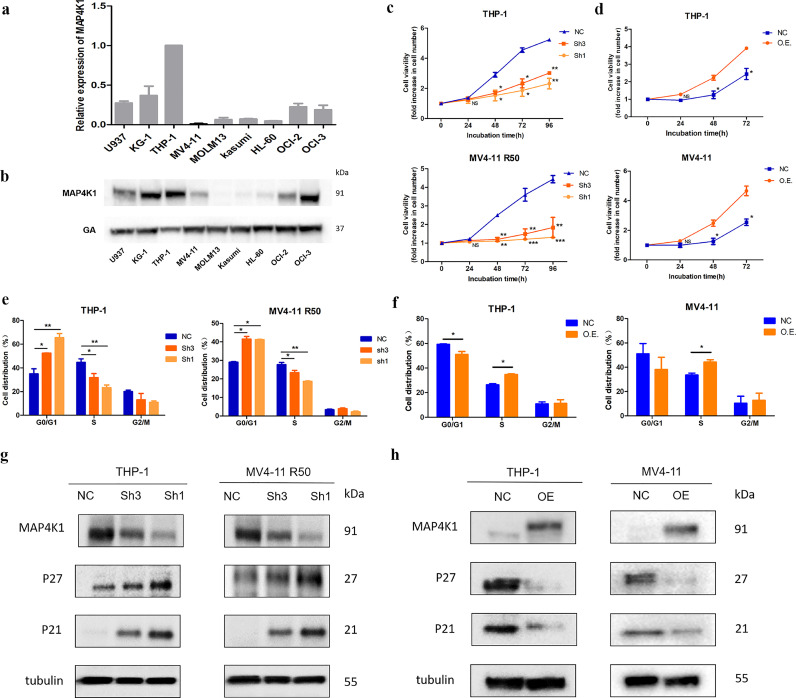
Expression levels of MAP4K1 in nine AML cell lines were determined using RT-qPCR and western blot. After analysis, we chose THP-1 and MV4-11 R50 cell lines for MAP4K1 knockdown and MV4-11 for MAP4K1 overexpression (Fig. 4a-b). To evaluate the role of MAP4K1 expression in AML progression, we altered MAP4K1 expression level by transfecting THP-1 and MV4-11 R50 with MAP4K1 short hairpin RNAs (shRNAs) for knockdown experiments or MAP4K1-PCDH lentivirus vector for overexpression experiments. We designed three shRNA sequences and selected the first and third sequences for subsequent experiments (Fig. S1a–d). As shown in the cell growth curve, cell proliferation showed a significant decrease in MAP4K1 knockdown cells compared with the cell proliferation levels in shScramble controls. On the contrary, elevated expression of MAP4K1 promoted proliferation of AML cells (Fig. 4c-d). Further, we performed flow cytometry to determine levels of cell apoptosis and cell cycle progression after MAP4K1 knockdown and overexpression in AML cell lines. MAP4K1 knockdown and overexpressed cells showed no significant change in apoptosis compared with the control group (Fig.S1e-f). MAP4K1 knockdown cells showed significant G0/G1 arrest and an increase in S phase was observed in MAP4K1 overexpressed cells (Fig. 4e-f). We determined the expression levels of cyclins by western blot. P21 and P27 were up-regulated in MAP4K1 knockdown group whereas in MAP4K1 overexpression group these two proteins were down-regulated (Fig. 4g-h). Notably, G0/G1 arrest can be induced by the upregulation of P21 and P27 [21]. Therefore, we concluded that MAP4K1 expression levels affected the growth of AML cells by blocking G0/G1 mainly via modulating expression levels of P21 and P27.
Fig. 4.
a Relative mRNA expression level of MAP4K1 in nine AML cell lines (expression of MAP4K1 in other cell lines compared with THP-1). b Westernblot analysis of the protein level of MAP4K1 in nine AML cell lines. c Cell viability after MAP4K1 knockdown in AML cell line THP-1 and MV4-11 R50 were measured by MTS assay [two-tailed Student's t-tests; *P < 0.05, **P < 0.01, ***P < 0.001]. d Cell viability after MAP4K1 overexpression in AML THP-1 and MV4-11 cell lines as determined by MTS assay [two-tailed Student's t-tests; *P < 0.05, **P < 0.01]. e The percentage of cells in the G1, S, or G2 phase in MAP4K1 knockdown cell lines for 48 h was detected by flow cytometry [two-tailed Student's t-tests; *P < 0.05, **P < 0.01]. f Percentage of cells in the G1, S, or G2 phase in MAP4K1 overexpressed cell lines for 48 h was detected by flow cytometry. g Cell lysates after MAP4K1 knockdown were collected, and levels of cell cycle-related proteins P21 and P27 were determined by Western blotting with the respective antibodies. h Cell lysates after MAP4K1 overexpression were collected, and the cell cycle-related proteins P21, P27 were examined by Western blotting with the respective antibodies. Western Blot bands were derived from three separate experiments but only one representative loading control is shown. Each value indicates the mean ± SEM of three or more independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
5 MAP4K1 modulates AML progression through MAPK & DNA damage/repair pathways
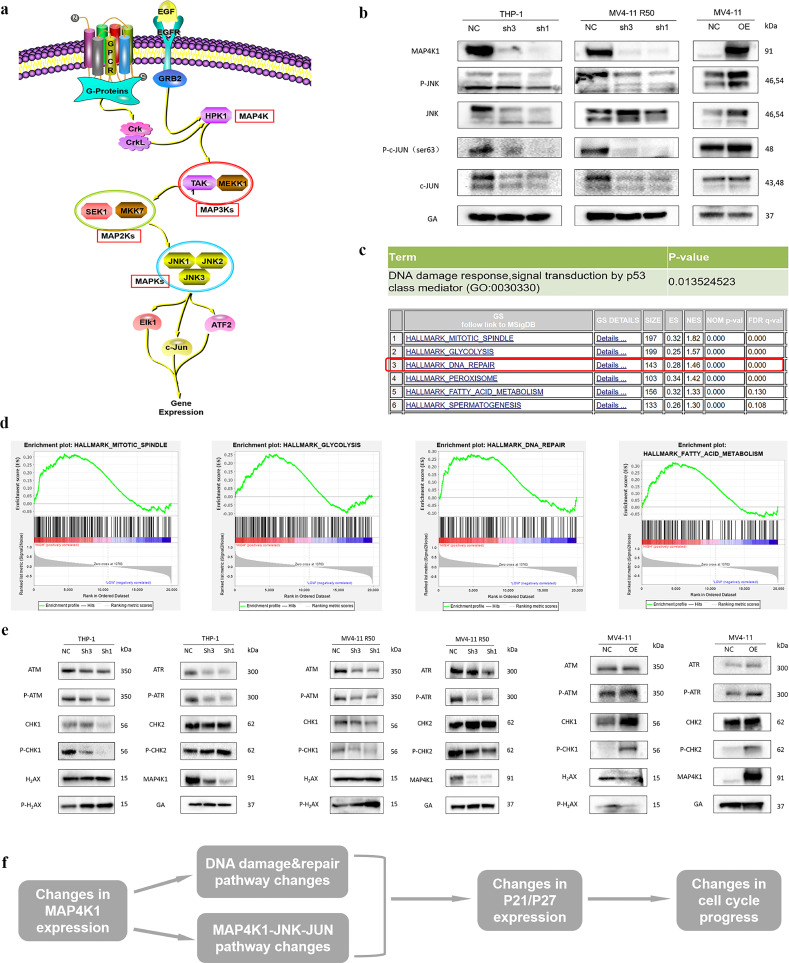
MAP4K1 is a MAP4 level kinase that transmits signals downstream to modulate the MAPK pathway. The cascade involves JNK and C-JUN genes which directly affect P21 functions (Fig. 5a) [22]. Western blot analysis showed that both total and phosphorylated JNK and C-JUN expression levels were greatly downregulated in MAP4K1 knockdown group. On the contrary, high levels of MAP4K1 upregulated total and phosphorylated JNK and C-JUN (Fig. 5b). Further, we performed GSEA and GO enrichment analysis to explore the specific regulatory mechanisms of MAP4K1 in AML. GSEA and GO enrichment analysis showed significant changes of DNA repair and damage pathways involved in the cell cycle after MAP4K1 overexpressed or knockdown. (Fig. 5c-d). Furthermore, genes coding for important DNA repair proteins like CHK1 were downregulated after MAP4K1 knockdown. In addition, DNA damage-inducible gene P-H2AX was induced after MAP4K1 knockdown. Conversely, overexpression of MAP4K1 showed enrichment of DNA repair pathways and reduction of DNA damage pathways in MV4-11 cell line (Fig. 5e). Moreover, DNA damage marker p-H2AX was positively correlated with molecule P21 [23]. In summary, MAP4K1 plays a role in AML progression through the MAPK & DNA damage/repair pathways. These pathways modulate the progression of cell cycle in AML through P21/P27 proteins and ultimately affect the progression of AML (Fig. 5f).
Fig. 5.
a Molecules in MAPK pathway (refer to ProteinLounge.com). b Protein levels of MAP4K1, JNK, C-JUN in AML cell lines. Total and phosphorylated JNK and C-JUN expression were downregulated after MAP4K1 knockdown while these two proteins were upregulated after MAP4K1 overexpression. c DNA repair and damage pathways were significantly dysregulated after MAP4K1 overexpression via bioinformatics analysis (UP panel: GO enrichment analysis, DOWN panel: GSEA analysis). d Several main pathways which had changed as shown by GSEA analysis. e Maker protein P-H2AX in DNA damage pathway up-regulated after MAP4K1 knockdown (e, left) and down-regulated after MAP4K1 overexpression (e, right) while DNA repair proteins have the opposite trend. f Possible mechanism of MAP4K1 in AML: MAP4K1 plays a biological role in AML through MAPK&DNA damage/repair pathways. These pathways modulated the progression of the cell cycle in AML through P21/P27 and ultimately influenced the progress of AML. Western Blot bands were derived from three separate experiments but only one representative loading control.
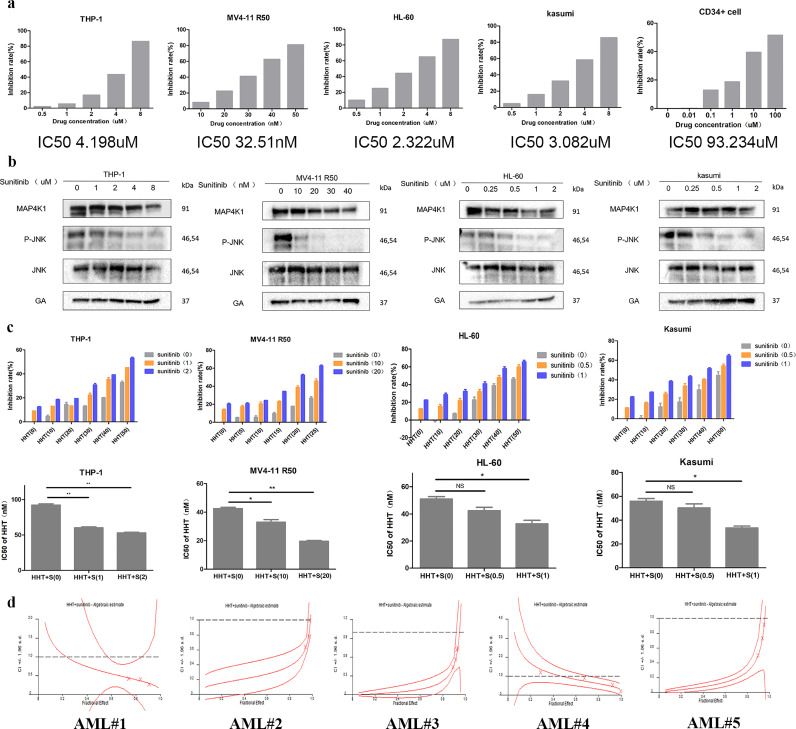
6 Sunitinib has a synergistic effect with HHT in AML
Sunitinib is a multi-receptor tyrosine kinase (RTK) inhibitor that binds to the kinase domain of MAP4K1 with high affinity [24]. First, we explored whether sunitinib also functions in AML and can regulate MAP4K1 in AML. We found an increase in sunitinib concentration decreased cell proliferation in four AML cell lines and, sunitinib showed good selectivity and less toxicity against normal human CD34+ cells (Fig. 6a). It has been reported that hematopoietic progenitor kinase 1 (HPK1 or MAP4K1) is a Ser/Thr kinase that operates via the c-Jun N-terminal kinase (JNK) and extracellular signal-regulated kinase (ERK) signaling pathways [24,25]. Firstly, the changes in downstream kinase JNK activity was examed after using MAP4K1 inhibitor (sunitinib) on four cell lines to select the appropriate drug concentration (The concentration can downregulate the activation of JNK but have a relatively minor effect on MAP4K1 expression) for downstream experiments (Fig. 6b). Following, we investigated the effect of sunitinib on sensitivity to HHT in AML. Firstly, THP-1 was treated with 0, 1, and 2uM sunitinib, MV4-11 R50 was treated with 0, 10, 20nM sunitinib, HL-60 was treated with 0, 0.5, 1uM sunitinib, Kasumi was treated with 0, 0.5, 1uM sunitinib for 24h. Then the IC50 values of these cells were determined after treatment with HHT (THP-1, MV4-11 R50, HL-60, and Kasumi were pretreated with sunitinib for 24h and then incubated with HHT for 24h). Treatment with sunitinib enhanced the inhibitory effect of HHT on cell viability (Fig. 6c). In addition, sunitinib and HHT had a synergistic effect on AML primary patient cells (Fig. 6d Tables 5-6) and AML cell lines (Fig. S2 Table 7).
Fig. 6.
a The AML cell proliferation slowed down after administration of MAP4K1 inhibitor sunitinib (left panel). Inhibition rate of sunitinib in CD34+ cell was detected by MTS and IC50 value and analysed using GraphPad prism (right panel). b Sunitinib binds to the kinase domain of MAP4K1 with high affinity and it can affect the main kinase activity downstream of the MAP4K1 in the four AML cell lines. c MTS assay was used to explore the sensitization of MAP4K1 inhibitor sunitinib to HHT in THP-1, MV4-11 R50, HL-60, and Kasumi cell lines (THP-1, MV4-11 R50, HL-60, Kasumi were pretreated with sunitinib for 24h and then incubated with HHT for 24h) [two-tailed Student's t-tests; *P < 0.05, **P < 0.01, ***P < 0.001]. d Combination effect of sunitinib and HHT on AML primary patient cells, MTS assay was performed once due to limited sample. The combination index (CI) was calculated using CalcuSyn software. Western Blot bands were derived from three separate experiments but only one representative loading control is shown. Each value indicates the mean ± SEM of three or more independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
Table 5.
Combination index value of HHT combined with sunitinib in AML patient cells.
| AML cell lines | Combination Index Values |
||
| ED50 ED75 ED90 | |||
| AML#1 | 0.63633 | 0.43349 | 0.30308 |
| AML#2 | 0.24064 | 0.34752 | 0.51013 |
| AML#3 | 0.06139 | 0.16814 | 0.48612 |
| AML#4 | 0.95910 | 0.66475 | 0.49205 |
| AML#5 | 0.10415 | 0.23024 | 0.51108 |
Table 6.
Characteristics of primary AML patients.
| Diagnose | Gender | Age(year) | FAB type | Cytogenetics | Molecular | |
| AML#1 | de novo | female | 33 | M4 | 46, XX | WT1 MLL-AF9 |
| AML#2 | de novo | male | 52 | M2 | 46, XY | BCR-ABL NPM1 |
| AML#3 | de novo | male | 50 | M5b | 46, XY | IDH2-R140Q |
| AML#4 | refractory | female | 59 | M4a | 46, XX | FLT3-ITD |
| AML#5 | refractory | male | 51 | M2a | 46, XY | IDH2 |
Table 7.
Combination index value of HHT combined with sunitinib in AML cell lines.
| AML cell lines | Combination Index Values |
||
| ED50 ED75 ED90 | |||
| THP-1 | 0.66106 | 0.57802 | 0.52817 |
| MV4-11 R50 | 0.82535 | 0.69637 | 0.58762 |
| MV4-11 | 0.82370 | 0.77211 | 0.75655 |
| HL-60 | 0.58978 | 0.47952 | 0.46336 |
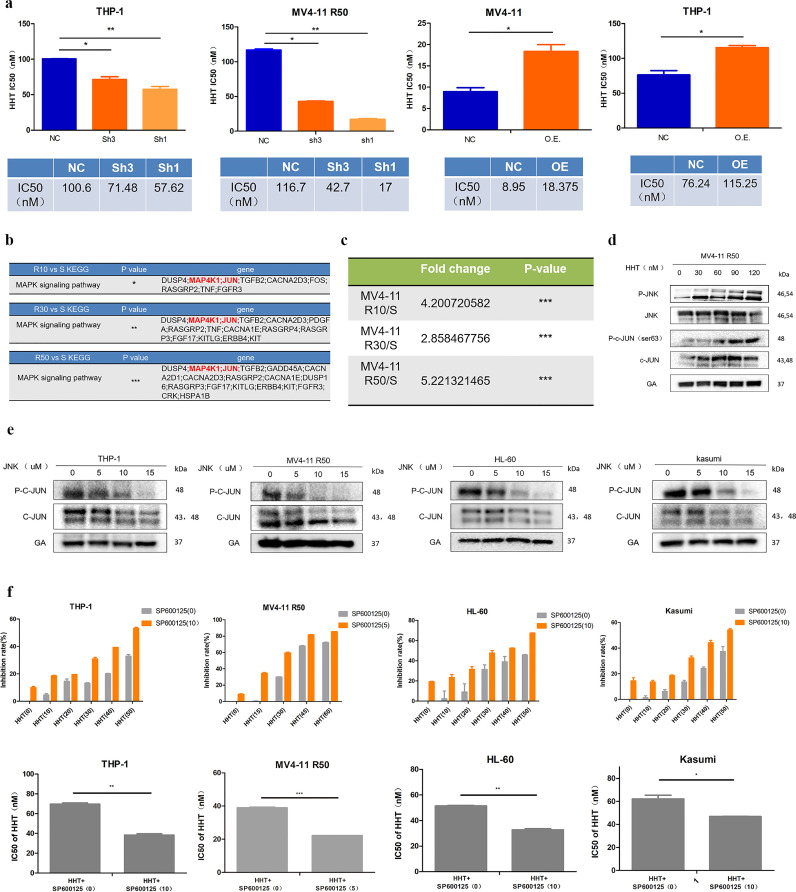
7 MAP4K1 modulates HHT resistance through regulation of MAPK pathway by JUN& JNK factors
MAP4K1 knockdown increased sensitivity of AML cells to HHT whereas overexpression of MAP4K1 increased resistance of AML cells to HHT (Fig. 7a). However, the change of MAP4K1 expression did not affect cell sensitivity to conventional chemotherapy drugs like Arac, IDA, and DNR (Fig. S3a-b). Subsequently, we explored the mechanism of MAP4K1 in modulating HHT resistance in AML. The KEGG analysis of RNA-seq data showed that the MAPK pathway including high expression levels of MAP4K1 and JUN was significantly up-regulated in the three HHT-resistant groups (Fig. 7b). In addition, JUN was significantly up-regulated in all three resistant strains (Fig. 7c). JUN is a transcription factor associated with tumorigenesis and drug resistance [26, 27]. This implied that MAP4K1 mediated HHT resistance in AML cells may mainly through the MAPK pathway by modulating JUN. Western blot showed an increase in JNK and JUN activation after the increase in HHT concentration (Fig. 7d). Thus, these two proteins may play a role in mediating HHT resistance. To explore the role of JNK and JUN in HHT resistance, a specific JNK inhibitor (SP600125) was used. First, we observed the changes in downstream JUN activity after using JNK inhibitor (SP600125) on four cell lines to select the appropriate drug concentration for downstream experiments (Fig. 7e). Next, 10, 5, 10, 10 uM JNK inhibitor (SP600125) were added to THP-1, MV4-11 R50, HL-60, and Kasumi cell lines separately and IC50 value of these cells were determined after treatment with HHT (THP-1, MV4-11 R50, HL-60, and Kasumi were pretreated with SP600125 for 24h and then incubated with HHT for 24h). Cell lines treated with JNK inhibitor were more sensitive to HHT and showed a lower HHT IC50 value (Fig. 7f). Drug combination experiments showed that JNK inhibitor and HHT had a high synergistic effect on resistant and sensitive cell lines (Fig. S3c). These results confirmed the importance of these two molecules in regulating HHT resistance. Combined with the previous finding that knockdown of MAP4K1 can down-regulate JNK and JUN (Fig. 5b), we concluded that MAP4K1 modulated HHT resistance through the regulation of JUN and JNK factors.
Fig. 7.
a THP-1, MV4-11 R50 AML cells became more sensitive to HHT after MAP4K1 knockdown whereas THP-1 and MV4-11 cell lines acquired resistance to HHT after MAP4K1 overexpression [two-tailed Student's t-tests; *P < 0.05, **P < 0.01]. b MAPK pathway was significantly up-regulated in the three groups (MV4-11 R10 vs MV4-11, MV4-11 R30 vs MV4-11, MV4-11 R50 vs MV4-11) [two-tailed Student's t-tests; *P < 0.05, **P < 0.01, ***P < 0.001]. c Fold change and p value of JUN expression in the three groups [two-tailed Student's t-tests; ***P < 0.001]. d JNK and C-JUN protein levels after treatment of MV4-11 R50 with 0, 30, 60, 90, 120 nM HHT for 48h. e Downstream JUN activity was detected by westernblot after administration of JNK inhibitor (SP600125) on the four AML cell lines. f MTS assay was used to evaluate sensitization of JNK inhibitor SP600125 to HHT in THP-1, MV4-11 R50, HL-60, and Kasumi cell lines (THP-1, MV4-11 R50, HL-60, and Kasumi were pretreated with sunitinib for 24h and then incubated with HHT for 24h) [two-tailed Student's t-tests; *P < 0.05, **P < 0.01, ***P < 0.001]. Western Blot bands were derived from three separate experiments but only one representative loading control is shown. Each value indicates the mean ± SEM of three or more independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
4. Discussion
The findings of this study showed that MAP4K1, an upstream MAPK modulator, regulated HHT resistance. In vitro experiments showed that overexpression of MAP4K1 decreased the sensitivity of AML cell lines to HHT. In addition, MAP4K1 was an independent predictor of poor AML outcome at the patient level and both in vitro and in vivo. These findings showed that MAP4K1 played a role as an independent risk factor and a mediator of HHT resistance.
MAP4K1, an upstream MAPK, is highly expressed in the lymphatic and myeloid lineage. Previous studies explored the function of MAP4K1 in lymphatic lineage, mainly focusing on the role of MAP4K1 on immune responses [28]. The associations between MAP4K1 expression and progression of several cancers have been discussed [10], however, the specific role of MAP4K1 in AML has not been explored. In this study, the prognostic role of MAP4K1 in AML at the patient level was explored for the first time. There are many factors that can affect the prognosis of the disease. However, in the prognostic part of our article, we only confirmed that MAP4K1 is indeed an independent prognostic risk factor, but we did not further explore whether the effect is related to drug resistance or disease relapse, or other factors. This part may need to be explored in the future. Furthermore, our results showed that downregulation of MAP4K1 slowed the proliferation of leukemia cells and lead to prolonged survival in vivo. MAP4K1 knockdown slowed AML cell progression through G0/G1 phase arrest via up-regulating P21, P27. (Fig. 4e,g). A recent study also showed that restoration of wild-type HPK1 in pancreatic ductal carcinoma cells increased p21 and p27 expression levels leading to cell cycle arrest [29].
MAP4K1 induces SAPK/JNK-JUN pathway in several systems [25,30]. Notably, knockdown of MAP4K1 inhibited activation of JNK and JUN in THP-1 and MV4-11 R50 cell line (Fig. 5b). In addition, MAP4K1 knockdown upregulated P21 and P27 resulting in cell cycle arrest (Fig. 4g). Transcriptional run-off experiments by Shaulian et, al. showed that c-Jun is a potent negative regulator of p21 transcription [21]. These findings implied that knockdown of MAP4K1 can down-regulate the activities of JNK and JUN, then up-regulate the activation of P21 and P27, and ultimately modulated the progression of AML. Further, to clarify the specific regulatory pathways associated with MAP4K1 in AML, we used GSEA and KEGG to analyze differential molecules in high and low MAP4K1 expression groups (Fig. 5c-d). Data showed that DNA damage and repair pathways involved in the cell cycle process were significantly changed after MAP4K1 overexpressed and knockdown (31). Changes in downstream pathways caused by changes in MAP4K1 expression levels were consistent with the final phenotype. Therefore, MAP4K1 modulates AML progression mainly through JNK-JUN and DNA damage/repair pathways thus ultimately regulating the cell cycle. Our study is the first study to report the mechanism of MAP4K1 in the modulation of AML progression.
Moreover, to promote clinical application, the role of sunitinib, a non-specific but high-affinity inhibitor of MAP4K1 was explored. Sunitinib selectively killed AML cells and gradually inhibited the downstream kinase JNK activity of MAP4K1 in a concentration-dependent manner. Results from drug rescue experiments showed that treatment with sunitinib increased the sensitivity of AML cells to HHT. In addition, sunitinib and HHT showed a high synergistic effect on primary patient cell lines and AML cell lines. This study is the first one to report a novel activity of sunitinib on AML cell lines. Due to the high homogeneity of the MAP4K family, the search for specific inhibitors of MAP4K1 is challenging. Further studies showed be carried out to explore specific inhibitors of MAP4K1 for the development of novel therapies.
Further, we explored the role of MAP4K1 expression levels on the sensitivity of AML cells to HHT. MAP4K1 knockdown increased sensitivity of AML cells to HHT whereas overexpression of MAP4K1 increased resistance of AML cells to HHT (Fig. 7a). The change of MAP4K1 expression did not affect cell sensitivity to conventional chemotherapy drugs like Arac, IDA, and DNR (Fig. S3a-b). However, our drug sensitivity test is limited to clinically commonly used drugs, while in clinical, the treatment is a combination of drugs and several schemes. So we can further discuss the function of MAP4K1 in other schemes in the future. Sequencing data revealed that the MAPK pathway included molecules MAP4K1 and c-JUN was significantly up-regulated in all three drug-resistant cell lines, and it became more obvious as the drug resistance index increased. Notably, an increase in HHT concentration on drug-resistant strain MV4-11 R50 increased the activation of JNK and c-JUN factors (Fig. 7d). Therefore, the activation of the MAPK pathway, especially downstream JNK and c-JUN factors played an important role in regulating HHT resistance. Similar findings were obtained in drug rescue and combination experiments (Figs. 7f & S3c). In this study, we identified an important pathway through which MAP4K1 regulated HHT resistance and reported important regulatory molecules in this pathway. Further studies should be carried out to explore the role of c-JUN in the regulation of HHT sensitivity and the corresponding regulatory network.
In this study, we reported that MAP4K1 can be an independent prognostic marker of AML and a mediator for regulating HHT resistance. In addition, the findings of this study showed that the MAPK signaling pathway modulated AML survival and drug resistance. In summary, the findings of this study provided information on the function and mechanisms of MAP4K1. In addition, sunitinib and HHT as well as JNK inhibitor and HHT can be used as a novel combination therapy for clinical treatment of AML.
Declaration of Competing Interest
Authors declare that they have no competing interests.
Acknowledgments
Acknowledgments
This work was supported by grants numbers 81800199, 81670124, 82070118 from the National Natural Science Foundation of China (NSFC); grant number LY20H080008 from the Natural Science Foundation of Zhejiang Province. We thank all the funding sources. We thank the patients who took part in donating leukemia specimens. We thank our laboratory members for their helpful discussions. The funders had no role in the study design, data collection, data analyses, interpretation, or writing of the manuscript.
Data sharing statement
The RNA sequencing data has been uploaded to NCBI's SRA, accession to cite for the SRA data: PRJNA664675. Data used for GSEA analysis were retrieved from the Cancer Genome Atlas (TCGA) (https://tcga-data.nci.nih.gov/).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103441.
Appendix. Supplementary materials
References
- 1.Burnett A., Wetzler M., Löwenberg B. Therapeutic advances in acute myeloid leukemia. J Clin Oncol. 2011;29(5):487–494. doi: 10.1200/JCO.2010.30.1820. [DOI] [PubMed] [Google Scholar]
- 2.Short N.J., Konopleva M., Kadia T.M., Borthakur G., Ravandi F., DiNardo C.D. Advances in the treatment of acute myeloid leukemia: new drugs and new challenges. Cancer Discov. 2020;10(4):506–525. doi: 10.1158/2159-8290.CD-19-1011. [DOI] [PubMed] [Google Scholar]
- 3.Kantarjian H.M., Talpaz M., Santini V., Murgo A., O'Brien SM. Homoharringtonine: history, current research, and future direction. Cancer. 2001;92(6):1591. doi: 10.1002/1097-0142(20010915)92:6<1591::aid-cncr1485>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 4.Jin J., Wang J.X., Chen F.F., Wu D.P., Hu J., Zhou J.F. Homoharringtonine-based induction regimens for patients with de-novo acute myeloid leukaemia: a multicentre, open-label, randomised, controlled phase 3 trial. Lancet Oncol. 2013;14(7):599–608. doi: 10.1016/S1470-2045(13)70152-9. [DOI] [PubMed] [Google Scholar]
- 5.Quan C., Xiao J., Liu L., Duan Q., Zhu F. Protein kinases as tumor biomarkers and therapeutic targets. Curr Pharm Des. 2017;23(29) doi: 10.2174/1381612823666170720113216. [DOI] [PubMed] [Google Scholar]
- 6.Sebolt-Leopold J.S., Herrera R. Targeting the mitogen-activated protein kinase cascade to treat cancer. Nat Rev Cancer. 2004;4(12):937–947. doi: 10.1038/nrc1503. [DOI] [PubMed] [Google Scholar]
- 7.Lee Jr J.T., Mccubrey J.A. The Raf/MEK/ERK signal transduction cascade as a target for chemotherapeutic intervention in leukemia. Leukemia. 2002;16(4):486–507. doi: 10.1038/sj.leu.2402460. [DOI] [PubMed] [Google Scholar]
- 8.Bosman M.C., Schepers H., Jaques J., Brouwers-Vos A.Z., Quax W.J., Schuringa J.J. The TAK1-NF-κB axis as therapeutic target for AML. Blood. 2014;124(20):3130–3140. doi: 10.1182/blood-2014-04-569780. [DOI] [PubMed] [Google Scholar]
- 9.Platanias LC. Map kinase signaling pathways and hematologic malignancies. Blood. 2003;101(12):4667–4679. doi: 10.1182/blood-2002-12-3647. [DOI] [PubMed] [Google Scholar]
- 10.Yang H.S., Matthews C.P., Clair T., Wang Q., Baker A.R., Li C.C.H. Tumorigenesis suppressor Pdcd4 down-regulates MAP4K1 expression to suppress colon carcinoma cell invasion. Mol Cell Biol. 2006;26(4):1297–1306. doi: 10.1128/MCB.26.4.1297-1306.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J., Song L., Yang S., Zhang W., Lu P., Li S. HPK1 positive expression associated with longer overall survival in patients with estrogen receptor-positive invasive ductal carcinoma-not otherwise specified. Mol Med Rep. 2017;16(4):4634–4642. doi: 10.3892/mmr.2017.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiangwen C.-.D., George P. Studzinski. dual role of hematopoietic progenitor kinase 1 (HPK1) as a positive regulator of 1α,25-dihydroxyvitamin D-induced differentiation and cell cycle arrest of AML cells and as a mediator of vitamin D resistance. Cell Cycle. 2014;11(7):1364–1373. doi: 10.4161/cc.19765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J.H., Chen W.L., Li J.M., Wu S.F., Chen T.L., Zhu Y.M. Prognostic significance of 2-hydroxyglutarate levels in acute myeloid leukemia in China. Proc Natl Acad Sci USA. 2013;110(42):17017–17022. doi: 10.1073/pnas.1315558110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Donnell M.R., Abboud C.N., Altman J., Appelbaum F.R., Arber D.A., Attar E. Acute myeloid leukemia. Hematol Am Soc Hematol Educ Program. 1987;149(29):1959–1960. [Google Scholar]
- 15.Ma Q.L., Wang J.H., Wang Y.G., Hu C., Jin J. High IDH1 expression is associated with a poor prognosis in cytogenetically normal acute myeloid leukemia. Int J Cancer. 2015;137(5) doi: 10.1002/ijc.29395. [DOI] [PubMed] [Google Scholar]
- 16.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;(W1):W1. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang Z., Kang B., Li C., Chen T., Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nuclc Acids Res. 2019;(W1):W1. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;(W1):W90–WW7. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A., Kuehn H., Gould J., Tamayo P., Mesirov JP. GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics. 2007;23(23):3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- 20.Shak Ed Y. The pro-tumorigenic host response to cancer therapies. Nat Rev Cancer. 2019;19(12):667–685. doi: 10.1038/s41568-019-0209-6. [DOI] [PubMed] [Google Scholar]
- 21.Bertoli C., Skotheim J.M., de Bruin RA. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol. 2013;14(8):518–528. doi: 10.1038/nrm3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaulian E., Schreiber M., Piu F., Beeche M., Wagner E.F., Karin M. The mammalian UV response: c-Jun induction is required for exit from p53-imposed growth arrest. Cell. 2000;103(6):897–908. doi: 10.1016/s0092-8674(00)00193-8. [DOI] [PubMed] [Google Scholar]
- 23.Fragkos M., Jurvansuu J., Beard P. H2AX is required for cell cycle arrest via the p53/p21 pathway. Mol Cell Biol. 2009;29(10):2828–2840. doi: 10.1128/MCB.01830-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson E., Mctigue M., Gallego R.A., Johnson T.W., Timofeevski S., Maestre M. Multiple conformational states of the HPK1 kinase domain in complex with sunitinib reveal the structural changes accompanying HPK1 trans-regulation. J Biol Chem. 2019 doi: 10.1074/jbc.AC119.007466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiefer F., Tibbles L.A., Anafi M., Janssen A., Zanke B.W., Lassam N. HPK1, a hematopoietic protein kinase activating the SAPK/JNK pathway. EMBO J. 1996;15(24):7013–7025. [PMC free article] [PubMed] [Google Scholar]
- 26.Shaulian E. AP-1-the Jun proteins: oncogenes or tumor suppressors in disguise? Cell Signal. 2010;22(6):894–899. doi: 10.1016/j.cellsig.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 27.Ozanne B.W., Spence H.J., Mcgarry L.C., Hennigan RF. Transcription factors control invasion: AP-1 the first among equals. Oncogene. 2007;26(1):1–10. doi: 10.1038/sj.onc.1209759. [DOI] [PubMed] [Google Scholar]
- 28.Chuang H.C., Wang X., Tan TH. MAP4K family kinases in immunity and inflammation. Adv Immun. 2016;129:277–314. doi: 10.1016/bs.ai.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 29.H. W., X. S., C. L. Proteasome-mediated degradation and functions of hematopoietic progenitor kinase 1 in pancreatic cancer. Cancer Res. 2009;69(3) doi: 10.1158/0008-5472.CAN-08-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen YR. The c-Jun N-terminal kinase pathway and apoptotic signaling (review) Int J Oncol. 2000;16(4):651–662. doi: 10.3892/ijo.16.4.651. [DOI] [PubMed] [Google Scholar]
- 31.Lanz M.C., Dibitetto D., Smolka MB. DNA damage kinase signaling: checkpoint and repair at 30 years. EMBO J. 2019;38(18) doi: 10.15252/embj.2019101801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.