Abstract
The severe acute respiratory syndrome – coronavirus 2 (SARS‐CoV‐2), the infectious agent responsible for COVID‐19 – has caused more than 2.5 million deaths worldwide and triggered a global pandemic. Even with successful vaccines being delivered, there is an urgent need for novel treatments to combat SARS‐CoV‐2, and other emerging viral diseases. While several organic small molecule drug candidates are in development, some effort has also been devoted towards the application of metal complexes as potential antiviral agents against SARS‐CoV‐2. Herein, the metal complexes that have been reported to show antiviral activity against SARS‐CoV‐2 or one of its target proteins are described and their proposed mechanisms of action are discussed.
Keywords: antiviral agents, bioinorganic chemistry, coronavirus, COVID-19, metallodrugs
Metal complexes for COVID‐19 targets: SARS‐CoV‐2 has caused a global pandemic that has affected millions of people worldwide. Herein, metal complexes that have been reported to inhibit various SARS‐CoV‐2 enzymes and their proposed mechanism of action are discussed.
Introduction
On March 11, 2020, the World Health Organization (WHO) officially declared the outbreak of the severe acute respiratory syndrome – coronavirus 2 (SARS‐CoV‐2), the causative agent of COVID‐19, a global pandemic. [1] Fortunately, a considerable number of efforts have produced effective vaccines from efforts in both the pharmaceutical industrial and academia. [2] However, there remains an urgent need for the development of additional therapeutics to treat this disease. While vaccinations provide prophylactic protection for healthy adults, they may be less effective for individuals with compromised immune systems or other underlying medical conditions. In addition, the emergence of viral variants could undermine the effectiveness of current vaccines. [3] Indeed, the recently identified E484 K mutation in the receptor binding domain of the spike protein of a new SARS‐CoV‐2 strain has been shown to reduce vaccine effectiveness. [4]
Throughout 2020, numerous drug discovery and development campaigns have been initiated, involving the repurposing of known drugs, as well as the development of novel drugs. Several of these efforts have led to compounds that are currently being evaluated in clinical trials. [5] Most of these efforts are focused on conventional organic small molecules or antibody‐based therapies. [6] As an alternative, inorganic complexes, which can exhibit biological activity, have also been explored. [7] Notably, Messori [8] and Bergamini [9] have recently reported on the potential of metal complexes as antiviral agents, including for COVID‐19. Herein, metal complexes investigated as antiviral agents active against SARS‐CoV‐2 are critically discussed with focus on their proposed mechanism of action.
Viral Life Cycle of SARS‐CoV‐2
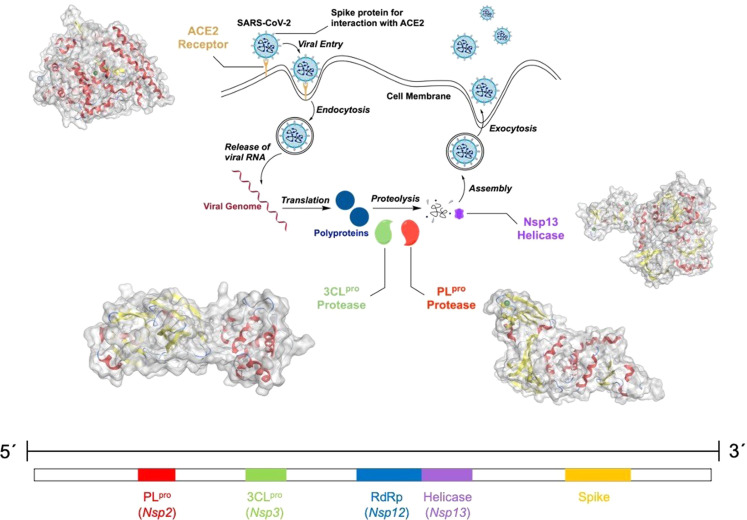
A simplified version of the viral life cycle and the viral genome of SARS‐CoV‐2 is presented in Figure 1. In the first step of viral infection of a cell, the receptor binding domain of the spike protein of SARS‐CoV‐2 binds at the cell surface protein angiotensin‐converting enzyme II (ACE2). Using the host cell transmembrane serine protease 2 (TMPRSS2), which is expressed in the human respiratory tract, the spike protein is primed and the virus internalized by clathrin‐mediated endocytosis. [10] Studies have indicated that the endosomal/lysosomal cysteine proteases cathepsin B and L can assist in this process, but are not essential. [11] The TMPRSS2 inhibitors Camostat Mesylate and Nafamostat Mesylate are currently being investigated in clinical trials to prevent viral cellular entry. [12]
Figure 1.
Top: Simplified viral life cycle of SARS‐CoV‐2. The therapeutic targets discussed here include the angiotensin‐converting enzyme II (ACE2, PDB: 2AJF), papain‐like protease (PLpro, PDB: 7NFV), the 3‐chymotrypsin‐like protease (3CLpro, PDB: 6Y2F) and helicase (Nsp13, PDB: 6ZSL) boxed in red. The secondary structure of the proteins is highlighted with α‐helices in red and β‐sheets in yellow. The Zn(II) ions in the crystal structures of ACE2, PLpro, and NSP13 are colored in green. Bottom: Single stranded RNA genome of SARS‐CoV‐2.
Following cellular uptake, the viral genome is released into the host cell cytoplasm. Upon translation of the open reading frames ORF1a and ORF1b from the genomic RNA, the polyproteins pp1a and pp1ab are generated and sixteen non‐structural proteins (Nsp) are co‐translationally released. Among these, Nsp3, also referred to as the papain‐like protease (PLpro), and Nsp5, also referred to as 3‐chymotrypsin‐like protease (3CLpro) or main protease, are used for proteolytic processing of the majority of the polyprotein cleavage sites. Interference with the activity of these proteases can disrupt the viral life cycle, providing additional targets for therapeutic intervention. [13] Clinical studies have demonstrated that coronavirus patients treated with protease inhibitors showed reduced symptoms and a decreased mortality rate. [14] The clinically‐approved compound Disulfiram has been proposed as a PLpro inhibitor and the compounds Lopinavir, Ritonavir, Darunavir, and Cobicistat are being investigated as 3CLpro inhibitors in ongoing clinical trials. [5a] Nsp2–11 were determined to be involved in the generation of the viral replication and transcription complexes by modulation of intracellular membranes, evasion of the host immune system, and providing cofactors for replication. In addition, Nsp12–16 are associated with RNA preparation, proofreading, and modification. [15] RNA synthesis is performed by Nsp12, the RNA‐dependent RNA polymerase (RdRp), and Nsp13 is a viral helicase. [16] RNA synthesis is crucial for viral replication and therefore presents yet another target for drug development. The compounds Favipiravir, Ribavirin, Penciclovir, Galidesivir, and Remdesivir are currently under investigation in clinical trials as inhibitors for the viral RNA synthesis machinery. [5a] Ultimately, the translated proteins assemble together and translocate to the endoplasmic reticulum and Golgi apparatus network. Following this, upon rearrangement of the viral content and sealing of the spherical capsid the virions mature and are released from the host cells by exocytosis. [17] Based on the different stages of the viral life cycle, several enzymes are considered possible therapeutic targets. In the following sections, metal complexes which have been studied as inhibitors for these enzymes are discussed.
Angiotensin‐Converting Enzyme II (ACE2)
ACE2 is a Zn‐dependent metalloenzyme that is attached to the membranes of cells found in the lungs, arteries, heart, kidneys, and intestines. [18] As a transmembrane protein, ACE2 was identified as a cellular entry receptor for SARS‐CoV‐2 via the viral spike protein. Consequently inhibition of the interaction of ACE2 with the spike proteins could potentially block viral entry into the host cells. [10] Consistent with this hypothesis, clinical studies on patients infected with SARS‐CoV‐2 and diagnosed with hypertension demonstrated a drop in mortality rate from 9.8 % to 3.7 % upon treatment with ACE2 inhibitors and angiotensin II receptor blockers, suggesting a connection between the blockage of viral cellular entry and therapeutic efficiency. [19]
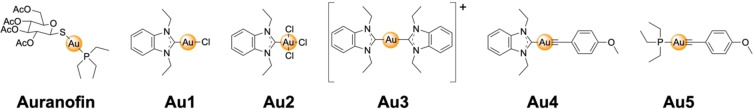
Au complexes functionalized with N‐heterocyclic carbene or alkynyl ligands Au1–Au5 as well as Auranofin (Figure 2) have been studied by Ott and co‐workers as inhibitors of ACE2. Using an enzyme‐linked immunosorbent assay (ELISA), the ability of the Au(I)/Au(III) compounds to inhibit binding of the spike protein receptor binding domain to the ACE receptor was quantified. The tested Au complexes were found to have a half maximal inhibitory concentration (IC50) in the micromolar range (IC50=16.2–25.0 μM). As described in the next section, these metal complexes were also inhibitors of the Papain‐like protease (PLpro) of SARS‐CoV‐2, presenting a possible multimodal mechanism of action. [20]
Figure 2.
Chemical structures of N‐heterocyclic carbene or alkynyl functionalized Au(I) (Auranofin, Au1, Au3–Au5) or Au(III) (Au2) complexes as angiotensin‐converting enzyme II (ACE2) or Papain‐like protease (PLpro) inhibitors.
Papain‐Like Protease (PLpro)
PLpro and 3CLpro are essential enzymes of SARS‐CoV‐2. These proteins are necessary for the cleavage of the viral polyproteins that regulate viral replication and transcription and influence viral maturation and infectivity. [13] In addition to the previsouly described inhibition of the ACE2 interaction with the spike proteins (Figure 1), Ott and co‐workers also tested the same Au complexes as inhibitors of the cysteine protease PLpro. Interestingly, while Au3 (IC50>100 μM) and Au4 (IC50>50 μM) did not inhibit PLpro in an enzymatic assay, the other Au complexes (Au1, Au2, Au5) demonstrated inhibition of PLpro in the low micromolar range (IC50=0.96–1.44 μM). Auranofin, a clinically approved antirheumatic agent, [21] showed slightly better inhibition in the high nanomolar range (IC50=0.75±0.13 μM). [20] Au(I) complexes are well known to be highly thiophilic and can form coordinate covalent adducts with thiol groups of cysteine residues. Some Au complexes have shown to bind to cysteine residues in metalloproteins, resulting in ejection of the metal ion from the coordination site. [22] Based on these findings, the authors investigated the ejection of Zn(II) ions from PLpro using a Zn‐selective fluorescent chelator. Although exposure to some of the Au(I)/Au(III) complexes did result in Zn(II) ejection from PLpro (as shown by a fluorescence assay using the Zn‐specific fluorophore FluoZin‐3), details of the amount of Zn(II) ejected (i. e., molar amount per PLpro protein) were not provided in this study. In addition, a substantial amount of Au(I)/Au(III) complex (50 μM), a 100‐fold excess over the concentration of PLpro protein (500 nM), was reported as necessary to achieve Zn(II) ejection. [20] The metal complexes with the strongest inhibitory activity were also found to result in the greatest amount of Zn ejection. Previous studies on Au1, Au3, and Au5 demonstrated that these compounds are cytotoxic, [23] which will likely prevent their further utility as SARS‐CoV‐2 therapeutics.
Beyond the ability to inhibit PLpro, Marzo and Messori proposed that Auranofin (Figure 2) could also act as a SARS‐CoV‐2 therapeutic [24] based on the known ability of this compound to inhibit redox enzymes such as thioredoxin reductase, the induction of endoplasmic reticulum stress, and the activation of the unfolded protein response in cells. [25] Studies have shown that endoplasmic reticulum stress and the activation of the unfolded protein response significantly contributes to viral replication and pathogenesis in coronaviruses. [26] Cells infected with coronaviruses were found to have an upregulated unfolded protein response. [27] It was hypothesized that Auranofin could potentially intervene by inhibition of redox enzymes and by causing endoplasmic reticulum stress, hampering SARS‐CoV‐2 protein synthesis. Indeed, a recent study by Rothan and Kumar showed a 95 % reduction of viral SARS‐CoV‐2 RNA (after 48 h) in human cells (Huh7) upon treatment with Auranofin at low micromolar concentrations (4 μM). [28] In light of these results, Auranofin may possess greater potential as an antiviral agent for SARS‐CoV‐2 when compared to other Au complexes.
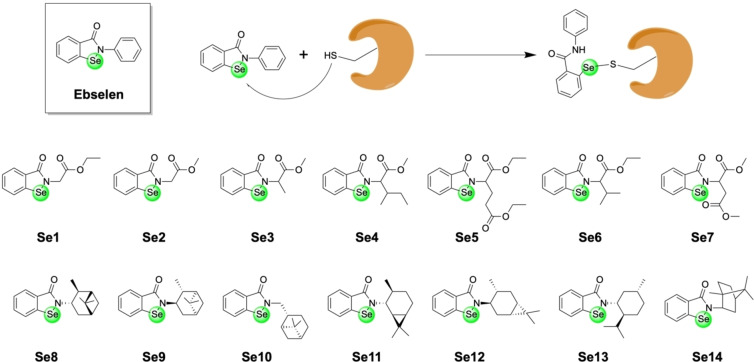
Ebselen (Figure 3) is an organoselenium drug with a wide range of reported biological activities, including anti‐inflammatory and anti‐oxidant activity. [29] It has not received FDA approval for use in humans for any indication, although it has been examined in a handful of Phase II and Phase III clinical trials. Although selenium is not a metal, it is classified as a metalloid and as such is included here as a compound of interest. Similar to the Au(I)/Au(III) compounds described above, Ebselen was also investigated as a potential Zn(II) ion ejector for PLpro. The compound showed activity against the PLpro enzyme (IC50 ∼2.4 μM), and even showed modest activity towards SARS‐CoV‐2 in infected kidney epithelial Vero E6 cells. Zn(II) ejection was monitored using the same Zn‐selective fluorophore (FluoZin‐3) with high equimolar concentrations of Ebselen and PLpro enzyme (5 μM each). Details of the amount of Zn(II) ejected per PLpro protein were not provided in this study. Using matrix‐assisted laser desorption ionization time‐of‐flight mass spectrometry (MALDI‐TOF), the covalent linkage of Ebselen to PLpro was investigated. Upon incubation with Ebselen, mass spectrometry experiments produced very broad peaks that shift towards higher molecular weights, which the authors attributed to the binding of one or even two molecules of Ebselen to PLpro; however, these data are not of sufficient resolution to definitively assign binding of Ebselen to the enzyme. [30] Computational experiments were presented that suggest that Ebselen could be covalently bound to the catalytic Cys111 residue in PLpro.[ 30 , 31 ] As shown below, Ebselen was also found to be an inhibitor of the 3‐chymotrypsin‐like protease (3CLpro) of SARS‐CoV‐2, presenting a possible multimodal mechanism of action. Despite these findings, the application of Ebselen as a potential SARS‐CoV‐2 therapeutic is limited as this compound is highly non‐specific and has been reported to bind to cysteine residues in many other enzymes, including chaperonin, heat shock protein 70, β‐tublin, vimentin, enolase I, and laminin receptor 1. [32]
Figure 3.
Top: Chemical structure of the organoselenium compound Ebselen and its proposed reaction with cysteine residues of proteins to form covalent adducts. If the cysteine residue is bound to a Zn(II) ion, reaction with Ebselen can result in ejection of the Zn(II) ion from the protein. Bottom: Amide functionalized derivatives of Ebselen investigated as inhibitors for PLpro and/or 3CLpro.
3‐Chymotrypsin‐Like Protease (3CLpro)
The 3CLpro protease, which is also referred to as the main protease (Mpro), is required for processing of the viral polyprotein and is therefore essential for viral replication and transcription. [13] While Ebselen was found to inhibit PLpro as described above, this compound was also identified in an enzymatic high‐throughput screening of a library of more than 10000 compounds to inhibit 3CLpro in the nanomolar range (IC50=0.67±0.09 μM). In an antiviral cellular assay using SARS‐CoV‐2 infected Vero E6 cells, Ebselen was shown to have a half‐maximal effective dose (EC50) in the micromolar range (EC50=4.67 μM). Using tandem protein mass spectrometry, the metal complex was found to form multiple adducts with the enzyme (with 1–2 Ebselen molecules bound); however, the majority of 3CLpro remained unmodified. [33] Computational modeling studies suggest that the compound could interact with the catalytic Cys145 residue.[ 30 , 34 ] Capitalizing on these findings, Santi and co‐workers made a series of Ebselen derivatives and screened them in an enzymatic 3CLpro assay (Figure 3). The majority of derivatives (Se1–Se3, Se5, Se7‐Se14) had similar or poorer activity than Ebselen (IC50=121.0±46.5 nM) against 3CLpro. Only compounds Se4 (IC50=149.5±18.1 nM) and Se6 (IC50=56.3±31.1 nM) were found to display modestly improved inhibition. A handful of the compounds in this study were further tested in a cell assay. While Se2 was found to have an IC50 value comparable to Ebselen, Se2 demonstrated a greater activity when compared to Ebselen in the cellular assay, suggesting that derivatization may be useful for obtaining more bioactive compounds in vivo. [35]
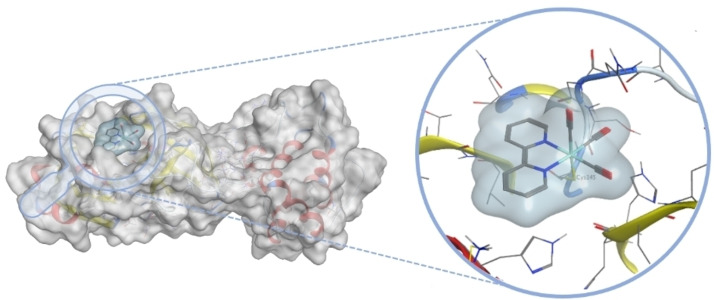
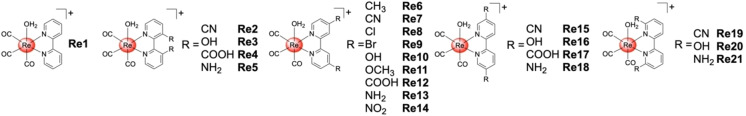
The Cohen group has recently reported Re(I) tricarbonyl complexes as inhibitors of 3CLpro. Using computational docking methods, 2,2'‐bipyridine Re(I) tricarbonyl complexes were identified as potential metal‐based fragments for binding to the catalytic Cys145 residue (Figure 4).Based on these docking studies, metal complexes with various functional groups on the 2,2'‐bipyridine scaffold were prepared with chloride or water molecules as labile ligands (Figure 5). To investigate the release of the labile chloride or water ligands in the presence of amino acids, the Re(I) tricarbonyl complexes were exposed to several different amino acids and their reactivity monitored. The metal complexes reacted completely with the thiol group of a cysteine, but did not react, or only slowly reacted with other polar (i. e., metal coordinating) amino acids. While the aqua complex was shown to react quickly with cysteine (within one hour), the analogous chloride compounds required days, demonstrating the importance of the labile ligand. Following this, the ability of such metal complexes to inhibit 3CLpro was investigated. All Re(I) tricarbonyl complexes Re1–Re21 showed activity at an inhibitor concentration of 50 μM. Further examination of the most potent compounds revealed that inhibition in the low micromolar range could be achieved (e. g., Re13, IC50=7.5±1.3 μM). Due to the reactivity of these compounds with sulfur containing biomolecules, the interaction with glutathione was investigated. Pre‐incubation of Re13 with glutathione (240 min), resulted in only a slight loss of activity (IC50=9.1±1.8 μM). The binding of these complexes to 3CLpro was investigated by electrospray ionization time‐of‐flight mass spectrometry (ESI‐TOF). While the native protein was found to have a well‐resolved mass of m/z 33797, upon incubation with Re1 for 2 h a new peak at m/z 34225 was observed, which precisely corresponds to a single attached Re(I) tricarbonyl complex. To identify the specific Cys residue modified by Re1, 3CLpro was first incubated with GC376, a known inhibitor that covalently modifies Cys145. Following incubation with GC376, the enzyme was then exposed to Re1. ESI‐TOF revealed a GC376‐protein adduct (m/z 34201) under these conditions, with no mixture of adducts and no addition of the Re1, suggesting that Re1 targets the same residue (Cys145). Finally, the selectivity of these Re(I) compounds towards the human proteases serine protease dipeptidyl peptidase‐4 (DPP4), aspartate protease beta‐secretase 1 (BACE1), and cysteine protease cathepsin B was evaluated. Lead compound Re13 did not inhibit DPP4 or cathepsin B (IC50>100 μM), while only weak inhibition of BACE1 (IC50=89.2±5.7 μM) was observed, indicative of some selectivity of the Re(I) tricarbonyl complexes for 3CLpro. [36] These findings are noteworthy, because previously reported organic covalent inhibitors of 3CLpro showed inhibition of cathepsins, particularly cathepsin B, resulting in potential off‐target activity and possible side effects. [37] This is particularly problematic as these enzymes are found in the respiratory cells that are infected by SARS‐CoV‐2. [38]
Figure 4.
Computationally predicted binding pose of the [Re(2,2'‐bipyridine)(CO)3]+ fragment bound to the thiol group of Cys145 of 3CLpro (PDB of 3CLpro: 6Y2F).
Figure 5.
Chemical structures of Re(I) tricarbonyl complexes as 3‐Chymotrypsin‐like protease (3CLpro) inhibitors. The metal complexes were isolated as triflate salts.
Helicase (Nsp13)
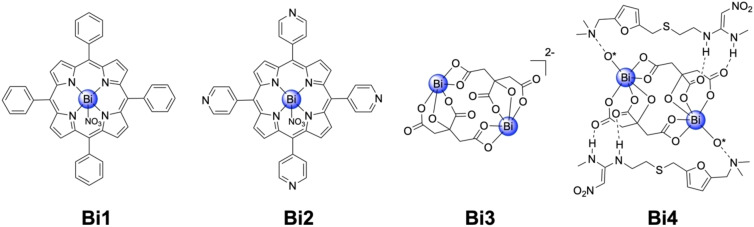
The RNA‐dependent RNA polymerase (RdRp, Nsp12) and the Helicase (Nsp13) are responsible for the viral replication of the genome. Studies have shown that the inhibition of the Zn‐dependent Nsp13 hampers these vital viral processes and as such presents a promising target for drug development. [16] Yuen and Sun demonstrated the potential of Bi(III) complexes Bi1–Bi4 (Figure 6) as replication inhibitors and therefore potential therapeutics for SARS‐CoV‐2. Using a fluorescence resonance energy transfer (FRET) based assay, the ability of several Bi(III) complexes to inhibit ATPase and to unwind DNA was investigated. The Bi(III) complexes were found to inhibit the activity of ATPase (IC50,ATPase=0.69–4.68 μM) and to unwind DNA (IC50,unwind=0.70–3.69 μM). Based on these results, the half maximal cytotoxic concentration (CC50) and EC50 values in infected Vero E6 cells was tested. All of the Bi(III) complexes demonstrated antiviral activity in infected Vero E6 cells in the low micromolar range (EC50=2.3–7.5 μM), with some achieving a high therapeutic index (707 for Bi3, 975 for Bi4, Figure 6). [39] As Bi(III) containing compounds have been previously shown to replace Zn(II) ions in metalloproteins and thereby reduce the activity of the corresponding metalloenzyme, [40] this was considered a possible mechanism of action for these compounds. Nsp13 was incubated with various concentration of Bi4 and the displacement of the three Zn(II) ions with Bi(III) measured using inductively coupled plasma mass spectrometry (ICP‐MS). Of note, several essential SARS‐CoV‐2 proteins are Zn(II)‐dependent, including Nsp10, Nsp12, Nsp13, Nsp14, and PLpro. In this case, the authors only studied Zn(II) displacement from Nsp13, but acknowledge that other Zn(II)‐dependent SARS‐CoV‐2 proteins could also undergo displacement by the Bi(III) complexes. Upon titration of Bi4 up to an excess of 10 equivalents compared to Nsp13, ∼2.9 Zn(II) ions were displaced, indicating that this may contribute to the mechanism of action. [39] The most potent compound was ranitidine bismuth citrate Bi4, which is a clinically approved drug for the treatment of Helicobacter pylori infections and peptic ulcers. [41] Bi4 was investigated in vivo for SARS‐CoV‐2 using a Syrian hamster model. The metal complex was intraperitoneally injected (15 mg/kg body weight) for four consecutive days in SARS‐CoV‐2 infected hamsters. While the animals in the control groups developed symptoms for the viral infection such as lethargy, ruffled fur, hunched back posture, and rapid breathing after 2 days, the animals treated with Bi4 did not show these symptoms. Consistent with this observation, hamsters treated with Bi4 showed an order of magnitude lower viral load than the untreated control group, confirming a reduction in SARS‐CoV‐2 viral load. [39]
Figure 6.
Chemical structures of Bi(III) complexes as helicase (Nsp13) inhibitors. The oxygen atoms marked with an asterisk in Bi4 belong to adjacent citrate molecules.
Summary and Outlook
Since its outbreak, the COVID‐19 infections have had a profound effect on all aspects of human life, health, and economic activity around the world. Based on the high infection and the mortality rates, there is an urgent need for novel therapeutics. While the majority of drug candidates under investigation are organic molecules and antibodies, some attention has also been devoted towards the use of metal complexes. Metal complexes offer distinct mechanisms of action when compared to organic compounds based on their rich molecular geometries, their ability to undergo ligand exchange reactions, and accessible redox processes. Based on the early results described in this review, it is expected that further efforts could lead to metal‐based inhibitors with improved affinity and selectivity, that ultimately may present an alternative to the more conventional therapeutics currently under investigation. Demonstration of potent and selective antiviral SARS‐CoV‐2 activity by a metal‐based compound would be an important validation for the viability of novel metallodrugs.
Conflict of interest
The authors declare no conflict of interest.
Biographical Information
Johannes Karges undertook his undergraduate studies at the Philipps‐University Marburg (Germany) where he was awarded a Deutschlandstipendium scholarship and the Imperial College London (United Kingdom) thanks to an Erasmus scholarship. He then joined the lab of Prof. Gilles Gasser at the École Nationale Supérieure de Chimie de Paris/Chimie ParisTech, Paris Sciences et Lettres University (France) to undertake a PhD thesis in bioinorganic chemistry. The focus of his work was the development of novel metal complexes as photosensitizers for photodynamic therapy and their selective delivery to tumor tissue. He did part of this work in the lab of Prof. Hui Chao at Sun Yat‐sen University (China). Currently, Johannes is a postdoctoral fellow in medicinal inorganic chemistry in the lab of Prof. Seth M. Cohen at the University of California, San Diego (USA). There, the focus of his work is the development of novel metal complexes as enzyme inhibitors.

Biographical Information
Seth M. Cohen studied at Stanford University (B.S., B.A.), the University of California, Berkeley (Ph.D.), and the Massachusetts Institute of Technology (postdoctoral fellowship). In 2001, he joined the Department of Chemistry and Biochemistry at the University of California, San Diego, where he has served on the faculty for ∼20 years, including three years as Chair of the Department (2012–2015). His research is focused on the development of metalloenzyme inhibitors, novel metal complexes as enzyme inhibitors, metal‐organic frameworks (MOFs), and MOF‐polymer hybrid materials.

Acknowledgements
This work was supported by National Institute of Health grant R21 AI138934.
J. Karges, S. M. Cohen, ChemBioChem 2021, 22, 2600.
Contributor Information
Johannes Karges, Email: jkarges@ucsd.edu.
Seth M. Cohen, Email: scohen@ucsd.edu.
References
- 1. https://www.who.int/director-general/speeches/detail/who-director-general–s-opening-remarks-at-the-media-briefing-on-covid-19-11-march-2020, last accessed on 10. 05. 2021.
- 2.
- 2a. Callaway E., Nature 2020, 588, 16–18; [DOI] [PubMed] [Google Scholar]
- 2b. Ita K., Arch. Med. Res. 2021, 52, 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.
- 3a. Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J. C. C., Muecksch F., Rutkowska M., Hoffmann H.-H., Michailidis E., Gaebler C., Agudelo M., Cho A., Wang Z., Gazumyan A., Cipolla M., Luchsinger L., Hillyer C. D., Caskey M., Robbiani D. F., Rice C. M., Nussenzweig M. C., Hatziioannou T., Bieniasz P. D., eLife 2020, 9, e61312; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3b. Greaney A. J., Loes A. N., Crawford K. H. D., Starr T. N., Malone K. D., Chu H. Y., Bloom J. D., Cell Host Microbe 2021, 29, 463–476.e466; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3c. Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C. O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J. A., Oliveira T. Y., Yang Z., Abernathy M. E., Huey-Tubman K. E., Hurley A., Turroja M., West K. A., Gordon K., Millard K. G., Ramos V., Da Silva J., Xu J., Colbert R. A., Patel R., Dizon J., Unson-O'Brien C., Shimeliovich I., Gazumyan A., Caskey M., Bjorkman P. J., Casellas R., Hatziioannou T., Bieniasz P. D., Nussenzweig M. C., Nature 2021, 592, 616-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jangra S., Ye C., Rathnasinghe R., Stadlbauer D., Krammer F., Simon V., Martinez-Sobrido L., García-Sastre A., Schotsaert M., MedRxiv 2021, 10.1101/2021.01.26.21250543. [DOI] [Google Scholar]
- 5.
- 5a. Li G., De Clercq E., Nat. Rev. Drug Discovery 2020, 19, 149–150; [DOI] [PubMed] [Google Scholar]
- 5b. Crosby J. C., Heimann M. A., Burleson S. L., Anzalone B. C., Swanson J. F., Wallace D. W., Greene C. J., JACEP Open 2020, 1, 231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.
- 6a. Morse J. S., Lalonde T., Xu S., Liu W. R., ChemBioChem 2020, 21, 730–738; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6b. Jawaid Akhtar M., Bioorg. Chem. 2020, 101, 104027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.
- 7a. Anthony E. J., Bolitho E. M., Bridgewater H. E., Carter O. W. L., Donnelly J. M., Imberti C., Lant E. C., Lermyte F., Needham R. J., Palau M., Sadler P. J., Shi H., Wang F.-X., Zhang W.-Y., Zhang Z., Chem. Sci. 2020, 11, 12888–12917; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7b. Boros E., Dyson P. J., Gasser G., Chem. 2020, 6, 41–60; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7c. Mjos K. D., Orvig C., Chem. Rev. 2014, 114, 4540–4563; [DOI] [PubMed] [Google Scholar]
- 7d. Karges J., ChemBioChem 2020, 21, 3044–3046; [DOI] [PubMed] [Google Scholar]
- 7e. Meier-Menches S. M., Gerner C., Berger W., Hartinger C. G., Keppler B. K., Chem. Soc. Rev. 2018, 47, 909–928; [DOI] [PubMed] [Google Scholar]
- 7f. Karges J., Stokes R. W., Cohen S. M., Trends Chem. 2021, 3, 523-534 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cirri D., Pratesi A., Marzo T., Messori L., Expert Opin. Drug Discovery 2021, 16, 39–46. [DOI] [PubMed] [Google Scholar]
- 9. de Paiva R. E. F., Marçal Neto A., Santos I. A., Jardim A. C. G., Corbi P. P., Bergamini F. R. G., Dalton Trans. 2020, 49, 16004–16033. [DOI] [PubMed] [Google Scholar]
- 10.
- 10a. Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F., Proc. Acad. Nat. Sci. 2020, 117, 11727–11734; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10b. Trougakos I. P., Stamatelopoulos K., Terpos E., Tsitsilonis O. E., Aivalioti E., Paraskevis D., Kastritis E., Pavlakis G. N., Dimopoulos M. A., J. Biomed. Sci. 2021, 28, 9; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10c. Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L., Nature 2020, 579, 270–273; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10d. Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., Hao P., Sci. China Life Sci. 2020, 63, 457–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N.-H., Nitsche A., Müller M. A., Drosten C., Pöhlmann S., Cell 2020, 181, 271–280.e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hoffmann M., Schroeder S., Kleine-Weber H., Müller M. A., Drosten C., Pöhlmann S., Antimicrob. Agents Chemother. 2020, 64, e00754–00720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.
- 13a. Chen Y., Liu Q., Guo D., J. Med. Virol. 2020, 92, 418–423; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13b. Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A., Schulz L., Widera M., Mehdipour A. R., Tascher G., Geurink P. P., Wilhelm A., van der Heden van Noort G. J., Ovaa H., Müller S., Knobeloch K.-P., Rajalingam K., Schulman B. A., Cinatl J., Hummer G., Ciesek S., Dikic I., Nature 2020, 587, 657–662; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13c. Sacco M. D., Ma C., Lagarias P., Gao A., Townsend J. A., Meng X., Dube P., Zhang X., Hu Y., Kitamura N., Hurst B., Tarbet B., Marty M. T., Kolocouris A., Xiang Y., Chen Y., Wang J., Sci. Adv. 2020, 6, eabe0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chu C. M., Cheng V. C. C., Hung I. F. N., Wong M. M. L., Chan K. H., Chan K. S., Kao R. Y. T., Poon L. L. M., Wong C. L. P., Guan Y., Peiris J. S. M., Yuen K. Y., Thorax 2004, 59, 252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.
- 15a.E. J. Snijder, E. Decroly, J. Ziebuhr, Adv. Virus Res., Vol. 96 (Ed.: J. Ziebuhr), Academic Press, 2016, pp. 59–126; [DOI] [PMC free article] [PubMed]
- 15b. V′Kovski P., Gerber M., Kelly J., Pfaender S., Ebert N., Braga Lagache S., Simillion C., Portmann J., Stalder H., Gaschen V., Bruggmann R., Stoffel M. H., Heller M., Dijkman R., Thiel V., eLife 2019, 8, e42037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.
- 16a. Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L. W., Wang Q., Lou Z., Rao Z., Science 2020, 368, 779–782; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16b. Lee N.-R., Kwon H.-M., Park K., Oh S., Jeong Y.-J., Kim D.-E., Nucleic Acids Res. 2010, 38, 7626–7636; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16c. Adedeji A. O., Singh K., Calcaterra N. E., DeDiego M. L., Enjuanes L., Weiss S., Sarafianos S. G., Antimicrob. Agents Chemother. 2012, 56, 4718–4728; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16d. White M. A., Lin W., Cheng X., J. Phys. Chem. Lett. 2020, 11, 9144–9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. V'kovski P., Kratzel A., Steiner S., Stalder H., Thiel V., Nat. Rev. Microbiol. 2021, 19, 155–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gheblawi M., Wang K., Viveiros A., Nguyen Q., Zhong J.-C., Turner A. J., Raizada M. K., Grant M. B., Oudit G. Y., Circ. Res. 2020, 126, 1456–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang P., Zhu L., Cai J., Lei F., Qin J.-J., Xie J., Liu Y.-M., Zhao Y.-C., Huang X., Lin L., Xia M., Chen M.-M., Cheng X., Zhang X., Guo D., Peng Y., Ji Y.-X., Chen J., She Z.-G., Wang Y., Xu Q., Tan R., Wang H., Lin J., Luo P., Fu S., Cai H., Ye P., Xiao B., Mao W., Liu L., Yan Y., Liu M., Chen M., Zhang X.-J., Wang X., Touyz R. M., Xia J., Zhang B.-H., Huang X., Yuan Y., Loomba R., Liu P. P., Li H., Circ. Res. 2020, 126, 1671–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gil-Moles M., Basu U., Büssing R., Hoffmeister H., Türck S., Varchmin A., Ott I., Chem. Eur. J. 2020, 26, 15140–15144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roder C., Thomson M. J., Drugs R&D 2015, 15, 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abbehausen C., Metallomics 2018, 11, 15–28. [DOI] [PubMed] [Google Scholar]
- 23.
- 23a. Rubbiani R., Can S., Kitanovic I., Alborzinia H., Stefanopoulou M., Kokoschka M., Mönchgesang S., Sheldrick W. S., Wölfl S., Ott I., J. Med. Chem. 2011, 54, 8646–8657; [DOI] [PubMed] [Google Scholar]
- 23b. Andermark V., Göke K., Kokoschka M., Abu el Maaty M. A., Lum C. T., Zou T., Sun R. W.-Y., Aguiló E., Oehninger L., Rodríguez L., Bunjes H., Wölfl S., Che C.-M., Ott I., J. Inorg. Biochem. 2016, 160, 140–148. [DOI] [PubMed] [Google Scholar]
- 24. Marzo T., Messori L., ACS Med. Chem. Lett. 2020, 11, 1067–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.
- 25a. Harbut M. B., Vilchèze C., Luo X., Hensler M. E., Guo H., Yang B., Chatterjee A. K., Nizet V., Jacobs W. R., Schultz P. G., Wang F., Proc. Acad. Nat. Sci. 2015, 112, 4453–4458; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25b. Thangamani S., Mohammad H., Abushahba M. F. N., Sobreira T. J. P., Seleem M. N., Int. J. Antimicrob. Agents 2016, 47, 195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.
- 26a. Fung T. S., Liu D. X., Front. Microbiol. 2014, 5, 296; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26b. Rothan H. A., Byrareddy S. N., J. Autoimmune Dis. 2020, 109, 102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sung S.-C., Chao C.-Y., Jeng K.-S., Yang J.-Y., Lai M. M. C., Virology 2009, 387, 402–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rothan H. A., Stone S., Natekar J., Kumari P., Arora K., Kumar M., Virology 2020, 547, 7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schewe T., Gen. Pharmacol. 1995, 26, 1153–1169. [DOI] [PubMed] [Google Scholar]
- 30. Sargsyan K., Lin C.-C., Chen T., Grauffel C., Chen Y.-P., Yang W.-Z., Yuan H. S., Lim C., Chem. Sci. 2020, 11, 9904–9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Węglarz-Tomczak E., Tomczak J. M., Talma M., Brul S., BioRxiv 2020, 10.1101/2020.05.17.100768. [DOI] [Google Scholar]
- 32. Sakurai T., Kanayama M., Shibata T., Itoh K., Kobayashi A., Yamamoto M., Uchida K., Chem. Res. Toxicol. 2006, 19, 1196–1204. [DOI] [PubMed] [Google Scholar]
- 33. Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L. W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H., Nature 2020, 582, 289–293. [DOI] [PubMed] [Google Scholar]
- 34. Ma C., Hu Y., Townsend J. A., Lagarias P. I., Marty M. T., Kolocouris A., Wang J., ACS Photonics 2020, 3, 1265–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sancineto L., Mangiavacchi F., Dąbrowska A., Pacuła A., Obieziurska-Fabisiak M., Scimmi C., Lei Y., Kong J., Zhao Y., dos Santos Machado K., Werhli A. V., Ciancaleoni G., Nascimento V., Kula-Pacurar A., Lenardao E. J., Yang H., Ścianowski J., Pyrc K., Santi C., ChemRxiv. 2020, 10.26434/chemrxiv.12994250.v1. [DOI] [Google Scholar]
- 36. Karges J., Kalaj M., Gembicky M., Cohen S. M., Angew. Chem. Int. Ed. 2021, 60, 10716–10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Steuten K., Kim H., Widen J. C., Babin B. M., Onguka O., Lovell S., Bolgi O., Cerikan B., Neufeldt C. J., Cortese M., Muir R. K., Bennett J. M., Geiss-Friedlander R., Peters C., Bartenschlager R., Bogyo M., ACS Infect. Dis. 2021, 10.1021/acsinfecdis.0c00815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lukassen S., Chua R. L., Trefzer T., Kahn N. C., Schneider M. A., Muley T., Winter H., Meister M., Veith C., Boots A. W., Hennig B. P., Kreuter M., Conrad C., Eils R., EMBO J. 2020, 39, e105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yuan S., Wang R., Chan J. F.-W., Zhang A. J., Cheng T., Chik K. K.-H., Ye Z.-W., Wang S., Lee A. C.-Y., Jin L., Li H., Jin D.-Y., Yuen K.-Y., Sun H., Nat. Microbiol. 2020, 5, 1439–1448. [DOI] [PubMed] [Google Scholar]
- 40.
- 40a. Wang R., Lai T.-P., Gao P., Zhang H., Ho P.-L., Woo P. C.-Y., Ma G., Kao R. Y.-T., Li H., Sun H., Nat. Commun. 2018, 9, 439; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40b. Cun S., Sun H., Proc. Acad. Nat. Sci. 2010, 107, 4943–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pipkin G. A., Mills J. G., Kler L., Dixon J. S., Wood J. R., Pharmacoepidemiol. Drug Saf. 1996, 5, 399–407. [DOI] [PubMed] [Google Scholar]