Abstract
Airborne spread of coronavirus disease 2019 (COVID‐19) by infectious aerosol is all but certain. However, easily implemented approaches to assess the actual environmental threat are currently unavailable. We present a simple approach with the potential to rapidly provide information about the prevalence of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) in the atmosphere at any location. We used a portable dehumidifier as a readily available and affordable tool to collect airborne virus in the condensate. The dehumidifiers were deployed in selected locations of a hospital ward with patients reporting flu‐like symptoms which could possibly be due to COVID‐19 over three separate periods of one week. Samples were analyzed frequently for both virus envelope protein and SARS‐CoV‐2 RNA. In several samples across separate deployments, condensate from dehumidifiers tested positive for the presence of SARS‐CoV‐2 antigens as confirmed using two independent assays. RNA was detected, but not attributable to SARS‐CoV‐2. We verified the ability of the dehumidifier to rapidly collect aerosolized sodium chloride. Our results point to a facile pool testing method to sample air in any location in the world and assess the presence and concentration of an infectious agent to obtain quantitative risk assessment of exposure, designate zones as “hot spots” and minimize the need for individual testing which may often be time consuming, expensive, and laborious.
Keywords: airborne, condensate, COVID‐19, dehumidifier, detection, SARS‐CoV‐2
Airborne spread of COVID‐19 by infectious aerosol is all but certain. This study presents a simple, elegant approach using compact dehumidifiers deployed in selected locations of a hospital ward to rapidly provide information about the prevalence of SARS‐CoV‐2 in the atmosphere. Our results point to a facile pool testing method to sample air in any location in the world. Coupled with better virus recovery, this approach may also be useful to track the emergence of virus variants.
1. INTRODUCTION
Since the emergence of the first case of coronavirus in Wuhan, China in December 2019, coronavirus disease 2019 (COVID‐19) has infected over 60 million people and claimed 1,412,328 lives worldwide with a staggering number of 12,754,013 affected individuals in the United States alone by November 24, 2020. The mortality rate is estimated to be around 1%, although these figures are not very accurate due to the lack of widespread testing and thereby under‐reported. The unavailability of rapid testing has severely hampered efforts to manage the disease and assess its risk of transmission. Furthermore, uncertainty about its mode of spreading has created much perplexity and resulted in incoherent and constantly changing guidelines (Lewis, 2020), creating public confusion and noncompliance. The case of mass infections from the Biogen conference, the Washington Choir (Hamner, 2020) and the Wuhan restaurant (Lu et al., 2020) are concrete evidence of the ease with which social contact can spread the virus. Even as the World Health Organization (2020) is evaluating the spread of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), understanding has evolved that the infection transmission mode is primarily respiratory through airborne transmission of aerosols (Prather et al., 2020). Due to the shared similarities between SARS‐CoV‐2 and other coronaviruses like the Middle East respiratory syndrome (MERS‐CoV) and severe acute respiratory syndrome (SARS‐CoV), both of which were found to be airborne and could be potentially transmitted to long distances, it is essential to investigate this feature of the new virus and mitigate any plausible risks. Based on aerodynamic analyses in hospital settings, there is growing evidence for airborne transmission of COVID‐19 that causes resurgence of infection in closed network topology (Chia et al., 2020; Guo et al., 2020; Liu et al., 2020; Ma et al., 2020; Morawska & Cao, 2020; Morawska, Tang, Bahnfleth, et al., 2020; Santarpia, Herrera, et al., 2020; Santarpia, Rivera, et al., 2020; Stadnytskyi et al., 2020). Recent findings from a study conducted in a hospital ward further confirmed aerosol‐based transmission of viable SARS‐CoV‐2 from air samples collected 2–4.8 m away from patients (Lednicky et al., 2020). This necessitates detection protocols to be in place for modeling strategic quarantining policies and to assess transmission dynamics. Hence, understanding of our day‐to‐day exposure risk to these lethal bioaerosols is vital to implement near real‐time interventions to prevent the spread of the virus as well as safeguard human health (Prussin & Marr, 2015). This is especially useful, as several reports indicating the spread of the virus through asymptotic and presymptomatic patients have surfaced (Furukawa et al., 2020; Lee et al., 2020). A testing device thus placed in areas of high footfall and capable of bypassing individual testing is an effective way of controlling the spread of the deadly disease. Therefore, a simple, robust method, capable of providing rapid and accurate results on SARS‐CoV‐2 exposure would be extremely impactful to slow the spread of the disease and cater to community health at large.
We hypothesized that collecting condensate from the atmosphere could provide a simple means of assessing viral load in the surroundings. To this end, we set up four portable dehumidifiers at various test locations around a hospital ward at the University of Maryland Medical Center in Baltimore and obtained condensate samples for viral load analysis on different dates (Figures 1 and S1). The condensate was sampled at three separate time periods between June 29–July 5, July 22–August 10, and September 3–10, with the last set of samples being collected in viral transport medium (VTM). VTM liquid Amies from Innovative Research was used for this study. This is a clear, colorless liquid and is negative for microbial growth after 48 h at 37°C. The VTM consists of 1× sterile Hanks balanced salt solution (HBSS) with calcium and magnesium ions, 2% heat‐inactivated fetal bovine serum, gentamicin sulfate (100 µg/ml) and amphotericin B (0.5 µg/ml).
Figure 1.
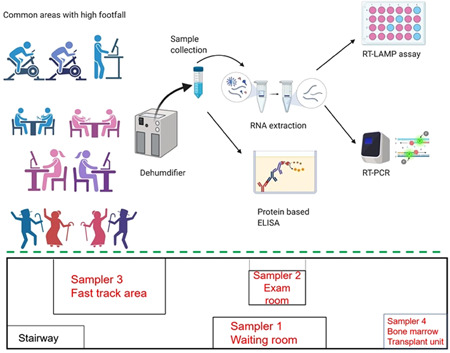
Schematic representation of sample collection and analysis for mass detection (top) and simplified layout of the hospital ward indicating the positions of various dehumidifiers during September 3–10, 2020 (bottom) [Color figure can be viewed at wileyonlinelibrary.com]
Our air sampling methodology can be a robust indicator of a potential contact pool of SARS‐CoV‐2 which can be used as a tool to implement strategies in a community bubble. Furthermore it may help in developing isolation strategies focusing on reducing disease burden thereby lowering morbidity and mortality. Thus, community mixing can be restricted through various social behavioral patterns by indirectly measuring the surge in COVID‐19 as opposed to observing confirmed cases (La Rosa et al., 2020), many of which could have been arrested beforehand. By employing this simple methodology (Figure 1) to monitor the presence of SARS‐CoV‐2 especially in areas with high human footfall or mass gatherings, appropriate preventive measures can be adopted to identify, track possible hotspots, and protect individuals from being infected. Among other uses, this type of monitoring can be used to enhance the effectiveness of vaccination strategies as they become available.
2. MATERIALS AND METHODS
Four identical 900 ml dehumidifiers (ICETEK B0863HNVNS from Amazon.com) were numbered 1–4 and deployed at the various sites indicated. These dehumidifiers use a muffin fan that draws room air past a Peltier‐cooled heat exchanger and deposits condensate into a tank underneath. While one dehumidifier was placed in the command center of the hospital to serve as a control, the other dehumidifiers were placed in staging areas involving the use of automated external defibrillators and powered air‐purifying respirator units. The condensate tanks were sampled at 24 or 48 h intervals and 50 ml samples were further processed for analysis. A three‐prong approach was followed for the identification of viral load in the collected condensate samples. All samples were deactivated in a water bath set at 65°C for 30 min, following which they were either stored at 4°C for protein detection using a protein enzyme‐linked immunosorbent assay (ELISA) kit or aliquoted and freeze‐dried for RNA‐based analysis employing commercially available RT‐LAMP and reverse‐transcription polymerase chain reaction (RT‐PCR) kits. It may be noted that the general targets towards the specific detection of SARS‐CoV‐2 are either the spike (S) or nucleocapsid (N) protein. We, therefore, used an ELISA assay targeted towards SARS‐CoV‐2 spike (S) protein and designed the primers for RT‐LAMP and RT‐PCR assay targeted towards SARS‐CoV‐2 N gene. Sampling lag, heat treatment, and 4°C storage time may well have impaired stability and hence the detection of S protein and N gene. As a parallel third detection technique, we employed a previously developed nano‐sensing platform from lanthanide‐doped carbon nanoparticles (LCNPs) which provide a distinct fluorescence response in presence of SARS‐CoV‐2 (Alafeef et al., 2019, 2020; Moitra et al., 2020). For samples collected in VTM, 50 ml of each sample was freeze dried and the residue redispersed in 2 ml of RNase free water and analyzed for the presence of RNA. Detailed procedures have been provided in the supporting information.
To further validate the ability of a dehumidifier to concentrate an aerosolized substance from the air, a cool‐mist humidifier (CVS Health) and an 1800 cubic feet dehumidifier (ICETEK) were first placed inside a small, sealed room. After the operation of only the dehumidifier overnight, the humidifier was turned on. After 2 h, 10 g of sodium chloride was added to the water inside the humidifier. Six hours later, the humidifier was turned off and removed from the room. Over the course of the entire experiment, a GSP‐6 temperature and humidity data logger (Elitech) was used to measure the humidity and temperature of the room and an Orion DuraProbe 4‐electrode conductivity probe (Thermo Fisher Scientific) connected to an Orion VersaStar Pro electrochemistry meter (Thermo Fisher Scientific) was used to measure the conductivity of the condensate collected inside the dehumidifier. The temperature was maintained at 22 ± 3°C.
3. RESULTS AND DISCUSSION
Water samples collected at the University of Maryland Medical Center between June 29–July 5, 2020 were first analyzed using a SARS‐CoV‐2 S‐protein ELISA kit. All the experiments were carried out at room temperature and samples were tested in duplicates. The mean value obtained was then utilized to determine the final S‐protein concentration. A calibration curve was initially generated using the known S‐protein concentrations (Figure S2) and S‐protein in the samples was then estimated (Table S1 and Figure 2). Only one sample presented a detectable dose of the virus SARS‐CoV‐2 S‐protein (dated July 5 from AED yellow zone by WGL214 door; S‐protein concentration 2.61 ng/ml) while others were well below the detection limit of the kit. Interestingly, the virus could be detected only when sampling was continued over the weekend in comparison with the daily sampling protocol. This indicated the requirement of concentrate sampling for the successful detection of the viral spike protein.
Figure 2.
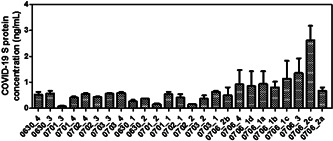
Concentration of SARS‐CoV‐2 S‐protein as determined by the ELISA assay for the samples collected over the period of June 29–July 5, 2020. The sample code starts with the date of sample collection from the hospital followed by the dehumidifier number, that is, 0630_4 indicates the water sample has been collected from dehumidifier number 4 on June 30, 2020. ELISA, enzyme‐linked immunosorbent assay; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
As mentioned earlier, we employed a nanosensing platform that was previously developed in our laboratory as a parallel detection technique (Alafeef et al., 2019, 2020; Moitra et al., 2020). This platform was applied to 17 condensate (water) samples collected as described previously. The sensor consists of lanthanide‐doped carbon nanoparticles (LCNPs) that provide a distinct fluorescence response towards the presence of SARS‐CoV‐2 specific viral protein (Table S2). The fluorescence responses obtained from the sensors were classified using a k‐mean clustering machine‐learning algorithm to identify the presence of SARS CoV‐2 (Alafeef et al., 2019). The clustered signature attributes were used to identify the pathogen type based on the commonalities in the data set (Moitra et al., 2017). The results obtained were also compared with those from the ELISA kit to confirm the reliability of the lanthanide sensor matrix.
Based on the promising results just described we next attempted to quantify the viral SARS‐CoV‐2 RNA in the water samples. Accordingly, RNA was extracted from all the samples (Table S3) and RT‐LAMP and RT‐PCR were performed to detect the presence of the viral SARS‐CoV‐2 RNA results obtained for the water samples indicated that no viral RNA was detected. This was attributed to either the low detection limit of the methods used or to deactivation or destabilization of SARS CoV‐2 RNA in the dehumidifier chamber. To remove the possibility of viral destabilization in the sampling method, we added 50 ml VTM to the dehumidifier chamber. This was done to ensure the stability of the viral RNA in the condensate.
Following sample collection in VTM, and although we were able to detect RNA in most of the samples (Table S4), both RT‐PCR and RT‐LAMP again did not detect viral RNA. Since sampling was performed at regular intervals and the condensate was collected as a whole instead of as fractions, this implies the presence of other detected RNAs alongside the viral RNA. Since VTM stabilizes RNA, we infer the cohabitation of all RNA types in the sampling chamber. This indicates that destabilization of viral RNA in the sampler is not the cause of the lack of viral RNA detection, but instead the cause is likely due to the relatively low sensitivity of the detection method used. To confirm this, we performed an experiment using gamma‐killed virions from BEI (sample NR‐52287, BEI Resources, NIAID, NIH, consists of a crude preparation of cell lysate and supernatant from Cercopithecus aethiops kidney epithelial cells (Vero E6; ATCC CRL‐1586) infected with SARS‐CoV‐2, isolate USA‐WA1/2020 that was gamma‐irradiated (5 × 106 RADs) on dry ice. The viral samples were diluted to similar concentrations as used for other samples obtained from the dehumidifier condensate. Two different concentrations were used and were spiked into the dehumidifier condensate. An RT‐LAMP experiment was then performed using these samples which showed an increase in emission at 520 nm confirming the presence of SARS‐CoV‐2 viral RNA (Figure S3). Hence, the discrepancy of RT‐LAMP results among the spiked and actual dehumidifier condensate samples can likely be attributed to the extensive dilution of the SARS‐CoV‐2 virus beyond the detection limit (0.75 copies/µL) of the RT‐LAMP assay used in case of the actual samples.
Interestingly, we were still able to detect the presence of S‐protein close to the minimum detectable limit (Figure 3). The sensitivity of the ELISA kit used (Protein ELISA from RayBiotech) is relatively lower than some of the recently available S‐protein based ELISA kits. However, at the time of conducting this study, which was in the early stages of the pandemic, the only commercially available kit was from RayBiotech. This kit, which was the one employed in this study, had a detection range of 2.7–2000 ng/ml and therefore a relatively low sensitivity of 2.7 ng/ml.
Figure 3.
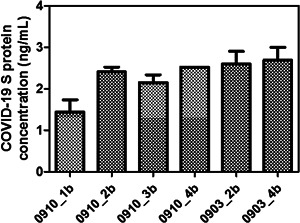
Determination of SARS‐CoV‐2 S‐protein concentration using protein‐based ELISA assay. The sample code starts with the date of sample collection from the hospital followed by the dehumidifier number, that is, 0910_1b indicates the water sample has been collected from dehumidifier number 1 on September 10, 2020. ELISA, enzyme‐linked immunosorbent assay; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
To further confirm our results, RT‐PCR, RT‐LAMP, and protein ELISA assays were performed with respect to positive and negative controls. For RT‐PCR and RT‐LAMP, quantitative PCR (qPCR) control RNA from heat‐inactivated SARS‐related coronavirus 2, isolate USA‐WA1/2020, NR 52347, obtained from BEI, was used as the positive control and RNAse free water was used as the negative control. For the protein ELISA assay, we used the SARS‐CoV‐2 spike protein provided with the kit as the positive control and assay buffer as the negative control. The standard curve, shown in Figure S2, was generated accordingly with the kit provided S‐protein. Thus, the positive results from the protein samples can be concluded to be true positives.
We are pursuing studies with different protein loads and contaminants generally encountered in a hospital environment. These samples will be aerosolized to assess interferences and to obtain data on false positives and negatives. Based on these studies, we will develop controls that will be used to assess accuracy and quantify false alarm rates. We are simultaneously working towards using Physics‐Informed Neural Networks (PINNs) (Raissi et al., 2019) and deep learning methods for error detection and standardization of the sampling protocol.
Furthermore, it can be expected that the risk of false positives and negatives depends upon a variety of diverse factors. For example, if the kit is used to perform either ELISA or RT‐PCR is not sensitive enough towards the target, then a false negative may occur. Pekosz et al. (2021), recently conducted a study that evaluated both RT‐PCR and antigen‐based COVID‐19 diagnosis using the conventional gold standard technique (i.e., virus culture in VeroE6TMPRSS2 cell). The study revealed that the antigen test demonstrated a higher positive predictive value (90%) than RT‐PCR (70%) when compared with the virus culture results. It is worth mentioning that this report supports the antigen tests over RT‐PCR, however, the RT‐PCR kit used in their study for comparison is not a kit with high sensitivity. Therefore, the choice of the kit could affect both sensitivity and specificity of the obtained results. In addition, if the RT‐PCR or ELISA technique is not performed following good molecular biology practices, carryover contamination might be observed in subsequent reactions resulting in false positive or false‐negative results.
The overall results of this study are summarized in Table 1. Most strikingly, SARS‐CoV‐2 viral protein was detected over some period in all the samplers. This could have implications for the efficacy of air filtration systems currently employed. Although airborne SARS‐CoV‐2 is widely implicated in the spread of COVID‐19, there is great uncertainty over the precise mechanisms of exposure and susceptibility. The viral load in the atmosphere presumably fluctuates depending on the actual shedding by the infected persons and their number. Our results cast a new light on this subject. We show that the simple technique of sampling condensate from a dehumidifier can provide evidence of the airborne virus. Given the widespread use of air‐conditioning equipment in homes and businesses worldwide, sampling their condensate provides a simple means of pool testing for virus presence analogous to those proposed for sewage monitoring. This approach also solves the major problem faced by conventional swab or saliva testing, where results can take several days. Antibody and point‐of‐care (POC) tests are more rapid but are geared towards individual patient testing and do not assess environmental airborne infection risk.
Table 1.
Summary of results
| Number of samples analyzed (Phase I without VTM) | 25 |
|---|---|
| Found positive using Protein ELISA | 1 (4% positive) |
| Found positive using Lanthanide Array | 5 (20% positive) |
| Found positive using RT‐LAMP | Not detected |
| Found positive using RT‐PCR | Not detected |
| Number of samples analyzed (Phase II with VTM) | 8 |
| Found positive using Protein ELISA | 5 (62.5% positive) |
| Found positive using RT‐LAMP | Not detected |
| Found positive using RT‐PCR | Not detected |
Note: Condensate samples collected during Phase I: June 29–July 5, July 22–August 10, and Phase II: September 3–10. Phase II samples included viral transport medium (VTM) in tank to stabilize any collected virus. RT‐LAMP and RT‐PCR analyses were performed on RNA isolated from samples; ELISA and Lanthanide array were performed directly on the samples.
Abbreviations: ELISA, enzyme‐linked immunosorbent assay; RT‐PCR, reverse‐transcription polymerase chain reaction; VTM, viral transport medium.
We further validated the ability of the dehumidifier to collect aerosolized sodium chloride. The results for the validation study on the collection of aerosolized substances from the air are shown in Figure 4. It can be seen that between the time sodium chloride was added to the water inside the humidifier and the time the humidifier was turned off, the conductivity of the condensate collected inside the dehumidifier increases with the humidity inside the test room. The correlation coefficient of the two variables was calculated to be 0.985, demonstrating that the dehumidifier is capable of collecting aerosolized components.
Figure 4.
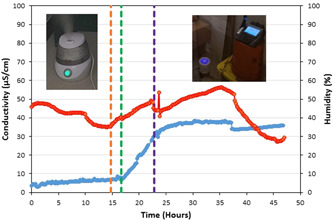
Plot of conductivity (blue line) and humidity (red line) inside the test room. The orange, green, and purple vertical dashed lines show the time the humidifier was turned on, the time sodium chloride was added to the water inside the humidifier, and the time the humidifier was turned off, respectively. Images showing this arrangement in the room can also be seen in this figure [Color figure can be viewed at wileyonlinelibrary.com]
Although RT‐LAMP and RT‐PCR‐based analyses did not detect the virus, as mentioned earlier this may be attributed to the dilution of the viral concentration in a large volume of media and inherent instability of the viral RNA in the further processing steps used. In support of this conclusion, past studies on wastewater sampling and detection indicate the low concentration of the virus to be a major limitation (La Rosa et al., 2020). The key to our approach is the ability to reliably integrate air sampling, virus capture, virus concentration, virus detection, and virus confirmation. By capturing virus from a known volume of air (specified by the room dimensions) and then measuring the amount of virus, it should be possible to determine the viral load and thereby assess infection risk in the hospital environment. There are three important parameters for this process: (1) the flow rate of air through the sampler; (2) the sampler run time; and (3) the amount of virus collected. The volume of air can be simply calculated by multiplying the flow rate through the sampler by the sampler run time. However, the capture efficiency is a function of not only the viral load but temperature, and humidity parameters in the sampling environment, in which case inferring the original amount of virus in the air from the amount of virus captured may be a source of a significant error in the method.
RT‐PCR has a limit of detection (LOD) of 6 copies/µl while RT‐LAMP has a corresponding value of 0.75 copies/µl. It may be presumed that these LOD values are above the detection limit required for analyses of the wastewater samples used here where the viruses are extensively diluted. Typical limits of detection required for wastewater analyses are in the range of 2 copies/100 ml–3×103 copies/ml (Foladori et al., 2020). In the current study, we did not have access to details concerning the persons in the hospital near our samplers. Instead, the focus of our study was on environmental monitoring of the viral load in different locations. In addition, any SARS‐CoV‐2 infected patients were possibly on closed‐circuit ventilators, and the efficiency of air exchanges in different locations of the hospital also varied. We intend to conduct further studies in the near future on aerosol collection from patient's breath, cough, and sneeze to confirm the viral load in those samples. Droplets naturally emanating from humans during respiration, speech and cough contain epithelial cells and immune system cells, inorganic ions (sodium, potassium, and chloride) present in mucous and saliva, and infectious load (bacteria, fungi, and virus). On the other hand, the droplets generated artificially in hospital settings have sterile water containing saline and pharmaceutical aerosols as the primary constituents. These factors too will be taken into account in our future studies (Atkinson et al., 2009).
Sampling in the current study was done at regular intervals of 24–72 h and the condensate stored at 4°C for further analyses. To ascertain the stability of the viral RNA, we used VTM for the latter phase of studies while maintaining the same sampling intervals. We were able to detect RNA in both phases of the study although the presence of SARS‐CoV‐2 viral RNA was not confirmed using both RT‐PCR and RT‐LAMP. We presume that this could be due to the extensive dilution of the viral RNA in the sample chamber and the limitations of the detection methods for wastewater samples. It is believed that the inherent instability of the viral RNA in different processing steps might not be the reason behind this failure in SARS‐CoV‐2 detection as is also supported by the recently published reports. The viral RNA remains detectable and does not degrade for up to 7 days or longer in VTM (Rogers et al., 2020). In fact, stability studies of the influenza virus A (H1N1) in a similar storage medium (PrimeStore MTM) indicate that viral RNA can be preserved and stabilized for up to 30 days under these conditions (Daum et al., 2011). Since the Coronavirus is an enveloped virus, its recovery rate from water samples is substantially lower than that of non‐enveloped viruses (Rusiñol et al., 2020). The major approaches to concentrate water samples include precipitation using polyethylene glycol (PEG), adsorption/elution, centrifugal ultrafiltration, aluminum hydroxide flocculation, and electronegative filtration (Ahmed et al., 2020; Hjelmsø et al., 2017). Recovery rates are also specific to the strain of the virus, their charge and hydrophobicity, and partition to solids. Despite these study limitations, our primary results present a novel method for air sampling in any resource‐limited settings across the globe. Coupled with sensitive and rapid assays that are being developed, there is the possibility of achieving near real‐time sensing of SARS‐CoV‐2 in the atmosphere, thereby providing an actionable threat assessment.
In light of the recent pandemic, most countries are struggling to strike a balance between protecting their residents and maintaining their economies. In such unprecedented times, the world has witnessed overburdening of healthcare facilities and increased risk of transmission via healthcare workers and in places with high human footfall. In an attempt to reduce the possibility of infection by adopting testing methods capable of producing effective and fast results in a cost‐effective manner, we have proposed herein a simple, facile, and affordable testing method for areas with high population density or footfall by avoiding laborious and time‐consuming individual testing. The use of dehumidifiers in designated areas would allow for analysis of the collected condensate in a rapid and facile manner, thus allowing authorities to designate zones as “hot spots” in case of a positive result. The method of sampling is both novel and effective, given the nature of transmission of coronaviruses and the unavailability of individual testing in many remote areas. As more sensitive tests are developed (Ogata et al., 2020), the approach outlined in this paper should lead to a novel, compact, and potentially wearable technology. It may also be useful for rapidly assessing the prevalence of variants in the sampled location, thereby assisting in epidemiological studies. Finally, there is still ongoing debate over the airborne nature of COVID‐19 transmission (Greenhalgh et al., 2021). This study, along with improved approaches for nucleic acid recovery may help resolve the issue.
CONFLICT OF INTERESTS
GR has filed a provisional patent application.
AUTHOR CONTRIBUTIONS
Study concept and design by Govind Rao, James Chang, Sai Sathish Ramamurthy, and Xudong Ge. Study design regarding the molecular testing of SARS‐CoV‐2 by Dipanjan Pan, Parikshit Moitra, Ketan Dighe, and Maha Alafeef. Product analysis and method development (protein and nucleic acid) by Dipanjan Pan, Parikshit Moitra, Maha Alafeef, Ketan Dighe, and Priyanka Ray. Sampling by James Chang, Parikshit Moitra, Ketan Dighe, and Priyanka Ray. Sample preparations by Ketan Dighe, and Priyanka Ray. Data analysis was done by Parikshit Moitra, Ketan Dighe, Priyanka Ray, Maha Alafeef, and Dipanjan Pan. Aerosolized salt collection experiments by Aaron Thole, Michael Tolosa, Benjamin Punshon‐Smith, and Douglas D. Frey. Manuscript written by Parikshit Moitra, Priyanka Ray, Maha Alafeef, Ketan Dighe, Dipanjan Pan, Sai Sathish Ramamurthy, and Govind Rao.
Supporting information
Supplementary information.
ACKNOWLEDGMENTS
We thank Dr. Bruce Jarrell for rapidly facilitating access to hospital sampling. The following reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: Quantitative PCR (qPCR) Control RNA from Heat‐Inactivated SARS‐Related Coronavirus 2, Isolate USA‐WA1/2020, NR 52347.
Moitra, P. , Alafeef, M. , Dighe, K. , Ray, P. , Chang, J. , Thole, A. , Punshon‐Smith, B. , Tolosa, M. , Ramamurthy, S. S. , Ge, X. , Frey, D. D. , Pan, D. , & Rao, G. (2021). Rapid and low‐cost sampling for detection of airborne SARS‐CoV‐2 in dehumidifier condensate. Biotechnology Bioengineering. 118, 3029–3036. 10.1002/bit.27812
Parikshit Moitra, Maha Alafeef, Ketan Dighe, and Priyanka Ray contributed equally to the study.
Contributor Information
Dipanjan Pan, Email: dipanjan@som.umaryland.edu.
Govind Rao, Email: grao@umbc.edu.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Ahmed, W. , Bertsch, P. M. , Bivins, A. , Bibby, K. , Farkas, K. , Gathercole, A. , & Kitajima, M. (2020). Comparison of virus concentration methods for the RT‐qPCR‐based recovery of murine hepatitis virus, a surrogate for SARS‐CoV‐2 from untreated wastewater. Science of the Total Environment, 739, 139960. 10.1016/j.scitotenv.2020.139960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alafeef, M. , Dighe, K. , & Pan, D. (2019). Label‐free pathogen detection based on yttrium‐doped carbon nanoparticles up to single‐cell resolution. ACS Applied Materials & Interfaces, 11(46), 42943–42955. 10.1021/acsami.9b14110 [DOI] [PubMed] [Google Scholar]
- Alafeef, M. , Moitra, P. , & Pan, D. (2020). Nano‐enabled sensing approaches for pathogenic bacterial detection. Biosensors and Bioelectronics, 165, 112276. 10.1016/j.bios.2020.112276 [DOI] [PubMed] [Google Scholar]
- Atkinson, J. , Chartier, Y. , Pessoa‐Silva, C. L. , Jensen, P. , Li, Y. , & Seto, W. (2009). Natural ventilation for infection control in health‐care settings. World Health Organization. https://www.who.int/water_sanitation_health/publications/natural_ventilation/en/ [PubMed] [Google Scholar]
- Chia, P. Y. , Coleman, K. K. , Tan, Y. K. , Ong, S. W. X , Gum, M. , Lau, S. K. , Lim, X. F. , Lim, A., S. , Sutjipto, S. , Lee, P. H. , Son, T. T. , Young, B. E. , Milton, D. K. , Gray, G. C. , Schuster, S. , Barkham, T. , De, P. P. , Vasoo, S. , Chan, M. , Ang, B. S. P. , Tan, B. H. , Leo, Y.‐S. , Ng, O.‐T. , Wong, M. S. Y. , & Marimuthu, K. (2020). Detection of air and surface contamination by SARS‐CoV‐2 in hospital rooms of infected patients. Nature Communications, 11(1), 1–7. 10.1038/s41467-020-16670-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum, L. , Worthy, S. , Yim, K. , Nogueras, M. , Schuman, R. , Choi, Y. , & Fischer, G. W. (2011). A clinical specimen collection and transport medium for molecular diagnostic and genomic applications. Epidemiology and Infection, 139(11), 1764–1773. 10.1017/S0950268810002384 [DOI] [PubMed] [Google Scholar]
- Foladori, P. , Cutrupi, F. , Segata, N. , Manara, S. , Pinto, F. , Malpei, F. , Bruni, L. , & La Rosa, G. (2020). SARS‐CoV‐2 from faeces to wastewater treatment: What do we know? A review. Science of The Total Environment, 743, 140444. 10.1016/j.scitotenv.2020.140444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa, N. W. , Brooks, J. T. , & Sobel, J. (2020). Evidence supporting transmission of severe acute respiratory syndrome coronavirus 2 while presymptomatic or asymptomatic. Emerging Infectious Diseases, 26(7), e1–e6. 10.3201/eid2607.201595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhalgh, T. , Jimenez, J. L. , Prather, K. A. , Tufekci, Z. , Fisman, D. , & Schooley, R. (2021). Ten scientific reasons in support of airborne transmission of SARS‐CoV‐2. The Lancet, 397(10285), 1603–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Z.‐D. , Wang, Z.‐Y. , Zhang, S.‐F. , Li, X. , Li, L. , Li, C. , & Chi, X.‐Y. (2020). Aerosol and surface distribution of severe acute respiratory syndrome coronavirus 2 in hospital wards, Wuhan, China, 2020. Emerging Infectious Diseases, 26(7), 1583–1591. 10.3201/eid2607.200885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamner, L. (2020). High SARS‐CoV‐2 attack rate following exposure at a choir practice‐Skagit County, Washington, March 2020. Morbidity and Mortality Weekly Report, 69(19), 606–610. 10.15585/mmwr.mm6919e6externalicon [DOI] [PubMed] [Google Scholar]
- Hjelmsø, M. H. , Hellmér, M. , Fernandez‐Cassi, X. , Timoneda, N. , Lukjancenko, O. , Seidel, M. , & Schultz, A. C. (2017). Evaluation of methods for the concentration and extraction of viruses from sewage in the context of metagenomic sequencing. PLoS One, 12(1), e0170199. 10.1371/journal.pone.0170199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lednicky, J. A. , Lauzardo, M. , Fan, Z. H. , Jutla, A. , Tilly, T. B. , Gangwar, M. , & Eiguren‐Fernandez, A. (2020). Viable SARS‐CoV‐2 in the air of a hospital room with COVID‐19 patients. International Journal of Infectious Diseases, 100, 476–482. 10.1016/j.ijid.2020.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. , Meyler, P. , Mozel, M. , Tauh, T. , & Merchant, R. (2020). Asymptomatic carriage and transmission of SARS‐CoV‐2: What do we know? [Patients asymptomatiques du SARS‐CoV‐2 et transmission du virus: Où en sont nos connaissances?]. Canadian Journal of Anaesthesia/Journal Canadien d' Anesthesie, 67(10), 1424–1430. 10.1007/s12630-020-01729-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, D. (2020). Is the coronavirus airborne? Experts can't agree. Nature, 580(7802), 175. 10.1038/d41586-020-00974-w [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Ning, Z. , Chen, Y. , Guo, M. , Liu, Y. , Gali, N. K. , & Westerdahl, D. (2020). Aerodynamic analysis of SARS‐CoV‐2 in two Wuhan hospitals. Nature, 582(7813), 557–560. 10.1038/s41586-020-2271-3 [DOI] [PubMed] [Google Scholar]
- Lu, J. , Gu, J. , Li, K. , Xu, C. , Su, W. , Lai, Z. , & Yang, Z. (2020). COVID‐19 outbreak associated with air conditioning in restaurant, Guangzhou, China, 2020. Emerging Infectious Diseases, 26(7), 1628–1631. 10.3201/eid2607.200764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J. , Qi, X. , Chen, H. , Li, X. , Zhan, Z. , Wang, H. , & Morawska, L. (2020). Exhaled breath is a significant source of SARS‐CoV‐2 emission. medRxiv. 10.1101/2020.05.31.20115154 [DOI] [Google Scholar]
- Moitra, P. , Alafeef, M. , Dighe, K. , Frieman, M. B. , Pan, D. (2020). Selective naked‐eye detection of SARS‐CoV‐2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano, 14, 7617–7627. 10.1021/acsnano.0c03822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitra, P. , Subramanian, Y. , & Bhattacharya, S. (2017). Concentration dependent self‐assembly of TrK‐NGF receptor derived tripeptide: new insights from experiment and computer simulations. The Journal of Physical Chemistry B, 121(4), 815–824. 10.1021/acs.jpcb.6b10511 [DOI] [PubMed] [Google Scholar]
- Morawska, L. , & Cao, J. (2020). Airborne transmission of SARS‐CoV‐2: The world should face the reality. Environment International, 139, 105730. 10.1016/j.envint.2020.105730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawska, L. , Tang, J. W. , Bahnfleth, W. , Bluyssen, P. M. , Boerstra, A. , Buonanno, G. , & Franchimon, F. (2020). How can airborne transmission of COVID‐19 indoors be minimised? Environment International, 142, 105832–105837. 10.1016/j.envint.2020.105832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata, A. F. , Maley, A. M. , Wu, C. , Gilboa, T. , Norman, M. , Lazarovits, R. , Mao, C. P. , Newton, G. , Chang, M. , Nguyen, K. , Kamkaew, M. , Zhu, Q. , Gibson, T. E. , Ryan, E. T. , Charles, R. C. , Marasco, W. A. , & Walt, D. R. (2020). Ultra‐sensitive serial profiling of SARS‐CoV‐2 antigens and antibodies in plasma to understand disease progression in COVID‐19 patients with severe disease. Clinical Chemistry, 66(12), 1562–1572. 10.1093/clinchem/hvaa213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekosz, A. , Parvu, V. , Li, M. , Andrews, J. C. , Manabe, Y. C. , Kodsi, S. , Gary, D. S. , Roger‐Dalbert, C. , Leitch, J. , & Cooper, C. K. (2021). Antigen‐based testing but not real‐time polymerase chain reaction correlates with severe acute respiratory syndrome coronavirus 2 viral culture. Clinical Infectious Diseases, ciaa1706. 10.1093/cid/ciaa1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prather, K. A. , Wang, C. C. , & Schooley, R. T. (2020). Reducing transmission of SARS‐COV‐2. Science, 368(6498), 1422–1424. 10.1126/science.abc6197 [DOI] [PubMed] [Google Scholar]
- Prussin, A. J. , & Marr, L. C. (2015). Sources of airborne microorganisms in the built environment. Microbiome, 3(1), 78. 10.1186/s40168-015-0144-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raissi, M. , Perdikaris, P. , & Karniadakis, G. E. (2019). Physics‐informed neural networks: A deep learning framework for solving forward and inverse problems involving nonlinear partial differential equations. Journal of Computational Physics, 378, 686–707. 10.1016/j.jcp.2018.10.045 [DOI] [Google Scholar]
- Rogers, A. A. , Baumann, R. E. , Borillo, G. A. , Kagan, R. M. , Batterman, H. J. , Galdzicka, M. M. , & Marlowe, E. M. (2020). Evaluation of transport media and specimen transport conditions for the detection of SARS‐CoV‐2 by use of real‐time reverse transcription‐PCR. Journal of Clinical Microbiology, 58(8), 1–5. 10.1128/JCM.00708-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa, G. , Iaconelli, M. , Mancini, P. , Bonanno Ferraro, G. , Veneri, C. , Bonadonna, L. , & Suffredini, E. (2020). First detection of SARS‐COV‐2 in untreated wastewaters in Italy. Science of the Total Environment, 736, 139652. 10.1016/j.scitotenv.2020.139652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiñol, M. , Martínez‐Puchol, S. , Forés, E. , Itarte, M. , Girones, R. , & Bofill‐Mas, S. (2020). Concentration methods for the quantification of coronavirus and other potentially pandemic enveloped virus from wastewater. Current Opinion in Environmental Science & Health, 17, 21–28. 10.1016/j.coesh.2020.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarpia, J. L. , Herrera, V. L. , Rivera, D. N. , Ratnesar‐Shumate, S. , Denton, P. W. , Martens, J. W. S. , & Lawler, J. V. (2020). The infectious nature of patient‐generated SARS‐CoV‐2 aerosol. MedRxiv. 10.1101/2020.07.13.20041632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarpia, J. L. , Rivera, D. N. , Herrera, V. L. , Morwitzer, M. J. , Creager, H. M. , Santarpia, G. W. , & Broadhurst, M. J. (2020). Aerosol and surface contamination of SARS‐CoV‐2 observed in quarantine and isolation care. Scientific Reports, 10(1), 1–8. 10.1038/s41598-020-69286-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadnytskyi, V. , Bax, C. E. , Bax, A. , & Anfinrud, P. (2020). The airborne lifetime of small speech droplets and their potential importance in SARS‐CoV‐2 transmission. Proceedings of the National Academy of Sciences of the United States of America, 117(22), 11875–11877. 10.1073/pnas.2006874117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . (2020). Modes of transmission of virus causing COVID‐19: implications for IPC precaution recommendations: scientific brief. WHO reference number: WHO/2019-nCoV/Sci_Brief/Transmission_modes/2020.2
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


