Abstract
The COVID‐19 pandemic has led to an unprecedented demand for real‐time surveillance data in order to inform critical decision makers regarding the management of the pandemic. The aim of this review was to describe how the Danish national microbiology database, MiBa, served as a cornerstone for providing data to the real‐time surveillance system by linkage to other nationwide health registries. The surveillance system was established on an existing IT health infrastructure and a close network between clinical microbiologists, information technology experts, and public health officials. In 2020, testing capacity for SARS‐CoV‐2 was ramped up from none to over 10,000 weekly PCR tests per 100,000 population. The crude incidence data mirrored this increase in testing. Real‐time access to denominator data and patient registries enabled adjustments for fluctuations testing activity, providing robust data on crude SARS‐CoV‐2 incidence during the changing diagnostic and management strategies. The use of the same data for different purposes, for example, final laboratory reports, information to the public, contact tracing, public health, and science, has been a critical asset for the pandemic response. It has also raised issues concerning data protection and critical capacity of the underlying technical systems and key resources. However, even with these limitations, the setup has enabled decision makers to adopt timely interventions. The experiences from COVID‐19 may motivate a transformation from traditional indicator‐based public health surveillance to an all‐encompassing information system based on access to a comprehensive set of data sources, including diagnostic and reference microbiology.
Keywords: electronic reporting, COVID‐19, microbiological test results, national surveillance
The severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) pandemic represents a challenge for nearly all parts of society, including the economy, trade, travel, health care, and social sectors, and has created an unprecedented need for real‐time health data. Healthcare managers, clinicians, clinical microbiologists, public health officials, researchers, and decision makers, including governments and international organizations, as well as media, all have expressed an urgent demand for surveillance data.
Infectious disease surveillance has been defined as “the ongoing systematic collection, analysis, interpretation, and dissemination of health data used for public health practice within a supporting legal framework” [1]. A well‐functioning surveillance system is representative of the population, comprehensive in scope, complete in content, timely, flexible, and economic and able to integrate information from a range of sources. Surveillance based on automated machine‐to‐machine (M2M) laboratory data reporting has been shown to be more effective, timely, and complete than conventional indicator‐based manual systems [2, 3]. A move toward M2M laboratory data reporting is in line with WHO and ECDC strategies and is taking place around the world. A recent European survey showed that electronic reporting is implemented in nearly half of the EU/EEA countries, including Denmark [2].
The Danish Microbiology Database (MiBa), established in 2010, is based on fully automated M2M laboratory data reporting. MiBa is unique as it provides nationwide access for individuals to their own microbiology test results and for clinicians to laboratory test reports for clinical purposes, while at the same time providing automated data transfer for surveillance purposes across all healthcare sectors in the country.
The aim of this review paper is to describe how MiBa is a cornerstone in the surveillance and control of the COVID‐19 pandemic in Denmark and in the provision of data and statistics for national and international stakeholders. By means of COVID‐19 data, generated from the MiBa‐based national surveillance system, we examine the course of the pandemic in Denmark to date and discuss how pandemic surveillance systems may be improved in the future.
Data Sources on SARS‐CoV‐2 Test Results in Denmark
In Denmark, persons with symptoms suggestive of COVID‐19, all patients requiring hospitalization or outpatient treatment for any reason and healthcare personnel, are tested in one of ten departments of clinical microbiology serving public and private hospitals and primary care. This workflow is referred to as the “Health care track”.
In addition, a centralized high throughput public COVID‐19 test laboratory, TestCenter Denmark (TCDK), was established by the end of April 2020. TCDK offers testing to asymptomatic persons and persons with mild symptoms. Test slots at the TCDK are made publicly available and can be booked online [4]. Furthermore, TCDK facilitates targeted testing efforts to specific population groups, for instance, healthcare staff working with vulnerable people and to employees in larger outbreaks at workplaces. TCDK is usually referred to as the “Community track” and was established due to an increased need of testing and contact tracing during the reopening of the country in the spring of 2020 (Table 1).
Table 1.
Management strategies and interventions to control the epidemic
| Time period 1 | Strategy | Comments |
|---|---|---|
| January to March 11, 2020 | Containment | The aim was to delay the introduction of SARS‐CoV‐2 with testing and isolation of possible cases and contact tracing. Some non‐pharmaceutical measures were adopted, including banning of large gatherings. |
| March 12 to April 2020 | Mitigation | A public lockdown in Denmark was instituted March 12. Schools, institutions for higher education, and daycare centers were closed. Workplaces were asked to introduce home working if possible. Restaurants, entertainment, sports facilities, and museums were closed. Adaptation of workplaces and public places to physical distancing and frequent use of hand hygiene have been an ongoing process ever since. |
| April to December 2020 | Suppression Or “Hammer & dance” |
Suppression of the epidemic with test, isolation and contact tracing and adapted non‐pharmaceutical interventions according to the situation. Late April: daycare centers and primary schools were reopened. May: Workplaces, worship places, secondary schools were reopened June: Entertainment and sport centers for sport were reopened in the beginning of June. Mid‐August schools and higher education institutions opened with eased restrictions, while most universities continued with online lectures. By the end of August, restaurants were again subjects to restrictions and one week later mandatory use of masks in closed places were introduced |
| December 2020 | Mitigation/suppression | Second wave: Due to increased transmission and increases in number of patients admitted to hospitals, a new lockdown was implemented gradually, starting with the capital area from December 7 to a national lockdown December 16, including closure of schools, shops and restaurants |
https://en.coronasmitte.dk/announcements/political‐agreements‐and‐initiatives [13].
Mobile sampling stations are also used to ensure coverage in places remote from permanent sampling stations and to assist in local outbreak responses by providing on‐site testing capacity.
Booking, testing, and reporting in both testing tracks are controlled and secured nationwide by using a unique personal identification number and a unique sample identifier, that permits data flow and integration across all healthcare sectors and the national surveillance systems at Statens Serum Institute (SSI) (Box 1).
Box 1. Principles of MiBa and data flows and nationwide access to test results.
During several decades, the healthcare system in Denmark has been developed to provide access to test results throughout the entire health sector, which is vital for the development of secure, efficient work processes and high standards of public health surveillance [32]. This includes facilitating the cooperation between authorities, organizations and private companies linked to the Danish healthcare sector, with special focus on solutions for electronic communication.
MiBa was launched in 2010 and is built on this framework and a close network between clinical microbiologists, information technology staff and public health officials. MiBa is established in collaboration between the regions and the state with funding from both sides and is governed within a framework for trans‐sectorial systems across the state, the regions and the municipalities.
Data from the 10 departments of clinical microbiology in Denmark, the national reference laboratory and TestCenter Denmark TCDK are transferred to and from MiBa via The Danish Health Data Network, which allows the health sector in Denmark to communicate and transfer data with connected organizations through one secure digital solution protecting sensitive personal information [3, 33].
The dramatic upscaling of the test capacity during the COVID‐19 pandemic was developed within this existing framework in which all test results are automatically transferred, processed, and accessible within already existing platforms.
The electronic requesting, testing, and reporting process, using PCR for Coronavirus SARS‐CoV‐2 as an example
Prior to testing a person for COVID‐19, an electronic requisition (ER) is created in the hospitals patient records or in the common Danish electronic laboratory request system WebReq (Fig. 1) [34]. The ER is created either by a general practitioner (GP) in the WebReq: Web‐based electronic request system (WebReq) or by a hospital physician. In the case of SARS‐CoV‐2 testing, individuals may also be tested in drive‐in clinics or at mobile units for this purpose, the ER is autogenerated in WebReq, allowing free access to testing for all persons.
During sampling, the specimen (e.g., a throat swab) is labeled with a unique identification number linking the sample to the ER. The ER is sent electronically to the laboratory information system (LIS). When the sample arrives at the laboratory, the matching ER is identified by the unique identification number and retrieved.
The samples are analyzed with PCR at either a department of clinical microbiology, at POCT‐equipment at the hospital, at an external laboratory connected with a department of clinical microbiology or at TCDK/SSI.
After analysis in the laboratory, the laboratory report is sent directly electronically from LIS to the electronic patient record of the requester, either the GP or the hospital. At the same time, a copy of the report is sent electronically to MiBa. Through system‐integrations to MiBa, the microbiology results can also be accessed from the electronic patient record at all hospitals and general practitioners throughout Denmark; people can access their own report through a secure website (www.sundhed.dk) or alternatively through a mobile app. All positive cases, not hospitalized or residents in a long‐time care facility is, in addition, contacted by the Danish Patient Safety Authority for contact tracing.
The system for requisition and reporting of laboratory analyses is based on a common national set of MedCom standards (EDIFACT REQ01 and OIO‐XML XREQ01 for requisition, EDIFACT RPT02 and OIO‐XML XRPT05 for reporting the results, and OIO‐XML XRPT06 for transfer of the results to MiBa) [35]. It is mandatory for all publicly funded hospitals and practitioners to communicate by this system. During the COVID epidemic, only minor changes in the process have been necessary to accommodate the huge escalation in testing activity.
MiBa holds highly structured and coded raw data from all diagnostic laboratory reports in Denmark. For use of data for surveillance purposes, new cases of a specified disease need to be identified from these unprocessed diagnostic microbiological data. This is done by disease specific data extraction and data processing algorithms. Each specified disease has its own diagnostic attributes and case definitions, so data algorithms need to be customized accordingly.
From the summer of 2020, private vendors have also offered COVID‐19 tests. Initially, a limited number of SARS‐CoV‐2 RNA PCRs and antigen tests for specific purposes, for example, for football teams or employers, have been performed in parallel with the free of cost, public testing system. To further increase the free of cost testing capacity, public funding was assigned to antigen testing provided by private vendors in December 2020. Test results from these private vendors were initially not captured by MiBa. From February 3, 2021, the first private vendors have been able to upload their test results, and from March 15, the 98 Danish municipalities have been able to upload tests result from screening in school settings. Antigen test results were included in the SSI surveillance system by March 10 [5].
The Danish Microbiology Database (MiBa) and the COVID‐19 Surveillance System
MiBa receives, in real‐time, copies of all laboratory test results from all clinical microbiology departments in Denmark, the national reference laboratories as well as TCDK (Box 1). A data upload module to MiBa for private laboratories has now been developed, and in early 2021, it became mandatory for private vendors to report electronically to MiBa.
Specific algorithms for retrieval of data from MiBa to identify new COVID‐19 cases were developed in March 2020 at the start of the epidemic. A COVID‐19 case was defined as “a person with at least one positive SARS‐CoV‐2 RNA test”. A test‐negative individual was defined as “a person with at least one negative and no positive SARS‐CoV‐2 RNA test”. New COVID‐19 cases were counted on the date of the first positive test result. Individuals tested negative were registered on every date the person is tested. Persons with more than a single test on the same date were counted only once.
For surveillance of COVID‐19, data on SARS‐CoV‐2 RNA tests are retrieved every hour from MiBa and processed to identify new cases of COVID‐19, as well as test‐negative individuals. Additional information on both positive cases and negative test is obtained by data linkage from a number of national registries, which all contain the unique identifier of all residents (see Fig. 1). These registries include the Civil Registration System, the National Patient Registry, the National Immunization Registry, the Register of Causes of Deaths, a registry of long‐term care facilities, and the National Database of work and Productivity as well as with whole genome sequencing data (WGS) of virus sequences from the Danish Covid‐19 Genome Consortium [6].
Fig. 1.
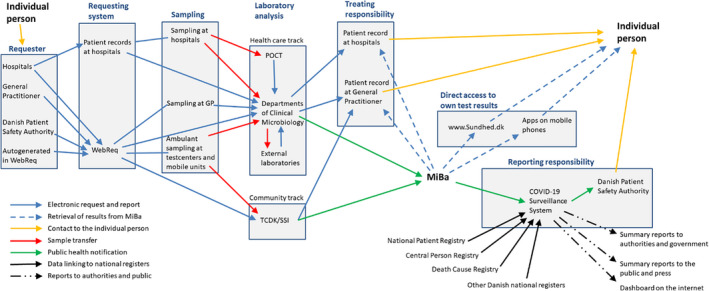
Dataflow from test request to reporting of the final result to the requesting doctor, to the tested person and to MiBa for the national surveillance and display of statistics as well as for reporting of positive cases to the Danish Patient Safety Authority. POCT, Point Of Care Testing; SSI, Statens Serum Institut; TCDK, Test Center Denmark; WebReq, Web‐based electronic request system [20]. Up load mechanisms for private vendors and school screening is not included in this outline of the system, for clarity reasons.
Data on all new positive cases, including information on hospitalization or residence in long‐term care facility, are transferred three times daily to the Danish Patient Safety Authority, who contacts the patient in order to ensure quarantine and instigate contact tracing. In addition, the SARS‐CoV‐2 test data are transferred every hour to the Danish COVID‐19 infection control app [7].
Data on all tested individuals are also transferred on a daily basis to research databases, available for researchers after application [8] and also to the expert group on mathematical modeling of COVID‐19 associated to SSI and public health officers at SSI [9].
All steps in processing of the data are automated and at close to real time provide a broad range of summary reports and autogenerated e‐mails to authorities and decision makers, which further disseminates situation reports [5]. A range of statistics on COVID‐19 is also accessible to the public at close to real time via www.ssi.dk, including the SSI COVID‐19 dashboard [10].
COVID‐19 in Denmark, 2020
In January 2020, COVID‐19 was assessed as an outbreak primarily occurring in Wuhan, China, with only a moderate likelihood of detecting cases imported into Europe. Testing was centralized to the national reference laboratory for virology at SSI [11].
When transmission of COVID‐19 at European ski resorts was evident, an increase in SARS‐CoV‐2 testing capacity was needed. Therefore, the clinical microbiology laboratories were asked to implement PCR analyses. The first COVID‐19 patient in Denmark was diagnosed on February 27, 2020 [12]. During the initial phase of the epidemic, mainly caused by imported cases, testing was restricted to symptomatic individuals meeting a case definition, including relevant travel history or exposure to a confirmed COVID‐19 case. When evidence of community transmission in Europe accumulated, the strategy was changed from containment to mitigation, with a focus on testing symptomatic patients requiring hospitalization to prevent health care‐associated transmission (Table 1) [13, 14, 15]. Targeted efforts to protect vulnerable populations, such as senior citizens in long‐term care facilities, were also implemented [16, 17]. During the first month of the epidemic, the testing capacity was restricted until the RT‐PCR test was implemented in all departments of clinical microbiology. Later on, basic reagents necessary for RT‐PCR were limited in supply [14].
In April, the testing strategy changed again, now focusing on easy access to free of cost COVID‐19 testing for all persons in Denmark, regardless of symptoms or exposure to persons with confirmed COVID‐19 [16, 18]. The nationwide access to free of cost testing was accomplished by building test facilities in all larger cities. The testing capacity has increased throughout the pandemic, and reached 704,700 total weekly tests by the second week of December 2020, corresponding to more than 10,000 tests per 100,000 inhabitants (Fig. 2B) [10]. The large testing capacity was considered a prerequisite for safe reopening of society and for the continuous suppression of the epidemic [19].
Fig. 2.
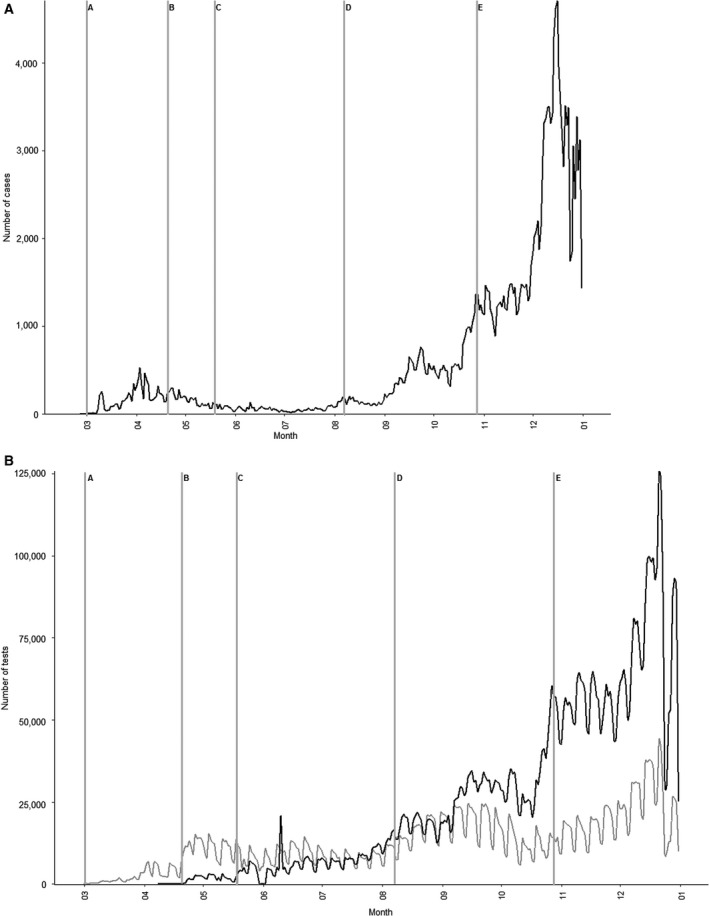
Case numbers and test activity in the Health care track and the Community track from March to December 2020. (A) Total number of COVID‐19 cases per day and (B) test activity in the Health care track and the Community track per day. Letters indicate changes in test strategy: A: Only test of hospitalized patients with symptoms or relevant exposure, B: Test of all hospitalized patients and patients with relevant symptoms, C: Access to test for all persons, D: Local outbreak in Aarhus with excessive testing, E: Systematic testing of staff in selected sectors.
Despite the huge increase in testing capacity, turnaround time from sampling to result has not increased. On the contrary, as shown in Fig. 3, turnaround time has slightly decreased over the period and 95% of samples were now reported in less than 24 h for prioritized samples within the Health care track, and in less than 48 h from sampling in the community track.
Fig. 3.
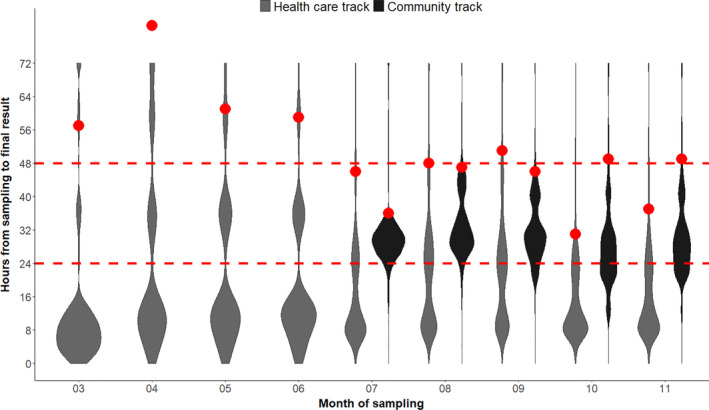
Distributions of turnaround times in hours. For the Health care track, turnaround times are shown for 1,936,468 samples analyzed at the regional clinical microbiology laboratories from March to November 2020. For the Community track, turnaround times are shown for 2,907,660 samples analyzed at TestCenter Denmark from July to November. The red points are the 95% quantiles. The plot is a so‐called violinplot showing the density distribution of the turnaround times (the areas are normalized to the same size). Samples with turnaround times below 4 h are due to point of care testing.
Reports in MiBa can be accessed through local IT systems for healthcare staff and through websites for the patients themselves (Box 1). The impact of increased testing capacity and the increasing need for access to test reports are illustrated by the increasing number of enquiries in MiBa per month (Fig. 4).
Fig. 4.
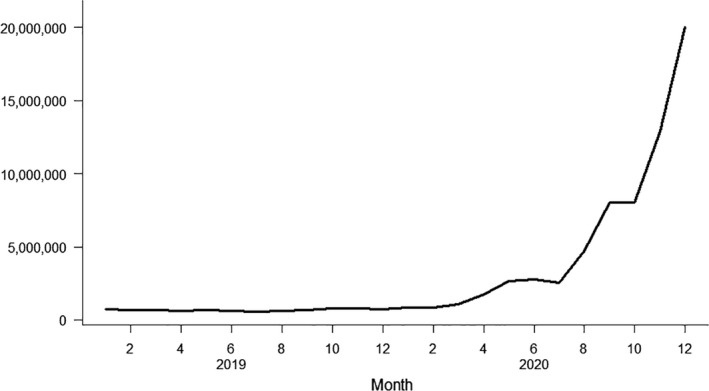
Number of all enquiries to MiBa per month, including enquiries for non‐COVID‐19‐related reports in the period from January 2019 to December 2020. During the year 2015, the monthly number of enquiries increased from on average x to y. Each time a person’s test results is accessed in MiBa, it is counted as one enquiry, regardless of the number of reports accessed.
The Epidemic Scenarios During 2020
Table 1 summarizes the management strategies during 2020, and Fig. 5 shows the crude incidence of COVID‐19 [13]. The first countrywide peak in the spring was associated with the initial travel‐associated importation and subsequent community transmission. Transmission was effectively curtailed with the lockdown in mid‐March with a subsequent decline in incidence. This resulted in a gradual reopening of society. In this period, the overall strategy was to suppress the epidemic, while at the same time to open the society and economy as much as possible after the initial lockdown [17]. A cross‐sectorial task force (Indsatsgruppen) became a key element after the summer of 2020. This group followed the number of new cases at national and local levels. The group proposed interventions and targeted testing to relevant authorities in response to local increases in occurrence and outbreaks of COVID‐19. Examples of outbreaks include social events at universities and other educational institutions, all types of local gatherings such as funerals, birthdays, and other celebrations, and in occupational settings, for example, health care, long‐term care facilities, and abattoirs. Some outbreaks were subject to considerable local containment efforts. One example is an outbreak in Region Midtjylland with epicenter in Aarhus, August 2020 [20] (Box 2). Another important event was the emergence of mink‐associated SARS‐CoV‐2 variants mainly in the Northern region of Denmark in October 2020, which resulted in lockdown and population screening in the most affected municipalities [21, 22]. Both outbreaks were mirrored as regional increases in incidence (Fig. 5). In Fall 2020, widespread community transmission was recognized with a corresponding increase in incidence and testing activity (Fig. 2). This necessitated after November 23, 2020, the gradual implementation of a number of new interventions culminating in a second national lockdown from December 16 [23].
Fig. 5.
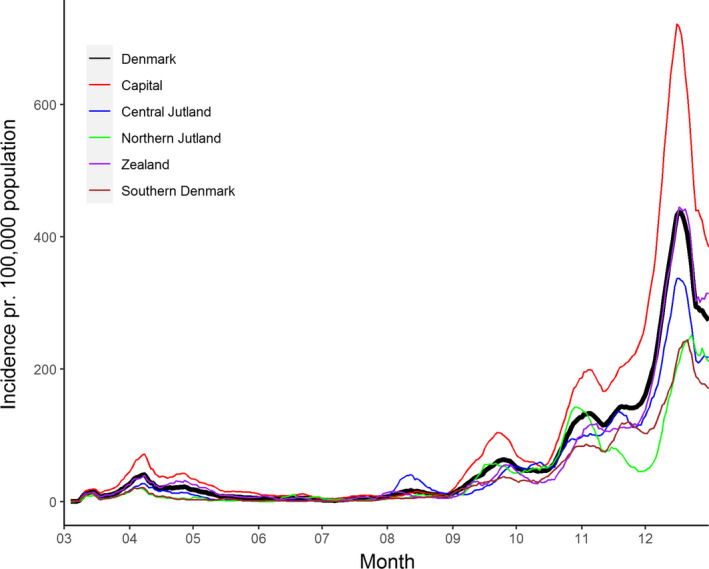
Rolling 7‐day incidences of COVID‐19 cases at national and regional levels from March to December 2020. The curves show the data from the whole country, Capital Region of Denmark; Central Jytland: Central Denmark Region; Northern Jutland: North Denmark Region; Zealand: Region Zealand; Southern Denmark: Region of Sothern Denmark.
Box 2. Controlling local outbreaks—the Aarhus outbreak as an example.
Surveillance data showed an increase in incidence in Central Denmark Region and the city of Aarhus by the beginning of August 2020 (Fig. 5). To control disease transmission, use of face masks in public transport became mandatory August 7. The public was encouraged to work from home and physical attendance in high schools was not allowed. Simultaneously, local testing efforts were massively increased including mobile test capacity in geographic hot spots and focus on communication within identified subpopulations. The case incidence normalized in the following weeks (Fig. 5).
The Rt estimated for Central Denmark Region adjusted for test activity (Fig. 7) shows that disease transmission accelerated by the end of June (R > 1). Transmission reached a peak at the end of July (R > 1.5). After the outbreak was recognized, the contact number reached a nadir (R = 0.8). It is likely that increased awareness and the reinforcement of general precautions contributed to control the outbreak. Control measures applied to the Region were suspended August 28. At that time Rt had stabilized at a level similar to the rest of Denmark (R = 1.1).
Besides the effect of management strategies on testing activity during the course of the epidemic, the decisions of governments and the media affect citizens’ behavior, compliance to regulations, and trust. This interrelationship has been followed by the HOPE project: “How Democracies Cope with COVID‐19: A Data‐Driven Approach” [24].
Challenges in the Interpretation of Numbers
The number and incidence of daily COVID‐19 cases are highly dependent on testing activity, which has changed profoundly during the period (Fig. 2B). The epidemiology of COVID‐19 cases may further be skewed, for example, if testing is performed in localized outbreak settings with intensified contact tracing. To obtain a more realistic estimate of the transmission dynamics underlying the epidemic, such variations must be adjusted for.
One strategy to control these confounders is to test a representative sample of the Danish population regularly. In the reopening period, after the early lockdown (Table 1), an unbiased representative surveillance system was established based on 1750 Danish citizens being invited each week to be tested. Unexpectedly, participation was low, declining from only 29.6% to 12.9% from the first to the third test round [25]. The coincidence of free testing opportunities outside the study possibly explains this. The power of the study was also challenged by a low prevalence of COVID‐19 during the reopening period studied.
Another more pragmatic approach to obtain estimates of how the changing testing activity influences estimates of COVID‐19 incidence was established by the expert group on mathematical modeling of COVID‐19 at SSI who introduced normalization of the daily data outputs by using a “reference population” [26]. In May, the National Health Authority decided to implement screening of all hospital patients (in and outpatients) for SARS‐CoV‐2 prior to their appointment. These individuals were obviously not representative for the Danish population or for the risk of contracting COVID‐19, but their indication for being tested can be assumed to be constant throughout the epidemic. For practical purposes, patients with respiratory infections were not included in this reference group, as COVID‐19 may be a differential diagnosis. Pregnant women were also excluded because they may avoid exposure for precautionary reasons. It may be assumed that if the general transmission risk changes because of increases in incidence, this will be reflected in this group, which therefore can be used as a reference group to adjust for changes in testing activity.
It was found that the incidence of COVID‐19 correlated with testing activity with a power function of 0.7, that is, a doubling in testing activity would result in an approximately 62% (20.7‐1) increase in the number of detected cases. Using this correction for testing activity, the COVID‐19 incidence appeared hugely underdiagnosed in early spring 2020, and the estimated incidence during spring before the lockdown was higher than in the fall (Fig. 6). The incidence adjusted for changes in testing activity correlates with the number of daily COVID‐19 hospitals admissions in all months except for December 2020, which is related to changes in testing patterns up to and during Christmas holiday [26].
Fig. 6.
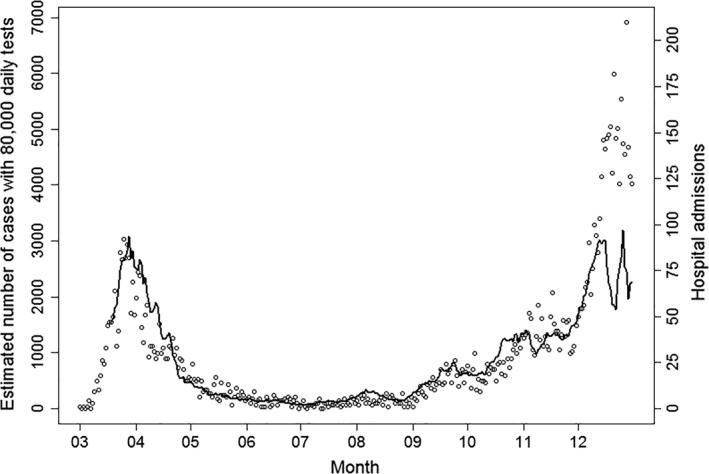
Estimated number of new cases using a correction factor for test activity (solíd black line), which empirically corrects for time‐varying testing behavior, test capacity, and targeted testing efforts. For ease of interpretation, the calculation is made relative to a fixed reference number of 80.000 daily tests. Points show the daily number of hospital admissions of cases with COVID‐19 for comparison. The estimated number of new cases follows hospital admission closely up to the middle of December, but the model fit was impacted by the substantial variation in testing activity in the last half of December due to seasonal changes in testing behavior around Christmas and the New Year.
There are caveats associated with using this reference group to estimate the relative changes in disease transmission. Firstly, changes in healthcare service, for example, elicited by the epidemic, can change the case‐mix of the reference group; secondly, behavioral changes, provision of testing by private vendors, and implementation of epidemic control measures are likely to have more immediate impact in the general society.
Calculation of the Effective Reproduction Number, Rt, During Changes in Testing Activity
The effective reproduction number “R” is the estimated number of secondary cases attributed to a primary case. Using the principles, described above, to adjust for variations in testing activity, a corrected time‐dependent reproduction number Rt was estimated (Fig. 7) [27].
Fig. 7.
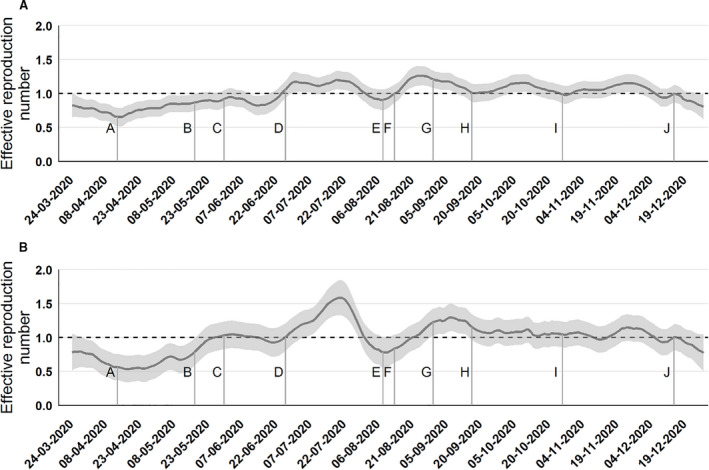
Effective reproduction number (Rt) corrected for variations in test activity at the national level (A) and the Central Denmark Region (B). The Rt calculations are based on the model also described in Fig. 6. For further details about these calculations (https://www.ssi.dk/‐/media/ssi‐files/ekspertrapport‐af‐den‐23‐oktober‐2020‐incidens‐og‐fremskrivning‐af‐covid19‐tilflde.pdf?la=da). In short, the model is a consideration of the complex testing behavior and uses a 7 day smoothing. The letters designate: A: Early limited reopening on April 15, B: Late reopening on May 19, C: Border reopening June 18, D: School summer holiday starts June 28, E: School summer holiday ends August 10, F: Mandatory use of masks in public transport August 15, G: Universities open September 1, H: Work from home recommendations September18, I: Mandatory use of masks in public areas (in doors) October 28, J: Second lockdown.
Figure 7 shows that Rt was <1 following the spring lockdown, indicating that the COVID‐19 epidemic was declining. The corrected Rt increased to >1 already in late June and had until mid‐December been around 1. This may indicate that the general precautionary measures in society deteriorated during the summer, which together with a relaxation of control measures, reignited a smoldering epidemic. This was largely unrecognized by authorities, who at the time focused mostly on the low incidence, as the estimate of the uncorrected Rt obscured by variations in testing activity did not appear alarming. The corrected Rt was used by the authorities from October 2020.
Discussion
The existing MiBa infrastructure has provided a robust linkage between diagnostic testing, reporting, and surveillance. The Danish electronic system for requesting and reporting of laboratory results was able to accommodate a ramp‐up in testing capacity. This infrastructure has enabled accumulation of test results in real time and direct communication of the results through daily updated publically available webpages and weekly surveillance reports. Sharing of reliable and timely data, supports communication, trust, and coherence in a country during a crisis.
Having this fundament of ample testing capacity and solid data, there are still lessons to be learned. A danger of real‐time reporting on daily basis is the associated perception of a need to act on fluctuations and increases in daily numbers sometimes associated with variations in the amount of testing done. Maybe also, the focus on the relative successful containment of local outbreaks contributed to a feeling of controlling the epidemic. It is the nature of epidemics to have exponential growth, and what may be a steady increase on a log scale seems at the start as a negligible increase in numbers associated with a comfortable, but false feeling of control. The same exponential increase will later in the epidemic, when daily numbers are high, be perceived as a lack of control, setting healthcare infrastructures at risk, even though the underlying transmission dynamics are the same. To focus on daily variations and local outbreaks may divert attention from the underlying changes in transmission dynamics that may have occurred earlier than perceived from absolute numbers. A parsimonious view on the Danish SARS‐CoV‐2 epidemic is that an initial surge in spring was curtailed by a lockdown period during which the reproduction number Rt was well below 1. This brought down the absolute number of infected people to a minimum. At the end of the gradual reopening of society in early summer, Rt increased to above 1 and has remained so in the second half of 2020.
A cornerstone for the Danish strategy to control the epidemic since the spring lockdown has been to provide ample testing capacity that would allow a fast identification of transmission chains including local outbreaks or other upsurges. This so‐called “hammer and dance” strategy was thought ideally to be able to suppress the epidemic and to ensure that the reopening of society following the lockdown was safe. This strategy was based on unlimited testing free of cost and accessible to all regardless of symptoms or exposure. Denmark is currently one of the world leaders in providing tests at a population level as reported by the John Hopkins Coronavirus Resource Center [28]. This strategy has been a partial success. Excess mortality has been nearly absent during the epidemic [29]. However, as a strategy to maintain an open society, it failed when the winter season appeared. Hence, the government decided to implement the second lockdown.
Although MiBa in many ways served as the key data source, surveillance faced a number of shortcomings and limitations. Data automatically captured from administrative databases have limited epidemiological information. For example, it is difficult to determine why a test was requested, or the symptoms that a patient experienced, which is essential for interpretation. Also, detailed exposure data are generally unavailable [22]. These data, which are collected in outbreak investigations, are valuable for understanding risk factors and transmission routes. Hence, electronic laboratory and administrative data will never be able to stand alone. We still need to undertake epidemiologic studies and surveys to understand transmission patterns and qualify interventions. It remains, for example, to determine the effectiveness of the contact tracing system and the mass testing, and to see how this can be improved for better management of the epidemic. Hence, whether the resurgence of the epidemic in the fall could have been curtailed by improved contact tracing remains to be answered. It is, however, clear that the timeliness of contact tracing is critical [30]. Turnaround time from test to result may obviously impact linkage to contact tracing and quarantine. With centralized testing supplemented with a limited number of PCR‐based POCT, it is difficult to bring down turnaround time of PCR testing to less than what we currently observe in Denmark. However, this may not be fast enough to provide efficient linkage to contact tracing and infection control. If testing is to some extent done for recreational use, that is, motivated by temporary non‐adherence to social distancing rules, a report of a negative test result 2 days old may provide false security. Targeted use of Antigen testing has recently been implemented in the national testing strategy.
In addition to the extensive SARS‐Cov2 testing, a gradual escalation of virus sequencing capacity has taken place, reaching approximately 50% of all positive samples in January 2021 and a goal of sequencing of all suitable samples by end of February 2021. This effort provides surveillance of the emerging and circulating variants of SARS‐Cov2, which will be of increasing importance when immunity from prior infection or vaccination increase in the populations in Denmark and abroad. Raw sequence data are complex and not suited for direct handling in MiBa, but typing results based on SARS‐CoV‐2 sequencing are recorded and have since February 2021 been available in MiBa for TCDK, microbiologists, and epidemiologists, but so far not for the patients and clinicians as they are rather difficult to interpret for non‐specialists [31].
An existing IT health infrastructure based on a close network between microbiologists, information technology experts, and public health officials enabled almost real‐time access to data for action and situation reports for the decision makers. The use of the same data for different purposes, for example, clinical reports, citizen information, contact tracing, public health, and science, has been a critical asset for the pandemic response. For example, real‐time access to high‐quality denominator data (e.g., number of tests undertaken and hospital capacity) and transmission in occupational settings were essential and would not have been available without this data fundament. This information has enabled timely containment outbreaks and enabled the decision makers to adopt timely interventions according to the situational awareness, and hitherto has navigated Denmark through the epidemic with only limited excess mortality.
However, the use of the same data for different purposes has raised questions regarding data protection and privacy. As mentioned in the introduction, the epidemic fueled an unprecedented demand for data, including person‐identifiable data to researchers. In particular, in a crisis, data should be regarded as a common good in order to provide evidence as timely as possible. This view stands against data protection. In fact, the EU General Data Protection Regulation law (GDPR), which was put in place May 2018, is the toughest privacy and security law in the world. The challenges imposed by GDPR have been one of the obstacles in sharing of detailed data at the individual level to researchers. In the future, it will be important to create robust solutions and governance models that on one hand respects privacy and GDPR, and on the other hand accommodates legitimate demands for data to public and private researchers. Other challenges were limitations in critical capacity of information technology, including hardware limitations, and limited number of key resources with in‐debt knowledge on the complexity of the infrastructure, coding, and the data. It is mission impossible to stabilize a somewhat fragile system built in haste with an exponential increase in dataload and at the same time accommodate the massive need to build new solutions to answer requests from a large number of stakeholders and the government. In a crisis, it is important to have a clear decision path and the ability to make difficult prioritizations, when all wishes and needs cannot be answered at the same time.
It is important to address these issues in the years to come. By the establishment of robust data solutions, COVID‐19 may at the end motivate a transformation from traditional indicator‐based public health surveillance to an all‐encompassing information systems based on access to a comprehensive set of data sources, including diagnostic and reference microbiology.
In conclusion, the MiBa system is integrated into the Danish health informatics infrastructure and facilitated sharing test results when upscaling testing capacity. The data could be used both for reporting of individual clinical results and in real time for statistics to support the national policy toward the COVID‐19 epidemic.
This is a truly collaborative effort between many stakeholders. Firstly, we thank the MiBa Board of Representatives and all departments of clinical microbiology in Denmark; we thank the managing people behind “Sundhedsjournal komplekset” and “Sundhed.dk” and The Danish Health Data Authority; we thank the expert group on mathematical modeling of COVID‐19 associated to SSI; and finally, we thank the Data Analysis and Integration Secretary, the Department of Infection Epidemiology and TestCenter Denmark, all at SSI.
Schønning K, Dessau RB, Jensen TG, Thorsen NM, Wiuff C, Nielsen L, Gubbels S, Denwood M, Thygesen UH, Christensen LE, Møller CH, Møller JK, Ellermann‐Eriksen S, Østergaard C, Lam JUH, Abushalleeh N, Meaidi M, Olsen S, Mølbak K, Voldstedlun M. Electronic reporting of diagnostic laboratory test results from all healthcare sectors is a cornerstone of national preparedness and control of COVID‐19 in Denmark. APMIS. 2021; 129: 438–451.
References
- 1. Thacker SB, Qualters JR, Lee LM. Public health surveillance in the United States: evolution and challenges. MMWR Suppl 2012;61:3–9. [PubMed] [Google Scholar]
- 2. Leitmeyer KC, Espinosa L, Broberg EK, Struelens MJ. Automated digital reporting of clinical laboratory information to national public health surveillance systems, results of a EU/EEA survey, 2018. Eurosurveillance 2020;25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Voldstedlund M, Haarh M, Mølbak K. The Danish Microbiology Database (MiBa) 2010 to 2013. Euro Surveill 2014;19. [DOI] [PubMed] [Google Scholar]
- 4. Coronaprover.dk. Available from: https://www.coronaprover.dk [25‐01‐2021]. [Google Scholar]
- 5. SSI . Daglige covid‐19‐tal på kommunalt, regional samt nationalt niveau. Available from: https://covid19.ssi.dk/overvagningsdata/daglige‐covid‐19‐tal [Google Scholar]
- 6. Danish Covid‐19 Genome Consortium. Available from: https://www.covid19genomics.dk/home [25‐01‐2021]. [Google Scholar]
- 7. SmitteIStop app. Available from: https://smittestop.dk/en [25‐01‐2021]. [Google Scholar]
- 8. Pottegård A, Kristensen KB, Reilev M, Lund LC, Ernst MT, Hallas J, et al. Existing data sources in clinical epidemiology: the Danish COVID‐19 cohort. Clin Epidemiol 2020;12:875–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hansen CH, Michlmayr D, Gubbels SM, Mølbak K, Ethelberg S, et al. Assessment of protection against reinfection with SARS‐CoV‐2 among 4 million PCR‐tested individuals in Denmark in 2020: a population‐level observational study. Lancet 2021;397:1204–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. SSI . Statens Serum Institut COVID‐19 dashboard. Available from: https://experience.arcgis.com/experience/aa41b29149f24e20a4007a0c4e13db1d [25‐01‐2021]. [Google Scholar]
- 11. ECDC . Rapid Risk Assessment: Outbreak of acute respiratory syndrome associated with a novel coronavirus, Wuhan, China; first update – 22 January 2020; 2020. [Google Scholar]
- 12. Sundhedsministeriet . En dansker er testet positiv for COVID‐19. 27‐02‐2020 – Pressemeddelelse COVID‐19. Available from: https://sum.dk/nyheder/2020/februar/en‐dansker‐er‐testet‐positiv‐for‐covid‐19 [Google Scholar]
- 13. Danish authorities joint website: Political agreements and initiatives. Available from: https://en.coronasmitte.dk/announcements/political‐agreements‐and‐initiatives [Google Scholar]
- 14. Sundhedsministeriet . Teststrategi skal sikre at flest muligt testes for COVID‐19. https://sum.dk/nyheder/2020/marts/teststrategi‐skal‐sikre‐at‐flest‐muligt‐testes‐for‐covid‐19 [Google Scholar]
- 15. Styrelsen for patientsikkerhed: COVID‐19: I denne uge er inddæmningsstrategien erstattet af en afbødningsstrategi. Available from: https://stps.dk/da/nyheder/2020/inddaemningsstrategi‐erstattes‐af‐afboedningsstrategi/
- 16. Sundhedsministeriet, Regeringen og regionerne øger antallet af test markant; 2020. Available from: https://sum.dk/nyheder/2020/april/regeringen‐og‐regionerne‐oeger‐antallet‐af‐test‐markant‐ [Google Scholar]
- 17. SSI . Uge 12 – 2020: Coronavirus pandemien – Status på udbruddet og håndtering af patienter; 2020. Available from: https://www.ssi.dk/aktuelt/nyhedsbreve/epi‐nyt/2020/uge‐12 [Google Scholar]
- 18. Sundhedsministeriet . Alle borgere får mulighed for at blive testet for COVID‐19; 2020. Available from: https://sum.dk/nyheder/2020/maj/alle‐borgere‐faar‐mulighed‐for‐at‐blive‐testet‐for‐covid‐19‐ [Google Scholar]
- 19. Imperial College COVID‐19 Response Team: Report 12: The Global Impact of COVID‐19 and Strategies for Mitigation and Suppression. London, United Kingdom; 2020. [Google Scholar]
- 20. Sundhedsministeriet . COVID‐19: Status på udbrud i Aarhus og Silkeborg; 2020. Available from: https://sum.dk/nyheder/2020/august/covid‐19‐status‐paa‐udbrud‐i‐aarhus‐og‐silkeborg [Google Scholar]
- 21. WHO . SARS‐CoV‐2 mink‐associated variant strain – Denmark. Disease Outbreak News. 2020; 6. [Google Scholar]
- 22. Sundhedsministeriet . Nye tiltag og restriktioner for at reducere smitteudviklingen i Nordjylland; 2020. Available from: https://sum.dk/nyheder/2020/november/nye‐tiltag‐og‐restriktioner‐for‐at‐reducere‐smitteudviklingen‐i‐nordjylland [05‐11‐2020]. [Google Scholar]
- 23. Sundhedsministeriet . COVID‐19‐nedlukning hen over jul og nytår; 2020. Available from: https://sum.dk/nyheder/2020/december/covid‐19‐nedlukning‐hen‐over‐jul‐og‐nytaar [16‐12‐2020]. [Google Scholar]
- 24. The Hope Project. Available from: https://hope‐project.dk/#/about [Google Scholar]
- 25. SSI . Notat: Resultater fra den repræsentative PCR‐undersøgelse af COVID‐19; 2020. Available from: https://files.ssi.dk/Notat_Resultater_PCR_praevalensundersoegelse_16juni2020.pdf [Google Scholar]
- 26. SSI . Ekspertrapport af d. 23. oktober 2020 – Incidens og fremskrivning af COVID‐19 tilfælde. Available from: https://covid19.ssi.dk/‐/media/ssi‐files/ekspertrapport‐af‐den‐23‐oktober‐2020‐incidens‐og‐fremskrivning‐af‐covid19‐tilflde.pdf?la=da [23‐10‐2020]. [Google Scholar]
- 27. Covid19RR – Estimation of reproduction rates for the COVID‐19 pandemic in Denmark based on publicly available data. Available from: https://github.com/ku‐awdc/Covid19RR [25‐01‐2021]. [Google Scholar]
- 28. John Hopkins University & Medicine . The Johns Hopkins Coronavirus Resource Center (CRC). Available from: https://coronavirus.jhu.edu/testing/international‐comparison [Google Scholar]
- 29. Euromomo Bulletins; 2020; Available from: https://euromomo.eu [Google Scholar]
- 30. SSI . Ekspertrapport af den 10. december 2020 – Effekten af kontaktopsporing. Available from: https://covid19.ssi.dk/‐/media/arkiv/subsites/covid19/modelberegninger/ekspertrapport‐effekten‐af‐kontaktopsporing‐10122020.pdf?la=da [10.12.2020]. [Google Scholar]
- 31. Bager P, Wohlfahrt J, Fonager J, Albertsen M, Yssing Michaelsen T, Holten Møller C, et al. Increased risk of hospitalisation associated with infection with SARS‐CoV‐2 lineage B.1.1.7 in Denmark. SSRN Electronic J. 2021. 10.2139/ssrn.3792894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. e‐health in Denmark. e‐health as a part of a coherent Danish health care system. Available from: https://www.medcom.dk/media/1211/ehealth‐in‐denmark‐ehealth‐as‐a‐part‐of‐a‐coherent‐danish‐health‐care‐system.pdf [Google Scholar]
- 33. MedCom – The Danish Health Data Network 1994‐2014 (MC‐S238); 2014. Available from: https://www.medcom.dk/media/1167/medcom‐20years.pdf [Google Scholar]
- 34. WebReq – off to a good start (MC‐S239) 2007 (July). Available from: https://www.medcom.dk/media/1157/webreq‐off‐to‐a‐good‐start.pdf [Google Scholar]
- 35. EDIFACT/XML standards; 2020. Available from: https://www.medcom.dk/standarder/edifactxml [Google Scholar]


